Abstract
The 300 kDa cation-independent mannose 6-phosphate receptor (CI-MPR) mediates the intracellular transport of newly synthesized lysosomal enzymes containing mannose 6-phosphate on their N-linked oligosaccharides. In addition to its role in lysosome biogenesis, the CI-MPR interacts with a number of different extracellular ligands at the cell surface, including latent transforming growth factor-β, insulin-like growth factor-II, plasminogen, and urokinase-type plasminogen activator receptor (uPAR), to regulate cell growth and motility. We have solved the crystal structure of the N-terminal 432 residues of the CI-MPR at 1.8 Å resolution, which encompass three out of the 15 repetitive domains of its extracytoplasmic region. The three domains, which exhibit similar topology to each other and to the 46 kDa cation-dependent mannose 6-phosphate receptor, assemble into a compact structure with the uPAR/plasminogen and the carbohydrate-binding sites situated on opposite faces of the molecule. Knowledge of the arrangement of these three domains has allowed us to propose a model of the entire extracytoplasmic region of the CI-MPR that provides a context with which to envision the numerous binding interactions carried out by this multi-faceted receptor.
Keywords: IGF-II receptor, lectin, lysosome biogenesis, mannose 6-phosphate receptor, protein trafficking
Introduction
The 300 kDa cation-independent mannose 6-phosphate receptor (CI-MPR) and the 46 kDa cation-dependent MPR (CD-MPR) are ubiquitously expressed type I transmembrane glycoproteins that cycle continuously between the cell surface and intracellular compartments. The best characterized function of the MPRs is their ability to direct the delivery of ∼50 different newly synthesized soluble acid hydrolases bearing mannose 6-phosphate (Man-6-P) on their N-linked oligosaccharides to lysosomes (Ghosh et al, 2003). In addition to its intracellular role in lysosome biogenesis, the CI-MPR, in contrast to the CD-MPR, has been implicated in numerous cellular processes, including cell growth, apoptosis, and cell migration, due to its ability to bind a wide range of Man-6-P-containing (e.g., latent transforming growth factor-β (TGF-β), granzyme B, CD26) and non-Man-6-P-containing (insulin-like growth factor II (IGF-II), retinoic acid, urokinase-type plasminogen activator receptor (uPAR), plasminogen) molecules at the cell surface (Dahms and Hancock, 2002). The ability of the CI-MPR to interact with many different proteins and a lipophilic molecule is facilitated by the receptor's large (∼2270 amino acids) extracytoplasmic region comprising 15 homologous domains, in which several distinct ligand-binding sites have been localized to individual domains.
Our crystal structure of the 154-residue extracytoplasmic region of the CD-MPR in the presence and absence of bound carbohydrate (Roberts et al, 1998; Olson et al, 1999, 2002) provides an insight into the mechanism of high-affinity (nM) phosphomonoester recognition by this homodimeric receptor. Unlike the CD-MPR, which contains a single carbohydrate recognition site per polypeptide, the CI-MPR contains two high-affinity Man-6-P binding sites (Tong et al, 1989; Westlund et al, 1991), with limited sequence identity to the CD-MPR (see Figure 2C), and essential residues for binding reside in domains 3 and 9 (Dahms et al, 1993; Hancock et al, 2002a). Surprisingly, our recent studies have revealed that the two carbohydrate-binding sites of the CI-MPR are structurally quite different. As with the CD-MPR, domain 9 can be expressed as an isolated domain that retains high-affinity (∼1 nM) carbohydrate binding. In contrast, expression of domain 3 alone results in a protein with ∼1000-fold reduction in affinity for a lysosomal enzyme compared to a construct encoding domains 1–3, which binds lysosomal enzymes with high affinity (Hancock et al, 2002b). These results suggest that residues in domain 1 and/or 2 either directly or indirectly influence carbohydrate recognition. The two Man-6-P-binding sites of the CI-MPR are also distinct with respect to their ligand specificities: domain 9 is similar to the CD-MPR in its specificity for phosphomonoesters, while domains 1–3 are more promiscuous in their recognition of related structures, including phosphodiesters and mannose 6-sulfate (Marron-Terada et al, 2000). These results are consistent with the observed greater efficiency displayed by the CI-MPR versus the CD-MPR in the delivery of acid hydrolases to the lysosome (Pohlmann et al, 1995).
Figure 2.
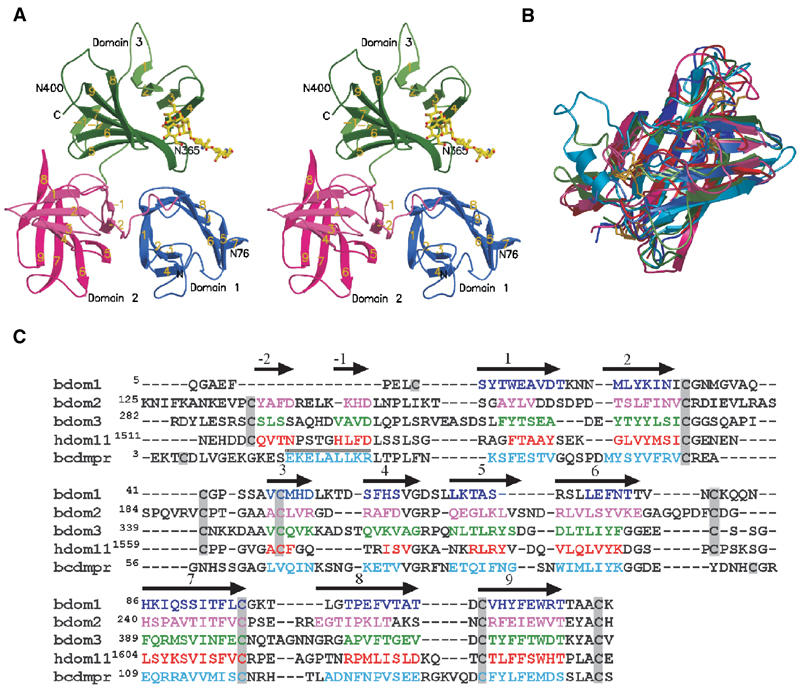
Structure of the three N-terminal domains of the bovine CI-MPR. (A) Stereo view of the ribbon diagram of the protein encoding domain 1 (blue), domain 2 (pink), and domain 3 (green). The oligosaccharide (yellow) attached to Asn76 of an adjacent crystallographic neighbor is shown. The position of the two glycosylated asparagine residues (Asn76 and Asn365) and that of the potential glycosylation site at Asn400 are indicated. The β-strands are labeled, showing the relative orientation of the domains to each other. (B) Overlay of the structures of the individual domains (domain 1, 2, and 3) along with those of the CD-MPR (cyan) and domain 11 (red) of the CI-MPR. Color schemes for domains 1–3 are the same as in panel A. The disulfide bridges are shown in gold. Note that the location of the first disulfide pair in domain 1 differs from that of domains 2, 3, and 11. (C) Structure-based sequence alignment of domains 1–3 of the bovine CI-MPR, domain 11 of the human CI-MPR, and the bovine CD-MPR. Secondary structure elements are indicated above the sequence, with arrows indicating β-strands and the cylinder indicating the single α-helix present in the CD-MPR. β-Strands of the β-barrel in each domain are sequentially numbered with strands in the preceding linker region as -1and -2. Residues found in the secondary structure elements are indicated by colored text. Cysteine residues are boxed in gray.
The designation of the CI-MPR (also referred to as the IGF-II receptor) as a putative tumor-suppressor gene is based on the ability of the receptor to divert hydrolytic enzymes from the secretory pathway (Ludwig et al, 1994; Pohlmann et al, 1995; Sohar et al, 1998) to facilitate the activation of the growth inhibitor TGF-β (Dennis and Rifkin, 1991; Ghahary et al, 2000) and to decrease serum levels of the mitogen IGF-II (Oka et al, 1985; Lau et al, 1994; Wang et al, 1994; Ludwig et al, 1996). Consistent with IGF-II clearance being an essential function of the receptor is the observation that mice deficient in the CI-MPR exhibit increased levels of serum IGF-II, enlarged organs, and perinatal lethality (Lau et al, 1994; Wang et al, 1994; Ludwig et al, 1996). Of the non-Man-6-P-containing ligands, the interaction between the CI-MPR and IGF-II has been characterized most extensively. In contrast, limited information is available concerning the retinoic acid-binding site of the CI-MPR: IGF-II and Man-6-P do not inhibit retinoic acid binding and the ∼40 kDa C-terminal region of the receptor appears to be essential for this interaction (Kang et al, 1997). The IGF-II-binding site has been mapped to domain 11 and sequences in domain 13 have been shown to contribute an ∼10-fold enhancement in affinity (Devi et al, 1998; Grimme et al, 2000; Linnell et al, 2001). Studies by Brown et al (2002) on a single isolated domain have provided the first structural view of the CI-MPR. Although these studies and a subsequent report (Uson et al, 2003) were performed in the absence of bound IGF-II, the crystal structure of domain 11 reveals that its overall fold is similar to that of the CD-MPR and a hydrophobic region corresponding to the location of the Man-6-P-binding site in the CD-MPR is predicted to bind IGF-II.
Recent additions to the growing list of ligands for the CI-MPR are plasminogen (Godar et al, 1999) and the glycosyl-phosphatidyl-inositol (GPI)-anchored uPAR (Nykjaer et al, 1998; Godar et al, 1999; Kreiling et al, 2003). uPAR is expressed on many cell types and functions in the regulation of cell adhesion, migration, and extracellular proteolysis via its interactions with vitronectin, integrins, and uPA (Preissner et al, 2000; Blasi and Carmeliet, 2002). uPAR binds to the CI-MPR with a Kd of ∼1–10 μM and involves domains DII and DIII of the three domain-containing uPAR (Nykjaer et al, 1998). Co-expression studies have shown that the CI-MPR can influence the subcellular distribution of uPAR by targeting uPAR for degradation in lysosomal compartments (Nykjaer et al, 1998). However, others have speculated that the CI-MPR augments the activity of uPAR by serving as a platform for the activation of pro-uPA and latent TGF-β on the cell surface (Godar et al, 1999). The proposed uPAR- and plasminogen-binding sites have recently been mapped to the N-terminal half of domain 1 of the CI-MPR (Leksa et al, 2002). Although the addition of the lysine analog tranexamic acid was shown to inhibit the interaction of the CI-MPR with either uPAR or plasminogen (Godar et al, 1999; Leksa et al, 2002), the structural determinants required for these interactions are not known. Here we report the crystal structure of the N-terminal three domains (residues 1–432) of the bovine CI-MPR at 1.8 Å resolution, which provides an insight into the mechanisms of carbohydrate, uPAR, and plasminogen recognition by the receptor.
Results and discussion
Overall structure of domains 1–3 of the CI-MPR
The structure of domains 1–3, which contains 24 cysteines (four disulfides per domain with no inter-domain disulfides), 362 water molecules, and one glycerol molecule, is a single polypeptide chain comprising residues Gln5 to Val432. Carbohydrates are detected at two out of the three potential N-glycosylation sites: Asn76 of domain 1 is glycosylated with two N-acetyl-D-glucosamine and three mannose residues (Figure 1), Asn365 in domain 3 has two N-acetyl-D-glucosamine residues with clear electron density, and Asn400 shows no visible electron density for the oligosaccharide, but there is ample room in the structure to accommodate several sugar residues.
Figure 1.
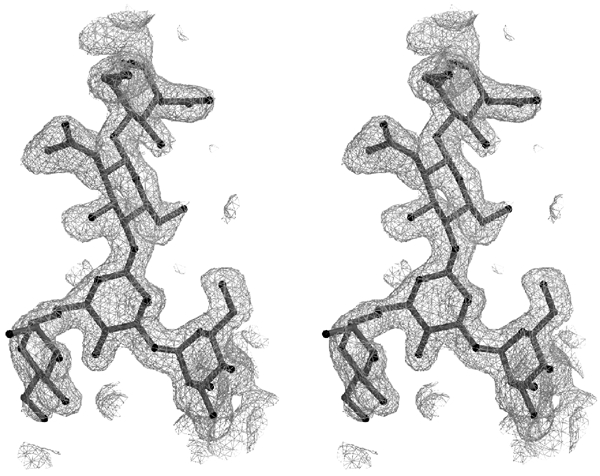
Electron density map. Stereo view of an Fo-Fc omit map in the region of the oligosaccharide Manα(1,3)[Manα(1,6)]Manβ(1,4)GlcNAcβ(1,4)GlcNAc attached to Asn76, contoured at 2σ level. The figures were generated with either MOLSCRIPT (Kraulis, 1991) and rendered with Raster3D (Merritt and Bacon, 1997) or with GRASP (Nicholls et al, 1991).
As predicted from sequence alignments of the CI-MPR (Lobel et al, 1988), the N-terminal 432 residues of the mature protein are folded into three distinct domains with a topology similar to each other (Figures 2A and B), as well as to the CD-MPR (Roberts et al, 1998; Olson et al, 1999) and domain 11 of the CI-MPR (Brown et al, 2002; Uson et al, 2003) (Figure 2B). The folding of the polypeptide for the three N-terminal domains of the CI-MPR appears to be unique: submission of the model coordinates to DALI (http://www.ebi.ac.uk/dali) for comparison with the database yielded no similar structures other than the CD-MPR and domain 11 of the CI-MPR. The molecule is wedge-shaped (approximate dimensions of 70 Å on each side and a height of 50 Å), with each domain having dimensions of 40 Å × 32 Å × 50 Å. The core structure of each domain consists of a flattened β-barrel comprising a four-stranded antiparallel β-sheet (β1–β4) with its strand direction orthogonally oriented over a five-stranded β-sheet (β5–β9), of which the fifth strand (β9) interjects between β7 and β8 (Figure 2A). Each domain contains four disulfide bridges, three of which are conserved with the CD-MPR, and all four are conserved with domain 11 of the CI-MPR (Figures 2B and C). The N-terminus of domain 2 (residues 125–147) and domain 3 (residues 282–302) each contains a linker region composed of a random coil followed by two anti-parallel β-strands (-2β and -1β), which functions to connect the core-flattened β-barrel structures together. Domains 1 and 2 are oriented such that the five-stranded second sheet (β5–β9) of domain 2 and the four-stranded first sheet (β1–β4) of domain 1 form a continuous surface making the ‘base' of the triangular wedge (Figures 2A and 5), while domain 3 sits on top of domains 1 and 2. The relative orientation between domains (Figure 2A) is not a simple repetitive symmetry operation as suggested by Brown et al (2002).
Domain interface interactions
Digestion of the CI-MPR with subtilisin, followed by N-terminal sequencing, showed that the smallest isolated fragment that retained carbohydrate-binding capabilities encompassed domains 1–3, indicating that this region of the receptor forms a stable functional unit (Westlund et al, 1991). Our current studies reveal that these domains do indeed fold into a compact structural unit. However, in comparison to the CD-MPR in which extensive contacts exist between the five-stranded second sheets (β5–β9) of the two monomers that form the homodimeric structure (Roberts et al, 1998; Olson et al, 1999, 2002), the contacts between the three N-terminal domains of the CI-MPR are quite different, reflecting the different functional roles carried out by these regions. Rather, the contacts between the three domains are mediated mainly by residues within the linker regions (domain 2, residues 125–147; domain 3, residues 282–302) and loops (Figure 2A).
Approximately 16% (3686 Å2) of the entire surface area of the three N-terminal domains of the CI-MPR is contained in interfaces. In comparison, the dimeric CD-MPR has a buried interface of 2063 Å2/dimer (20%) in the ligand-bound state (Roberts et al, 1998; Olson et al, 1999) and 1536 Å2 (15%) in the unbound state (Olson et al, 2002). Domains 1 and 2 share the largest interface at 1720 Å2 (22%). The N-terminal strand β1 of domain 1 interacts with the linker strand -2β of domain 2, and β8 of domain 1 interacts with -1β of domain 2. Domains 2 and 3 share 1426 Å2 (16%) in solvent-inaccessible area between them, whereas domains 1 and 3 share the smallest interface (712 Å2, 8%). Thus, these extensive interactions, along with relatively low B-factors of the linker regions, strongly suggest that the flexibility in the linker regions between domains is very limited and that the three-domain arrangement observed in the current structure is likely to form an independent structural entity within the entire CI-MPR molecule. Furthermore, the domain interactions that we observe in the N-terminal region of the CI-MPR are significantly more extensive than in other multi-domain lectins, such as wheat germ agglutinin-3 in which there are only three contacts less than 3.3 Å between adjacent domains and no contacts between non-adjacent domains (Harata et al, 1995). In addition, the recently reported structure of the LDL receptor (Rudenko et al, 2002) shows that the multiple cysteine-rich repeats, R2–R7, do not interact with each other directly, but rather exist in an extended conformation relative to each other. Thus, in comparison, the multiple domains of the CI-MPR exhibit significant contacts with each other to form a compact structure.
Oligosaccharide-binding site
Although the crystallization medium contained Man-6-P, the structure reveals that the predicted ligand-binding site of domain 3 is occupied by a nonphosphorylated oligosaccharide of a crystallographically related neighboring molecule: the binding site residues interact with the three mannose residues of the branched oligosaccharide (Manα(1,3)[Manα(1,6)]Manβ(1,4)GlcNAcβ(1,4)GlcNAc) attached to Asn76 from an adjacent molecule (Figure 3A and B). Inhibition studies confirmed that the amino-terminal binding site of the CI-MPR is able to recognize nonphosphorylated mannose-containing disaccharides of various linkages, although at an approximately 150–400-fold lower affinity than that observed for Man-6-P (Figure 4). The terminal α1-3-linked mannose (M3) is in the predicted binding site, with its 6-OH linked to the hydroxyl group of Ser386 through two water molecules (Figure 3A). The residues involved in the binding of the oligosaccharide are consistent with previous mutagenesis studies that identified Gln348, Arg391, Glu416, and Tyr421 as essential for high-affinity Man-6-P binding (Dahms et al, 1993; Hancock et al, 2002a), and are also conserved in the CD-MPR (Gln66, Arg11, Glu133, and Tyr143) (see Figure 2C). In addition, comparison of the structures of the two binding pockets reveals that: (1) the terminal α1-3-linked mannose (M3) of domains 1–3 is in the same position as the terminal Man-6-P of the bound ligand in CD-MPR, and (2) the four residues (Gln, Arg, Glu, Tyr) involved in Man-6-P binding are topologically conserved (Figure 3B and C). Ser387, also shown to be essential for Man-6-P binding (Hancock et al, 2002a), does not make direct contact with the oligosaccharide, but is near the binding pocket and close (7.3 Å) to the 6-OH group of the terminal α1–3-linked mannose moiety (Figure 3B). It is possible that Ser387 would make a hydrogen bond with the phosphate of the terminal α1–3-linked mannose, if the latter were phosphorylated. Clearly, additional studies are needed to reveal the role of the 6-phosphate moiety in carbohydrate recognition by the CI-MPR. In addition to the α1–3-linked mannose, the receptor makes several hydrogen-bonding interactions with the remaining two mannose residues. Glu323 is hydrogen bonded to the 4-OH and Tyr324 to the 6-OH, respectively, of the α1–6-linked mannose (M2, Figure 3A), while the 4-OH of the penultimate mannose moiety (M1, Figure 3A) forms hydrogen bonds with both Lys350 and Tyr324. These additional interactions between the mannose residues and the polypeptide are most likely responsible for the ability of the receptor to bind the oligosaccharide attached to Asn76 of the neighboring molecule, rather than to Man-6-P present in the crystallization medium.
Figure 3.
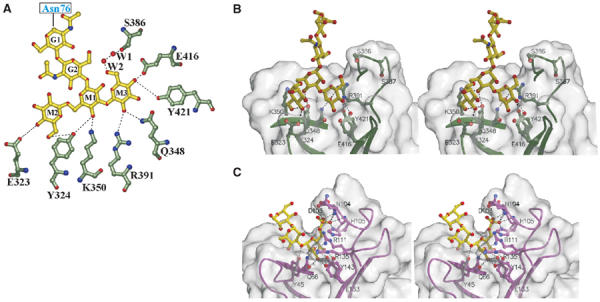
Man3GlcNAc2-binding site. (A) Flattened view of the ligand-binding site. Oligosaccharide residues are shown with the single letter code G for GlcNAc and M for mannose. M3 is joined to M1 via an α1–3 linkage, whereas M2 is joined to M1 via an α1–6 linkage. The two water molecules (W1 and W2) are bridging between the 6-hydroxyl of M3 and Ser386. Gln348, Arg391, Glu416, and Tyr421 correspond to conserved residues found in the Man-6-P-binding pocket of the CD-MPR crystal structure (Roberts et al, 1998; Olson et al, 1999), and previous mutagenesis studies demonstrate the essential nature of these residues for Man-6-P binding by a construct encoding domains 1–3 of the CI-MPR (Dahms et al, 1993; Hancock et al, 2002a). (B) Stereo diagram of the ligand-binding pocket of domains 1–3 showing the molecular surface of the protein. Potential hydrogen bonds between side chains and ligand are shown by the dotted lines. (C) Stereo diagram of the ligand-binding pocket of the CD-MPR in complex with pentamannosyl phosphate (PDB code, 1C39), showing the molecular surface of the protein. Potential hydrogen bonds between side chains and ligand are shown by the dotted lines.
Figure 4.
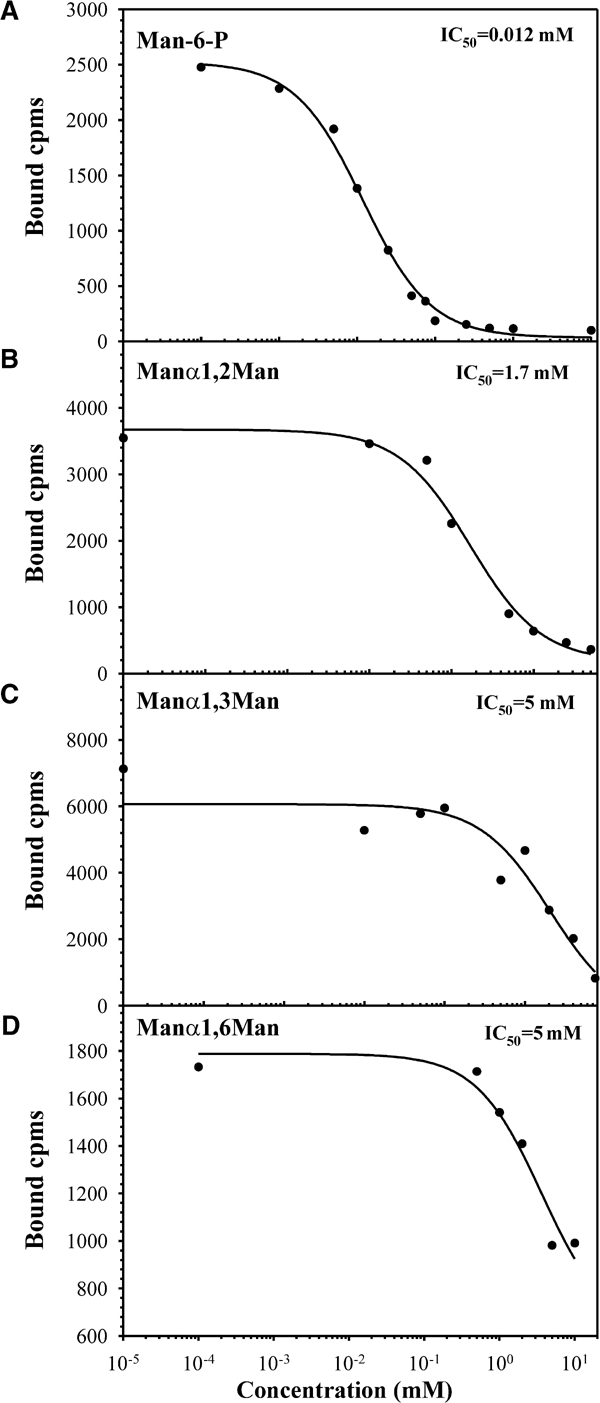
Inhibition of β-glucuronidase binding to domains 1–3His by Man-6-P or nonphosphorylated disaccharides. Purified domains 1–3His were incubated with 125I-β-glucuronidase (0.5 nM) in the presence of increasing concentrations of Man-6-P (A), Manα1,2Man (B), Manα1,3Man (C), or Manα1,6Man (D). The receptor and bound ligand were immunoprecipitated with bovine CI-MPR-specific antiserum pre-bound to protein A-sepharose beads and washed, and the bound β-glucuronidase was eluted specifically with 5 mM Man-6-P.
The binding pocket appears to be wide and shallow, with only ∼16% of the solvent-accessible surface area of the three mannose residues of the oligosaccharide being buried in the protein (Figure 3B). In contrast, the CD-MPR, which is specific for phosphomonoesters, has a narrow and deep binding pocket, with 97, 62, and 38% of the terminal, penultimate, and prepenultimate mannose residues, respectively, of the linear pentamannosyl phosphate oligosaccharide being buried in the protein (Olson et al, 1999) (Figure 3C). Taken together, the shallow binding pocket of the N-terminal carbohydrate recognition site of the CI-MPR likely accounts for the relative promiscuity observed in its recognition of related carbohydrate structures that include mannose 6-sulfate and phosphodiesters (Marron-Terada et al, 2000).
The current structure also reveals the existence of close interactions between the regions containing the carbohydrate-binding residues of domain 3 and segments of domain 1: the loop between β6 and β7 containing Ser386Ser387 and the region encompassing Glu416 and Tyr421 of domain 3 make salt bridges and hydrogen bonds with both the loop between β7 and β8 of domain 1 and the connecting region between domains 1 and 2 (Ala122–Phe128). These interactions contribute to the formation and stabilization of the carbohydrate-binding cavity of domain 3, and are most likely responsible for the reduced Man-6-P-binding affinity observed for a construct encoding domain 3 alone (Kd=500 nM) compared to domains 1–3 (Kd=0.5 nM) (Hancock et al, 2002b).
Proposed uPAR and plasminogen-binding site
Expression of various truncated versions of the CI-MPR has localized the proposed uPAR and plasminogen-binding sites to the N-terminal half of domain 1 (Leksa et al, 2002). The corresponding region (residues 1–73) in our model encompasses the N-terminus through a part of β6, which consists of the first sheet of the flattened β-barrel plus two β-strands of the second sheet. β1 of domain 1 interacts with -1β of the linker region of domain 2, forming a hydrophobic base to the crevice with approximate overall dimensions of 20 Å parallel to β1, 10 Å between domains 1 and 2, and 8 Å deep (Figure 5). The sides of the crevice are formed by β2 of domain 1, which has an overall positive electrostatic potential, as does the other side of the crevice, which is formed by β5 of domain 2. Recent studies by Stockinger and co-workers (Godar et al, 1999; Leksa et al, 2002) demonstrated that the binding of plasminogen or uPAR to the CI-MPR can be abolished in the presence of tranexamic acid, a lysine analog. An 18-residue synthetic peptide encompassing Lys29 of the CI-MPR was also shown to inhibit the interaction of the CI-MPR with uPAR (Leksa et al, 2002). In addition, previous studies have shown that the kringle domains of plasminogen interact with lysine residues of fibrin and cell surface receptors (Plow et al, 1995). The current structure demonstrates the presence of Lys29 located on β1 of domain 1 of the CI-MPR as well as two nearby lysines (Lys132 and Lys215) in domain 2 that are situated on the same face of the molecule (Figure 5), and it is likely that this entire surface participates in interactions with uPAR and plasminogen. Additional studies will be required to probe whether these lysine residues serve as critical determinants for plasminogen and/or uPAR binding. Of note is the location of the proposed uPAR/plasminogen-binding site relative to the carbohydrate recognition site: the two ligand-binding sites are situated on opposite faces of the molecule (Figure 5A). Thus, it is possible that the CI-MPR is able to bind simultaneously both uPAR/plasminogen and an oligosaccharide, as evidenced by the observation that the binding of membrane-associated uPAR is not inhibited by the presence of Man-6-P (Nykjaer et al, 1998).
Figure 5.
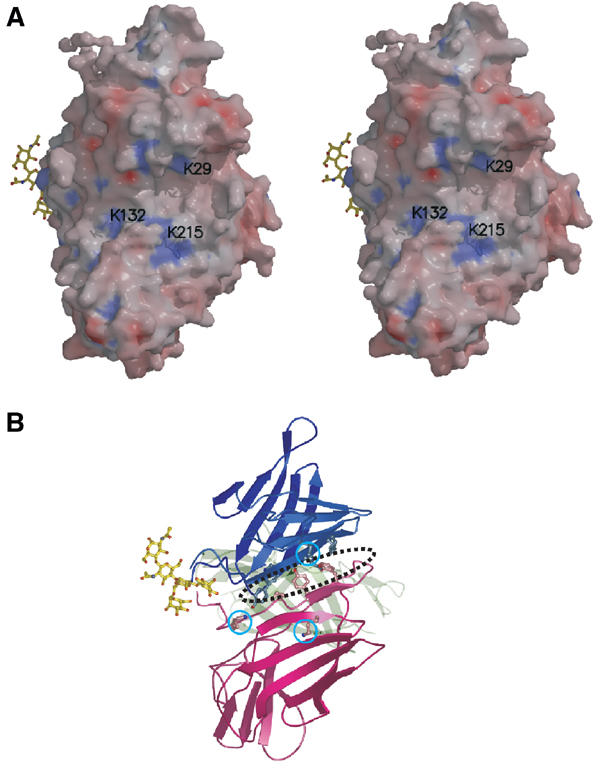
uPAR/plasminogen-binding site. The view (rotated 90° left along the y-axis and 90° up along the x-axis relative to the one shown in Figure 2A) shows the surface containing the first β-sheet of domain 1 and the second β-sheet of domain 2, forming the ‘base' of the wedge-shaped domains 1–3. (A) Stereo diagram of the molecular surface colored by electrostatic potential. Values range from −15 kBT (red) to +15 kBT (blue), with zero being neutral, where kB is Bolzmann's constant and T the absolute temperature. Solvent-exposed lysine residues in the vicinity of the proposed uPAR/plasminogen-binding site are labeled. The oligosaccharide from a crystallographic neighbor is shown in the binding site of domain 3 for orientation purposes. (B) Corresponding ribbon diagram showing the aromatic residues located at the bottom of the crevice between domains 1 and 2, along with the three lysine residues (cyan circles) shown in panel A. A dotted ellipse indicates the position of the surface crevice.
Proposed model for domains 1–15 of the CI-MPR
The CI-MPR is a multifunctional protein that binds carbohydrate (Man-6-P), several proteins (IGF-II, uPAR, plasminogen), and a lipophilic molecule (retinoic acid) at distinct sites. To date, all but the retinoic acid-binding site have been identified and involve sequences within domains 1, 3, 9, 11, and 13. However, the question remains as to how do the 15 domains of the receptor's extracellular region assemble to accomplish these various functions. The packing of the molecules in the orthorhombic crystal allows us to propose a simple model for the extracellular portion of the receptor. The current structure of domains 1–3 shows that the interdomain arrangement among the three domains is rather rigid and therefore can be assumed to be the structural unit. In the orthorhombic space group, the molecules progress along each of the three two-fold screw axes of the unit cell, thus forming an alternating front-to-back pattern of domains 1–3 molecules. Among the three possible stacking arrangements, the one along the y-axis places the N-terminal cysteine and the adjacent neighbor's C-terminal cysteine 16 Å apart, compared to the other two arrangements that are 42 or 57 Å apart. The number of residues between these two points varies from seven residues between domains 9 and 10, to 11 residues between domains 6 and 7. Therefore, even the shortest linker is long enough to span this region. For this reason, we choose the stacking arrangement along the y-axis to generate the 15-domain-containing model representing the entire extracytoplasmic region of the CI-MPR. In our proposed model, five units of the three-domain unit of domains 1–3 were used to stack up in a back-to-front manner to produce a long stalk approximately 210 Å in length (Figure 6A). Digestion with subtilisin (see below) supports domains 1–3 and domains 4–6 existing as stable units. In addition, a structure-based alignment comparing domains 1–3, 4–6, 7–9, 10–12, and 13–15 of the CI-MPR was performed along with the CD-MPR to evaluate whether the residues involved in the contacts among domains 1–3 are conserved in the other proposed 3-domain units. Analysis of the sequence alignment reveals that, while not absolutely conserved, there is a general conservation of residue type in the proposed contact regions between domains (data not shown), indicating that domains 4–6, 7–9, 10–12, and 13–15 can each assemble into a 3-domain unit similar to that of the domains 1–3 structure. Therefore, the proposed model is consistent with the available data and provides a basis for further experimental testing. Brown et al (2002) proposed a model of domains 10–13 of the CI-MPR based on the crystal packing of domain 11. In their model, a single domain was the repeating unit along the two-fold screw axis, which had the effect of orienting the odd domains on one side and even domains rotated 180° on the other side. Unlike the model proposed by Brown et al (2002), the alternating domains in our model are not oriented by a 180° rotation, but rather domains 1 and 3 are rotated relative to each other by approximately 180° (Figure 2A).
Figure 6.
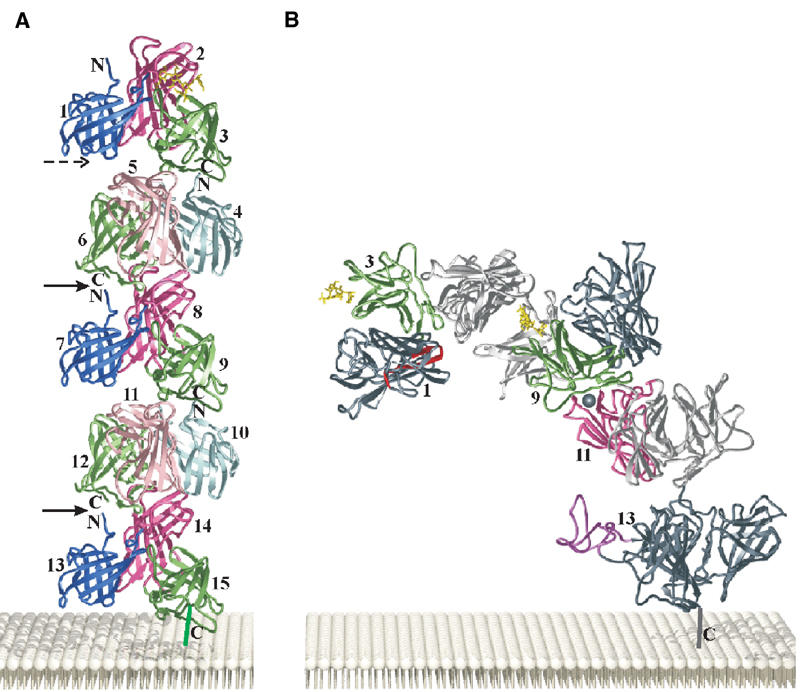
Proposed model of the entire extracytoplasmic region (domains 1–15) of the CI-MPR. (A) Five domain 1–3 structural units are arranged along a crystallographic two-fold screw axis (y-axis) forming the entire extracytoplasmic portion of the CI-MPR. Domains are numbered from the N- to C-termini. The two points where the model has been bent in panel B are indicated by the solid arrows and the dotted arrow indicates another site of flexibility. (B) Generation of a bent model of the CI-MPR to accommodate biochemical data. The proposed uPAR/plasminogen-binding crevice is shown in red. Two oligosaccharides (yellow) bound to domains 3 and 9 (shown in green) are located on the same side of the molecule (front of the page), opposite to the side of the molecule encompassing the uPAR/plasminogen-binding site (back of the page). The proposed IGF-II-binding site (shown as a gray ball) located in domain 11 (pink) is oriented on the same side of the molecule as the 43-residue fibronectin type II repeat of domain 13 (purple) that has been implicated in enhancing the affinity of the CI-MPR for IGF-II. The fibronectin type II structure from human fibronectin (PDB code 2FN2) was used to model this region of the receptor. The 25-residue region of the receptor that connects the C-terminus of domain 15 to the transmembrane region is shown as a vertical line. The 23-residue transmembrane region and the 163-residue cytoplasmic region of the CI-MPR are not shown.
Data from several laboratories indicate that the CI-MPR is capable of forming oligomeric structures, most likely dimers, through its extracytoplasmic domain. Recent studies suggest that the binding of a lysosomal enzyme stabilizes the oligomeric state (York et al, 1999) and that residues in or near domain 12 may play a role in the formation of oligomers (Byrd et al, 2000). However, our current model does not reveal an obvious mechanism for receptor oligomerization.
To accommodate biochemical data on the functional properties of the CI-MPR, a minimal number of bends between the three-domain units were introduced into the model (Figure 6B). Tong et al (1989) reported that an oligosaccharide with two phosphomonoesters bound with high affinity to the CI-MPR. The authors proposed that the two Man-6-P's on the single oligosaccharide (maximum distance between two phosphorylated mannose residues is ∼30 Å) bind to domains 3 and 9 located on the same polypeptide of the CI-MPR. One or two bends were incorporated into our model in an attempt to bring the two carbohydrate-binding sites into close proximity. A bend was applied to the model in the linker region between domains 3 and 4 and/or domains 6 and 7. The flexible nature of these linkers is consistent with proteolytic data indicating that these regions are exposed and susceptible to cleavage by subtilisin (Westlund et al, 1991). The closest distance that could be obtained without steric hindrance was ∼70 Å with one bend (Figure 6B) and ∼45 Å with two bends, indicating that a single diphosphorylated oligosaccharide cannot bind to both domains 3 and 9 located within the same polypeptide. Thus, our model suggests that the observed high-affinity binding is due to the diphosphorylated oligosaccharide spanning two binding sites, with each binding site located on a different polypeptide of the CI-MPR dimer. An intriguing possibility is that the receptor exists in a dynamic state, in which the spacing between the two carbohydrate-binding sites flexes between ∼45 Å (fully bent) and 85 Å (fully extended), thus enhancing the ability of the receptor to interact with a single protein containing multiple oligosaccharides.
The model has one additional bend inserted between domains 12 and 13 to move the fibronectin type II-like region located on domain 13 into the vicinity of domain 11, as sequences in domain 13 have been shown to contribute an ∼10-fold enhancement of IGF-II binding by the receptor (Devi et al, 1998; Grimme et al, 2000; Linnell et al, 2001). Introducing these two bends reduces the height of the entire extracytoplasmic region of the CI-MPR such that the proposed uPAR-binding site of the receptor is located less than 90 Å from the membrane surface, a reasonable height for the three domain-containing uPAR molecule. It has been reported that the binding of IGF-II can inhibit the binding and cellular uptake of a lysosomal enzyme by the CI-MPR (Kiess et al, 1989). In our proposed model, the binding sites for IGF-II and Man-6-P are situated on opposite faces of the molecule. Thus, our model indicates that the binding of the small 7.5 kDa IGF-II polypeptide cannot prevent the binding of a lysosomal enzyme by steric hindrance. However, our model does not rule out the possibility that the binding of IGF-II could induce conformational changes that would perturb carbohydrate-binding affinity. In addition to these three sites (i.e., domains 3/4, 6/7, and 12/13), the linker region between other domains may exhibit some flexibility. The possibility of the CI-MPR adopting multiple conformations is attractive as it could allow the receptor to modulate the spacing of its various ligand-binding sites. This flexibility would clearly be advantageous in the intracellular transport of newly synthesized lysosomal enzymes where the receptor needs to interact with ∼50 different lysosomal enzymes that are likely to exhibit a wide range of spacing of their phosphorylated N-linked oligosaccharides.
Unlike many other receptors, the MPRs travel between numerous organelles and are capable of binding their ligands in the trans-Golgi network and at the plasma membrane. However, only the CI-MPR functions efficiently at the near neutrality of the cell surface. The ability of the MPRs to undergo pH-dependent dissociation with their cargo in the acidic environment (pH<6) of late endosomal compartments is crucial for the proper targeting of lysosomal enzymes, as inhibition of this process results in the excessive secretion of these enzymes (von Figura and Hasilik, 1986). The unique orientation of the three homologous domains revealed in the structure of the N-terminal region of the CI-MPR can now serve as a foundation for future investigations into the structural basis of uPAR and plasminogen recognition and the mechanisms underlying the pH dependence of ligand binding and release.
Materials and methods
Protein expression, purification, and crystallization
Recombinant protein encoding residues 1–432 (domains 1 (residues 1–124), 2 (residues 125–281), and 3 (residues 282–432); domains 1–3) of the mature bovine CI-MPR was cloned and expressed in Trichoplusia ni 5B1-4 cells and purified to near homogeneity by pentamannosyl phosphate-agarose affinity chromatography as described previously (Marron-Terada et al, 2000). Protein crystals were grown at 19°C by vapor diffusion in sitting drops using a 1:1 ratio of well buffer (0.1 M sodium cacodylate, pH 6.35, 25% PEG 4000) and protein solution (8 mg/ml domains 1–3 in 50 mM imidazole, pH 6.75, 150 mM NaCl, 10 mM MnCl2, 5 mM β-glycerophosphate, 10 mM mannose 6-phosphate). Rectangular bar-shaped crystals appeared in approximately 5 weeks. Heavy atom derivatives were generated by soaking crystals in heavy metal-containing solution (0.1 M sodium cacodylate (pH 6.35) and 28% PEG 4000) for the times indicated in Table I, immediately prior to soaking in a cryoprotectant solution. Crystals were incubated for 15 min in the above soaking buffer supplemented with 20% glycerol prior to flash freezing in liquid nitrogen.
Table 1.
Data collection, phase determination, and refinement statistics
| Space group | P212121 | |||||||
|---|---|---|---|---|---|---|---|---|
| Unit cell dimensions (Nativea), Å | 61.2 | 84.8 | 96.1 | |||||
| # monomers/asymmetric unit | 1 | |||||||
| Crystal | Nativeb | Nativea | K2OsCl6b | PIPb | Thimersolb | SmCl3b | K2PtCl6b | K2IrCl6b |
| Resolution (Å) | 30–2.2 | 40–1.8 | 30–2.8 | 30–2.2 | 30–2.5 | 30–2.5 | 30–2.2 | 30–2.2 |
| Rsym (%) | 5.0 (29.4) | 5.9 (37.3) | 7.8 (24.0) | 5.2 (22.5) | 6.0 (26.6) | 8.4 (28.8) | 6.3 (50.9) | 6.3 (50.9) |
| Total observations | 175686 | 184236 | 72292 | 206508 | 91049 | 40992 | 177763 | 136977 |
| Unique reflections | 24298 | 43050 | 12383 | 24117 | 16988 | 15856 | 23894 | 22037 |
| Completenessc (%) | 92.4 (75.9) | 91.7 (52.6) | 93.0 (71.8) | 91.6 (79.8) | 94.6 (84.6) | 87.8 (75.7) | 91.6 (64.4) | 83.8 (58.2) |
| Riso (%) | 10.5 | 15.7 | 7.4 | 8.7 | 8.9 | 8.7 | ||
| Overall figure of merit | 0.53 for 30–3.2 Å resolution data (for initial MIRAS phases) | |||||||
| Phasing power | 0.54 | 0.36 | 0.41 | 0.88 | 0.46 | 0.39 | ||
| Concentration (mM) | 5 | 5 | 5 | 2 | 5 | 1 | ||
| Soaking time (h) | 16 | 5 | 5 | 0.5 | 5 | 2.5 | ||
| No. of sites | 3 | 2 | 2 | 1 | 1 | 2 | ||
| Refinement (with APS data set) | ||||||||
| Rfree (%, 30–1.8 Å) | 28.1 | |||||||
| Rcryst (%, 30–1.8 Å) | 23.2 | |||||||
| R.m.s.d. |
||||||||
| # of residues |
Average B (Å2) |
Bond length (Å) |
Bond angles (deg) |
|||||
| Protein | 427 | 33.9 | 0.006 | 1.4 | ||||
| Water | 362 | 38.0 | ||||||
| Asn76-carbohydrate | 5 | 28.5 | ||||||
| Asn365-carbohydrate | 2 | 46.1 | ||||||
| Glycerol |
|
1 |
49.1 |
|
|
|
|
|
| aData collected at the Advanced Photon Source, beamline BioCARS 14BM-C at 0.9000 Å at −175°C. | ||||||||
| bData collected in-house at −175°C on an R-AXIS IIC image plate detector system with a Rigaku RU200 rotating anode generator operating at 50 kV and 100 mA with Osmic mirrors. | ||||||||
| cValues in parentheses are for the highest resolution shell. | ||||||||
Data collection and structure determination
A native data set of domains 1–3 was collected using an R-AXIS IIC image plate system equipped with a Rigaku RU200 generator, a Rigaku/MSC X-stream cooling system operating at −175°C, and Osmic blue confocal mirrors. The crystals belong to the space group P212121, and the cell dimensions are a=61.2 Å, b=84.8 Å, and c=96.1 Å. Assuming one molecule of domains 1–3 per asymmetric unit, the calculated Matthew's coefficient is 2.47 Å3/Da, which corresponds to ∼50% solvent content in the crystal. High-resolution data were collected at the Advanced Photon Source (Argonne, IL), beam-line 14-BM-C. All data sets were processed using the HKL suite of programs (Otwinowski and Minor, 1997). The structure was solved using multiple isomorphous replacement with anomalous scattering (MIRAS) (Table I). Difference Patterson map analysis was performed in XtalView (McRee, 1999). Initial phases were obtained using SOLVE followed by density modification, and automated model building was performed using RESOLVE (154 alanines placed out of 432 total residues) (Terwilliger and Berendzen, 1999). MIRAS phases from SOLVE were combined with native anomalous data (Native 1) and were used to calculate an anomalous difference Fourier map. The anomalous map verified the 12 disulfide positions in our model as well as the positions of two methionine residues (Met26 and Met49). The sulfur positions were included in further phase refinement, together with the original heavy atom sites using SHARP (La Fortelle and Bricogne, 1997), followed by density modification to improve the map quality. The resulting sulfur positions were used to aid in model building. All model building and manual refinements were carried out using Turbo-Frodo (Roussel and Cambillau, 1994). Model refinement and water addition were carried out using CNS (Brunger et al, 1998). The final Rcryst and Rfree (30–1.8 Å) are 23.2 and 28.1%, respectively. Table I shows data collection and refinement statistics.
Inhibition studies
Inhibition studies were performed as previously described, using a construct encoding domains 1–3 that contains six His residues at its C-terminus (Hancock et al, 2002b). Briefly, the purified receptor was incubated with 125I-β-glucuronidase (0.5 nM) in the presence of increasing concentrations of Man-6-P, or the disaccharides Manα1,2Man, Manα1,3Man, or Manα1,6Man (Dextra Laboratories). The receptor and bound ligand were immunoprecipitated with bovine CI-MPR-specific antiserum pre-bound to protein A-Sepharose beads and the bound β-glucuronidase was eluted specifically from the receptor with 5 mM Man-6-P. The results were analyzed by nonlinear regression (SigmaPlot version 5.05, SPSS Science) to obtain IC50 values.
Acknowledgments
We thank the staff at the APS beamline BioCARS 14BM-C for excellent assistance in data collection. NMD and J-JPK acknowledge the support of NIH grant DK42667. Use of the Advance Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by NCRR grant RR07707 from the NIH. The atomic coordinates have been deposited to the Protein Data Bank, accession code 1Q25.
References
- Blasi F, Carmeliet P (2002) uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol 3: 932–943 [DOI] [PubMed] [Google Scholar]
- Brown J, Esnouf RM, Jones MA, Linnell J, Harlos K, Hassan AB, Jones EY (2002) Structure of a functional IGF2R fragment determined from the anomalous scattering of sulfur. EMBO J 21: 1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Byrd JC, Park JH, Schaffer BS, Garmroudi F, MacDonald RG (2000) Dimerization of the insulin-like growth factor II/mannose 6-phosphate receptor. J Biol Chem 275: 18647–18656 [DOI] [PubMed] [Google Scholar]
- Dahms NM, Hancock MK (2002) P-type lectins. Biochim Biophys Acta 1572: 317–340 [DOI] [PubMed] [Google Scholar]
- Dahms NM, Rose PA, Molkentin JD, Zhang Y, Brzycki MA (1993) The bovine mannose 6-phosphate/insulin-like growth factor II receptor. The role of arginine residues in mannose 6-phosphate binding. J Biol Chem 268: 5457–5463 [PubMed] [Google Scholar]
- Dennis PA, Rifkin DB (1991) Cellular activation of latent transforming growth factor beta requires binding to the cation-independent mannose 6-phosphate/insulin-like growth factor type II receptor. Proc Natl Acad Sci USA 88: 580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi GR, Byrd JC, Slentz DH, MacDonald RG (1998) An insulin-like growth factor II (IGF-II) affinity-enhancing domain localized within extracytoplasmic repeat 13 of the IGF-II/mannose 6- phosphate receptor. Mol Endocrinol 12: 1661–1672 [DOI] [PubMed] [Google Scholar]
- Ghahary A, Tredget EE, Shen Q, Kilani RT, Scott PG, Houle Y (2000) Mannose-6-phosphate/IGF-II receptors mediate the effects of IGF-1-induced latent transforming growth factor beta 1 on expression of type I collagen and collagenase in dermal fibroblasts. Growth Factors 17: 167–176 [DOI] [PubMed] [Google Scholar]
- Ghosh P, Dahms NM, Kornfeld S (2003) Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol 4: 202–213 [DOI] [PubMed] [Google Scholar]
- Godar S, Horejsi V, Weidle UH, Binder BR, Hansmann C, Stockinger H (1999) M6P/IGFII-receptor complexes urokinase receptor and plasminogen for activation of transforming growth factor-beta1. Eur J Immunol 29: 1004–1013 [DOI] [PubMed] [Google Scholar]
- Grimme S, Honing S, von Figura K, Schmidt B (2000) Endocytosis of insulin-like growth factor II by a mini-receptor based on repeat 11 of the mannose 6-phosphate/insulin-like growth factor II receptor. J Biol Chem 275: 33697–33703 [DOI] [PubMed] [Google Scholar]
- Hancock MK, Haskins DJ, Sun G, Dahms NM (2002a) Identification of residues essential for carbohydrate recognition by the insulin-like growth factor II/mannose 6-phosphate receptor. J Biol Chem 277: 11255–11264 [DOI] [PubMed] [Google Scholar]
- Hancock MK, Yammani RD, Dahms NM (2002b) Localization of the carbohydrate recognition sites of the insulin-like growth factor II/mannose 6-phosphate receptor to domains 3 and 9 of the extracytoplasmic region. J Biol Chem 277: 47205–47212 [DOI] [PubMed] [Google Scholar]
- Harata K, Nagahora H, Jigami Y (1995) X-ray structure of wheat germ agglutinin isolectin 3. Acta Crystallogr D 51: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Kang JX, Li Y, Leaf A (1997) Mannose-6-phosphate/insulin-like growth factor-II receptor is a receptor for retinoic acid. Proc Natl Acad Sci USA 94: 13671–13676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiess W, Thomas CL, Greenstein LA, Lee L, Sklar MM, Rechler MM, Sahagian GG, Nissley SP (1989) Insulin-like growth factor-II (IGF-II) inhibits both the cellular uptake of β-galactosidase and the binding of β-galactosidase to purified IGF-II/mannose 6-phosphate receptor. J Biol Chem 264: 4710–4714 [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Kreiling JL, Byrd JC, Deisz RJ, Mizukami IF, Todd RF III, MacDonald RG (2003) Binding of urokinase-type plasminogen activator receptor (uPAR) to the mannose 6-phosphate/insulin-like growth factor II receptor: contrasting interactions of full-length and soluble forms of uPAR. J Biol Chem 278: 20628–20637 [DOI] [PubMed] [Google Scholar]
- La Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- Lau MM, Stewart CE, Liu Z, Bhatt H, Rotwein P, Stewart CL (1994) Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev 8: 2953–2963 [DOI] [PubMed] [Google Scholar]
- Leksa V, Godar S, Cebecauer M, Hilgert I, Breuss J, Weidle UH, Horejsi V, Binder BR, Stockinger H (2002) The N terminus of mannose 6-phosphate/insulin-like growth factor 2 receptor in regulation of fibrinolysis and cell migration. J Biol Chem 277: 40575–40582 [DOI] [PubMed] [Google Scholar]
- Linnell J, Groeger G, Hassan AB (2001) Real time kinetics of insulin-like growth factor II (IGF-II) interaction with the IGF-II/mannose 6-phosphate receptor. The effects of domain 13 and pH. J Biol Chem 276: 23986–23991 [DOI] [PubMed] [Google Scholar]
- Lobel P, Dahms NM, Kornfeld S (1988) Cloning and sequence analysis of the cation-independent mannose 6-phosphate receptor. J Biol Chem 263: 2563–2570 [PubMed] [Google Scholar]
- Ludwig T, Eggenschwiler J, Fisher P, D'Ercole AJ, Davenport ML, Efstratiadis A (1996) Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol 177: 517–535 [DOI] [PubMed] [Google Scholar]
- Ludwig T, Munier-Lehmann H, Bauer U, Hollinshead M, Ovitt C, Lobel P, Hoflack B (1994) Differential sorting of lysosomal enzymes in mannose 6-phosphate receptor-deficient fibroblasts. EMBO J 13: 3430–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marron-Terada PG, Hancock MK, Haskins DJ, Dahms NM (2000) Recognition of Dictyostelium discoideum lysosomal enzymes is conferred by the amino-terminal carbohydrate binding site of the insulin-like growth factor II/mannose 6-phosphate receptor. Biochemistry 39: 2243–2253 [DOI] [PubMed] [Google Scholar]
- McRee DE (1999) Practical Protein Crystallography, 2nd edn. San Diego: Academic Press [Google Scholar]
- Merritt EA, Bacon DJ (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol 277: 505–524 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Christensen EI, Vorum H, Hager H, Petersen CM, Roigaard H, Min HY, Vilhardt F, Moller LB, Kornfeld S, Gliemann J (1998) Mannose 6-phosphate/insulin-like growth factor-II receptor targets the urokinase receptor to lysosomes via a novel binding interaction. J Cell Biol 141: 815–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Rozek LM, Czech MP (1985) Direct demonstration of rapid insulin-like growth factor II. Receptor internalization and recycling in rat adipocytes. Insulin stimulates 125I-insulin-like growth factor II degradation by modulating the IGF-II receptor recycling process. J Biol Chem 260: 9435–9442 [PubMed] [Google Scholar]
- Olson LJ, Zhang J, Dahms NM, Kim J-JP (2002) Twists and turns of the CD-MPR: ligand-bound versus ligand-free receptor. J Biol Chem 277: 10156–10161 [DOI] [PubMed] [Google Scholar]
- Olson LJ, Zhang J, Lee YC, Dahms NM, Kim J-JP (1999) Structural basis for recognition of phosphorylated high mannose oligosaccharides by the cation-dependent mannose 6-phosphate receptor. J Biol Chem 274: 29889–29896 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Plow EF, Herren T, Redlitz A, Miles LA, Hoover-Plow JL (1995) The cell biology of the plasminogen system. FASEB J 9: 939–945 [DOI] [PubMed] [Google Scholar]
- Pohlmann R, Boeker MW, von Figura K (1995) The two mannose 6-phosphate receptors transport distinct complements of lysosomal proteins. J Biol Chem 270: 27311–27318 [DOI] [PubMed] [Google Scholar]
- Preissner KT, Kanse SM, May AE (2000) Urokinase receptor: a molecular organizer in cellular communication. Curr Opin Cell Biol 12: 621–628 [DOI] [PubMed] [Google Scholar]
- Roberts DL, Weix DJ, Dahms NM, Kim J-JP (1998) Molecular basis of lysosomal enzyme recognition: three-dimensional structure of the cation-dependent mannose 6-phosphate receptor. Cell 93: 639–648 [DOI] [PubMed] [Google Scholar]
- Roussel A, Cambillau C (1994) TURBO-FRODO. The Manual (Marseille, France, Biographics, LCCMB)
- Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J (2002) Structure of the LDL receptor extracellular domain at endosomal pH. Science 298: 2353–2358 [DOI] [PubMed] [Google Scholar]
- Sohar I, Sleat D, Gong Liu C, Ludwig T, Lobel P (1998) Mouse mutants lacking the cation-independent mannose 6- phosphate/insulin-like growth factor II receptor are impaired in lysosomal enzyme transport: comparison of cation-independent and cation-dependent mannose 6-phosphate receptor-deficient mice. Biochem J 330: 903–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated structure solution for MIR and MAD. Acta Crystallogr D 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong PY, Gregory W, Kornfeld S (1989) Ligand interactions of the cation-independent mannose 6-phosphate receptor. The stoichiometry of mannose 6-phosphate binding. J Biol Chem 264: 7962–7969 [PubMed] [Google Scholar]
- Uson I, Schmidt B, von Bulow R, Grimme S, von Figura K, Dauter M, Rajashankar KR, Dauter Z, Sheldrick GM (2003) Locating the anomolous scatterer substructures in halide and sulfur phasing. Acta Crystallogr D 59: 57–66 [DOI] [PubMed] [Google Scholar]
- von Figura K, Hasilik A (1986) Lysosomal enzymes and their receptors. Annu Rev Biochem 55: 167–193 [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Fung MR, Barlow DP, Wagner EF (1994) Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature 372: 464–467 [DOI] [PubMed] [Google Scholar]
- Westlund B, Dahms NM, Kornfeld S (1991) The bovine mannose 6-phosphate/insulin-like growth factor II receptor. Localization of mannose 6-phosphate binding sites to domains 1–3 and 7–11 of the extracytoplasmic region. J Biol Chem 266: 23233–23239 [PubMed] [Google Scholar]
- York SJ, Arneson LS, Gregory WT, Dahms NM, Kornfeld S (1999) The rate of internalization of the mannose 6-phosphate/insulin-like growth factor II receptor is enhanced by multivalent ligand binding. J Biol Chem 274: 1164–1171 [DOI] [PubMed] [Google Scholar]


