Abstract
Recurrent deletions of chromosome 15q13.3 associate with intellectual disability, schizophrenia, autism and epilepsy. To gain insight into its instability, we sequenced the region in patients, normal individuals and nonhuman primates. We discovered five structural configurations of the human chromosome 15q13.3 region ranging in size from 2 to 3 Mbp. These configurations arose recently (~0.5–0.9 million years ago) as a result of human-specific expansions of segmental duplications and two independent inversion events. All inversion breakpoints map near GOLGA8 core duplicons—a ~14 kbp primate-specific chromosome 15 repeat that became organized into larger palindromic structures. GOLGA8-flanked palindromes also demarcate the breakpoints of recurrent 15q13.3 microdeletions, the expansion of chromosome 15 segmental duplications in the human lineage, and independent structural changes in apes. The significant clustering (p=0.002) of breakpoints provides mechanistic evidence for the role of this core duplicon and its palindromic architecture in promoting evolutionary and disease-related instability of chromosome 15.
INTRODUCTION
A ~2.5 Mbp region on human chromosome 15q13.3, distal to the Prader-Willi/Angelman locus represents one of the most genetically unstable regions of the human genome1,2. Rare recurrent microdeletions between blocks of segmental duplications (SDs) (BP4 and BP5) are strongly associated with intellectual disability, schizophrenia, autism and other neurodevelopmental disorders3–7. The deletion is, in fact, now recognized as one of the most prevalent major risk factors for idiopathic generalized epilepsy (~1% of all cases)4. The reciprocal duplication as well as smaller internal deletions that encompass the entire CHRNA7 gene have also been described in patients with a range of neurodevelopmental phenotypes8,9. Numerous additional structural variants, including common copy number and an inversion polymorphism, have been reported within the 15q13.3 region3,8,10,11. The majority of the common and rare 15q13.3 structural polymorphisms are associated with complex, high-identity blocks of SDs that arose recently in primate evolution12–17. Owing to the genomic complexity of the region, neither the extent of human structural diversity nor the breakpoints of most rearrangement events are understood at the molecular genetic level.
In this study, we sought to better understand the mechanisms leading to genomic instability of the 15q13.3 locus by characterizing breakpoints of evolutionary and contemporary rearrangements. We used an integrated comparative genomics approach to sequence characterize structural haplotypes from multiple human and ape genomes. This entailed the construction of BAC libraries, high-quality finished sequencing using single-molecule real-time (SMRT) sequencing technology to resolve structural haplotypes18, and cytogenetic-based assays to characterize the organization, orientation and SD architecture of the 15q13.3 region. We performed detailed sequence-based analysis of 80 15q13.3 microdeletions. Our results suggest a molecular convergence on specific repeat sequences as the potential source for genetic instability of these regions.
RESULTS
Copy number polymorphism
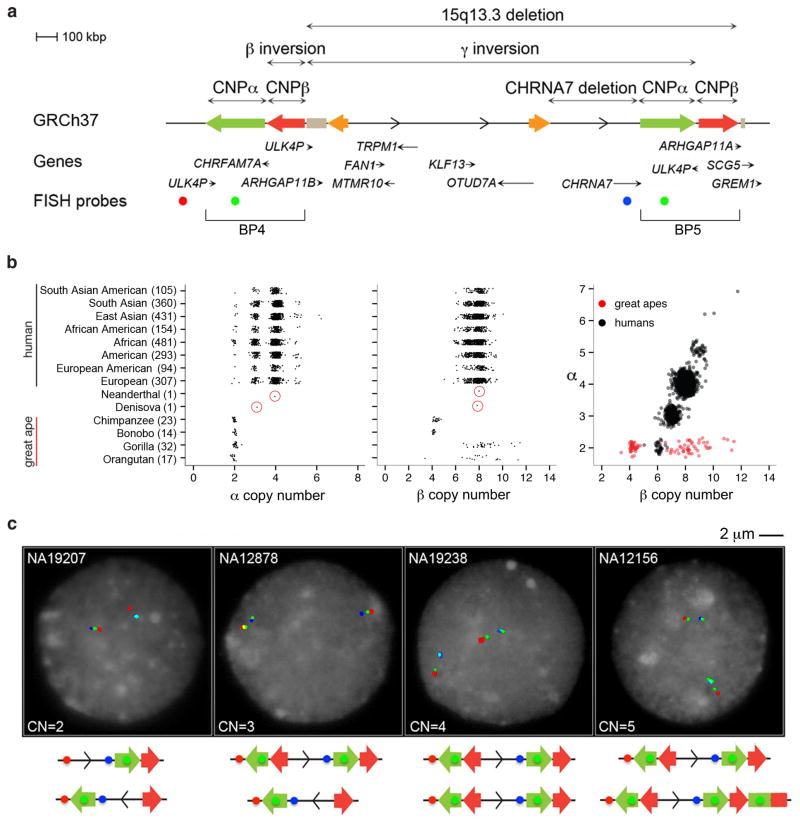
Since most breakpoints map to the large blocks of SDs at BP4 and BP5 (Figure 1a; Table 1), we first assessed the extent of copy number polymorphism of these regions using sequence read-depth approaches19 applied to 2313 human, ape and archaic hominin genomes (Supplementary Tables 1–3). We identified two large copy number polymorphic (CNP) regions of ~300 kbp and ~210 kbp referred to here as CNPα and CNPβ, respectively. These two copy number variable regions are separated by a GOLGA8 repeat and correspond to two SDs, each with >99.5% identity, in which the breakpoints of the recurrent 2 Mbp deletions were originally predicted to occur3. CNPα is a human-specific SD whose diploid copy number (CN) ranges from 2–7, with 77% of humans apparently fixed for the duplication (diploid CN=4) (Figure 1b; Supplementary Figure 1). In contrast, copy number states for CNPβ range from 5–12 with 72% of humans showing a diploid copy number of 8 with four of these copies mapping elsewhere on chromosome 15. A strong correlation (r=0.82, Pearson correlation) in copy number is observed between CNPα and CNPβ suggesting that in the human lineage (but not in the ape lineage) the two SDs have expanded in concert as part of a larger 510 kbp cassette.
Figure 1. 15q13.3 structural variation.
(a) Different structural rearrangements at the 15q13.3 region include a 2 Mbp microdeletion between BP4 and BP53, a 430 kbp microdeletion involving the CHRNA7 gene8, a 1.8 Mbp polymorphic inversion of the same region (γ inversion)3,10,11, two CNP SDs (CNPα and CNPβ) mapping at BP4 and BP5 of the 15q13.3 microdeletion, and a small inversion (β inversion) overlapping CNPβ at BP4. (b) Read-depth-based copy number estimates of CNPα and CNPβ in 2225 HapMap individuals from the 1000 Genome Project and 86 nonhuman ape, Neanderthal and Denisova genomes (circled in red). The number of individuals from each population is indicated in parentheses. A strong correlation (r=0.82, Pearson correlation which is significant using an F test) in copy number is observed between CNPα and CNPβ in humans but not apes. (c) FISH analysis using a probe mapping at CNPα (WIBR2-1388I24, green) and two probes mapping in the unique sequence (WIBR2-1462O20, red; WIBR2-3158E16, blue) shows a variable copy number between 0 and 1 at BP4 and between 0 and 2 at BP5.
Table 1. 15q13.3 structural variant events.
Different structural rearrangements at the 15q13.3 region and the frequency of each rearrangement are shown.
| Species | Structural variant | Size | GRCh37 coordinates | Disease/Predisposition | Frequency |
|---|---|---|---|---|---|
| Human | 15q13.3 microdeletion | 2 Mbp | Chr15: 30.7–32.7 Mbp; Chr15: 30.9–32.9 Mbp | intellectual disability, epilepsy, autism and schizophrenia | 0.27% (42/15,767) cases; 0% (0/8,329) controls * |
| Human | 15q13.3 microduplication | 2 Mbp | Chr15: 30.7–32.7 Mbp; Chr15: 30.9–32.9 Mbp | intellectual disability, epilepsy and autism | 0.13% (20/15,767) cases; 0.3% (3/8,329) controls * |
| Human | γ inversion (BP4-BP5) | 1.8 Mbp | Chr15: 30.80–32.70 Mbp | predisposition to CHRNA7 microdeletion | 6% (haplotype freq.) |
| Human | β inversion (BP4) | 130 kbp | Chr15: 30.70–30.84 Mbp | - | 10% (haplotype freq.) |
| Human | CNPα duplication | 300 kbp | Chr15: 30.37–30.67 Mbp Chr15: 32.45–32.75 Mbp |
- | CN=4 77%; CN=3 19%; CN=2 2%; CN=5 2% |
| Human | CNPβ duplication | 210 kbp | Chr15: 30.70–30.91 Mbp Chr15: 32.68–32.89 Mbp |
- | CN=8 72%; CN=7 21%; CN=9 5%; CN=6 2% |
| Human | CHRNA7-adj duplication | 124 kbp | Chr15: 30.97–31.09 Mbp | predisposition to CHRNA7 microdeletion | fixed |
| Human | ARHGAP11B duplication | 39 kbp | Chr15: 30.90–30.93 Mbp | - | fixed |
| Chimpanzee | β inversion (BP5) | 120 kbp | Chr15: 32.7–32.8 Mbp | - | ND |
| Gorilla | γ inversion | 1.9 Mbp | Chr15: 30.40–32.90 Mbp | - | fixed |
| Gorilla | inversion of a portion of α(BP5) | 80 kbp | Chr15: 32.61–32.69 Mbp | - | ND |
| Gorilla | partial duplication of α (BP4) | 80 kbp | Chr15: 30.37–30.45 Mbp | - | fixed |
Cooper et al., 2011
Based on the extremes observed in this study, the data suggest that individuals in the human population may differ by as much as 1 Mbp with respect to SD content between BP4 and BP5. We designed a series of three-color interphase fluorescence in situ hybridization (FISH) experiments to investigate the location of the copy number differences of CNPα among different individuals. The FISH analysis indicates copy number polymorphism at both breakpoints of the 15q13.3 microdeletion. At a chromosomal level, we estimate a haploid variable CN between 0 and 1 for BP4 and between 0 and 2 at BP5 (Figure 1c; Supplementary Table 4). Note: due to additional copies of CNPβ mapping to chromosome 15, BP4 and BP5 signals could not be clearly resolved by FISH for CNPβ.
Discovery and characterization of the β inversion
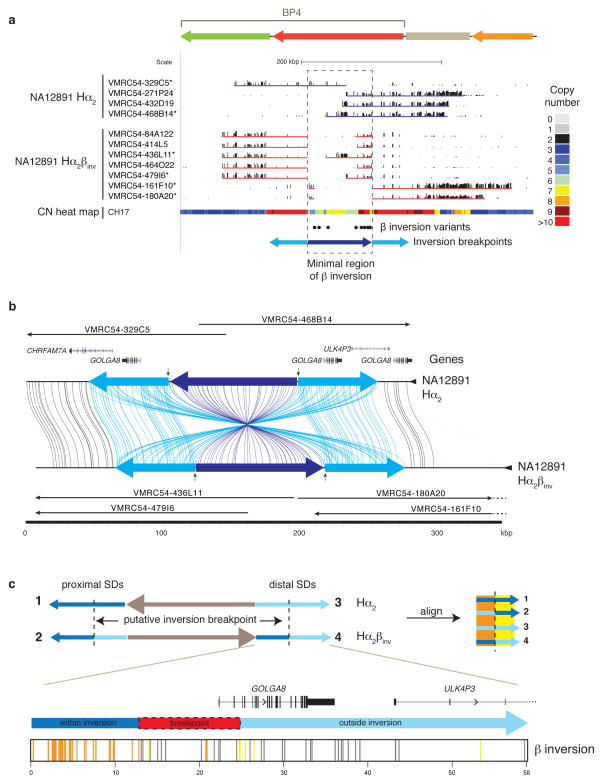
Due to the potential for assembly errors within SD regions20–23, we established an alternate reference assembly for 15q13.3 from a hydatidiform (haploid) mole source (CHM1hTERT). We constructed a map of 23 contiguous BAC clones (CH17) and sequenced 21 of these using SMRT and capillary sequencing methods to establish a 4 Mbp high-quality alternate reference assembly (Supplementary Figure 2a; Supplementary Table 5). The new reference differed structurally from GRCh37 by a 130 kbp inversion corresponding to CNPβ at BP4 (Supplementary Figure 3). To ensure the β inversion was not a hydatidiform cell line artifact, we identified a set of eight single nucleotide variants that distinguished it from GRCh37 (Supplementary Tables 6 and 7) and screened additional DNA samples from the 1000 Genomes Project24, identifying a European individual, NA12891, that was heterozygous for the β inversion. We constructed and arrayed a large-insert genomic BAC library from this DNA sample (VMRC54) as well as the two other members of the NA12878 trio (Methods; Supplementary Table 8). We recovered and sequenced VMRC54 BAC clones from the BP4 region, independently validating the sequence structure of the β inversion and the GRCh37 configuration (Figure 2a and b; Supplementary Table 9). For simplicity, we refer to the CH17 haplotype as Hα2βinv because it carries the β inversion and two haploid copies of CNPα compared to the directly oriented structural configuration (Hα2) of the reference assembly.
Figure 2. Sequence refinement of β inversion breakpoints.
(a) A 210 kbp β inversion was identified, validated, and sequenced using the VMRC54 BAC library (NA12891 individual). Illumina-generated sequences of clones spanning the BP4 CNPβ were mapped to human reference GRCh37. Clones sequenced using PacBio are indicated with asterisks. The copy number (CN) heat map shows the total diploid CN of a region in the CH17 hydatidiform mole cell line. The locations of the β inversion haplotype-tagging variants are pictured as dots. The blue arrows represent the BP4 CNPβ (dark blue) with the flanking 58 kbp inverted SDs (light blue). (b) Homologous sequences of clones, generated using PacBio and assembled into sequence contigs, are connected with colored lines between the direct (Hα2) and inverted (Hα2βinv) haplotypes from NA12891 using Miropeats55. Vertical arrows indicate the minimal inversion breakpoints. (c) Homologous sequences (58 kbp) from the BP4 CNPβ flanking inverted SDs were aligned from multiple individuals (NA12891 and CH17) and haplotypes (β direct: SDs 1 and 3, and β inverse: SDs 2 and 4; see Supplementary Figure 5 for a more detailed alignment) and variant sites compared. Variant positions showing signatures of being within or outside of the β inversion breakpoints are indicated as colored lines under the picture of the distal β inverse SD including: within the inversion (orange; consensus of SDs 1 & 4 and SDs 2 & 3), outside the inversion (yellow; consensus of SDs 1 & 2 and SDs 3 & 4), and gene conversion (gray; consensus of SDs 1 & 3 and SDs 2& 4). The inversion breakpoint, refined to a region in which we observe a transition from orange to yellow lines, is highlighted with a dash-outlined red box.
The β inversion consists of three SDs (Supplementary Figure 2b): a pair of two highly identical (58 kbp, 99.6% identity) inversely oriented SDs flanking a 95 kbp duplication. The flanking 58 kbp palindrome corresponds to the GOLGA8 gene family15,16, one of the core duplicons found to be associated with most of the interspersed SD blocks across chromosome 1515,16. The β inversion configuration increases the length of the largest contiguous tract of directly oriented SDs between BP4 and BP5 from 58 to ~188 kbp of near perfect sequence (99.4% identity) (Supplementary Figure 4) in principle creating a better substrate for unequal crossover and instability associated with disease. To assess the breakpoints of the β inversion, we constructed a multiple sequence alignment (MSA) from three distinct haplotypes (Figure 2c; Supplementary Figure 5) and used unique sequence differences in the duplicated regions to define the most likely breakpoint transition region. We narrowed the inversion breakpoint to a ~12 kbp region spanning from intron 2 of the GOLGA8 repeat to 9.6 kbp upstream of the gene (Figure 2c; Supplementary Table 10).
Sequence structure of the γ inversion
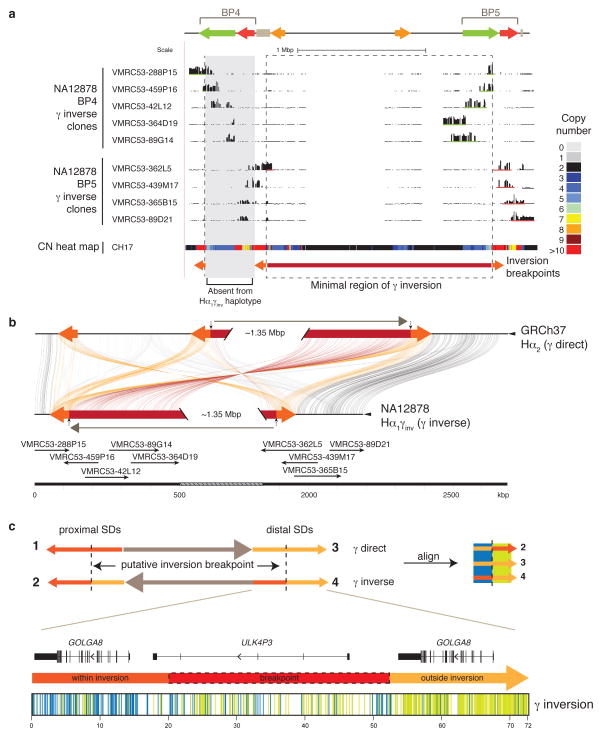
We sequence resolved the larger human inversion polymorphism spanning the entire BP4-BP5 region (referred to as the γ inversion)3,10,11 (Figure 1). This entailed sequencing of 21 clones from a BAC library (VMRC53) constructed from a heterozygous individual (NA12878); single nucleotide polymorphism (SNP) genotyping to assign maternal and paternal haplotypes; and high-quality sequencing of 11 nonredundant clones to generate an alternate reference assembly at the breakpoint region (Figure 3a and b; Supplementary Figure 2c; Supplementary Figures 6 and 7; Supplementary Table 9). Compared to the reference, the γ inversion spans ~1.844 Mbp from BP4 to BP5 and is flanked by palindromic SDs containing two GOLGA8 genes and a ULK4P3 gene, (~71 kbp, 98.5% identity; Figure 3c; Supplementary Figure 2d). The Hα1γinv assembly contains a single copy of CNPα at BP4 and CNPβ at BP5 suggesting that it arose from a simpler human haplotype Hα1 where CNPα was moved from BP5 to BP4 by the inversion (Supplementary Figure 8). Based on sequence alignment, we refined the γ inversion breakpoints to a ~32 kbp region within the palindrome containing the ULK4P3 gene and flanked on either side by GOLGA8 core duplicons (Figure 3c; Supplementary Figure 9; Supplementary Table 10). The high sequence identity of the duplications as well as alternative sequence signatures consistent with historical gene conversion events made it impossible to refine the breakpoint with any further precision (Supplementary Table 11).
Figure 3. Sequence refinement of γ inversion breakpoints.
(a) The γ inversion was identified, validated, and sequenced using the VMRC53 BAC library (NA12878 individual). Illumina-generated sequences of clones spanning the 15q13.3 BP4 (green bars) and BP5 (red bars) loci were mapped to the human reference GRCh37. The nine clones pictured were sequenced using PacBio. The copy number (CN) heat map shows total diploid CN of a region in the CH17 hydatidiform mole cell line. The minimal region of the inversion spans ~1.8 Mbp (highlighted with a dashed box and a red bar). The orange arrows represent the flanking 72 kbp flanking inverted SDs that mediate the γ inversion. The Hα1γinv haplotype likely arose from the Hα1 haplotype, which does not harbor CNPα and CNPβ at BP4. (b) Homologous sequences of clones, generated using PacBio and assembled into contigs, and the human reference are connected with colored lines between γ direct (Hα2) and inverse (Hα1γinv) haplotypes using Miropeats55. Vertical arrows indicate the minimal inversion breakpoints. (c) Homologous sequences (72 kbp) from the orange flanking inverted SDs were aligned from multiple individuals (NA12878, CH17, and GRCh37) and haplotypes (γ direct: SD 3, and γ inverse: SDs 2 and 4; see Supplementary Figure 9 for a more detailed alignment) and variant sites compared. Variant positions showing signatures of being within or outside of the γ inversion breakpoints are indicated as colored lines under the picture of the distal γ inverse SD including: within the inversion (blue; consensus of SDs 2 & 3), and outside the inversion (green; consensus of SDs 3 & 4). The inversion breakpoint, refined to a region in which we observe a transition from blue to green lines, is highlighted with a dash-lined red box.
Population frequency of β and γ inversion polymorphisms
To estimate the frequency of the γ inversion, we initially tested lymphoblastoid cell lines from 20 diverse HapMap individuals using a three-color interphase FISH assay (Supplementary Figure 10). Without exception, all chromosomal haplotypes (n=16/16) with higher copy number of CNPα (n=2–3) were directly configured similar to the reference genome (Supplementary Table 4). In contrast, all γ inversion haplotypes showed a single copy of CNPα consistent with our BAC sequencing results. Note: 10/24 of the chromosomes with a single copy of CNPα carried the γ inversion. An additional series of three-color FISH experiments confirmed that when there is a single haploid copy number of CNPα and it carries the γ inversion (Hα1γinv configuration) (Supplementary Figure 11), we always observe an absence of CNPα at BP5 consistent with a single structural haplotype for this inversion. Thus, CNPα varies between 1 and 3 copies only in the directly oriented configurations for the BP4-BP5 region (Hα1, Hα2, Hα3) (Supplementary Figure 8) with 0 and 1 copies at BP4 and between 1 and 2 copies at BP5 (Supplementary Table 4). Combining this cytogenetic inference with copy number data from 1311 human genomes with ethnicities matching our original FISH survey, we estimate an allele frequency of 6% for the γ inversion (Supplementary Table 12) with slightly elevated frequency of the inversion in Tuscany and African populations. Notably, our inversion frequency estimate is lower than previously reported3. In the previous study, a two-probe FISH assay was used to genotype the γ inversion resulting in a higher error of detection compared to our study, which used a three-probe assay.
Since the β inversion is smaller and embedded within a complex region flanked by high-identity duplications, FISH could not be used to assess its frequency. Instead, we leveraged the unique tag SNPs used to recover and sequence the inversion in NA12891. Using these SNPs as a surrogate, we designed molecular inversion probes (MIPs)25,26 to capture, sequence (Illumina), and genotype the eight haplotype-tagging variants across 904 individuals from diverse human populations from the 1000 Genomes Project (Supplementary Tables 13, 14, and 15). We estimate a haplotype frequency of ~38% across European populations (n=275, CEU, TSI, and GBR) with reduced frequencies of ~10% in African populations (n=299, LWK, MKK, YRI, ESN, and GWD) and ~4% in Asian populations (n=221, CHB, CDX, KHV, and JPT). No β inversion haplotypes were observed in either Chinese Dai (CDX) or Cambodian (KHV) populations. These data suggest considerable stratification especially between Europeans and Asians (average Fst=0.28), with a maximum Fst of 0.36 between Toscani (TSI) and Chinese Dai (CDX) populations (Supplementary Table 16).
Evolution of chromosome 15q13.3
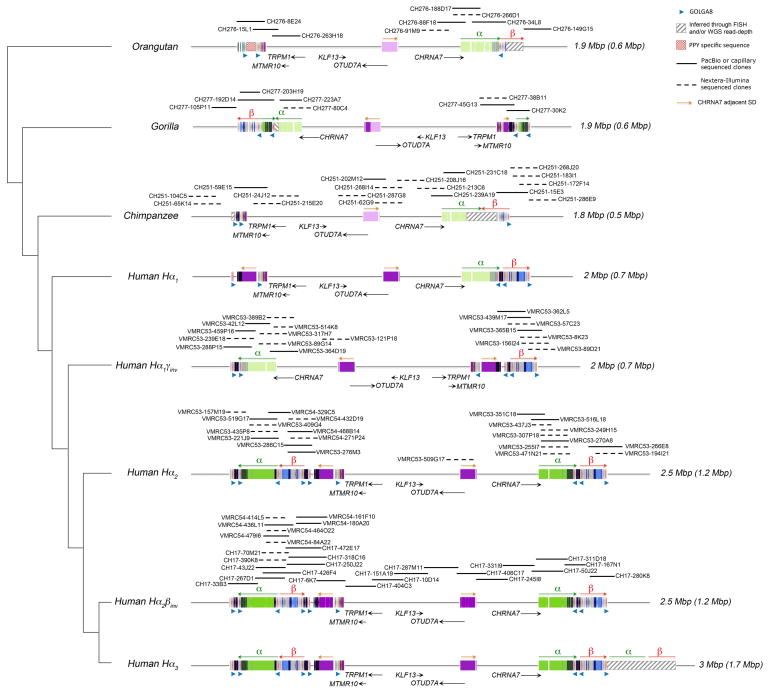
We examined the organization of the region in multiple nonhuman ape samples by FISH and found that chimpanzee and orangutan show a direct orientation between BP4 and BP5, while gorilla is in inverted orientation compared to the human reference genome (Supplementary Figure 12; Supplementary Table 17). Next, we sequenced 48 BAC clones from chimpanzee, gorilla and orangutan in order to reconstruct the most likely ancestral sequence structure of the breakpoint regions (Supplementary Figure 13; Supplementary Table 18). Sequencing data show that both gorilla and orangutan lack the ULK4P3 gene where the γ inversion breakpoints map in humans, indicating that the 1.8 Mbp γ inversion likely occurred as two independent events in human and gorilla lineages. Recurrences of large inversion events across primate species have been reported for other regions, including the 17q21.31 and 16p12.1 microdeletion regions23,27.
Our sequence analysis reveals a much simpler organization of the 15q13.3 orthologous region in nonhuman primates when compared to human (Figure 4). The sequenced chimpanzee, gorilla and orangutan haplotypes, for example, lack the large SDs found at most human BP4 regions predicting that BP5 was the ancestral source (Supplementary Figures 14 and 15). Phylogenetic analysis confirms this and predicts that the proximal GOLGA8 repeats at BP4 are orthologous among apes and humans and, thus, pre-existed the duplicative transpositions of CNPα and CNPβ to the region (Supplementary Figure 16; Supplementary Table 19). This duplication also includes ARHGAP11, a gene that was previously described to have undergone a human-specific expansion compared to other primate lineages19. In this case, we observe that the proximal breakpoint of the ARHGAP11 duplication maps within a GOLGA8 repeat (13.8 kbp resolution) (Supplementary Figure 17).
Figure 4. Comparative sequence analysis of the 15q13.3 region among apes.
The genomic structure is schematized within the context of a generally accepted phylogeny of orangutan, gorilla, chimpanzee and human. A tiling path of BAC clones was sequenced for each haplotype (dashed lines Illumina/solid lines PacBio or capillary finished sequence). A total of 66 BACs were completely sequenced and used to determine the SD organization (colored boxes). Colored boxes with lighter shades indicate segments that are single copy but duplicated in other species. Nonhuman primates lack most of the larger duplications (including CNPα and CNPβ) observed in humans but do carry ancestral GOLGA8 repeats. The region has expanded from 1.8–1.9 Mbp in nonhuman apes to 2–3 Mbp in humans as a result of SD accumulation (colored rectangles). The size of each haplotype is indicated on the right, with the size of the duplicated bases in parentheses. The addition of a polymorphic 500 kbp at BP4 occurred specifically in the human lineage, associated with an expansion of the GOLGA8 repeats at BP4 (CN=6 compared to CN=2 in human simpler haplotypes and nonhuman primates). Sequence and FISH data indicate that chimpanzee and orangutan were found to be in direct orientation while gorilla was found to be in inverse orientation for the γ inversion suggesting separate inversion events occurred at this locus across primate species.
There are numerous additional structural differences between apes and humans in this region. Our analysis shows that CNPβ maps in an inverted orientation in chimpanzee at BP5 (120 kbp inversion) with a GOLGA8 repeat defining at least one boundary of this chimpanzee-specific event (Figure 4; Supplementary Figures 13 and 14). A ~80 kbp inversion of the distal portion of CNPα is identified in gorilla at BP5. This particular segment is also partially duplicated at BP4 in gorilla, and in both instances the rearrangement (duplication at BP4 and inversion at BP5) is flanked by the GOLGA8 repeats. Finally, the CHRNA7-adjacent SD (purple block with orange arrow in Figure 4) is completely absent at BP4 in all analyzed primates with the exception of a partial duplication in gorilla. Interestingly, the distal breakpoint of the CHRNA7-adjacent duplication at BP4 in humans maps within a GOLGA8 repeat.
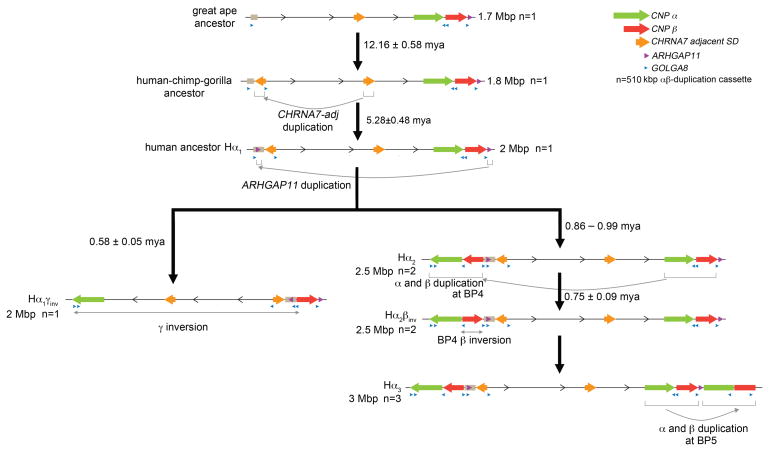
To estimate the order and timing of the major structural changes during human evolution, we constructed a series of phylogenetic trees and estimated the coalescence/divergence time using locally calibrated molecular clocks and predicted divergence time of 6 million years between human and chimpanzee. The earliest events in restructuring this region include the duplicative transposition of the adjacent CHRNA7 segment to the proximal 15q13.3 region before divergence of the African apes (12.16 ± 0.58 mya) (Figure 5; Supplementary Figure 18). This was followed by the human-specific ARHGAP11 duplication from BP5 (ARHGAP11A) to BP4 (ARHGAP11B), which occurred soon after humans and chimpanzees diverged (5.28 ± 0.48 mya) (Supplementary Figure 19). We estimate that the largest CNPα and CNPβ duplications from BP5 to BP4 occurred in close succession or concurrently at 995 ± 61 and 862 ± 99 thousand years ago, respectively (Supplementary Figure 20). These estimates are consistent with the finding that both duplications were already present before the split of Denisova and Neanderthal from the Homo sapiens lineage (Figure 1b). Further, by comparing sequences between the human CH17 contig (CNPβinv) and GRCh37 (CNPβ), we predict the β inversion to have occurred shortly thereafter 748 ± 92 thousand years ago (Supplementary Figure 20b). We calculate that the time to most recent common ancestor of the inverted NA12878 Hα1γinv and the Hα1 haplotype (direct for the γ inversion) is 578 ± 47 thousand years ago (Supplementary Figure 21).
Figure 5. Model of chromosomal 15q13.3 evolution.
Based on comparisons to outgroup primates, we propose a simpler human ancestral organization (Hα1)—a configuration that is found enriched in contemporary African populations. A 510 kbp duplicative transposition from BP5 to BP4 (α and β duplications) occurred potentially in a palindromic configuration (Hα2), followed by an inversion of β at BP4 (Hα2βinv) between 700–900 thousand years ago. NAHR within BP5 leads to tandemization of the 510 kbp duplication (Hα3) and larger configurations primarily in East Asian populations. Approximately 500 thousand years ago, the 1.8 Mbp γ inversion independently rearranged to the Hα1γinv inverted haplotype.
Overall, these data suggest radical restructuring of this region in the Homo lineage over a short epoch of evolutionary time and a clear polarity of duplicative transposition events moving segments from BP4 to BP5 in association with GOLGA8 repeats. These events have led to the emergence of at least five alternate chromosomal configurations in the human population ranging from ~2 to >3 Mbp in size.
15q13.3 microdeletion patient breakpoint analysis
We analyzed 80 total DNA samples from children with autism, intellectual disability, and/or developmental delay that were previously identified as carrying 15q13.3 microdeletions by clinical array comparative genomic hybridization (CGH). These include 77 cases with intellectual disability and developmental delay referred to Signature Genomic Laboratories (24 unpublished and 53 previously reported)1 and three cases with idiopathic autism from the Simons Simplex Collection (SSC)28. We screened the 80 patients using complementary methods targeted to the 15q13.3 region: (1) a higher density customized microarray and (2) sequencing via MIP-capture of singly unique nucleotide (SUN) k-mers (SUNKs). Both methods mapped the breakpoints of the disease-critical region to a ~500 kbp region spanned by the CNPα and CNPβ SDs (Supplementary Figures 22 and 23; Supplementary Tables 20 and 21).
Since the β inversion configuration creates a potentially more competent substrate for non-allelic homologous recombination (NAHR) because of its longer stretch (188 kbp) of directly oriented sequence, we tested whether this particular configuration was enriched in patients as has been observed for other microdeletion regions23,29,30. We compared the frequency of this configuration in patients and controls of European ancestry using sequence markers specific for the β inversion (Figure 2a). We found that the frequency of the β inversion does not differ significantly in 15q13 microdeletion patients [~28% (n=40) when compared to the European average (38%) (p=0.27, Fisher’s exact two-tailed test); Supplementary Table 15]. These data suggest that factors other than simply the length of homology promote the instability of this locus.
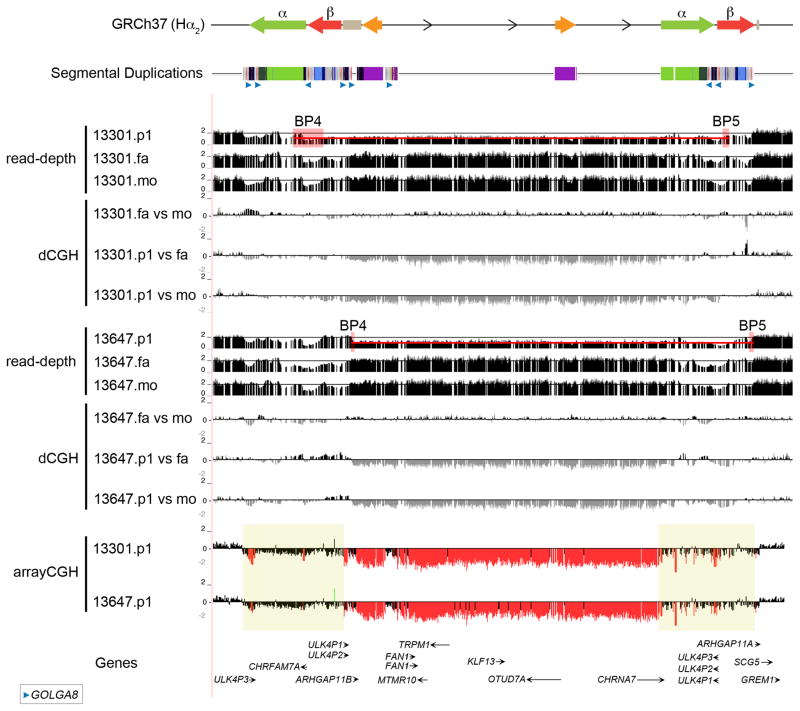
To refine the breakpoints with greater precision, we performed whole-genome sequencing of two idiopathic autism patients from the SSC carrying de novo 15q13.3 microdeletions along with their unaffected parents using the Illumina HiSeq 2000 (101 bp PE reads) (Supplementary Table 22). The generated sequences were aligned to the human GRCh37 reference and the alternate CH17 Hα2βinv assembly. We investigated paralog-specific read-depth over 1 bp windows in each trio at all sites where both parents had the expected copy number of 2. Using SUN variants that allowed us to discriminate between the paralogous copies19, we narrowed proband 13647.p1 breakpoints to a 14 kbp segment at BP4 and a 22 kbp segment at BP5 and proband 13301.p1 breakpoints to a 155 kbp segment at BP4 and a 30 kbp at BP5 (Figure 6; Supplementary Table 10). The two probands have different breakpoints but in both cases the breakpoints map at or adjacent to directly oriented copies of GOLGA8 (Figure 7).
Figure 6. 15q13.3 microdeletion breakpoints analysis.
Array CGH data for two 15q13.3 microdeletion patient samples are mapped against the GRCh37 human reference. The microdeletion breakpoints map within a 500 kbp region (yellow boxes) where both α and β SDs are mapping. Digital comparative genomic hybridization (dCGH)17 was used to detect regions of gain or loss in probands (p1) compared to their parents (mo, mother; fa, father). The method measures differences in Illumina sequence read-depth compared to a reference genome to define sites of copy number variation. Paralog-specific read-depth analysis in each proband and their parents was performed at all sites where both parents had the expected copy number of 2. This allowed us to refine proband 13647.p1 breakpoints to a 13 kbp segment and proband 13301.p1 breakpoints to a 30 kbp between BP4 and BP5 (red boxes). The two probands have different breakpoints but in both cases the breakpoints map at or adjacent to the GOLGA8 repeats.
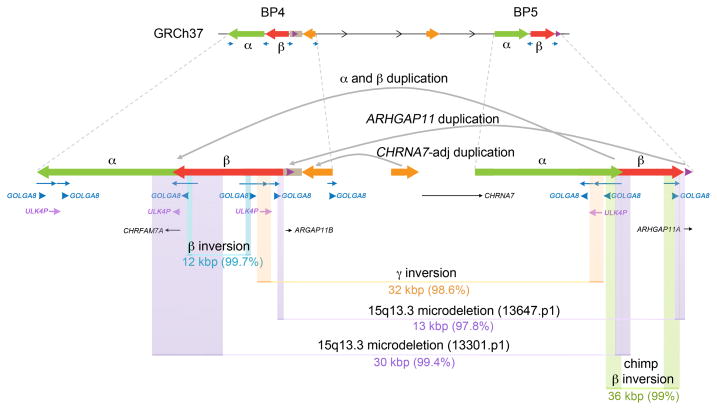
Figure 7. Summary of 15q13.3 rearrangements mediated by GOLGA8 repeats.
Shown are eight independent rearrangements at the 15q13.3 region. Colored boxes indicate the breakpoints identified for each rearrangement (Supplementary Table 10). The size and the percent of similarity of the paralogous sequences at the rearrangement breakpoints are shown.
We tested by simulation to determine if the apparent clustering of evolutionary and disease breakpoints within or near GOLGA8 sequences was significant. We identified the positions of all GOLGA8 sequences (Supplementary Table 23) within the BP4 and BP5 regions and created a null model by randomly distributing the breakpoint intervals to the SDs mapping to this portion of 15q13.3 (chr15:30,362,914–31,196,467 and chr15:32,442,314–32,927,877; Supplementary Table 24). We computed the number of times the mean distance of sampled breakpoints from the null distribution was less than or equal to the mean of the observed distances between 15q13.3 breakpoints and GOLGA8 repeats (66,801 bp). The results suggest that the clustering of breakpoints with GOLGA8 sequences is significant (empirical p=0.002, n=100,000 permutations).
DISCUSSION
Our comparative sequence analysis of human and primate genomes reveals that the 15q13.3 region has become increasingly complex over the course of human evolution, with an expansion in size from 1.8 Mbp in apes to 2–3.5 Mbp in humans. There has been a clear polarity with most duplicative transpositions occurring from BP5 to BP4. Most of the largest structural changes, including large-scale inversion polymorphisms, arose over a narrow evolutionary period (500–900 thousand years ago)—a time when ancestral Homo sapiens was diverging from archaic hominins31,32. We have resolved five distinct structural configurations in humans that differ radically in organization and SD content. Our results suggest that human chromosomal 15q13.3 haplotypes can vary by as much as 75% of their euchromatic length and are stratified among different populations. The simplest ancestral configurations (e.g., Hα1) show elevated frequency among African populations while some of the largest and potentially disease-prone configurations are enriched in out-of-Africa populations (e.g., Hα2βinv in Europeans and Hα3 in East Asians).
At least nine 15q13.3 rearrangement breakpoints (six human, one chimpanzee, and two gorilla rearrangements) map at or adjacent to GOLGA8 core duplicons (Table 1; Figures 4, 6, and 7). Although our breakpoint precision ranges from 12–155 kbp and cannot be further refined due to the presence of virtually identical sequence within these regions (Supplementary Table 10), our simulations strongly suggest that this association is significant. The GOLGA repeat encodes a primate-specific chromosome 15 gene family of 14 kbp15 that expanded over the last 20 million years of primate evolution12,13. It has become dispersed to multiple locations across the long arm of chromosome 15 and is the most enriched sequence associated with SD blocks promoting disease instability, including Prader-Willi/Angelman syndromes, 15q24 microdeletions and 15q25.2 microdeletions33–36 (Supplementary Figure 24a and b). GOLGA is one of fourteen “core duplicons” associated with the burst of interspersed SDs in human–great ape ancestral lineage17,37.
We propose that the GOLGA core duplicons are preferential sites of genomic instability that have driven both disease and evolutionary instability of chromosome 15. In addition to the clustering of breakpoints on chromosome 15q13.3, other data are supportive of a more global association. We note, for example, that this same GOLGA repeat demarcates a pericentric inversion breakpoint between human and chimpanzee 15q11-q1338 and a more ancient inversion in the Catarrhini ancestor39. Analysis of the SDs mapping at other chromosome 15 microdeletion regions (e.g., 15q24 and 15q25) show the breakpoints often occur in directly orientated duplications that are short and have a low percentage of identity (Supplementary Figures 24c and 25) but contain multiple copies of the GOLGA repeats. Array CGH experiments on ten previously published 15q24 microdeletion cases confirm that the GOLGA repeat maps at or near most rearrangement breakpoints (Supplementary Figures 24c and 26)33,40. These findings are also consistent with our observation that we find no evidence of an enrichment of the Hα2βinv haplotype among 15q13.3 deletion patients even though this configuration expands the directly orientated segment from 58 to 188 kbp in length. Although orientation, length and degree of sequence identity between duplicated sequences are frequently deemed the most important parameters for NAHR1,41, the presence of a GOLGA repeat may bias the actual position of the unequal crossover (i.e., an NAHR hotspot).
These results also bear striking similarities to the microdeletion encompassing the neurofibromatosis type-1 (NF1) gene and its flanking regions at 17q11.2. The most common NF1 microdeletions (type-1) span 1.4 Mbp and have breakpoints located within SDs containing LRRC37 core duplicons42. The same LRRC37 core duplicons at 17q21.31 are known to have mediated the 970 kbp polymorphic inversions of the MAPT locus that also underlies the syndromes associated with recurrent 17q21.31 microdeletions27. The presence of core duplicons at multiple evolutionary breakpoints as well as at a variety of recurring disease-associated rearrangements are indicative of the high degree of genomic instability driven by these sequences.
Our evolutionary reconstruction suggests that the GOLGA8 core, in particular, has promoted both inversions and the formation of large palindromic SD structures. Palindromic sequences, or inverted repeats, have been known to be unstable and represent hotspots for deletion or recombination in bacteria, yeast, and mammals43–46. This genetic instability has generally been related to DNA replication: slow replication was observed in an inverted repeat sequence in Escherichia coli44, and inverted repeats lead to chromosomal rearrangements more frequently in yeast that are deficient in DNA polymerase activity47,48. In the events discussed here, the presence of palindromic structures might have promoted stalling of the replication fork, creating an opportunity for the chromosome to break, and recombination might have occurred in a non-allelic fashion using the homology of the GOLGA repeats. In humans, short palindromic AT-rich repeats (or PATRR) have been implicated in chromosomal aberrations via non-homologous end joining leading to gross transchromosomal events49 and instability in cancer cells50. Most experimental demonstrations of palindrome formation and instability have involved smaller structures. The putative palindromes here are massive—for instance, ~210 kbp in length with 58 kbp inverted arms flanking a 95 kbp spacer for CNPβ—and attempts to detect its formation by in vitro snapback assays50 were inconclusive.
The recurrent use of GOLGA core duplicons suggests a fundamental role in the cycles of chromosomal rearrangement that have intertwined large-scale inversions and SD expansions in this region. We note that most of the largest interspersed SDs have been transposed in an inverted orientation. Similar inverted configurations also occur for contemporary rearrangements such as the PLP1 locus, which is known to be associated with inverted repeats51,52. Microhomology-mediated break-induced replication (MMBIR) mechanisms may be responsible for initial SD formation53,54, and sequences such as GOLGA may also represent preferred or “fragile” sites for MMBIR. It is intriguing that the GOLGA repeats corresponding to sites of rearrangement and conversion maintain an open reading frame while those at the periphery are disrupted (see Supplementary Note). We previously showed that core sequences are generally more transcriptionally active than unique or flanking duplicated sequence16. Thus, transcription and maintenance of an open reading frame may be a critical feature of core duplicons in order to serve as seeds of genomic instability and punctuated SD in the human genome. The mechanism by which these elements promote evolutionary and disease instability during replication will require future experimental investigation.
METHODS
FISH analysis
Interphase nuclei and metaphase spreads were obtained from lymphoblast and fibroblast cell lines from 20 human HapMap individuals (Coriell Cell Repository, Camden, NJ), four chimpanzees (Katie; Veronica; Cochise; PTR8), two gorillas (GGO5; GGO8) and three orangutans (PPY9; PPY16; PPY13). All cell lines were tested for mycoplasma contamination. Primate cell lines were previously collected at the University of Washington and at the University of Bari (Supplementary Table 17) and have not been authenticated. FISH experiments were performed using fosmid clones directly labeled by nick-translation with Cy3-dUTP (PerkinElmer), Cy5-dUTP (PerkinElmer), and fluorescein-dUTP (Enzo) as described previously23. A minimum of 50 interphase cells were scored for each inversion to statistically determine the orientation of the examined region.
Copy number variation analysis
Array CGH was performed on 80 samples with 15q13.3 microdeletions using custom, high-density oligonucleotide 4×180K Agilent chips targeted to with a density of 1 probe per 100 bp. Labeling, hybridization, scanning, and data processing were performed as directed by the manufacturer. DNA sample NA19240 was used as reference. We estimated the copy number of 15q13.3 SDs among 2225 HapMap individuals of different ethnicity24 using a sequence read-depth method19. The duplication content of human, chimpanzee, gorilla, orangutan, and macaque was determined using the whole-genome shotgun sequence detection (WSSD) method as described in Marques-Bonet, et al.37.
BAC library construction and screening
We constructed individual BAC libraries from each member of the NA12878 parent-child trio, namely: NA12878 (VMRC53), NA12891 (VMRC54) and NA12892 (VMRC57). High molecular weight DNA was isolated, partially EcoRI digested, and subcloned into pCC1BAC vector (Epicentre) to create >150 kbp insert libraries using previously described protocols 56. Clones were plated into 384 microtiter plates and were transferred to high-density nylon filters for library screening.
Illumina sequencing of BAC clones
DNA from CH17, VMRC53, VMRC54, CH251, CH276 and CH277 BAC clone libraries was isolated, prepped into barcoded genomic libraries and sequenced (PE101) on an Illumina HiSeq 2000 using a Nextera protocol29. Sequencing data (~300-fold coverage) were mapped with mrsFAST57 to the reference genome and SUN identifiers were used to discriminate between highly identical SDs19.
PacBio clone sequencing and assembly
DNA was isolated from CH17, VMRC53, VMRC54, CH251, CH277 and CH276 BAC clones, PacBio SMRTbell libraries were prepared and sequenced using RSII C2P4 chemistry (one SMRT cell/BAC sample with two 45-minute movies). Inserts were assembled using Quiver and HGAP as described18. Alternate human genome assemblies, including PacBio and capillary sequenced clones from CH17, RP11 and VMRC53 BAC libraries, were assembled with Sequencher and compared to the human reference genome using Miropeats55 and BLAST58.
Sequence analyses
Multiple sequence alignments (MSAs) of representative human haplotypes, paralogs, and/or orthologs from human, chimpanzee, gorilla and orangutan were generated using Clustal W59. We constructed a series of phylogenetic trees using the neighbor-joining method with a complete deletion option (MEGA5)60. Genetic distances were calculated using the Kimura 2-parameter with standard error estimates (an interior branch test of phylogeny; N=500 boot straps replicates); Tajima’s relative rate test was used to assess validity of the molecular clock. We then estimated the coalescence/divergence time using the equation T=K/2R and an estimated divergence time of 6 million years between human and chimpanzee and 15 million years between human and orangutan.
Whole-genome sequencing of 15q13.3 microdeletion samples
Using SSC autism trios (proband, father, and mother) 13301 and 13647, 3 μg of genomic DNA were sheared, end-repaired, an A-tail added, and adaptors ligated to the fragments as described61. Afterwards, ligation samples were run on a 6% pre-cast polyacrylamide gel (Invitrogen, Cat. No. EC6265BOX). The band at 400–550 bp was excised, diced, and incubated as described above. Size-selected fragments were amplified with 0.5 μL of primers, 25 μL of 2X iProof, 0.25 μL of SYBR green, and 8.25 μL of dH2O under the following conditions: 98°C for 30 sec, 30 cycles of 98°C for 10 sec, 60°C for 30 sec, 72°C for 30 sec, 72°C for 15 sec followed by 72°C for 2 min. Fluorescence was assessed between the 30 and 15 sec 72°C step. Amplified, size-selected libraries were quantified using an Agilent 2100 Bioanalyzer and paired-end sequenced (101 bp reads) on an Illumina HiSeq 2000. Sequence read-depth corresponding to SUNs was used to refine the breakpoints as previously described19. Digital comparative genomic hybridization (dCGH) was performed using the sequences from these samples using previously described methods17.
Molecular inversion probe (MIP) genotyping
We used 70 bp MIPs to capture and sequence the β inversion haplotype-tagging variants (n=8) and SUNKs (n=235) spanning the 15q13.3 region. The β inversion haplotype-tagging variants were identified from an MSA of CNPβ at BP4 and BP5 from our CH17-derived assembly and the human reference (Supplementary Table 6). We identified 3544 SUNKs across the 15q13.3 region (chr15:30,350,000–32,950,000; GRCh37) using previously described methods19. MIP design, capture, and sequencing were performed as previously described26,62. MIP sequences are listed in Supplementary Table 13. Any individual with less than 5000 reads mapping was removed from subsequent analyses. In the case of the β inversion haplotype-tagging variants, we genotyped an individual as carrying the β inversion if they had at least one read mapping to seven out of the eight variants (Supplementary Table 14).
Human subjects
The human samples included in this study do not meet the federal definitions for human subjects research. All samples were publicly available or encoded with no individual identifiers available to the study authors. Samples were collected at respective institutions after receiving informed consent and approval by the appropriate institutional review boards. There are no new health risks to participants. Samples that fall within this category include autism probands and parents from the SSC, probands with intellectual disability and developmental delay referred to Signature Genomic Laboratories, and individuals from representative human populations from the 1000 Genomes Project.
Supplementary Material
Acknowledgments
We thank M. Ventura, C. Campbell and H.C. Mefford for useful discussions and T. Brown for critical review of the manuscript. We also thank S. Diede, H. Tanaka, B. Brewer, C. Payen, L. Harshman, and K. Penewit for experimental advice and support for the palindromic snapback assay. This work was supported, in part, by U.S. National Institutes of Health (NIH) grants HG002385 and HG004120 to E.E.E. M.Y.D. is supported by the National Institute of Neurological Disorder and Stroke of the U.S. National Institutes of Health (award K99NS083627). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
BAC clone sequences generated using capillary and SMRT sequencing have been deposited into GenBank with accession numbers listed in Supplementary Tables 5, 9, and 18.
AUTHOR CONTRIBUTIONS
This study was designed by F.A., M.Y.D. and E.E.E. F.A. performed FISH experiments, Illumina sequencing libraries construction, array CGH experiments and sequence analysis. M.Y.D. performed MIPs experiments, Illumina sequencing libraries construction, array CGH experiments and sequence analysis. J.H. performed PacBio sequence analysis and haplotypes reconstruction. P.H.S. and K.M.S. performed sequencing data analysis. T.A.G. and R.K.W. performed CH17 and nonhuman primate BAC clones capillary sequencing and analysis. L.V. and M.M. performed FISH experiments. M.M. performed array CGH experiments. B.M. performed PacBio sequencing libraries construction. L.D performed MIP experiments and PacBio sequencing libraries construction. A.R. performed PacBio sequence analysis. C.T.A., A.S. and J.T. performed VMRC53, VMRC54 and VMRC57 BAC libraries construction. J.A.R. and L.G.S. contributed to 15q13.3 microdeletion data collection. F.A., M.Y.D. and E.E.E. contributed to data interpretation. F.A., M.Y.D. and E.E.E. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and was an SAB member of Pacific Biosciences, Inc. (2009–2013) and SynapDx Corp. (2011–2013). J.A.R. is an employee of Signature Genomic Laboratories, a subsidiary of PerkinElmer, Inc. L.G.S. was an employee of Signature Genomic Laboratories and is now an employee of Genetic Veterinary Sciences, Inc.
References
- 1.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaminsky EB, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in medicine: official journal of the American College of Medical Genetics. 2011;13:777–84. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp AJ, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–8. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helbig I, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–2. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller DT, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–8. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinawi M, et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet. 2009;41:1269–71. doi: 10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams NM, et al. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. The Am J Psychiatry. 2012;169:195–204. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonacci F, et al. Characterization of six human disease-associated inversion polymorphisms. Hum Mol Gens. 2009;18:2555–66. doi: 10.1093/hmg/ddp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pujana MA, et al. Additional complexity on human chromosome 15q: identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26. Genome Res. 2001;11:98–111. doi: 10.1101/gr.155601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pujana MA, et al. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur J Hum Genet. 2002;10:26–35. doi: 10.1038/sj.ejhg.5200760. [DOI] [PubMed] [Google Scholar]
- 14.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 15.Zody MC, et al. Analysis of the DNA sequence and duplication history of human chromosome 15. Nature. 2006;440:671–5. doi: 10.1038/nature04601. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–8. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 17.Sudmant PH, et al. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 2013;23:1373–82. doi: 10.1101/gr.158543.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huddleston J, et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 2014 doi: 10.1101/gr.168450.113. [DOI] [PMC free article] [PubMed]
- 19.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–6. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis MY, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–22. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itsara A, et al. Resolving the breakpoints of the 17q21.31 microdeletion syndrome with next-generation sequencing. Am J Hum Genet. 2012;90:599–613. doi: 10.1016/j.ajhg.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.She X, et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–30. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- 23.Antonacci F, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat Genet. 2010;42:745–50. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardenbol P, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–8. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 26.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–83. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girirajan S, et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am J Hum Genet. 2013;92:221–37. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinberg KM, et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet. 2012 doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–42. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 31.Meyer M, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2014;505:403–6. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 32.Prufer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–9. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mefford HC, et al. Further clinical and molecular delineation of the 15q24 microdeletion syndrome. J Med Genet. 2012;49:110–8. doi: 10.1136/jmedgenet-2011-100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wat MJ, et al. Recurrent microdeletions of 15q25.2 are associated with increased risk of congenital diaphragmatic hernia, cognitive deficits and possibly Diamond--Blackfan anaemia. J Med Genet. 2010;47:777–81. doi: 10.1136/jmg.2009.075903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amos-Landgraf JM, et al. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–86. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Hattab AW, et al. Redefined genomic architecture in 15q24 directed by patient deletion/duplication breakpoint mapping. Hum Genet. 2009;126:589–602. doi: 10.1007/s00439-009-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marques-Bonet T, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–81. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Locke DP, et al. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol. 2003;4:R50. doi: 10.1186/gb-2003-4-8-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannuzzi G, et al. Hominoid fission of chromosome 14/15 and the role of segmental duplications. Genome Res. 2013;23:1763–73. doi: 10.1101/gr.156240.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp AJ, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet. 2007;16:567–72. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 41.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 42.Bengesser K, et al. A novel third type of recurrent NF1 microdeletion mediated by nonallelic homologous recombination between LRRC37B-containing low-copy repeats in 17q11.2. Hum Mutat. 2010;31:742–51. doi: 10.1002/humu.21254. [DOI] [PubMed] [Google Scholar]
- 43.Gordenin DA, et al. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–22. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays: news and reviews in molecular, cellular and developmental biology. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 45.Collick A, et al. Instability of long inverted repeats within mouse transgenes. The EMBO J. 1996;15:1163–71. [PMC free article] [PubMed] [Google Scholar]
- 46.Akgun E, et al. Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol. 1997;17:5559–70. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruskin B, Fink GR. Mutations in POL1 increase the mitotic instability of tandem inverted repeats in Saccharomyces cerevisiae. Genetics. 1993;134:43–56. doi: 10.1093/genetics/134.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–98. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 49.Inagaki H, et al. Two sequential cleavage reactions on cruciform DNA structures cause palindrome-mediated chromosomal translocations. Nat Comm. 2013;4:1592. doi: 10.1038/ncomms2595. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat Genet. 2005;37:320–7. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 51.Carvalho CM, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074–81. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 53.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLOS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLOS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons JD. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–9. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 56.Smith JJ, Stuart AB, Sauka-Spengler T, Clifton SW, Amemiya CT. Development and analysis of a germline BAC resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma. 2010;119:381–9. doi: 10.1007/s00412-010-0263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–7. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 59.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 60.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 61.Igartua C, et al. Targeted enrichment of specific regions in the human genome by array hybridization. In: Haines Jonathan L, et al., editors. Curr Protoc Hum Genet. Unit 18. Chapter 18. 2010. p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nuttle X, et al. Rapid and accurate large-scale genotyping of duplicated genes and discovery of interlocus gene conversions. Nature Methods. 2013;10:903–9. doi: 10.1038/nmeth.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.