Abstract
The heme-copper oxidase (HCO) superfamily includes HCOs in aerobic respiratory chains and nitric oxide reductases (NORs) in the denitrification pathway. The HCO/NOR catalytic subunit has a core structure consisting of 12 transmembrane helices (TMHs) arranged in three-fold rotational pseudosymmetry, with six conserved histidines for heme and metal binding. Using sensitive sequence similarity searches, we detected a number of novel HCO/NOR homologs and named them HCO Homology (HCOH) proteins. Several HCOH families possess only four TMHs that exhibit the most pronounced similarity to the last four TMHs (TMHs 9–12) of HCOs/NORs. Encoded by independent genes, four-TMH HCOH proteins represent a single evolutionary unit (EU) that relates to each of the three homologous EUs of HCOs/NORs comprising TMHs 1–4, TMHs 5–8, and TMHs 9–12. Single-EU HCOH proteins could form homotrimers or heterotrimers to maintain the general structure and ligand-binding sites defined by the HCO/NOR catalytic subunit fold. The remaining HCOH families, including NnrS, have 12-TMHs and three EUs. Most three-EU HCOH proteins possess two conserved histidines and could bind a single heme. Limited experimental studies and genomic context analysis suggest that many HCOH proteins could function in the denitrification pathway and in detoxification of reactive molecules such as nitric oxide. HCO/NOR catalytic subunits exhibit remarkable structural similarity to the homotrimers of MAPEG (membrane-associated proteins in eicosanoid and glutathione metabolism) proteins. Gene duplication, fusion, and fission likely play important roles in the evolution of HCOs/NORs and HCOH proteins.
Keywords: Heme-copper oxidase superfamily, HCO homology proteins, denitrification, detoxification, MAPEG proteins, gene duplication, gene fusion and fission
Introduction
Aerobic respiration has evolved to use oxygen, which produces about 16 times more adenosine triphosphates (ATPs) than anaerobic respiration.1 This advantage likely facilitated some critical evolutionary steps, such as the origin of eukaryotes and the increase of body size.2,3 Heme-copper oxidases (HCOs) are membrane-bound enzyme complexes functioning in the terminal step of aerobic respiratory chains.4–6 They catalyze the reduction of dioxygen to water using electrons transferred from cytochrome c or a quinol derivative. The released energy is coupled to the translocation of protons across the membrane to generate an electrochemical gradient that can be used for ATP synthesis. All HCOs possess a catalytic subunit, an integral membrane protein with 12 core transmembrane helices (TMHs). Six conserved histidines in the TMHs of the catalytic subunit coordinate three co-factors: a high-spin heme and a copper ion in the binuclear catalytic site, and an additional low-spin heme functioning in the electron transfer pathway.7,8 Two of the six histidines function as axial ligands to coordinate the low-spin heme, while the rest participate in the catalytic site, with three histidines positioning the copper on one side of the high-spin heme and one histidine serving as the axial ligand on the other side. The 12-TMH core structure of the HCO catalytic subunit displays three-fold rotational pseudosymmetry and distributes the two heme groups into two of the three proposed pseudosymmetric units,9,10 each of which consists of four TMHs.
HCOs from various organisms have been discovered. They differ in heme types, electron donors (such as cytochrome c and ubiquinol), proton transfer pathways, and subunit composition. Three major types of HCOs (A, B, and C) have been defined based on sequence and structural analyses.5,11 A-type HCOs include cytochrome c oxidases in mitochondria, and cytochrome c oxidases and quinol oxidases in many bacteria and some archaea.12 B-type HCOs are mainly found in the archaeal phylum of Crenarchaeota and appear to use one proton pathway compared to two proton pathways in A-type HCOs. A- and B-type HCOs share a conserved tyrosine residue residing in the sixth TMH. This tyrosine is covalently linked to a copper-binding histidine and was proposed to donate a fourth electron to the binuclear center in the catalytic process.13 C-type HCOs, mainly from Proteobacteria, use a catalytic tyrosine residue located in a structurally different position (the seventh TMH) than that of A- and B-type HCOs.14,15
Sequence and structural analyses revealed that the catalytic subunit of nitric oxide reductases (NORs) is homologous to that of the HCOs.6,16 NOR catalytic subunit also has 12 core TMHs sharing the same topology of HCO catalytic subunit and binds two hemes and a non-heme iron (instead of copper in HCOs) in a similar fashion by using six conserved histidines. The HCO superfamily thus includes both HCOs and NORs. NORs catalyze the reduction of nitric oxide (NO) to nitrous oxide (N2O) with the help of the heme groups and the non-heme iron in the denitrification pathway of the nitrogen cycle.6,17,18 As NO is a toxic reactive agent, NORs in some pathogenic bacteria also play important roles in detoxifying exogenous NO generated by hosts.19 Besides substrate preference, NORs differ from HCOs in that they do not translocate protons across the membrane and do not have a catalytic tyrosine due to fewer electrons required in one catalytic cycle. Two major subgroups of NORs have been described: the cytochrome c-dependent cNOR and the quinol-dependent qNOR.20 NORs appear to be more closely related to C-type HCOs than A- and B-type HCOs in terms of sequence similarity and subunit composition.21
Although NORs and the three types of HCOs each form well-separated clades in the phylogeny reconstructed for the HCO superfamily, the position of the root remains controversial. Different evolutionary scenarios have been proposed for the origin and evolutionary order of HCOs and NORs. Several studies18,22–25 suggested that NORs may be more ancient than HCOs, consistent with the assumption that aerobic respiration evolved from denitrification after the emergence of atmospheric oxygen. Other researchers have proposed that the widely distributed A-type HCOs were present before the split of bacteria and archaea and are ancestors to B- and C-type HCOs and NORs.12 These hypotheses, still in debate, explain how the 12 core TMHs developed into various types of oxidases. However, the origin of this pseudosymmetric helical architecture, an ancient event, is rarely discussed.
In this study, we used sensitive sequence similarity search methods such as transitive PSI-BLAST26 searches and HHpred27 to detect proteins homologous to the catalytic subunits of the HCO superfamily members. We called the newly found homologs HCO homology (HCOH) proteins. Interestingly, we discovered HCOH proteins with only four TMHs. These four-TMH proteins exhibit the highest similarity to the last four TMHs of HCOs (TMHs 9–12). They are considered to correspond to one evolutionary unit (EU) and are called single-EU HCOH proteins. Single-EU HCOH proteins may form homotrimers or heterotrimers to maintain the general structure and the ligand-binding sites defined by the fold of HCO/NOR catalytic subunits. HCO/NOR catalytic subunits are proposed to contain three homologous EUs made of TMHs 1–4, TMHs 5–8, and TMHs 9–12. We also discovered several groups of 12-TMH HCOH proteins that, like HCOs/NORs, contain three EUs. The majority of these three-EU HCOH proteins possess two conserved histidines that are predicted to bind a single heme. Most of the newly found remote homologs of HCOs/NORs are hypothetical proteins without experimental characterization. Only two of the seven major groups of HCOH proteins have been defined in current domain databases (DUF2871 and NnrS). Limited experimental studies and genomic context analysis suggest that they could function in the denitrification pathway and in the detoxification of reactive agents such as NO. Remarkably, the structural core of the three-EU assembly of HCOs/NORs resembles that of a diverse family of trimeric membrane-associated proteins in eicosanoid and glutathione metabolism (MAPEG).28,29 We propose the potential evolutionary scenarios linking existing families, as well as the early evolutionary events of HCOs/NORs in aerobic respiration.
Results and Discussion
Transitive PSI-BLAST26 searches (see Materials and Methods) and HHpred27 were used to detect proteins homologous to the catalytic subunits of the HCO superfamily members. A number of remote homologs of HCOs/NORs with different patterns of conserved histidines were discovered. We called these newly found superfamily members HCOH proteins and divided them into groups based on the number of TMHs in the homologous regions, patterns of conserved histidines, and the CLANS30 sequence clustering results.
Four-TMH proteins homologous to HCOs/NORs help define EUs
The catalytic subunits of HCOs/NORs exhibit an approximate three-fold structural symmetry and are considered as a result of duplications of four-TMH units.9,10 In previous structure studies, three pseudosymmetric structural units (SUs) have been defined as TMHs 11/12/1/2, TMHs 3/4/5/6, and TMHs 7/8/9/10 [Fig. 1(A)] (TMHs 1–12 correspond to previously defined TMHs I–XII).9,10 We detected a set of four-TMH proteins homologous to the catalytic subunits of HCOs/NORs. Most of these four-TMH proteins possess the HxH motif at the beginning of the second TMH. These proteins exhibit the highest sequence similarity to the last four TMHs of HCOs/NORs (TMHs 9/10/11/12). The HxH motif of these four-TMH proteins aligns to the HxH motif in the tenth TMH of the catalytic subunits of HCOs/NORs. We consider that these four TMHs correspond to an evolutionarily conserved unit and define them as one EU. Each of the four-TMH HCOH proteins possesses one EU and is thus called a single-EU HCOH protein. On the other hand, HCOs/NORs contain three EUs: TMHs 1/2/3/4 (EU1), TMHs 5/6/7/8 (EU2), and TMHs 9/10/11/12 (EU3) [Fig. 1(B,C)].
Figure 1.
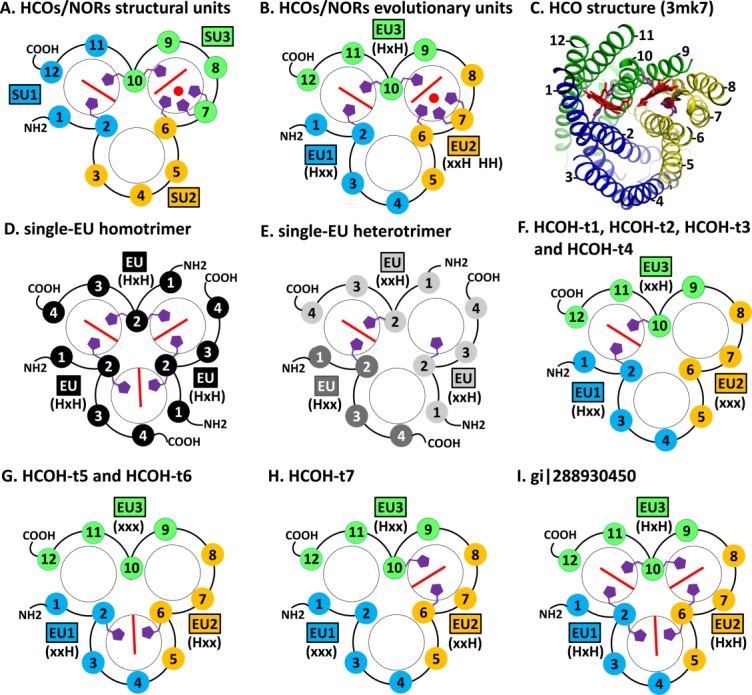
TMH topology diagrams of HCOs/NORs and HCOH proteins. TMHs are shown in numbered circles. TMHs in the same structure unit (marked by SU) or the same evolutionary unit (marked by EU) are filled with the same color. The histidine patterns of the EUs are shown in parentheses. Hemes are shown as red lines. Copper or non-heme irons in HCOs/NORs are shown as red spheres. Conserved histidine sidechains are shown in purple. N- and C-termini of each modeled protein are marked by NH2 and COOH, respectively. (A). Previously defined structural units of HCOs/NORs. (B). Newly defined EUs of HCOs/NORs. (C). Structure of a C-type HCO catalytic subunit (pdb: 3mk7). (D). Model of homotrimer for single-EU proteins with the HxH motif. (E). Model of heterotrimer for single-EU proteins with xxH and Hxx motifs. (F). Model for HCOH-t1, HCOH-t2, HCOH-t3 and HCOH-t4. (G). Model for HCOH-t5 and HCOH-t6. (H). Model for HCOH-t7. (I). Model for the protein (gi|288930450) with the HxH motif in all three EUs.
The second TMH in each of the three EUs in HCOs/NORs harbors conserved histidine(s) for heme or metal-binding, with characteristic three-residue motifs of Hxx, xxH, and HxH in EU1, EU2, and EU3, respectively (x: a variable residue) (Fig. 2). These motifs are homologous and occupy structurally equivalent positions in the superposition of EU1, EU2, and EU3. The third TMH of HCO/NOR EU2 additionally harbors a conserved HH motif (Fig. 2). The histidine in the Hxx motif of EU1 and the second histidine in the HxH motif of EU3 coordinate the low-spin heme in the ligand-binding pocket between EU1 and EU3 [Fig. 1(B,C)]. The histidines in the xxH motif and the HH motif of EU2 as well as the first histidine in the HxH motif of EU3 contribute to the binding of high-spin heme and copper/non-heme iron in the pocket between EU2 and EU3 [Fig. 1(B,C)].
Figure 2.
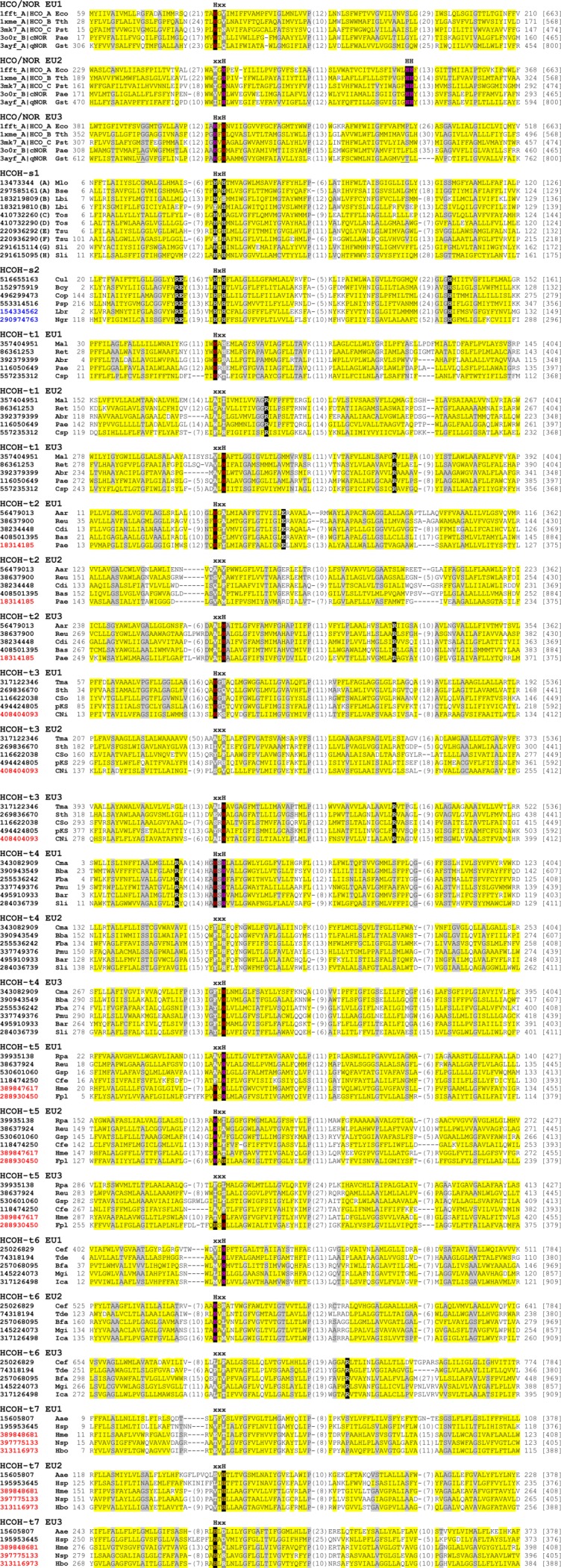
Multiple sequence alignment of EUs of HCOs/NORs and HCOH proteins. For HCOs/NORs, the sequences are denoted by their pdb id and chain id, followed by type name. For HCOH proteins, each sequence is denoted by its NCBI gene identification (gi) number. The cluster type for HCOH-s1 is shown in parentheses after gi numbers. Starting and ending residues numbers are shown before and after the sequences, respectively. Protein lengths are shown in brackets at the end. Positions with conserved motifs in the second TMHs of these EUs are marked with HxH, xxH, Hxx, or xxx. The conserved HH motif in the second EU of HCOs/NORs is also marked. Conserved histidines in these positions are in black background. For three-EU proteins, conserved histidines at the interface between EU1 and EU3 are colored red, those at the interface between EU2 and EU3 are colored orange, and those at the interface between EU1 and EU2 are colored magenta. Other family-specific conserved residues are also highlighted in black background. Non-charged residues in positions with mainly hydrophobic residues are shaded in yellow. Small residues (G,A,S,C,T,P, and V) in positions with mainly small residues are shaded in grey. Insertion regions are replaced by the number of inserted residues in parentheses or omitted in between underscored letters. Three-letter species name abbreviations shown before the residue starting numbers are as follows: Aae, Aquifex aeolicus; Aar, Aromatoleum aromaticum; Abr, Azospirillum brasilense; Bcy, Bacillus cytotoxicus; Bse, Bacillus selenitireducens; Bba, Belliella baltica; Bas, Bifidobacterium asteroides; Bar, Bizionia argentinensis; Bfa, Brachybacterium faecium; Cfe, Campylobacter fetus; Csp, Campylobacter sp.; CNi, Candidatus Nitrososphaera gargensis; CSo, Candidatus Solibacter usitatus; Cop, Coprobacillus; Cdi, Corynebacterium diphtheriae; Cef, Corynebacterium efficiens; Cul, Corynebacterium ulceribovis; Cma, Cyclobacterium marinum; Eco, Escherichia coli; Fpl, Ferroglobus placidus; Fba, Flavobacteriaceae bacterium; Gsp, Geobacillus sp.; Gst, Geobacillus stearothermophilus; Hme, Haloferax mediterranei; Hbo, Halogeometricum borinquense; Hsp, Hydrogenobaculum sp.; Ica, Intrasporangium calvum; Lbr, Leishmania braziliensis; Lbi, Leptospira biflexa; Mlo, Mesorhizobium loti; Mal, Methylomicrobium alcaliphilum; Mgi, Mycobacterium gilvum; Ngr, Naegleria gruberi; Nsp, Natrinema sp.; pKS, planctomycete KSU-1; Pmu, Paenibacillus mucilaginosus; Psp, Peptoniphilus sp.; Pae, Pseudomonas aeruginosa; Pst, Pseudomonas stutzeri; Pae, Pyrobaculum aerophilum; Reu, Ralstonia eutropha; Ret, Rhizobium etli; Rpa, Rhodopseudomonas palustris; Sli, Sideroxydans lithotrophicus; Sth, Sphaerobacter thermophilus; Sli, Spirosoma linguale; Tma, Thermaerobacter marianensis; Tos, Thermus oshimai; Tth, Thermus thermophilus; Tsu, Thioalkalivibrio sulfidophilus; Tde, Thiobacillus denitrificans. Gi numbers of bacterial, archaeal, and eukaryotic proteins are in black, red, and blue colors, respectively.
Single-EU proteins with four TMHs may form homotrimers or heterotrimers
The single-EU proteins can be roughly divided into two major groups: HCOH-s1 (HCO homology proteins with a single EU, group 1) and HCOH-s2, according to CLANS sequence clustering results (Fig. 3) and sequence conservation.
Figure 3.
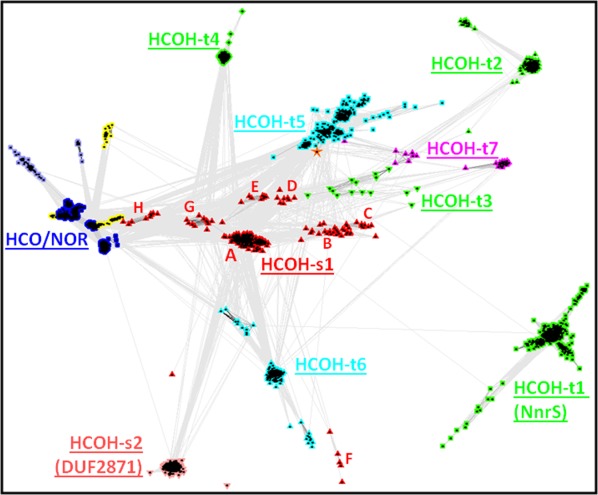
CLANS diagram of HCOs/NORs and HCOH proteins. Connections between proteins indicate BLAST P-values less than 1e-10. Single-EU HCOH proteins are shown as red up triangles (HCOH-s1) and pink low down triangles (HCOH-s2). Members of HCOH-t1, HCOH-t2, HCOH-t3, and HCOH-t4, the four groups with the Hxx.xxx.xxH motif pattern, are shown in green squares, green up triangles, green down triangles, and green diamonds, respectively. HCOH-t5 and HCOH-t6 members, both with the xxH.Hxx.xxx motif pattern, are shown in cyan square and cyan up triangles, respectively. The sequence in the HCOH-t5 group with the HxH.HxH.HxH motif pattern is marked by an orange star. HCOH-t7 members with the xxx.xxH.Hxx motif pattern are shown in magenta. Underlined group names are shown. For the HCOH-s1 group, the A to H clusters are marked in red letters. HCOs/NORs are shown as blue dots. Two small groups of proteins shown as light blue dots are closely related to HCOs/NORs. They do not contain the HH motif in the third helix of EU2, while maintaining all the other conserved histidines (Hxx, xxH, and HxH in EU1, EU2, and EU3, respectively). Some HCO-related sequences with all conserved histidines deteriorated are shown as yellow dots.
HCOH-s1 (Fig. 3, red up triangles) consists of a main cluster (marked by A in Fig. 3) and several nearby small clusters (marked by letters B, C, D, E, F, G, and H in Fig. 3). HCOH-s1 proteins of the main cluster (cluster A) are mostly from Proteobacteria and Firmicutes (Supporting Information Fig. S1). To maintain the structural compactness in the general fold of HCOs/NORs, these single-domain proteins most likely assemble as trimers. They could form homotrimers, since genomes of the main cluster of HCOH-s1 proteins contain only one single-EU protein. As this cluster of HCOH-s1 contains the HxH motif, three symmetric heme-binding sites at the interfaces of EUs can be inferred for the homotrimers [Fig. 1(D)], similar to the fashion of coordination of the low-spin heme by HCOs/NORs.
The B cluster (Fig. 3) of HCOH-s1 consists of closely related proteins encoded by gene pairs that are chromosomal neighbors [see three gene structure examples in Fig. 4(A–C)]. Interestingly, two proteins from the same species have the xxH and Hxx motifs, respectively (e.g., gi|183219809 and gi|183219810 in Fig. 2, also see Supporting Information Fig. S1 for other B cluster proteins). These neighboring gene products could from heterotrimers, with the histidines contributing to the heme-binding site similar to the way the low-spin heme is coordinated in HCOs/NORs. Figure 1(E) depicts one possible way of forming a heterotrimer consisting of two proteins with the xxH motif and one protein with the Hxx motif. A single heme-binding site can be inferred for such a heterotrimer.
Figure 4.
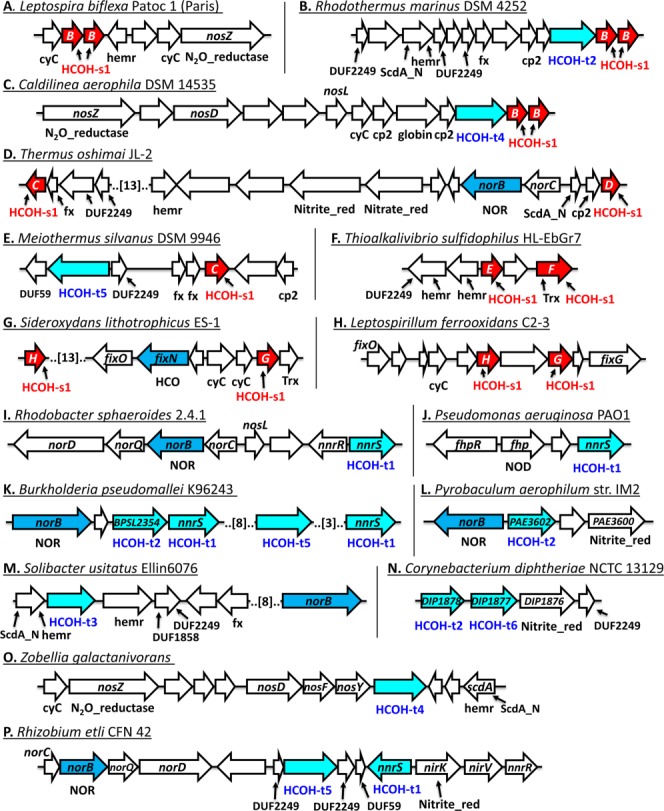
Gene structure diagrams of selected HCOH proteins. Genes are shown as arrows. HCOH-s1 genes, three-EU HCOH genes, and HCO/NOR genes are shown as red arrows, cyan arrows, and blue arrows, respectively. Names of frequently occurring domains are shown below the genes. Numbers of omitted genes are shown in brackets. Domain name abbreviations are as follows: cp2, cupin_2 domain; cyC, cytochrome c; fx, iron-sulfur ferredoxin domain; HCO: heme-copper oxidase; hemr, hemerythrin domain; N2O_reductase, nitrous oxide reductase; Nitrate_red, nitrate reductase; Nitrite_red, nitrite reductase; NOD, nitric oxide dioxygenase; NOR, nitric oxide reductase; and Trx, thioredoxin.
The remaining six small clusters (C–H) of the HCOH-s1 each have limited species distribution. HCOH-s1 proteins of clusters C and D contain the HxH motif and are from the Thermales order of the Deinococcus-Thermus phylum. In several species of the Thermus genus, a cluster C member and a cluster D member are products of genes not far away from each other, separated by gene clusters containing denitrification enzymes [see one example of Fig. 4(D)]. These proteins may form homotrimers or heterotrimers for genomes containing both C and D cluster members. On the other hand, two species (Meiothermus silvanus DSM 9946 and Oceanithermus profundus DSM 14977) have cluster C members and do not have cluster D members [see the example of Fig. 4(E)], suggesting that cluster C members can form homotrimers. HCOH-s1 proteins of clusters E and F, mostly from the Thioalkalivibrio genus, are also largely encoded by neighboring gene pairs [one example shown in Fig. 4(F)]. Although cluster E proteins have the HxH motif, cluster F proteins are characterized by the xxH motif (Supporting Information Fig. S1). Products of these neighboring gene pairs may form heterotrimers. HCOH-s1 proteins of clusters G and H all possess the HxH motif (Supporting Information Fig. S1) and are encoded by pairs of genes in chromosomal vicinity [see two examples in Fig. 4(G,H)].
Members of the HCOH-s1 group have not been classified in publicly available domain databases, while the HCOH-s2 group corresponds to Pfam family DUF2871 and consists of proteins with unknown function. HCOH-s2 members have the conserved HxH motif in their second TMH. They likely form homotrimers with three symmetric sites that can coordinate three hemes [Fig. 1(D)]. Compared to HCOH-s1 proteins, HCOH-s2 proteins additionally possess a conserved “RE” motif at the end of the first TMH and a conserved histidine in the fourth TMH (Fig. 2 and Supporting Information Fig. S2). HCOH-s2 proteins are mostly from bacterial phyla Firmicutes and Actinobacteria (Supporting Information Fig. S2). Interestingly, several single-cell eukaryotes also possess HCOH-s2 proteins, including some species in the order of Trypanosomatida such as those of the Leishmania genus, Angomonas deanei, and Strigomonas culicis. Manual inspection of weak PSI-BLAST hits also revealed a divergent HCOH-s2 protein from Naegleria gruberi (gi|290974763, Fig. 2), a free-living single-cell eukaryotic species of the Heterolobosea class. HCOH-s2 may be present in the ancestor of eukaryotes, and its patchy phylogenetic distribution suggests that it may be lost independently in most eukaryotic lineages. Leishmania species are parasites for leishmaniasis, a disease that causes skin sores and visceral failure.31,32 Leishmania species only include the last three enzymes in the heme biosynthetic pathway.33 Although Leishmania may be able to synthesize heme from heme precursors, it is thought to transport heme with an unknown mechanism34,35 and is uniquely dependent on the acquisition of exogenous heme for survival.36,37 Considering the membrane localization and potential heme-binding capability of HCOH-s2 proteins, Leishmania HCOH-s2 proteins might be involved in the maintenance of heme homeostasis in these parasites. One hypothesis about their function is that Leishmania HCOH-s2 proteins sequester hemes in the membrane to reduce heme toxicity38 to the cell and increase heme accessibility to other membrane proteins.
The majority of HCOH-s1 and HCOH-s2 proteins consist of a single domain corresponding to one EU of four TMHs. As exceptions, one HCOH-s1 protein has an N-terminal divergent cupin_2 domain as suggested by HHpred (gi|91786010, Fig. 5), and all F-cluster HCOH-s1 proteins contain an N-terminal thioredoxin domain (e.g., gi|220936290, Fig. 5). A small number of HCOH-s2 members have several additional TMHs (four or seven) (Fig. 5) in their N-termini that do not show detectable sequence similarity to the EUs of HCOs/NORs and HCOH proteins (based on PSI-BLAST and HHpred results).
Figure 5.
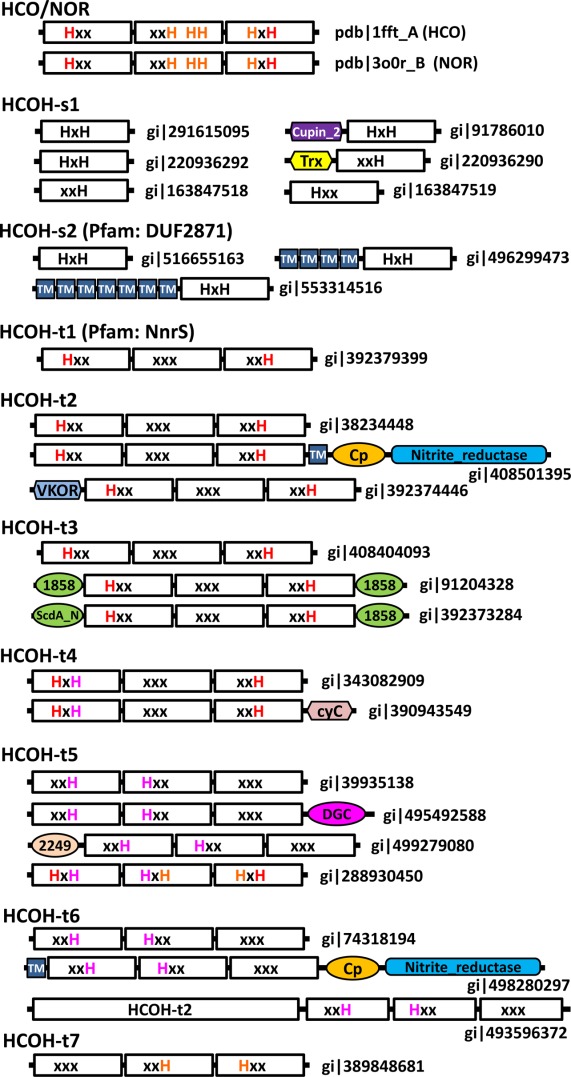
Domain structure diagrams of selected HCO/NOR and HCOH proteins. NCBI gi number or pdb/chain id is shown for each protein. EUs are shown in white rectangular boxes with motifs (HxH, xxH, Hxx, xxx, and HH). For three-EU proteins, conserved histidines at the interface of EU1 and EU3 are shown in red letters, those at the interface of EU2 and EU3 are shown in orange letters, and those at the interface of EU1 and EU2 are shown in magenta letters. Domain or module name abbreviations are as follows: cupin_2, cupin 2 domain; cyC, cytochrome c domain, Trx: thioredoxin domain; Cp, cupredoxin domain; 1858, DUF1858; 2249, DUF2249; TM, predicted transmembrane helix; and VKOR, vitamin K epoxide reductase domain.
HCOH groups with three EUs
We found a number of HCOH proteins with 12 TMHs (about 1500 proteins in the nre90 database). These HCOH proteins, like HCOs/NORs, consist of three EUs. However, they usually have fewer conserved histidines than HCOs/NORs. They form several clusters in the CLANS protein clustering diagram (Fig. 3). We divided them into seven groups: HCOH-t1 (HCO homology proteins with three EUs, group 1), HCOH-t2, HCOH-t3, HCOH-t4, HCOH-t5, HCOH-t6, and HCOH-t7 (see Supporting Information Figs. S3–S9 for their sequence information and alignments).
HCOH-t1, HCOH-t2, and HCOH-t3 proteins possess two conserved histidine residues in motifs Hxx of EU1 and xxH of EU3 and do not have conserved histidines in EU2 [Figs. 1(F) and 2]. These proteins also possess a conserved arginine in the third TMH of EU3 (Fig. 2). The two conserved histidines correspond to the two residues in HCOs/NORs that bind the low-spin heme. HCOH-t1 proteins, mainly from Proteobacteria (Supporting Information Fig. S3), correspond to the previously classified NnrS family (PF05940) in the Pfam database. The nnrS gene was identified as the neighboring gene of nnrR in Rhodobacter sphaeroides 2.4.1 and R. sphaeroides 2.4.3.39 The nnrR gene encodes a transcriptional regulator that responses to nitric oxide (NO) to activate the expression of the NOR gene norB. The expression of the nnrS gene is also dependent on the nnrR gene.39 The STRING functional association server40 suggests that the nnrR and nnrS genes co-occur with the norB-containing nor operon in various bacterial genomes, and they are often chromosomal neighbors, such as in the genome of R. sphaeroides 2.4.1 [Fig. 4(I)]. The purified NnrS protein appears to contain heme and copper.41 Disruption of the nnrS gene affected taxis towards nitrate and nitrite, suggesting a role of NnrS in the denitrification process.41
Recent studies showed that NnrS contributes to NO resistance in the bacterial pathogen Vibrio cholerae.42 NO is a host-generated reactive nitrogen species toxic to many bacterial pathogens such as V. cholerae. The nnrS gene in V. cholerae is up-regulated by the NorR transcriptional regulator in response to NO.42 NorR also activates the expression of the hmpA gene that encodes a protein with nitric oxide dioxygenase (NOD) activity that turns NO to less toxic nitrogen oxides.42 Gene disruption experiments suggest that nnrS and hmpA are important for V. cholerae colonization of intestines under the NO conditions, suggesting their roles in NO detoxification.42 Unlike HmpA, V. cholerae NnrS does not remove NO, but it may protect cellular iron pool from NO damage.43 The STRING server revealed that genes encoding NorR, HmpA, and NnrS are chromosomal neighbors in some bacteria, such as the opportunistic pathogen Pseudomonas aeruginosa [Fig. 4(J), fhpR and fhp encoding orthologous genes of norR and hmpA in V. cholerae, respectively), further supporting their functional association. The nnrS gene was also identified in a transposon mutagenesis screen to be important in host colonization of Neisseria meningitidis, a bacterial pathogen that causes meningitis.44
The majority of HCOH-t2 proteins are from Actinobacteria and Proteobacteria (Supporting Information Fig. S4). They also include some archaeal members mainly from the Crenarchaeota phylum. Most of the HCOH-t2 proteins are annotated as hypothetical proteins. A few HCOH-t2 proteins harbor additional domains. For example, the protein gi|408501395 has an additional TMH, a cupredoxin domain, and a copper-containing nitrite reductase domain C-terminal to the HCOH domain (Fig. 5). Such a domain composition suggests that HCOH-t2 may function in the denitrification process. Another protein, gi|392374446, has a different oxidoreductase domain (VKOR, vitamin K epoxide reductase) located N-terminally to the HCOH domain (Fig. 3). Genes encoding HCOH-t2 proteins are frequently found as neighbors of reductases in the denitrification process. For example, the HCOH-t2 gene is the neighbor to a NOR gene (norB) in Burkholderia pseudomallei K96243 [Fig. 4(K)] and to a copper-containing nitrite reductase gene in Corynebacterium diphtheriae NCTC 13129 [Fig. 4(N)]. In the hyperthermophilic crenarchaeon Pyrobaculum aerophilum str. IM2, the HCOH-t2 gene (PAE3602) is adjacent to a NOR gene (norB) and a nitrite reductase subunit (cytochrome D1) [Fig. 4(L)]. These three genes were all up-regulated after induction with nitrate.45 The association with these reductase genes also suggests that HCOH-t2 may function in the denitrification process.
The HCOH-t3 group consists of 14 bacterial sequences and one archaeal sequence forming a loosely connected cluster in the CLANS diagram (Fig. 3). A few HCOH-t3 proteins also possess DUF1858 (Pfam: PF08984) and ScdA_N (Pfam: PF04405) domains (Fig. 5). HHpred results suggest that DUF1858, a domain of unknown function, is distantly related to the ScdA_N domain. ScdA_N domain is named after the N-terminal domain of Staphylococcus aureus protein ScdA, which also contains the hemerythrin domain (Pfam: PF01814) that binds non-heme diirons.46 The bacterial hemerythrin domain-containing RIC (repair of iron centers) family proteins, including ScdA from S. aureus, DnrN from Neisseria gonorrhoeae and YtfE from Escherichia coli, confer resistance to reactive nitrogen and oxygen molecules such as NO and H2O2 by repairing their damages to iron-sulfur centers.47 The presence of the ScdA_N domain in many RIC proteins suggests that ScdA_N could aid in oxidative or nitrosative stress response. Such a domain in a few HCOH-t3 proteins indicates that they may also be involved in resistance to reactive nitrogen molecules such as NO, like some HCOH-t1 (NnrS) members. In Solibacter usitatus Ellin6076, the HCOH-t3 gene (Acid_2923) is in the neighborhood of genes of the RIC family and genes containing ScdA_N and DUF1858 domains [Fig. 4(M)], further supporting their functional associations.
HCOH-t4 proteins exhibit the histidine patterns of HxH, xxx, xxH in EU1, EU2, and EU3, respectively (Fig. 2). Similar to HCOH-t1, HCOH-t2, and HCOH-t3, such a pattern allows coordination of a heme group at the interface of EU1 and EU3. HCOH-t4 proteins are mainly from the Bacteroidetes phylum (Supporting Information Fig. S6). A few of them contain a cytochrome c domain at the C-terminus (Fig. 5), suggesting that they may be involved in cytochrome c-dependent electron transfer. The HCOH-t4 gene is often located near the nos operon [one example shown in Fig. 4(O)] that encodes nitrous oxide (N2O) reductase, which catalyzes the final step of the denitrification pathway: conversion of N2O to dinitrogen. A gene encoding the RIC family protein ScdA is also frequently found to be close to the HCOH-t4 gene [e.g., Fig. 4(O)], suggesting that HCOH-t4 could be involved in denitrification and detoxification of reactive molecules.
The majority of HCOH-t5 and HCOH-t6 proteins possess the xxH and Hxx motifs in the second TMH of EU1 and the second TMH of EU2, respectively, and lack conserved histidines in EU3 (Fig. 2 and Supporting Information Figs. S7 and S8). Such a pattern allows coordination of one heme group at the interface between EU1 and EU2 [Fig. 1(G)]. HCOH-t5 members are present in both bacteria and archaea. The bacterial members of HCOH-t5 are mainly from Proteobacteria and Firmicutes, while the archaeal members are all from the Halobacteria class of the Euryarchaeota phylum (Supporting Information Fig. S7). The majority of HCOH-t5 proteins are annotated as hypothetical proteins and do not contain additional domains. As exceptions, a few HCOH-t5 proteins possess domains of unknown function such as DUF2249 (Pfam: PF10006) and DGC (Pfam: PF08859, a domain with four conserved cysteines that likely coordinate zinc). The STRING server revealed strong association of HCOH-t5 proteins with proteins containing DUF2249 domain, proteins with DUF59 domain, and RIC proteins based on evidence of gene neighborhood, gene fusion (in the case of DUF2249 domain), and gene co-occurrence. For example, the HCOH-t5 gene from Rhizobium etli CFN 42 (RHE_PF00521) is predicted to be functionally associated with two genes containing DUF2249 domains (RHE_PF00520 and RHE_PF00522) and a gene with DUF59 domain (RHE_PF00523) [Fig. 4(P)]. These genes are also neighbors to nnrS (a HCOH-s1 gene), nnrR, and gene clusters encoding denitrification enzymes such as NOR and nitrite reductase [Fig. 4(O)].
HCOH-t6 proteins are mainly from Actinobacteria (Supporting Information Fig. S8). Some HCOH-t6 proteins are annotated as “multicopper oxidase” or “nitrite reductase,” as they also possess a cupredoxin domain and a copper-containing nitrite reductase domain. Such a domain composition is similar to a few HCOH-t2 proteins described above (Fig. 5). Interestingly, HCOH-t6 and HCOH-t2 genes are often chromosomal neighbors, such as DIP1877 (a HCOH-t6 gene) and DIP1878 (a HCOH-t2 gene) of Corynebacterium diphtheriae NCTC 13129 [Fig. 4(N)]. In one case, HCOH-t2 and HCOH-t6 are fused together in one open reading frame (gi|493596372 from Actinomyces urogenitalis, Fig. 5). These observations suggest that HCOH-t2 and HCOH-t6 may have related functions.
HCOH-t7 proteins exhibit yet another pattern of histidine motifs with xxH in EU2 and Hxx in EU3, while lacking conserved histidines in EU1 (Fig. 2 and Supporting Information Fig. S9). Such a pattern would allow coordination of one heme group at the interface between EU2 and EU3 [Fig. 1(H)]. HCOH-t7 proteins form two small clusters in the CLANS diagram (Fig. 3). Each cluster has restricted phylogenetic distribution. They are from the bacterial phylum Aquificae and from the archaeal class Halobacteria of the Euryarchaeota phylum, respectively (Supporting Information Fig. S9). These proteins are annotated as hypothetical proteins and do not have additional domains.
In summary, three-EU HCOH genes are often neighbors to genes involved in the denitrification process and/or detoxification of small reactive molecules such as NO (Fig. 4). Many of these neighboring genes encode various denitrification enzymes such as nitrate reductase, nitrite reductase, NOR, and nitrous oxide reductase (Fig. 4). The detoxification genes include those that encode NOD and the RIC family proteins with hemerythrin domains. In addition, genes encoding a few domains of unknown functions, such as ScdA_N, DUF2249, DUF1858, and DUF59, are also frequently found in the neighborhood of three-EU HCOH genes. Three-EU HCOH proteins from different groups are sometimes gene neighbors (Fig. 4), suggesting that they are involved in the same biological process. The gene neighborhood associations are consistent with the domain contents of limited multi-domain three-EU HCOH proteins, as some of these proteins contain nitrite reductase domain, DUF2249 domain and DUF1858 domain (Fig. 5).
For some single-EU HCOH proteins such as HCOH-s2 (DUF2871) proteins and cluster A HCOH-s1 proteins, we did not find strong functional associations to denitrification/detoxification genes according to the results of the STRING server. On the other hand, genes encoding HCOH-s1 proteins of clusters B, C, D, E, and F are frequently neighbors to denitrification genes and potential detoxification genes such as those with the hemerythrin domain and the globin domain48 (Fig. 4(A–F)]. Like many three-EU HCOH genes, these single-EU HCOH genes often have neighbors with domains of unknown function such as ScdA_N, DUF2249 and DUF59 [Fig. 4(A–F)]. Other genes frequently in the vicinity of HCOH-s1 genes include those encoding proteins with cytochrome c domain (cyC), ferredoxin domain (fx), thioredoxin domain (Trx), and cupin_2 domain (cp2) [Fig. 4(A–F)]. Some three-EU HCOH genes are also found to be neighbors of HCOH-s1 genes [e.g., Fig. 4(B,C,E)].
Limited experimental studies on NnrS proteins, gene context analysis and domain content analysis indicate the involvement of many HCOH proteins in denitrification and detoxification. The molecular mechanisms of their functions remain unknown and await further experimental investigations. As putative heme-binding proteins, HCOH proteins could contribute to denitrification and detoxification in several ways, such as maintenance of cellular iron and heme homeostasis, being part of the electron transfer pathways in various denitrification enzymes, binding/sequestering/exporting reactive nitrogen species, and possessing enzymatic activities that convert reactive nitrogen species to less toxic molecules.
Structure similarity between the catalytic subunits of HCOs/NORs and the MAPEG family proteins
After establishing the EUs for the HCO superfamily, we sought to explore its relationship to known structures. We queried the structures of HCO/NOR EUs against the SCOP49 database using the HorA server,50 which not only reports structural similarity, but also evaluates if the similarity represents homology or analogy. HorA identified a MAPEG structure as the top hit following the structures from the HCO superfamily, with relatively good scores for both structure comparison (Dali Z-score 7.6) and sequence comparison (HHpred probability 0.52), resulting in an overall score (5.042) that is consistent with scores derived for distant homologs.51 MAPEG proteins are a group of membrane-bound enzymes with diverse functions, including glutathione transferase activity that provides protection from oxidative stress in the membrane.28 The MAPEG fold, consisting of four TMHs, forms a homotrimer that binds three substrates in a symmetric manner. The four TMHs of a MAPEG subunit adopt the same topology as the four TMHs in each EU of HCOs/NORs, showing right-handed connections between them. In addition, an unexpected structural similarity lies in the same arrangement of the three subunits of the MAPEG homotrimer compared to the three EUs in HCOs/NORs, as noticed before.29 A superposition of a trimeric MAPEG structure (pdb id: 4al0) onto an HCO structure (pdb id: 3mk7) (Fig. 6) reveals a striking similarity (Dali Z-score: 20.6, RMSD: 3.3Å) covering all 12 TMHs. Each of the three EUs of HCO structure corresponds to one monomer of the MAPEG homotrimer (Fig. 6). Interestingly, the hemes from the HCO structure (red sticks, Fig. 6) overlap with the MAPEG substrate (glutathione, magenta sticks, Fig. 6), which is located at the interfaces of the monomers. Combined with the trimeric state required for MAPEG enzyme function,52,53 the structural similarity and similar active site position suggest that MAPEG proteins and HCOs/NORs are evolutionarily related.29
Figure 6.
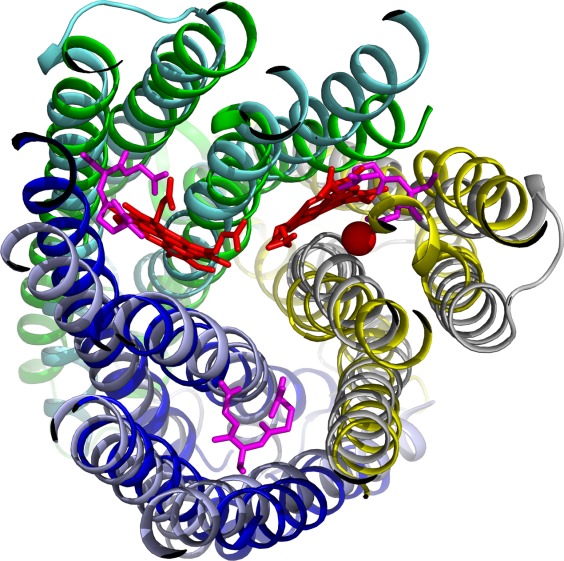
Structure superposition of an HCO superfamily member (pdb id: 3mk7) and a MAPEG member (pdb id: 4al0). EU1, EU2, and EU3 of 3mk7 are shown in blue, yellow, and green, respectively. The heme groups and the non-heme iron are shown as red sticks and a red sphere, respectively. The three chains of 4al0 aligned to EU1, EU2, and EU3 of 3mk7 are shown in light blue, grey, and cyan, respectively. The ligands of 4al0 are shown as magenta sticks.
Evolutionary scenarios of HCOs/NORs and the HCOH proteins
Gene duplication, divergence, fusion, and fission are the main driving forces in the evolution of proteins with novel functions.54–56 It is estimated that a large fraction of proteins form oligomers with functional importance.57 Evolution of oligomeric protein complexes has drawn interest in both theoretic and experimental studies.58,59 Gene duplication is considered as the cause to generate heteromers (hetero-oligomers) from homomers (homo-oligomers), for example, in the evolution of proteasomes60 and chaperonins.61 Subunits in a heteromer have different sequences and structures that break the symmetry of homomers and allow more versatile functions.62 It is estimated that a large fraction of membrane proteins form oligomers or are internally pseudosymmetric,63,64 like the catalytic subunits of HCOs/NORs.
The structural similarity between the catalytic subunits of HCOs/NORs and the MAPEG trimers suggests that HCOs/NORs could have evolved from a four-TMH ancestral protein that forms MAPEG-like homotrimers (Fig. 7). It is likely that a fortuitous sequence divergence event of this ancestor [Fig. 7(A)] gave rise to the HxH motif in the second TMH that enabled binding of heme groups. The discovery of single-EU HCOH proteins with four TMHs and the HxH motif supports this hypothesis. The homotrimers of four-TMH single-EU proteins with the HxH motif would have three symmetric heme-binding sites located at the interfaces of the monomers, similar to those of the MAPEG proteins. Single-EU HCOH genes could have undergone gene duplications [Fig. 7(B)] followed by sequence divergence to generate multiple copies of single-EU HCOH genes encoding proteins that form heterotrimers. If the HxH motif is kept in all duplicated genes, a heterotrimer with three heme-binding sites could be formed. In case some histidines are mutated [Fig. 7(C)], like the HCOH-s1 cluster B proteins, heterotrimers that bind less than three hemes could evolve. The fusion of three single-EU genes would result in an open reading frame with three EUs [Fig. 7(D,F)]. Three-EU proteins could bind three hemes with six histidines if the HxH motif is kept in all three EUs [Fig. 7(D)]. Interestingly, we identified one archaeal protein in the HCOH-t5 group (gi|288930450 in Fig. 2, marked by a star in Fig. 3) that has three HxH motifs and could bind three hemes. Deterioration of some histidines, either in the stage of single-EU ancestors [Fig. 7(C)] or three-EU ancestors [Fig. 7(E)], could lead to three-EU HCOH proteins with fewer than six conserved histidines and thus less than three heme-binding sites.
Figure 7.
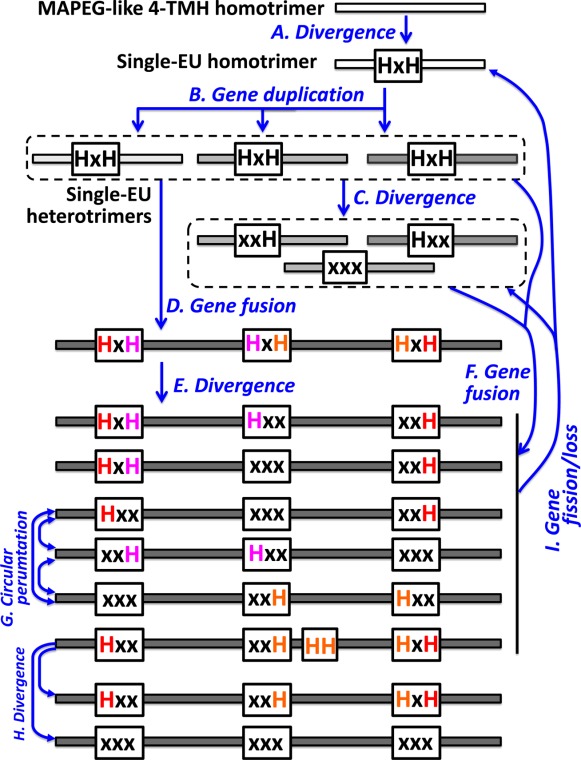
Possible evolutionary events in HCOs/NORs and HCOH proteins.
Most of extant three-EU HCOH proteins possess two conserved histidines and are inferred to bind a single heme. They mostly follow one of three histidine patterns: Hxx.xxx.xxH (HCOH-t1, HCOH-t2, HCOH-t3, and HCOH-t4), xxH.Hxx.xxx (HCOH-t5 and HCOH-t6) and xxx.xxH.Hxx (HCOH-t7) (Fig. 7). These three patterns can also be related by circular permutations of the three EUs [Fig. 7(G)]. We also identified a small number of archaeal HCOH-t5 proteins (e.g., gi|389847617 in Fig. 2) that contain a combination of patterns of HCOH-t1 and HCOH-t5 and have four histidines in the motif pattern of HxH.Hxx.xxH (Fig. 7). These proteins could thus bind two hemes.
HCOs/NORs follow the histidine pattern of Hxx.xxH.HH.HxH. The HH motif in the seventh TMH (the third TMH in EU2) is a unique feature of HCOs/NORs that is not present in HCOH proteins. Such an addition allows HCOs/NORs to coordinate a copper or non-heme iron in addition to binding two hemes. We also identified several small groups of proteins closely related to HCOs/NORs that have some or all of the conserved histidines deteriorated. Two small groups (light blue dots in Fig. 3 and also see the close-up view in Supporting Information Fig. S10) have the HH motif deteriorated (see Supporting Information Figs. S14 and S15 for their alignments). Four small groups (yellow dots in Fig. 3 and Supporting Information Fig. S10) have all of the six histidines deteriorated (see Supporting Information Figs. S16–S19 for their alignments). These groups could have evolved from the HCOs/NORs by sequence divergence [Fig. 7(H)].
Although it is likely that three-EU HCOH proteins have evolved by gene duplication and fusion of single-EU HCOH ancestors, the opposite evolutionary scenario, gene split (or fission) of three-EU HCOH genes to generate single-EU HCOH open reading frames [Fig. 7(I)], is also plausible. For example, HCOH-s1 proteins of clusters H and G, especially cluster H proteins, are close to the HCOs/NORs in the CLANS diagram (Fig. 3). Their top BLAST hits include C-type HCOs. In the genome of Sideroxydans lithotrophicus ES-1, one cluster G HCOH-s1 gene and one cluster H HCOH-s1 gene are close to each other and neighboring to a C-type HCO (Fig. 4). It is likely that HCOH-s1 proteins of clusters G and H are derived from C-type HCO proteins by gene fission.
Conclusions
Comparative sequence-structure analysis revealed novel homology between a number of HCOH proteins and the catalytic subunits of HCOs/NORs. HCOH proteins form groups of four-TMH single-EU proteins (HCOH-s1 and HCOH-s2) and groups of 12-TMH three-EU proteins (HCOH-t1, HCOH-t2, HCOH-t3, HCOH-t4, HCOH-t5, HCOH-t6, and HCOH-t7). Among these groups, only HCOH-s2 and HCOH-t1 correspond to known domains (DUF2871 and NnrS, respectively), while the majority of other HCOH members are currently annotated as hypothetical proteins without known domains. Gene context and domain content analyses, coupled with limited experimental studies of NnrS, suggest that most HCOH proteins are involved in the denitrification process and/or detoxification of reactive small molecules. Based on the structures of HCOs/NORs, single-EU HCOH proteins could form homotrimers or heterotrimers with active sites located at the interfaces between monomers. Conserved histidines in HCOH proteins indicate that they can bind heme. Strong structural similarity was observed between the homotrimers of the MAPEG family membrane enzymes and the catalytic subunits of HCOs/NORs. Such a similarity, together with the discovery of single-EU HCOH proteins, suggests that HCOs/NORs and three-EU HCOH proteins could have evolved from four-TMH ancestors that form homotrimers similar to MAPEG proteins. Gene duplication, sequence divergence, and gene fusion of ancestral single-EU HCOH proteins could give rise to three-EU HCOH proteins and HCOs/NORs. Conversely, gene fission of three-EU HCOH proteins or HCOs/NORs may have produced some extant single-EU HCOH proteins.
Materials and Methods
Sequence similarity searches
PSI-BLAST26 iterations were conducted to search for homologs of the HCO superfamily proteins starting from one representative with known structure (protein databank (PDB65) id: 3o0r, chain B)16 against a database composed of NCBI non-redundant proteins and environmental sequences with maximal 90% identity (nre90) protein database (e-value inclusion cutoff: 1e-4). To perform transitive searches, PSI-BLAST hits were grouped by BLASTCLUST (with the score coverage threshold [−S, defined as the bit score divided by alignment length) set to 1, length coverage threshold (−L) set to 0.5, and no requirement of length coverage on both sequences (−b F)], and a representative sequence from each group was used to initiate new PSI-BLAST searches. Such an iterative procedure was repeated until convergence. HHpred27 was used for profile-profile-based similarity searches to identify distant homologous relationships for (1) HCOs/NORs (gi|315583520), (2) the HCOH-s1 group (gi|499132825), (3) the HCOH-s2 group (gi|316941303), (4) the HCOH-t1 group (gi|110347088), (5) the HCOH-t5 group (gi|292656262), and (6) the HCOH-t6 group (gi|256378768) (profile databases used: Pfam,66 PDB,65 and CDD67). Detections of conserved domains are performed by the CDD server67 and the HMMER3 package.68 We also employed the HorA server50 to detect structural homologs for the pseudosymmetric units of HCO proteins, using the C terminus of a NOR structure (pdb: 3o0r, chain B, residue 302–458) as input.
Sequence clustering and multiple sequence alignment
Sequence clustering was performed and visualized by the CLANS program.30 Several cutoffs of P-values were tried. The P-value cutoff 1e-10 was chosen since it gave the best separation between clusters according to manual inspections. We extracted the sequences in each manually defined group of CLANS results and performed multiple sequence alignments by PROMALS3D69 for each group. These alignments as well as information about their sequences such as species and domain ranges are available in the Supporting Information. Representative sequences for HCOs/NORs and the newly defined HCOH groups were selected and split into individual EUs of four TMHs. A multiple sequence alignment was constructed for the EUs of these representatives by PROMALS3D. This alignment was manually adjusted by taking into account sequence conservation, structural superposition of known structures of HCOs/NORs by DaliLite70 and MUSTANG,71 hydrophobicity and small residue patterns, and transmembrane regions predictions by Phobius72 and TMHMM.73
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
References
- 1.Catling DC, Glein CR, Zahnle KJ, McKay CP. Why O2 is required by complex life on habitable planets and the concept of planetary “oxygenation time”. Astrobiology. 2005;5:415–438. doi: 10.1089/ast.2005.5.415. [DOI] [PubMed] [Google Scholar]
- 2.Payne JL, Boyer AG, Brown JH, Finnegan S, Kowalewski M, Krause RA, Jr, Lyons SK, McClain CR, McShea DW, Novack-Gottshall PM, Smith FA, Stempien JA, Wang SC. Two-phase increase in the maximum size of life over 3.5 billion years reflects biological innovation and environmental opportunity. Proc Natl Acad Sci USA. 2009;106:24–27. doi: 10.1073/pnas.0806314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahl TW, Hammarlund EU, Anbar AD, Bond DP, Gill BC, Gordon GW, Knoll AH, Nielsen AT, Schovsbo NH, Canfield DE. Devonian rise in atmospheric oxygen correlated to the radiations of terrestrial plants and large predatory fish. Proc Natl Acad Sci USA. 2010;107:17911–17915. doi: 10.1073/pnas.1011287107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gribaldo S, Talla E, Brochier-Armanet C. Evolution of the haem copper oxidases superfamily: a rooting tale. Trends Biochem Sci. 2009;34:375–381. doi: 10.1016/j.tibs.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 6.van der Oost J, de Boer AP, de Gier JW, Zumft WG, Stouthamer AH, van Spanning RJ. The heme-copper oxidase family consists of three distinct types of terminal oxidases and is related to nitric oxide reductase. FEMS Microbiol Lett. 121:1–9. doi: 10.1111/j.1574-6968.1994.tb07067.x. [DOI] [PubMed] [Google Scholar]
- 7.Richter OM, Ludwig B. Cytochrome c oxidase—structure, function, and physiology of a redox-driven molecular machine. Rev Physiol Biochem Pharmacol. 2003;147:47–74. doi: 10.1007/s10254-003-0006-0. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson-Miller S, Babcock GT. Heme/copper terminal oxidases. Chem Rev. 1996;96:2889–2908. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 9.Michel H, Behr J, Harrenga A, Kannt A. Cytochrome c oxidase: structure and spectroscopy. Annu Rev Biophys Biomol Struct. 1998;27:329–356. doi: 10.1146/annurev.biophys.27.1.329. [DOI] [PubMed] [Google Scholar]
- 10.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 11.Sousa FL, Alves RJ, Ribeiro MA, Pereira-Leal JB, Teixeira M, Pereira MM. The superfamily of heme-copper oxygen reductases: types and evolutionary considerations. Biochim Biophys Acta. 2012;1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Brochier-Armanet C, Talla E, Gribaldo S. The multiple evolutionary histories of dioxygen reductases: Implications for the origin and evolution of aerobic respiration. Mol Biol Evol. 2009;26:285–297. doi: 10.1093/molbev/msn246. [DOI] [PubMed] [Google Scholar]
- 13.Gennis RB. Multiple proton-conducting pathways in cytochrome oxidase and a proposed role for the active-site tyrosine. Biochim Biophys Acta. 1998;1365:241–248. [Google Scholar]
- 14.Rauhamaki V, Baumann M, Soliymani R, Puustinen A, Wikstrom M. Identification of a histidine-tyrosine cross-link in the active site of the cbb3-type cytochrome c oxidase from Rhodobacter sphaeroides. Proc Natl Acad Sci USA. 2006;103:16135–16140. doi: 10.1073/pnas.0606254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemp J, Christian C, Barquera B, Gennis RB, Martinez TJ. Helix switching of a key active-site residue in the cytochrome cbb3 oxidases. Biochemistry. 2005;44:10766–10775. doi: 10.1021/bi050464f. [DOI] [PubMed] [Google Scholar]
- 16.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 17.Moenne-Loccoz P, Fee JA. Biochemistry. Catalyzing NO to N2O in the nitrogen cycle. Science. 2010;330:1632–1633. doi: 10.1126/science.1200247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendriks J, Gohlke U, Saraste M. From NO to OO: nitric oxide and dioxygen in bacterial respiration. J Bioenerg Biomembr. 1998;30:15–24. doi: 10.1023/a:1020547225398. [DOI] [PubMed] [Google Scholar]
- 19.Stevanin TM, Laver JR, Poole RK, Moir JW, Read RC. Metabolism of nitric oxide by Neisseria meningitidis modifies release of NO-regulated cytokines and chemokines by human macrophages. Microbes Infect. 2007;9:981–987. doi: 10.1016/j.micinf.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Hendriks J, Oubrie A, Castresana J, Urbani A, Gemeinhardt S, Saraste M. Nitric oxide reductases in bacteria. Biochim Biophys Acta. 2000;1459:266–273. doi: 10.1016/s0005-2728(00)00161-4. [DOI] [PubMed] [Google Scholar]
- 21.Sousa FL, Alves RJ, Pereira-Leal JB, Teixeira M, Pereira MM. A bioinformatics classifier and database for heme-copper oxygen reductases. PLoS One. 2011;6:e19117. doi: 10.1371/journal.pone.0019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castresana J, Lubben M, Saraste M, Higgins DG. Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J. 1994;13:2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Vries S, Schroder I. Comparison between the nitric oxide reductase family and its aerobic relatives, the cytochrome oxidases. Biochem Soc Trans. 2002;30:662–667. doi: 10.1042/bst0300662. [DOI] [PubMed] [Google Scholar]
- 24.Ducluzeau AL, van Lis R, Duval S, Schoepp-Cothenet B, Russell MJ, Nitschke W. Was nitric oxide the first deep electron sink? Trends Biochem Sci. 2009;34:9–15. doi: 10.1016/j.tibs.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Castresana J, Saraste M. Evolution of energetic metabolism: the respiration-early hypothesis. Trends Biochem Sci. 1995;20:443–448. doi: 10.1016/s0968-0004(00)89098-2. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsson PJ, Morgenstern R, Mancini J, Ford-Hutchinson A, Persson B. Common structural features of MAPEG—a widespread superfamily of membrane associated proteins with highly divergent functions in eicosanoid and glutathione metabolism. Protein Sci. 1999;8:689–692. doi: 10.1110/ps.8.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm PJ, Bhakat P, Jegerschold C, Gyobu N, Mitsuoka K, Fujiyoshi Y, Morgenstern R, Hebert H. Structural basis for detoxification and oxidative stress protection in membranes. J Mol Biol. 2006;360:934–945. doi: 10.1016/j.jmb.2006.05.056. [DOI] [PubMed] [Google Scholar]
- 30.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 31.Oliveira F, de Carvalho AM, de Oliveira CI. Sand-fly saliva—man: the trigger trio. Frontiers Immunol. 2013;4:375. doi: 10.3389/fimmu.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vannier-Santos MA, Martiny A, de Souza W. Cell biology of Leishmania spp.: invading and evading. Curr Pharma Des. 2002;8:297–318. doi: 10.2174/1381612023396230. [DOI] [PubMed] [Google Scholar]
- 33.Koreny L, Lukes J, Obornik M. Evolution of the haem synthetic pathway in kinetoplastid flagellates: an essential pathway that is not essential after all? Intl J Parasitol. 2010;40:149–156. doi: 10.1016/j.ijpara.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Campos-Salinas J, Cabello-Donayre M, Garcia-Hernandez R, Perez-Victoria I, Castanys S, Gamarro F, Perez-Victoria JM. A new ATP-binding cassette protein is involved in intracellular haem trafficking in Leishmania. Mol Microbiol. 2011;79:1430–1444. doi: 10.1111/j.1365-2958.2010.07531.x. [DOI] [PubMed] [Google Scholar]
- 35.Huynh C, Yuan X, Miguel DC, Renberg RL, Protchenko O, Philpott CC, Hamza I, Andrews NW. Heme uptake by Leishmania amazonensis is mediated by the transmembrane protein LHR1. PLoS Pathog. 2012;8:e1002795. doi: 10.1371/journal.ppat.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dutta S, Furuyama K, Sassa S, Chang KP. Leishmania spp.: delta-aminolevulinate-inducible neogenesis of porphyria by genetic complementation of incomplete heme biosynthesis pathway. Exp Parasitol. 2008;118:629–636. doi: 10.1016/j.exppara.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang CS, Chang KP. Heme requirement and acquisition by extracellular and intracellular stages of Leishmania mexicana amazonensis. Mol Biochem Parasitol. 1985;16:267–276. doi: 10.1016/0166-6851(85)90069-6. [DOI] [PubMed] [Google Scholar]
- 38.Anzaldi LL, Skaar EP. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun. 2010;78:4977–4989. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwiatkowski AV, Laratta WP, Toffanin A, Shapleigh JP. Analysis of the role of the nnrR gene product in the response of Rhodobacter sphaeroides 2.4.1 to exogenous nitric oxide. J Bacteriol. 1997;179:5618–5620. doi: 10.1128/jb.179.17.5618-5620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartnikas TB, Wang Y, Bobo T, Veselov A, Scholes CP, Shapleigh JP. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a haem-copper protein. Microbiology. 2002;148:825–833. doi: 10.1099/00221287-148-3-825. [DOI] [PubMed] [Google Scholar]
- 42.Stern AM, Hay AJ, Liu Z, Desland FA, Zhang J, Zhong Z, Zhu J. The NorR regulon is critical for Vibrio cholerae resistance to nitric oxide and sustained colonization of the intestines. MBio. 2012;3:e00013–00012. doi: 10.1128/mBio.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern AM, Liu B, Bakken LR, Shapleigh JP, Zhu J. A novel protein protects bacterial iron-dependent metabolism from nitric oxide. J Bacteriol. 2013;195:4702–4708. doi: 10.1128/JB.00836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamet A, Euphrasie D, Martin P, Nassif X. Identification of genes involved in Neisseria meningitidis colonization. Infect Immun. 2013;81:3375–3381. doi: 10.1128/IAI.00421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cozen AE, Weirauch MT, Pollard KS, Bernick DL, Stuart JM, Lowe TM. Transcriptional map of respiratory versatility in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. J Bacteriol. 2009;191:782–794. doi: 10.1128/JB.00965-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.French CE, Bell JM, Ward FB. Diversity and distribution of hemerythrin-like proteins in prokaryotes. FEMS Microbiol Lett. 2008;279:131–145. doi: 10.1111/j.1574-6968.2007.01011.x. [DOI] [PubMed] [Google Scholar]
- 47.Overton TW, Justino MC, Li Y, Baptista JM, Melo AM, Cole JA, Saraiva LM. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J Bacteriol. 2008;190:2004–2013. doi: 10.1128/JB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frey AD, Kallio PT. Nitric oxide detoxification—a new era for bacterial globins in biotechnology? Trends Biotechnol. 2005;23:69–73. doi: 10.1016/j.tibtech.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. SCOP: a structural classification of proteins database. Nucleic Acids Res. 2000;28:257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim BH, Cheng H, Grishin NV. HorA web server to infer homology between proteins using sequence and structural similarity. Nucleic Acids Res. 2009;37:W532–W538. doi: 10.1093/nar/gkp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng H, Kim BH, Grishin NV. Discrimination between distant homologs and structural analogs: lessons from manually constructed, reliable data sets. J Mol Biol. 2008;377:1265–1278. doi: 10.1016/j.jmb.2007.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez Molina D, Wetterholm A, Kohl A, McCarthy AA, Niegowski D, Ohlson E, Hammarberg T, Eshaghi S, Haeggstrom JZ, Nordlund P. Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase. Nature. 2007;448:613–616. doi: 10.1038/nature06009. [DOI] [PubMed] [Google Scholar]
- 53.Martinez Molina D, Eshaghi S, Nordlund P. Catalysis within the lipid bilayer-structure and mechanism of the MAPEG family of integral membrane proteins. Curr Opin Struct Biol. 2008;18:442–449. doi: 10.1016/j.sbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 54.Ohno S. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- 55.Kummerfeld SK, Teichmann SA. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21:25–30. doi: 10.1016/j.tig.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Pasek S, Risler JL, Brezellec P. Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins. Bioinformatics. 2006;22:1418–1423. doi: 10.1093/bioinformatics/btl135. [DOI] [PubMed] [Google Scholar]
- 57.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 58.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynch M. The evolution of multimeric protein assemblages. Mol Biol Evol. 2012;29:1353–1366. doi: 10.1093/molbev/msr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouzat JL, McNeil LK, Robertson HM, Solter LF, Nixon JE, Beever JE, Gaskins HR, Olsen G, Subramaniam S, Sogin ML, Lewin HA. Phylogenomic analysis of the alpha proteasome gene family from early-diverging eukaryotes. J Mol Evol. 2000;51:532–543. doi: 10.1007/s002390010117. [DOI] [PubMed] [Google Scholar]
- 61.Archibald JM, Logsdon JM, Jr, Doolittle WF. Origin and evolution of eukaryotic chaperonins: phylogenetic evidence for ancient duplications in CCT genes. Mol Biol Evol. 2000;17:1456–1466. doi: 10.1093/oxfordjournals.molbev.a026246. [DOI] [PubMed] [Google Scholar]
- 62.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi S, Jeon J, Yang JS, Kim S. Common occurrence of internal repeat symmetry in membrane proteins. Proteins. 2008;71:68–80. doi: 10.1002/prot.21656. [DOI] [PubMed] [Google Scholar]
- 64.Duran AM, Meiler J. Inverted topologies in membrane proteins: a mini-review. Comput Struct Biotechnol J. 2013;8:e201308004. doi: 10.5936/csbj.201308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rose PW, Bi C, Bluhm WF, Christie CH, Dimitropoulos D, Dutta S, Green RK, Goodsell DS, Prlic A, Quesada M, Quinn GB, Ramos AG, Westbrook JD, Young J, Zardecki C, Berman HM, Bourne PE. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 2013;41:D475–482. doi: 10.1093/nar/gks1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 71.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 72.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35:W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Intl Conf Intellig Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


