Abstract
NKG2D is an activating receptor expressed by all NK cells and subsets of T cells. It serves as a major recognition receptor for detection and elimination of transformed and infected cells and participates in the genesis of several inflammatory diseases. The ligands for NKG2D are self-proteins that are induced by pathways that are active in certain pathophysiological states. NKG2D ligands are regulated transcriptionally, at the level of mRNA and protein stability, and by cleavage from the cell surface. In some cases, ligand induction can be attributed to pathways that are activated specifically in cancer cells or infected cells. We review the numerous pathways that have been implicated in the regulation of NKG2D ligands, discuss the pathologic states in which those pathways are likely to act, and attempt to synthesize the findings into general schemes of NKG2D ligand regulation in NK cell responses to cancer and infection.
INTRODUCTION
Natural killer (NK) cells were first discovered based on their capacity to lyse tumor cells without prior sensitization (1). Early studies also demonstrated a role for NK cells in limiting certain viral infections. It soon became clear that NK cells do not express T cell or B cell antigen receptors. Consequently, the mechanism of specific recognition of tumor cells and virus-infected cells remained a mystery for many years, until numerous inhibitory and activating receptors were eventually discovered. Each NK cell expresses several different activating receptors and a few different inhibitory receptors. These receptors and the corresponding modes of recognition are mentioned only in passing below. This review focuses on the best-characterized activating NK receptor, called NKG2D, and specifically on the regulation of the ligands recognized by NKG2D. NKG2D is one of the most important activating receptors expressed by NK cells in terms of tumor cell recognition (2, 3), although NKp46, NKp44, NKp30, DNAM1, SLAM-family ligands, and others also play important roles (1). Notably, NKG2D binds to several different ligands that are encoded by distinct genes in the host's own genome, i.e., the ligands are self-proteins, as opposed to foreign antigens. Several different ligands, encoded by distinct genes, exist in each individual. Most importantly, NKG2D ligands are expressed poorly or not at all by most normal cells but are upregulated in cancer cells and virus-infected cells. This type of recognition process, in which self-encoded ligands for activating receptors are induced on unhealthy cells, has been called “induced self-recognition” (4), distinct from “missing self-recognition,” the phenomenon in which loss of MHC ligands for NK inhibitory receptors sensitizes cells for elimination by NK cells (5). As we describe below, various cellular pathways activated as a result of cellular stress, infection, or tumorigenesis regulate expression of the NKG2D ligands. These findings underlie the concept that NK cells recognize unhealthy or distressed cells, though there are clearly other modes of NK recognition, such as missing self-recognition and recognition of certain foreign ligands (1). The purpose of this review is to describe the current understanding of the pathways that regulate the display of NKG2D ligands on cells and that, therefore, regulate the sensitivity of target cells to elimination by NK cells. This information informs a broader understanding of the role of NK cells in immune recognition.
PROPERTIES OF NKG2D
NKG2D is a lectin-like, type 2 transmembrane receptor (2, 6, 7). It functions as an activating receptor by virtue of its interactions with the signaling adapter molecule DAP10 in humans and with DAP10 and DAP12 in mice (7, 8). When the receptor is ligated, DAP10 provides signals that recruit the p85 subunit of phosphatidylinositol 3-kinase (PI3K) and a complex of GRB2 and VAV1, whereas DAP12 activates protein tyrosine kinases Syk and ZAP70. Engagement of NKG2D on NK cells induces degranulation and cytokine production. Earlier analyses of transgenic target cells indicated that expression of NKG2D ligands was sufficient to convert normal cells (lymphocytes, at least) into target cells for NK cells, as tested in vitro and in vivo (9, 10). Those results further suggested that the sensitivity of ligand-expressing cells to NK cells did not depend on the induction of other types of activating ligands in conjunction with NKG2D ligands or on the loss of inhibitory MHC molecules by the cells. However, naive human NK cells failed to respond well when stimulated through NKG2D alone, but did respond well when NKG2D was stimulated along with other receptors such as 2B4, a SLAM family receptor whose ligand is broadly expressed by hematopoietic cells (11). In this case, however, the coactivating ligand is broadly expressed even in normal hematopoietic cells, though not in most nonhematopoietic cells. Therefore, in humans, as well as in mice, induced expression of NKG2D ligands by otherwise normal cells is likely a sufficient alteration for converting many cell types into NK-sensitive target cells.
NKG2D is expressed by all NK cells but is not limited to NK cells, as it is also expressed by many T cells, including all CD8+ T cells in humans (all activated CD8+ T cells in mice), subsets of γδ T cells, and subsets of NKT cells (2). Expression by certain CD4+ T cells has also been reported, at least in humans, although in mice it is difficult to detect expression by conventional CD4+ T cells (12, 13). In T cells, NKG2D may have several functions. In specialized T cells, such as the subset of γδ T cells resident in the skin of mice, NKG2D provides potent costimulatory signals for T cell activation (14, 15). In conventional CD8+ T cells, NKG2D may also provide costimulatory function (16, 17), although this is most evident for the subset of highly activated CD8+ T cells that lacks expression of CD28, the conventional costimulatory receptor for T cells (16). In some scenarios, such as after potent activation of human CD8+ T cells with IL-15 and CD3 engagement, subsequent NKG2D engagement in the absence of T cell receptor activation is sufficient to trigger target cell killing (18). Hence, NKG2D provides signals that activate, or in some cases coactivate, killing and cytokine production by NK cells and certain T cells.
NK cell activation as a result of NKG2D engagement can modify, or be modified by, engagement of other NK receptors. For naive human NK cells, synergistic activation occurs when NKG2D is coengaged with 2B4, as already mentioned, or with NKp46, another NK activating receptor (11). Conversely, NKG2D-induced NK cell activation can be inhibited (albeit not necessarily completely) if the target cell expresses MHC class I molecules that engage inhibitory receptors on NK cells, such as Ly49 receptors in mice or KIRs (killer cell immunoglobulin-like receptors) in humans (13, 19). We emphasize this point to highlight the possibility that unhealthy cells may simultaneously alter the expression of various activating and inhibitory ligands for NK cells; the NK cell is thought to integrate these various signals, ultimately balancing activating signals against inhibitory signals and responding accordingly.
NKG2D LIGANDS
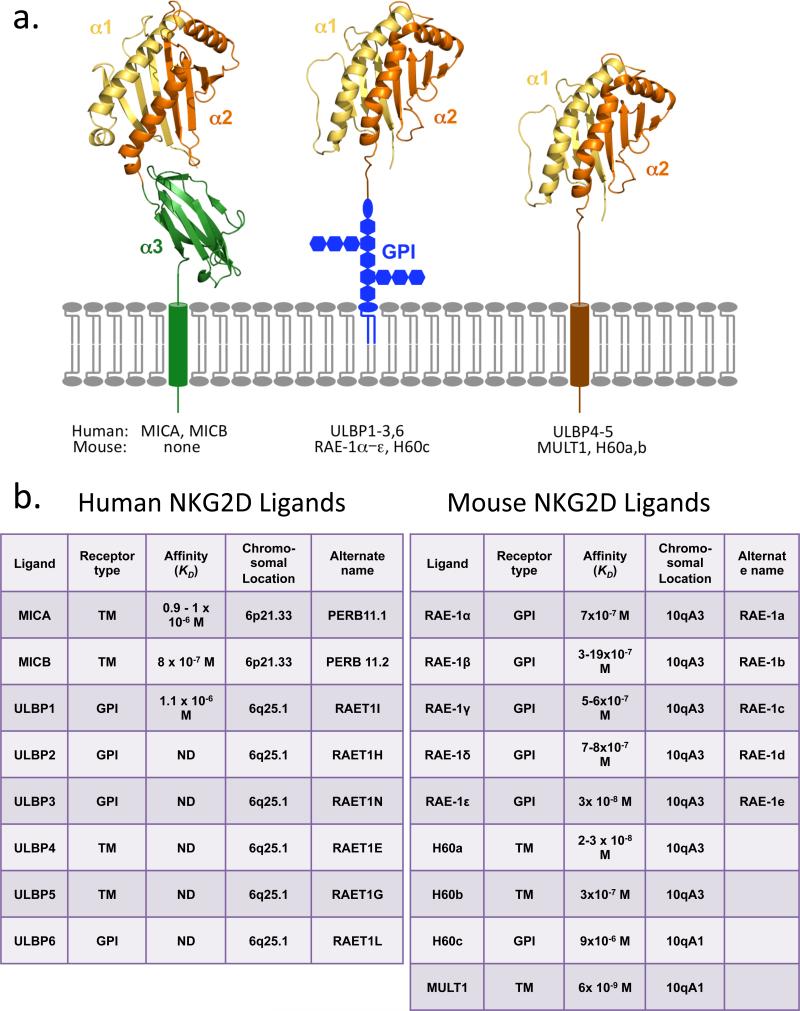
Multiple NKG2D ligands have been identified in humans and mice, all of which are homologous to MHC class I molecules (2, 7, 20) (Figure 1). Like MHC proteins, they exhibit considerable allelic variation. In humans, the NKG2D ligands include MICA and MICB (MHC class I chain–related proteins A and B), both encoded by genes in the MHC, and up to six different proteins called ULBPs (UL16-binding proteins), also known as RAET1 proteins. The latter group of genes is clustered on human chromosome 6. In mice, there are no orthologs of the MICA and MICBgenes, but a family of genes orthologous to the human ULPB/RAET1 family is present on chromosome 10. These genes encode proteins that fall into three subgroups of NKG2D ligands, including five different isoforms of RAE-1 (retinoic acid early inducible-1) proteins, one MULT1 (murine UL16-binding protein-like transcript 1) protein, and three different isoforms of H60 proteins (though not all mouse strains express all the isoforms).
Figure 1.
NKG2D ligands in humans and mice. (a) The ligands fall into three general structures. MICA and MICB are transmembrane proteins with three domains analogous to the α1–α3 domains of MHC Ia proteins (MICB structure shown on left; Reference 21). The remaining ligands contain two domains analogous to α1 and α2 of MHC Ia proteins but no α3-like domain (the RAE-1β α1 and α2 domains are depicted in both the middle and right structures; Reference 21). Human ULBP1–3 and 6 and mouse RAE1α–ε and H60c are GPI-linked, whereas human ULBP4–5 and mouse MULT1 and H60a and b are transmembrane proteins. In some cases (e.g., ULBP2 and possibly others), both GPI-linked and transmembrane forms of the protein are found on the same cell (22). The original image was kindly provided by Dr. Roland Strong of the Fred Hutchinson Cancer Research Center. (b) Properties of NKG2D ligands and genes are summarized, including transmembrane versus GPI linkage, affinity for NKG2D, and chromosomal location based on Ensembl (http://www.ensembl.org) and the UC Santa Cruz genome browser (http://genome.ucsc.edu). The affinities are from equilibrium determinations (23, 24, 25, 26, 27, 28). Note that MICA and MICB are within the human MHC, whereas the remaining human ligand genes are located on the same chromosome but on the other side of the centromere. The mouse NKG2D ligands are located on chromosome 10, separate from the mouse MHC on chromosome 17. (ND, not determined.)
Different NKG2D ligands vary considerably in sequence and bind NKG2D with a wide range of affinities, with KDs ranging from 9 × 10−6 to 6 × 10−9 M (2, 7, 29) (Figure 1). Furthermore, some of the ligands are transmembrane proteins, while others are glycosylphosphatidylinositol (GPI)-linked (2, 7, 30) (Figure 1). Despite these variations, the topography of binding is similar, though not identical, in each of the NKG2D-ligand structures that have been solved by X-ray diffraction analysis (29, 31). Moreover, the various ligands trigger NK cell and T cell functions similarly, as tested using cell lines transfected with different ligands.
Why there are so many distinct NKG2D ligands remains a subject of speculation, but several possibilities exist, many of which are not mutually exclusive: (a) As is discussed in sections below, the ligands are likely regulated somewhat differently by stress pathways. This feature presumably allows the same receptor to stimulate a response in different contexts, including responses to cells undergoing different forms of stress, because these different contexts result in upregulation of distinct ligands. (b) Pathogen infections are likely to have exerted selection for diversity and a degree of redundancy of NKG2D ligands. It is common for viruses to encode proteins that destroy or inhibit NKG2D ligands and other immune receptor ligands in order to evade the corresponding responses, and the existence of a multitude of ligands regulated by common mechanisms confers fitness to the host by making such evasion more difficult. (c) The different ligands may vary somewhat in how they trigger or engage NKG2D, in such a way that different outcomes occur. Such differences have not been well documented in functional studies. Nevertheless, as already mentioned, the ligands do vary considerably in affinity for NKG2D and are likely to reside in different membrane compartments because some are GPI-linked and some are not. They may also differ in how well they are shed or secreted from cells. (d) The different ligands may engage other receptors in addition to NKG2D, as was suggested in one report (32). (e) The various ligands may exert different effects on the cells that express them, independent of NKG2D engagement. One report suggested that RAE-1 expression is required for neural cell proliferation, for example, making this idea plausible (33).
In addition to being displayed on the cell surface, some, or perhaps all, NKG2D ligands can be shed or excreted from cells. In some cases, the ligands are cleaved from the plasma membrane by proteinases, but in others, the ligands are found associated with membrane vesicles that are excreted from cells, such as exosomes, or secreted from cells. Shedding and excretion of NKG2D ligands are discussed in detail in a later section of this review.
REGULATION OF THE EXPRESSION OF NKG2D LIGANDS: OVERVIEW AND BACKGROUND
It is well accepted that cellular stress pathways play a role in regulating NKG2D ligands. The term “stress” is difficult to define, however, and a more meaningful description would specify the underlying molecular pathways and the conditions under which they are active. Some of the relevant pathways may be activated normally in development, as suggested by the finding that NKG2D ligands are expressed in early embryonic tissues, so it is arguable whether all such pathways should be defined as stress pathways. The following sections summarize current knowledge of how the cell surface expression of various NKG2D ligands is regulated and the potential significance of the corresponding pathways in the context of disease. In particular, these findings are considered in the context of events that accompany cellular infections and tumorigenesis. Furthermore, NKG2D ligands are regulated at several stages of biogenesis, including (at least) transcription, RNA stabilization, protein stabilization, and cleavage from the cell membrane. In the following sections, we consider the stage in ligand biogenesis at which each of the regulatory mechanisms acts. Finally, the totality of the information is integrated in an attempt to develop a unifying synthesis of how NKG2D ligands are regulated in the service of host defense.
Before delving into the themes of regulation, we summarize relevant events that accompany tumorigenesis, infection, and injury, to provide a context for the findings. Some of this information is relevant for understanding the specific mechanisms that regulate NKG2D ligands, which are discussed in subsequent sections. But these summaries are most pertinent for underpinning the closing sections of the review, which attempt to synthesize the findings concerning specific regulatory mechanisms, from the broader perspectives of the processes of tumorigenesis, infection, and injury.
However, some of the key pathways discussed here have not been implicated in regulation of NKG2D ligands. They are mentioned here for the sake of completeness.
Cellular Pathways Activated in Tumorigenesis
Cell transformation and tumorigenesis are associated with the activation of numerous stress pathways in the affected cells. Early in tumorigenesis, rapid, poorly regulated cell proliferation (hyperplasia) is thought to activate a stress pathway called the DNA damage response. Poorly regulated DNA replication (termed replication stress), characterized by collapsed replication forks, can result in DNA breaks (34, 35). The replication stress itself, as well as the resulting DNA breaks, are each thought to independently activate the DNA damage response, a protein kinase cascade that regulates numerous aspects of cell physiology (36). The following underlying mechanisms of induction have been discerned: Collapsed replication forks that accompany replication stress are detected by the DNA damage sensor kinase ATR (Ataxia telangiectasia and Rad3 related), and the accompanying DNA breaks are detected by the related protein kinase ATM (Ataxia telangiectasia, mutated). Activated ATR and/or ATM initiates a protein kinase cascade in which many downstream mediators are activated post-transcriptionally, including the checkpoint kinases CHK1 and CHK2 and the key tumor suppressor p53. Studies show that various DNA damage response proteins such as ATM, CHK2, and p53 are activated early in tumorigenesis, in some cases in precancerous lesions (34, 35). ATM or ATR activation can cause cell cycle arrest, either through the action of p53 or by a p53-independent mechanism (37). The p19ARF tumor suppressor is also induced as a result of strong proliferative signals in developing tumors, associated with highly active c-MYC or RAS (38, 39). Similar to the DNA damage response, induced p19ARF can activate p53. p53 activation by the DNA damage response or p19ARF imposes a strong cell cycle arrest and can also induce apoptosis or cellular senescence, depending on the cell type and other factors (40).
The p16INK4A tumor suppressor, encoded by a gene that overlaps thep19Arf gene, represents another means of feedback inhibition of cell proliferation (41). p16INK4A is induced by oncogenic signals as well by the E2F transcription factor, inhibits cyclin-dependent kinase 4 (CDK4) and CDK6, and thereby suppresses cell proliferation.
Among the most common alterations in cancer cells are those that activate the PI3K pathway, which promotes survival and proliferation (42,43). Mutations in PI3K itself, receptors that activate PI3K, or downstream mediators of PI3K signaling are commonly detected in cancers.
In addition to the pathways described above, developing tumors have been reported to show activation of other stress pathways that promote cell survival, such as the hypoxia response, the unfolded protein response, and the heat shock response. For example, the heat shock proteins (HSP), which are frequently induced in cancer cells (44), inhibit multiple apoptotic signaling components and therefore promote survival.
The tumor suppressive mechanisms and stress pathways described here operate to some extent intrinsically in the cancer cell, but as we discuss below, some of them also play roles in promoting or modulating immune reactions, suggesting a role in immune surveillance of cancer. Ultimately, many developing tumors acquire mutations in genes encoding key regulatory proteins, including p53, p19ARF, p16INK4A, and pRb, and such mutations enable the tumors to bypass tumor suppression. In some cases, these mutations are believed also to play a role in escape from immune surveillance.
Cellular Pathways Activated During Infections
Infections can induce numerous pathways associated with immune responses as well as various stress pathways. Among the innate signaling pathways are those triggered by Toll-like receptors (TLRs) and intracellular sensors of microbial molecules such as NOD-like receptors (NLRs), RNA sensors, DNA sensors, and sensors of microbe-derived cyclic dinucleotides (45). Many of these receptors and sensors are expressed not only by immune cells such as dendritic cells, but also by many other cell types including epithelial cells and fibroblasts. When a cell is exposed to microbes or viruses via intracellular or extracellular routes, one or more of these innate receptors are typically activated and can induce various intracellular signaling pathways. Among these pathways are those involving activation of the signaling molecule TANK-binding kinase-1 (TBK1) and the transcription factors IRF3, IRF5, IRF7, or NF-κB, which regulate chemokine and cytokine genes, including type I interferon (IFN) genes, as well as genes encoding costimulatory ligands such as CD40, CD80, and CD86 (46). Some of the innate receptors instead activate the inflammasome, typically resulting in caspase-1 activation, processing of certain cytokines such as IL-1 and IL-18, and cell death.
Infections can also induce one or more stress pathways. Various infections induce the DNA damage response, described above, in some cases because of the mode or rapid pace of viral DNA replication (47, 48,49) and in other cases because viral products induce DNA damage or specifically activate the ATR or ATM kinases that initiate the DNA damage response (50, 51). Some viruses encode proteins that suppress downstream components of the DNA damage response, apparently to enable their host cells to evade the consequences of having activated the pathway (47, 48).
Infections can also activate other major stress pathways. Among these are, as mentioned above for tumorigenesis, the heat shock pathway and the unfolded protein response, as well as the oxidative stress pathway (52, 53, 54). In addition, many viral infections cause the activation of the PI3K and/or mitogen-activated protein kinase (MAPK) pathways (55). The activation of these stress and signaling pathways may benefit the pathogen by enhancing replication processes and enabling the cells to withstand the stress accompanying infections, but the activated state of the pathways may at the same time provide cues that indicate to the immune system that the cells are potentially infected or otherwise distressed.
Injury and Inflammatory Disease
In the absence of infection or transformation, tissue injury can induce numerous stress responses. Injury that damages DNA induces the DNA damage response, as already discussed, and unhealthy cells often establish an endoplasmic reticulum stress response. Certain forms of injury induce senescence (40). In some models of injury, p53 and/or p16/INK4A are activated, which is necessary for the senescent state to be established (56, 57). Senescent cells can accumulate in conditions of chronic injury and exert pathological effects, as occurs in mouse models of liver fibrosis, a precursor of cirrhosis. The removal of senescent cells in injured tissues can therefore promote resolution of the injury. Indeed, induced expression of NKG2D ligands may enable NK cells to eliminate senescent cells and thereby aid in resolving the injury (56, 58).
In other disease states, inflammation results in the accumulation of aberrant cells in tissues. One example is metabolic syndrome, associated with type 2 diabetes, where the liver and other organs are highly infiltrated by immune cells (59, 60, 61). This condition is often accompanied by atherosclerosis, which is the constriction of blood vessels due to the accumulation of plaque, consisting largely of activated so-called foamy macrophages. The underlying cause of inflammation in such diseases varies, but in the case of metabolic syndrome, a significant role is believed to be played by metabolites that accumulate in patients with metabolic syndrome. Evidence has been provided of a role for oxidized low-density lipoproteins (Ox-LDLs), which are believed to engage a TLR/CD36 complex (62), and advanced glycation endproduct (AGE), which engages the receptor for AGE (RAGE) (63, 64). Stimulation of cell types expressing these innate receptors may underlie inflammation in metabolic syndrome.
REGULATION OF NKG2D LIGANDS
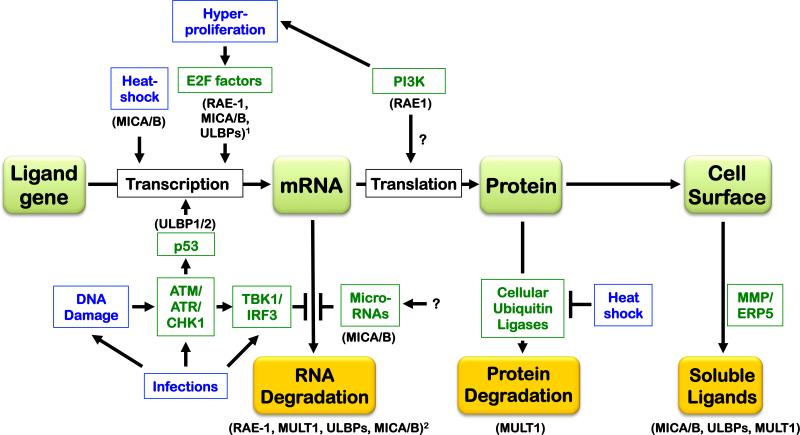
We describe the pathways and signals known to regulate NKG2D ligands initially in the order of the stage in ligand biogenesis that is regulated: transcription, RNA stabilization, translation, protein stabilization, cell surface egress, and excretion/shedding of ligands from cells. The reader is referred to Figure 2 to put the published findings in perspective. The specific ligands that undergo different types of regulation are specified in the text and the figures to emphasize the point that some forms of regulation impact one subgroup of ligands and not others.
Figure 2.
Regulation of NKG2D ligands at different stages of ligand biogenesis (gene→mRNA→protein, depicted in green boxes) and degradation (orange boxes). The regulatory signals and pathways that regulate NKG2D ligands are depicted, as well as whether they act transcriptionally, by stabilizing or targeting ligand mRNAs, by stabilizing ligand proteins, or by cleavage of the ligand from the cell surface. The blue text boxes specify stress or pathological states, whereas the green text boxes specify different mediators, with the affected ligands indicated parenthetically in black. 1Proliferative signals regulate mouse Raet1 and human RAET1/ULBP and possibly MICA/B expression, but the role of E2F was demonstrated specifically in the case of Raet1 genes. 2The activated DNA damage response induces mouse Raet1 and Mult1 expression as well as human RAET1/ULBP and possibly MICA/B expression, but the role of mRNA stabilization was demonstrated specifically for Raet1 transcripts. See the text for additional details.
TRANSCRIPTIONAL REGULATION
Although it is generally assumed that most regulation of NKG2D ligands occurs at the transcriptional stage, some of the mechanisms that have been discovered work primarily at post-transcriptional stages. The pathways thought to regulate ligand transcription directly are discussed in this section.
Heat Shock Pathway
The expression of MICA and MICB, two of the human NKG2D ligands, is regulated in some conditions by the heat shock stress pathway. For example, in confluent cells that become quiescent, imparting heat shock resulted in increased MICA and MICB mRNA accumulation and cell surface expression (65). Heat shock response elements (HSE) were defined in the corresponding promoter elements. Chromatin immunoprecipitation (ChIP) experiments showed that the promoter was occupied by the heat shock transcription factor 1 (HSF1) in heat-shocked cells (66). Reporter plasmids driven by the MICA promoter were induced by heat shock, and mutations in the HSE that bind HSF1 caused a reduction in reporter activity in heat-shocked cells. Transcriptional regulation directed at HSE in the MICA promoter was not necessary for induction of MICA or MICB in response to viral infections or in proliferating cells, however (66), suggesting the existence of independent modes of stress-induced transcriptional regulation. On the basis of sequence analysis, investigators suggested that HSE may also exist in some of the ULBP genes (67), but it has not been demonstrated that these genes are regulated by heat shock. HSE have not been implicated in the transcriptional regulation of any mouse NKG2D ligand genes.
E2F Transcription Factors
Transcription of the mouse Raet1 family of ligands is regulated by the E2F family of transcription factors (68). This discovery was based on the findings that expression of RAE-1 was highly correlated with cell proliferation in fibroblasts and in tumor cell lines in vitro. Inhibitors of transcription rapidly suppressed Raet1 mRNA and RAE-1 protein in proliferating cells in culture. Conversely, even some normal cell types that were actively proliferating in vivo, such as embryonic brain cells and cells in wounded skin, upregulated RAE-1 (68). Nuclear run-on experiments showed that proliferative conditions were associated with increased transcription of the Raet1e gene. An investigation of the roles of factors known to regulate the cell cycle showed that E2F1–3, the activating E2F transcription factors, bind to the Raet1e promoter in vivo and transactivate the endogenous Raet1e gene as well as the Raet1epromoter in a reporter plasmid. Mutations in two consensus E2F sites in the promoter impaired the activation of the Raet1e promoter reporter plasmid.
The available data in mice suggest that proliferation-associated signals induce transcription of Raet1e, Raet1d, and one or more of the Raet1a, b, and g genes, but do not appreciably induce transcription of Mult1 orH60b. Hence, E2Fs may regulate only a subset of NKG2D ligands in mice, specifically most or all Raet1 genes. In the human HCT116 cell line, proliferation was associated with increased cell surface expression of MICA, MICB, ULBP2, ULBP3, and possibly ULBP1 (68, 69), although the role of E2F transcription factors has not been directly addressed experimentally. Analysis indicated that proliferation was associated with higher activity of the MICA and MICB promoters in these cells (66). These data suggest that regulation associated with proliferative signals is a common feature of numerous NKG2D ligands.
p53 Transcription Factor
The possible role of the p53 tumor suppressor in regulating NKG2D ligands has been investigated in numerous studies. Early work showed that expression of NKG2D ligands occurs in many tumor cell lines, even when the p53 gene was deleted, indicating that p53 is not essential for ligand expression (70, 71). Additional studies in murine systems have failed to detect a major role for p53 in ligand expression (A. Iannello & D.H. Raulet, unpublished data). However, analysis of human ligands ULBP1 and 2 suggests that p53 can reinforce higher expression of these ligands. p53 response elements were localized in the first introns of the genes, and in a cell line p53 was associated with the ULBP1 and 2genes, based on the results of ChIP experiments (72, 73). Cell lines treated with drugs that induce p53 showed a higher expression of ULBP1and 2. These data suggested that p53 action amplifies transcription of certain human NKG2D ligands. Countering this effect, another report indicated that microRNAs induced by p53, miR34a, and miR34c can downregulate ULBP2, as is discussed again later in this review (74). No role of p53 in regulating NKG2D ligands in mouse cells has yet been documented. Furthermore, although the DNA damage response induces NKG2D ligands in mouse cells, and p53 is activated downstream of the DNA damage response, p53 was dispensable for induction of NKG2D ligands by DNA-damaging agents (70). Hence, the DNA damage response induces NKG2D ligands at least in part by a p53-independent process. Together, these data suggest selective, and in some cases opposing, regulatory effects of p53 on the expression of NKG2D ligands.
NF-κB Transcription Factors
Investigators proposed several years ago that NF-κB transcription factors induced in activated T cells may stimulate MICA transcription in human cells (75). Consistent with a role for NF-κB, treating human cell lines with TNF-α, which induces NF-κB activity, caused a modest induction of MICA, and a binding site for NF-κB was found to overlap the HSE in the MICA promoter (76). Hence, inflammatory signals and NF-κB may amplify MICA expression. Attempts to induce NKG2D ligands in mouse cells with TNF-α have been unsuccessful, however (S. Gasser & D.H. Raulet, unpublished data).
Sp Family Transcription Factors
Studies of the MICA, MICB, and ULBP1 genes identified potential binding sites for Sp family transcription factors (66, 77). Binding of the promoter to Sp1 and in some cases to Sp3 was demonstrated in the case of all three promoters, and mutations of the binding sites resulted in reduced transcription in reporter assays. The Sp site was necessary for optimal transcriptional activation in both heat-shocked and proliferating cells, but not in cells infected with human cytomegalovirus (66). Transactivation assays suggested that the long form of the Sp3 factor plays a particularly important role, at least in the case of the ULBP1 promoter. In general, binding of Sp transcription factors to the promoters of ligand genes was constitutive (66), and it has not been shown that regulatory changes in ligand expression are due to alterations in Sp factors or binding.
AP-1 and AP-2a
Evidence has also emerged that the murine Raet1e transcripts are much more abundant in cells lacking JunB, a subunit of the activator protein (AP)-1 transcription complex, resulting in substantially higher cell surface display of RAE-1ε (78). It remains unclear, however, whether AP-1 acts directly on the Raet1e gene to repress transcription or by an indirect mechanism that alters Raet1e transcription or mRNA stability.
In a similar vein, AP-2a may negatively regulate the ULBP1 gene. It was reported that the ULBP1 promoter contains an AP-2 site that partially overlaps the binding site for Sp family transcription factors (77). Mutating the AP-1 site resulted in modestly increased expression of a reporter plasmid driven by the ULBP1 promoter. It was proposed that AP-2a inhibits ULBP1 expression by blocking binding of Sp family transcription factors.
REGULATION AT THE mRNA LEVEL
Evidence abounds that NKG2D ligands are regulated post-transcriptionally, and a prominent level of regulation occurs at the RNA stage. Stabilization of NKG2D ligand mRNA is particularly important for induction of ligands in cells that sustain DNA damage.
The DNA Damage Response
The finding that DNA damage is associated with induction of NKG2D ligands came in studies of cultured cell lines (70). Analysis of transformed cell lines, a cultured fibroblast cell line, and numerous other cell lines showed that drugs or irradiation that damaged DNA consistently induced several NKG2D ligands in mouse cells, including RAE-1, MULT1, and H60 (70, 79). In the case of human cell lines, DNA-damaging agents induced the expression of ULBP1, 2, and 3 and, in some studies, MICA and MICB (70, 80, 81). In addition, ligands were induced at the cell surface by treating cells with aphidicolin, a drug that inhibits DNA replication by inhibiting DNA polymerase (70). By disrupting DNA replication, aphidicolin causes replication stress and can also cause DNA breaks. Cells subjected to DNA damage exhibited enhanced sensitivity to NK lysis in vitro.
As described above, DNA-damaging agents activate ATM and ATR and therefore activate the DNA damage response. Induction of ligands in cells subjected to DNA damage was inhibited by conditional deletion of the ATR gene; by small hairpin RNA (shRNA) knockdowns of ATR, ATM, or a downstream kinase, CHK1; or by treating the cells with drugs that inhibit ATR, ATM, or CHK1 (70). The outcome depended on how the DNA damage was inflicted, consistent with findings that ATM and ATR are activated by different types of DNA damage. These findings indicated that DNA damage induces NKG2D ligands via the DNA damage response. Importantly, although p53 is activated by the DNA damage response, p53 played little or no role in ligand induction in cell lines subjected to DNA damage (70, 71).
The finding that the DNA damage response induces NKG2D ligands took on added significance in light of evidence that tumor cells in situ, including precancerous lesions, exhibit constitutive activation of ATM and other components of the DNA damage response (34, 35). Studies with tumor cell lines showed that inhibiting the DNA damage response caused a decrease in the display of NKG2D ligands on the cell surface (70, 79). Hence, the DNA damage response has the potential to serve as a sentinel for tumor formation by inducing NKG2D ligands and therefore mobilizing NK cells and T cells against the tumors. In a related scenario, chemotherapy drugs may work in part by intensifying the DNA damage response, resulting in enhanced expression of NKG2D ligands (79). Evidence consistent with such a role was obtained in the case of multiple myelomas and Ewing sarcomas in humans (81).
The DNA damage response can also be activated in rapidly proliferating cells as a result of DNA replication stress. Accordingly, cultured human T cells stimulated with mitogens, superantigens, or antigens were induced to express NKG2D ligands in vitro, and the expression of the ligands was decreased by treating the cells with drugs or siRNAs that inhibit the DNA damage response (80). In contrast to these results with human T cells, NKG2D ligands were not strongly induced in mouse T cells stimulated to proliferate in vitro, although a modest upregulation of ligands was observed in stimulated B cells in culture (30, 70). Because it could be counterproductive to upregulate NKG2D ligands during immune responses in a manner that causes proliferating lymphocytes to be eliminated by NK cells, it might be expected that NKG2D ligand induction would be suppressed in order to prevent such events. Whether and how NKG2D ligands are regulated differentially in proliferating lymphocytes versus other cell types remain to be established. It has been proposed, however, that killing of activated T cells that express NKG2D ligands could serve as part of a mechanism to limit undesirable immune responses (69, 80, 82); if this is the case, it seems unlikely that proliferation could be the sole determinant of ligand expression in lymphocytes because such a mechanism would not readily discriminate undesirable activated lymphocytes from desirable ones.
The DNA damage response is also activated in certain infections (see the Introduction). Induction of NKG2D ligands by this pathway may therefore serve as a mechanism to enable NK cells and T cells to eliminate infected cells. Evidence in favor of this idea has emerged in two instances. In one study, pre-B cells infected with Abelson murine leukemia virus were induced to display NKG2D ligands on the cell surface (50). Abelson virus infection inappropriately induces the expression of activation-induced cytidine deaminase (AID) in infected pre-B cells. AID is a mutagen in vivo that deaminates bases in DNA, and these researchers (50) proposed that DNA damage inflicted inappropriately by AID in infected cells was responsible for ligand induction.
A second example in which the DNA damage response was implicated in the induction of NKG2D ligands in infected cells comes from studies of HIV-infected cells. HIV encodes the Vpr protein, which activates the ATR kinase and thereby activates the DNA damage response. Studies showed that HIV infection of cells in culture results in induction of ULBP1 and 2 (51). Induction depends on Vpr expression by HIV and is ATR dependent. A subsequent study showed that the HIV Vif protein also impacts NKG2D ligand expression, in this case by inhibiting it (83). The role of Vif is related to its activity in targeting the degradation of the antiviral host protein APOBEC3G. APOBEC3G deaminates cytosine residues in the HIV genome, causing mutations that inactivate the viral genome. Analysis suggested that DNA damage created while repairing these mutations induces the DNA damage response and hence the expression of NKG2D ligands on infected cells (83). HIV Vif partly counteracts this effect by decreasing the amounts of APOBEC3G in infected cells.
How the DNA damage response regulates the expression of NKG2D ligands remains poorly understood. An important issue is to define the stage of biogenesis of NKG2D ligands that is regulated by the DNA damage response. Treating cell lines with agents that activate the DNA damage response induces transcripts for NKG2D ligands, but evidence suggests that much of the regulation is post-transcriptional. Thus, the results of nuclear run-on experiments showed that in cells treated with agents that induce the DNA damage response, there was no increase in the rate of Raet1 gene transcription. However, analysis showed that the rate of degradation of preexisting Raet1 transcripts is significantly inhibited in cells with an activated DNA damage response (B. Hsiung & D.H. Raulet, unpublished data). Therefore, a principal effect of the DNA damage response is to stabilize the normally labile transcripts encoding RAE-1 proteins, and possibly other NKG2D ligands, instead of to induce transcription. This distinction is important because it implies that the DNA damage response by itself does not induce NKG2D ligands in cells where transcription of NKG2D ligands is not enabled by another mechanism.
The induction of NKG2D ligands in cells afflicted with DNA damage was impaired in cells with deficient function of signaling molecules TBK1 and IRF3 (A.R. Lam, N. Le Bert, S.S.W. Ho, Y.J. Shen, L.F.M. Tang, G.M. Xiong, J.L. Croxford, M.F. Pan, C.W. Huang, C.X. Koo, K.J. Ishii, S. Akira, D.H. Raulet & S. Gasser, unpublished data), which mediate signaling downstream of cytoplasmic sensors for DNA and RNA, as well as TLRs (46). Lam and colleagues (unpublished) propose that DNA damage results in the accumulation of cytosolic DNA, activating the DNA sensor and thereby inducing NKG2D ligands.
microRNAs
Several reports have provided evidence that certain NKG2D ligand genes are regulated by microRNAs, which are noncoding RNAs that bind the 3′-untranslated regions of target genes and induce mRNA degradation or impair translation. The initial relevant findings were that the human cytomegalovirus (84) as well as other viruses (85, 86) encode microRNAs that downregulate the expression of MICA, MICB, or ULBP3. Presumably, these microRNAs aid in virus replication by reducing the susceptibility of infected cells to NK attack. However, it was subsequently shown that certain cellular microRNAs also target MICA and MICB transcripts. Among those initially identified were miR-17-5p, miR-20a, miR-93, miR-106b, miR-373, and miR-520 (87). Subsequent studies identified additional microRNAs that target NKG2D ligands, including miR-20a, miR-34a, and miR-34c (74, 88).
The cellular processes that regulate microRNA-mediated suppression of NKG2D ligands remain unclear. Originally, Stern-Ginossar et al. (87) proposed that the micro-RNAs are expressed constitutively and serve to suppress basal NKG2D ligand expression; transcriptional induction of NKG2D ligand genes might then overcome this buffering effect and induce cell surface expression of NKG2D ligands.
However, the expression of several micro-RNAs that target NKG2D ligands is regulated by immune stimuli or activated p53, suggesting a regulatory role for the microRNAs (74). The logic of the regulation varies depending on the microRNA. Lipopolysaccharide activation decreased the expression of miR-17-5, miR-20a, and miR-93, which were shown to target MICA mRNA in human macrophages (88). These data suggested that suppression of these microRNAs is one mechanism by which NKG2D ligands are induced by infections or inflammatory signals.
In contrast, miR-520b, which inhibits MICA expression by direct as well as indirect mechanisms, is upregulated by IFN-γ (89). This finding is consistent with an earlier report that IFN-γ signaling can downregulate certain NKG2D ligands (90). These researchers (90) proposed that IFN-γ's effect may be part of a switching mechanism that acts during the progression of an immune response to suppress NK susceptibility while at the same time promoting sensitivity to cytotoxic T cells.
Yet another type of microRNA regulation is suggested by evidence that miR-34a and miR-34c, which are upregulated in response to p53 activation, interact with the 3′-untranslated regions of ULBP2 mRNA and suppress ULBP2 expression (74). Accordingly, induction of p53 activation with the drug Nutlin-3a caused a reduction in ULBP2 expression. Such a mechanism is predicted to lead to higher expression of ULBP2 in cells lacking p53, and thus this mechanism may serve to increase the efficiency of NK-mediated surveillance of such p53-deficient tumors. However, these results seem to be at odds with the finding, mentioned earlier in this review, that p53 enhances transcription of ULBP1 and 2, and that p53 activation has the effect of increasing ULBP2 expression rather than decreasing it (73). In mouse studies, effects of p53 on expression of NKG2D ligands have not been detected (70; A. Iannello & D.H. Raulet, unpublished data). Further studies are needed to understand the roles of p53 in regulating NKG2D ligands.
OTHER REGULATORY PATHWAYS THAT IMPACT LIGAND mRNA ABUNDANCE
In addition to the aforementioned transcriptional regulatory events, some studies have documented pathways that regulate NKG2D ligands that may have a transcriptional component, although this has not been documented directly. Four examples follow.
Toll-Like Receptors
A connection between innate immune receptor engagement and NKG2D ligand expression was indicated by evidence that various agonists for TLRs induced Raet1 transcripts and cell surface RAE-1 protein in cultured macrophages (91). In other studies, induction of cell surface expression of RAE-1 protein was not observed, despite clear induction ofRaet1 mRNA (A.M. Jamieson & D.H. Raulet, unpublished data). Not yet determined is whether induction of Raet1 transcripts reflects transcriptional induction or post-transcriptional events such as stabilization of the mRNA. The induction of RAE-1 by TLR ligands was abrogated in mice lacking MYD88, a major signaling adapter for TLRs (91).
PI3K
A study of how mouse cytomegalovirus infections induce expression of NKG2D ligands uncovered a role of PI3K in the process (92). Infections of mouse cells including fibroblasts or macrophages with mouse cytomegalovirus resulted in a sharp increase in mRNAs encoding several ligands, but the virus counters this effect by directing the synthesis of several viral proteins that prevent NKG2D ligand surface expression (93,94). By using a virus containing a deletion of a viral gene necessary for suppressing RAE-1 expression, investigators showed that the induction of RAE-1 expression in infected cells was associated with the induction of PI3K activity and was blocked by inhibitors of PI3K (92). Various PI3K isoforms have been identified, and the studies implicated the p110α subunit in the process of ligand induction. That isoform is also frequently mutated in cancer cells, and experiments confirmed that inhibitors of the p110α isoform also cause a significant reduction in the expression of RAE-1 in tumor cell lines (92). Therefore, PI3K function was linked to RAE-1 expression in both virus-infected cells and tumor cells. Inhibiting PI3K caused a significant reduction in Raet1 mRNA but an even larger reduction in RAE-1 protein expression at the cell surface (92), suggesting that it may act at multiple steps in RAE-1 biogenesis, including potentially transcriptional and post-transcriptional stages. A study examining the role of activated RAS in RAE-1 induction also implicated PI3K as a downstream mediator of the effect and provided evidence that the regulation occurs in part at the level of translation of Raet1 mRNAs (95).
Viral or Cellular Oncogenes
A report showed that mouse cell lines or primary cells transformed with the E1A oncogene of Adenovirus serotype 5 contained larger amounts ofRaet1 mRNAs and of RAE-1, but not of MULT1, on the cell surface (96). It remains unclear whether ligand induction resulted from a direct effect of E1A on the Raet1 gene or mRNA or, alternatively, was an indirect consequence of E1A-dependent transformation of the cells.
Another study examined RAE-1ε expression on B cells in Eμ-Myc transgenic mice, a mouse model of B lymphoma (97). In these mice, MYC is expressed in all B cells, and virtually all the mice eventually develop B lymphomas. It was reported that RAE-1ε is often expressed on the B lymphomas that arise in Eμ-Myc transgenic mice. ChIP analysis indicated that c-MYC binds to a site within the promoter of the Raet1e gene. Interestingly, however, although MYC expression occurs in all B cells inEμ-Myc mice, RAE-1 expression was restricted to lymphomas that were clonal with respect to the Ig gene rearrangements they contained. This finding suggested that RAE-1 expression occurs as tumors grow out and not before. It is difficult to ascertain whether the effect of MYC expression in this system is to directly transactivate the Raet1e gene or is instead indirect. One possibility is that enforced MYC expression promotes the formation of B lymphomas, which express RAE-1ε for other reasons. Notably, a separate study showed that the MYC binding site in theRaet1e promoter was not necessary for transcriptional activation of theRaet1e gene in proliferating cells or even in cells transfected with a c-MYC expression vector, supporting an indirect role (68). One function of c-MYC is to transactivate E2f genes, so an interesting possibility is that MYC acts in part by inducing the expression of activating E2F transcription factors, which act directly on the Raet1e promoter.
As noted in the previous section, RAS activation is also associated with induction of NKG2D ligands, specifically RAE-1α and RAE-1β in mouse cells and ULBP1–3 in human cells (95). Induction of NKG2D ligands by activated RAS depended on downstream pathways, including PI3K, MAPK/MEK, and RAF, but did not depend on the DNA damage response. RAS activation was associated with a modest increase in Raet1 mRNA, but accompanying evidence suggested that regulation may also be exerted by increasing the efficiency of translation of Raet1 mRNAs (95).
Type I and Type II Interferons
In one study, human dendritic cells and tumor cells treated with IFN-α displayed greater amounts of cell surface MICA (98). Treatment of cells with IFN-α increased MICA promoter activity, as tested using reporter constructs (99). In contrast, IFN-α and IFN-γ inhibited the expression of the mouse NKG2D ligand H60 on sarcoma cell lines, and IFN-γ inhibited MICA expression in human melanoma cells. In contrast, no effects of IFN on ligand expression were discerned in analyses of mouse fibroblast, carcinoma, or lymphoma cell lines (70).
REGULATION AT THE PROTEIN LEVEL
Protein Stabilization
Numerous instances have been reported in which NKG2D ligand proteins are targeted by virally encoded proteins in a manner that prevents their expression on the cell surface and presumably enables viruses to evade NKG2D-dependent detection by NK cells and T cells (100). But host cells also employ post-translational mechanisms to regulate NKG2D ligand expression, presumably in the service of host defense.
As one example, a stress-induced mechanism stabilizes the murine MULT1 protein by preventing degradation (101). Whereas some NKG2D ligands are GPI-linked proteins, MULT1 is a membrane-spanning type 1 glycoprotein (Figure 1) (102). Expression vectors directing the synthesis of wild-type MULT1 resulted in poor surface expression in several unstressed cell lines, but deleting the long cytoplasmic tail of MULT1 or mutating the lysines in the cytoplasmic tail to arginine residues resulted in a high level of MULT1 expression on the cell surface (101). The necessity for lysine residues suggested the role of protein ubiquitination in the degradation of MULT1 in unstressed cells, a conclusion supported by evidence that MULT1 is polyubiquitinated and degraded in unstressed cells.
In cells subjected to heat shock or UV irradiation, in contrast, the wild-type protein was less ubiquitinated and was induced on the cell surface, and in parallel, the half-life of the protein was extended (101). Genotoxic agents other than UV irradiation failed to induce MULT1 stabilization, consistent with additional evidence that the effect was independent of the DNA damage response. A subsequent study demonstrated that MULT1 degradation could be mediated by MARCH4 and MARCH9, members of the MARCH family of transmembrane E3 ubiquitin ligases (103). However, other E3 ubiquitin ligases presumably participate in this process, given that depletion of MARCH4 and MARCH9 failed to prevent rapid MULT1 degradation in unstressed cells (103).
Several other NKG2D ligands are also transmembrane proteins, including H60a and H60b in mice (30) and MICA, MICB, ULBP4/RAET1E, and ULBP5 in humans (65, 104, 105). A study of human tumor cells suggested that MICA may be sequestered inside of tumor cells, perhaps by a similar or related mechanism (106). These findings therefore suggest that, in addition to being regulated at the transcriptional and mRNA levels, NKG2D ligands can be regulated by stress pathways at the post-translational level.
Shedding, Secretion, and Exosomal Excretion of NKG2D Ligands
Another level of potential regulation is represented by instances in which ligands are cleaved from the cell surface, excreted in vesicles such as exosomes, or secreted from the cell. Such events may exert effects by decreasing expression of NKG2D ligands by the affected cell, and they may also exert distinct regulatory effects because of functional activities mediated by the cleaved, excreted, or secreted ligands. Furthermore, it is plausible that different and even opposing activities could be mediated by a cleaved form of a ligand as opposed to a membrane vesicle– bound form. Some confusion may attach to the literature because in many studies the shed form was detected by ELISA and investigators did not clarify whether the detected material corresponded to the cleaved form, the vesicle form, or both. Although many uncertainties remain concerning how cleavage or excretion of NKG2D ligands is regulated and how these cell-free forms of NKG2D ligands regulate immune responses, the present understanding of these processes is summarized here.
Initially, soluble MICA was detected in the sera of patients with lung, breast, and gastrointestinal malignancies (107, 108), and cell-free forms of MICA and MICB were subsequently detected in patients with a variety of cancers (109, 110, 111, 112, 113, 114, 115). Soluble forms of ULBP1, ULBP2, and ULBP3 have also been reported in cancer patients or cancer cell culture supernatants (116, 117, 118, 119, 120). In the mouse, both RAE-1 (7) and MULT1 (W. Deng & D.H. Raulet, unpublished data) ligands have been detected in soluble form. The presence of soluble ligands in the sera of cancer patients may in some cases serve as indicators of prognosis. For example, the presence of soluble ULBP2 in melanoma patients was correlated with poor prognosis (119).
Shedding of Ligands as a Result of Proteolytic Cleavage
Cleavage of the extracellular domain by matrix metalloproteases (MMPs) is responsible for shedding of several human ligands, including MICA (108, 121), MICB (115), and ULBP2 (116). Hence, both transmembrane (MICA and MICB) and predominantly GPI-linked (ULBP2) NKG2D ligands may be shed as result of protein cleavage. In the mouse, the MULT1 ligand, at least, is also cleaved from the cell surface by MMPs (W. Deng & D.H. Raulet, unpublished data). Among the MMPs, members of the ADAM (a disintegrin and metalloproteinase) family, including ADAM10 and ADAM17, as well as MMP14, have been specifically implicated in ligand cleavage (122, 123, 124). In the case of MICA, cleavage requires the participation of a membrane-associated disulphide isomerase endoplasmic reticulum protein 5 (ERP5) (121), which is hypothesized to cause conformational changes in the extracellular stem of the protein that favor proteolytic cleavage. ERP5 may be upregulated in certain cancers, such as multiple myelomas, increasing the rate of ligand cleavage (125). Interestingly, in cleavage dependent on ERP5, MMP inhibitors did not prevent MICA cleavage, suggesting the participation of other types of proteases (121).
Membrane Vesicle–Mediated Excretion of NKG2D Ligands
Although soluble forms of ligands were initially assumed to be generated exclusively by proteolytic cleavage, evidence has accumulated that ligands are sometimes excreted in vesicles such as exosomes. Exosomes are small vesicles formed in multivesicular endosomes and released by fusion with the plasma membrane. In one study, ULBP3 from tumor cell culture supernatants was associated with exosomes, whereas ULBP2 was primarily in a cleaved form (120). Another report demonstrated that the form of MICA encoded by the most common MICA allele is primarily excreted in exosomes, whereas MICA encoded by other alleles is released primarily by membrane cleavage (126). There are indications that some ligands can be released by both proteolytic cleavage and exosome excretion (120; W. Deng & D.H. Raulet, unpublished data), suggesting that care must be taken to characterize the mode of ligand excretion that occurs in different circumstances.
Secretion of NKG2D Ligands
In addition to shedding and exosomal excretion, NKG2D ligand transcripts can in some cases, through alternative processing, generate a secreted form of the ligands. ULBP4 (RAET1E) and ULBP5 (RAET1G) can be alternatively spliced to generate soluble forms (RAET1E2 and RAET1G2, respectively) (118, 127). RAET1E2, at least, can inhibit NK cell–target cell interactions in vitro, resulting in reduced cytotoxicity (118).
Biological Effects of Ligand Shedding and Secretion
Shedding of NKG2D ligands can result in sharply lower cell surface levels on the affected cells, such as tumor cells, and these lower ligand levels are associated with a reduced susceptibility of tumor cells to NK cells and T cells (116). It is likely that by reducing ligand densities at the cell surface, proteolytic shedding has a substantial role in reducing the capacity of affected cells to be killed by NK cells or to stimulate them to secrete cytokines. Whether exosome shedding influences the concentration of cell surface NKG2D ligands has not been determined.
Another potential consequence of ligand excretion is that shed ligands may interact with NKG2D on the surface of NK cells and T cells, even those some distance from the source (128). Soluble ligands, at least those with high affinity, could thus block the receptors on the cells in a manner that inhibits their interactions with target cells. Alternatively, if the soluble ligands can transmit signals through NKG2D, these interactions have the potential to either activate or desensitize the NK cells or T cells.
Numerous reports have suggested that binding to cells of excreted NKG2D ligands present in patient samples or cell culture supernatants can cause downregulation of NKG2D from the cell surface and that receptor downregulation is associated with desensitization of the cells (107, 120). In some cases, for example, cancer patients with elevated soluble MICA in their serum exhibited strongly reduced NKG2D staining of their peripheral blood CD8+ T cells (107). In a mouse MICB transgenic prostate tumor model, shed MICB was reported to promote tumor formation, presumably by inhibiting NKG2D-dependent tumor surveillance mechanisms (129). However, numerous conflicting reports have failed to confirm receptor downregulation in the presence of soluble NKG2D ligands, even with samples containing the same soluble ligand (115, 116,130). Furthermore, NKG2D was not appreciably downregulated on cells in patients with rheumatoid arthritis or celiac disease (131, 132) or in mice with a ubiquitously expressed MICA transgene (130), all of which have high levels of serum MICA. Several explanations can be considered for the conflicting results. One is that the outcome depends on the nature of the excreted ligands; in contrast to monovalent ligands, multivalent ligands, such as found in exosomes, are predicted to cross-link receptors and convey signals that are generally necessary for receptor downregulation (120). Another possible explanation is that receptor downregulation observed in some reports is caused not by soluble ligands but by other components that may be present in patient samples, such as TGF-β (133, 134). Finally, in some studies, reduced receptor staining after incubation with soluble ligands may reflect blockade of the epitope recognized by NKG2D-specific antibodies, rather than receptor downregulation.
Regulation Related to Ligand Shedding and Excretion
Shedding and exosome excretion of NKG2D ligands are likely regulated processes, but little is known at present concerning such regulation. MMPs are known to be upregulated on many cancer cells in association with increased tumor invasiveness (135, 136), so it will be interesting to investigate whether ligand shedding is correlated with the invasive stage of tumorigenesis. In addition, ERP5, which promotes MICA shedding, is associated with metastasis (137) and is upregulated in certain cancers, such as multiple myelomas, increasing the rate of ligand cleavage (125). Furthermore, it has been reported that MICA is subject to palmitoylation and that this modification causes the protein to be recruited to membrane microdomains and promotes shedding (138). How this process may be regulated is unknown.
External agents may also modulate ligand cleavage. One report showed that IFN-γ induces increased amounts of MMP, resulting in more cleavage and therefore lower surface expression of MICA in tumor cell lines (99). Another report suggested that the MMP-dependent shedding of MICA from cell lines was induced by hypoxic conditions and was inhibited by nitric oxide (139). In addition, it was reported that the surface expression of ERP5 in a cell line was inhibited by histamine, but this inhibition was accompanied by a decrease in cell surface MICA expression rather than by the increase that might be expected if proteolytic cleavage was blocked (140).
The regulation of exosome excretion of NKG2D ligands is even less well understood than the regulation of ligand cleavage. However, one report provided evidence that oxidative and thermal stress modestly boost the rate of exosome excretion of MICA and MICB (141).
As noted above, extracellular forms of NKG2D ligands can inhibit the cell surface expression and functional activities of NK cells and T cells. Interestingly, reports suggest that exposure of the lymphocytes to cytokines, including IL-15 and IL-12, can increase NKG2D expression and restore the functionality of NK cells (142). Hence, downregulation of NKG2D expression by soluble ligands probably varies depending not only on the form of the soluble ligands but also on the cytokine milieu.
SYNTHESIS: COORDINATED REGULATION BY MULTIPLE DISEASE-ASSOCIATED SIGNALS
The previous discussion makes clear that NKG2D ligands are regulated by numerous pathways and signals. Here, we attempt to synthesize this information and propose schemes in which the various changes accompanying transformation, infection, and other pathological states work together to induce or amplify NKG2D ligand expression. Inherent in this analysis is the notion that regulation acts at different levels of biogenesis (Figure 2). Clearly, for example, a mechanism that stabilizes ligand transcripts will be ineffective at inducing NKG2D ligands in cells that do not transcribe the ligand genes. Regulation at multiple levels of biogenesis may also result in a potentially wide range of ligand expression on cells, depending on which combination of pathways is active in a given cell. Finally, some mechanisms are likely to work in opposition, as, for example, a cell that produces large amounts of a ligand protein but is also hyperactive at shedding the ligand from the cell surface. This discussion is designed to provide ideas for future studies, but it is also necessarily speculative because the specific roles of different mechanisms working in concert have not yet been systematically evaluated experimentally.
Synthesis: Cancer
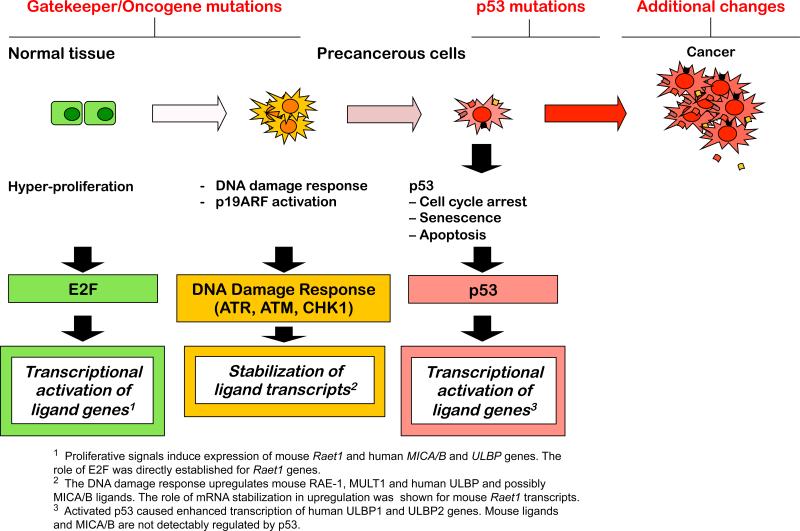
In the case of cancer, numerous independent pathways that are activated in cancer cells are likely to cooperate in the induction of NKG2D ligands. In Figure 3, some of these have been arranged in a scheme that emphasizes the relationship of these pathways and signals to the events that are thought to accompany the onset of tumorigenesis.
Figure 3.
Regulation of NKG2D ligands in the context of the multistep process of tumorigenesis. The figure maps various regulators of NKG2D ligand expression onto a scheme depicting the multistep process of tumorigenesis. Tumorigenesis may initiate with loss of gatekeeper tumor suppressors and activation of oncogenes, resulting in hyperproliferation. Strong proliferative signals result in DNA damage, and consequent activation of ATM, ATR and the DNA damage response, as well as p19ARF activation. Both of these pathways activate p53, which imposes a major barrier to transformation. An activated DNA damage response and activated p53 each impart blocks in cell cycle progression, and p53 can also trigger apoptosis as well as senescence. Loss of p53 as a result of mutation enables tumor progression. See Figure 1 for more detailed information concerning the specific ligands and the mediators and pathways that regulate them.
An early event in tumorigenesis is a hyperproliferative state that occurs in association with the activation of oncogenes or the loss of gatekeeper tumor suppressors. Cell cycle entry typically requires active E2F transcription factors, which also transcriptionally activate various NKG2D ligands, including RAE-1 in mouse cells. Proliferation is also associated with induction of MICA, MICB, and several ULBPs in human cells (66, 68). These findings suggest a direct linkage between the proliferative state of tumor cells and transcription of NKG2D ligands. PI3K, by enhancing proliferation, may also enhance transcription of NKG2D ligands (92, 95). Additionally, however, several oncogenes have been proposed to transcriptionally activate NKG2D ligand genes.
The hyperproliferative state often results in activation of p19ARF and/or the DNA damage response, both of which activate the p53 tumor suppressor, which serves as a key barrier to tumorigenesis. Activated p53 can enhance transcription of certain NKG2D ligands, such as ULBP1 and 2 (72, 73), although upregulation of mouse NKG2D ligands has not been linked to p53. Finally, the heat shock stress response, which is reportedly often activated in tumor cells (44), has been implicated in transcription of MICA and MICB ligands in human cells.
Induced transcription of NKG2D ligand genes may suffice to induce cell surface expression of the ligands in some contexts, but in others the relatively rapid degradation of the ligand transcripts appears to limit the amount of ligand mRNA found in the cells and therefore the amount of the corresponding protein displayed on the cell surface. The early activation of the DNA damage response, which occurs often in tumorigenesis (34,35, 71), activates IRF3 and TBK1 via the action of cytosolic DNA sensors, ultimately inducing conditions that stabilize the ligand transcripts and therefore induce greater expression of ligands (Figure 3). Accordingly, expression of cell surface ligands in tumor cell lines was depressed when the DNA damage response was inhibited with drugs or shRNAs that deplete critical mediators in the pathway (70, 79).
Regulation may also occur at the level of ligand mRNA translation. PI3K activation, which supports expression of NKG2D ligands in tumor cell lines and cell lines with activated RAS, regulates translation of numerous genes, for example, and may also play a role in translational regulation of RAE-1 family NKG2D ligands (95).
NKG2D ligands are also regulated at the protein level. At least one NKG2D ligand, MULT1 in mice, is upregulated in heat-shocked cells by a mechanism that prevents ubiquitin-dependent degradation of the protein (101). Whether this pathway plays a role in tumor cells remains to be determined. Events that occur in tumor cells are also likely to impact shedding and possibly exosome excretion of NKG2D ligands, although in this case the effect may be to reduce the amount of ligand displayed on the cells. For example, expression of MMPs and the ERP5 protein occurs in invasive stages of tumorigenesis (135, 136, 137), and these molecules are expected to facilitate shedding and therefore decrease the amount of NKG2D ligands available at the cell surface.
Synthesis: Infections
Ligand induction in infected cells likely also involves the coordinated effects of several pathways. Little is known concerning transcriptional induction of NKG2D ligands in infected cells. However, induction of PI3K, as occurs in certain viral infections (55, 92), may support activation of E2F and NF-κB transcription factors, both of which have been implicated in transcriptional regulation of NKG2D ligands (68, 75, 76). The heat shock pathway, shown to play a role in the induction of MICA and MICB at the transcriptional level, is also induced in certain infections (53, 75,76). Engagement of TLRs on macrophages induces transcripts for NKG2D ligands, especially Raet1 family transcripts in mice (91). Induction is dependent on MYD88, which acts upstream of NF-κB. Indeed, many other innate receptors also induce NF-κB activation, suggesting another possible mechanism whereby infections induce transcription of NKG2D ligands. However, this connection has not been tested directly, and additional studies will be necessary to determine whether NF-κB plays a role in ligand induction downstream of innate receptor stimulation.
A second signaling module downstream of TLRs acts via the TRIF signaling molecule, resulting in activation of TBK1 and IRF3. As noted in an earlier section (The DNA Damage Response), TBK1 and IRF3 also play important roles in induction of NKG2D ligands in cells subjected to genotoxic stress. In these cells, Raet1 transcripts were induced, but analysis showed that this was primarily the result of stabilization of Raet1transcripts, rather than increases in Raet1 transcription. Taken together, these findings suggest that the TBK1/IRF3 pathway plays a role primarily in stabilizing ligand transcripts as opposed to inducing ligand gene transcription. Presumably, IRF3 induces mediators that interact directly with NKG2D ligand transcripts, rather than doing so itself.
One implication of these findings is that innate sensors other than TLRs that act via TBK1 and IRF3 may also stabilize transcripts for NKG2D ligands, leading to greater cell surface expression of the corresponding proteins. These other sensors include cytosolic RNA sensors RIG-I and MDA-5 and various cytosolic DNA sensors (46). In this way, various different sensors of pathogen-associated molecular patterns may act to induce NKG2D ligands in infected cells that express those sensors. In certain other cases, such as in HIV (51) or Abelson virus–infected cells (50), events occurring during the infection induce the DNA damage response, also resulting in increased cell surface display of NKG2D ligands.
Synthesis: Injury
Induction of NKG2D ligands may also occur under conditions of injury. Little is known concerning the underlying mechanisms, but some of the same pathways described above may play a role. Excessive cell proliferation in injured tissue may act to induce ligand gene transcription via E2F transcription factors (68). p53 activation has been documented in fibrotic livers and healing cutaneous wounds (56, 57), and it may also enhance transcription of NKG2D ligand genes (72, 73) in the context of injury. The DNA damage response may also be activated in injured tissue in some contexts (57), resulting in stabilization of NKG2D ligand transcripts.
Induction of NKG2D ligands also occurs in other pathological states such as inflammatory diseases, but little is known concerning mechanisms for ligand induction in these syndromes. In one example, ligand induction was documented in atherosclerotic plaques and in inflamed liver in animals with metabolic syndrome. Evidence suggested a role of abnormal metabolites that accumulate under these conditions, such as Ox-LDL and AGE (61). Both of these metabolites reportedly interact with immune receptors, TLRs, and CD36 in the case of Ox-LDL (62) and RAGE and other receptors in the case of AGE (63, 64). In vitro studies suggested that both metabolites induce expression of NKG2D ligands in cultured macrophages (61). TLRs may function as already discussed, by activating TBK1/IRF3 and NF-κB, stabilizing ligand transcripts, and supporting ligand transcription, respectively. How AGE enhances NKG2D ligand expression remains to be determined. Given the potential role of these specialized metabolites in atherosclerosis and metabolic syndrome, it will be interesting to determine whether other abnormal metabolites or, alternatively, environmental stimuli play a role in ligand induction that underlies NKG2D activation in various other inflammatory and autoimmune diseases such as colitis and diabetes.
CONCLUSIONS
The available data suggest that NKG2D ligands are regulated at several levels of biogenesis by various types of signals that are elaborated in distressed, infected, or transformed cells. Regulation has been discerned at the transcriptional, post-transcriptional, translational, and post-translational levels (Figure 2). Many basic questions remain to be explored regarding regulation of NKG2D ligand expression, particularly at the transcriptional and post-translational levels. The molecular mechanisms of secretion, shedding, and exosome excretion and their regulation and consequences are currently poorly understood. Furthermore, there are several examples in which a given stimulus is effective at inducing an NKG2D ligand in one cell type but not another, such as the induction of RAE-1 on proliferating fibroblasts but much less so on proliferating mouse T cells. Examining these differences will be essential for a comprehensive understanding of NKG2D function.
NKG2D Ligand Regulation: A Bar Code for Defining Unhealthy Cells?
The signals that regulate the various steps of ligand biogenesis range from those induced by stress pathways to those that are commonly elaborated in at least some normal cells, such as signals associated with cell proliferation. An attractive idea is that optimal ligand induction requires a collection of different signals that regulate different stages of ligand biogenesis, acting contemporaneously. Each set of signals that are associated with a given disease state, and act together to induce ligands, could be considered a bar code that the cell uses to define various unhealthy states. Cancer cells, or at least some subtypes of cancer cells, might display a unique bar code consisting of several elements, including proliferation to induce Raet1 transcription, the DNA damage response to stabilize the transcripts, and a stress response to stabilize the corresponding ligand protein. Infected cells might rely on a different bar code, such as innate receptor stimulation (via NF-κB) or PI3K activation to enhance transcription, TBK1/IRF3 signals to stabilize the transcripts, etc. Such a scheme includes certain fail-safe mechanisms, in that activation of one relevant signal in otherwise normal cells may not be enough to trigger abundant ligand expression. A signal that stabilizes a ligand transcript or protein will have no effect if signals for transcriptional activation are absent. The requirement for a coordination of different signals for ligand induction would also provide the system with the specificity necessary to discriminate normal cells from unhealthy ones.
Given that numerous unhealthy states are accompanied by the induction of NKG2D ligands, a variety of different effective bar codes may be necessary to accomplish this task. A given step in ligand biosynthesis may therefore be regulated independently by different signals, associated with different disease states. An alternative strategy is to provide for a specialization of NKG2D ligands, such that different sets of signals preferentially regulate different ligands. There are several indications of such specialization, including the findings that MICA/B ligands but not most other NKG2D ligands are transcriptionally regulated by heat shock, that RAE-1 ligands but not other mouse ligands are regulated by proliferative signals, and that MULT1, but not other murine NKG2D ligands, is regulated at the protein stabilization level by heat shock.
Or, Do the Various Regulatory Steps Constitute a System to Quantify the Threat Level?
The bar code concept is attractive in some respects, but a caveat is that it tends to imply that ligand expression is an on/off state. The published data provide many examples of partial effects in which activating a pathway boosts ligand expression above a significant, basal level. In light of these data, we might consider that the multiplicity of functions involved in regulating NKG2D ligands serves to quantitatively define a threat level, as used for warnings of possible terrorism. Activation of some of the relevant pathways may induce modest expression of ligands, whereas high expression may require participation of several signals, providing evidence of multiple dysregulated pathways in the cell. Increased amounts of NKG2D ligands would increase the sensitivity of the cells to elimination by immune cells (17). NKG2D ligand regulation might incorporate elements of both the bar code and threat level concepts.
Clearly much work remains to discern the logic whereby different sets of signals regulate distinct ligands. The number of stress stimuli that induce NKG2D ligands may also continue to grow with additional research. A better and more comprehensive understanding of the regulation of NKG2D ligand expression will not only aid in understanding the biological roles of NKG2D and NK cells, but will also provide insights that may inform efforts to target the NKG2D system therapeutically. The use of stress inducers of NKG2D ligand expression may provide new methods for enhancing NKG2D-mediated immune responses in vivo. Conversely, it may be possible to selectively target specific inducers of NKG2D ligands relevant in inflammatory disease, while avoiding those relevant for host defense.
ACKNOWLEDGMENTS
We thank the members of our laboratories for contributing to the original studies reviewed here and our colleagues in the field for their important contributions. We thank Dr. Roland Strong for providing the image that is the basis of Figure 1a. Work in the Raulet laboratory is supported by the National Institutes of Health (RO1AI039642, RO1CA093678) and the Prostate Cancer Foundation. The Gasser laboratory is supported by the Biomedical Research Council grant 07/1/21/19/513 and the National Research Foundation grant HUJ-CREATE—Cellular and Molecular Mechanisms of Inflammation. Weiwen Deng was the recipient of a Cancer Research Institute Postdoctoral Fellowship. Heiyoun Jung was the recipient of a predoctoral scholarship from the Samsung Scholarship Foundation, and Benjamin Gowen was the recipient of a National Science Foundation Predoctoral Fellowship.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1•.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2•.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 2003;3:781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 3•.Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat. Rev. Immunol. 2009;9:568–80. doi: 10.1038/nri2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•.Diefenbach A, Raulet DH. Strategies for target cell recognition by natural killer cells. Immunol. Rev. 2001;181:170–84. doi: 10.1034/j.1600-065x.2001.1810114.x. [DOI] [PubMed] [Google Scholar]
- 5•.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today. 1990;11:237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 6•.Houchins JP, Yabe T, McSherry C, Bach FH. DNA sequence analysis of NKG2, a family of related cDNA clones encoding type II integral membrane proteins on human natural killer cells. J. Exp. Med. 1991;173:1017–20. doi: 10.1084/jem.173.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol. Rev. 2010;235:267–85. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Wu J, Song Y, Bakker AB, Bauer S, Spies T, et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–32. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 9•.Oppenheim DE, Roberts SJ, Clarke SL, Filler R, Lewis JM, et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat. Immunol. 2005;6:928–37. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
- 10•.Champsaur M, Beilke JN, Ogasawara K, Koszinowski UH, Jonjic S, Lanier LL. Intact NKG2D-independent function of NK cells chronically stimulated with the NKG2D ligand Rae-1. J. Immunol. 2010;185:157–65. doi: 10.4049/jimmunol.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107:159–66. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Groh V, Smythe K, Dai Z, Spies T. Fas ligand-mediated paracrine T cell regulation by the receptor NKG2D in tumor immunity. Nat. Immunol. 2006;7:755–62. doi: 10.1038/ni1350. [DOI] [PubMed] [Google Scholar]
- 13•.Jamieson AM, Diefenbach A, McMahon CW, Xiong N, Carlyle JR, Raulet DH. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17:19–29. doi: 10.1016/s1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 14•.Whang MI, Guerra N, Raulet DH. Costimulation of dendritic epidermal γδ T cells by a new NKG2D ligand expressed specifically in the skin. J. Immunol. 2009;182:4557–64. doi: 10.4049/jimmunol.0802439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, et al. Regulation of cutaneous malignancy by γδ T cells. Science. 2001;294:605–9. [PubMed] [Google Scholar]
- 16•.Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8 αβ T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat. Immunol. 2001;2:255–60. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 17•.Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–71. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–66. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 19•.Regunathan J, Chen Y, Wang D, Malarkannan S. NKG2D receptor-mediated NK cell function is regulated by inhibitory Ly49 receptors. Blood. 2005;105:233–40. doi: 10.1182/blood-2004-03-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Eagle RA, Trowsdale J. Promiscuity and the single receptor: NKG2D. Nat. Rev. Immunol. 2007;7:737–44. doi: 10.1038/nri2144. [DOI] [PubMed] [Google Scholar]
- 21•.Li P, McDermott G, Strong RK. Crystal structures of RAE-1β and its complex with the activating immunoreceptor NKG2D. Immunity. 2002;16:77–86. doi: 10.1016/s1074-7613(02)00258-3. [DOI] [PubMed] [Google Scholar]
- 22•.Fernandez-Messina L, Ashiru O, Aguera-Gonzalez S, Reyburn HT, Vales-Gomez M. The human NKG2D ligand ULBP2 can be expressed at the cell surface with or without a GPI anchor and both forms can activate NK cells. J. Cell Sci. 2011;124:321–27. doi: 10.1242/jcs.076042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Carayannopoulos LN, Naidenko OV, Kinder J, Ho EL, Fremont DH, Yokoyama WM. Ligands for murine NKG2D display heterogeneous binding behavior. Eur. J. Immunol. 2002;32:597–605. doi: 10.1002/1521-4141(200203)32:3<597::aid-immu597>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- 24•.O'Callaghan CA, Cerwenka A, Willcox BE, Lanier LL, Bjorkman PJ. Molecular competition for NKG2D: H60 and RAE1 compete unequally for NKG2D with dominance of H60. Immunity. 2001;15:201–11. doi: 10.1016/s1074-7613(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 25•.Takada A, Yoshida S, Kajikawa M, Miyatake Y, Tomaru U, et al. Two novel NKG2D ligands of the mouse H60 family with differential expression patterns and binding affinities to NKG2D. J. Immunol. 2008;180:1678–85. doi: 10.4049/jimmunol.180.3.1678. [DOI] [PubMed] [Google Scholar]
- 26•.McFarland BJ, Strong RK. Thermodynamic analysis of degenerate recognition by the NKG2D immunoreceptor: not induced fit but rigid adaptation. Immunity. 2003;19:803–12. doi: 10.1016/s1074-7613(03)00320-0. [DOI] [PubMed] [Google Scholar]
- 27•.Carayannopoulos LN, Naidenko OV, Fremont DH, Yokoyama WM. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 2002;169:4079–83. doi: 10.4049/jimmunol.169.8.4079. [DOI] [PubMed] [Google Scholar]
- 28•.Li PW, Morris DL, Willcox BE, Steinle A, Spies T, Strong RK. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat. Immunol. 2001;2:443–51. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 29•.Strong RK, McFarland BJ. NKG2D and related immunoreceptors. Adv. Protein Chem. 2004;68:281–312. doi: 10.1016/S0065-3233(04)68008-9. [DOI] [PubMed] [Google Scholar]
- 30•.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 2000;1:119–26. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 31•.Radaev S, Sun PD. Structure and function of natural killer cell surface receptors. Annu. Rev. Biophys. Biomol. Struct. 2003;32:93–114. doi: 10.1146/annurev.biophys.32.110601.142347. [DOI] [PubMed] [Google Scholar]
- 32•.Kriegeskorte AK, Gebhardt FE, Porcellini S, Schiemann M, Stemberger C, et al. NKG2D-independent suppression of T cell proliferation by H60 and MICA. Proc. Natl. Acad. Sci. USA. 2005;102:11805–10. doi: 10.1073/pnas.0502026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Popa N, Cedile O, Pollet-Villard X, Bagnis C, Durbec P, Boucraut J. RAE-1 is expressed in the adult subventricular zone and controls cell proliferation of neurospheres. Glia. 2011;59:35–44. doi: 10.1002/glia.21074. [DOI] [PubMed] [Google Scholar]
- 34•.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 35•.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 36•.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–94. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 37•.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–47. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 38•.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–33. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–26. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 40•.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 41•.Romagosa C, Simonetti S, Lopez-Vicente L, Mazo A, Lleonart ME, et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30:2087–97. doi: 10.1038/onc.2010.614. [DOI] [PubMed] [Google Scholar]
- 42•.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 43•.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–96. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5:2592–601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- 45•.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 46•.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–84. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 47•.Weitzman MD, Carson CT, Schwartz RA, Lilley CE. Interactions of viruses with the cellular DNA repair machinery. DNA Repair. 2004;3:1165–73. doi: 10.1016/j.dnarep.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 48•.Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 2010;64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- 49•.Nikitin PA, Luftig MA. The DNA damage response in viral-induced cellular transformation. Br. J. Cancer. 2012;106:429–35. doi: 10.1038/bjc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Gourzi P, Leonova T, Papavasiliou FN. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 2006;24:779–86. doi: 10.1016/j.immuni.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 51•.Ward J, Davis Z, DeHart J, Zimmerman E, Bosque A, et al. HIV-1 Vpr triggers natural killer cell-mediated lysis of infected cells through activation of the ATR-mediated DNA damage response. PLoS Pathog. 2009;5:e1000613. doi: 10.1371/journal.ppat.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Jackson AC, Kammouni W, Fernyhough P. Role of oxidative stress in rabies virus infection. Adv. Virus Res. 2011;79:127–38. doi: 10.1016/B978-0-12-387040-7.00008-1. [DOI] [PubMed] [Google Scholar]
- 53•.LaThangue NB, Latchman DS. Nuclear accumulation of a heat-shock 70-like protein during herpes simplex virus replication. Biosci. Rep. 1987;7:475–83. doi: 10.1007/BF01116504. [DOI] [PubMed] [Google Scholar]
- 54•.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 2005;79:6890–99. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Buchkovich NJ, Yu Y, Zampieri CA, Alwine JC. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 2008;6:266–75. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–67. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Jun JI, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat. Cell Biol. 2010;12:676–85. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Radaeva S, Sun R, Jaruga B, Nguyen VT, Tian Z, Gao B. Natural killer cells ameliorate liver fibrosis by killing activated stellate cells in NKG2D-dependent and tumor necrosis factor-related apoptosis-inducing ligand-dependent manners. Gastroenterology. 2006;130:435–52. doi: 10.1053/j.gastro.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 59•.Ludwig J, Viggiano TR, McGill DB, Oh BJ. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin. Proc. 1980;55:434–38. [PubMed] [Google Scholar]
- 60•.Tous M, Ferré N, Camps J, Riu F, Joven J. Feeding apolipoprotein E-knockout mice with cholesterol and fat enriched diets may be a model of non-alcoholic steatohepatitis. Mol. Cell. Biochem. 2005;268:53–58. doi: 10.1007/s11010-005-2997-0. [DOI] [PubMed] [Google Scholar]
- 61•.Xia M, Guerra N, Sukhova GK, Yang K, Miller CK, et al. Immune activation resulting from NKG2D/ligand interaction promotes atherosclerosis. Circulation. 2011;124:2933–43. doi: 10.1161/CIRCULATIONAHA.111.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 2010;11:155–61. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Park L, Raman KG, Lee KJ, Lu Y, Ferran LJ, Jr, et al. Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat. Med. 1998;4:1025–31. doi: 10.1038/2012. [DOI] [PubMed] [Google Scholar]
- 64•.Bucciarelli LG, Wendt T, Qu W, Lu Y, Lalla E, et al. RAGE blockade stabilizes established atherosclerosis in diabetic apolipoprotein E-null mice. Circulation. 2002;106:2827–35. doi: 10.1161/01.cir.0000039325.03698.36. [DOI] [PubMed] [Google Scholar]
- 65•.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl. Acad. Sci. USA. 1996;93:12445–50. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Venkataraman GM, Suciu D, Groh V, Boss JM, Spies T. Promoter region architecture and transcriptional regulation of the genes for the MHC class I-related chain A and B ligands of NKG2D. J. Immunol. 2007;178:961–69. doi: 10.4049/jimmunol.178.2.961. [DOI] [PubMed] [Google Scholar]
- 67•.Eagle RA, Traherne JA, Ashiru O, Wills MR, Trowsdale J. Regulation of NKG2D ligand gene expression. Hum. Immunol. 2006;67:159–69. doi: 10.1016/j.humimm.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 68•.Jung H, Hsiung B, Procyk E, Raulet DH. E2F transcription factors regulate expression of RAE1 ligands for NKG2D, an immune cell receptor implicated in tumor surveillance. J. Exp. Med. 2012;209:2409–22. doi: 10.1084/jem.20120565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Rabinovich BA, Li J, Shannon J, Hurren R, Chalupny J, et al. Activated, but not resting, T cells can be recognized and killed by syngeneic NK cells. J. Immunol. 2003;170:3572–76. doi: 10.4049/jimmunol.170.7.3572. [DOI] [PubMed] [Google Scholar]
- 70•.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–90. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71•.Gasser S, Raulet DH. The DNA damage response arouses the immune system. Cancer Res. 2006;66:3959–62. doi: 10.1158/0008-5472.CAN-05-4603. [DOI] [PubMed] [Google Scholar]
- 72•.Li H, Lakshmikanth T, Garofalo C, Enge M, Spinnler C, et al. Pharmacological activation of p53 triggers anticancer innate immune response through induction of ULBP2. Cell Cycle. 2011;10:3346–58. doi: 10.4161/cc.10.19.17630. [DOI] [PubMed] [Google Scholar]
- 73•.Textor S, Fiegler N, Arnold A, Porgador A, Hofmann TG, Cerwenka A. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 2011;71:5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- 74•.Heinemann A, Zhao F, Pechlivanis S, Eberle J, Steinle A, et al. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012;72:460–71. doi: 10.1158/0008-5472.CAN-11-1977. [DOI] [PubMed] [Google Scholar]
- 75•.Molinero LL, Fuertes MB, Girart MV, Fainboim L, Rabinovich GA, et al. NF-κB regulates expression of the MHC class I-related chain A gene in activated T lymphocytes. J. Immunol. 2004;173:5583–90. doi: 10.4049/jimmunol.173.9.5583. [DOI] [PubMed] [Google Scholar]
- 76•.Lin D, Lavender H, Soilleux EJ, O'Callaghan CA. NF-κB regulates MICA gene transcription in endothelial cell through a genetically inhibitable control site. J. Biol. Chem. 2012;287:4299–310. doi: 10.1074/jbc.M111.282152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77•.López-Soto A, Quiñones-Lombraña A, López-Arbesú R, López-Larrea C, González S. Transcriptional regulation of ULBP1, a human ligand of the NKG2D receptor. J. Biol. Chem. 2006;281:30412–30. doi: 10.1074/jbc.M604868200. [DOI] [PubMed] [Google Scholar]
- 78•.Nausch N, Florin L, Hartenstein B, Angel P, Schorpp-Kistner M, Cerwenka A. Cutting edge: The AP-1 subunit JunB determines NK cell-mediated target cell killing by regulation of the NKG2D-ligand RAE-1ε. J. Immunol. 2006;176:7–11. doi: 10.4049/jimmunol.176.1.7. [DOI] [PubMed] [Google Scholar]
- 79•.Gasser S, Raulet DH. Activation and self-tolerance of natural killer cells. Immunol. Rev. 2006;214:130–42. doi: 10.1111/j.1600-065X.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- 80•.Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK-cell lysis. Blood. 2007;110:606–15. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- 81•.Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113:3503–11. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- 82•.Dai Z, Turtle CJ, Booth GC, Riddell SR, Gooley TA, et al. Normally occurring NKG2D+CD4+ T cells are immunosuppressive and inversely correlated with disease activity in juvenile-onset lupus. J. Exp. Med. 2009;206:793–805. doi: 10.1084/jem.20081648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83•.Norman JM, Mashiba M, McNamara LA, Onafuwa-Nuga A, Chiari-Fort E, et al. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat. Immunol. 2011;12:975–83. doi: 10.1038/ni.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84•.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85•.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–85. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 86•.Bauman Y, Nachmani D, Vitenshtein A, Tsukerman P, Drayman N, et al. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9:93–102. doi: 10.1016/j.chom.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 87•.Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, et al. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat. Immunol. 2008;9:1065–73. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- 88•.Eissmann P, Evans JH, Mehrabi M, Rose EL, Nedvetzki S, Davis DM. Multiple mechanisms downstream of TLR-4 stimulation allow expression of NKG2D ligands to facilitate macrophage/NK cell crosstalk. J. Immunol. 2010;184:6901–9. doi: 10.4049/jimmunol.0903985. [DOI] [PubMed] [Google Scholar]
- 89•.Yadav D, Ngolab J, Lim RS, Krishnamurthy S, Bui JD. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-γ-induced microRNA. J. Immunol. 2009;182:39–43. doi: 10.4049/jimmunol.182.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90•.Bui JD, Carayannopoulos LN, Lanier LL, Yokoyama WM, Schreiber RD. IFN-dependent down-regulation of the NKG2D ligand H60 on tumors. J. Immunol. 2006;176:905–13. doi: 10.4049/jimmunol.176.2.905. [DOI] [PubMed] [Google Scholar]
- 91•.Hamerman JA, Ogasawara K, Lanier LL. Cutting edge: Toll-like receptor signaling in macrophages induces ligands for the NKG2D receptor. J. Immunol. 2004;172:2001–5. doi: 10.4049/jimmunol.172.4.2001. [DOI] [PubMed] [Google Scholar]
- 92•.Tokuyama M, Lorin C, Delebecque F, Jung H, Raulet DH, Coscoy L. Expression of the RAE-1 family of stimulatory NK-cell ligands requires activation of the PI3K pathway during viral infection and transformation. PLoS Pathog. 2011;7:e1002265. doi: 10.1371/journal.ppat.1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93•.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat. Rev. Immunol. 2008;8:259–68. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94•.Jonjic S, Babic M, Polic B, Krmpotic A. Immune evasion of natural killer cells by viruses. Curr. Opin. Immunol. 2008;20:30–38. doi: 10.1016/j.coi.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95•.Liu XV, Ho SS, Tan JJ, Kamran N, Gasser S. Ras activation induces expression of Raet1 family NK receptor ligands. J. Immunol. 2012;189:1826–34. doi: 10.4049/jimmunol.1200965. [DOI] [PubMed] [Google Scholar]
- 96•.Routes JM, Ryan S, Morris K, Takaki R, Cerwenka A, Lanier LL. Adenovirus serotype 5 E1A sensitizes tumor cells to NKG2D-dependent NK cell lysis and tumor rejection. J. Exp. Med. 2005;202:1477–82. doi: 10.1084/jem.20050240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97•.Unni AM, Bondar T, Medzhitov R. Intrinsic sensor of oncogenic transformation induces a signal for innate immunosurveillance. Proc. Natl. Acad. Sci. USA. 2008;105:1686–91. doi: 10.1073/pnas.0701675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98•.Jinushi M, Takehara T, Kanto T, Tatsumi T, Groh V, et al. Critical role of MHC class I-related chain A and B expression on IFN-α-stimulated dendritic cells in NK cell activation: impairment in chronic hepatitis C virus infection. J. Immunol. 2003;170:1249–56. doi: 10.4049/jimmunol.170.3.1249. [DOI] [PubMed] [Google Scholar]
- 99•.Zhang C, Niu J, Zhang J, Wang Y, Zhou Z, et al. Opposing effects of interferon-α and interferon-γ on the expression of major histocompatibility complex class I chain-related A in tumors. Cancer Sci. 2008;99:1279–86. doi: 10.1111/j.1349-7006.2008.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100•.Jonjic S, Polic B, Krmpotic A. Viral inhibitors of NKG2D ligands: friends or foes of immune surveillance? Eur. J. Immunol. 2008;38:2952–56. doi: 10.1002/eji.200838823. [DOI] [PubMed] [Google Scholar]
- 101•.Nice TJ, Coscoy L, Raulet DH. Posttranslational regulation of the NKG2D ligand Mult1 in response to cell stress. J. Exp. Med. 2009;206:287–98. doi: 10.1084/jem.20081335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102•.Diefenbach A, Hsia JK, Hsiung MY, Raulet DH. A novel ligand for the NKG2D receptor activates NK cells and macrophages and induces tumor immunity. Eur. J. Immunol. 2003;33:381–91. doi: 10.1002/immu.200310012. [DOI] [PubMed] [Google Scholar]
- 103•.Nice TJ, Deng W, Coscoy L, Raulet DH. Stress-regulated targeting of the NKG2D ligand Mult1 by a membrane-associated RING-CH family E3 ligase. J. Immunol. 2010;185:5369–76. doi: 10.4049/jimmunol.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104•.Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–33. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 105•.Eagle RA, Traherne JA, Hair JR, Jafferji I, Trowsdale J. ULBP6/RAET1L is an additional human NKG2D ligand. Eur J. Immunol. 2009;39:3207–16. doi: 10.1002/eji.200939502. [DOI] [PubMed] [Google Scholar]
- 106•.Fuertes MB, Girart MV, Molinero LL, Domaica CI, Rossi LE, et al. Intracellular retention of the NKG2D ligand MHC class I chain-related gene A in human melanomas confers immune privilege and prevents NK cell-mediated cytotoxicity. J. Immunol. 2008;180:4606–14. doi: 10.4049/jimmunol.180.7.4606. [DOI] [PubMed] [Google Scholar]
- 107•.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–38. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 108•.Salih HR, Rammensee HG, Steinle A. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J. Immunol. 2002;169:4098–102. doi: 10.4049/jimmunol.169.8.4098. [DOI] [PubMed] [Google Scholar]
- 109•.Salih HR, Antropius H, Gieseke F, Lutz SZ, Kanz L, et al. Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood. 2003;102:1389–96. doi: 10.1182/blood-2003-01-0019. [DOI] [PubMed] [Google Scholar]
- 110•.Doubrovina ES, Doubrovin MM, Vider E, Sisson RB, O'Reilly RJ, et al. Evasion from NK cell immunity by MHC class I chain-related molecules expressing colon adenocarcinoma. J. Immunol. 2003;171:6891–99. doi: 10.4049/jimmunol.171.12.6891. [DOI] [PubMed] [Google Scholar]
- 111•.Jinushi M, Takehara T, Tatsumi T, Hiramatsu N, Sakamori R, et al. Impairment of natural killer cell and dendritic cell functions by the soluble form of MHC class I-related chain A in advanced human hepatocellular carcinomas. J. Hepatol. 2005;43:1013–20. doi: 10.1016/j.jhep.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 112•.Boissel N, Rea D, Tieng V, Dulphy N, Brun M, et al. BCR/ABL oncogene directly controls MHC class I chain-related molecule A expression in chronic myelogenous leukemia. J. Immunol. 2006;176:5108–16. doi: 10.4049/jimmunol.176.8.5108. [DOI] [PubMed] [Google Scholar]
- 113•.Marten A, von Lilienfeld-Toal M, Buchler MW, Schmidt J. Soluble MIC is elevated in the serum of patients with pancreatic carcinoma diminishing γδ T cell cytotoxicity. Int. J. Cancer. 2006;119:2359–65. doi: 10.1002/ijc.22186. [DOI] [PubMed] [Google Scholar]
- 114•.Jinushi M, Hodi FS, Dranoff G. Therapy-induced antibodies to MHC class I chain-related protein A antagonize immune suppression and stimulate antitumor cytotoxicity. Proc. Natl. Acad. Sci. USA. 2006;103:9190–95. doi: 10.1073/pnas.0603503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115•.Salih HR, Goehlsdorf D, Steinle A. Release of MICB molecules by tumor cells: mechanism and soluble MICB in sera of cancer patients. Hum. Immunol. 2006;67:188–95. doi: 10.1016/j.humimm.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 116•.Waldhauer I, Steinle A. Proteolytic release of soluble UL16-binding protein 2 from tumor cells. Cancer Res. 2006;66:2520–26. doi: 10.1158/0008-5472.CAN-05-2520. [DOI] [PubMed] [Google Scholar]
- 117•.Song H, Kim J, Cosman D, Choi I. Soluble ULBP suppresses natural killer cell activity via down-regulating NKG2D expression. Cell. Immunol. 2006;239:22–30. doi: 10.1016/j.cellimm.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 118•.Cao W, Xi X, Hao Z, Li W, Kong Y, et al. RAET1E2, a soluble isoform of the UL16-binding protein RAET1E produced by tumor cells, inhibits NKG2D-mediated NK cytotoxicity. J. Biol. Chem. 2007;282:18922–28. doi: 10.1074/jbc.M702504200. [DOI] [PubMed] [Google Scholar]
- 119•.Paschen A, Sucker A, Hill B, Moll I, Zapatka M, et al. Differential clinical significance of individual NKG2D ligands in melanoma: soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin. Cancer Res. 2009;15:5208–15. doi: 10.1158/1078-0432.CCR-09-0886. [DOI] [PubMed] [Google Scholar]
- 120•.Fernandez-Messina L, Ashiru O, Boutet P, Aguera-Gonzalez S, Skepper JN, et al. Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J. Biol. Chem. 2010;285:8543–51. doi: 10.1074/jbc.M109.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121•.Kaiser BK, Yim D, Chow IT, Gonzalez S, Dai Z, et al. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature. 2007;447:482–86. doi: 10.1038/nature05768. [DOI] [PubMed] [Google Scholar]
- 122•.Waldhauer I, Goehlsdorf D, Gieseke F, Weinschenk T, Wittenbrink M, et al. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 2008;68:6368–76. doi: 10.1158/0008-5472.CAN-07-6768. [DOI] [PubMed] [Google Scholar]
- 123•.Boutet P, Aguera-Gonzalez S, Atkinson S, Pennington CJ, Edwards DR, et al. Cutting edge: The metalloproteinase ADAM17/TNF-α-converting enzyme regulates proteolytic shedding of the MHC class I-related chain B protein. J. Immunol. 2009;182:49–53. doi: 10.4049/jimmunol.182.1.49. [DOI] [PubMed] [Google Scholar]
- 124•.Liu G, Atteridge CL, Wang X, Lundgren AD, Wu JD. The membrane type matrix metalloproteinase MMP14 mediates constitutive shedding of MHC class I chain-related molecule A independent of A disintegrin and metalloproteinases. J. Immunol. 2010;184:3346–50. doi: 10.4049/jimmunol.0903789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125•.Jinushi M, Vanneman M, Munshi NC, Tai YT, Prabhala RH, et al. MHC class I chain-related protein A antibodies and shedding are associated with the progression of multiple myeloma. Proc. Natl. Acad. Sci. USA. 2008;105:1285–90. doi: 10.1073/pnas.0711293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126•.Ashiru O, Boutet P, Fernandez-Messina L, Aguera-Gonzalez S, Skepper JN, et al. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. 2010;70:481–89. doi: 10.1158/0008-5472.CAN-09-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127•.Bacon L, Eagle RA, Meyer M, Easom N, Young NT, Trowsdale J. Two human ULBP/RAET1 molecules with transmembrane regions are ligands for NKG2D. J. Immunol. 2004;173:1078–84. doi: 10.4049/jimmunol.173.2.1078. [DOI] [PubMed] [Google Scholar]
- 128•.Chauveau A, Aucher A, Eissmann P, Vivier E, Davis DM. Membrane nanotubes facilitate long-distance interactions between natural killer cells and target cells. Proc. Natl. Acad. Sci. USA. 2010;107:5545–50. doi: 10.1073/pnas.0910074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129•.Wu JD, Atteridge CL, Wang X, Seya T, Plymate SR. Obstructing shedding of the immunostimulatory MHC class I chain-related gene B prevents tumor formation. Clin. Cancer Res. 2009;15:632–40. doi: 10.1158/1078-0432.CCR-08-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130•.Wiemann K, Mittrucker HW, Feger U, Welte SA, Yokoyama WM, et al. Systemic NKG2D down-regulation impairs NK and CD8 T cell responses in vivo. J. Immunol. 2005;175:720–29. doi: 10.4049/jimmunol.175.2.720. [DOI] [PubMed] [Google Scholar]
- 131•.Groh V, Bruhl A, El-Gabalawy H, Nelson JL, Spies T. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl. Acad. Sci. USA. 2003;100:9452–57. doi: 10.1073/pnas.1632807100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132•.Hue S, Mention JJ, Monteiro RC, Zhang S, Cellier C, et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 2004;21:367–77. doi: 10.1016/j.immuni.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 133•.Dasgupta S, Bhattacharya-Chatterjee M, O'Malley BW, Jr, Chatterjee SK. Inhibition of NK cell activity through TGF-β1 by down-regulation of NKG2D in a murine model of head and neck cancer. J. Immunol. 2005;175:5541–50. doi: 10.4049/jimmunol.175.8.5541. [DOI] [PubMed] [Google Scholar]
- 134•.Lee JC, Lee KM, Kim DW, Heo DS. Elevated TGF-β1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J. Immunol. 2004;172:7335–40. doi: 10.4049/jimmunol.172.12.7335. [DOI] [PubMed] [Google Scholar]
- 135•.Stetler-Stevenson WG, Aznavoorian S, Liotta LA. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu. Rev. Cell Biol. 1993;9:541–73. doi: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- 136•.Sato H, Takino T. Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances pericellular proteolysis and invasion. Cancer Sci. 2010;101:843–47. doi: 10.1111/j.1349-7006.2010.01498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137•.Gumireddy K, Sun F, Klein-Szanto AJ, Gibbins JM, Gimotty PA, et al. In vivo selection for metastasis promoting genes in the mouse. Proc. Natl. Acad. Sci. USA. 2007;104:6696–701. doi: 10.1073/pnas.0701145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138•.Aguera-Gonzalez S, Gross CC, Fernandez-Messina L, Ashiru O, Esteso G, et al. Palmitoylation of MICA, a ligand for NKG2D, mediates its recruitment to membrane microdomains and promotes its shedding. Eur. J. Immunol. 2011;41:3667–76. doi: 10.1002/eji.201141645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139•.Siemens DR, Hu N, Sheikhi AK, Chung E, Frederiksen LJ, et al. Hypoxia increases tumor cell shedding of MHC class I chain-related molecule: role of nitric oxide. Cancer Res. 2008;68:4746–53. doi: 10.1158/0008-5472.CAN-08-0054. [DOI] [PubMed] [Google Scholar]
- 140•.Nagai Y, Tanaka Y, Kuroishi T, Sato R, Endo Y, Sugawara S. Histamine reduces susceptibility to natural killer cells via down-regulation of NKG2D ligands on human monocytic leukaemia THP-1 cells. Immunology. 2012;136:103–14. doi: 10.1111/j.1365-2567.2012.03565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141•.Hedlund M, Nagaeva O, Kargl D, Baranov V, Mincheva-Nilsson L. Thermal- and oxidative stress causes enhanced release of NKG2D ligand-bearing immunosuppressive exosomes in leukemia/lymphoma T and B cells. PLoS ONE. 2011;6:e16899. doi: 10.1371/journal.pone.0016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142•.Wu JD, Higgins LM, Steinle A, Cosman D, Haugk K, Plymate SR. Prevalent expression of the immunostimulatory MHC class I chain-related molecule is counteracted by shedding in prostate cancer. J. Clin. Investig. 2004;114:560–68. doi: 10.1172/JCI22206. [DOI] [PMC free article] [PubMed] [Google Scholar]