Abstract
To investigate the binding specificity of turnip mosaic virus (TuMV) viral protein-genome linked (VPg) with translation initiation factor 4E, we evaluated here the kinetic parameters for the interactions of human eIF4E, Caenorhabditis elegans IFE-3 and IFE-5 and Arabidopsis eIFiso4E, by surface plasmon resonance (SPR). The results indicated that TuMV VPg does not show a binding preference for Arabidopsis eIFiso4E, even though it is from a host species whereas the other eIF4E orthologues are not. Surprisingly, the effect of m7GTP on both the rate constants and equilibrium binding constants for the interactions of VPg differed for the four eIF4E orthologues. In the case of eIFiso4E and IFE-3, m7GTP increased kon, but for eIF4E and IFE-5, it decreased kon. To provide insight into the structural basis for these differences in VPg binding, tertiary structures of the eIF4E orthologues were predicted on the basis of the previously determined crystal structure of m7GpppA-bound human eIF4E. The results suggested that in cap-bound eIF4E orthologues, the VPg binds to the C-terminal region, which constitutes one side of the entrance to the cap-binding pocket, whereas in the cap-free state, VPg binds to the widely opened cap-binding pocket and its surrounding region. The binding of VPg to the C-terminal region was confirmed by the SPR analyses of N- or C-terminal residues-deleted eIF4E orthologues.
Keywords: 3D structure prediction, eIF4E, surface plasmon resonance, turnip mosaic virus, VPg
Positive-stranded RNA viruses have evolved to use a wide variety of mechanisms for translation initiation (1). Several viruses use a 5′-cap dependent mechanism for initiation, whereas other viruses use a cap-independent IRES (2). In recent years, it has been reported that members of the families Picornaviridae, Potyviridae, Luteoviridae, Comoviridae and Caliciviridae are not capped, but are instead covalently linked at the 5′-end of the RNA genome to a small protein called the VPg (viral protein linked to the genome) (3) through either tyrosine or serine residues (4–8). The general structural similarity between their RNAs and eukaryotic mRNAs suggests that the VPg acts functionally like a cap. Indeed, an interaction between the VPg and the cap-binding protein eIF4E and other initiation factors has been demonstrated for several viruses (9–14), suggesting that the VPg plays a role in initiating translation of viral RNA through protein–protein interactions with the translation machinery.
Turnip mosaic virus (TuMV) is a member of the genus Potyvirus, in the family Potyviridae (15), and is known to infect cruciferous plants including Arabidopsis thaliana. The VPg of TuMV is a factor in virus infectivity because of its interaction with eIF4E (16). Interaction with both eIFiso4E and eIFiso4F has been reported (17,18). Recently, the detailed binding properties and kinetics of these interactions have been investigated by biophysical methods (13, 14). In particular, it was shown by SPR that the TuMV VPg binds to eIFiso4E but not to eIF4E or nCBP, the two other family members of eIF4E in A. thaliana (13).
This striking finding led us to investigate the interaction between TuMV VPg and other eIF4E orthologues, because we are interesting in clarifying whether the VPg of each virus has any preference for the interaction with the host cellular initiation factor 4E variant. Also, we are interested in clarifying the 4E orthologue-dependent binding region and mode of VPg, because such studies may lead insight into the structural basis for VPg binding, which will be critical for understanding the biological function of VPg. Multiple eIF4E family members have been found in plants, mammals, insects, amphibians, nemotodes, certain protists and fission yeast (19, 20). These provide an opportunity to study eIF4E molecules varying widely in amino-acid sequence yet still retaining the ability to specifically interact with the cap. In the present work, we studied the interaction of the TuMV VPg with four eIF4E orthologues, A. thaliana eIFiso4E, human eIF4E, Caenorhabditis elegans IFE-3 and C. elegans IFE-5 by SPR in both the presence and absence of m7GTP. In order to interpret the results, we also constructed models for the tertiary structures of these orthologues based on the known structure of human eIF4E bound to m7GpppA (21).
MATERIALS AND METHODS
Preparation of His-Tagged TuMV VPg
The DNA encoding the TuMV VPg was prepared according to the method of Miyoshi et al. (13). The expression plasmid was constructed from the pET-21a vector (Novagene Co.). The construct was confirmed by DNA sequencing. His-tagged VPg protein was then expressed in Escherichia coli BL21(DE3)pLysS cells (Novagene Co.). Transformed cells were grown in LA broth (Nacalai). After addition of 100 μM IPTG, cells were cultured for 3 h at 37°C, harvested, suspended in 15 ml of buffer A (50 mM Tris–HCl pH 7.5, 100 mM NaCl) at 0°C, and lysed by sonication.
The supernatant was then purified on Ni–Sepharose (Pharmacia Biotech) by gravity flow. The column was eluted step-wise with buffer A containing 10–500 mM imidazole, the VPg being eluted at 200 mM. The sample was dialysed against buffer B (20mM Tris–HCl pH 8.5, 2 mM DTT), applied to an anion exchange column (HiTrap™ Q-HP), and eluted with a linear gradient of buffer B containing 50–500 mM NaCl. The protein exhibited a single band on SDS–PAGE.
Preparation of m7GTP–Free and -Bound eIF4E Orthologues
Expression of recombinant human eIF4E (wild and an N-terminal 33 residues-deleted mutant) in E. coli and its purification by the cap affinity chromatography were carried out as described previously (22). Expressions of His-tagged C. elegans IFE-3 (wild and a C-terminal 45 residues-deleted mutant) and IFE-5 and purifications by Ni-chelating chromatography were performed as previously described (23, 24). A. thaliana eIFiso4E was expressed as a fusion protein with TF by using the pCold TF vector (Novagen) in E. coli BL21(DE3). The transformed cells were grown in LA broth. After addition of 100 μM IPTG, cells were cultured for 24 h at 15°C, harvested, suspended in 15 ml of buffer A at 0°C, and lysed by sonication. Proteins were purified from the supernatants by Ni–Sepharose chromatography by step-wise elution with buffer A containing 10–500 mM imidazole, TF-fused eIFiso4E being eluted at 150 mM. The cleavage of TF was performed with factor Xa [1 U per 500 μg for eIFiso4E] for 20 h at 4°C. The purification of eIFiso4E was performed in the same manner as IFE-3 and IFE-5. The recombinant samples of human eIF4E, IFE-3, IFE-5 and eIFiso4E migrated as single bands on SDS–PAGE analyses.
The m7GTP-free and -bound forms of the eIF4E orthologues were prepared as follows. The purified samples were applied to an m7GTP-Sepharose 4B affinity column equilibrated with buffer C [50 mM HEPES–NaOH (pH 7.0), 1 mM EDTA, 100 mM NaCl]. After washing the column with buffer C, the m7GTP-free and -bound eIF4E orthologues were prepared by elution with buffer C containing 1 M NaCl or 100 μM m7GTP, respectively, followed by gel filtration. The protein solutions were concentrated with a Centricon 10 (Amicon Co.) to the desired concentration, which was determined by the Bradford method (25). Because of the relatively rapid proteolysis of m7GTP-free eIF4E preparations at room temperature (21), the SPR experiments were all performed within 8 h after the sample preparation.
SPR-Binding Analysis
Binding characteristics were performed using BIAcore X instrument (Biacore, Inc.) with a CM-dextran sensor chip. The VPg was immobilized on the sensor chip by the conventional amine coupling method at a concentration of 400–500 RU, which is the range recommended for accurate measurement of the kinetic parameters (26). M7GTP-free or -bound eIF4E orthologues were injected into the chip with the running buffer D (10 mM HEPES–NaOH pH 7.4, 200 mM NaCl, 0.005% surfactant P-20, 1 mM DTT). All measurements were performed at a flow rate of 20 μl/min at 25°C. Sensorgrams were evaluated using the BIA evaluation software package. The responses from blank chips were subtracted from the sample responses to obtain the sensorgram for a specific interaction. The constants for association rate, kon, dissociation rate, koff, and the equilibrium constant for dissociation, KD (= koff/kon), were obtained using the Langmuir(1:1)-binding model. These kinetic parameters were checked for consistency using local fitting for each analyte concentration and were judged to be reliable based on χ2 values less than 10 and plots of residuals.
3D Structure Prediction of eIF4E Orthologues
Attempt to crystallize the eIF4E orthologues used in this study other than human eIF4E has not yet been successful. We therefore made 3D structure predictions of cap-bound IFE-3, IFE-5 and eIFiso4E using the crystal structure of m7GpppA-bound human eIF4E [PDBcode: 1IPB] (21) as a template for the homology modelling. The amino-acid sequences of IFE-3 (23), IFE-55 (24) and eIFiso4E (27) are 51%, 43% and 40% identical to human eIF4E, respectively. Sequence alignments were performed with Sequence Analysis and Protein Families modules in Discovery Studio (28). A multiple-sequence alignment technique was adopted based on the structural similarity between the template and target molecules with the aid of BLAST sequence alignment software.
The 3D structure of each 4E orthologue was generated from the results of the alignment procedure by Modeler module within Discovery Studio. An initial 3D structure was obtained by transferring the backbone coordinates of human eIF4E (21) to the corresponding residues of the aligned eIF4E orthologue. For mismatched residues only the Cα atom coordinates were copied from the template protein while the remaining atomic coordinates were constructed using internal coordinates derived from a CHARMM topology library. The refinement of the homology model was then performed by the Protein Refine module within Discovery Studio, which is based on CHARMM energy minimization and is not dependent on the initial structure (ab initio approach). The steepest descent algorithm was used for 5,000 iterations and then the conjugated gradient calculation was carried out to 0.1 kcal mol−1 Å−1 of convergence. After this simulation, the homology model was further refined by molecular dynamics calculation. Finally a conjugate gradient energy minimization was performed until the root-mean square gradient energy was lower than 0.001 kcal mol−1 Å−1.
RESULTS
SPR Analyses
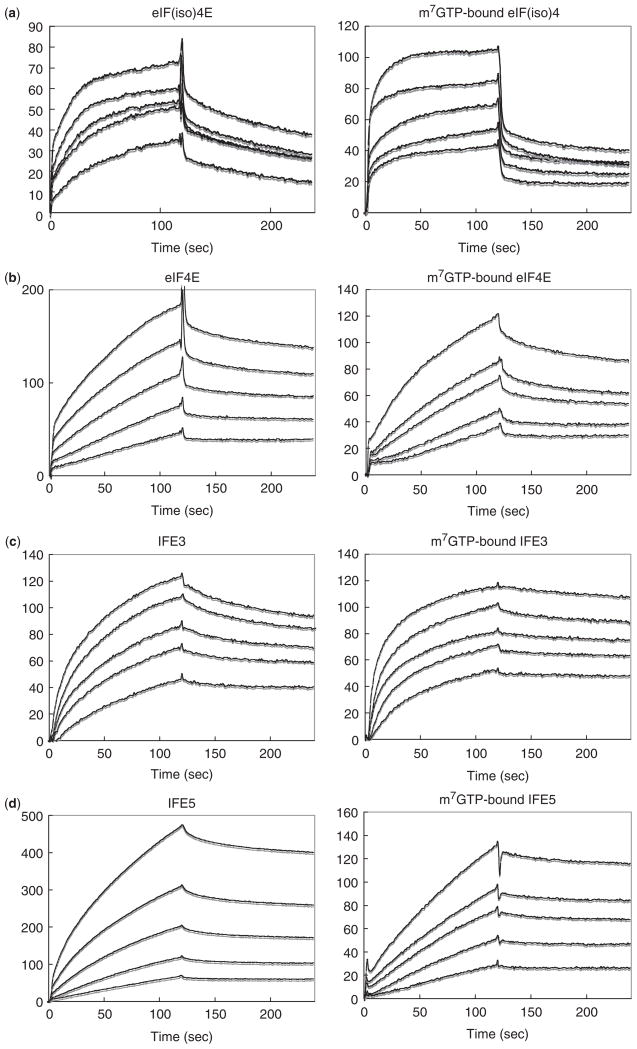
We previously reported that TuMV VPg binds to eIFiso4E from the host plant species A. thaliana, but not to eIF4E or nCBP, the two other family members in A. thaliana (13), probably due to the different binding specificity of TuMV VPg for eIF4E orthologues in the same host plant. To gain insight into this specificity, we examined the interaction kinetics of the same VPg with three other eIF4E orthologues, human eIF4E, C. elegans IFE-3 and C. elegans IFE-5, and compared that with its host A. thaliana eIF(iso)4E: these four orthologues are subsequently referred to as eIFiso4E, eIF4E, IFE-3 and IFE-5, respectively. The SPR sensorgrams for interactions with m7GTP-free and -bound eIF4E orthologues are shown in Fig. 1, and the kon, koff and KD values are collected in Table 1. kon for the binding to eIFiso4E was increased 6.4-fold by the presence of m7GTP. Similarly, kon for binding to IFE-3 was accelerated 17.7-fold by m7GTP. Interestingly, the opposite behaviour was seen with eIF4E and IFE-5; kon was decreased 1.7- and 4.4-fold by m7GTP, respectively. koff for the dissociation of VPg from eIFiso4E, eIF4E and IFE-5 was essentially unaffected by m7GTP, but koff for IFE-3 was decreased 2.4-fold by m7GTP.
Fig. 1. SPR sensorgrams for the interaction of immobilized TuMV VPg with (a) A. thaliana eIFiso4E, (b) human eIF4E, (c) C. elegans IFE-3, and (d) C. elegans IFE-5.
The left and right parts correspond to the m7GTP-free and -bound eIF4E orthologues, respectively. Each SPR signal reflects the response after buffer containing the eIF4E orthologue (analyte) begins to flow over a CM dextran sensor chip to which the VPg is coupled through amino groups. The concentrations of analytes, corresponding to curves of increasing RU, are: 5.0–20.0 μM and 1.3–5.0 μM for m7GTP-free and -bound forms for (a), 0.2–2.5 μM and 1.3–30.0 μM for (b), 0.6–7.5 μM and 0.1–0.9 μM for (c) and 0.3–5.0 μM and 0.6–10.0 μM for (d). Each sensorgram consist of a formation portion (injection of analyte) and a dissociation portion (injection of buffer alone).
Table 1.
Kinetic and equilibrium parameters for the interaction of TuMV VPg with four eIF4E orthologues.
| Ligand | Analyte | kon (M−1s−1) | koff (s−1) | KD (M) |
|---|---|---|---|---|
| m7GTP-free state | ||||
| VPg | eIFiso4E | 3.25 ×103 | 4.59 ×10−3 | 1.41 ×10−6 |
| eIF4E | 7.29 ×103 | 1.29 ×10−3 | 1.76 ×10−7 | |
| IFE-3 | 3.84 ×103 | 1.55 ×10−3 | 4.03 ×10−7 | |
| IFE-5 | 3.08 ×103 | 8.88 ×10−4 | 2.88 ×10−7 | |
| m7GTP-bound state | ||||
| VPg | eIFiso4E | 2.08 ×104 | 4.32 ×10−3 | 2.08 ×10−7 |
| eIF4E | 4.33 ×102 | 1.63 ×10−3 | 3.68 ×10−6 | |
| IFE-3 | 6.78 ×104 | 6.40 ×10−4 | 9.44 ×10−9 | |
| IFE-5 | 7.03 ×102 | 4.64 ×10−4 | 6.60 ×10−7 | |
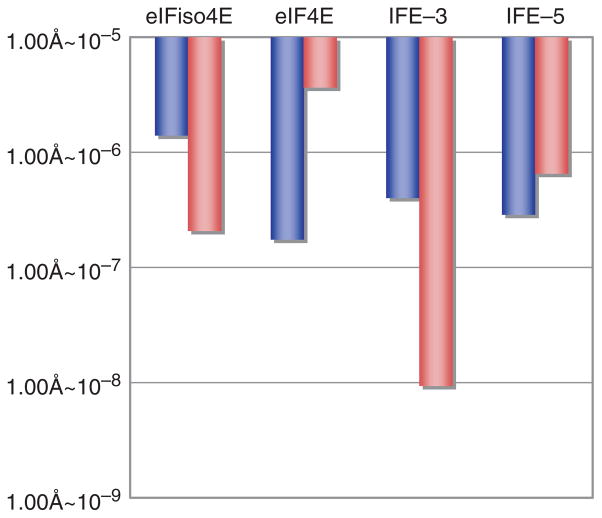
The results indicate that there is no selective binding of the TuMV VPg to eIFiso4E of A. thaliana, the host species for this virus. The KD values indicate that, in the absence of m7GTP, the VPg-binding strength is in the order of IFE-3 ≈IFE-5 ≈eIF4E > eIFiso4E. In the presence of m7GTP, the order is IFE-3 > IFE-5 ≈eIFiso4E > eIF4E. Three patterns are apparent: KD is lower in the presence of m7GTP (IFE-3 and eIFiso4E); KD is higher in the presence of m7GTP (IFE-5); KD is essentially unchanged in the presence of m7GTP (eIF4E) (Fig. 2). The KD values for VPg binding are about 10-fold different between the m7GTP-free and -bound states of eIF4E orthologues, except for that of IFE-5. As it is generally considered that the difference of 10-fold is significant (26), the three different patterns suggest that there are different mechanism of binding.
Fig. 2. Comparative histogram of KD values of m7GTP-free (blue) and -bound (red) eIF4E orthologues for interaction with TuMV VPg.
Each bar represents mean of KD values measured in this work and is within the deviation of ±5%.
3D Structure Prediction of eIF4E Orthologues
Although the tertiary structures of the MuMV VPg, eIFiso4E, IFE-3 and IFE-5 have not yet been established, that of human eIF4E has been solved (21). As an aid to better understand the differences in the sites and modes of VPg binding to these various eIF4E orthologues, we predicted the 3D structures of eIFiso4E, IFE-3 and IFE-5 from the X-ray structure of m7GpppA-bound human eIF4E (MATERIALS AND METHODS section). The secondary structure of each eIF4E orthologue was sequentially aligned on the basis of the secondary structure of human eIF4E, i.e. the major sheet and helix structures were imposed on the amino-acid sequences of each orthologue. Less homology was observed at both N- and C-terminal regions. The sequence alignments for eIFiso4E, IFE-3 and IFE-5 with respect to eIF4E are shown in Fig. 3. A common feature is that the N-terminal portions of eIFiso4E, IFE-3 and IFE-5 are shorter than that of eIF4E. The other feature is that the most sequence divergence occurs in the C-terminal portions; compared with eIF4E, eight residues are inserted for IFE-5 (residues 165–172) and 36 residues are added to the end of IFE-3 (residues 215–251). No insertions or additions occur for eIFiso4E but the sequence of the C-terminal portion is nonetheless more divergent than other parts of the protein.
Fig. 3. Sequence alignments of eIFiso4E, IFE-3 and IFE-5 based on 3D structure of human m7GpppA-bound eIF4E.

Two Trp and one Glu residues participating in the recognition of mRNA cap-structure are shown in green. The α-helix and β-sheet structures are indicated with red and blue arrows above the sequences, respectively. Three loop structures (Phe48–Gln57, Asp96–Gly110 and Val156–Lys162) and a C-terminal region (Ala201–Val217), which make up the cap-binding pocket, are shown with green, blue, purple and red broken lines, respectively.
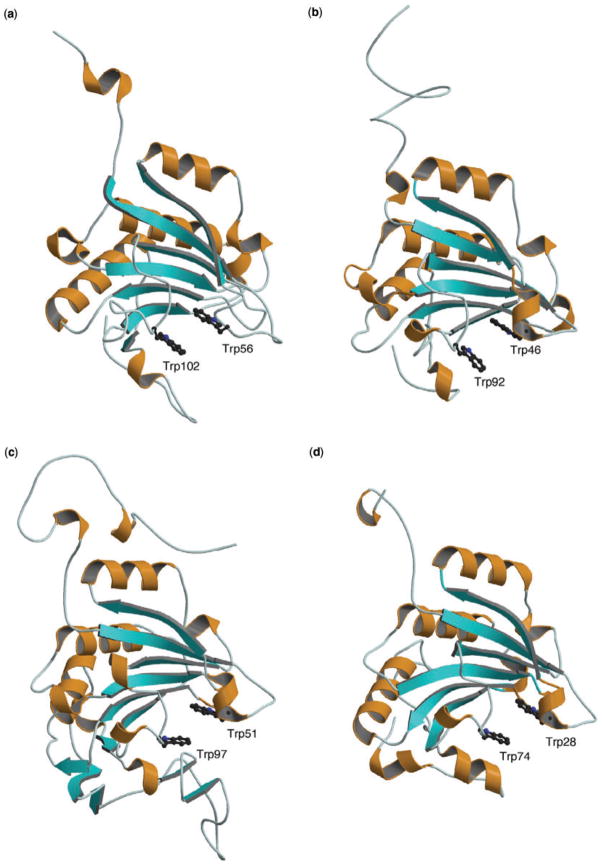
The predicted 3D structures of eIFiso4E, IFE-3 and IFE-5 are shown in Fig. 4b, c and d, respectively; the result of eIFiso4E was already reported in a previous article (13). For comparison, the previously published X-ray structure of human eIF4E (21) is shown as well (Fig. 4a). Since the latter structure was the cap-bound state, the predicted structures simulate the same cap-binding state. In all four eIF4E orthologues the orientations of α-helix and β-sheet secondary structures are nearly the same. This similarity in secondary structure is also suggested by similar circular dichroism (data not shown). It can also be seen that the cap-binding Trp and Glu residues are completely conserved among the eIF4E orthologues (Fig. 3). Previous studies with several eIF4E orthologues (29–31) have shown that the cap-binding to human eIF4E involves π–π stacking interactions with Trp56 and Trp102 residues and H-bond pairing with Glu103. These correspond to Trp46, Trp92 and Glu93 in eIFiso4E, Trp51, Trp97 and Glu98 in IFE-3, and Trp28, Trp74 and Glu75 in IFE-5 (Fig. 4); these cap-binding residues for IFE-3 and IFE-5 have been reported by the other 3D modelling approach (32). The same spatial positions of these conserved residues in the predicted structures suggest that all four eIF4E orthologues utilize a similar mode of cap-recognition.
Fig. 4. Drawing of the X-ray structure of eIF4E (21) (a) and the predicted 3D structures of (b) eIFiso4E, (c) IFE-3 and (d) IFE-5.
The α-helix and β-sheet structures are shown with the ribbon model. Two Trp residues participating in cap binding are shown with ball and stick models.
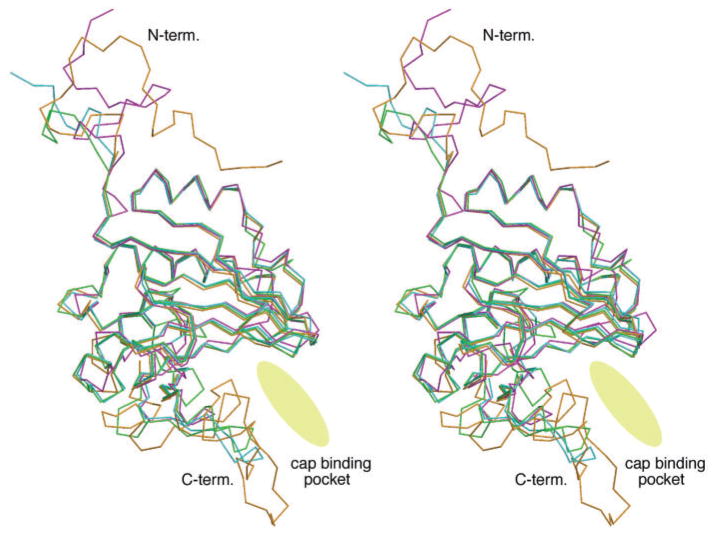
In the C-terminal regions, however, there are differences among the four orthologues. These differences can be seen more clearly in a stereoscopic depiction in which all four structures are superimposed (Fig. 5). The most obvious difference among the eIF4E orthologues is at the periphery of the C-terminal region, which creates one side of the entrance to the cap-binding pocket. The N-terminal region, though divergent among the four orthologues, does not assume a defined conformation, although in yeast eIF4E it has been shown to become folded upon eIF4G binding (33). This suggests that the binding site of VPg is at the C-terminal region, but not the N-terminal one, of eIF4E orthologue. To confirm this suggestion, the SPR interaction kinetics of the VPg with N-terminal 33 residues-deleted human eIF4E (m7GTP-bound state) and C-terminal 45 residues-deleted IFE-3 (m7GTP-bound state) were measured. Consequently the N-terminal truncated eIF4E showed the nearly same values as those with the full-length eIF4E: kon (M−1 s−1) = 1.36 ×103, koff (s−1) = 4.47 ×10−3, KD (M) = 3.29 ×10−6. In contrast, the SPR signals of the VPg with the C-terminal truncated IFE-3 were too weak to estimate the kinetic parameters by the BIA evaluation software.
Fig. 5. Stereoscopic drawing of superimposition of 3D backbone structures of eIF4E (pink), eIFiso4E (purple), IFE-3 (orange) and IFE-5 (green).
The cap-binding entrance and pocket is indicated with an ellipsoidal disk.
DISCUSSION
A cause of virus infectivity of cruciferous plant by TuMV correlates with the VPg covalently linked at the genome RNA 5′-end of TuMV, where it inhibits the protein biosynthesis of host cell through the interaction with the initiation factor 4E. To clarify the infectious mechanism of A. thaliana, the interaction between TuMV VPg and its host cellular initiation factor 4E has been investigated by various physicochemical methods (13, 14, 17, 18). The present work was performed to determine (i) whether the VPg of TuMV has a binding specificity for the eIFiso4E of one of its host plant species, A. thaliana and (ii) which structural moiety of eIF4E isoform participates in VPg binding. The SPR results indicated that there is no specific preference for TuMV VPg binding to its host cellular eIFiso4E among the eIF4E orthologues used in the study (Table 1). This is in contrast with the previous results, i.e. the TuMV VPg binds to eIFiso4E but not to eIF4E or nCBP in A. thaliana (13). Although the reason for the lack of binding to the eIF4E or nCBP is not explainable at present, the difference of buffers used for SPR measurement may be probable, i.e. the reducing agent was included in the present buffer D but not in the previous buffer (20 mM Tris–HCl pH 7.6 and 150 mM NaCl), because a recent paper (34) reported that eIF4E from plant such as wheat causes the large conformational change as well as the formation of an intramolecular disulfide bridge in its oxidized form.
A characteristic difference among the four orthologues is in the effect of m7GTP on the binding of VPg (Fig. 2). The 3D structure prediction suggests that this largely results from the different structural situation at either or both of the N- and C-terminal regions of each eIF4E orthologue, because the structural difference could be mainly observed at these regions (Fig. 5).
Concerning the binding site of VPg on eIF4E ortholo-gue, two models have been reported: one is at or near the cap-binding pocket (13,18), and the other is a region in which the cap and VPg do not interfere with each other, implying a site distant from the cap-binding pocket (10). It is unlikely that the potyvirus VPg interacts with the N-terminal region of eIF4E, because the SPR interaction kinetics of the VPg with N-terminal truncated human eIF4E were nearly the same as those with the full-length eIF4E. As the N-terminal regions of the eIF4E orthologues used in this work are all shorter than that of human eIF4E, the following would be applicable, i.e. no notable affect of the N-terminal region to the cap-binding pocket of each eIF4E orthologue. Thus, it would be reasonable to consider that the difference between the association constants of VPg is mainly dependent on the binding situation at the C-terminal regions of the m7GTP-free and -bound eIF4E orthologues, because the deletion of IFE-3 C-terminal 45 residues, which constitute one side of the entrance to the cap-binding pocket, caused the complete loss of the binding ability. The predicted 3D structures also show that the largest differences are found in the C-terminal regions.
As is shown in Fig. 6, three loop structures and a C-terminal region construct the cap-binding entrance and pocket in human eIF4E, i.e. Phe48–Gln57 (green), Asp96–Gly110 (blue), Val156–Lys162 (purple) and Ala201–Val217 (red) (also refer Fig. 3). These regions would be highly flexible in the cap-free 4E orthologue due to the lack of interaction with cap. In fact, the NMR solution structure of the cap-free eIF4E showed the large fluctuation of these regions, in particular Phe48–Gln57, Asp96–Gly110 loops (35). This makes us imagine that the defined cap-binding pocket is not constructed in the cap-free eIF4E orthologue and VPg interacts with the widespread flexible region including the Phe48–Gln57, Asp96–Gly110 loops, consequently leading to the relatively small difference among the KD values (Fig. 2).
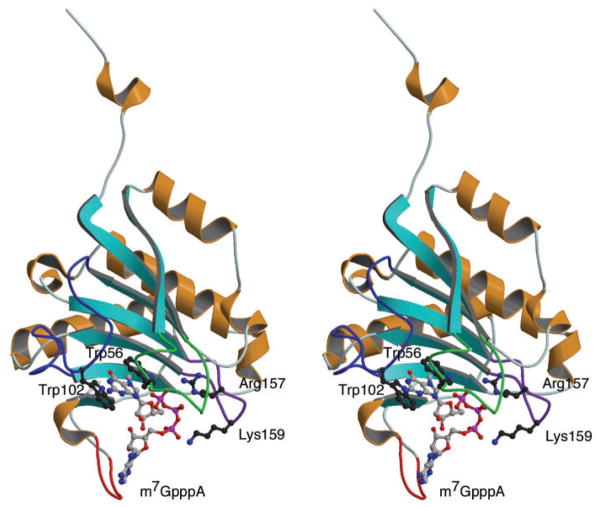
Fig. 6. Stereoscopic drawing of X-ray structure of m7GpppA-bound eIF4E (21).
The m7GpppA bound to the cap-binding pocket is shown (ball and stick model). The sequences of Phe48–Gln57, Asp96–Gly110, Val156–Lys162 and Ala201–Val217 are shown with green, blue, purple and red ropes, respectively.
In the case of m7GTP-bound eIF4E, on the other hand, flexibility of the Phe48–Gln57 and Asp96–Gly110 loops would be restricted because of the π–π stacking formation of Trp56 and Trp102 with m7GTP and H-bonding with Glu103. These loops may thus be less available for interaction with the VPg, leaving only the Val156–Lys162 loop and C-terminal Ala201–Val217 sequence. The X-ray and NMR structures (21, 30–32) showed that Arg157 and Lys159 on the Val156–Lys162 loop stabilize the cap structure through the H-bonds with the phosphate group of m7GTP. Since these two polar residues are also conserved in the eIF4E orthologues under study, it would be reasonable to consider that the association of VPg is largely influenced by the C-terminal conformation of each eIF4E orthologue (Fig. 5).
The relation of the predicted 3D structures of cap-bound human eIF4E and C. elegens IFE-3 with their KD values indicates that the C-terminal conformation of IFE-3 contributes to the increased association with VPg; this extra loop may be wide and flexible enough to wrap the VPg, consequently increasing the association. The predicted 3D structure showed the C-terminal conformation characteristic for each eIF4E orthologue. Although the detailed discussion on the relation between this conformation and KD value is impossible, it may say that the binding affinity of VPg to the cap-bound eIF4E orthologue is controlled by the length and conformation of the C-terminal region.
Abbreviations
- 4EBP
eIF4E-binding protein
- eIF
eukaryotic initiation factor
- CM
carboxymethyled
- eIF4E
eukaryotic initiation factor 4E
- IRES
internal ribosome entry site
- GST
glutathione-S-transferase
- IPTG
isopropyl-β-D-thiogalactopyranoside
- m7GTP
7-methylguanosine 5′-triphosphate
- m7GpppA
P1-7-methylguanosine-P3-adenosine- 5′,5′-triphosphate
- LB
Luria-Bertani
- nCBP
novel cap-binding protein
- PCR
polymerase chain reaction
- P-20
poly(oxyethylene)sorbitan monolaulate (Tween 20)
- RU
resonance unit
- SDS–PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SPR
surface plasmon resonance
- TF
trigger factor
- TuMV
turnip mosaic virus
- VPg
viral protein linked to the genome
Footnotes
CONFLICT OF INTEREST
None declared.
References
- 1.Pe’ery T, Mathews M. Viral translational strategies and host defense mechanism. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- 2.Belsham GJ, Jackson RJ. In: Translation initiation on picornavirus RNA in Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 869–900. [Google Scholar]
- 3.Sadowy E, Milner AD, Haenni AL. Proteins attached to viral genomes are multifunctional. Adv Virus Res. 2001;57:185–262. doi: 10.1016/s0065-3527(01)57004-9. [DOI] [PubMed] [Google Scholar]
- 4.Rothberg PG, Harris TJR, Nomoto A, Wimmer E. O4-(5′-Uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of Poliovirus. Proc Natl Acad Sci USA. 1978;75:4868–4872. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vartapetian AB, Drygen YU, Chumakov KM, Bogdanov AA. The structure of the covalent linkage between proteins and RNA in encephalomyocarditis virus. Nucleic Acids Res. 1980;8:3729–3742. doi: 10.1093/nar/8.16.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaegle M, Wellink J, Goldbach R. The genome-linked protein of Cowpea mosaic virus is bound to the 5′ terminus of virus RNA by a phosphodiester linkage to serine. J Gen Virol. 1987;68:627–632. [Google Scholar]
- 7.Murphy JF, Rychlik W, Rhoads RE, Hunt AG, Shaw JG. A tyrosine residue in the small nuclear inclusion protein of tobacco vein mottling virus links the VPg to the viral RNA. J Virol. 1991;65:511–513. doi: 10.1128/jvi.65.1.511-513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy J, Klein P, Hunt A, Shaw J. Replacement of the tyrosine residue that links a potyviral VPg to the viral RNA is lethal. Virology. 1996;220:535–538. doi: 10.1006/viro.1996.0344. [DOI] [PubMed] [Google Scholar]
- 9.Daughenbaugh KF, Fraser CS, Hershey JWB, Hardy ME. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodfellow I, Chaudhry Y, Gioldasi I, Gerendopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michon T, Estevez Y, Walter J, Germon-Retana S, Gall OL. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 2006;273:1312–1322. doi: 10.1111/j.1742-4658.2006.05156.x. [DOI] [PubMed] [Google Scholar]
- 12.Grzela R, Strokovska L, Andrieu JP, Dublet B, Zagorski W, Chroboczek J. Potyvirus terminal protein VPg, effector of host eukaryotic initiation factor eIF4E. Biochimie. 2006;88:887–896. doi: 10.1016/j.biochi.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Miyoshi H, Suehiro N, Tomoo K, Muto S, Takahashi T, Tsukamoto T, Ohmori T, Natsuaki T. Binding analyses for the interaction between plant virus genome-linked protein (VPg) and plant translational initiation factors. Biochimie. 2006;88:329–340. doi: 10.1016/j.biochi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Khan MA, Miyoshi H, Ray S, Natsuaki T, Suehiro N, Goss DJ. Interaction of genome-linked protein (VPg) of turnip mosaic virus with whear germ translation initiation factors eIFiso4E and eIFiso4F. J Biol Chem. 2006;281:28002–28010. doi: 10.1074/jbc.M605479200. [DOI] [PubMed] [Google Scholar]
- 15.Walsh JA, Jenner CE. Turnip mosaic virus and the quest for durable resistance. Mol Plant Pathol. 2002;3:289–300. doi: 10.1046/j.1364-3703.2002.00132.x. [DOI] [PubMed] [Google Scholar]
- 16.Leonard S, Viel C, Beauchemin C, Daigneault N, Fortin MG, Laliberte JF. Interaction of VPg-Pro of Turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J Gen Virol. 2004;85:1055–1063. doi: 10.1099/vir.0.19706-0. [DOI] [PubMed] [Google Scholar]
- 17.Wittman S, Chatel H, Fortin MG, Laliberte JF. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology. 1997;234:84–92. doi: 10.1006/viro.1997.8634. [DOI] [PubMed] [Google Scholar]
- 18.Leonard S, Plante D, Wittmann S, Daigneault N, Fortin MG, Laliberte JF. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlate with virus infectivity. J Virol. 2000;74:7730–7737. doi: 10.1128/jvi.74.17.7730-7737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi B, Lee K, Maeder D, Jagus R. Phylogenetic analysis of eIF4E-family members. BMC Evol Biol. 2005;5:48. doi: 10.1186/1471-2148-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhoads RE, Dinkova TD, Jagus R. Approaches for analyzing the differential activities and functions of eIF4E family members. In: Lorsch J, editor. Methods in Enzymology. Vol. 429. Elsevier; 2007. pp. 261–297. [DOI] [PubMed] [Google Scholar]
- 21.Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Sasaki M, Taniguchi T, Miyagawa H, Kitamura K, Miura K, Ishida T. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J Mol Biol. 2003;328:365–383. doi: 10.1016/s0022-2836(03)00314-0. [DOI] [PubMed] [Google Scholar]
- 22.Morino S, Hazama H, Ozaki M, Teraoka Y, Shibata S, Doi M, Ueda H, Ishida T, Uesugi S. Analysis of the mRNA cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur J Biochem. 1996;239:597–601. doi: 10.1111/j.1432-1033.1996.0597u.x. [DOI] [PubMed] [Google Scholar]
- 23.Jankowska-Anyszka M, Lamphear BJ, Aamodt EJ, Harrington T, Darzynkiewicz E, Stolarski R, Rhoads RE. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated mRNA cap structures. J Biol Chem. 1998;273:10538–10542. doi: 10.1074/jbc.273.17.10538. [DOI] [PubMed] [Google Scholar]
- 24.Keiper BD, Lamphear BJ, Deshpande AM, Jankowska-Anyszka M, Aamodt EJ, Blumenthal T, Rhoads RE. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J Biol Chem. 2000;275:10590–10596. doi: 10.1074/jbc.275.14.10590. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 26.Nagata K, Handa H. Real Time Analysis of Biomolecular Interactions. Springer-Verlag; Tokyo: 1998. [Google Scholar]
- 27.Allen ML, Metz AM, Timmer RT, Rhoads RE, Browning KS. Isolation and sequence of the cDNAs encoding the subunits of the isozyme form of wheat protein synthesis initiation factor 4F. J Biol Chem. 1992;267:23248–23252. [PubMed] [Google Scholar]
- 28.Discovery Studio Version 1.5.1. Accerlys Inc; San diego, CA: 2005. [Google Scholar]
- 29.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 30.Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 31.Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Ishida T, Taniguchi T, Hasegawa H, Terashima A, Sasaki M, Katsuya Y, Kitamura K, Miyoshi H, Ishikawa M, Miura K. Crystal structure of 7-methylguanosine 5′-triphosphate (m7GTP)-and P1-7-methylguanosine-P3-adenosine-5′,5′-triphosphate (m7GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem J. 2002;362:539–544. doi: 10.1042/0264-6021:3620539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyoshi H, Dwyer DS, Keiper B, Jankowska-Anyszka M, Darzynkiewicz E, Rhoads RE. Discrimination between mono- and trimetylated cap structures by two isoforms of Caenorhabditis elegans eIF4E. EMBO J. 2002;21:4680–4690. doi: 10.1093/emboj/cdf473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hershey PEC, McWhirter SM, Gross JD, Wagner G, Alber T, Sachs AB. The cap-binding protein eIF4E promotes folding of a functional domain of yeast translation intiation factor eIF4G1. J Biol Chem. 1999;274:21279–21304. doi: 10.1074/jbc.274.30.21297. [DOI] [PubMed] [Google Scholar]
- 34.Monzingo AF, Dhaliwal S, Dutt-Chaudhuri A, Lyon A, Sadow JH, Hoffman DW, Robertus JD, Browning KS. The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol. 2007;143:1504–1518. doi: 10.1104/pp.106.093146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volpon L, Osborne MJ, Topisirovic I, Siddiqui N, Borden KLB. Cap-free structure of eIF4E suggests a basis for conformational regulation by its ligands. EMBO J. 2006;25:5138–5149. doi: 10.1038/sj.emboj.7601380. [DOI] [PMC free article] [PubMed] [Google Scholar]