Abstract
The F-box protein Skp2 mediates c-Myc ubiquitylation by binding to the MB2 domain. However, the turnover of c-Myc is largely dependent on phosphorylation of threonine-58 and serine-62 in MB1, residues that are often mutated in cancer. We now show that the F-box protein Fbw7 interacts with and thereby destabilizes c-Myc in a manner dependent on phosphorylation of MB1. Whereas wild-type Fbw7 promoted c-Myc turnover in cells, an Fbw7 mutant lacking the F-box domain delayed it. Furthermore, depletion of Fbw7 by RNA interference increased both the abundance and transactivation activity of c-Myc. Accumulation of c-Myc was also apparent in mouse Fbw7−/− embryonic stem cells. These observations suggest that two F-box proteins, Fbw7 and Skp2, differentially regulate c-Myc stability by targeting MB1 and MB2, respectively.
Keywords: c-Myc, Fbw7, SCF complex, Skp2, ubiquitin ligase
Introduction
The oncoprotein c-Myc, a basic helix–loop–helix/leucine zipper (bHLH/Zip)-type transcription factor, is a master regulator of cell proliferation. c-Myc forms a heterodimer with the bHLH/Zip protein Max, and this complex binds to the CACGTG sequence, known as the E-box motif (Grandori et al, 2000), present in target genes, such as those for lactate dehydrogenase (LDH) and heat shock protein 60 (Hsp60), and thereby activates their transcription. The transcription of other genes, including those for cyclin D1 and carboxypeptidase D, is repressed by the c-Myc–Max complex or by c-Myc alone (Philipp et al, 1994; Guo et al, 2000). These positive and negative effects on gene transcription are thought to contribute to the promotion of cell proliferation by c-Myc.
Whereas Max is expressed constitutively, the expression of c-Myc is transient and is directly related to the proliferative potential of cells. Whereas c-Myc is virtually undetectable in quiescent cells, its expression is rapidly induced as cells enter the G1 phase of the cell cycle in response to stimulation with serum or specific mitogens. The abundance of c-Myc subsequently decreases gradually to a low steady-state level at which it remains for as long as the cells continue to proliferate. The expression level of c-Myc is increased in many malignant tumors as a result of amplification or mutation of the c-Myc gene. Given that many c-MYC mutations affect the stability of c-Myc (Bahram et al, 2000; Grandori et al, 2000), its turnover is thought to be a critical determinant of carcinogenesis.
The half-life of c-Myc is extremely short (∼30 min) in proliferating cells (Hann and Eisenman, 1984), and the protein has been shown to undergo ubiquitylation and degradation by the proteasome (Ciechanover et al, 1991; Salghetti et al, 1999). The region of c-Myc that signals its ubiquitylation (the degron) overlaps with the transactivation domain (TAD) (Salghetti et al, 2000). Two highly conserved sequence elements, Myc box 1 (MB1) and MB2, in the TAD have been implicated not only in the proteolysis of c-Myc but also in its transactivation and oncogenic activities (Flinn et al, 1998; Grandori et al, 2000). In particular, phosphorylation of Thr-58 and Ser-62 in MB1 is an important determinant of c-Myc stability (Lutterbach and Hann, 1994; Sears et al, 1999, 2000). Consistent with their effect on c-Myc stability, these two residues are frequently mutated in various tumors (Bahram et al, 2000). Phosphorylation of Thr-58 appears to be both mediated by glycogen synthase kinase 3 (GSK3) and dependent on the prior phosphorylation of Ser-62, which is likely mediated by the Ras–ERK (extracellular signal-regulated kinase) pathway. Whereas phosphorylation of Ser-62 alone seems to stabilize c-Myc, subsequent phosphorylation of Thr-58 promotes c-Myc ubiquitylation and degradation (Sears et al, 1999, 2000).
The ubiquitin ligase (E3) component of the enzyme cascade that mediates ubiquitin–protein conjugation is responsible for target specificity (Hershko and Ciechanover, 1998). Two E3s, the SCF complex and the anaphase-promoting complex or cyclosome (APC/C), are thought to regulate cell cycle progression predominantly at G1–S and M–G1 phases, respectively (Elledge and Harper, 1998; Zachariae and Nasmyth, 1999). The SCF complex consists of four components: the invariable subunits Skp1, Cul1 (also known as Cdc53) and Rbx1 (Roc1, Hrt1) and a variable F-box protein that serves as a receptor for target proteins and thereby determines target specificity (Elledge and Harper, 1998; Kamura et al, 1999; Ohta et al, 1999; Seol et al, 1999). Among the many F-box proteins that have been identified, Skp2 and Fbw7 have been well characterized and shown to control the abundance of proteins important in cell cycle regulation. Skp2, which contains leucine-rich repeats in addition to its F box, ubiquitylates various cell cycle regulators, including p27Kip1 (Carrano et al, 1999; Sutterluty et al, 1999; Nakayama et al, 2000), p21Cip1 (Bornstein et al, 2003), and p57Kip2 (Kamura et al, 2003) as well as free cyclin E (Nakayama et al, 2000), E2F-1 (Marti et al, 1999), Orc1 (Mendez et al, 2002), B-Myb (Charrasse et al, 2000), Rb-related protein p130 (Tedesco et al, 2002), and Cdt1 (Li et al, 2003). Fbw7 (hCdc4, Sel-10, hAgo) contains WD40 repeats in addition to its F box and ubiquitylates cyclin E (Koepp et al, 2001; Moberg et al, 2001; Strohmaier et al, 2001), Notch1, and Notch4 (Gupta-Rossi et al, 2001; Oberg et al, 2001).
We and others recently showed that Skp2 binds to c-Myc via its MB2 and HLH-Zip domains and thereby mediates its ubiquitylation and degradation. Skp2 also increases the transactivation activity of c-Myc, suggesting that Skp2 is a transcriptional cofactor (Kim et al, 2003; von der Lehr et al, 2003). Given that Skp2 is itself an oncoprotein with growth-promoting properties (Gstaiger et al, 2001; Latres et al, 2001), we hypothesized the existence of another regulator that controls c-Myc stability in a manner dependent on the phosphorylation of Thr-58 and Ser-62. We now show that Fbw7 interacts with and promotes the degradation of c-Myc in such a manner. Furthermore, suppression of Fbw7 by RNA interference (RNAi) resulted in stabilization of c-Myc as well as consequent transcriptional activation of its target genes and promotion of cell proliferation. These results suggest that c-Myc undergoes dual regulation by two F-box proteins, Fbw7 and Skp2, that target its MB1 and MB2 domains, respectively.
Results
Interaction of Fbw7 with c-Myc
Skp2 associates with c-Myc in a manner independent of MB1. However, c-Myc stability is largely dependent on the phosphorylation of Thr-58 and Ser-62 in MB1. We therefore investigated whether another regulator might control c-Myc stability in a manner dependent on the phosphorylation of these residues. The peptide sequence surrounding Thr-58 of c-Myc conforms to a motif known as the Cdc4 phospho-degron (CPD) (Nash et al, 2001), which is also present in the Fbw7 substrate cyclin E (Figure 1A). We thus tested whether Fbw7 interacts with c-Myc. We expressed the hemagglutinin epitope (HA)-tagged F-box proteins Fbw1a, Fbw2, Fbw4, and Fbw7 together with FLAG-tagged c-Myc in HEK293T cells. Cell lysates were subjected to immunoprecipitation with antibodies to FLAG, and the resulting precipitates were subjected to immunoblot analysis with antibodies to HA or FLAG. HA-Fbw7 was co-precipitated by the antibodies to FLAG in the presence of FLAG-c-Myc, whereas HA-tagged Fbw1a, Fbw2, and Fbw4 (F-box proteins that, like Fbw7, contain WD40 repeats) were not (Figure 1B). These results suggested that Fbw7 interacts with c-Myc and might target it for ubiquitylation.
Figure 1.
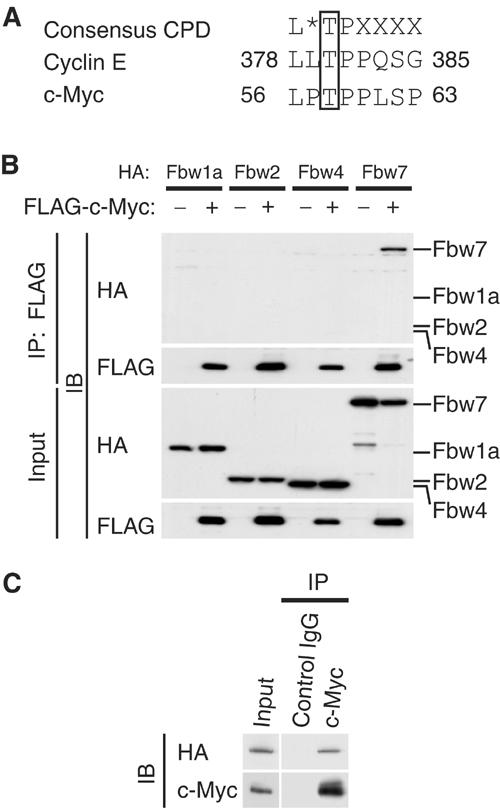
Interaction of Fbw7 with c-Myc in vivo. (A) The Cdc4 phospho-degron (CPD) sequences of human cyclin E and c-Myc. The asterisk indicates L, I, or P; X indicates any residue other than K or R. (B) HEK293T cells were transfected with vectors for FLAG-c-Myc and either HA-Fbw1a, HA-Fbw2, HA-Fbw4, or HA-Fbw7, as indicated, and were then incubated with MG132 for 6 h. Cell lysates were subjected to immunoprecipitation (IP) with antibodies to FLAG, and the resulting precipitates as well as the original cell lysates (input) were subjected to immunoblot analysis (IB) with antibodies to HA or FLAG. (C) HEK293T cells were transfected with a vector for HA-Fbw7, subjected to serum deprivation, stimulated by re-exposure to serum for 2 h, and then incubated for 6 h in the additional presence of MG132. Cell lysates were then subjected to immunoprecipitation with antibodies to c-Myc or with control mouse IgG, and the resulting precipitates were subjected to immunoblot analysis with antibodies to HA or c-Myc.
To determine whether HA-Fbw7 interacts with endogenous c-Myc, we deprived transfected HEK293T cells of serum and then stimulated them by re-exposure to serum in order to induce expression of endogenous c-Myc. Cell lysates were subjected to immunoprecipitation with antibodies to c-Myc or with control mouse immunoglobulin G (IgG). HA-Fbw7 was co-precipitated by the antibodies to c-Myc but not by the control IgG (Figure 1C), suggesting that endogenous c-Myc associates with HA-Fbw7 in these cells.
Purified recombinant SCFFbw7 ubiquitylates c-Myc in vitro
To determine whether the SCFFbw7 complex ubiquitylates c-Myc, we purified recombinant SCFFbw7 from insect cells infected with baculoviruses encoding the four components of this complex: Fbw7 (fused with an NH2-terminal hexahistidine tag; His6-Fbw7), Rbx1, Cul1, and Skp1. We also prepared a His6-c-Myc substrate with this system; this protein was phosphorylated on both Thr-58 and Ser-62 by the insect cells (see Figure 3C). We then assayed the ubiquitylation activity of the recombinant SCFFbw7 complex in vitro with the His6-c-Myc substrate. Immunoblot analysis of the reaction mixtures with antibodies to c-Myc detected the ubiquitylation of His6-c-Myc only in the presence of Uba1 (E1), UbcH5A (E2), ubiquitin, ATP, and SCFFbw7 (E3) (Figure 2A). Lack of any of these components prevented c-Myc ubiquitylation. The SCFFbw7 complex was thus shown to ubiquitylate c-Myc in an ATP-dependent manner.
Figure 3.
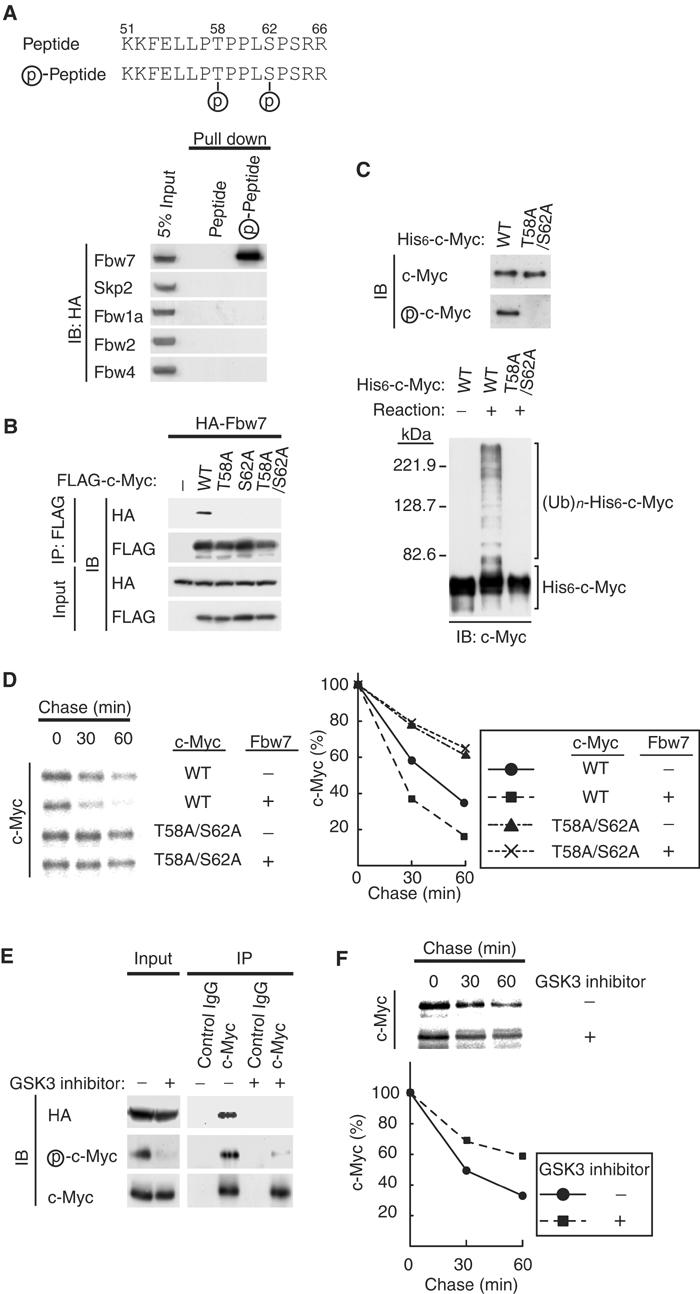
Phosphorylation of c-Myc on Thr-58 and Ser-62 is required for its recognition by the SCFFbw7complex. (A) Interaction of Fbw7 with a synthetic CPD peptide in vitro. HEK293T cells were transfected with vectors for HA-Fbw7, HA-Skp2, HA-Fbw1a, HA-Fbw2, or HA-Fbw4. Cell lysates were subsequently subjected to a ‘pull-down' assay with beads linked to nonphosphorylated or phosphorylated peptides corresponding to the CPD of c-Myc (upper panel), and the resulting precipitates (or 5% of the input cell lysates) were subjected to immunoblot analysis with antibodies to HA. (B) In vivo association of Fbw7 with c-Myc derivatives. HEK293T cells were transfected with vectors for HA-Fbw7 and either wild type (WT) or the indicated Thr-58 or Ser-62 mutants of FLAG-c-Myc. They were then subjected to in vivo binding analysis as described in Figure 1B. (C) Ubiquitylation of phosphorylated but not nonphosphorylated c-Myc by recombinant SCFFbw7 in vitro. His6-c-Myc and the His6-c-Myc(T58A/S62A) mutant purified from Sf21 cells were subjected to immunoblot analysis with antibodies to c-Myc or phospho-c-Myc (upper panel). The purified His6-c-Myc and His6-c-Myc(T58A/S62A) proteins were also tested as substrates in the in vitro ubiquitylation assay, performed with all reaction components (lower panel), as described in Figure 2A. (D) HEK293T cells were transfected with the indicated combinations of FLAG-c-Myc and HA-Fbw7 vectors and then subjected to pulse-chase analysis as described in Figure 2B. (E) HEK293T cells transfected with a vector for HA-Fbw7 were deprived of serum and then stimulated with serum in the absence or presence of a GSK3 inhibitor as described in Materials and methods. Cell lysates were then subjected to immunoprecipitation and immunoblot analysis as described in Figure 1B. (F) HEK293T cells transfected with a vector for FLAG-c-Myc were subjected to pulse-chase analysis in the absence or presence of a GSK3 inhibitor.
Figure 2.
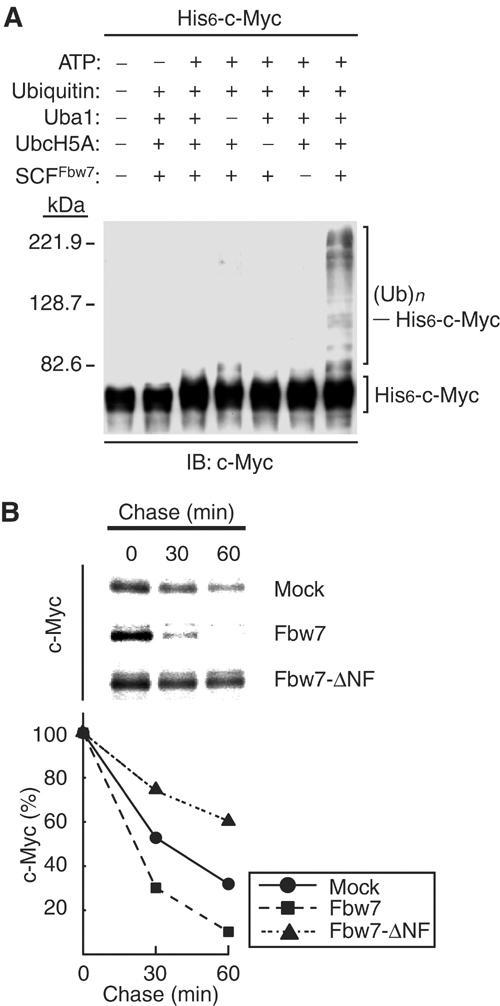
Promotion of the ubiquitylation and degradation of c-Myc by Fbw7. (A) Ubiquitylation of c-Myc by the recombinant SCFFbw7 complex in vitro. Recombinant SCFFbw7 was assayed for ubiquitylation activity with His6-c-Myc as substrate in the absence or presence of the indicated reaction mixture components. The reaction mixtures were then subjected to immunoblot analysis with antibodies to c-Myc. The positions of unmodified His6-c-Myc and of His6-c-Myc conjugated with ubiquitin ((Ub)n) are indicated. (B) Promotion of c-Myc degradation by Fbw7 in vivo. HEK293T cells were transfected with vectors for FLAG-c-Myc and either HA-Fbw7 or HA-Fbw7-ΔNF (or the corresponding empty vector; mock). The cells were then subjected to pulse-chase analysis by metabolic labeling with [35S]methionine, and cell lysates were prepared at the indicated times of the chase incubation and subjected to immunoprecipitation with antibodies to FLAG. The precipitates were subjected to SDS–polyacrylamide gel electrophoresis and autoradiography (upper panel). The percentage of FLAG-c-Myc remaining after the various chase times was quantitated by image analysis (lower panel).
Promotion of c-Myc degradation by Fbw7 in vivo
To examine the possible effect of Fbw7 on the degradation of c-Myc in vivo, we transfected HEK293T cells with vectors for c-Myc and for either wild-type Fbw7 or an Fbw7 mutant (Fbw7-ΔNF) that lacks the F-box domain and is therefore unable to associate with Skp1. Pulse-chase analysis revealed that expression of wild-type Fbw7 markedly promoted the degradation of c-Myc, whereas Fbw7-ΔNF delayed it (Figure 2B). These results thus suggested that Fbw7 contributes to the turnover of c-Myc in intact cells.
Phosphorylation of Thr-58 and Ser-62 is required for c-Myc degradation mediated by SCFFbw7
To determine whether phosphorylation of Thr-58 and Ser-62 in MB1 of c-Myc is required for the binding of Fbw7, we tested the ability of phosphorylated and nonphosphorylated forms of a synthetic peptide encompassing the c-Myc CPD (amino acids 51–66) to interact with Fbw7 in vitro. Recombinant Fbw7 interacted only with the phosphorylated form of the peptide, whereas Skp2, Fbw1a, Fbw2, or Fbw4 did not interact with either form (Figure 3A). To investigate the possible effect of phosphorylation of these residues of c-Myc on its degradation by the SCFFbw7 complex, we first performed in vivo analysis of the interaction between Fbw7 and c-Myc mutants (T58A, S62A, T58A/S62A) in which either or both Thr-58 and Ser-62 were replaced by alanine. In transfected HEK293T cells, wild-type c-Myc interacted with Fbw7, whereas the T58A, S62A, and T58A/S62A mutants did not (Figure 3B), suggesting that phosphorylation of c-Myc on both Thr-58 and Ser-62 is required for interaction with Fbw7. We next examined the possible requirement for phosphorylation of Thr-58 and Ser-62 in the ubiquitylation of c-Myc in vitro. The His6-c-Myc substrate was shown to be phosphorylated by immunoblot analysis with antibodies that specifically recognize c-Myc phosphorylated on Thr-58 and Ser-62 (Figure 3C); the mutant His6-c-Myc(T58A/S62A) was not phosphorylated. The in vitro ubiquitylation assay revealed that, unlike the wild-type protein, the c-Myc mutant was not ubiquitylated by purified SCFFbw7 (Figure 3C). These data suggest that phosphorylation of c-Myc on Thr-58 and Ser-62 is required for its ubiquitylation.
We next performed pulse-chase analysis to examine the effect of Fbw7 on the stability of the T58A/S62A mutant of c-Myc. Whereas expression of Fbw7 promoted the degradation of wild-type c-Myc, it had no effect on the stability of the T58A/S62A mutant (Figure 3D). These observations thus suggested that phosphorylation of c-Myc on Thr-58 and Ser-62 is also essential for its ubiquitylation by Fbw7 and its degradation in vivo.
Given that phosphorylation of c-Myc on Thr-58 appears to be mediated primarily by GSK3 (Sears et al, 1999, 2000), we examined the effects of treatment of HEK293T cells expressing HA-Fbw7 with a GSK3 inhibitor during induction of c-Myc (after serum deprivation). Immunoblot analysis with antibodies specific for phospho-c-Myc revealed that the inhibitor markedly reduced the extent of c-Myc phosphorylation on Thr-58 (or Ser-62) (Figure 3E). Immunoprecipitation with antibodies to c-Myc and subsequent immunoblot analysis with antibodies to HA, c-Myc, or phospho-c-Myc revealed that HA-Fbw7 was co-precipitated only with c-Myc that was phosphorylated by GSK3 (Figure 3E). Phosphorylation of c-Myc on Thr-58 by GSK3 thus appears to be essential for recognition by Fbw7. Moreover, pulse-chase analysis showed that the GSK3 inhibitor delayed the turnover of FLAG-c-Myc in HEK293T cells (Figure 3F).
Depletion of Fbw7 results in accumulation of c-Myc
To examine whether the abundance of endogenous Fbw7 affects that of c-Myc, we used RNAi to deplete HeLa cells of Fbw7. Reverse transcription (RT) and polymerase chain reaction (PCR) analysis revealed that transfection of HeLa cells with small interfering RNAs (siRNAs) for Fbw7 or Skp2, or with the combination thereof, resulted in downregulation of the corresponding mRNAs (Figure 4A). Depletion of Fbw7 mRNA induced the accumulation of c-Myc and, consistent with previous observations (Koepp et al, 2001), that of cyclin E (Figure 4B). Similar experiments by another group revealed no effect of an Fbw7 siRNA on c-Myc abundance (Kim et al, 2003); this previous study, however, failed to demonstrate the effectiveness of the Fbw7 siRNA by RT–PCR analysis. As previously described (Nakayama et al, 2000; Kim et al, 2003), depletion of Skp2 led to the accumulation of c-Myc, cyclin E, and p27Kip1 (Figure 4B). Moreover, the combination of siRNAs for Fbw7 and Skp2 resulted in an additive effect on the abundance of c-Myc and cyclin E (Figure 4B).
Figure 4.
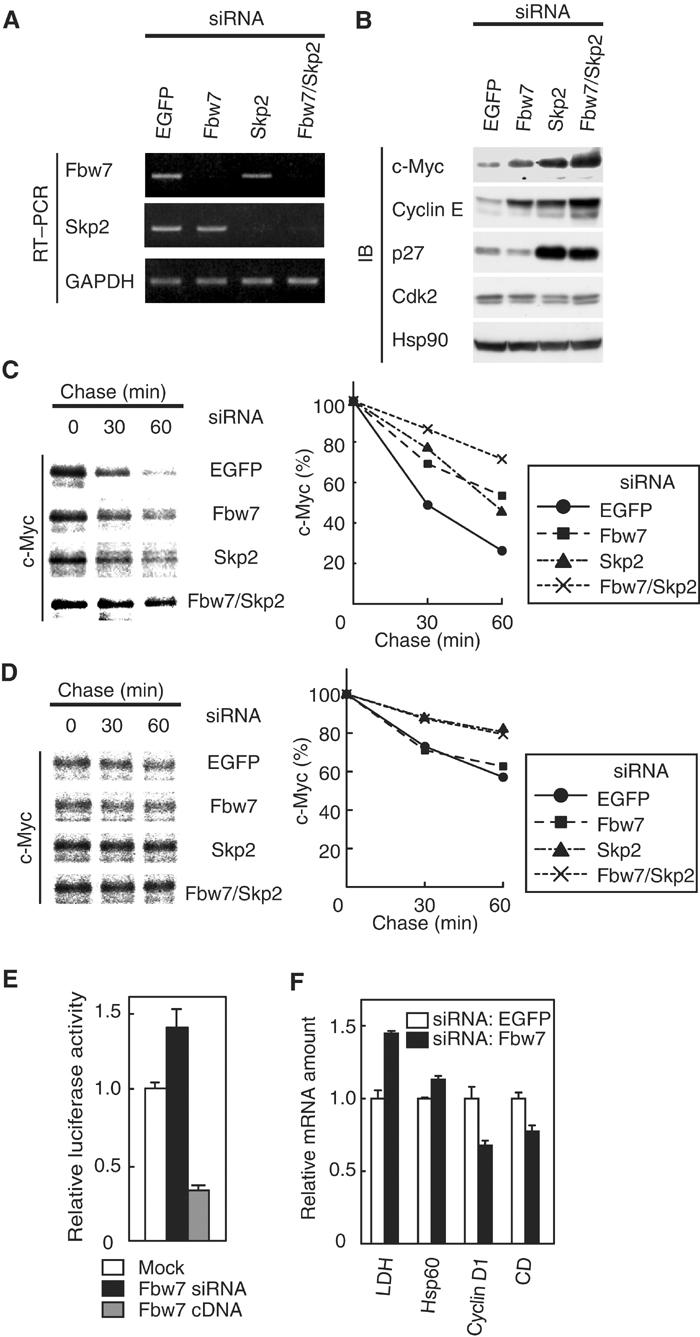
Depletion of Fbw7 by RNAi induces accumulation of c-Myc and promotes c-Myc-dependent transactivation. (A) At 48 h after transfection of HeLa cells with the indicated siRNAs, the abundance of mRNAs for Fbw7, Skp2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; internal standard) was determined by RT–PCR. (B) At 48 h after siRNA transfection, HeLa cell lysates were subjected to immunoblot analysis with antibodies to c-Myc, cyclin E, p27, Cdk2, or Hsp90 (internal standard). (C) At 48 h after siRNA transfection, HeLa cells were transfected with a vector for FLAG-c-Myc and then subjected to pulse-chase analysis. (D) At 48 h after siRNA transfection, HeLa cells were transfected with a vector for FLAG-c-Myc(T58A/S62A) and then subjected to pulse-chase analysis. (E) Activity of a c-Myc-dependent luciferase reporter gene. At 48 h after mock, Fbw7 siRNA, or Fbw7 cDNA transfection, HeLa cells were transfected with a c-Myc-dependent luciferase reporter plasmid (p4 × E-SVP-Luc). The cells were incubated for an additional 24 h and then assayed for relative luciferase activity. Data are means±s.d. of triplicates from a representative experiment. (F) Transcriptional activity of c-Myc target genes. The abundance of transcripts derived from the indicated genes (CD, carboxypeptidase D) was determined by RT and real-time PCR 72 h after siRNA transfection. Data are means±s.d. of triplicates from a representative experiment.
To evaluate further the effect of Fbw7 or Skp2 depletion on the turnover of c-Myc, we performed pulse-chase analysis. The degradation of c-Myc in cells transfected with the siRNAs for Fbw7 or Skp2 was delayed compared with that apparent in cells transfected with a control siRNA. Moreover, the combination of the siRNAs for Fbw7 and Skp2 delayed the turnover of c-Myc in an additive manner (Figure 4C). These data suggested that endogenous Fbw7 and Skp2 both participate in regulation of the turnover of c-Myc in vivo. We next investigated the effect of Fbw7 or Skp2 depletion on the stability of the T58A/S62A mutant of c-Myc for further understanding the relative contribution of Fbw7 and Skp2 to the degradation of c-Myc. Whereas the siRNA for Fbw7 had no effect on the stability of the T58A/S62A mutant, the siRNA for Skp2 delayed its degradation (Figure 4D). These observations suggest that Fbw7 ubiquitylates c-Myc in a manner dependent on phosphorylation at Thr-58 and Ser-62, whereas Skp2 ubiquitylates it in a manner independent of the phosphorylation.
Depletion of Fbw7 promotes c-Myc-dependent transactivation
To examine the effect of Fbw7 deficiency on c-Myc-dependent transactivation, we measured the relative luciferase activity of cells transfected with a c-Myc-responsive reporter construct (p4 × E-SVP-Luc) (Mori et al, 1998). The relative luciferase activity of cells transfected with the Fbw7 siRNA was increased compared with that apparent in cells transfected with the control siRNA (Figure 4E). Conversely, we found that overexpression of Fbw7 reduced the extent of c-Myc-dependent transactivation (Figure 4E). In addition, RT and real-time PCR analysis revealed that depletion of Fbw7 by RNAi resulted in an increase in the abundance of transcripts derived from the LDH and Hsp60 genes, both of which are positively regulated by c-Myc (Guo et al, 2000). In contrast, the abundance of transcripts derived from the cyclin D1 and carboxypeptidase D genes, both of which are negatively regulated by c-Myc (Philipp et al, 1994; Guo et al, 2000), was reduced in cells transfected with the Fbw7 siRNA relative to that in cells transfected with the control siRNA (Figure 4F). These observations thus suggested that the accumulation of c-Myc induced by depletion of Fbw7 also resulted in an increase in the transactivation activity of c-Myc.
Stabilization of c-Myc in Fbw7−/− ES cells
We have generated Fbw7-deficient mice and found that these animals die in utero at embryonic day 10.5 as a result of impaired vascular development (Tsunematsu et al, 2004). Attempts to isolate mouse embryonic fibroblasts (MEFs) from such immature embryos have not been successful. Instead, we have generated mouse embryonic stem (ES) cells with deletions in both Fbw7 alleles (Tsunematsu et al, 2004). Marked accumulation of c-Myc was apparent in these Fbw7−/− ES cells compared with the low concentration of this protein detected in Fbw7+/− ES cells (Figure 5A). Furthermore, pulse-chase analysis showed that the stability of c-Myc was greatly increased in Fbw7−/− ES cells compared with that in Fbw7+/− ES cells (Figure 5B). Accumulation of cyclin E was not apparent in the Fbw7−/− ES cells (Figure 5A) or in Fbw7−/− embryos at embryonic day 9.5 (Tsunematsu et al, 2004), excluding the possibility that the increased abundance of c-Myc in the Fbw7−/− cells is a secondary effect of cyclin E accumulation. Skp2−/− MEFs also exhibited a substantial accumulation of c-Myc as well as of p27 compared with the abundance of these proteins in Skp2+/− cells (Figure 5C). The degradation of c-Myc in Skp2−/− MEFs appeared to be especially impaired at early time points (20 min) of cycloheximide treatment (Figure 5D), suggesting that Skp2 is important for the early phase of c-Myc degradation, whereas Fbw7 might contribute to a later phase. The difference in c-Myc stability between the control cells in Figure 5B and those in Figure 5D may be attributable to the difference in cell type (ES cells versus MEFs). Together with previous biochemical data (Kim et al, 2003; von der Lehr et al, 2003), our genetic evidence indicates that both Fbw7 and Skp2 participate in the degradation of c-Myc.
Figure 5.
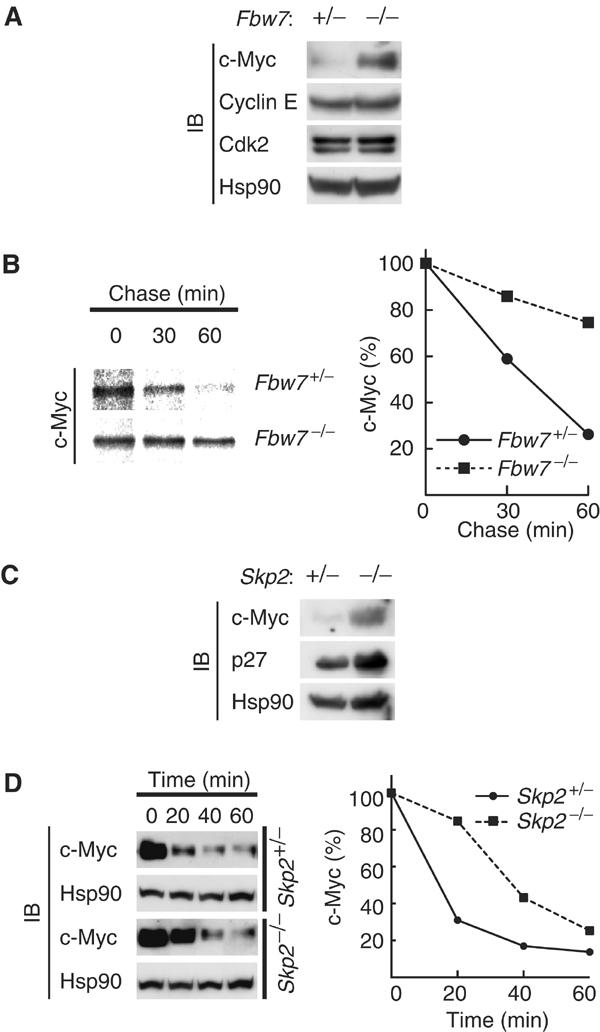
Stabilization of c-Myc in Fbw7−/− ES cells. (A) Lysates of asynchronous Fbw7+/− or Fbw7−/− ES cells were subjected to immunoblot analysis with antibodies to the indicated proteins. (B) ES cells were transfected with a vector for FLAG-c-Myc controlled by the phosphoglycerate kinase gene promoter and then subjected to pulse-chase analysis. (C) Lysates of asynchronous Skp2+/− or Skp2−/− MEFs were subjected to immunoblot analysis with antibodies to c-Myc, p27, or Hsp90. (D) MEFs were deprived of serum, stimulated by re-exposure to serum for 2 h, and then incubated for the indicated times in the additional presence of cycloheximide (50 μg/ml). Cell lysates were subjected to immunoblot analysis with antibodies to c-Myc or Hsp90.
Discussion
The ubiquitin–proteasome pathway plays important roles in many cell functions by determining the abundance of regulatory proteins (Hershko and Ciechanover, 1998). c-Myc, a bHLH/Zip-type transcription factor, was previously shown to be ubiquitylated and degraded by the proteasome (Ciechanover et al, 1991; Salghetti et al, 1999; Gregory and Hann, 2000). Given that many mutations of c-MYC associated with cancer affect the stability of c-Myc (Bahram et al, 2000), characterization of the mechanism of c-Myc ubiquitylation is important for understanding the oncogenic process mediated by c-Myc.
We and others have recently shown that Skp2 participates in c-Myc degradation through interaction with MB2 (and the HLH-Zip region) (Kim et al, 2003; von der Lehr et al, 2003). However, MB1 has also been implicated in the degradation of c-Myc, and phosphorylation of Thr-58 and Ser-62 in MB1 has been thought to be primarily responsible for the regulation of c-Myc stability (Lutterbach and Hann, 1994; Sears et al, 1999, 2000). Indeed, most c-MYC mutations associated with cancer affect these or neighboring residues (Bahram et al, 2000). Skp2 does not appear to contribute to the regulation of c-Myc stability mediated at the level of MB1 (Kim et al, 2003; von der Lehr et al, 2003). Furthermore, Skp2 is itself an oncoprotein with growth-promoting properties (Gstaiger et al, 2001; Latres et al, 2001), which renders its potential role as the E3 that targets c-Myc for degradation inconsistent with the notion that such a molecule is likely to be a tumor suppressor. We thus hypothesized that another E3 might also mediate c-Myc degradation in a manner dependent on the phosphorylation of Thr-58 and Ser-62.
The c-Myc degron in MB1 (LPTPPLSP) conforms to the CPD sequence recognized by Fbw7. We now show that Fbw7 binds to c-Myc in a manner dependent on the phosphorylation of Thr-58 and Ser-62. We therefore propose that ubiquitylation of c-Myc is mediated by two F-box proteins, Fbw7 and Skp2 (Figure 6). Given that Fbw7 recognizes c-Myc in a phosphorylation-dependent manner, the Fbw7-mediated degradation of c-Myc is likely triggered by activation of GSK3 and the Ras–ERK signaling pathway. In contrast, Skp2-mediated degradation of c-Myc is not dependent on the phosphorylation of Thr-58, although it is not clear whether Skp2 requires phosphorylation at other sites in c-Myc for its recognition of this protein.
Figure 6.
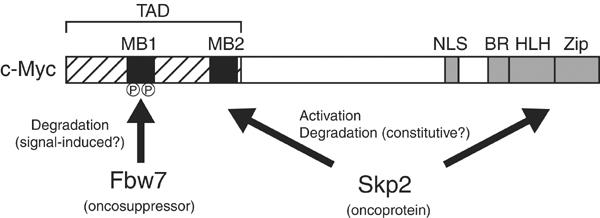
A model for c-Myc ubiquitylation. Two F-box proteins, Fbw7 and Skp2, regulate the turnover of c-Myc. The putative oncosuppressor Fbw7 recognizes c-Myc molecules that are phosphorylated in the MB1 region of the transcriptional activation domain (TAD). The oncoprotein Skp2 recognizes the MB2 and HLH-Zip domains. Skp2 both enhances the transactivation activity of c-Myc and promotes its degradation. NLS, nuclear localization signal; BR, basic region.
c-Myc is virtually undetectable in resting cells, but its abundance increases on entry of cells into G1 triggered by exposure to mitogens, during which time it activates the transcription of various genes important for progression into S phase. Thereafter, c-Myc is phosphorylated on Thr-58 and Ser-62, probably by GSK3 and ERK, respectively (Sears et al, 2000), and is then immediately degraded by the ubiquitin–proteasome pathway. Our present data indicate that phosphorylation of c-Myc at these sites is essential for its binding to Fbw7, its Fbw7-mediated ubiquitylation, and its Fbw7-dependent turnover. Consistent with this notion, a GSK3 inhibitor reduced the extent of phosphorylation of c-Myc on Thr-58 (or Ser-62) as well as that of the interaction between c-Myc and Fbw7. In contrast, Skp2 binds to a c-Myc mutant that lacks MB1 (von der Lehr et al, 2003) and to the T58A mutant of c-Myc but not to a phosphorylated peptide corresponding to c-Myc(51–66), with which Fbw 7 interacts. The role of Skp2-mediated ubiquitylation of c-Myc has been unclear. However, given that Skp2 is expressed predominantly in S phase (Lisztwan et al, 1998; Hara et al, 2001), Skp2-mediated degradation of c-Myc must occur in this phase of the cell cycle. In contrast, Fbw7 is expressed throughout the cell cycle (Spruck et al, 2002). The timing of the actions of Fbw7 and Skp2 in late G1–S phase thus appears too close to readily allow their delineation. c-Myc, Skp2, and Fbw7 are also all localized predominantly in the nucleus (von der Lehr et al, 2003), suggesting that the intracellular sites of c-Myc ubiquitylation mediated by Fbw7 and Skp2 are similar or identical. The degradation of c-Myc in Fbw7−/− ES cells was impaired for up to 60 min in the chase incubation of pulse-chase analysis, whereas that in Skp2−/− MEFs appeared to be impaired in particular at early times (20 min) of cycloheximide treatment, suggesting that Skp2 is important for an early phase of degradation and that Fbw7 is required for a later phase. However, we cannot exclude the possibility that this difference in the time course of c-Myc degradation is attributable to the difference in the cell types studied. Overall, although the timing and subcellular site of c-Myc ubiquitylation mediated by Fbw7 or Skp2 appear to be similar, Fbw7-mediated ubiquitylation is regulated by phosphorylation of c-Myc on Thr-58 and Ser-62, whereas Skp2-mediated ubiquitylation is dependent on the time course of Skp2 expression.
The stability of IκBα, a relatively well-characterized ubiquitylated protein, is also determined by two molecular regions: an NH2-terminal domain that is phosphorylated by the IκB kinase complex in response to extracellular signals (Regnier et al, 1997), and a COOH-terminal PEST domain that is important in the basal turnover of the protein. The F-box protein Fbw1 recognizes the phospho-degron (DpSGXXpS) that is present in the NH2-terminal domain of IκBα and conserved in all members of the IκB and β-catenin families, and it mediates the ubiquitylation of these proteins (Hatakeyama et al, 1999; Kitagawa et al, 1999). We propose that the turnover of c-Myc is also determined by signal-induced and basal degradation mediated by Fbw7 and Skp2, respectively.
Whereas overexpression of Fbw7 reduced the transactivation activity of c-Myc, overexpression of Skp2 had an opposite effect (von der Lehr et al, 2003). The decrease in the transactivation activity of c-Myc induced by Fbw7 likely results from the degradation of c-Myc mediated by the ubiquitin–proteasome pathway. In contrast, Skp2 appears to induce both the degradation and the activation of c-Myc. This latter effect of Skp2 might result from its also targeting a negative regulator of c-Myc, such as MM-1 (Mori et al, 1998), for ubiquitin-mediated proteolysis.
The turnover of cyclin E is also regulated by both Fbw7 (Koepp et al, 2001; Moberg et al, 2001; Strohmaier et al, 2001) and Skp2 (Nakayama et al, 2000). Fbw7 recognizes cyclin E phosphorylated on Thr-380. In contrast, Skp2 interacts only with free cyclin E in a phosphorylation-independent manner; the association of cyclin E with Cdk2 prevents its interaction with Skp2. We thus propose that Fbw7 plays an important role in termination of the action of the cyclin E–Cdk2 complex, whereas Skp2 controls the abundance of the cyclin E pool during the cell cycle. Mice lacking Skp2 manifest cellular accumulation of cyclin E and p27Kip1 as well as over-replication of chromosomes and centrosomes (Nakayama et al, 2000). The increased abundance of cyclin E in Skp2−/− mice is not a secondary effect of p27Kip1 accumulation, given that it was also apparent in Skp2−/− p27−/− double-mutant mice (Nakayama et al, 2004) and that cyclin E ubiquitylation was markedly increased by recombinant Skp2 in vitro (Nakayama et al, 2000). The collaboration of two cell cycle-related F-box proteins thus appears to be a common feature of the mechanisms by which the turnover of cyclin E and c-Myc, both of which promote cell proliferation and have oncogenic potential, is regulated.
Given that Fbw7 targets cyclin E, Notch1, Notch4, and c-Myc, all of which are thought to be oncoproteins, for degradation, it is likely that Fbw7 functions as an oncosuppressor. Indeed, FBW7 has been shown to be mutated in at least 16% of human endometrial tumors (Spruck et al, 2002). In contrast, overexpression of Skp2 has been detected in many types of cancer, and mice transgenic for Skp2 in the lymphoid lineage are predisposed to malignant lymphoma (Gstaiger et al, 2001; Latres et al, 2001). It is thus possible that the accumulation of c-Myc and cyclin E due to Fbw7 deficiency contributes to tumorigenesis, in which case restoration of FBW7 expression by gene therapy may prove beneficial in individuals with malignant tumors.
Materials and methods
Antibodies
Rabbit polyclonal antibodies to c-Myc (sc-764), cyclin E (sc-198, sc-481), Cdk2 (sc-163), or HA (sc-805) as well as mouse monoclonal antibodies to c-Myc (sc-42) were obtained from Santa Cruz Biotechnology. Rabbit polyclonal antibodies to phospho-c-Myc (M8433) and mouse monoclonal antibodies to FLAG (M2) or HA (HA11) were from Sigma and Babco, respectively. Mouse monoclonal antibodies to p27Kip1 or Hsp90 were from BD Biosciences.
Plasmid construction and mutagenesis
Complementary DNAs encoding HA-Fbw7, HA-Fbw7-ΔNF (human), and HA-Fbw4 (zebrafish) were cloned into pCGN (Kitagawa et al, 1999); cDNAs encoding HA-Fbw1a, HA-Fbw2, and HA-Skp2 (mouse) were cloned into pcDNA3 (Invitrogen); and a cDNA encoding FLAG-c-Myc (human) was cloned into pCI (Promega). The pCI vectors encoding the FLAG-c-Myc substitution mutants T58A, S62A, and T58A/S62A were generated with the use of a QuickChange site-directed mutagenesis kit (Stratagene). For ectopic expression in ES cells, a vector for FLAG-c-Myc controlled by the phosphoglycerate kinase gene promoter was constructed.
Complementary DNAs encoding either a glutathione S-transferase (GST) fusion protein of Myc epitope-tagged Cul1 (mouse) or T7-tagged Skp1 (human) were cloned into pBacPAK9 or pBacPAK8 (BD Biosciences). Complementary DNAs encoding c-Myc, c-Myc(T58A/S62A), or Fbw7 were cloned into pFASTBacHTa (Invitrogen).
Transfection, immunoprecipitation, and immunoblot analysis
HEK293T cells were transfected with vectors with the use of FuGENE6 (Roche). Cells were incubated for 6 h with the proteasome inhibitor MG132 (10 μM) (Peptide Institute) beginning 24 h after transfection. Cell lysis and immunoprecipitation were performed as described (Kitagawa et al, 1999). The immunoprecipitates were subjected to immunoblot analysis as described (Kamura et al, 2003).
In vitro ‘pull-down' assay
Nonphosphorylated and phosphorylated peptides corresponding to the CPD region of c-Myc (residues 51–66) were chemically synthesized and conjugated to NHS-activated beads (Pharmacia). Cell lysates (200 μg of protein) were incubated for 30 min at 4°C with the peptide-conjugated beads. The resulting precipitates were washed four times with lysis buffer and subjected to immunoblot analysis.
Serum deprivation and stimulation
HEK293T cells and MEFs were deprived of serum for 48 h by culture in Dulbecco's modified Eagle's medium (DMEM) supplemented with 0.1% fetal bovine serum (FBS). They were then stimulated for various times by incubation with DMEM supplemented with 30% FBS.
Production of recombinant proteins
Saccharomyces cerevisiae Uba1, human UbcH5A, and human ubiquitin were expressed in and purified from Escherichia coli as described (Hara et al, 2001). Recombinant baculoviruses were generated with the BacPAK (BD Biosciences) or Bac-To-BacHT (Invitrogen) baculovirus systems. The recombinant SCFFbw7 complex was purified as described (Kamura et al, 2003). His6-c-Myc and His6-c-Myc(T58A/S62A) were purified similarly.
Assay of ubiquitylation in vitro
The ability of the purified recombinant SCFFbw7 complex to ubiquitylate c-Myc was assayed by incubation of 50 ng of His6-c-Myc and 200 ng of the complex as described (Kamura et al, 2003). The reaction mixtures were subjected to immunoblot analysis with antibodies to c-Myc (sc-764).
Pulse-chase analysis
Pulse-chase analysis was performed with the use of antibodies to FLAG as described (Kitagawa et al, 1999). All pulse-chase data are from experiments that were repeated three times with similar results.
GSK3 inhibitor treatment
After transfection and serum deprivation, HEK293T cells were stimulated by re-exposure to serum in the presence of 20 μM 2-thio(3-iodobenzyl)-5-(1-pyridyl)-[1,3,4]-oxadiazole (GSK3 inhibitor II, Calbiochem) or 0.1% dimethyl sulfoxide (vehicle) for 2 h. The cells were then incubated in the additional presence of 10 μM MG132 for a further 6 h before immunoprecipitation and immunoblot analysis.
RNAi
RNAi was performed as described (Elbashir et al, 2001), with the exception that Oligofectamine (Invitrogen) was used for transfection. The siRNA duplexes corresponding to Fbw7, Skp2, or enhanced green fluorescent protein (EGFP) were synthesized by Dharmacon Research; sequences are available on request.
Luciferase assay
At 48 h after siRNA transfection, HeLa cells were transfected with both p4 × E-SVP-Luc (Mori et al, 1998), provided by Y Ariga, and pRL-Tk (Promega), as an internal control, with the use of FuGENE6. After incubation for an additional 24 h, the cells were assayed with a Dual-Luciferase Reporter Assay System (Promega).
RT and real-time PCR
Total RNA (1 μg) isolated from HeLa cells with the use of an RNAssay kit (Qiagen) was subjected to RT with a RevaTra Ace kit (Toyobo). The resulting cDNA was subjected to PCR with SYBR Green PCR Master Mix in an ABI-PRISM 7000 Sequence Detection System (Applied Biosystems). The relative amounts of each target mRNA were calculated as described (Pfaffl, 2001).
Acknowledgments
We thank H Ariga (Hokkaido University) for p4 × E-SVP-Luc, R Yasukochi, S Matsushita, N Nishimura, and K Shimoharada for technical assistance, and C Sugita and M Kimura for help in preparation of the manuscript. This work was supported in part by a grant from the Ministry of Education, Science, Sports, and Culture of Japan, and by a research grant from the Human Frontier Science Program.
References
- Bahram F, von der Lehr N, Cetinkaya C, Larsson LG (2000) c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 95: 2104–2110 [PubMed] [Google Scholar]
- Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A (2003) Role of SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 during S-phase. J Biol Chem 278: 25752–25757 [DOI] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M (1999) SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1: 193–199 [DOI] [PubMed] [Google Scholar]
- Charrasse S, Carena I, Brondani V, Klempnauer KH, Ferrari S (2000) Degradation of B-Myb by ubiquitin-mediated proteolysis: involvement of the Cdc34–SCF(p45Skp2) pathway. Oncogene 19: 2986–2995 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, DiGiuseppe JA, Schwartz AL, Brodeur GM (1991) Degradation of MYCN oncoprotein by the ubiquitin system. Prog Clin Biol Res 366: 37–43 [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Harper JW (1998) The role of protein stability in the cell cycle and cancer. Biochim Biophys Acta 1377: M61–M70 [DOI] [PubMed] [Google Scholar]
- Flinn EM, Busch CM, Wright AP (1998) myc boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Mol Cell Biol 18: 5961–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16: 653–699 [DOI] [PubMed] [Google Scholar]
- Gregory MA, Hann SR (2000) c-Myc proteolysis by the ubiquitin–proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol 20: 2423–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W (2001) Skp2 is oncogenic and overexpressed in human cancers. Proc Natl Acad Sci USA 98: 5043–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET (2000) Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res 60: 5922–5928 [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A (2001) Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem 276: 34371–34378 [DOI] [PubMed] [Google Scholar]
- Hann SR, Eisenman RN (1984) Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol 4: 2486–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Kamura T, Nakayama K, Oshikawa K, Hatakeyama S, Nakayama KI (2001) Degradation of p27(Kip1) at the G(0)–G(1) transition mediated by a Skp2-independent ubiquitination pathway. J Biol Chem 276: 48937–48943 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Kitagawa M, Nakayama K, Shirane M, Matsumoto M, Hattori K, Higashi H, Nakano H, Okumura K, Onoe K, Good RA, Nakayama KI (1999) Ubiquitin-dependent degradation of IκBα is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proc Natl Acad Sci USA 96: 3859–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI (2003) Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci USA 100: 10231–10236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW (1999) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284: 657–661 [DOI] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP (2003) Skp2 regulates myc protein stability and activity. Mol Cell 11: 1177–1188 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama KI, Nakayama K (1999) An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J 18: 2401–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ (2001) Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294: 173–177 [DOI] [PubMed] [Google Scholar]
- Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, Pagano M (2001) Role of the F-box protein Skp2 in lymphomagenesis. Proc Natl Acad Sci USA 98: 2515–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao Q, Liao R, Sun P, Wu X (2003) The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem 278: 30854–30858 [DOI] [PubMed] [Google Scholar]
- Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W (1998) Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): evidence for evolutionary conservation in the subunit composition of the CDC34–SCF pathway. EMBO J 17: 368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B, Hann SR (1994) Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol 14: 5510–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti A, Wirbelauer C, Scheffner M, Krek W (1999) Interaction between ubiquitin–protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol 1: 14–19 [DOI] [PubMed] [Google Scholar]
- Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell 9: 481–491 [DOI] [PubMed] [Google Scholar]
- Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK (2001) Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413: 311–316 [DOI] [PubMed] [Google Scholar]
- Mori K, Maeda Y, Kitaura H, Taira T, Iguchi-Ariga SM, Ariga H (1998) MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc. J Biol Chem 273: 29794–29800 [DOI] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, Kitagawa M, Nakayama KI, Hatakeyama S (2000) Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J 19: 2069–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T, Nakayama KI (2004) Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell, in press [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [DOI] [PubMed] [Google Scholar]
- Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U (2001) The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem 276: 35847–35853 [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y (1999) ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell 3: 535–541 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, Alitalo K, Eilers M (1994) Repression of cyclin D1: a novel function of MYC. Mol Cell Biol 14: 4032–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M (1997) Identification and characterization of an IkappaB kinase. Cell 90: 373–383 [DOI] [PubMed] [Google Scholar]
- Salghetti SE, Kim SY, Tansey WP (1999) Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J 18: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti SE, Muratani M, Wijnen H, Futcher B, Tansey WP (2000) Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc Natl Acad Sci USA 97: 3118–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Leone G, DeGregori J, Nevins JR (1999) Ras enhances Myc protein stability. Mol Cell 3: 169–179 [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR (2000) Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14: 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ (1999) Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev 13: 1614–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck CH, Strohmaier H, Sangfelt O, Muller HM, Hubalek M, Muller-Holzner E, Marth C, Widschwendter M, Reed SI (2002) hCDC4 gene mutations in endometrial cancer. Cancer Res 62: 4535–4539 [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI (2001) Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413: 316–322 [DOI] [PubMed] [Google Scholar]
- Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W (1999) p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol 1: 207–214 [DOI] [PubMed] [Google Scholar]
- Tedesco D, Lukas J, Reed SI (2002) The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein–ubiquitin ligase SCF(Skp2). Genes Dev 16: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI (2004) Mouse Fbw7/Sel-10/Cdc4 is required for Notch degradation during vascular development. J Biol Chem 279: 9417–9423 [DOI] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, Soderberg O, Kerppola TK, Larsson LG (2003) The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 11: 1189–1200 [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev 13: 2039–2058 [DOI] [PubMed] [Google Scholar]


