Abstract
Click Chemistry is a set of rapid, selective and robust reactions that give near-quantitative yield of the desired product in aqueous solutions. The Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) that forms 1,4-disubstituted triazoles is a prototypical example of click chemistry that features exquisite selectivity and bioorthogonality—that is, non-interacting with biological components while proceeding under physiological conditions. Over the past ten years, CuAAC has found extensive applications in the field of chemical biology. In this chapter, we describe the discovery of Cu(I) catalysts for this transformation and the recent development of the strain-promoted azide-alkyne cycloaddition that eliminate the use of copper. We also highlight several recent applications toward conjugating biomolecules, including proteins, nucleic acids, lipids and glycans, with biophysical probes for both in vitro and in vivo studies.
Keywords: Bioconjugation, Bioorthogonal, Click chemistry
1 Introduction
Over the past forty years, a growing synergy has developed between the fieldd of organic chemistry and the modern biosciences. The discovery of chemoselective transformations has enabled the incorporation of probes and tags onto biomolecules to create new therapeutic agents or research tools. For example, covalent attachment of polyethylene glycol (PEG) chains, a process termed PEGylation, is routinely practiced in pharmaceutical companies to improve pharmacokinetics of therapeutic proteins. This modification is often realized by exploiting the electrophilic reactivity of succinimidyl carbonate-functionalized PEGs for ε-amino groups of lysine residues or the N-terminal amino acid of target proteins. However, since proteins often possess multiple copies of lysine residues, site-specific labeling can be difficult to achieve. PEG molecules randomly introduced to a target protein may interfere with its desired biological function, hampering its therapeutic application. To address this challenge, researchers have developed genetically encoded unnatural amino acids [1] and peptide tags [2, 3] for introducing novel functional groups beyond those found in the 20 canonical amino acids. These new tagging methods, combined with tools from the emerging field of bioorthogonal click chemistry [4], have conferred site specificity to protein modification manipulations. Indeed, orthogonality and selectivity, two central factors that govern the success of these approaches, have been increasingly adopted by chemical biologists in recent years. Today, bioorthogonal click chemistry has found applications beyond protein science and offers reliable methods for bioconjugation in the covalent labeling of biological macromolecules, including proteins, nucleic acids, glycans, and lipids [5]. Featuring benign reaction conditions and unprecedented selectivity, bioorthogonal click chemical transformations are the first choice for in situ labeling of biomolecules in living systems.
2 CuAAC and Copper-Free Click Chemistry
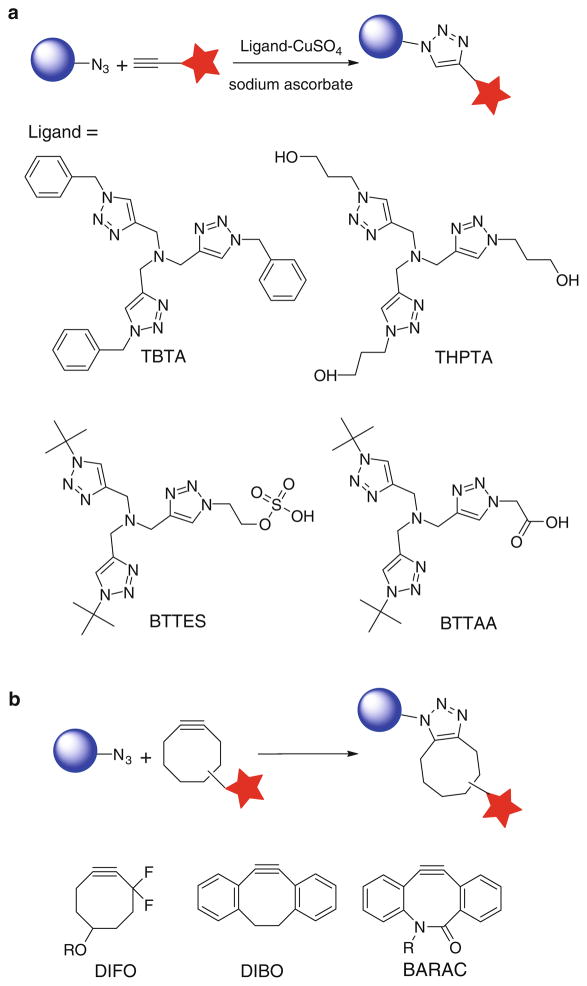
Discovered by Sharpless-Fokin and Meldal in 2002, the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) that forms 1,4-disubstituted triazoles is a prototypical example of bioorthogonal click chemistry (Fig. 1a) [6, 7]. This cycloaddition reaction is accelerated by Cu(I) approximately seven orders of magnitude compared to the uncatalyzed version [8, 9]. As a ligand-assisted process, the reaction is further accelerated by Cu(I)-stabilizing ligands, such as tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) and tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) [10, 11]. Small size and inertness of azides and terminal alkynes make them popular tags to be incorporated into biomolecules, and their detections are realized by using probes functionalized in a complementary fashion via the CuAAC. The simple procedure and exquisite selectivity of this transformation has gained it widespread utilization in bioorthgonal conjugation [12, 13].
Fig. 1.
(a) Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC)- and (b) copper-free click chemistry
In the past few years, several protocols have been developed for the CuAAC-mediated bioconjugation, in which CuSO4 or CuBr is used as the copper source, sodium ascorbate or tris(2-carboxyethyl)phosphine (TCEP) as the reducing agent, and TBTA as the Cu(I)-stabilizing ligand [5]. However, these formulations have two major problems: toxicity, hindering the use of CuAAC in living systems [5, 14], and slow kinetics in aqueous solutions at micromolar substrate concentrations [15], resulting in incomplete cycloaddition reaction [16] and hampering the modification of biomolecules of limited quantities. TBTA, the ligand utilized in the canonical CuAAC conditions to stabilize the Cu(I) oxidation state, has very poor water solubility, which mandates the use of high Cu loading (0.2–1 mM) to achieve reasonable reaction rates. Free Cu(I) ions that escape from the coordination sphere of TBTA promote the generation of reactive oxygen and nitrogen species and induce detrimental consequences to cellular metabolism [17]. For example, Escherichia coli that incorporated azidohomoalanine into their outer membrane protein C survived the initial treatment with 100 μM CuBr for 16 h, but were no longer able to divide [18]. Similarly, greater than 90% of mammalian cells underwent apoptosis and cell lysis within 20 min when treated with 1 mM Cu(I) under optimized CuAAC conditions [5]. Zebrafish embryos exhibited a similar sensitivity to Cu(I). Following treatment with 1 mM CuSO4, 1.5 mM sodium ascorbate, and 0.1 mM TBTA ligand, none of the embryos survived beyond 15 minutes [5].
To improve upon the biocompatibility of the CuACC and extend its application into living systems, our laboratory has developed a new generation of tris (triazolylmethyl)amine-based ligands for CuAAC, such as 2-(4-((bis((1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl) ethyl hydrogen sulfate (BTTES) [15] and 2-[4-({bis[(1-tert-butyl-1H-1,2,3-triazol-4-yl) methyl]amino}methyl)-1H-1,2,3-triazol-1-yl]acetic acid (BTTAA) (Fig. 1a) [19]. These ligands not only dramatically accelerate the rate of the azide-alkyne cycloaddition by coordinating with the in situ generated Cu(I), but they also rendered the CuAAC biocompatible. At physiological conditions (pH = 7.4), the hydrogen sulfate group in BTTES and acetic acid in BTTAA ionize to negatively charged groups, i.e., sulfate and acetate, respectively, securing the solubility of the ligand–Cu(I) complex and preventing cellular internalization of the coordinated copper ions.
Alternatively, the Bertozzi laboratory pioneers the development of a copper-free [3+2] cycloaddition reaction by employing ring strain as a driving force for alkyne activation (Fig. 1b) [20, 21]. Among the cyclooctyne derivatives developed by the Bertozzi and the Boons groups, 3,3-difluorocyclooct-1-yne, DIFO [22]; 4-dibenzocyclooctynol (DIBO) [23]; and biarylazacyclooctynone, BARAC [24], showed rapid kinetics in biomolecular labeling experiments. Particularly, fluorescently labeled DIFO reagents have been elegantly used for imaging azide-tagged glycans within complex biological systems, including live cells [22], C. elegans [25], and zebrafish embryos [26, 27], with very low background fluorescence. However, recent in vivo studies revealed that DIFO-based probes have nonspecific reactivity toward mouse serum albumin, presumably via covalent-bond formation between the cyclooctyne and cysteine residues [28].
Today, CuAAC and copper-free click chemistry are serving as complementary approaches to modify biomolecules for both in vitro and in vivo applications. In this chapter, we will highlight the recent applications of these triazole-forming reactions to the imaging and profiling of biomolecules. Comprehensive reviews of click chemistry-based approaches for bioconjugation have been published recently [12, 29].
3 Triazole Formation-Based Bioconjugation for Imaging Cell-Surface Proteins and Newly Synthesized Proteins
The homeostasis of cellular proteins is tightly regulated by synthesis and degradation machineries in response to their ever-changing microenvironment. Inside a live cell, proteins are continuously synthesized to replenish those that have been naturally metabolized or degraded in response to stimuli. Techniques that enable the visualization of proteins in their cellular environment are powerful tools for analyzing proteins’ cellular dynamics and functions. Popular methods for visualizing proteins in their native environments rely on genetically encoded fluorescent proteins (e.g., GFP). By fusing a target protein to GFP, a protein of interest can be visualized in live cells or organisms. However, the large size of GFP may interfere with the structure and function of the fused protein.
Alternatively, a consensus peptide sequence that harbors a specific site for bioorthogonal conjugation can be exploited to modify a target protein for in vivo imaging. By treating the tagged protein on the cell surface with the enzyme that adds a synthetic prosthetic group analog bearing the desired orthogonal functionality, the protein can then be labeled with an imaging probe via bioorthogonal click reactions. Biotin ligase [30], formylglycine-generating enzyme [31, 32], and transglutaminase [33] have all been employed for this purpose. For example, Ting and coworkers redirected a microbial lipoic acid ligase (LplA) [34] to specifically attach an alkyl azide onto an engineered LplA acceptor peptide (LAP) fused to cell-surface proteins in live mammalian cells. The alkyl azide was then selectively labeled with fluorescently conjugated cyclooctyne probes to image the dynamics of the labeled proteins (Fig. 2) [35]. Such methods are ideal for studying the dynamics and distribution of one or a few preselected proteins. Since genetic manipulation is required for tagging a fluorescent protein or a peptide tag to the protein of interest, it is difficult to apply these methods to monitor the newly synthesized proteins at a global level.
Fig. 2.
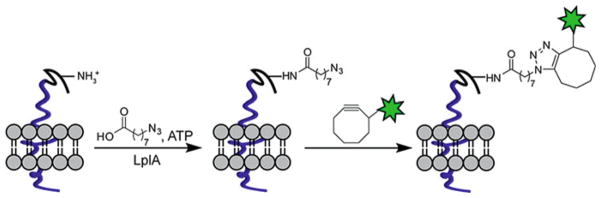
Site-specific labeling of cell-surface proteins using lipoic acid ligase
In order to analyze the global protein synthesis, researchers in the Tirrell group have developed a method called BONCAT (bioorthogonal noncanonical amino acid tagging), a two-step labeling approach. In the first step, the methionine analogue azidohomoalanine (AHA) or homopropargylglycine (HPG) (Fig. 3) is metabolically incorporated into the newly synthesized proteins. In the second step, an alkyne- or azide-bearing fluorophore is used to detect these proteins in fixed cells via TBTA-mediated CuAAC [36].
Fig. 3.
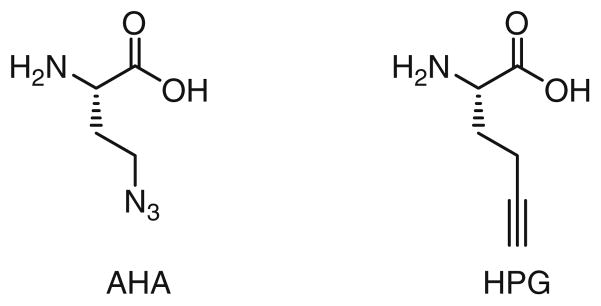
Methionine analogues used for metabolically labeling of newly synthesized proteins
They successfully applied this method for visualizing newly synthesized proteins in rat hippocampal neurons. After a 1-h incubation with either AHA or HPG, newly synthesized proteins can be detected in both the somata and dendrites. Newly synthesized proteins in spine-like protrusions from dendrites can be detected after a 2-hour exposure to AHA or HPG.
In order to monitor the newly synthesized proteins inside live cells, the Tirrell lab has developed a set of cell-permeable coumarin-functionalized cyclooctynes. These coumarin cyclooctynes would ligate to the cellular azide-bearing proteins, allowing visualizing the newly synthesized proteins dynamically [37].
4 Triazole Formation-Based Bioconjugation for Imaging and Profiling Protein Posttranslational Modifications
We have come a long way in the journey toward understanding the molecular basis of life. Half a century ago, Francis Crick proposed the central dogma as a working model that credits the three-dimensional structure and function of proteins to their one-dimensional progenitors: DNAs and RNAs [38]. However, the discovery of posttranslational modification (PTM) makes it clear that the structure and function of proteins and polypeptides can be dictated by factors other than their encoding nucleic acids and amino acid building blocks [39].
A major task in the postgenomic era is to assign specific functions of individual proteins [40]. Genetic tools such as gene microarrays and RNA interference have played major roles in revealing the expression patterns and functions of target genes [41]. However, these techniques are insensitive to most PTM that are under spatiotemporal control—processes that modulate protein folding [42], functional states [43], and their subcellular localizations [41].
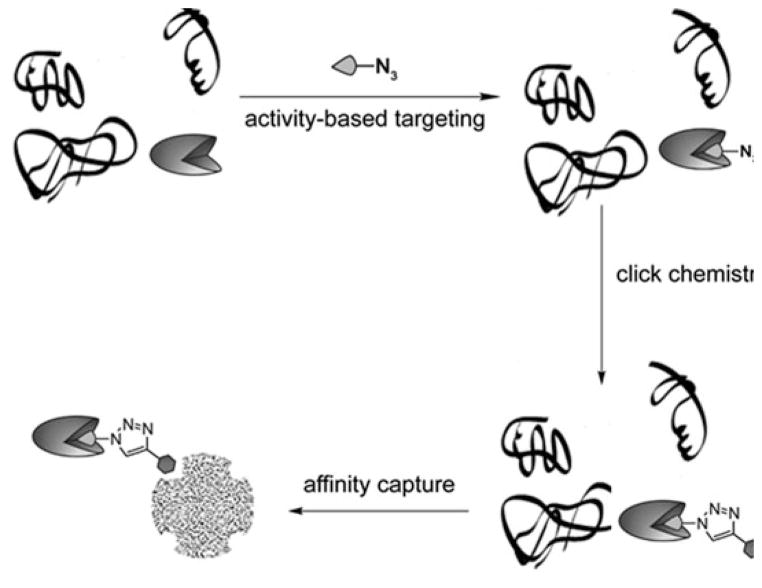
In the past decade, bioconjugation techniques based on CuAAC have emerged as new tools for profiling protein PTMs, including poly(ADP-ribosyl)ation, methylation, acylation, and glycosylation. In these methods, an azide or an alkyne moiety is first introduced to a natural substrate (e.g., cofactor monosaccharide, etc.) of a PTM enzyme. The modified substrate is then processed by its respective endogenous PTM enzymes to modify the substrate proteins in their native environment or by a recombinant PTM enzyme in crude cell lysates. In a subsequent step, an imaging probe can be introduced, allowing the visualization of the labeled biomolecules in living systems or in sodium dodecyl sulfate-polyacrylamide gel. Alternatively, the tagged proteins can be conjugated with an affinity probe via CuAAC and isolated for proteomic analysis and functional studies.
4.1 Profiling Protein Poly(ADP-ribosyl)ation
In 2010, the Lin group created a clickable analogue of nicotinamide adenine dinucleotide (NAD), an ADP-ribose donor, to identify novel substrates of poly (ADP-ribose) polymerases (PARPs). PARPs mediate protein poly(ADP-ribosyl) ation by adding one or more ADP-ribose moieties posttranslationally to target proteins. Seventeen different variants of PARP have been identified in humans, and they are generally known to play an important role in DNA repair, transcription, and mitosis. In order to understand the functions of individual PARPs, it is necessary to know which substrate proteins each PARP is modifying. Using an alkyne-tagged NAD analogue specific to PARP-1 in a PARP-catalyzed reaction, Lin and coworkers enabled the conjugation of affinity tags to the substrates of PARP-1 and performed affinity purification under denaturing conditions in order to minimize nonspecific interactions within the cell lysate and give fewer false-positive results than the alternative methods (Fig. 4). Following affinity purification, they were able to identify over 70 candidate substrates for PARP-1 by tandem mass spectrometry, 40% of which had been previously identified by other methods [44].
Fig. 4.
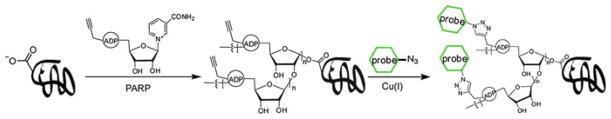
Identifying substrates of poly(ADP-ribose) polymerases using a CuAAC-based strategy
4.2 Profiling Protein Fatty Acylation
Protein fatty acylation plays an important role in normal eukaryotic cell function and signaling as well as in human disease. Conventional approaches for detecting protein fatty acylation involve metabolic labeling with radiolabeled fatty acids and subsequent analysis using autoradiography. Such methods are hazardous and not compatible with downstream proteomic studies. To circumvent these limitations, the Hang group developed an alternative approach to metabolically treat mammalian cells with alkyne-modified fatty acids and labeled the alkyne-incorporated proteins with a biotin-azide probe by CuAAC for affinity capture and MS identification (Fig. 5). Using this approach, Hang and coworkers were able to identify from Jurkat cells 57 known fatty-aceylated proteins and 109 new candidate proteins with high confidence, including histone H3 variants [45]. Recently, the same approach was applied to profile palmitoylated proteins in dendritic cells, a type of phagocytic cells in mammalian immune system. Protein S-palmitoylation is a PTM that occurs on cysteine residues that facilitates targeting and function of membrane-associated proteins [46]. Due to the diversity of S-palmitoylation in eukaryotic cells, it was suggested that multiple cellular pathways could be regulated by this PTM. Hang et al. performed large-scale profiling of S-palmitoylated proteins in dendritic cell line DC2.4 and found that interferon-induced transmembrane protein 3 (IFITM3), a protein associated with the host immune response to microbial infection, was palmitoylated. Further study revealed that S-palmitoylation of IFITM3 is responsible for the clustering of this protein in the ER membrane, and this PTM is also critical for IFITM3’s antiviral activity against influenza viruses [47].
Fig. 5.
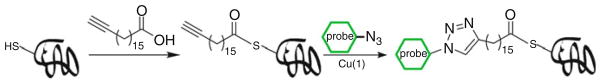
Two-step labeling strategy for detecting S-palmitoylated proteins
4.3 Imaging Cellular Glycans
In nature, nine monosaccharides are used as building blocks to produce a variety of polysaccharides that decorate the surface of eukaryotic cells. These glycans are critical mediators of many biological processes, including angiogenesis, fertilization, cell–cell communication, and neuronal development [48]. Inside the cell, glycans regulate fundamental cellular processes including transcription, translation, and protein trafficking [49]. Glycosylation is a dynamic process that reports on a cell’s physiological state. When a cell undergoes a transformation of state, the glycans on its cell surface will change. In many cases, altered glycosylation, i.e., over- and down-expression of naturally occurring glycans and the appearance of glycans normally restricted to embryonic expression, is a hallmark of tumor phenotype [50]. Therefore, the ability to visualize the dynamic changes of glycans in vivo will augment our understanding of glycans’ physiological roles in developmental processes and in malignant transformation.
Glycosylation is a posttranslational modification, not under direct genetic control. Therefore, genetic reporters (e.g., GFP) used for in vivo protein imaging cannot be exploited for visualizing glycans. Instead, imaging of glycans is rendered possible by integrating bioorthogonal click chemistry with metabolic oligosaccharide engineering. By hijacking a cell’s biosynthetic machinery, monosaccharide building blocks functionalized with bioorthogonal chemical tags can be incorporated into target glycans. In a subsequent step, a complementary imaging probe is conjugated via a bioorthogonal click reaction, enabling the visualization of the tagged glycoconjugates (Fig. 6) [51].
Fig. 6.
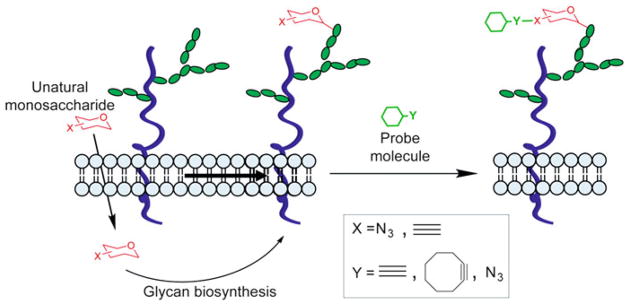
Detection of cell-surface glycans using metabolic labeling and triazole-forming click chemistry
Since the breakthrough discovery by the Bertozzi group in 1997 that bioorthogonal ligation can take place on the surface of live cells [52], several different classes of glycans have been detected using this method by employing the azide or the alkyne as the chemical reporter (tag) (Table 1) [51]. Detection of the azide-bearing glycans in live cells or living organism is realized using the Bertozzi–Staudinger ligation [53] or the triazole-forming reactions [51]. Likewise, detection of the alkyne-bearing glycans in live cells can be accomplished using the biocompatible CuAAC (Table 1) [15].
Table 1.
Azide- and alkyne-bearing monosaccharides used for metabolic labeling of glycans
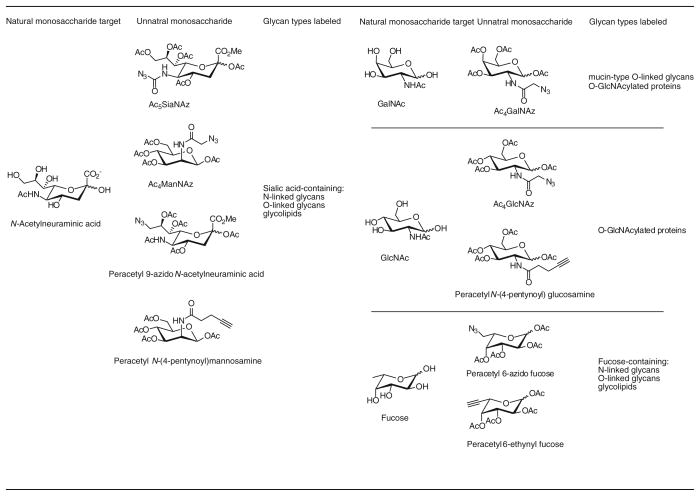
|
The first example of triazole formation-enabled glycan imaging in a living organism was reported by the Bertozzi group in 2007, in which the dynamic glycosylation in zebrafish was revealed by fluorescence imaging [26]. They cultured zebrafish embryos in media supplemented with peracetylated N-azidoacetylgalactosamine (Ac4GalNAz) to metabolically label O-linked glycans. The embryos were then reacted with fluorescently labeled DIFO, allowing the visualization of the O-linked glycans tagged by the azide. They further demonstrated that dynamic O-linked glycosylation can be visualized by quenching unreacted azide groups on the cell surface of the labeled embryos with TCEP, pulsing labeling the embryos with additional GalNAz, and probing the embryos with a blue-shifted DIFO conjugate [26]. Using this method, they discovered that newly synthesized O-glycans are primarily located in the fins, jaw, and olfactory organs. One obvious advantage of this technique is better tissue accessibility of the DIFO-based imaging probes compared to what can be achieved by using bulky lectins or antibodies. However, robust labeling requires 1-hour incubation time, limiting its application to follow a fast biological process taking place in minute time scale [26].
The discovery of the BTTES- and BTTAA-mediated CuAAC overcomes this obstacle and paves the way for rapid imaging of glycans in vivo. This method allowed the visualization of fucosylated glycans in developing zebrafish for the first time [15]. Our lab showed that by using guanidine 5′-diphosphate-β-6-ethynyl fucose (GDP-FucAl) directly as a metabolic precursor to bypass the guanidine 5′-diphosphate-β-L-fucose (GDP-fucose) salvage pathway, fucosylated glycans in zebrafish embryos can be labeled with an alkyne tag. Upon a 3-minute click reaction with a fluorophore-conjugated azide, the fucosylated glycans in enveloping layer, the embryos’ outermost layer of cells, were lightened up for fluorescence visualization, and the labeled glycans were detectable as early as 2.5 hour postfertilization (hpf) (Fig. 7) [15]. Notably, a single dose of GDP-FucAl could yield a detectable fluorescent signal up to 96 hpf.
Fig. 7.
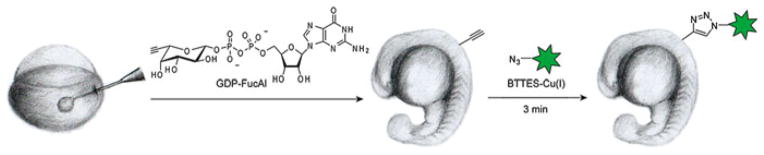
Imaging fucosylated glycans in developing zebrafish embryos
However, the metabolic method is only applicable for monosaccharide labeling. Since each monosaccharide is usually found in a variety of cell-surface polysaccharide glycans, higher-order glycans, such as disaccharides or trisaccharides, of specific composition cannot be uniquely labeled by hijacking their biosynthetic pathways with unnatural monosaccharides. As the first step to overcome this challenge, our lab developed a chemoenzymatic approach that has been successfully applied to label cell-surface polysaccharides bearing N-acetyllactosamine (LacNAc; Galβ1,4GlcNAc) building blocks.
This method employs a recombinant Helicobacter pylori α(1,3)fucosyltransferase [54] to transfer a C-6 azide- or alkyne-tagged fucose residue to the 3-OH of N-acetylglucosamine unit of the LacNAc disaccharide. The tagged LacNAc can then be selectively derivatized with probes via triazole-forming click reactions. Using this method, we discovered that murine lymphocytes with an activated memory phenotype (CD44highCD62Llow, CD25+) exhibited higher levels of LacNAc compared to their naïve counterparts (CD44lowCD62Lhigh, CD25−) [55]. Thus, this chemoenzymatic approach may serve as a valuable tool to discriminate differentially activated cell subsets ex vivo (Fig. 8).
Fig. 8.
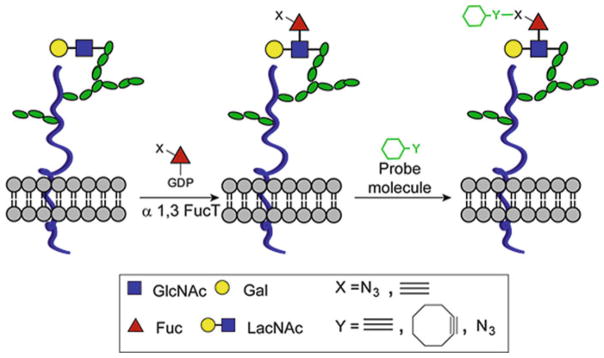
A chemoenzymatic method for the detection of LacNAc disaccharides
4.4 Profiling Sialylated Glycoproteins
The metabolic labeling and chemical reporter strategy also found applications in targeted glycoproteomics. Based on the pioneering work of Bertozzi and coworkers, the Wong laboratory designed a glycoproteomic method for identifying sialylated glycoproteins from crude cell lysates [56]. In this method, mammalian cells are metabolically treated with Ac4ManNAl to label sialylated glycoproteins with alkyne tags. These proteins are then biotinylated via CuAAC and enriched by affinity capture on streptavidin beads. Following on-bead trypsin digestion and PNGase F treatment, N-linked glycoproteins that are sialylated are selectively released and identified by LC-MS2. Adopting this method to profile sialylated glycoproteins in syngeneic prostate cancer cell lines PC3-N2 (nonmetastatic) and PC3-ML2 (highly metastatic), Semmes and coworkers identified and annotated 64 (26% of total nonredundant proteins) and 72 (29%) extracellular or membrane-bound glycoproteins from N2 and ML2 cells, respectively. Using ingenuity pathway analysis, they discovered that the majority of glycoproteins unique to the nonmetastatic PC3-N2 cell line were mediators of basic cell functions, such as vesicular transport, lipid metabolism, and mRNA processing and splicing. By contrast, the majority of glycoproteins overexpressed in the metastatic ML2 cell line were involved in cell motility, migration, and invasion [57].
4.5 Profiling O-Linked-β-N-Acetylglucosamine (O-GlcNAc) Glycosylation
O-GlcNAcylation is a dynamic glycosylation that is found on cytosolic and nuclear proteins. Today, more than 500 proteins are found to be O-GlcNAcylated [58]. These proteins participate in numerous cellular processes, including transcription, apoptosis, signal transduction, nutrient sensing, and proteasomal degradation. To identify O-GlcNAcylated proteins at the proteome level, Hsieh-Wilson and coworkers developed a chemoenzymatic approach, which involves transferring N-azidoacetyl galactosamine (GalNAz) to O-GlcNAc residues of O-GlcNAcylated proteins in crude cell lysates. The azido-tagged proteins can then be modified with biotin tags via CuAAC and be purified for mass spectrometry identification (Fig. 9) [59].
Fig. 9.
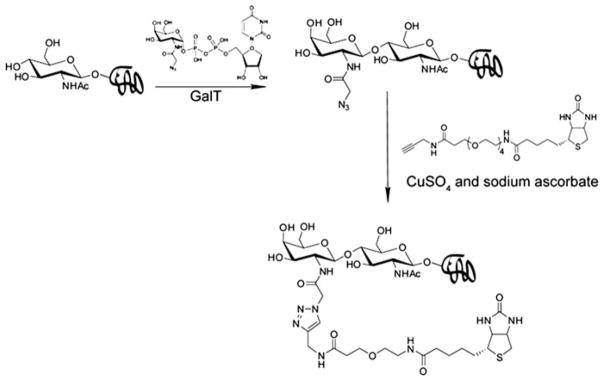
A chemoenzymatic method for the detection of protein O-GlcNAcylation
Using this method, the Hart lab discovered that histones, nuclear proteins in eukaryotic cells that assemble DNAs into nucleosomes, are O-GlcNAcylated [60]. Histones isolated from Hela cells were first subjected to tryptic digestion, followed by the chemoenzymatic labeling and mass spectrometry identification. Among the four core histones, three were found to be modified by O-GlcNAc, i.e., H2A, H2B, and H4. Threonine101 of H2A, serine36 of H2B, and serine47 of H4 were identified as the sites of O-GlcNAcylation. Interestingly, O-GlcNAcylation of serine36 of H2B and serine47 of H4 regulate histone-tail dynamics and may induce changes in the nucleosomal structure. It was discovered that histone O-GlcNAcylation level increased with heat shock and this increase accompanies chromatin condensation.
5 Triazole Formation-Based Bioconjugation for Activity-Based Protein Profiling
Another area in chemical biology that the triazole-forming click reactions have revolutionized is activity-based protein profiling (ABPP). ABPP is a chemical biology platform that uses active site-directed probes [61] to study the dynamic expression and function of mechanistically similar enzymes in their various physiological complexes [62, 63]. ABPP complements conventional proteomic approaches, which measure protein variations based on relative abundance [64]. By design, activity-based probes (ABPs) label only the catalytically active enzymes to exclude – their inactive counterparts, irrespective of transcript or protein expression level of the targets [41]. Generally, ABPs contain three integral pieces: an electrophilic reactive group (RG) that selectively binds and covalently modifies catalytically important and reactive nucleophiles, a high-affinity binding group that can guide and direct the ABP to structurally conserved features in the active sites of the target enzymes, and an analytical handle for visualization and isolation of probe-labeled targets [65, 66].
As an early success story, ABPP, integrated with CuAAC, led to the identification of the target protein of FR182877, a natural product isolated from a fermentation broth of Streptomyces species. FR182877 shows activities to induce microtubule assembly in vitro and antitumor activity in vivo [67].
Since this natural product contains two electrophilic sites, its biological activity is predicted to be derived from covalent modifications of specific proteins in vivo. An affinity tag-conjugated FR182877 derivative is ideal to verify this hypothesis. Due to the structural complexity of FR182877 and its sensitivity toward nucleophiles, conventional methods for its functionalization were complicated by side reactions. Using the Cu(I)-catalyzed triazole formation, a short peptide containing both a rhodamine tag and a biotin tag was introduced to FR182877 to generate a trifunctional probe (Fig. 10) [68]. Incubating the probe with a mouse heart proteome in vitro, followed by isolation of the biotin-labeled proteins by avidin chromatography, carboxylesterase-1 (CE-1) was identified as a specific target of this natural product. Competition experiments with the serine hydrolase-directed probe fluorophosphonate rhodamine determined that (−)FR182877 is a highly potent (IC50 = 34 nM) and selective inhibitor of CE-1.
Fig. 10.
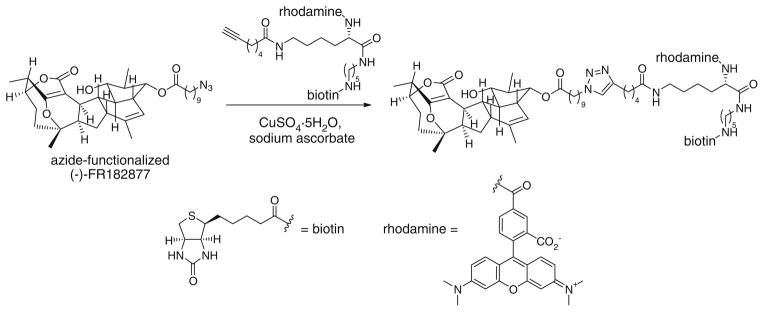
Synthesis of reporter-tagged (−)-FR182877 via CuAAC
Despite the successful development of ABPP probes for several enzyme classes, the large reporter tags, e.g., fluorophore or biotin, prevents the cellular uptake and distribution of many ABPP probes in living organisms. To solve this problem, Cravatt [69] and Overkleeft [70] independently designed a second generation probes, in which the large reporter tag is replaced with a small, bioorthogonal chemical handle (i.e., an alkyne or an azide). After covalent modification of target proteins in crude cell lysates, the alkyne or azide tag can be further conjugated to affinity probes via CuAAC or Bertozzi–Staudinger ligation [53], allowing the enrichment of tagged proteins for further analysis (Fig. 11).
Fig. 11.
The workflow of activity-based protein profiling
Using an azide-bearing active-site probe, azidohexyl benzenesulfonate and a rhodamine-alkyne detection probe and this two-step labeling strategy, Cravatt and coworkers were able to identify a number of serine hydrolases, including ECH-1, in the human breast cancer cell line MDA-MB-435 [69]. Azidohexyl benzenesulfonate was also active in live mice, enabling the capture of ECH-1 and its subsequent detection via rhodamine-alkyne and mass spectrometry analyses ex vivo in mice homogenes [69]. Further studies revealed that background noise in proteome labeling can be significantly reduced by replacing the azidohexyl benzenesulfonate/rhodamine-alkyne cycloaddition pair by the hex-5-ynyl benzenesulfonate/rhodamine-azide pair [66]. The higher sensitivity of the latter enabled the detection of low-abundance enzyme activity that had eluded detection in azidohexyl benzenesulfonate/rhodamine-alkyne reaction. Since 2004, the optimized ABPP conditions have been successfully applied to profile glycosidases [71], cytochrome P450 [72], and other enzyme classes [73].
Combined with photocrossing linkers, ABPP has also been utilized to profile enzymes that do not form covalent bond with activity-based probes, including metallohydrolases [74] and histone deacetylases [75].
6 Triazole Formation-Based Bioconjugation for Labeling Nucleic Acids
Likewise, the remarkable selectivity of Triazole forming bioconjugation reactions have been expoited to modify nucleic acids for a number of applications such as detecting newly synthesized DNA and determining genomic locations of 5-hydroxymethylcytosine.
A cell’s ability to proliferate reflects its healthy conditions and is influenced by metabolic and environmental cues. The most accurate method of measuring this property is directly measuring a cell’s DNA synthesis. Traditional methods for detecting and quantitating newly synthesized DNA utilize 5-bromo-2′-deoxyuridine (BrdU) and a corresponding antibody, which requires harsh DNA denaturation procedures that can disrupt dsDNA integrity and complicate the downstream analysis. In 2007, Mitchison reported that 5-ethynyl-2′-deoxyuridine (EdU) is metabolically incorporated into DNA during replication. After cell fixation, EdU in dsDNA can be detected with a fluorescent azide probe via CuAAC [76]. By using this procedure, newly synthesized DNA can be visualized with excellent sensitivity under milder conditions than the BrdU-based method.
5-Hydroxymethylcytosine (5-hmc), an oxidized form of 5-methylcytosine (5-mc), is an epigenetic mark discovered recently in certain mammalian tissues [77, 78]. The function of 5-hmC in epigenetic regulation is thought to be different from 5-methylcytosine (5-mC) and currently remains a mystery. Determination of genomic locations of 5-hmc serves as the first step to dissecting its functional roles. He and coworkers developed a chemoenzymatic method for site-specific labeling of 5-hmc [79]. This method utilizes β-glucosyltransferase to transfer an azido-containing glucose from the corresponding uridine 5′-diphospho-α-D-glucopyranoside to the 5-hydroxymethyl group of 5-hmc. The resulting azide-modified 5-hmc can then be conjugated to biotin probes via the triazole-formation reactions and quantified using avidin-based assays. Using this method, they discovered that the expression of 5-hmc in mouse cerebellum is developmentally regulated and is increased accompanying neuronal maturation (Fig. 12).
Fig. 12.
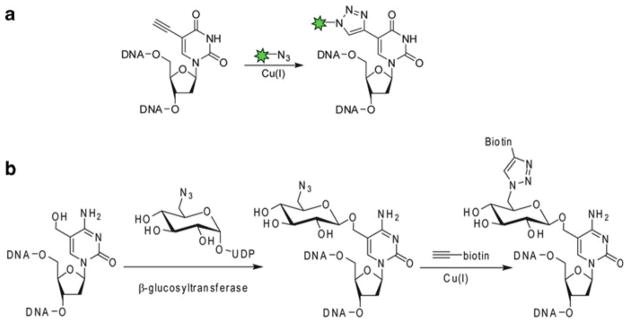
Detecting newly synthesized DNA (a) and determining genomic locations of 5-hydroxymethylcytosine (b) via CuAAC
7 Summary and Outlook
In the past ten years, the discovery of the triazole-forming click reactions has paved the way for a number of breakthroughs in the field of chemical biology. However, several technique challenges are waiting to be overcome. First, CuAAC, as presently formulated, cannot be applied to label biomolecules in the cytoplasm of live cells. To mitigate the toxicity associated with CuAAC, we incorporated functional groups that bear negative charges (at physiological pH) into Cu(I)-coordinating ligands to minimize the membrane permeability of the coordinated Cu(I). Accordingly, only cell-surface biomolecules are efficiently labeled. In a few studies, copper-free click chemistry has been adopted to modify azide-containing cytosolic proteins. However, high background noise is associated with this methodology [37]. Second, the invention of cyclooctyne probes eliminates the use of Cu(I) catalysts. Nevertheless, elevated ground-state energy of the cyclooctynes by ring strain inevitably increases their nonspecific reactivity. Through the joint efforts of chemists and biologists, we believe that these hurdles will be overcome in the near future by the discovery of new accelerating ligands for Cu(I) or new bioorthogonal reactions with better kinetics and selectivity than the triazole-forming reactions.
Acknowledgments
The author’s work that was highlighted in this book chapter was supported by the National Institutes of Health to P.W. (GM080585 and GM093282), the Mizutani Foundation for Glycoscience, and DuPont (DuPont Young Professor Award).
References
- 1.Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 2.O’Hare HM, Johnsson K, Gautier A. Chemical probes shed light on protein function. Curr Opin Struct Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Rabuka D. Chemoenzymatic methods for site-specific protein modification. Curr Opin Chem Biol. 2010;14:790–796. doi: 10.1016/j.cbpa.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baskin JM, Bertozzi CR. Bioorthogonal click chemistry: covalent labeling in living systems. Qsar Comb Sci. 2007;26:1211–1219. [Google Scholar]
- 5.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 7.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 8.Wu P, Fokin VV. Catalytic azide-alkyne cycloaddition: reactivity and applications. Aldrichimica Acta. 2007;40:7–17. [Google Scholar]
- 9.Meldal M, Tornoe CW. Cu-catalyzed azide-alkyne cycloaddition. Chem Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 10.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 11.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 13.Lutz JF. 1,3-dipolar cycloadditions of azides and alkynes: a universal ligation tool in polymer and materials science. Angew Chem Int Ed. 2007;46:1018–1025. doi: 10.1002/anie.200604050. [DOI] [PubMed] [Google Scholar]
- 14.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 15.Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan A, Levy M, Liu Y, Marlow FL, Wu P. Biocompatible copper(I) catalysts for in vivo imaging of glycans. J Am Chem Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaltgrad E, Sen Gupta S, Punna S, Huang CY, Chang A, Wong CH, Finn MG, Blixt O. Anti-carbohydrate antibodies elicited by polyvalent display on a viral scaffold. Chembiochem. 2007;8:1455–1462. doi: 10.1002/cbic.200700225. [DOI] [PubMed] [Google Scholar]
- 17.Gaetke LM, Chow CK. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology. 2003;189:147–163. doi: 10.1016/s0300-483x(03)00159-8. [DOI] [PubMed] [Google Scholar]
- 18.Link AJ, Vink MK, Tirrell DA. Presentation and detection of azide functionality in bacterial cell surface proteins. J Am Chem Soc. 2004;126:10598–10602. doi: 10.1021/ja047629c. [DOI] [PubMed] [Google Scholar]
- 19.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano Del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: a comparative study. Angew Chem Int Ed. 2011;50:8051–8056. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 21.Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ning X, Guo J, Wolfert MA, Boons GJ. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast Huisgen cycloadditions. Angew Chem Int Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jewett JC, Sletten EM, Bertozzi CR. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J Am Chem Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laughlin ST, Bertozzi CR. In vivo imaging of Caenorhabditis elegans glycans. ACS Chem Biol. 2009;4:1068–1072. doi: 10.1021/cb900254y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proc Natl Acad Sci USA. 2010;107:10360–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Copper-free click chemistry in living animals. Proc Natl Acad Sci USA. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lallana E, Riguera R, Fernandez-Megia E. Reliable and efficient procedures for the conjugation of biomolecules through Huisgen azide-alkyne cycloadditions. Angew Chem Int Ed. 2011;50:8794–8804. doi: 10.1002/anie.201101019. [DOI] [PubMed] [Google Scholar]
- 30.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 31.Carrico IS, Carlson BL, Bertozzi CR. Introducing genetically encoded aldehydes into proteins. Nat Chem Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 32.Wu P, Shui W, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR. Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proc Natl Acad Sci USA. 2009;106:3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin CW, Ting AY. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J Am Chem Soc. 2006;128:4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green DE, Morris TW, Green J, Cronan JE, Jr, Guest JR. Purification and properties of the lipoate protein ligase of Escherichia coli. Biochem J. 1995;309(Pt 3):853–862. doi: 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beatty KE, Fisk JD, Smart BP, Lu YY, Szychowski J, Hangauer MJ, Baskin JM, Bertozzi CR, Tirrell DA. Live-cell imaging of cellular proteins by a strain-promoted azide-alkyne cycloaddition. Chembiochem. 2010;11:2092–2095. doi: 10.1002/cbic.201000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 39.Dove A. Proteomics: translating genomics into products? Nat Biotech. 1999;17:233–236. doi: 10.1038/6972. [DOI] [PubMed] [Google Scholar]
- 40.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 41.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seo J, Lee K-J. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J Biochem Mol Biol. 2004;37:35–44. doi: 10.5483/bmbrep.2004.37.1.035. [DOI] [PubMed] [Google Scholar]
- 43.Kobe B, Kemp BE. Active site-directed protein regulation. Nature. 1999;402:373–376. doi: 10.1038/46478. [DOI] [PubMed] [Google Scholar]
- 44.Jiang H, Kim JH, Frizzell KM, Kraus WL, Lin H. Clickable NAD analogues for labeling substrate proteins of poly(ADP-ribose) polymerases. J Am Chem Soc. 2010;132:9363–9372. doi: 10.1021/ja101588r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson JP, Raghavan AS, Yang YY, Charron G, Hang HC. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.001198. 001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 47.Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, Lopez CB, Hang HC. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Eseentials of glycobiology. 2. Cold Spring Harbor; New York: 2008. [PubMed] [Google Scholar]
- 49.Zachara NE, Hart GW. The emerging significance of O-GlcNAc in cellular regulation. Chem Rev. 2002;102:431–438. doi: 10.1021/cr000406u. [DOI] [PubMed] [Google Scholar]
- 50.Taylor-Papadimitriou J, Epenetos AA. Exploiting altered glycosylation patterns in cancer: progress and challenges in diagnosis and therapy. Trends Biotechnol. 1994;12:227–233. doi: 10.1016/0167-7799(94)90121-X. [DOI] [PubMed] [Google Scholar]
- 51.Laughlin ST, Bertozzi CR. Imaging the glycome. Proc Natl Acad Sci USA. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 53.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 54.Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, Del Amo DS, Garret S, Seidel RD, III, Wu P. Chemoenzymatic synthesis of GDP-L-fucose and the Lewis X glycan derivatives. Proc Natl Acad Sci USA. 2009;106:16096–16101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng T, Jiang H, Gros M, del Amo DS, Sundaram S, Lauvau G, Marlow F, Liu Y, Stanley P, Wu P. Tracking N-acetyllactosamine on cell-surface glycans in vivo. Angew Chem Int Ed Engl. 2011;50:4113–4118. doi: 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanson SR, Hsu TL, Weerapana E, Kishikawa K, Simon GM, Cravatt BF, Wong CH. Tailored glycoproteomics and glycan site mapping using saccharide-selective bioorthogonal probes. J Am Chem Soc. 2007;129:7266–7267. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang L, Nyalwidhe JO, Guo S, Drake RR, Semmes OJ. Targeted identification of metastasis-associated cell-surface sialoglycoproteins in prostate cancer. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.007294. 007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 59.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cravatt BF, Sorensen EJ. Chemical strategies for the global analysis of protein function. Curr Opin Chem Biol. 2000;4:663–668. doi: 10.1016/s1367-5931(00)00147-2. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shields DJ, Niessen S, Murphy EA, Mielgo A, Desgrosellier JS, Lau SKM, Barnes LA, Lesperance J, Bouvet M, Tarin D, Cravatt BF, Cheresh DA. RBBP9: a tumor-associated serine hydrolase activity required for pancreatic neoplasia. Proc Natl Acad Sci. 2010;107:2189–2194. doi: 10.1073/pnas.0911646107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez J-C. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 65.Jessani N, Cravatt BF. The development and application of methods for activity-based protein profiling. Curr Opin Chem Biol. 2004;8:54–59. doi: 10.1016/j.cbpa.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 67.Sato B, Muramatsu H, Miyauchi M, Hori Y, Takase S, Hino M, Hashimoto S, Terano H. A new antimitotic substance, FR182877. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot (Tokyo) 2000;53:123–130. doi: 10.7164/antibiotics.53.123. [DOI] [PubMed] [Google Scholar]
- 68.Adam GC, Vanderwal CD, Sorensen EJ, Cravatt BF. (−)-FR182877 is a potent and selective inhibitor of carboxylesterase-1. Angew Chem Int Ed Engl. 2003;42:5480–5484. doi: 10.1002/anie.200352576. [DOI] [PubMed] [Google Scholar]
- 69.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 70.Ovaa H, van Swieten PF, Kessler BM, Leeuwenburgh MA, Fiebiger E, van den Nieuwendijk AM, Galardy PJ, van der Marel GA, Ploegh HL, Overkleeft HS. Chemistry in living cells: detection of active proteasomes by a two-step labeling strategy. Angew Chem Int Ed. 2003;42:3626–3629. doi: 10.1002/anie.200351314. [DOI] [PubMed] [Google Scholar]
- 71.Vocadlo DJ, Bertozzi CR. A strategy for functional proteomic analysis of glycosidase activity from cell lysates. Angew Chem Int Ed Engl. 2004;43:5338–5342. doi: 10.1002/anie.200454235. [DOI] [PubMed] [Google Scholar]
- 72.Wright AT, Cravatt BF. Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem Biol. 2007;14:1043–1051. doi: 10.1016/j.chembiol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 74.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci USA. 2004;101:10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci USA. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]