Abstract
Telocytes (TCs) were identified as a distinct cellular type of the interstitial tissue and defined as cells with extremely long telopodes (Tps). Our previous data demonstrated patterns of mouse TC-specific gene profiles on chromosome 1. The present study focuses on the identification of characters and patterns of TC-specific or TC-dominated gene expression profiles in chromosome 2 and 3, the network of principle genes and potential functional association. We compared gene expression profiles of pulmonary TCs, mesenchymal stem cells, fibroblasts, alveolar type II cells, airway basal cells, proximal airway cells, CD8+T cells from bronchial lymph nodes (T-BL), and CD8+ T cells from lungs (T-LL). We identified that 26 or 80 genes of TCs in chromosome 2 and 13 or 59 genes of TCs up-or down-regulated in chromosome 3, as compared with other cells respectively. Obvious overexpression of Myl9 in chromosome 2 of TCs different from other cells, indicates that biological functions of TCs are mainly associated with tissue/organ injury and ageing, while down-expression of Pltp implies that TCs may be associated with inhibition or reduction of inflammation in the lung. Dominant overexpression of Sh3glb1, Tm4sf1 or Csf1 in chromosome 3 of TCs is mainly associated with tumour promotion in lung cancer, while most down-expression of Pde5 may be involved in the development of pulmonary fibrosis and other acute and chronic interstitial lung disease.
Keywords: TCs, mesenchymal stem cells, fibroblasts, chromosome 2, chromosome 3, lung
Introduction
Telocytes (TCs) were first described as a distinct cell type in 2010 1,2, followed by a significant growing of research globally 3,4, as detailed in www.telocytes.com. TCs were found in multiple tissues and organs, such as heart 5–7, kidney 8 and urinary tract 9,10, skin 11,12 eye 13, mammary gland 14, digestive tract 15,16, skeletal muscles 17 and neuromuscular spindles 18, uterus 19–22 and placenta 2, liver 23 and gall bladder 24,25, pleura 26, trachea 27 and lungs 28. TCs are mainly recognized and characterized by electron microscopy, the only technique able to highlight their characteristic extensions – the telopodes (Tps), consisting of thin segments – podomers, alternating with dilated regions – podoms 1. Other characteristics of Tps include: (i) the unusual and varied length, between tens and thousands of micrometres; (ii) the branching network pattern, forming a labyrinthine system; (iii) the communications through homo-and heterocellular junctions exosome and ectosome release 6,20,22. TCs were found to link nerve fibres, blood vessels, secretory acini and exocrine epithelial ducts 29–31, and different cell types, e.g. macrophages, lymphocytes, mast cells, stem cells 32–34. TCs form 3-dimensional networks within organs/tissues 20,35. Networks integrity may be affected in many pathological conditions, such as systemic sclerosis 36, skin basal and squamous cell carcinomas 37 and Crohn's disease 38.
Telocytes differ from fibroblasts (Fbs) and mesenchymal stem cells (MSCs) as demonstrated by miR signatures and genetic profiles 15,39,40. Proteomic signatures of the TCs are also supportive for the uniqueness and helpful in understanding of the functions. The data from omics studies demonstrated that elements within TCs are involved in (i) intercellular signalling, (ii) mechanical sensing and mechanochemical conversion task, (iii) tissue homoeostasis and remodelling/renewal, (iv) anti-oxidative stress and anti-ageing cellular mechanisms, (v) cancer cell proliferation through the inhibition of apoptosis 40,41. Our recent work explored patterns of mouse TC-specific gene profiles on chromosome 1 and showed important roles for TCs in the prevention of tissue inflammation and fibrogenesis, development of lung inflammatory diseases or modulation of immune cell responses 42. However, dominant patterns and specificity of gene and protein profiles of TCs which are different from other cells existed in the lung is still not completed and unclear.
The present study undertakes an in-depth analysis to find out the characters and patterns of TC-specific or TC-dominated gene expression profiles in chromosome 2 and 3, investigate the network of principle genes, and explore potential functional association. Comparisons are made among pulmonary TCs, MSCs, Fbs, alveolar type II cells (ATII), airway basal cells (ABCs), proximal airway cells (PACs), CD8+T cells from bronchial lymph nodes (T-BL) and CD8+ T cells from lung (T-L), which may interact with TCs in the lung and trachea. Furthermore, we applied the most complete reference library of the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database to identify key functional genes, and characteristic networks by bioinformatics tools.
Material and methods
Isolation and culture
Telocytes were isolated from the lung tissues of mice, primary cultured in a concentration of 1 × 105 cells/cm2, and harvested on days 5 (TC D5) and on days 10 (TC D10), as described previously 43. RNA isolation, preparation, labelling, and hybridization were performed for DNA microarray (The Mouse 4 × 44K Gene Expression Array, Agilent, Shanghai, China). About 39,000+ mouse genes and transcripts represented with public domain annotations were gained, according to the protocol of One-Color Microarray-Based Gene Expression Analysis. The hybridized arrays were washed, fixed and scanned by the Agilent DNA Microarray Scanner (part number G2505B).
Data collection and mining
We selectively collected gene expression profiles of pulmonary TCs on days 5 (TC D5) and 10 (TC D10), Fbs, MSCs, from our study 43, ATII, ABCs, PACs, T-BL and T-L, from the NCBI Gene Expression Omnibus database (GSE6846 44, GSE27379 45, GSE28651 46). The microarray was composed of 45,101 probes. We eliminated the probe sets without corresponding official symbol, leaving 39,417 probes and 21,680 genes.
The gene expression profiles are from our earlier study, which are composed of 23,861 probes, of pulmonary TCs on days 5 and 10, Fbs and MSCs are composed of 23,861 probes 43. There were 13,236 probes and 11,545 genes after further eliminating the probes without corresponding official symbol, which we focused on in the present study. From the total of 11,545 genes, 917 genes of the chromosome 2 and 567 genes of the chromosome 3 were analysed.
Identification of differentially expressed genes
There are about 20,000–25,000 genes in mouse, of which about 85% are similar with humans, and the propensity of functional changes was reflected in different levels of the gene expression in particular cell types. We used gene expression profiles between mouse lung cells to seek for the specific regulated and identify genes specific to TCs and their function. The fold change was utilized to identify differentially expressed genes or simply differential genes. Up-or down-regulated folds of TCs genes were calculated as compared with other cells and subtracted its own multiple of TCs, after the average of gene expression in each cell was obtained from the raw data of multi-databases, as shown in Data S1.
Results
The final data analysis by bioinformatics tools showed that in chromosome 2, 26 genes were overexpressed in TCs, as compared with those in other cells (Table1). Among them, 20 genes (1110008F13Rik, 2310003F16Rik, 2900064A13Rik, Abl1, Ass1, Commd3, Commd7, Creb3l1, Dlgap4, Edf1, Id1, Manbal, Mocs3, Psmc3, Sdccag3, Slc39a13, Snai1, Spc25, Tubb2c, Srxn1) were overexpressed between 0 and 1 folds (Table1A). Four genes, Dbndd2 (Dysbindin domain-containing protein 2), Fbn1 (fibrillin 1), Tfpi (tissue factor pathway inhibitor) and Ak1 (adenylate kinase 1) genes, were overexpressed one-to-fourfold, in both TC D5 and TC D10, as compared with other cells (Table1B). Highest overexpressed gene, Myl9 (myosin, light chain 9), was over fourfold up-regulated in both TC D5 and TC D10 compared with other cells (Table1C). 80 genes in TCs were down-regulated, as compared with other cells (Table2). Of them, Gzf1, Pltp, Polr1b, Tasp1, Zbtb34 and Zfp120 were down-regulated more than onefold in TCs compared with other cells.
Table 1.
Summary of genes expressed preferentially in TCs, as compared with others
| Compared pairs/fold up-regulated | >0 | >1 | >4 | ||||
|---|---|---|---|---|---|---|---|
| TC5 versus others | 64 | 18 | 2 | ||||
| TC10 versus others | 56 | 13 | 4 | ||||
| TCs versus others | 26 | 6 | 2 | ||||
| Gene symbol | Folds (TC5 versus others/TC10 versus others) | ||||||
| Fibroblast | Stem | ATII | CD8_T_BL | CD8_T_LL | Basal_cell | Duct_cell | |
| (A) Genes up-regulated between 0-and 1-folds in TCs as compared with others | |||||||
| 1110008F13Rik | −0.41/−0.18 | −0.46/−0.25 | −0.73/−0.72 | −0.79/−0.80 | −0.85/−0.85 | −0.91/−0.91 | −0.91/−0.92 |
| 2310003F16Rik | −0.40/−0.04 | −0.49/−0.19 | −0.97/−0.97 | −0.52/−0.46 | −0.56/−0.50 | −0.82/−0.80 | −0.86/−0.84 |
| 2900064A13Rik | −0.13/−0.21 | −0.41/−0.46 | −0.37/−0.58 | −0.29/−0.54 | −0.30/−0.54 | −0.24/−0.51 | −0.19/−0.47 |
| Abl1 | −0.75/−0.67 | −0.43/−0.23 | −0.76/−0.76 | −0.29/−0.32 | −0.70/−0.71 | −0.82/−0.83 | −0.89/−0.90 |
| Ass1 | −0.44/−0.34 | −0.96/−0.95 | −0.82/−0.84 | −0.94/−0.95 | −0.80/−0.83 | −0.90/−0.92 | −0.80/−0.84 |
| Commd3 | −0.63/−0.13 | −0.63/−0.54 | −0.56/−0.62 | −0.78/−0.59 | −0.76/−0.60 | −0.13/−0.34 | −0.24/−0.65 |
| Commd7 | −0.29/−0.56 | −0.62/−0.91 | −0.58/−0.49 | −0.53/−0.99 | −0.54/−0.99 | −0.24/−0.87 | −0.60/−0.89 |
| Creb3l1 | −0.65/−0.66 | −0.93/−0.44 | −0.46/−0.35 | −0.99/−0.18 | −0.99/−0.52 | −0.86/−0.25 | −0.88/−0.29 |
| Dlgap4 | −0.79/−0.98 | −0.65/−0.96 | −0.44/−0.77 | −0.28/−1.00 | −0.58/−0.96 | −0.34/−0.37 | −0.38/−0.71 |
| Edf1 | −0.44/−0.17 | −0.40/−0.08 | −0.51/−0.73 | −0.78/−0.76 | −0.81/−0.61 | −0.26/−0.28 | −0.26/−0.66 |
| Id1 | −0.98/−0.70 | −0.97/−0.03 | −0.74/−0.20 | −0.99/−0.50 | −0.95/−0.45 | −0.27/−0.63 | −0.66/−0.59 |
| Manbal | −0.49/−0.51 | −0.43/−0.43 | −0.77/−0.34 | −0.79/−0.19 | −0.66/−0.19 | −0.37/−0.20 | −0.71/−0.25 |
| Mocs3 | −0.81/−0.53 | −0.38/−0.02 | −0.30/−0.90 | −0.55/−0.95 | −0.51/−0.95 | −0.67/−0.55 | −0.63/−0.57 |
| Psmc3 | −0.64/−0.46 | −0.58/−0.20 | −0.34/−0.29 | −0.17/−0.58 | −0.18/−0.36 | −0.18/−0.67 | −0.23/−0.93 |
| Sdccag3 | −0.44/−0.46 | −0.48/−0.60 | −0.76/−0.37 | −0.77/−0.98 | −0.74/−0.94 | −0.73/−0.45 | −0.67/−0.18 |
| Slc39a13 | −0.51/−0.59 | −0.28/−0.77 | −0.13/−0.42 | −0.46/−0.73 | −0.20/−0.75 | −0.58/−0.72 | −0.91/−0.80 |
| Snai1 | −0.84/−0.26 | −0.37/−0.15 | −0.92/−0.67 | −0.93/−0.74 | −0.95/−0.58 | −0.95/−0.40 | −0.97/−0.59 |
| Spc25 | −0.41/−0.37 | −0.24/−0.40 | −0.79/−0.91 | −0.61/−0.91 | −0.41/−0.91 | −0.19/−0.58 | −0.07/−0.85 |
| Tubb2c | −0.73/−0.59 | −0.84/−0.48 | −0.47/−0.40 | −0.74/−0.90 | −0.77/−0.89 | −0.73/−0.57 | −0.81/−0.74 |
| Srxn1 | −0.49/−0.68 | −0.71/−0.30 | −0.32/−0.86 | −0.96/−0.85 | −0.93/−0.83 | −0.69/−0.86 | −0.41/−0.93 |
| (B) Genes up-regulated between 1-and 4-folds in TCs as compared with other | |||||||
| Dbndd2 | −0.61/−1.00 | −0.66/−0.98 | −0.90/−0.73 | −0.96/−0.96 | −0.95/−0.96 | −0.83/−0.87 | −0.81/−0.90 |
| Fbn1 | −0.95/−0.98 | −0.84/−0.53 | −0.89/−0.84 | −0.95/−0.82 | −0.95/−0.84 | −0.66/−0.87 | −0.89/−0.76 |
| Tfpi | −0.60/−0.72 | −0.94/−0.58 | −0.85/−0.85 | −0.66/−0.95 | −0.73/−0.96 | −0.84/−0.78 | −0.82/−0.86 |
| Ak1 | −0.80/−0.74 | −0.84/−0.96 | −0.69/−0.93 | −1.00/−0.85 | −0.97/−0.87 | −0.94/−0.93 | −0.97/−0.92 |
| (C) Genes up-regulated between >4-folds in TCs as compared with others | |||||||
| Myl9 | −1.00/−0.96 | −0.92/−0.91 | −0.98/−0.92 | −1.00/−0.96 | −1.00/−0.98 | −0.98/−0.88 | −1.00/−0.94 |
Table 2.
Summary of genes expressed preferentially in TCs, as compared with others
| Compared pairs/fold down-regulated | >0 | >1 | >4 | ||||
|---|---|---|---|---|---|---|---|
| TC5 versus others | 140 | 14 | 0 | ||||
| TC10 versus others | 236 | 38 | 0 | ||||
| TCs versus others | 80 | 6 | 0 | ||||
| Gene symbol | Folds (TC5 versus others/TC10 versus others) | ||||||
| Fibroblast | Stem | ATII | CD8_T_BL | CD8_T_LL | Basal_cell | Duct_cell | |
| (A) Genes down-regulated between 0-and 1-folds in TCs as compared with others | |||||||
| 1500012F01Rik | 1.00/2.24 | 2.51/4.68 | 0.09/0.29 | 0.83/1.11 | 1.30/1.68 | 2.50/3.01 | 4.17/4.95 |
| 1600027N09Rik | 0.02/0.26 | 0.24/0.53 | 3.26/2.85 | 5.59/4.77 | 6.52/5.68 | 6.23/5.32 | 6.80/5.85 |
| 1700058C13Rik | 0.01/0.29 | 0.03/0.31 | 6.00/5.54 | 6.63/5.93 | 15.2/13.91 | 7.23/6.46 | 4.82/4.30 |
| 2010317E24Rik | 0.71/2.34 | 1.58/4.04 | 0.03/0.47 | 0.23/0.70 | 1.38/2.34 | 10.45/14.81 | 10.14/14.46 |
| 2810408M09Rik | 1.13/1.13 | 0.66/0.66 | 20.58/14.77 | 1.23/0.58 | 14.91/10.44 | 65.85/46.34 | 29.46/20.69 |
| 4921504E06Rik | 0.16/0.37 | 0.31/0.55 | 6.25/5.27 | 0.87/0.57 | 8.86/7.40 | 11.6/9.56 | 9.10/7.51 |
| 6820408C15Rik | 0.02/0.06 | 0.54/0.61 | 99.87/75.87 | 8.76/6.22 | 28.01/20.77 | 26.11/19.03 | 17.2/12.51 |
| Abca2 | 0.77/1.04 | 0.51/0.73 | 11.84/9.81 | 5.21/4.07 | 6.05/4.84 | 8.82/7.01 | 6.99/5.55 |
| Acvr2a | 1.31/1.33 | 0.42/0.43 | 23.28/16.86 | 13.98/9.7 | 25.29/18.05 | 84.04/59.65 | 70.93/50.56 |
| Angptl2 | 0.52/0.45 | 1.97/0.31 | 16.78/0.65 | 9.96/1.22 | 5.33/2.03 | 14.94/3.34 | 20.00/1.38 |
| Api5 | 0.06/0.28 | 0.40/1.50 | 3.39/9.94 | 6.09/5.56 | 4.92/2.84 | 3.10/8.51 | 3.11/11.60 |
| Arhgap1 | 0.30/0.39 | 0.05/0.84 | 1.45/3.21 | 1.27/5.61 | 0.44/4.59 | 4.67/2.81 | 3.64/2.84 |
| Arpc5l | 1.84/1.17 | 0.11/0.75 | 1.81/1.99 | 9.73/1.70 | 7.56/0.73 | 9.55/5.73 | 5.43/4.53 |
| Atf2 | 0.15/2.57 | 0.26/0.39 | 2.39/1.58 | 7.92/8.55 | 3.92/6.73 | 8.87/8.38 | 5.91/4.74 |
| B2m | 1.13/0.44 | 0.57/1.06 | 2.50/1.63 | 4.18/3.06 | 4.50/2.19 | 2.32/1.70 | 1.22/1.32 |
| Catsper2 | 1.51/0.83 | 0.31/1.01 | 14.79/2.94 | 24.41/9.06 | 27.92/4.63 | 19.7/10.11 | 15.59/6.82 |
| Cbfa2t2 | 0.21/0.82 | 0.43/0.34 | 0.27/1.18 | 0.97/2.13 | 0.86/2.37 | 0.60/1.00 | 0.28/0.35 |
| Cdca7 | 1.89/1.49 | 6.28/0.30 | 1.05/10.46 | 1.2/16.91 | 0.10/19.66 | 2.18/13.55 | 2.49/10.73 |
| Cep110 | 1.61/0.62 | 0.28/0.92 | 0.84/0.24 | 7.14/0.87 | 4.90/0.79 | 5.53/0.51 | 2.91/0.22 |
| Chchd5 | 0.45/3.93 | 0.13/11.43 | 6.34/1.55 | 3.16/1.67 | 2.13/0.35 | 12.54/2.84 | 10.66/3.24 |
| Ciz1 | 0.30/2.69 | 0.21/0.81 | 1.53/0.90 | 0.84/7.16 | 0.68/4.99 | 7.8/5.53 | 4.74/2.93 |
| Cry2 | 0.11/1.18 | 0.15/0.69 | 13.7/7.05 | 4.83/3.43 | 2.42/2.38 | 10.9/13.38 | 11.47/11.46 |
| Ddx27 | 0.42/0.74 | 0.44/0.62 | 0.20/1.47 | 1.27/0.74 | 1.09/0.61 | 5.47/7.32 | 6.52/4.46 |
| Ddx31 | 0.31/0.36 | 0.93/0.91 | 0.51/2.65 | 1.79/8.07 | 0.75/9.21 | 5.42/17.28 | 5.09/13.41 |
| Depdc7 | 1.34/0.33 | 0.42/0.38 | 13.2/11.89 | 0.67/3.96 | 2.85/1.96 | 2.96/9.11 | 33.03/9.65 |
| Dsn1 | 0.61/0.26 | 1.33/0.31 | 6.39/6.62 | 39.29/12.48 | 47.16/10.54 | 12.99/6.54 | 23.88/5.26 |
| Elp4 | 0.76/1.09 | 0.25/1.12 | 5.62/0.30 | 1.29/1.37 | 1.03/1.21 | 14.19/5.74 | 10.08/6.88 |
| Ext2 | 0.44/0.46 | 0.68/1.16 | 23.82/0.23 | 31.23/1.21 | 21.21/0.41 | 20.92/4.07 | 11.20/3.83 |
| Fmnl2 | 0.18/1.37 | 0.15/0.44 | 11.91/9.50 | 5.60/0.20 | 12.75/1.80 | 15.11/1.84 | 21.28/23.52 |
| Fubp3 | 0.04/0.99 | 0.42/1.88 | 14.72/5.69 | 34.28/34.4 | 32.79/41.90 | 93.41/11.27 | 73.83/20.93 |
| Gapvd1 | 0.04/0.18 | 0.09/3.47 | 3.56/5.04 | 5.04/13.37 | 3.84/8.91 | 3.82/4.01 | 3.24/2.43 |
| Gmeb2 | 0.30/1.39 | 0.70/0.70 | 15.73/5.58 | 17.11/1.21 | 7.65/0.98 | 2.14/13.63 | 2.63/9.73 |
| Hat1 | 0.56/0.99 | 0.09/1.32 | 12.93/24.06 | 49.03/30.61 | 46.29/21.08 | 26.19/20.45 | 29.7/11.00 |
| Il15ra | 1.42/0.59 | 0.07/0.54 | 2.25/11.67 | 0.64/5.29 | 2.21/12.29 | 4.80/14.33 | 1.58/20.32 |
| Mapkbp1 | 0.03/0.18 | 0.13/2.25 | 96.09/4.23 | 25.15/33.87 | 22.48/30.68 | 123.55/25.01 | 263.85/15.93 |
| Mga | 0.68/0.24 | 0.34/0.68 | 4.24/12.63 | 4.57/28.72 | 5.14/27.86 | 8.89/78.35 | 7.84/62.23 |
| Mkks | 0.26/0.64 | 0.25/0.71 | 1.66/4.26 | 3.04/5.77 | 2.84/4.51 | 6.40/4.39 | 6.51/3.77 |
| Mllt10 | 0.11/1.18 | 0.06/0.54 | 5.21/2.72 | 32.27/10.63 | 27.76/16.87 | 15.03/12.57 | 12.67/9.62 |
| Mrps5 | 0.30/0.37 | 0.29/0.79 | 2.85/11.87 | 3.12/12.53 | 1.15/5.55 | 16.43/1.34 | 8.73/1.72 |
| Necab3 | 3.74/0.72 | 0.49/0.20 | 3.72/10.21 | 2.23/38.12 | 6.28/36.48 | 11.85/20.21 | 3.01/23.07 |
| Nr6a1 | 0.87/2.71 | 0.40/0.63 | 18.84/2.64 | 14.87/0.78 | 18.42/2.54 | 41.84/5.30 | 67.71/1.82 |
| Ntng2 | 0.19/0.21 | 0.25/0.33 | 0.4/82.62 | 10.52/20.87 | 12.47/18.91 | 4.3/102.97 | 1.66/221.24 |
| Nusap1 | 0.59/1.31 | 1.62/0.83 | 0.56/4.26 | 0.21/4.42 | 0.4/5.07 | 2.96/8.61 | 4.62/7.64 |
| Olfr73 | 0.10/0.04 | 0.27/0.02 | 1.12/0.60 | 4.73/1.36 | 1.03/1.28 | 20.44/3.31 | 5.83/3.40 |
| P2rx3 | 0.88/0.59 | 0.47/0.52 | 0.77/5.49 | 5.71/32.77 | 1.29/28.59 | 5.36/15.24 | 12.31/12.91 |
| Pdhx | 1.09/0.82 | 0.10/0.80 | 11.01/2.93 | 7.06/3.08 | 5.76/1.16 | 5.97/16.23 | 6.61/8.67 |
| Phf20 | 0.77/3.35 | 0.87/0.37 | 0.88/2.17 | 5.00/1.11 | 3.20/3.81 | 1.03/7.36 | 0.67/1.62 |
| Polr3f | 0.83/1.25 | 0.21/0.69 | 10.77/16.44 | 10.86/12.56 | 14.35/15.82 | 15.73/35.52 | 7.5/57.87 |
| Rae1 | 0.71/0.82 | 0.58/0.91 | 0.59/0.56 | 1.34/11.47 | 1.61/13.79 | 4.08/4.73 | 1.55/1.89 |
| Rbm38 | 1.52/0.58 | 0.11/0.70 | 18.93/2.24 | 5.43/1.14 | 12.22/1.26 | 10.19/9.83 | 8.49/7.36 |
| Scn1a | 9.47/0.74 | 2.38/0.87 | 0.73/1.13 | 0.61/3.84 | 2.46/3.60 | 7.56/0.30 | 4.58/0.57 |
| Slc12a5 | 4.24/0.92 | 0.59/2.15 | 2.84/0.38 | 3.26/0.04 | 3.41/0.21 | 4.93/2.38 | 5.08/3.83 |
| Slc27a4 | 1.12/0.22 | 0.50/0.41 | 28.79/0.72 | 13.26/3.52 | 6.48/0.62 | 6.51/15.87 | 6.71/4.40 |
| Slc34a3 | 0.33/1.15 | 0.38/0.69 | 10.17/0.48 | 10.98/4.46 | 19.71/0.89 | 20.08/4.17 | 17.79/9.87 |
| Spata2 | 0.54/1.15 | 0.59/0.13 | 0.65/8.00 | 1.75/4.87 | 0.78/3.99 | 4.44/4.06 | 3.95/4.56 |
| Sptlc3 | 0.42/0.74 | 0.31/1.10 | 26.19/5.08 | 2.31/7.18 | 13.32/8.05 | 179.36/10.23 | 250.13/8.40 |
| Ss18l1 | 0.33/1.71 | 1.15/1.85 | 24.07/1.10 | 70.3/5.51 | 106.73/3.61 | 142.18/1.19 | 87.02/0.81 |
| Surf6 | 0.14/2.78 | 0.03/0.71 | 0.36/0.10 | 1.56/1.44 | 0.98/0.59 | 4.86/3.33 | 5.65/2.12 |
| Timm10 | 1.11/0.80 | 0.16/0.19 | 0.79/7.50 | 0.44/7.32 | 0.44/9.91 | 3.52/10.71 | 2.82/4.98 |
| Trub2 | 0.41/1.52 | 0.58/1.33 | 0.34/0.72 | 1.71/1.45 | 0.55/1.77 | 4.38/4.31 | 1.61/1.68 |
| Ttll9 | 0.06/2.02 | 0.03/0.34 | 13.59/16.46 | 4.75/4.47 | 8.44/10.41 | 63.39/8.51 | 39.11/7.1 |
| Yme1l1 | 1.17/14.07 | 0.17/3.87 | 4.23/0.82 | 10.71/0.64 | 12.14/2.59 | 21.28/7.73 | 18.6/4.72 |
| Arl6ip6 | 0.33/5.28 | 0.27/0.91 | 0.09/2.37 | 1.77/2.62 | 1.30/2.81 | 3.37/4.03 | 2.27/4.19 |
| Cep152 | 0.58/2.53 | 1.26/5.71 | 1.95/0.32 | 28.84/1.17 | 18.63/0.56 | 5.03/3.14 | 2.63/3.27 |
| Chd6 | 0.54/0.90 | 1.20/0.97 | 11.69/10.67 | 70.65/11.16 | 61.55/20.31 | 58.35/20.35 | 34.4/18.12 |
| Ddb2 | 3.10/1.00 | 0.72/1.06 | 10.74/0.56 | 81.94/1.53 | 91.16/0.66 | 88.59/3.99 | 61.48/3.57 |
| Dnmt3b | 1.85/0.69 | 0.05/0.56 | 1.45/22.66 | 16.44/1.80 | 14.99/11.27 | 3.61/151.12 | 1.68/211.91 |
| Dut | 0.04/2.30 | 0.36/0.50 | 0.80/1.54 | 2.32/5.41 | 1.35/3.55 | 14.44/1.97 | 9.05/1.33 |
| Emilin3 | 0.83/0.44 | 0.86/0.29 | 26.92/0.25 | 42.07/1.28 | 59.95/0.79 | 7.44/4.22 | 12.92/4.95 |
| Entpd6 | 1.61/1.68 | 0.30/0.48 | 41.36/0.66 | 38.04/0.30 | 17.21/0.32 | 17.79/3.07 | 12.23/2.46 |
| Mettl5 | 0.61/0.10 | 0.37/0.06 | 2.72/10.00 | 6.88/3.21 | 4.9/6.00 | 6.01/46.04 | 5.83/28.46 |
| Myef2 | 0.58/0.69 | 0.11/0.33 | 0.74/1.38 | 0.91/3.04 | 2.12/3.57 | 4.87/9.78 | 3.94/8.22 |
| Rif1 | 0.94/1.19 | 1.76/3.64 | 0.98/14.02 | 5.85/10.48 | 7.01/7.97 | 3.27/6.11 | 2.11/0.65 |
| Sfmbt2 | 2.56/2.25 | 3.80/1.98 | 1.09/1.49 | 7.35/0.88 | 9.00/1.40 | 2.16/7.25 | 0.94/5.39 |
| (B) Genes down-regulated between 1-and 4-folds in TCs as compared with others | |||||||
| Gzf1 | 1/3.12 | 3.24/1.05 | 17.78/47.88 | 13.78/42.76 | 10.39/19.69 | 8.17/20.02 | 1.12/13.88 |
| Pltp | 9.94/3.83 | 2.17/3.01 | 189.9/16.60 | 7.67/31.99 | 8.30/21.33 | 110.19/13.79 | 96.66/22.44 |
| Polr1b | 1.83/1.59 | 1.33/1.47 | 7.01/5.03 | 6.45/10.25 | 3.06/18.41 | 10.71/37.36 | 8.50/46.25 |
| Tasp1 | 2.19/7.78 | 1.12/8.13 | 1.96/2.59 | 15.91/2.34 | 12.47/4.49 | 20.97/8.59 | 8.31/9.50 |
| Zbtb34 | 2.82/4.42 | 1.78/2.60 | 58.06/2.67 | 131.6/19.40 | 78.44/15.48 | 69.44/25.45 | 58.72/10.27 |
| Zfp120 | 1.92/1.70 | 1.84/1.62 | 17.79/11.67 | 21.29/13.60 | 18.08/11.68 | 11.75/7.33 | 8.68/5.36 |
A set of genes are specifically up-or down-regulated in pulmonary TCs, as compared with other cells in chromosome 2 (Table3), up-or down-regulated genes more than 0-fold of TCs D5 were 576 or 341, 559 or 358, 228 or 689, 287 or 630, 277 or 640, 181 or 736, or 210 or 707, respectively, as compared with MSCs, Fbs, ATII, T-BL, T-L, ABCs, or PACs. Up-or down-regulated genes more than 0-fold of TCs D10 were 431 or 486, 408 or 509, 238 or 679, 294 or 623, 288 or 629, 182 or 735, or 222 or 695, as compared with MSCs, Fbs, ATII, T-BL, T-L, ABCs or PACs respectively. Up-and down-regulated genes more than 0-fold of TCs were 406 or 316, 388 or 338, 204 or 655, 262 or 598, 251 or 603, 158 or 712, or 180 or 665, as compared with MSCs, Fbs, ATII, T-BL, T-L, ABCs or PACs respectively.
Table 3.
The number of genes specifically up-or down-regulated in pulmonary telocytes, as compared with other cells respectively
| Compared pairs | Up>0 | Up>1 | Up>4 | Down>0 | Down>1 | Down>4 |
|---|---|---|---|---|---|---|
| TC5 versus stem | 576 | 194 | 50 | 341 | 80 | 14 |
| TC10 versus stem | 431 | 136 | 41 | 486 | 152 | 27 |
| TCs versus stem | 406 | 116 | 30 | 316 | 73 | 14 |
| TC5 versus fibroblast | 559 | 201 | 79 | 358 | 107 | 17 |
| TC10 versus fibroblast | 408 | 166 | 61 | 509 | 178 | 33 |
| TCs versus fibroblast | 388 | 140 | 56 | 338 | 93 | 14 |
| TC5 versus ATII | 228 | 102 | 36 | 689 | 504 | 289 |
| TC10 versus ATII | 238 | 104 | 35 | 679 | 516 | 296 |
| TCs versus ATII | 204 | 86 | 30 | 655 | 476 | 268 |
| TC5 versus CD8BL | 287 | 174 | 89 | 630 | 689 | 303 |
| TC10 versus CD8BL | 294 | 196 | 97 | 623 | 485 | 284 |
| TCs versus CD8BL | 262 | 160 | 81 | 598 | 461 | 262 |
| TC5 versus CD8LL | 277 | 178 | 92 | 640 | 487 | 306 |
| TC10 versus CD8LL | 288 | 187 | 97 | 629 | 482 | 289 |
| TCs versus CD8LL | 251 | 162 | 84 | 603 | 458 | 263 |
| TC5 versus basal cell | 181 | 89 | 42 | 736 | 612 | 414 |
| TC10 versus basal cell | 182 | 101 | 41 | 735 | 601 | 406 |
| TCs versus basal cell | 158 | 79 | 36 | 712 | 573 | 375 |
| TC5 versus duct cell | 210 | 118 | 50 | 707 | 552 | 358 |
| TC10 versus duct cell | 222 | 117 | 51 | 695 | 548 | 345 |
| TCs versus duct cell | 180 | 103 | 42 | 665 | 522 | 320 |
In chromosome 3, 13 genes were higher than 0-fold in TCs, as compared with those in other cells (Table4), of which 10 genes (Agl, Ecm1, Golim4, Kcnab1, Lce1a2, Nexn, Pde4dip, Plekho1, Psrc1, Rhoc, Rit1, Scamp3, Sec22b) were overexpressed 0-to 1-fold (Table4A). Three genes Sh3glb1 (SH3-domain GRB2-like B1 – endophilin), Tm4sf1 (transmembrane 4 superfamily member 1) and Csf1 (colony stimulating factor 1) were overexpressed more than onefold, in both TC D5 and TC D10, as compared with other cells (Table4B). 59 genes in TCs were down-regulated, as compared with other cells (Table5). Of them, 1700013F07Rik, Amy1, Anp32e, Dnase2b, Fmo5, Pde5a, Phf17, Rwdd3 and Trim33 were down-regulated more than onefold, in both TC D5 and TC D10, as compared with other cells.
Table 4.
Summary of genes expressed preferentially in TCs, as compared with others
| Compared pairs/fold up-regulated | >0 | >1 | >4 | ||||
|---|---|---|---|---|---|---|---|
| TC5 versus others | 42 | 10 | 0 | ||||
| TC10 versus others | 30 | 7 | 2 | ||||
| TCs versus others | 13 | 3 | 0 | ||||
| Gene symbol | Folds (TC5 versus others/TC10 versus others) | ||||||
| Fibroblast | Stem | ATII | CD8_T_BL | CD8_T_LL | Basal_cell | Duct_cell | |
| (A) Genes up-regulated between 0-and 1-folds in TCs as compared with others | |||||||
| Agl | −0.31/−0.25 | −0.42/−0.37 | −0.64/−0.6 | −0.54/−0.5 | −0.81/−0.79 | −0.63/−0.6 | −0.79/−0.77 |
| Ecm1 | −0.96/−0.96 | −0.9/−0.9 | −0.91/−0.91 | −0.96/−0.96 | −0.85/−0.85 | −0.48/−0.47 | −0.24/−0.23 |
| Golim4 | −0.65/−0.63 | −0.24/−0.2 | −0.86/−0.85 | −0.8/−0.79 | −0.86/−0.86 | −0.65/−0.63 | −0.48/−0.45 |
| Kcnab1 | −0.94/−0.95 | −0.39/−0.54 | −0.73/−0.8 | −0.62/−0.71 | −0.9/−0.93 | −0.24/−0.42 | −0.65/−0.73 |
| Lce1a2 | −0.16/−0.3 | −0.41/−0.52 | −0.01/−0.18 | −0.03/−0.2 | −0.24/−0.37 | −0.83/−0.86 | −0.93/−0.94 |
| Nexn | −0.59/−0.47 | −0.3/−0.08 | −0.82/−0.76 | −0.9/−0.87 | −0.96/−0.95 | −0.64/−0.52 | −0.78/−0.71 |
| Pde4dip | −0.65/−0.69 | −0.52/−0.58 | −0.19/−0.29 | −0.11/−0.22 | −0.4/−0.47 | −0.71/−0.75 | −0.68/−0.72 |
| Plekho1 | −0.07/−0.3 | −0.8/−0.85 | −0.8/−0.85 | −0.85/−0.89 | −0.97/−0.98 | −0.95/−0.97 | −0.98/−0.99 |
| Psrc1 | −0.8/−0.7 | −0.78/−0.67 | −0.81/−0.73 | −0.72/−0.59 | −0.62/−0.45 | −0.32/0 | −0.38/−0.1 |
| Rhoc | −0.57/−0.52 | −0.5/−0.43 | −0.85/−0.83 | −1/−1 | −0.92/−0.91 | −0.6/−0.55 | −0.73/−0.69 |
| Rit1 | −0.54/−0.53 | −0.66/−0.65 | −0.58/−0.57 | −0.69/−0.68 | −0.66/−0.65 | −−0.25/−0.22 | −0.57/−0.55 |
| Scamp3 | −0.34/−0.34 | −0.55/−0.55 | −0.79/−0.79 | −0.61/−0.61 | −0.72/−0.72 | −0.82/−0.82 | −0.83/−0.83 |
| Sec22b | −0.41/−0.08 | −0.5/−0.23 | −0.49/−0.21 | −0.55/−0.3 | −0.63/−0.43 | −0.56/−0.32 | −0.65/−0.46 |
| (B) Genes up-regulated between 1-and 4-folds in TCs as compared with other | |||||||
| Sh3glb1 | −0.73/−0.68 | −0.62/−0.54 | −0.81/−0.77 | −0.79/−0.75 | −0.8/−0.76 | −0.69/−0.62 | −0.7/−0.63 |
| Tm4sf1 | −1/−1 | −0.6/−0.67 | −0.85/−0.88 | −1/−1 | −1/−1 | −0.99/−0.99 | −0.99/−0.99 |
| Csf1 | −0.71/−0.65 | −0.73/−0.68 | −0.93/−0.92 | −0.98/−0.98 | −0.98/−0.97 | −0.91/−0.9 | −0.92/−0.91 |
Table 5.
Summary of genes expressed preferentially in TCs, as compared with others
| Compared pairs/fold down-regulated | >0 | >1 | >4 | ||||
|---|---|---|---|---|---|---|---|
| TC5 versus others | 79 | 12 | 0 | ||||
| TC10 versus others | 137 | 22 | 1 | ||||
| TCs versus others | 59 | 9 | 0 | ||||
| Gene symbol | Folds (TC5 versus others/TC10 versus others) | ||||||
| Fibroblast | Stem | ATII | CD8_T_BL | CD8_T_LL | Basal_cell | Duct_cell | |
| (A) Genes down-regulated between 0-and 1-folds in TCs as compared with others | |||||||
| 1700027A23Rik | 0.9/1.57 | 0.08/0.46 | 162.04/220.27 | 4.95/7.07 | 0.43/0.94 | 28.1/38.49 | 8.2/11.48 |
| 2810403A07Rik | 0.81/1.11 | 0.7/0.99 | 0.47/0.72 | 3.33/4.07 | 4.16/5.04 | 1.31/1.7 | 0.96/1.3 |
| 4932438A13Rik | 1.12/1.11 | 0.9/0.9 | 4.4/4.39 | 76.24/75.99 | 78.16/77.9 | 9.48/9.44 | 8.12/8.09 |
| 4933421E11Rik | 1.61/2.74 | 0.22/0.74 | 0.3/0.87 | 5.03/7.64 | 4.6/7.02 | 2.41/3.89 | 0.14/0.64 |
| A530020G20Rik | 0.13/0.92 | 0.02/0.73 | 6.3/11.39 | 1.91/3.94 | 2.91/5.64 | 2.4/4.77 | 1.14/2.63 |
| Acadm | 5.66/5.92 | 0.22/0.27 | 4.5/4.72 | 6.8/7.1 | 4.87/5.09 | 6.25/6.53 | 5.1/5.33 |
| Adh6a | 0.46/0.67 | 0.44/0.65 | 5.68/6.63 | 8.32/9.65 | 11.57/13.37 | 50.97/58.39 | 25.27/29.03 |
| Ahcyl1 | 0.62/0.88 | 0.14/0.32 | 1.5/1.91 | 1.14/1.49 | 0.51/0.76 | 2.4/2.95 | 1.9/2.38 |
| Alx3 | 1.08/1.53 | 1.35/1.87 | 0.45/0.77 | 0.95/1.38 | 5.61/7.05 | 0.82/1.22 | 1.64/2.21 |
| Atp11b | 0.44/0.93 | 0.99/1.66 | 2.2/3.29 | 28.57/38.63 | 22.24/30.15 | 10.08/13.85 | 15.11/20.59 |
| Car3 | 0.25/0.29 | 0.04/0.07 | 3.72/3.87 | 4.07/4.23 | 6.87/7.12 | 40.74/42.05 | 22.72/23.47 |
| Clcc1 | 1.62/2.56 | 0.03/0.39 | 0.08/0.47 | 0.81/1.45 | 0.7/1.31 | 3.29/4.83 | 3.25/4.77 |
| Cryz | 3.65/6.13 | 3.67/6.15 | 0.91/1.92 | 0.17/0.79 | 0/0.53 | 0.51/1.32 | 2.14/3.8 |
| Ctso | 0.22/0.15 | 1.18/1.05 | 151.36/142.4 | 336.91/317.03 | 271.25/255.24 | 245.94/231.42 | 92.66/87.15 |
| Gnat2 | 1.35/1.57 | 0.19/0.3 | 1.09/1.29 | 2.4/2.72 | 4.24/4.73 | 8.71/9.63 | 1.89/2.17 |
| Gpsm2 | 1.89/2.66 | 1.39/2.03 | 0.36/0.72 | 3.42/4.59 | 5.15/6.78 | 14.01/17.99 | 18.95/24.24 |
| Hax1 | 0.16/0.6 | 0.66/1.3 | 1.25/2.1 | 0.9/1.62 | 0.94/1.68 | 0.6/1.21 | 0.84/1.54 |
| Hltf | 0.31/0.45 | 0.45/0.61 | 3.26/3.75 | 18.55/20.78 | 18.24/20.44 | 7.82/8.83 | 3.57/4.09 |
| Hps3 | 1.39/1.42 | 0.56/0.58 | 0.8/0.82 | 8.25/8.37 | 6.09/6.18 | 2.97/3.02 | 0.26/0.27 |
| Ints3 | 0.83/0.94 | 0.23/0.3 | 27.92/29.65 | 13.8/14.68 | 6.12/6.54 | 23.06/24.5 | 26.27/27.9 |
| Isg20l2 | 0.09/0.62 | 0.06/0.57 | 2.61/4.35 | 4.77/7.55 | 3.23/5.27 | 5.9/9.22 | 6.17/9.62 |
| Lass2 | 0.06/0.12 | 0.01/0.07 | 5.54/5.9 | 2.13/2.3 | 1.64/1.79 | 1.12/1.23 | 0.41/0.49 |
| Lrrc40 | 0.4/0.16 | 0.42/0.18 | 102.03/84.89 | 256.01/213.26 | 257.71/214.68 | 537.73/448.12 | 349.78/291.43 |
| Lrrcc1 | 0.1/0.63 | 0.19/0.77 | 0.6/1.37 | 2.23/3.8 | 1.26/2.35 | 5.2/8.21 | 2.45/4.13 |
| Mfsd8 | 0.71/0.3 | 0.45/0.11 | 11.71/8.67 | 24.91/18.72 | 19.66/14.72 | 32.59/24.56 | 33.42/25.19 |
| Mrpl24 | 0.64/1.57 | 0.06/0.66 | 1.1/2.27 | 3.11/5.41 | 1.92/3.56 | 1.85/3.45 | 1.82/3.41 |
| Mrpl9 | 0.29/0.47 | 0.44/0.64 | 3.16/3.73 | 7.18/8.3 | 5.33/6.19 | 8.23/9.48 | 7.87/9.08 |
| Ndufb5 | 1/1.86 | 0.01/0.44 | 2.17/3.55 | 2.34/3.79 | 0.88/1.69 | 2.78/4.42 | 2.95/4.66 |
| Odf2l | 1.26/1.48 | 0.17/0.29 | 5.61/6.26 | 11.82/13.08 | 7.95/8.83 | 44.13/48.56 | 37.95/41.78 |
| Papss1 | 1.13/1.25 | 0.44/0.52 | 2.44/2.62 | 1.53/1.66 | 0.28/0.34 | 1.73/1.88 | 1.64/1.78 |
| Pgrmc2 | 1.11/1.55 | 0.08/0.31 | 4.75/5.96 | 1.45/1.97 | 2/2.62 | 29.73/36.17 | 15.37/18.8 |
| Plk4 | 0.46/0.88 | 0.71/1.2 | 2.33/3.29 | 11.64/15.29 | 12.8/16.79 | 7.3/9.69 | 12.53/16.43 |
| Prpf38b | 1.18/0.93 | 0.68/0.49 | 11.25/9.81 | 50/44 | 66.2/58.3 | 44.75/39.37 | 34.78/30.58 |
| Rabggtb | 0.81/0.52 | 1.09/0.75 | 8.12/6.64 | 53.93/45.01 | 57.59/48.07 | 37.82/31.52 | 48.92/40.81 |
| Rapgef2 | 0.29/0.68 | 0.63/1.11 | 2.8/3.92 | 10.47/13.85 | 8.15/10.86 | 0.24/0.6 | 0.61/1.09 |
| Rps4x | 0.11/0.67 | 0.62/1.45 | 0.39/1.1 | 0.85/1.79 | 1.99/3.51 | 0.67/1.53 | 0.54/1.32 |
| Sars | 3.16/4.85 | 1.32/2.27 | 1.03/1.86 | 0.4/0.98 | 0.09/0.54 | 0.41/0.99 | 0.3/0.83 |
| Setdb1 | 0.25/0.87 | 0.6/1.39 | 0.03/0.55 | 1.29/2.42 | 1.07/2.1 | 2.73/4.58 | 1.8/3.19 |
| Siah2 | 1.29/1.68 | 1.79/2.27 | 3.92/4.77 | 0.21/0.42 | 0.87/1.19 | 3.28/4.02 | 3.03/3.72 |
| Slc33a1 | 0.75/0.58 | 0.72/0.55 | 5.56/4.91 | 2.19/1.87 | 1.75/1.48 | 3.02/2.62 | 0.75/0.58 |
| Smc4 | 0.07/0.85 | 0.89/2.26 | 0.12/0.94 | 14.46/25.7 | 14.77/26.25 | 2.22/4.56 | 6.28/11.58 |
| Sohlh2 | 0.02/0.11 | 0.08/0.17 | 1.69/1.93 | 0.91/1.07 | 1.86/2.11 | 1.58/1.8 | 1.85/2.1 |
| Spata5 | 0.06/0.88 | 0.21/1.16 | 0.19/1.12 | 0.32/1.36 | 0.4/1.49 | 1.15/2.83 | 1.51/3.47 |
| Syt6 | 0.23/0.6 | 0.21/0.57 | 0.15/0.5 | 0.55/1.02 | 0.43/0.87 | 1.27/1.96 | 0.29/0.69 |
| Tbl1xr1 | 2.36/1.6 | 0.89/0.47 | 2.59/1.79 | 4.02/2.9 | 3.66/2.61 | 16.91/12.89 | 13.56/10.29 |
| Txnip | 0.77/0.21 | 1.35/0.6 | 54.04/36.58 | 107.86/73.33 | 65.79/44.6 | 36.39/24.53 | 26.15/17.53 |
| Ubqln4 | 0.18/0.58 | 0.21/0.63 | 11.94/16.36 | 5.82/8.15 | 2.66/3.91 | 28.73/38.87 | 40.6/54.79 |
| Wdr77 | 0.14/0.7 | 0.97/1.92 | 3.67/5.93 | 1.52/2.74 | 0.79/1.66 | 2.28/3.88 | 2.96/4.88 |
| Ythdf3 | 0.22/0.54 | 0.64/1.07 | 3.08/4.17 | 6.8/8.87 | 6.44/8.41 | 3.68/4.92 | 3/4.06 |
| Zzz3 | 0.1/0.64 | 0.73/1.58 | 0.1/0.64 | 3.91/6.33 | 3.67/5.96 | 1.29/2.42 | 0.6/1.39 |
| (B) Genes down-regulated between 1-and 4-folds in TCs as compared with others | |||||||
| 1700013F07Rik | 6.5/7.08 | 1.64/1.84 | 34.73/37.52 | 2.1/2.34 | 7.8/8.48 | 29.96/32.38 | 47.84/51.65 |
| Amy1 | 2.81/3.86 | 8.06/10.55 | 9.44/12.32 | 7.37/9.67 | 14.79/19.13 | 31.07/39.88 | 24.64/31.68 |
| Anp32e | 8.69/11.5 | 2.79/3.89 | 121.76/157.32 | 245.2/316.52 | 243.57/314.42 | 356.1/459.55 | 464.63/599.52 |
| Dnase2b | 2.08/2.96 | 1.85/2.67 | 1.08/1.68 | 1.08/1.67 | 9.48/12.48 | 10.22/13.44 | 6.68/8.87 |
| Fmo5 | 3.8/6 | 3.36/5.35 | 3.52/5.58 | 4.08/6.4 | 1.75/3.01 | 21.33/31.55 | 5.4/8.33 |
| Pde5a | 4.09/2.99 | 3.94/2.87 | 4.58/3.37 | 39.25/30.54 | 12.26/9.39 | 44.99/35.04 | 15.96/12.29 |
| Phf17 | 1.24/1.76 | 3.48/4.52 | 1.04/1.52 | 3.4/4.42 | 3.67/4.75 | 11.3/14.14 | 4.48/5.75 |
| Rwdd3 | 2.98/4.43 | 2.52/3.8 | 2.98/4.43 | 14.07/19.53 | 10.16/14.21 | 28.88/39.73 | 8.77/12.31 |
| Trim33 | 1.93/5.56 | 1.06/3.62 | 4.2/10.64 | 26.9/61.48 | 17.78/41.07 | 3.1/8.17 | 2.63/7.13 |
In chromosome 3 (Table6), up-or down-regulated genes more than 0-fold of TCs D5 were 345 or 222, 352 or 215, 377 or 190, 214 or 353, 201 or 366, 130 or 437, or 137 or 430, as compared with Fbs, MSCs, ATII, T-BL, T-L, ABCs or PACs respectively. Up-or down-regulated genes more than 0-fold of TCs D5 were 265 or 302, 263 or 304, 138 or 429, 188 or 379, 168 or 399, 95 or 472, or 120 or 447, as compared with Fbs, MSCs, ATII, T-BL, T-L, ABCs or PACs respectively. Up-and down-regulated genes more than 0-fold of TCs were 255 or 212, 247 or 199, 367 or 128, 181 or 346, 164 or 362, 87 or 429, or 110 or 420, as compared with Fbs, MSCs, ATII, T-BL, T-L, ABCs or PACs respectively. Details of up-or down gene variations of chromosome 2 and 3, including the number and names of up-or down-regulated genes more than 0-fold among different cells, were listed in Data S2.
Table 6.
The number of genes specifically up-or down-regulated in pulmonary telocytes, as compared with other cells respectively
| Compared pairs | Up>0 | Up>1 | Up>4 | Down>0 | Down>1 | Down>4 |
|---|---|---|---|---|---|---|
| TC10 versus fibroblast | 265 | 116 | 40 | 302 | 126 | 28 |
| TC5 versus fibroblast | 345 | 161 | 51 | 222 | 87 | 17 |
| TCs versus fibroblast | 255 | 100 | 33 | 212 | 78 | 12 |
| TC10 versus stem | 263 | 109 | 35 | 304 | 115 | 23 |
| TC5 versus stem | 352 | 134 | 41 | 215 | 63 | 12 |
| TCs versus stem | 247 | 85 | 27 | 199 | 58 | 12 |
| TC10 versus ATII | 138 | 67 | 22 | 429 | 306 | 177 |
| TC5 versus ATII | 377 | 278 | 137 | 190 | 93 | 32 |
| TCs versus ATII | 367 | 268 | 124 | 128 | 62 | 20 |
| TC10 versus CD8BL | 188 | 117 | 59 | 379 | 302 | 186 |
| TC5 versus CD8BL | 214 | 143 | 81 | 353 | 265 | 162 |
| TCs versus CD8BL | 181 | 113 | 57 | 346 | 254 | 153 |
| TC10 versus CD8LL | 168 | 99 | 51 | 399 | 307 | 197 |
| TC5 versus CD8LL | 201 | 138 | 66 | 366 | 278 | 163 |
| TCs versus CD8LL | 164 | 96 | 47 | 362 | 269 | 154 |
| TC10 versus basal cell | 95 | 55 | 19 | 472 | 388 | 261 |
| TC5 versus basal cell | 130 | 65 | 26 | 437 | 345 | 222 |
| TCs versus basal cell | 87 | 45 | 16 | 429 | 339 | 218 |
| TC10 versus duct cell | 120 | 56 | 26 | 447 | 373 | 234 |
| TC5 versus duct cell | 137 | 81 | 29 | 430 | 321 | 199 |
| TCs versus duct cell | 110 | 51 | 21 | 420 | 312 | 187 |
The relationships of the more than 0-fold up-regulated genes of chromosome 2 and 3 in TC D5 and/or TC D10 were analysed by String Network analysis (www.string-db.org), as compared with other cells, to identify direct (physical) and indirect (functional) associations between selected genes of TCs. TC-specific or dominating genes in TC D5 and TC D10 were selected by up-or down-expression more than 0-fold, as compared with other cells. Figure1A and C demonstrated the distribution of such active gene group in chromosome 2 and 3 of all cells, and interactions or potential functional links of those genes of TCs.
Figure 1.
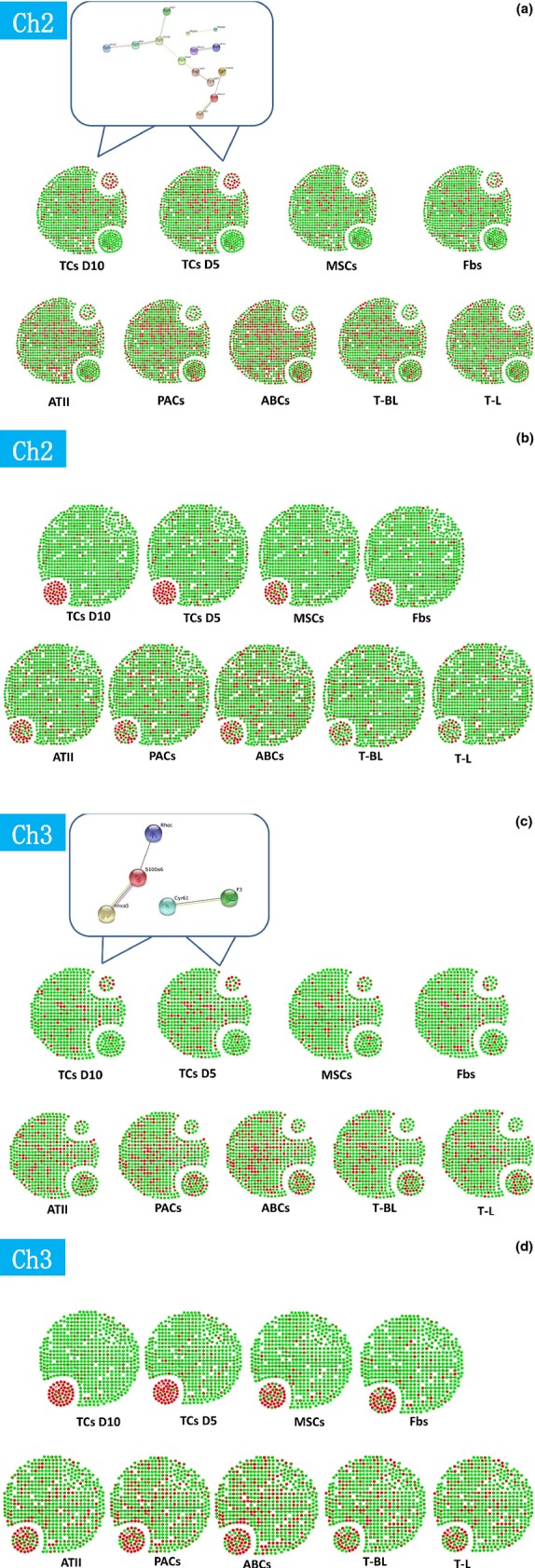
Expression profiles of the selected genes as an active group of chromosome 2 and 3 of TCs (TCs) isolated and cultured from mouse lungs on days 5 (D5) and 10 (D10), as compared with fibroblasts (Fbs), mesenchymal stem cells (MSCs), alveolar type II cells (ATII), airway basal cells (ABCs), proximal airway cells (PACs), CD8+ T cells come from bronchial lymph nodes (T-BL), and CD8+ T cells from lung (T-L) respectively (A and C). The profiles for entire genes are described in Data S1. The selected core network and whole mouse network are linked by the documented functional interactions from various databases (see Methods). Genes in each network are indicated in red and some of their nearest neighbours are indicated by dark grey nodes. A group of telocyte genes up-regulated and down-regulated more than 0-fold as compared with all other cells and existed in TCs on days 5 and 10 were selected as telocyte-specific or dominated genes in chromosome 2 and 3 (A and C). Top 50 up-or down-regulated genes of each cells were also evaluated and their distribution within chromosome 2 and 3 genes showed the difference between cells (B and D). Details of the selected network in each cell type are in Figure S1–S18.
In chromosome 2, about 30-50% of TCs genes showed similar patterns of gene expression in MSCs, Fbs or ATII, while 5-15% of TCs genes showed similarities with ABCs, PACs, T-BL or T-L. Top 50 up-or down-regulated genes of each cell were also evaluated and their distribution within chromosome 2 genes showed the difference between them, as shown in Figure1B. High expressed genes of each cell within chromosome 2 were evaluated and distributed as red colour (Fig.1B). The distribution of the high expressed genes and low expressed genes both in TC D5 and TC D10 indicates that they are in the centre of the small cluster and different from the other cells. Among the 26 co-up-expressed genes (Table1A–C), 7 genes were found to have certain interactions (Fig.1A).
In chromosome 3, about 50–60% of TCs genes showed similar patterns of gene expression in Fbs, MSCs, PACs or ABCs, while 0–20% of TCs genes showed similarities with ABCs, PACs, T-BL or T-L. Top 50 up-or down-regulated genes of each cell were also evaluated and their distribution within chromosome 3 genes showed the difference between them, as shown in Figure1D. High expressed genes of each cell within chromosome 3 were evaluated and distributed as red colour (Fig.1D). The distribution of up-expressed genes and down-expressed genes in TC D5 and TC D10 indicates that they are in the centre of the small cluster and different from the other cells. Among the 16 co-up-expressed genes (Table4A and B), no clear or certain interactions (Fig.1C) were found. The hierarchical cluster analysis of the differentially expressed genes (Fig. 2) clearly shows that TCs are poorly related to the other cell lines.
Figure 2.
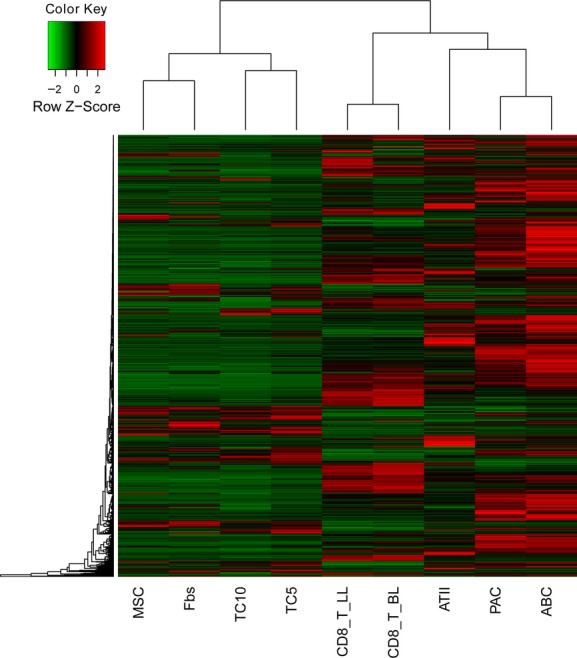
Hierarchical cluster analysis of the differentially expressed genes on chromosomes 2 and 3 among telocytes (TCs), mesenchymal stem cells (MSCs), fibroblasts (Fbs), lymphocytes from lungs (T-LL) and from bronchial lymph nodes (T-BL), alveolar type II cells (ATII), proximal airway cells (PAC) and airway basal cells (ABC).
Discussion
Mouse chromosomes are the best studied mammalian chromosomes and are considered as gold standard of human comparative map, although genomic rearrangements occur during evolution. Certain human disease genes were discovered by comparative genomics using the information derived from mapped mouse mutations, although they are not the simplest model for human comparison. In humans, chromosome 2 has the largest sequenced base pairs (237, 712, 649) 47, working with all of the autosomes in humans, spanning the second largest amount of total base pairs (242, 751, 149) and representing 16.2% of the total DNA 48–51. Over 31 exactly known diseases were proposed to be associated with genes on chromosome 2. In mouse, chromosome 2 is entirely sequenced and has 3146 genes encoding 1780 proteins 47, of which 917 genes were measured by bioinformatics tools in the present study. Our data demonstrated that there were 26 or 80 up-or down-regulated genes of chromosome 2 in TCs, as compared with MSCs, Fbs, ATII, T-BL, T-LL, ABCs or PACs.
One gene Myl9 (myosin, light polypeptide 9) was overexpressed most in TCs, different from other cells. Myl9 regulatory gene encodes the regulatory light chains of myosin II molecule, known to play a central role in cell adhesion, migration and division. Recent results showed Myl9 as the only gene differentially expressed in the aged versus young injured arteries 52 implying that it may be related to tissue/organ injury and ageing. Therefore, it is possible that the overexpression of Myl9 in pulmonary TCs may play an important role in lung injury and ageing. There were six genes, e.g. Pltp, Gzf1, Polr1b, Tasp1, Zbtb34 and Zfp120, down-expressed most in TCs, different from other cells. The Pltp (phospholipid transfer protein) gene is widely expressed in the body, and plays an important role in lipid metabolism, immune modulation, lipopolysaccharide binding or neurodegenerative disease 53. Pltp is highly expressed within the lung epithelium, in chronic obstructive pulmonary disease or pulmonary inflammation 54. TCs may play an important role of inhibiting inflammation in the lung. Roles of Gzf1 (GDNF-inducible zinc finger protein 1), Polr1b (polymerase (RNA) I polypeptide B), Tasp1 (taspase threonine aspartase 1), Zbtb34 (zinc finger and BTB domain containing 34) or Zfp120 (zinc finger protein 120) genes or proteins in the lung remain unclear. Thus, there is a further need to clarify the exact mechanisms and functions of these genes in TCs.
Mouse chromosome 3 has a total number of genes of 993 which encode a total of 669 proteins 47. Human chromosome 3 has ∼7% of the human genome probably related with, at least, 121 diseases that are associated with 105 genes 55 and also spanning the third largest amount of total base pairs (199, 446, 827) and represented about 6.5% of the total DNA in cells 56–59. The chromosome 3 has 1550 genes, of which 567 genes of chromosome 3 were measured by bioinformatics tools in the present study. We showed that there were 13 or 59 up-or down-regulated genes of chromosome 3 in TCs, as compared with Fbs, MSCs, ATII, T-BL, T-L, ABCs or PACs. There were three genes, e.g. Sh3glb1, Tm4sf1 and Csf1, overexpressed in TCs.
Sh3glb1 gene encodes SH3-domain GRB2-like B1 or endophilin, known to have an extremely close relationship with Bax-interacting factor-1 (bif-1) 60,61, involved in cell survival and proliferation under metabolic stress and evasion of apoptosis. SH3glb1 is a membrane curvature-inducing protein interact with BECN1 though UVRAG and regulates the post-Golgi trafficking of membrane-integrated ATG9A for autophagy. At the premalignant stage, allelic loss of Sh3glb1 could enhance Myc-induced chromosomal instability and result in the up-regulation of anti-apoptotic proteins, including MCL1 and BCL2L1 61, being responsible for enabling cells to survive and proliferate under metabolic stress and evasion of apoptosis. Endophilin is a membrane curvature-inducing protein that interacts with autophagy related beclin 1, although UV radiation resistance associated gene (Uvrag) and regulates the post-Golgi trafficking of membrane-integrated autophagy related 9A (Atg9A) protein. At the premalignant stage, allelic loss of Sh3glb1 enhances Myc-induced chromosomal instability and results in the up-regulation of anti-apoptotic proteins, including MCL1 and BCL2L1 61. So far, there is no reported association with any lung disease, however, we cannot exclude a role for TCs as having pro-proliferative effects through inhibition of apoptosis as showed in a previous study 41.
Tm4sf1 (transmembrane 4 superfamily member 1) is a distant member of the transmembrane 4 superfamily of cell-surface proteins characterized by the presence of four hydrophobic domains 62. It is highly expressed in different carcinomas, e.g. in lung cancer 62, and lowly expressed in normal tissues 63. Colony stimulating factor 1 (macrophage) (Csf1) plays an important role in cancer metastasis and invasion. It is highly expressed in different carcinomas and expressed at relatively low levels (if at all) in many normal tissues 63. High expression of Csf1 can increase metastasis and invasion of pulmonary adenocarcinomas 64. For example, Tm4sf1 it was up-regulated in human adenocarcinoma A549 cell line, suggesting a poor prognosis for anticancer therapy 65. Overexpression of TM4SF1 and Csf1 in lung TCs may have a role in the development of lung cancer. Among down-expressed genes in TCs, Pde5a (cGMP-specific phosphodiesterase 5A) has an obvious association with acute and chronic interstitial lung disease. Overexpression of Pde5a may accelerate the formation of pulmonary fibrosis, while down-expression of Pde5a has important roles and effects in pulmonary fibrosis-associated pulmonary hypertension 66,67. Therefore, we concluded that Tm4sf1 and Csf1 found to be overexpressed in lung TCs may have a role in tumour promotion. There were nine genes, e.g. 1700013F07Rik, Amy1, Anp32e, Dnase2b, Fmo5, Pde5a, Phf17, Rwdd3 and Trim33, down-expressed most in TCs, different from other cells.
Among them, only phosphodiesterase 5a (Pde5a) cGMP-specific gene is obviously associated with acute and chronic interstitial lung disease. Its high expression promotes the pulmonary fibrosis, while the inhibition of Pde5a expression ameliorates right ventricular failure and pulmonary, when is associated with bleomicin, through a reduction in reactive oxygen species 68. Therefore, Pde5a low expression in lung TCs may have therapeutic effect on pulmonary fibrosis and other acute and chronic interstitial lung disease, probably by modulation of oxidative stress levels, as previously shown 41.
In conclusion, the present study compared genetic variations of chromosome 2 and 3 of pulmonary TCs with other related cells, e.g. Fbs, MSCs, ATII, T-BL, T-L, ABCs or PACs. Our data showed a number of TCs-specific or dominant genes in chromosomes 2 and 3, different from other lung tissue resident cells or infiltrated cells. The TCs signatures of chromosome 2 and 3 genes indicate TCs may be mainly associated with anti-inflammatory responses, the prevention of lung cancer formation and development or protective effects on pulmonary fibrosis or acute and chronic interstitial lung diseases.
Acknowledgments
The work was supported by Shanghai Leading Academic Discipline Project (B115), Zhongshan Distinguished Professor Grant (XDW), the National Nature Science Foundation of China (91230204, 81270099, 81320108001, 81270131), the Shanghai Committee of Science and Technology (12JC1402200, 12431900207, 11410708600), Zhejiang Provincial Natural Science Foundation (Z2080988), Zhejiang Provincial Science Technology Department Foundation (2010C14011) and Ministry of Education, Academic Special Science and Research Foundation for PhD Education (20130071110043). This work was partially supported (for DC) by the Sectorial Operational Programme Human Resources Development (SOP HRD), financed from the European Social Fund and by the Romanian Government under the contract number POSDRU/89/1.5/S/141531.
Conflicts of interest
The authors confirm that there are no conflicts of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data profiles for all genes.
Details of up-or down regulated gene expression variations of chromosome 2 and 3.
Details of the selected core network genes in TCs isolated from the mouse lung and cultured for 5 days in chromosome 2.
Details of the selected core network genes in TCs isolated from the mouse lung and cultured for 10 days in chromosome 2.
Details of the selected core network genes in mouse mesenchymal stem cells in chromosome 2.
Details of the selected core network genes in mouse fibroblasts in chromosome 2.
Details of the selected core network genes in mouse alveolar type II cells in chromosome 2.
Details of the selected core network genes in mouse airway basal cells in chromosome 2.
Details of the selected core network genes in mouse proximal airway cells in chromosome 2.
Details of the selected core network genes in mouse CD8+ T cells come from bronchial lymph nodes in chromosome 2.
Details of the selected core network genes in mouse CD8+ T cells from lung in chromosome 2.
Details of the selected core network genes in TCs isolated from the mouse lung and cultured for 10 days in chromosome 3.
Details of the selected core network genes in TCs isolated from the mouse lung and cultured for 5 days in chromosome 3.
Details of the selected core network genes in mouse mesenchymal stem cells in chromosome 3.
Details of the selected core network genes in mouse fibroblasts in chromosome 3.
Details of the selected core network genes in mouse alveolar type II cells in chromosome 3.
Details of the selected core network genes in mouse airway basal cells in chromosome 3.
Details of the selected core network genes in mouse proximal airway cells in chromosome 3.
Details of the selected core network genes in mouse CD8+ T cells come from bronchial lymph nodes in chromosome 3.
Details of the selected core network genes in mouse CD8+ T cells from lung in chromosome 3.
References
- Popescu LM, Faussone-Pellegrini MS. TELOCYTES-a case of serendipity: the winding way from Interstitial Cells of Cajal (ICC), via Interstitial Cajal-Like Cells (ICLC) to TELOCYTES. J Cell Mol Med. 2010;14:729–40. doi: 10.1111/j.1582-4934.2010.01059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suciu L, Popescu LM, Gherghiceanu M, et al. Telocytes in human term placenta: morphology and phenotype. Cells Tissues Organs. 2010;192:325–39. doi: 10.1159/000319467. [DOI] [PubMed] [Google Scholar]
- Smythies J, Edelstein L. Telocytes, exosomes, gap junctions and the cytoskeleton: the makings of a primitive nervous system? Front Cell Neurosci. 2014;7:278. doi: 10.3389/fncel.2013.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Flores L, Gutierrez R, Garcia MP, et al. CD34+ stromal cells/fibroblasts/fibrocytes/telocytes as a tissue reserve and a principal source of mesenchymal cells. Location, morphology, function and role in pathology. Histol Histopathol. 2014;29:831–70. doi: 10.14670/HH-29.831. [DOI] [PubMed] [Google Scholar]
- Popescu LM, Manole CG, Gherghiceanu M, et al. Telocytes in human epicardium. J Cell Mol Med. 2010;14:2085–93. doi: 10.1111/j.1582-4934.2010.01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghiceanu M, Popescu LM. Cardiac telocytes-their junctions and functional implications. Cell Tissue Res. 2012;348:265–79. doi: 10.1007/s00441-012-1333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sun W, Wu SM, et al. Telocytes in human heart valves. J Cell Mol Med. 2014;18:759–65. doi: 10.1111/jcmm.12285. Doi: 10.1111/jcmm.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G, Lin M, Xu M, et al. Telocytes in the human kidney cortex. J Cell Mol Med. 2012;16:3116–22. doi: 10.1111/j.1582-4934.2012.01582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhu T, Lin M, et al. Telocytes in the urinary system. J Transl Med. 2012;10:188. doi: 10.1186/1479-5876-10-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert T, De Vos R, Van Der Aa F, et al. Identification of telocytes in the upper lamina propria of the human urinary tract. J Cell Mol Med. 2012;16:2085–93. doi: 10.1111/j.1582-4934.2011.01504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceafalan L, Gherghiceanu M, Popescu LM, et al. Telocytes in human skin–are they involved in skin regeneration? J Cell Mol Med. 2012;16:1405–20. doi: 10.1111/j.1582-4934.2012.01580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusu MC, Pop F, Hostiuc S, et al. Telocytes form networks in normal cardiac tissues. Histol Histopathol. 2012;27:807–16. doi: 10.14670/HH-27.807. [DOI] [PubMed] [Google Scholar]
- Luesma MJ, Gherghiceanu M, Popescu LM. Telocytes and stem cells in limbus and uvea of mouse eye. J Cell Mol Med. 2013;17:1016–24. doi: 10.1111/jcmm.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Y, Wang Y, Li J, et al. Immunohistochemical characterization and functional identification of mammary gland telocytes in the self-assembly of reconstituted breast cancer tissue in vitro. J Cell Mol Med. 2013;17:65–75. doi: 10.1111/j.1582-4934.2012.01646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zheng Y, Manole CG, et al. Telocytes in human oesophagus. J Cell Mol Med. 2013;17:1506–12. doi: 10.1111/jcmm.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannucchi MG, Traini C, Manetti M, et al. Telocytes express PDGFRalpha in the human gastrointestinal tract. J Cell Mol Med. 2013;17:1099–108. doi: 10.1111/jcmm.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manole CG, Cismasiu V, Gherghiceanu M, et al. Experimental acute myocardial infarction: telocytes involvement in neo-angiogenesis. J Cell Mol Med. 2011;15:2284–96. doi: 10.1111/j.1582-4934.2011.01449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Flores L, Gutierrez R, Saez FJ, et al. Telocytes in neuromuscular spindles. J Cell Mol Med. 2013;17:457–65. doi: 10.1111/jcmm.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretoiu SM, Simionescu AA, Caravia L, et al. Complex effects of imatinib on spontaneous and oxytocin-induced contractions in human non-pregnant myometrium. Acta Physiol Hung. 2011;98:329–38. doi: 10.1556/APhysiol.98.2011.3.10. [DOI] [PubMed] [Google Scholar]
- Cretoiu SM, Cretoiu D, Popescu LM. Human myometrium-the ultrastructural 3D network of telocytes. J Cell Mol Med. 2012;16:2844–9. doi: 10.1111/j.1582-4934.2012.01651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum ST, Svalo J, Nielsen K, et al. Immunolocalization and expression of small-conductance calcium-activated potassium channels in human myometrium. J Cell Mol Med. 2012;16:3001–8. doi: 10.1111/j.1582-4934.2012.01627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretoiu SM, Cretoiu D, Marin A, et al. Telocytes: ultrastructural, immunohistochemical and electrophysiological characteristics in human myometrium. Reproduction. 2013;145:357–70. doi: 10.1530/REP-12-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Wang F, Liu Z, et al. Telocytes in liver: electron microscopic and immunofluorescent evidence. J Cell Mol Med. 2013;17:1537–42. doi: 10.1111/jcmm.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinescu ME, Ardeleanu C, Gherghiceanu M, et al. Interstitial Cajal-like cells in human gallbladder. J Mol Histol. 2007;38:275–84. doi: 10.1007/s10735-007-9099-0. [DOI] [PubMed] [Google Scholar]
- Matyja A, Gil K, Pasternak A, et al. Telocytes: new insight into the pathogenesis of gallstone disease. J Cell Mol Med. 2013;17:734–42. doi: 10.1111/jcmm.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinescu ME, Gherghiceanu M, Suciu L, et al. Telocytes in pleura: two-and three-dimensional imaging by transmission electron microscopy. Cell Tissue Res. 2011;343:389–97. doi: 10.1007/s00441-010-1095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Li H, Manole CG, et al. Telocytes in trachea and lungs. J Cell Mol Med. 2011;15:2262–8. doi: 10.1111/j.1582-4934.2011.01404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu LM, Gherghiceanu M, Suciu LC, et al. Telocytes and putative stem cells in the lungs: electron microscopy, electron tomography and laser scanning microscopy. Cell Tissue Res. 2011;345:391–403. doi: 10.1007/s00441-011-1229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolescu MI, Bucur A, Dinca O, et al. Telocytes in parotid glands. Anat Rec (Hoboken) 2012;295:378–85. doi: 10.1002/ar.21540. [DOI] [PubMed] [Google Scholar]
- Bosco C, Diaz E, Gutierrez R, et al. Ganglionar nervous cells and telocytes in the pancreas of Octodon degus: extra and intrapancreatic ganglionar cells and telocytes in the degus. Auton Neurosci. 2013;177:224–30. doi: 10.1016/j.autneu.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Popescu BO, Gherghiceanu M, Kostin S, et al. Telocytes in meninges and choroid plexus. Neurosci Lett. 2012;516:265–9. doi: 10.1016/j.neulet.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Gherghiceanu M, Popescu LM. Cardiomyocyte precursors and telocytes in epicardial stem cell niche: electron microscope images. J Cell Mol Med. 2010;14:871–7. doi: 10.1111/j.1582-4934.2010.01060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faussone-Pellegrini MS, Bani D. Relationships between telocytes and cardiomyocytes during pre-and post-natal life. J Cell Mol Med. 2010;14:1061–3. doi: 10.1111/j.1582-4934.2010.01074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretoiu D, Cretoiu SM, Simionescu AA, et al. Telocytes, a distinct type of cell among the stromal cells present in the lamina propria of jejunum. Histol Histopathol. 2012;27:1067–78. doi: 10.14670/HH-27.1067. [DOI] [PubMed] [Google Scholar]
- Rusu MC, Loreto C, Manoiu VS. Network of telocytes in the temporomandibular joint disc of rats. Acta Histochem. 2014;116:663–8. doi: 10.1016/j.acthis.2013.12.005. Doi: 10.1016/j.acthis.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Manetti M, Guiducci S, Ruffo M, et al. Evidence for progressive reduction and loss of telocytes in the dermal cellular network of systemic sclerosis. J Cell Mol Med. 2013;17:482–96. doi: 10.1111/jcmm.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirancea N, Morosanu AM, Mirancea GV, et al. Infrastructure of the telocytes from tumor stroma in the skin basal and squamous cell carcinomas. Rom J Morphol Embryol. 2013;54:1025–37. [PubMed] [Google Scholar]
- Milia AF, Ruffo M, Manetti M, et al. Telocytes in Crohn's disease. J Cell Mol Med. 2013;17:1525–36. doi: 10.1111/jcmm.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu VB, Radu E, Popescu LM. miR-193 expression differentiates telocytes from other stromal cells. J Cell Mol Med. 2011;15:1071–4. doi: 10.1111/j.1582-4934.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Cretoiu D, Yan G, et al. Comparative proteomic analysis of human lung telocytes with fibroblasts. J Cell Mol Med. 2014;18:568–89. doi: 10.1111/jcmm.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Cretoiu D, Yan G, Cretoiu SM, et al. Protein profiling of human lung telocytes and microvascular endothelial cells using iTRAQ quantitative proteomics. J Cell Mol Med. 2014;18:1035–59. doi: 10.1111/jcmm.12350. Doi: 10.1111/jcmm.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zheng M, Zhang M, et al. Differences in the expression of chromosome 1 genes between lung telocytes and other cells: mesenchymal stem cells, fibroblasts, alveolar type II cells, airway epithelial cells and lymphocytes. J Cell Mol Med. 2014;18:801–10. doi: 10.1111/jcmm.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhang M, Qian M, et al. Genetic comparison of mouse lung telocytes with mesenchymal stem cells and fibroblasts. J Cell Mol Med. 2013;17:567–77. doi: 10.1111/jcmm.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ikegami M, Wang Y, et al. Gene expression and biological processes influenced by deletion of Stat3 in pulmonary type II epithelial cells. BMC Genomics. 2007;8:455. doi: 10.1186/1471-2164-8-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosiek MJ, Gruber AD, Bader SR, et al. CD4+CD25+Foxp3+ regulatory T cells are dispensable for controlling CD8+ T cell-mediated lung inflammation. J Immunol. 2011;186:6106–18. doi: 10.4049/jimmunol.1000632. [DOI] [PubMed] [Google Scholar]
- Hegab AE, Ha VL, Gilbert JL, et al. Novel stem/progenitor cell population from murine tracheal submucosal gland ducts with multipotent regenerative potential. Stem Cells. 2011;29:1283–93. doi: 10.1002/stem.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2008. All data in this table was derived from Vega Genome Browser database. Available at: http://vega.sanger.ac.uk/ (accessed Aug 15, 2014)
- Hillier LW, Graves TA, Fulton RS, et al. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005;434:724–31. doi: 10.1038/nature03466. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–6. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jw IJ, Baldini A, Ward DC, et al. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci USA. 1991;88:9051–5. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarello R, Pedicini A, Caiulo A, et al. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet. 1992;89:247–9. doi: 10.1007/BF00217134. [DOI] [PubMed] [Google Scholar]
- Shehadeh LA, Webster KA, Hare JM, et al. Dynamic regulation of vascular myosin light chain (MYL9) with injury and aging. PLoS ONE. 2011;6:e25855. doi: 10.1371/journal.pone.0025855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T, Lagrost L. Plasma PLTP (phospholipid-transfer protein): an emerging role in ‘reverse lipopolysaccharide transport’ and innate immunity. Biochem Soc Trans. 2011;39:984–8. doi: 10.1042/BST0390984. [DOI] [PubMed] [Google Scholar]
- Brehm A, Geraghty P, Campos M, et al. Cathepsin G degradation of phospholipid transfer protein (PLTP) augments pulmonary inflammation. FASEB J. 2014;28:2318–31. doi: 10.1096/fj.13-246843. Doi: 10.1096/fj.13-246843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor SL, Garcia DK. Chromosome 3. Hoboken, NJ, USA: eLS, John Wiley & Sons, Inc; 2006. DOI: 10.1038/npg.els.0005812. [Google Scholar]
- Pradella S, Hans A, Sproer C, et al. Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56. Arch Microbiol. 2002;178:484–92. doi: 10.1007/s00203-002-0479-2. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Paux E, Sourdille P, Salse J, et al. A physical map of the 1-gigabase bread wheat chromosome 3B. Science. 2008;322:101–4. doi: 10.1126/science.1161847. [DOI] [PubMed] [Google Scholar]
- Hergenreider E, Heydt S, Treguer K, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- Khan MM, Strack S, Wild F, et al. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy. 2014;10:123–36. doi: 10.4161/auto.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Young MM, Serfass JM, et al. Sh3glb1/Bif-1 and mitophagy: acquisition of apoptosis resistance during Myc-driven lymphomagenesis. Autophagy. 2013;9:1107–9. doi: 10.4161/auto.24817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GJ, Bueno R, Sugarbaker DJ. Genes associated with prognosis after surgery for malignant pleural mesothelioma promote tumor cell survival in vitro. BMC Cancer. 2011;11:169. doi: 10.1186/1471-2407-11-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriuchi N, Higuchi T, Hanaoka H, et al. Current status of cancer therapy with radiolabeled monoclonal antibody. Ann Nucl Med. 2005;19:355–65. doi: 10.1007/BF03027399. [DOI] [PubMed] [Google Scholar]
- Hung JY, Horn D, Woodruff K, et al. Colony-stimulating factor 1 potentiates lung cancer bone metastasis. Lab Invest. 2014;94:371–81. doi: 10.1038/labinvest.2014.1. Doi: 10.1038/labinvest.2014.1. [DOI] [PubMed] [Google Scholar]
- Seo DC, Sung JM, Cho HJ, et al. Gene expression profiling of cancer stem cell in human lung adenocarcinoma A549 cells. Mol Cancer. 2007;6:75. doi: 10.1186/1476-4598-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Escamilla J, Mok S, et al. CSF1R signaling blockade stanches tumor-infiltrating myeloid cells and improves the efficacy of radiotherapy in prostate cancer. Cancer Res. 2013;73:2782–94. doi: 10.1158/0008-5472.CAN-12-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino D, De Pitta C, Orso F, et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2013;27:1223–35. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- Hemnes AR, Zaiman A, Champion HC. PDE5A inhibition attenuates bleomycin-induced pulmonary fibrosis and pulmonary hypertension through inhibition of ROS generation and RhoA/Rho kinase activation. Am J Physiol Lung Cell Mol Physiol. 2008;294:L24–33. doi: 10.1152/ajplung.00245.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data profiles for all genes.
Details of up-or down regulated gene expression variations of chromosome 2 and 3.
Details of the selected core network genes in TCs isolated from the mouse lung and cultured for 5 days in chromosome 2.
Details of the selected core network genes in TCs isolated from the mouse lung and cultured for 10 days in chromosome 2.
Details of the selected core network genes in mouse mesenchymal stem cells in chromosome 2.
Details of the selected core network genes in mouse fibroblasts in chromosome 2.
Details of the selected core network genes in mouse alveolar type II cells in chromosome 2.
Details of the selected core network genes in mouse airway basal cells in chromosome 2.
Details of the selected core network genes in mouse proximal airway cells in chromosome 2.
Details of the selected core network genes in mouse CD8+ T cells come from bronchial lymph nodes in chromosome 2.
Details of the selected core network genes in mouse CD8+ T cells from lung in chromosome 2.
Details of the selected core network genes in TCs isolated from the mouse lung and cultured for 10 days in chromosome 3.
Details of the selected core network genes in TCs isolated from the mouse lung and cultured for 5 days in chromosome 3.
Details of the selected core network genes in mouse mesenchymal stem cells in chromosome 3.
Details of the selected core network genes in mouse fibroblasts in chromosome 3.
Details of the selected core network genes in mouse alveolar type II cells in chromosome 3.
Details of the selected core network genes in mouse airway basal cells in chromosome 3.
Details of the selected core network genes in mouse proximal airway cells in chromosome 3.
Details of the selected core network genes in mouse CD8+ T cells come from bronchial lymph nodes in chromosome 3.
Details of the selected core network genes in mouse CD8+ T cells from lung in chromosome 3.


