A novel gene, csi2+ (chromosome segregation impaired 2), is reported. It localizes to the spindle pole body, and its deletion leads to a transient monopolar spindle and subsequent chromosome lagging. It is proposed that csi2p regulates mitotic microtubule length, defects in which may cause kinetochore–microtubule attachment problems.
Abstract
Proper chromosome segregation is of paramount importance for proper genetic inheritance. Defects in chromosome segregation can lead to aneuploidy, which is a hallmark of cancer cells. Eukaryotic chromosome segregation is accomplished by the bipolar spindle. Additional mechanisms, such as the spindle assembly checkpoint and centromere positioning, further help to ensure complete segregation fidelity. Here we present the fission yeast csi2+. csi2p localizes to the spindle poles, where it regulates mitotic microtubule dynamics, bipolar spindle formation, and subsequent chromosome segregation. csi2 deletion (csi2Δ) results in abnormally long mitotic microtubules, high rate of transient monopolar spindles, and subsequent high rate of chromosome segregation defects. Because csi2Δ has multiple phenotypes, it enables estimates of the relative contribution of the different mechanisms to the overall chromosome segregation process. Centromere positioning, microtubule dynamics, and bipolar spindle formation can all contribute to chromosome segregation. However, the major determinant of chromosome segregation defects in fission yeast may be microtubule dynamic defects.
INTRODUCTION
Absolute fidelity of chromosome segregation is essential for proper cellular development (Siegel and Amon, 2012). Eukaryotic chromosome segregation is achieved by the bipolar spindle, a dynamic structure composed of microtubules, microtubule-associated proteins (MAPs), motors, and other regulatory proteins (Walczak and Heald, 2008; Tanenbaum and Medema, 2010; Meunier and Vernos, 2012). Of key importance is the correct attachment of the chromosome, via its kinetochore, to the microtubules responsible for subsequent chromosome separation to the opposite spindle poles (Verdaasdonk and Bloom, 2011; Foley and Kapoor, 2013). Indeed, cells have evolved the spindle assembly checkpoint (SAC), active at the kinetochore–microtubule interface, to ensure correct chromosome-to-microtubule attachment (Musacchio and Salmon, 2007; Lara-Gonzalez et al., 2012; Vleugel et al., 2012).
The fission yeast Schizosaccharomyces pombe serves as a good model system with which to dissect different aspects of the chromosome segregation pathway, from spindle formation (Hagan and Yanagida, 1992, 1995), to kinetochore structure (Goshima et al., 1999), to checkpoint regulators (He et al., 1997). It is reported that the fission yeast csi1+ regulates chromosome segregation by positioning the centromeres at the spindle pole body (SPB) during interphase, so that when mitosis starts, the chromosomes are efficiently captured by the spindle microtubules (Hou et al., 2012). Alternatively, csi1+ also organizes the bipolar spindle, which is required for proper chromosome segregation (Zheng et al., 2014). Of interest, it was reported in mammalian cells that the precise timing of bipolar spindle formation at mitosis onset is required for proper chromosome segregation, as induced transient monopolar spindles and/or delays in bipolar spindle formation lead to kinetochore–microtubule misattachment and chromosome segregation defects (McHedlishvili et al., 2012; Silkworth et al., 2012). In addition, regulation of microtubule lengths and dynamics also affects chromosome segregation, as depletion of the mitotic kinesin-8 MCAK, which depolymerizes microtubules, and depletion of the microtubule-associated proteins EB1, which stabilizes microtubules, both lead to chromosome segregation defects (Kline-Smith et al., 2004; Lan et al., 2004; Green et al., 2005). Thus proper centromere positioning, timing of bipolar spindle formation, and regulation of microtubule dynamics are all important for subsequent chromosome segregation. However, what are the relative contributions of these different mechanisms?
We report here the fission yeast csi2+. csi2p localizes to the SPB, similar to the reported localization of csi1p, and their localization is interdependent. csi2 deletion (csi2Δ) has similar chromosome segregation defects as reported for csi1Δ. However, csi2Δ does not have the centromere-positioning defects of csi1Δ (Hou et al., 2012), although it does have microtubule and transient monopolar spindle defects (Zheng et al., 2014). Thus csi2Δ uncouples the relative contribution of centromere positioning from microtubule dynamics and bipolar spindle formation, enabling establishment of their respective and relative contributions to chromosome segregation.
RESULTS
We used the S. pombe genome-wide yellow fluorescent protein (YFP)–tagged collection (Matsuyama et al., 2006) and the haploid deletion collection (Kim et al., 2010) to visually identify novel genes whose products localize to the SPB and whose deletions lead to spindle defects. We identified SPBC2G2.14 and SPAC4D7.07C. Consistent with the logic of our screen, we confirmed that the protein products of these genes localize to the SPB, and their deletions lead to transient monopolar spindles and chromosome missegregation (see later description). We thus named SPBC2G2.14 and SPAC4D7.07C as csi1+ and csi2+ (chromosome segregation impaired 1 and 2), respectively. The role of csi1+ in centromere–SPB anchoring has been described (Hou et al., 2012), as has its role in bipolar spindle formation (Zheng et al., 2014). Here we focus on the function of csi2+.
csi2Δ has spindle assembly defects
csi2Δ yielded viable cells. However, csi2Δ cells were sensitive to the microtubule-depolymerizing drug methyl benzimidazol-2-yl carbamate (MBC; Supplemental Figure S1A), suggesting that csi2Δ cells have defects in the microtubule cytoskeleton. We thus examined microtubule organization and dynamics in csi2Δ and wild-type cells expressing mCherry-atb2p (tubulin). No striking differences in interphase microtubules were observed in csi2Δ and wild-type cells. However, spindle microtubule organization and dynamics were markedly different in csi2Δ cells compared with wild type. In wild type, entry into mitosis is concurrent with the disassembly of cytoplasmic interphase microtubules (Hagan, 1998). To standardize measurements of mitosis time, we define the start of mitosis, time 0 min, as the complete disassembly of cytoplasmic interphase microtubules (Figure 1A). In wild type, time 0 min coincides with the assembly of a microtubule “bar” (83% of cells) or a “dot” (17% of cells) that quickly transitions into a bar (n = 18), representing the bipolar spindle (Figure 1, A and B). In contrast, only 18% of csi2Δ cells exhibited bars at time 0 min. The rest exhibited delayed bipolar spindle formation (Figure 1A), where the spindle dot occurred more frequently (60% of cells) and took longer to form bars (n = 51; Figure 1, A and B). Of interest, 22% of csi2Δ cells formed transient microtubule protrusions defined as monopolar spindle (mono; Figure 1, A and B). These microtubule protrusions emanated from both mother and daughter SPB (Figure 1D). Whereas wild-type microtubule dots quickly transitioned into bars (<1 min), the csi2Δ dots took significantly longer (2.8 ± 2.1 min; n = 43; p < 10−5); and the csi2Δ mono spindles persisted 5.3 ± 4.2 min (n = 11) before becoming the bipolar bar (Figure 1C). No wild-type cells exhibited monopolar spindles.
FIGURE 1:

csi2p organizes the prophase bipolar spindle. (A) Time-lapse images of wild-type and csi2Δ mitotic cells expressing mCherry-atb2p (tubulin). Wild-type cells typically show a stable bipolar spindle (bar) within 1 min after the start of mitosis, when interphase microtubules have completely depolymerized (time = 0 min). In contrast, some csi2Δ cells exhibit a transient prophase monopolar spindle (mono) or a faint dot (dot) instead of the bar. Other csi2Δ cells show no microtubule signal at mitotic entry (yellow box). Scale bar, 5μm. (B) Comparison of different spindle structures (bar, mono, dot) for wild-type and csi2Δ cells. Eighty-three percent of wild-type cells (n = 18) exhibit the bipolar spindle (bar) compared with 18% of csi2Δ cells (n = 51). Wild-type cells have zero monopolar spindles (mono), compared with 22% of csi2Δ cells. Wild-type cells have 17% of dot spindles (dot) that persisted <1min, compared with csi2Δ cells having 60% of dot spindles that persisted up to 10 min (p < 10−62; see C). (C) Box-and-dot plot comparison of duration of mono and dot spindle persistence in wild-type and csi2Δ cells. Wild-type cells exhibit zero mono spindles (n = 18), and the dot spindles are transient and persist for <1 min (n = 3). In contrast, csi2Δ cell mono spindles persist for 5.3 ± 4.2 min (n = 11), and dot spindles persist for 2.8 ± 2.1 min (n = 43; p < 10−5). (D) Images of wild-type and csi2Δ prophase mitotic cells expressing mCherry-atb2p and alp4p-GFP (γ-tubulin complex protein, marking the SPB). The wild-type cell shows a well-organized microtubule bar spindle between the two SPBs. In contrast, the csi2Δ cell, in addition to having the bar spindle, has microtubules protruding from both SPBs, suggesting that both mother and daughter SPBs are competent microtubule nucleators. Scale bar, 2 μm. (E) Time-lapse images of wild-type and csi2Δ mitotic cells expressing mCherry-atb2p and cut7p-GFP (kinesin-5; Hagan and Yanagida, 1992). For both wild type and csi2Δ, cut7p is recruited to the spindle approximately at the same time before the onset of mitosis (time = 0 min). Scale bar, 5 μm.
We note that csi1 deletion (csi1Δ) cells also yielded delay in bipolar spindle formation similar to csi2Δ (Supplemental Figure S1B), with 95% of cells exhibiting the transient monopolar microtubule protrusion phenotype and 5% exhibiting the transient dot phenotype. Monopolar spindle defects were recently observed in csi1Δ (Zheng et al., 2014). Here we highlight only similar roles of csi1p and csi2p in bipolar spindle formation.
It is known that the kinesin-5 cut7p localizes to the spindle and is essential for bipolar spindle formation (Hagan and Yanagida, 1992; Fu et al., 2009). We compared the recruitment of cut7–green fluorescent protein (GFP) to the spindle in wild-type and csi2Δ cells. Both wild-type and csi2Δ cells exhibited similar recruitment time of cut7p to the spindle, approximately 4 min before mitosis onset or t = 0 min (Figure 1E and Supplemental Figure S1C). Nevertheless, wild type took 5.1 ± 1.4 min (n = 32) after cut7p arrival to form a bipolar spindle bar, in contrast to csi2Δ, which took 7.4 ± 2.0 min (n = 15; p < 10−3; Supplemental Figure S1D). Taking the results together, we conclude that csi2p (and csi1p) functions in bipolar spindle formation. The observed defects in the bipolar spindle are not due to lack or delay of kinesin-5 recruitment to the spindle at the onset of mitosis.
csi2Δ has chromosome segregation defects
In wild-type cells, once spindle bipolarity has been achieved, the spindle elongates to its steady-state metaphase spindle length (Syrovatkina et al., 2013). Using cdc13-GFP (cyclin) signal degradation as a marker for the metaphase-to-anaphase transition (Tatebe et al., 2001), we compared the final metaphase spindle lengths of wild-type and csi2Δ cells (Figure 2, A and C). Whereas the total duration of mitosis was relatively similar between wild type (35.5 ± 4.2 min, n = 12) and csi2Δ (36.5 ± 5.8 min, n = 12, p = 0.65; Figure 2C), metaphase spindle lengths were different. Wild type had metaphase spindle length of 2.93 ± 0.37 μm (n = 16), significantly shorter than csi2Δ length of 4.30 ± 0.52 μm (n = 14, p < 10−6; Figure 2, B and C). We also observed that the csi2Δ metaphase spindles were not stable in length, but continued to slowly elongate (Figure 2C).
FIGURE 2:
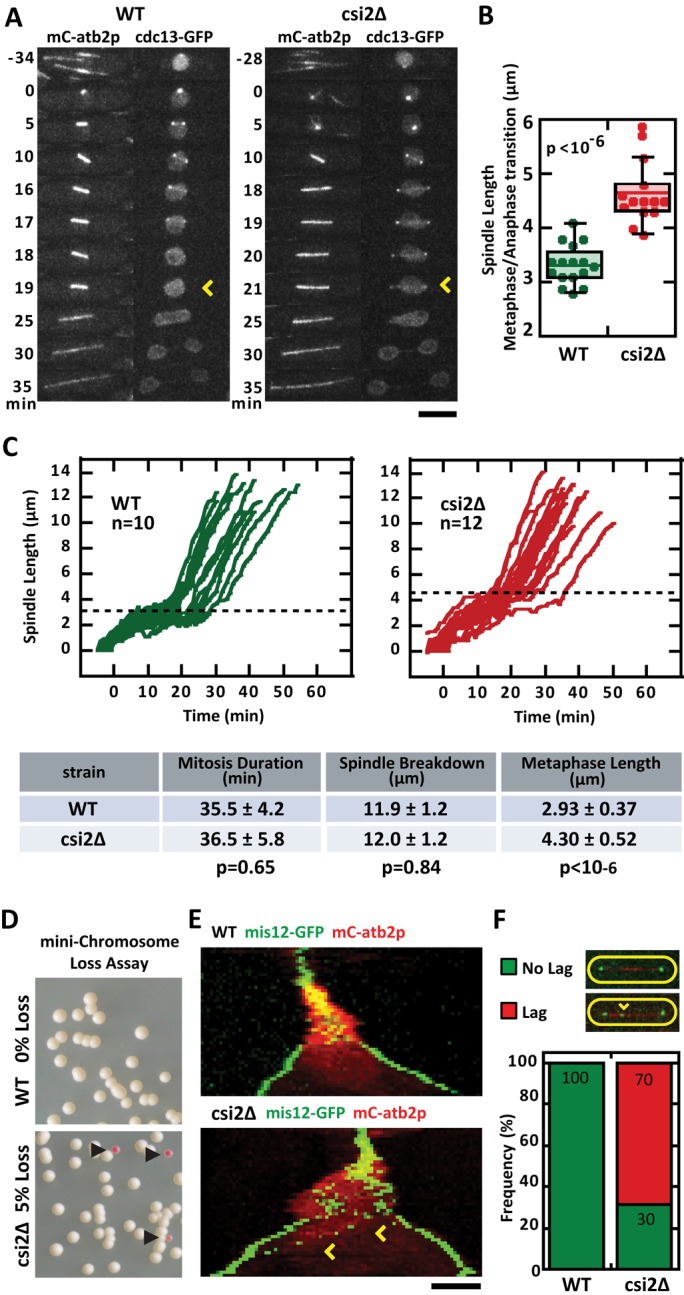
csi2p regulates metaphase spindle length and chromosome segregation. (A) Time-lapse images of wild-type and csi2Δ mitotic cells expressing mCherry-atb2p and cdc13p-GFP (cyclin B; Tatebe et al., 2001). cdc13p is degraded at the metaphase–anaphase transition (yellow arrow), marking the final length of the metaphase spindle. In the wild-type cell the final metaphase spindle (time = 18 min) was shorter than that of the csi2Δ cell (time = 20 min). Scale bar, 5 μm. (B) Box-and-dot plot comparison of final metaphase spindle length for wild-type and csi2Δ cells. Wild-type spindle length was 3.3 ± 0.4 μm (n = 16), compared with 4.7 ± 0.6 μm (n = 14) in csi2Δ (p < 10−6). A longer metaphase spindle suggests spindle checkpoint delay and/or misregulation of microtubule dynamics. (C) Spindle length vs. time for wild-type and csi2Δ cells. The proceeding table summarizes mitosis duration, spindle final length at end of mitosis breakdown, and metaphase spindle length. The duration of mitosis was the same for wild-type and csi2Δ cells (p = 0.65), suggesting that the spindle checkpoint is satisfied relatively quickly in csi2Δ cells. (D) Minichromosome loss assay (Niwa et al., 1989) for wild-type and csi2Δ cells. Wild-type cells exhibited all-white colonies on selection plates (n > 300), suggesting none or very low (<0.3%) minichromosome loss. In contrast, 5% (n > 300) of csi2Δ colonies were pink, indicating chromosome segregation defects (p < 0.05). (E) Kymograph comparison of wild-type and csi2Δ cells expressing mCherry-atb2p and mis12p-GFP (kinetochore marker; Goshima et al., 1999). By the start of anaphase B, when the spindle exhibits fast elongation, all kinetochores are properly segregated to the spindle poles in the wild-type cell. In contrast, the csi2Δ cell exhibits lagging kinetochores (yellow arrow) during anaphase B, suggesting improper kinetochore–microtubule attachment during mitosis. (F) Bar plot comparison of lagging kinetochores in wild-type and csi2Δ cells. Lagging kinetochores were not observed in wild-type cells (n = 19). In contrast, 70% (n = 32) of csi2Δ cells exhibited lag kinetochores (p < 10−49).
Spindle defects are known to correlate with chromosome segregation defects (Goshima and Scholey, 2010). We thus probed for chromosome segregation defects in wild-type and csi2Δ cells. First, using the artificial minichromosome loss color assay (Niwa et al., 1989), we observed that wild type had zero minichromosome loss (<0.003%, n = 300), represented by the white colonies, compared with 5% (n = 300, p < 0.02) of csi2Δ cells that had minichromosome loss, represented by the pink colonies (Figure 2D). Second, using either the kinetochore marker mis12-GFP (Goshima et al., 1999) or the centromere marker CEN1-GFP (Nabeshima et al., 1998), we observed kinetochore “lagging” in 70% of csi2Δ cells during anaphase B when the spindle length increased dramatically, compared with zero lagging in wild-type cells (Figure 2, E and F, and Supplemental Figure S2A).
Lagging kinetochores are indicative of microtubule–kinetochore attachment defects, which suggests that the spindle assembly checkpoint would have been activated (May and Hardwick, 2006). We thus monitored for checkpoint time delay before anaphase. In wild type, the duration from mitotic onset to the end of metaphase was 16.7 ± 3.0 min (n = 19), which has no statistical significance compared with 19.1 ± 4.6 min (n = 20; p = 0.06) for csi2Δ (Supplemental Figure S2B). This is consistent with total mitosis duration being similar between wild type and csi2Δ (Figure 2C). Nevertheless, in the absence of either of the three core SAC proteins mad2p, bub3p, and mph1p (May and Hardwick, 2006), csi2Δ cells exhibited cell death at progressively higher temperature (Supplemental Figure S2C), indicating that in the absence of the SAC, csi2Δ cells failed to segregate their chromosomes.
sad1p and csi1p are required for csi2p localization to the spindle pole body
We next examined csi2p localization throughout the cell cycle. Fluorescent tagging of csi2p at its native locus revealed that csi2p localizes to the SPB during interphase and mitosis (Figure 3A and Supplemental Figure S3A), consistent with the previous genome-wide YFP-tagged overexpression study (Matsuyama et al., 2006). csi2p localization at the SPB was further confirmed by its colocalization with other known SPB-localized proteins, such as sad1p (Hagan and Yanagida, 1995) and sid4p (Chang and Gould, 2000; Figure 3B and Supplemental Figure S3A). The conserved essential SUN-domain inner nuclear membrane protein sad1p has been proposed to act as a scaffold for the recruitment of other proteins to the SPB (Hiraoka and Dernburg, 2009). Using the temperature-sensitive mutant sad1.1ts, in which sad1p is inactivated at the nonpermissive temperature of 37°C, we observed no csi2p-mCherry signal at the SPB (Figure 3C). However, sad1-YFP signal was still present at the SPB in csi2Δ cells (Figure 3D). The result suggests that sad1p directly or indirectly recruits csi2p to the SPB.
FIGURE 3:
csi2p localizes to the SPB throughout the cell cycle. (A) Kymograph of a wild-type mitotic cell expressing mCherry-atb2p and csi2-GFP. csi2p localizes to the SPB throughout mitosis. Scale bar, 2 μm. (B) Images of a wild-type cell expressing sad1-YFP and csi2-mCherry. csi2p colocalizes with sad1p, the SUN-domain protein at the SPB. Scale bar, 5 μm. (C) Images of wild-type and temperature-sensitive sad1.1ts cells expressing csi2-mCherry. At the nonpermissive temperature (37°C), sad1p is inactivated (Hagan and Yanagida, 1995), and csi2p is not detected at the SPB, suggesting that sad1p recruits csi2p to the SPB. Scale bar, 5 μm. (D) Images of csi2∆ cells expressing sad1-YFP. sad1p localization to the SPB does not require csi2p. Scale bar, 5 μm. (E) Images of wild-type and csi1∆ cells expressing csi2-GFP. csi2p localization to the SPB is dependent on csi1p. Scale bar, 5 μm. (F) Time-lapse images of wild-type and csi2∆ cells expressing mCherry-atb2p and csi1-GFP. csi1p localization to the SPB is dependent on csi2p only at early mitosis. In contrast, during interphase and late anaphase, csi1p can localize to the SPB in the absence of csi2p. Scale bar, 5 μm. (G) Images of wild-type, csi1∆, and csi2∆ cells expressing mCherry-atb2p and mis12-GFP (kinetochore marker; Goshima et al., 1999). In csi1∆ interphase cells, kinetochores are declustered from the SPB (yellow arrowhead), represented by >1 dot of signal. In contrast, in both wild-type and csi2∆ cells, kinetochores are clustered at the SPB, represented by 1 one dot of signal. Scale bar, 5 μm. (H) Bar plot quantification of kinetochore clustering in interphase wild-type, csi1∆, and csi2∆ cells. In wild type, 96% of cells exhibit proper kinetochore clustering (1 dot) at the SPB (n = 51), and 4% have declustered kinetochore (>1 dot). In csi1∆, 62% of cells have proper kinetochore clustering, whereas 38% have declustered kinetochores (n = 139; p < 10−69). In csi2Δ, 98% of cells have proper kinetochore clustering, whereas only 2% have declustered kinetochores (n = 129), which is similar to wild type (p = 0.23).
It was shown that csi1p localizes to the SPB throughout the cell cycle, and its deletion leads to chromosome segregation defects (Hou et al., 2012; Zheng et al., 2014), very similar to the localization and function of csi2p. We thus checked SPB-localization dependence between csi1p and csi2p. In the absence of csi1+, no csi2-GFP signal was observed at the SPB (Figure 3E). Of interest, in the absence of csi2+, csi1-GFP failed to localize to the spindle SPB precisely at mitosis onset until anaphase, after which time, csi1-GFP signal gradually returned to the SPB (Figure 3F). Thus csi2p requires csi1p to be recruited to the SPB throughout the cell cycle. However, csi1p is only dependent on csi2p for recruitment to the SPB specifically during mitosis. Accordingly, we observed that the monopolar spindle defect in csi1Δ is dominant over csi2Δ cells. Whereas csi2Δ cells showed 21% transient monopolar spindles, the double-deletion csi1Δcsi2Δ cells showed 88%, which is similar to csi1Δ cells, at 95% (Supplemental Figure S3B).
In fission yeast, the three chromosomes are clustered at the interphase SPB via direct coupling between the centromere and the SPB (Kniola et al., 2001). csi1p was shown to be a coupler of centromere to SPB, as csi1Δ cells exhibited declustered centromeres (Hou et al., 2012). Probable direct or indirect interaction between csi1p and csi2p prompted us to test for centromere declustering in csi2Δ cells using the centromere (kinetochore) marker mis12-GFP (Goshima et al., 1999). Surprisingly, whereas 38% of csi1Δ interphase cells showed declustered centromeres, represented by >1 dot of mis12-GFP, only 2% (n = 129) of csi2Δ interphase cells showed declustered centromeres, which is similar to the 4% seen in wild-type cells (n = 51, p = 0.3; Figure 3, G and H). This suggests that both csi1Δ and csi2Δ have monopolar spindles and chromosome segregation defects, but only csi1Δ has the additional phenotype of centromere declustering. Thus the csi2Δ mutant becomes a useful tool to address the relative contributions of centromere declustering and/or monopolar spindle to chromosome segregation defects.
Centromere declustering and monopolar spindle phenotypes need not correlate with chromosome segregation defects
It was proposed that the centromere-declustering phenotype observed in csi1Δ cells would attenuate efficient kinetochore–microtubule attachment, resulting in chromosome segregation defects (Hou et al., 2012). Alternatively, transient monopolar spindle defects, such as seen in csi1Δ cells, have also been proposed to result in chromosome segregation defects (McHedlishvili et al., 2012; Silkworth et al., 2012; Zheng et al., 2014). To determine the relative contributions of these two phenotypes to the observed chromosome segregation defects would require uncoupling the two phenotypes. csi2Δ cells maintained csi1-GFP at the interphase SPB (Figure 3F), and therefore centromeres also remained clustered at the SPB (Figure 3G). However, csi2Δ exhibited monopolar spindles (Figure 1, A– D), similar to csi1Δ (Supplemental Figure S1B). Thus csi2Δ uncouples the interphase centromere declustering from the monopolar spindle phenotype. That we observed lagging kinetochores in csi2Δ cells (Figure 2, E and F, and Supplemental Figure S2B), which have properly clustered centromeres (Figure 3, G and H), indicates that centromere declustering, such as observed in csi1Δ cells (Figure 3G), is not the sole contributor to chromosome segregation defects.
We next determined whether the transient monopolar spindle phenotype observed in csi2Δ cells can lead to chromosome segregation defects. We tracked individual csi2Δ mitotic spindles and found no correlation between monopolar spindles and chromosome segregation defects. A csi2Δ cell with starting monopolar spindle can still have subsequent proper kinetochore segregation at anaphase (Figure 4A). In contrast, a csi2Δ cell with seemingly normal bipolar spindle may still exhibit kinetochore lagging (Figure 4A). We observed that csi2Δ defective spindles, either dot or monopolar, were equally likely to have lagging kinetochores or no lagging kinetochores at anaphase. In a population of mitotic csi2Δ cells, of 17 spindles that were initially dots, 11 showed lagging kinetochores and six did not; of 18 spindles that were monopolar, eight showed lagging kinetochores and 10 did not (Figure 4B). Of note, of the 5 csi2Δ spindles that had relatively “normal” bipolar spindles, represented by bars, all exhibited lagging kinetochores (Figure 4B). Taking the results together, we conclude that individually neither the interphase centromere declustering (Hou et al., 2012) nor the monopolar spindle (McHedlishvili et al., 2012; Silkworth et al., 2012; Zheng et al., 2014) hypothesis can completely account for the chromosome segregation defects seen in csi2Δ cells. There have to be additional mechanisms.
FIGURE 4:
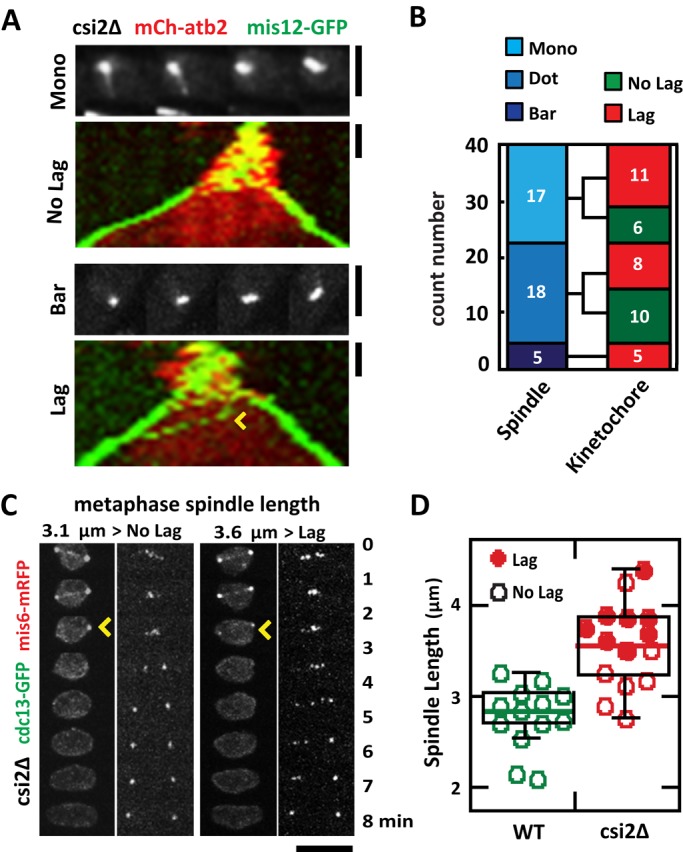
Abnormally long metaphase spindle length in csi2Δ correlates positively with chromosome segregation defects. (A) Initial monopolar spindle defects do not correlate with subsequent chromosome segregation defects. Shown are csi2∆ cells expressing mCherry-atb2p and mis12-GFP. Images show the initial state of the spindle, and kymographs show spindle and kinetochore dynamics. Top, an initial monopolar spindle, which subsequently exhibits no kinetochore lagging at anaphase B. Bottom, in contrast, a seemingly “normal” initial bipolar spindle, which subsequently exhibits kinetochore lagging (yellow arrowhead). Scale bar, 2 μm. (B) Bar plot quantification of the initial spindle structure (mono, dot, and bar) and subsequent anaphase B kinetochore dynamics (NO Lag, Lag) in csi2∆ cells. No correlation exists between the initial state of the spindle and subsequent kinetochore lagging (n = 40). (C) Time-lapse images of csi2Δ cells expressing cdc13-GFP and mis6-2mRFP (kinetochore marker). cdc13p (cyclin B) is degraded at the metaphase/anaphase transition (yellow arrowhead). In the left csi2Δ cell, the final metaphase pole-to-pole distance is 3.1 μm, and the cell has no kinetochore lag. In contrast, the right csi2Δ cell has pole-to-pole distance of 3.6 μm, and it has kinetochore lag. Scale bar, 5 μm. (D) Box-and-dot plot comparison between final metaphase spindle length and kinetochore lagging at anaphase B for wild-type and csi2Δ cells. Wild-type final metaphase spindle length is 2.8 ± 0.4 μm (n = 13), with no cell exhibiting kinetochore lagging. In contrast, csi2Δ final metaphase spindle length is 3.3 ± 0.5 μm (n = 15). Of these, the shorter spindles tend to have no kinetochore lag, and the longer spindles tend to have kinetochore lag.
Abnormally long metaphase spindles correlate with chromosome segregation defect
We noted that csi2Δ cells have significantly longer metaphase spindles compared with wild type (Figure 2, A– C). Abnormally long metaphase spindles are often a consequence of a defective force-balance mechanism for maintaining spindle length, resulting from defects of kinetochore–microtubule attachment and leading to chromosome segregation defects (Goshima and Scholey, 2010; Syrovatkina et al., 2013). We thus tracked wild-type and csi2Δ metaphase spindle lengths using cdc13-GFP, whose degradation marks the start of anaphase (Tatebe et al., 2001), and centromere lagging using the kinetochore marker mis6-RFP (Goshima et al., 1999). We found a strong correlation between abnormally long metaphase spindle and kinetochore lagging at anaphase (Figure 4, C and D). Wild-type metaphase spindle lengths ranged from 2 to 3 μm, with a median length of 2.8 μm (Figure 4D). None of the 13 observed wild-type spindles showed lagging kinetochores (Figure 4D). In contrast, csi2Δ final metaphase spindle lengths ranged from 3 to 4.5 μm, with a median length of 3.5 μm (Figure 4D). In the csi2Δ spindles, eight of 15 showed lagging kinetochores, and all of these spindles are equal to or above the median length (Figure 4D). Only one spindle above the median length did not exhibit centromere lagging (Figure 4D). Thus, abnormally long metaphase spindle length positively correlates with lagging kinetochores.
Abnormally long metaphase spindle lengths observed in csi2Δ indicate that spindle microtubules are also longer than wild type, which suggests that microtubule dynamics are perturbed in csi2Δ cells. Because the fission yeast spindle contains many individual microtubules (Ding et al., 1993), it is currently not possible to monitor individual microtubule dynamics within the bipolar spindle structure using optical microscopy. However, we reasoned that the monopolar spindle resulting from kinesin-5 cut7.24ts inactivation, which is not expected to alter microtubule dynamics (Hagan and Yanagida, 1992; Fu et al., 2009), would enable measurements of individual microtubules emanating from the spindle poles (Costa et al., 2013). We thus compared individual microtubule dynamics between cut7.24ts (control) and cut7.24ts csi2Δ cells expressing mCherry-atb2 (tubulin). At the nonpermissive temperature 37°C, control and csi2Δ cells exhibited the expected monopolar spindles, with microtubule protrusions likely composed of multiple microtubules (p = 0.7; Figure 5A and Supplemental Figure S4A). The number of microtubule protrusions was similar between control and csi2Δ monopolar spindles (Figure 5B), with control cells having 3 ± 1 (n = 18) microtubule bundles and csi2Δ cells also having 3 ± 1 (n = 19) microtubule bundles. However, the microtubule protrusions are longer in the csi2Δ cells compared with control (Figure 5C). Whereas the control cells have mitotic microtubule length of 0.7 ± 0.3 μm (n = 53), csi2Δ cells have mitotic microtubule length of 1.2 ± 0.3 μm (n = 59), ∼40% longer than control (p < 10−10).
FIGURE 5:
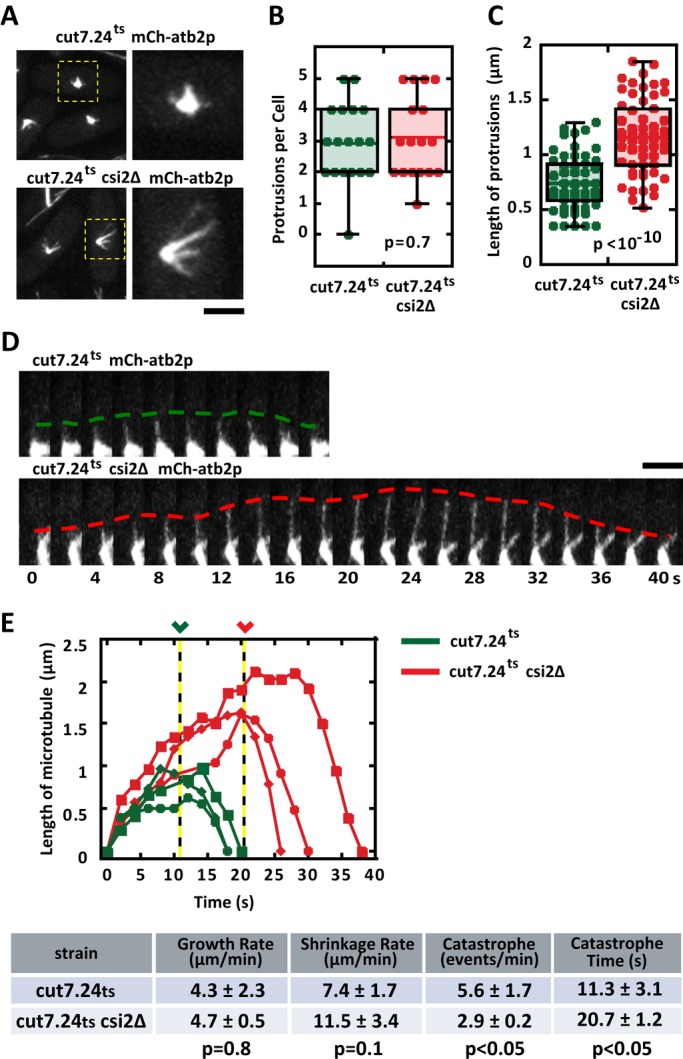
csi2p regulates mitotic microtubule length. (A) Images of cut7.24ts (control) and cut7.24tscsi2Δ cells expressing mCherry-atb2p. At the nonpermissive temperature (37°C), cut7.24ts cells fail to form bipolar spindles, but instead make monopolar spindles, which enable measurements of individual mitotic microtubule dynamics (Costa et al., 2013). Control cut7.24ts monopolar spindles exhibit relatively shorter microtubules than cut7.24tscsi2Δ. Scale bar, 2 μm. (B) Box-and-dot plot comparison of the number of mitotic microtubule bundles protruding from the monopolar spindles of cut7.24ts and cut7.24tscsi2Δ cells at 37°C. Control cut7.24ts cells have 3 ± 1 (n = 18) microtubule protrusions, and cut7.24tscsi2Δ cells also have 3 ± 1 (n = 19) microtubule protrusions (p = 0.7). (C) Box-and-dot plot comparison of the lengths of the microtubule protrusions from cut7.24ts and cut7.24tscsi2Δ cells at 37°C. Control cut7.24ts microtubule protrusion length is 0.7 ± 0.3 μm (n = 53). In contrast, cut7.24tscsi2Δ microtubule protrusion length is 1.2 ± 0.3 μm (n = 59; p < 10−10), suggesting that csi2p regulates mitotic microtubule lengths. (D) Time-lapse images of single microtubule dynamics emanating from the monopolar spindles of cut7.24ts and cut7.24tscsi2Δ cells at 37°C. In the control cut7.24ts cell, the mitotic microtubule elongates up to 1 μm in length before undergoing catastrophe after 10 s of growth and shrinks back to the spindle pole. In contrast, the cut7.24tscsi2Δ mitotic microtubule elongates up to 2 μm in length before undergoing catastrophe after 20 s of growth. Scale bar, 2 μm. (E) Length vs. time of individual microtubule dynamics of control cut7.24ts (green) and cut7.24tscsi2Δ cells (red). In control cut7.24ts cells, the mitotic microtubules grow for 11.3 ± 3.1 s (n = 3) before undergoing catastrophe. In contrast, cut7.24tscsi2Δ mitotic microtubules grow for 20.7 ± 1.2 s (n = 3), approximately twice as long (p < 0.05). The proceeding table summarizes the growth rate, shrinkage rate, catastrophe frequency, and time before catastrophe of control and csi2Δ cells.
When we could unambiguously determine a single microtubule, based on homogeneous fluorescence intensity along the entire length of the microtubule, we measured its length over time (Figure 5D). Although microtubule growth and shrinkage velocities were similar for control and csi2Δ cells (Figure 5E), spindle microtubules of csi2Δ cells consistently grew to longer lengths (∼2 μm) compared with control (∼1 μm; Figure 5, D and E). Of importance, the time until microtubule catastrophe was twice as long for csi2Δ cells (21 ± 1 s, n = 3) as for control (11 ± 3 s, n = 3; p < 0.05; Figure 5E), and the frequency of microtubule catastrophe was reduced by half in csi2Δ cells (2.9 ± 0.2 min−1, n = 3) compared with control (5.6 ± 1.7 min−1, n = 3; p < 0.05; Figure 5E). Of note, we observed that csi2Δ monopolar spindles had less microtubule fluorescence signal and less signal area than control cells (Supplemental Figure S4A). Whereas control spindle fluorescence signal was 2951 ± 615 a.u. (n = 24), for csi2Δ it was 2009 ± 526 a.u. (n = 16), or 32% less than control (p < 10−5; Supplemental Figure S4B). Similarly, whereas control spindle area was 158 ± 22 pixels squared (n = 24), for csi2Δ it was 114 ± 39 pixels squared (n = 16), or 28% less than control (p < 10−3). We conclude that csi2Δ cells have defects in spindle microtubule dynamics, leading to the abnormally long metaphase spindle and resulting, in large part, in the observed chromosome segregation defects.
DISCUSSION
csi2+ is a new gene involved in microtubule dynamics, bipolar spindle formation, and chromosome segregation. Our results suggest that sad1p recruits both csi2p and csi1p to the SPB, and csi2p and csi1p localizations at the SPB are partially interdependent. We attempted to show probable physical interaction between csi1p and csi2p by coimmunoprecipitation but without success (unpublished data), likely due to the fact that csi2p is a nuclear protein with a predicted transmembrane domain.
Although both csi2Δ and csi1Δ have transient monopolar spindle defects (Zheng et al., 2014) and chromosome segregation defects (Hou et al., 2012), csi2Δ does not have centromere-positioning defects exhibited by csi1Δ. Thus csi2Δ uncouples the two different phenotypes observed in csi1Δ. This enabled us to probe the respective contributions of centromere positioning (Hou et al., 2012) and bipolar spindle formation (McHedlishvili et al., 2012; Silkworth et al., 2012; Zheng et al., 2014) to chromosome segregation. That we observed chromosome segregation defects in csi2Δ, where centromere positioning is normal, suggests that centromere declustering seen in csi1Δ may not be the only cause of subsequent chromosome segregation defects.
Surprisingly, our results show that there is no strong correlation between the transient monopolar spindle defects observed in csi2Δ and subsequent chromosome segregation defects. Indeed, transient monopolar spindles in csi2Δ can have no chromosome segregation defects, and “normal” bipolar spindles can have high chromosome segregation defects. Thus, in contrast to mammalian cells (McHedlishvili et al., 2012; Silkworth et al., 2012), fission yeast transient monopolar spindle defects may not directly cause chromosome segregation defects. However, we cannot currently rule out the possibility that transient monopolar spindle defects partially contribute to chromosome segregation defects. Of interest, unlike mammalian cells, the centromeres of fission yeast are normally clustered at the SPB (Kniola et al., 2001), close proximity to the nascent spindle to facilitate being captured. Declustered centromeres may therefore delay or make defective chromosome capture by microtubules.
Our results show a strong correlation between the abnormally long metaphase spindle length and chromosome segregation defects observed in csi2Δ. Failure to control the metaphase spindle length is known to lead to chromosome segregation defects (Goshima and Scholey, 2010). It is also believed that the metaphase spindle length is control by a force-balance mechanism by which opposing forces produced by motors and MAPs located at the spindle midzone and the kinetochore dictate the final length (Goshima and Scholey, 2010; Syrovatkina et al., 2013). That we observed longer mitotic microtubules in csi2Δ suggests that csi2p regulates microtubule dynamics. That csi2p localizes to the SPB suggests that its microtubule length regulation occurs at the microtubule minus ends. How csi2p regulates microtubule length is not known. However, our observation that monopolar spindles of csi2Δ cells have fewer microtubule signals than wild-type cells suggests that there may be less microtubule nucleation in csi2Δ. Further, it is reported that mutations in fission yeast γ-tubulin, which is a nucleator for microtubules, results in abnormally long microtubules (Paluh et al., 2000). In this context, csi2p may act in the same pathway as γ-tubulin to ensure proper microtubule nucleation or proper microtubule lattice structure. Thus the longer metaphase spindle length observed in csi2Δ, in addition to being regulated by the force-balance mechanism, may also be regulated by microtubule dynamics itself. How do csi2Δ's abnormally long prophase and metaphase microtubules lead to chromosome segregation defects? It may be that the long microtubules are able to reach both sister kinetochores, resulting in merotelic attachment, which would satisfy the spindle assembly checkpoint and explain a lack of spindle delay and the numerous lagging kinetochores.
Finally, our results suggest a model in which multiple mechanisms contribute to ensure proper chromosome segregation. csi2p and csi1p may act as a complex that performs multiple functions, with centromere positioning, microtubule dynamics regulation, and bipolar spindle formation all contributing to chromosome segregation. Our analyses revealed that 40% of csi1Δ cells have declustered centromeres at interphase, yet 95% of cells have lagging chromosomes at anaphase B (Zheng et al., 2014). This implies that centromere declustering only partially contributes to chromosome segregation defects. Of interest, csi2Δ cells have no declustered centromeres but still have 70% of cells with lagging chromosomes. This implies that the declustered centromeres seen in csi1Δ cells contribute 25% of the chromosome segregation defects and that transient monopolar spindle and/or longer metaphase microtubules seen in both csi1Δ and csi2Δ cells account for the remaining 70% chromosome segregation defects. We do not see a strong correlation between monopolar spindles and subsequent chromosome lagging. Instead, we see a very strong correlation between abnormal spindle lengths, a consequence of defective mitotic microtubule dynamics, and subsequent chromosome lagging. Thus we favor a model in which defective mitotic microtubule dynamics seen in csi1Δ and csi2Δ accounts for 70% of chromosome segregation defects. Sequence homology revealed that csi1+ and csi2+ are both unique to fission yeast. However, given its important and diverse roles during mitosis, functional homology seems likely in higher eukaryotes.
MATERIALS AND METHODS
S. pombe strains and plasmid construction
Standard yeast media and genetic methods were used to create yeast strains, as previously described (Moreno et al., 1991; Forsburg and Rhind, 2006). Strains of csi2 deletion and GFP/mCherry tagging were carried out by a previously described PCR-based method (Bahler et al., 1998).
Screen for csi2+
We used the S. pombe genome-wide YFP-tagged overexpression collection (Matsuyama et al., 2006) and the haploid deletion collection (Kim et al., 2010) to identify novel genes whose products localize to the SPB and whose deletions lead to spindle defects. The novel gene SPAC4D7.07C was found to have spindle defects and chromosome segregation defects. We thus named this gene psr2+ (poles separation regulator 2), but subsequently renamed it csi2+ (chromosome segregation impaired 2) to be consistent with published convention.
Minichromosome loss assay
A strain containing the artificial minichromosome Ch16 was introduced into wild-type and csi2Δ cells by mating and random spore analysis, and selection was carried out on minimum media EMM plates lacking the selection marker adenine, according to the published protocol (Niwa et al., 1989). Equal amount of wild-type and csi2Δ cells carrying the minichromosome were plated on selective EMM plates lacking adenine. The plates were incubated at 30°C for 4 d, and colonies were examined for the color red, which indicated minichromosome loss.
Microtubule drug sensitivity assay
Wild-type and csi2∆ cells were grown in YE5S (yeast extract + 5 amino acid supplements) medium to mid log phase (OD600 nm = 0.5), and then a series of fourfold cell dilutions was spotted onto agar plates containing YE5S plus 4 μg/ml MBC (Sigma-Aldrich, St. Louis, MO). These plates were incubated at 30°C for 3 d and then assayed for colony growth.
Microscopy
Live-cell imaging was carried out at either room temperature (21°C) or 37°C, when temperature-sensitive mutants were used. We used a spinning-disk confocal microscope equipped with a PlanApo 100×/1.40 numerical aperture objective (Nikon, Melville, NY) and an ORCA charge-coupled device (CCD) camera (Hamamatsu, Hamamatsu, Japan) or ImagEM electron-multiplying CCD camera (Hamamatsu) as previously described (Tran et al., 2004). MetaMorph 7.5 (Molecular Devices, Sunnyvale, CA) was used to acquire and process all images.
For high temporal resolution, images were acquired at 200-ms exposures for mCherry at 2-s intervals, with each stack comprising four optical sections of 0.5-μm spacing. For longer time scale, images were acquired at 500- or 1000-ms exposures for GFP and mCherry at 1-min intervals, with each stack comprising 11 optical sections of 0.5-μm spacing.
The temperature-sensitive strain sad1-1ts (Hagan and Yanagida, 1995) was incubated in YE5S medium for 3 h at 37°C before imaging. The temperature-sensitive strain cut7.24ts (Hagan and Yanagida, 1992) was incubated in YE5S medium for 20 min at 37°C before imaging.
Data analysis
Data are presented as mean ± SD or as frequency. Statistical analyses on means were performed using the Student's t test. Statistical analyses on frequencies were performed using the χ2 test. All analyses were performed using Excel 2010 (Microsoft, Redmond, WA). All plots were created using KaleidaGraph 4.0 (Synergy Software, Reading, PA). Dot-and-box plots show all individual data points, and the plots enclose 50% of the data in the box, with the median value displayed as a line. The lines extending from the top and bottom of each box mark the minimum and maximum values within the data set that fall within an acceptable range. Outliers are displayed as individual points.
Temperature sensitivity assay
Wild-type, csi2∆, mad2∆, bub3∆, mph1∆, csi2∆.mad2∆, csi2∆.bub3∆, and csi2∆.mph1∆ cells were grown in YE5S medium to mid log phase (OD600 nm = 0.5), and then a series of fivefold cell dilutions was spotted onto YE5S agar plates. These plates were incubated at 25°C for 5 d, or at 30 or 37°C for 3 d, and then assayed for colony growth.
Supplementary Material
Acknowledgments
We thank A. Stout of the University of Pennsylvania CDB Microscopy Core for technical assistance; the laboratories of I. Hagan (University of Manchester, Manchester, United Kingdom), J. R. McIntosh (University of Colorado, Boulder, CO), T. Toda (Cancer Research UK, London, United Kingdom), and M. Sato (Waseda University, Tokyo, Japan) and the Japan National Bio Resource Project for generously providing reagents. We thank E. Gomes, M. Piel, A. Paoletti, and E. Bi for helpful discussions throughout this work. J.C. was supported by a predoctoral fellowship from the Fundação para a Ciência e a Tecnologia, Portugal. This work was supported by grants from the National Institutes of Health and the Agence Nationale de la Recherche.
Abbreviations used:
- MAP
microtubule-associated protein
- MBC
carbendazim
- SAC
spindle assembly checkpoint
- SPB
spindle pole body.
Footnotes
J.C. and C.F. created reagents; J.C. and V.M.K. performed experiments; J.C. and P.T.T. analyzed data and wrote the manuscript.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-09-1370) on September 24, 2014.
REFERENCES
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Chang L, Gould KL. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc Natl Acad Sci USA. 2000;97:5249–5254. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J, Fu C, Syrovatkina V, Tran PT. Imaging individual spindle microtubule dynamics in fission yeast. Methods Cell Biol. 2013;115:385–394. doi: 10.1016/B978-0-12-407757-7.00024-4. [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Ward JJ, Loiodice I, Velve-Casquillas G, Nedelec FJ, Tran PT. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev Cell. 2009;17:257–267. doi: 10.1016/j.devcel.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Green RA, Wollman R, Kaplan KB. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol Biol Cell. 2005;16:4609–4622. doi: 10.1091/mbc.E05-03-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM. The fission yeast microtubule cytoskeleton. J Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, Miller EA, Chang F, Jia S. Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol. 2012;199:735–744. doi: 10.1083/jcb.201208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola B, O'Toole E, McIntosh JR, Mellone B, Allshire R, Mengarelli S, Hultenby K, Ekwall K. The domain structure of centromeres is conserved from fission yeast to humans. Mol Biol Cell. 2001;12:2767–2775. doi: 10.1091/mbc.12.9.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- May KM, Hardwick KG. The spindle checkpoint. J Cell Sci. 2006;119:4139–4142. doi: 10.1242/jcs.03165. [DOI] [PubMed] [Google Scholar]
- McHedlishvili N, Wieser S, Holtackers R, Mouysset J, Belwal M, Amaro AC, Meraldi P. Kinetochores accelerate centrosome separation to ensure faithful chromosome segregation. J Cell Sci. 2012;125:906–918. doi: 10.1242/jcs.091967. [DOI] [PubMed] [Google Scholar]
- Meunier S, Vernos I. Microtubule assembly during mitosis—from distinct origins to distinct functions. J Cell Sci. 2012;125:2805–2814. doi: 10.1242/jcs.092429. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 1989;8:3045–3052. doi: 10.1002/j.1460-2075.1989.tb08455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh JL, Nogales E, Oakley BR, McDonald K, Pidoux AL, Cande WZ. A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol Biol Cell. 2000;11:1225–1239. doi: 10.1091/mbc.11.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JJ, Amon A. New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol. 2012;28:189–214. doi: 10.1146/annurev-cellbio-101011-155807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23:401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrovatkina V, Fu C, Tran PT. Antagonistic spindle motors and MAPs regulate metaphase spindle length and chromosome segregation. Curr Biol. 2013;23:2423–2429. doi: 10.1016/j.cub.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Tatebe H, Goshima G, Takeda K, Nakagawa T, Kinoshita K, Yanagida M. Fission yeast living mitosis visualized by GFP-tagged gene products. Micron. 2001;32:67–74. doi: 10.1016/s0968-4328(00)00023-8. [DOI] [PubMed] [Google Scholar]
- Tran PT, Paoletti A, Chang F. Imaging green fluorescent protein fusions in living fission yeast cells. Methods. 2004;33:220–225. doi: 10.1016/j.ymeth.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleugel M, Hoogendoorn E, Snel B, Kops GJ. Evolution and function of the mitotic checkpoint. Dev Cell. 2012;23:239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Zheng F, Li T, Jin DY, Syrovatkina V, Scheffler K, Tran PT, Fu C. Csi1p recruits alp7p/TACC to the spindle pole bodies for bipolar spindle formation. Mol Biol Cell. 2014;25:2750–2760. doi: 10.1091/mbc.E14-03-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



