Abstract
Previous studies have implicated autophagy in osteoclast differentiation. The aim of this study was to investigate the potential role of p62, a characterized adaptor protein for autophagy, in RANKL-induced osteoclastogenesis. Real-time quantitative PCR and western blot analyses were used to evaluate the expression levels of autophagy-related markers during RANKL-induced osteoclastogenesis in mouse macrophage-like RAW264.7 cells. Meanwhile, the potential relationship between p62/LC3 localization and F-actin ring formation was tested using double-labeling immunofluorescence. Then, the expression of p62 in RAW264.7 cells was knocked down using small-interfering RNA (siRNA), followed by detecting its influence on RANKL-induced autophagy activation, osteoclast differentiation, and F-actin ring formation. The data showed that several key autophagy-related markers including p62 were significantly altered during RANKL-induced osteoclast differentiation. In addition, the expression and localization of p62 showed negative correlation with LC3 accumulation and F-actin ring formation, as demonstrated by western blot and immunofluorescence analyses, respectively. Importantly, the knockdown of p62 obviously attenuated RANKL-induced expression of autophagy- and osteoclastogenesis-related genes, formation of TRAP-positive multinuclear cells, accumulation of LC3, as well as formation of F-actin ring. Our study indicates that p62 may play essential roles in RANKL-induced autophagy and osteoclastogenesis, which may help to develop a novel therapeutic strategy against osteoclastogenesis-related diseases.
Keywords: p62, RANKL, RAW264.7 cells, autophagy, osteoclastogenesis
Introduction
Throughout life, homeostasis of bone is elaborately maintained by a balance involving bone resorption and bone formation. Osteoclasts, which are confirmed as monocyte-macrophage derived multinucleated cells, are uniquely specialized to carry out bone resorption (Katagiri and Takahashi 2002; Lee et al. 2009). During the process of osteoclastogenesis, the finally differentiated cells are characterized as tartrate-resistant acid phosphatase (TRAP)-positive and multi-nucleated, and these two aspects are widely regarded as biological markers of mature osteoclasts (Bloemen et al. 2010).
Osteoclasts are derived from hematopoietic progenitors, which fuse to form multinucleated cells by an orchestrated process (Xu et al. 2009). In the presence of receptor activator of nuclear factor-κB ligand (RANKL), mouse RAW264.7 cells have been explained to possess the ability to differentiate into multinucleated, mature osteoclasts (Kim et al. 2008; Asai et al. 2014). RANKL as a member of the tumor necrosis factor (TNF) superfamily binds to the RANK on osteoclasts precursors to stimulate their differentiation and fusion into mature osteoclasts (Takayanagi 2009). After treatment with RANKL, mouse RAW264.7 cells have been shown to express high levels of osteoclast-related genes, including TRAP, Nuclear factor of activated T-cells, cytoplasmic 1 (NFATc1), Cathepsin K (CatK), and Fra-2 (Kim et al. 2009; Zhao et al. 2012).
The orchestrated process of osteoclast differentiation mainly involves rearrangement of the cytoskeleton, change of the organelle types, and the degradation and renewal of intracellular proteins (Chen and Olson 2005; Oberley et al. 2008; Oikawa et al. 2013). Autophagy, which has been implicated in numerous physiological and pathological processes in eukaryotic cells, possesses pivotal roles in regulating the degradation of cellular proteins and organelles (Mazure and Pouyssegur 2010; Mizushima and Levine 2010; Maruyama et al. 2014; Ryter et al. 2014). Moreover, several studies, including those from our laboratory, have implicated autophagy activation in osteoclast differentiation (Zhao et al. 2012; NY Lin et al. 2013; Xiu et al. 2014). Specifically, our previous study unmasked the precise molecular mechanisms underlying the activation of autophagy in mouse RAW264.7 cells under hypoxic conditions (Zhao et al. 2012). Generally, the activation of autophagy starts with the formation of characteristic double- or multi-membrane vesicles called autophagosomes, which wrap around parts of the cytosol or organelles. After that, autophagosomes fuse with lysosomal vesicles resulting in the formation of autolysosomes and completion of the maturation process (Liu et al. 2012). So far, more than 30 autophagy-related genes (Atg), including Atg5, microtubule-associated protein 1 light chain 3 (LC3), Beclin 1, Atg7, Atg12, and p62 (also known as SQSTM1/sequestome 1), have been verified to participate in the different stages of the autophagy activation (Mizushima et al. 2010; Rogov et al. 2014).
Recently, increasing evidence demonstrates that autophagy can degrade specific proteins and organelles under physiological conditions in order to complete the cell self-renewal process (Orvedahl et al. 2011; Rogov et al. 2014). Also, mounting evidence has identified various substrates of autophagy, including p62, neighbor of BRCA1 (NBR1), NDP52 and Nix. Among these substrates, p62, which binds ubiquitin and LC3, is the highest specific adaptor protein for autophagy, and it can regulate the formation of protein aggregates and is continuously degraded by autophagy (Komatsu et al. 2010; Johansen and Lamark 2011). Previous studies revealed that the adaptor protein p62 interacts with atypical protein kinase C (PKC), which is necessary for nuclear factor-kappa B (NF-κB) activation and osteoclast differentiation (Duran et al. 2004; Chamoux et al. 2013; Chung et al. 2014). These findings suggest a pivotal role for atypical PKC in p62-involved autophagy and osteoclastogenesis. Furthermore, p62 may interact with atypical PKC to affect a variety of cellular differentiation processes involved in the autophagy-lysosome pathway (Wang et al. 2011; Chamoux et al. 2013).
The aim of the present study was to investigate the potential role of p62 in soluble RANKL-induced osteoclastogenesis, which commonly occurs under both pathological and physiological conditions. Our results provide the first direct evidence to support p62 as an important bridging protein between RANKL-induced autophagy and osteoclastogenesis, and also reveal the potential involvement of autophagic degradation of p62 in the formation of F-actin rings. Our findings may shed new light on the complexity of osteoclastogenesis-related diseases, where autophagy may develop a new therapeutic strategy.
Materials & Methods
Reagents and Antibodies
Recombinant murine soluble RANKL (sRANKL) and macrophage colony-stimulating factor (M-CSF) were obtained from Peprotech (London, United Kingdom). Primary antibodies against LC3 and Atg5 were obtained from Cell Signaling Technology (Danvers, MA) (Zhao et al. 2012). The primary antibodies against p62 were obtained from Epitomics (Burlingame, CA) (Meng et al. 2014). Primary antibodies to GAPDH from Santa Cruz Biotechnology (Dallas, TX) served as a loading control (Yu et al. 2010). Tetramethyl rhodamine isothiocyanate (TRITC)- and fluorescein isothiocyanate (FITC)-conjugated secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA). FITC-labeled phalloidin was obtained from Sigma-Aldrich (St. Louis, MO). Both the p62 siRNA with lentivirus and the corresponding empty vectors were provided by Genechem (Shanghai, China). Culture media and fetal bovine serum (FBS) were provided by Gibco-BRL, Life Technologies (Gaithersburg, MD). All other reagents were from Sigma-Aldrich.
In vitro Osteoclastogenesis Assay
Mouse macrophage-like RAW264.7 cells were purchased from the China Center for Type Culture Collection (Wuhan, China). RAW264.7 cells were incubated (2.0×104 cells/well) overnight in 24-well plates containing α-minimal essential medium (α-MEM) with 10% FBS. After stimulation with 50 ng/ml sRANKL for 5 days, the cells were fixed with 4% paraformaldehyde for 10 min and then TRAP staining was performed using an Acid Phosphatase Kit (Sigma-Aldrich).
For bone marrow-derived macrophage (BMM) cultures, the femurs and tibiae of C57/BL6 male mice (4-6 weeks old, 18-20 g; purchased from the Experimental Animal Center of Wuhan University, China) were flushed using α-MEM with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 100 ng/ml M-CSF to obtain bone marrow cells. These cells were cultured for 20 hr, the non-adherent cells were collected and used as BMMs. BMMs were then seeded (6.0×104 cells/well) into 24-well plates and stimulated with 30 ng/ml M-CSF and 100 ng/ml RANKL. After 6 days of culture, cells were fixed and stained for TRAP. TRAP-positive, multinucleated cells containing three or more nuclei were considered as osteoclasts. Osteoclasts were observed and counted using a Leica light microscope (Solms, Germany). These animal experiments were approved and supervised by the Institutional Animal Care and Use Committee of Wuhan University, China.
Animal Models
All animal experiments were approved and supervised by the Institutional Animal Care and Use Committee of Wuhan University, China. Sprague-Dawley rats, weighing ~230g, were purchased from the Experimental Animal Center of Wuhan University. Rats were housed four or five per cage in temperature-controlled rooms. Rats were anesthetized by intraperitoneal administration of ketamine (80 mg/kg). Rats without additional treatment were used as normal controls. For induction of periapical lesions, the pulp chamber of the mandibular first molars was exposed with a round bur. The exposed teeth remained open to the oral environment throughout the experiment. Rats were euthanized at 21 days after lesion induction, as described elsewhere (Sun et al. 2014).
An experimental periodontitis model was induced by ligature placement around the cervix of the mandibular first molars. The thread was carefully pressed into the gingival sulcus and ligated at almost the same position relative to the gingival margin. Rats with ligatures were euthanized at 14 days after lesion induction, as described previously (Holzhausen et al. 2002). After establishment of the above models, the rat mandibles were fixed in 4% paraformaldehyde for 1 day, and then demineralized in 10% EDTA solution for 6 weeks. After that, the samples were embedded in paraffin using standard laboratory procedures, and 4-µm-thick sections were cut along a specific direction.
Lentivirus Transfection and Cell Viability Assay
Cells were cultured in 3.5-cm dishes and then transfected with p62 siRNA containing lentivirus or corresponding vector with 4 µg/ml polybrene. The lentivirus infection efficiency was always higher than 95%, as determined by ZsGreen1 expression.
For cell viability, the number of total and viable cells was determined using the Vi-CELL cell viability analyzer (Beckman Coulter; Fullerton, CA).
Detection of F-actin Ring Formation
For detection of F-actin ring formation, the pretreated RAW264.7 cells were fixed in 4% paraformaldehyde after washing in phosphate-buffered saline (PBS). After that, the cells were permeabilized with 0.1% Triton X-100 for 10 min, blocked with bovine serum albumin buffer, and then incubated with FITC-labeled phalloidin for 45 min. Cell nuclei were stained with 4’, 6-diamidino-2-phenylindole (DAPI), and a Leica fluorescence microscope was used to observe the F-actin ring formation.
Immunofluorescence Analysis
For immunofluorescence analyses, the pretreated RAW264.7 cells (2.0×105) were fixed and permeabilized with 0.1% Triton X-100 after washing with PBS. After that, a blocking solution of 10% non-immune goat serum was used for 1 hr at 37C. Then the cells were treated with rabbit anti-mouse LC3 or p62 antibody (1:100) overnight at 4C, following treatment with TRITC-conjugated goat anti-rabbit antibody for 1 hr. The colocalization of LC3 or p62 with F-actin ring was detected by double-labeling immunofluorescence. Nuclei were counterstained with DAPI, followed by observation under a Leica fluorescence microscope. For negative controls, the primary antibody was substituted with nonimmune isotype antibodies.
Real-time Quantitative PCR
Total RNA was isolated for the synthesis of cDNA, which was used for real-time quantitative PCR (qPCR), according to our previous study (Zhao et al. 2012). The mRNA expression is normalized using β-actin expression in the analyzed samples. The specific nucleotide primer sequences for PCR are shown in Table 1.
Table 1.
Primer Sequences Used for Real Time Quantitative PCR.
| Gene | Accession No. | Sequence (5’ to 3’) | Amplicon size, bp |
|---|---|---|---|
| GAPDH | NM_008084.3 | 5’-CCTGGAGAAACCTGCCAAGTAT | 133 |
| 5’-AGAGTGGGAGTTGCTGTTGAAG | |||
| TRAP | NM_001111036.1 | 5’-TGACAAGAGGTTCCAGGA | 318 |
| 5’-AGCCAGGACAGCTGAGTG | |||
| NFATc1 | NM_001164111.1 | 5’-GCATCACAGGGAAGACCGTGTC | 153 |
| 5’-GAAGTTCAATGTCGGAGTTTCTGAG | |||
| CatK | NM_007802.4 | 5’-ACCGGGGTATTGACTCTGAA | 190 |
| 5’-GAGGTCAGGCTTGCATCAAT | |||
| Fra-2 | NM_008037.4 | 5’-GATCAAGACCATTGGCACCAC | 79 |
| 5’-GCGACGCTTCTCCTCCTCT | |||
| Beclin1 | NM_019584.3 | 5’-AGCTCAGTACCAGCGGGAGT | 147 |
| 5’-TGGAAGGTGGCATTGAAGAC | |||
| Atg5 | NM_053069.5 | 5’-GCCTATATGTACTGCTTCATCCA | 267 |
| 5’-CATTTCAGGGGTGTGCCTTCA | |||
| Atg7 | NM_028835.4 | 5’-GCTGTGGAGCTGATGGTCTC | 166 |
| 5’-CCAGGCTGACAGGAAGAACA | |||
| Atg12 | NM_026217.3 | 5’-AACAAAGAAATGGGCTGTGG | 198 |
| 5’-TTGCAGTAATGCAGGACCAG | |||
| LC3 | NM_026160.4 | 5’-CGGAGCTTTGAACAAAGAGTG | 279 |
| 5’-TCTCTCACTCTCGTACACTTC |
Atg, autophagy-related genes; CatK, Cathepsin K; LC3, microtubule-associated protein 1 light chain 3; NFATc1, nuclear factor of activated T-cells, cytoplasmic 1 (NFATc1); TRAP, tartrate-resistant acid phosphatase.
Western Blot Analysis
Western blot analysis was carried out according to our previous described method (Zhang et al. 2013). Briefly, total proteins from RAW264.7 cells were collected, 25 µg of protein was separated on 10% SDS-polyacrylamide gels, and the gels were electroblotted onto polyvinylidene fluoride membranes (Roche Applied Science, Basel, Switzerland). Blots were then blocked overnight using 5% non-fat milk and probed with the corresponding primary antibodies at the following concentrations: 1:1000 for LC3, 1:800 for Atg5, 1:1000 for p62, and 1:800 for GAPDH. Membranes were then washed and incubated with the horseradish peroxidase-conjugated secondary antibody at a dilution of 1:2000, followed by washing and chemiluminescence using a chemiluminescence substrate kit (both from Pierce Biotechnology; Rockford, IL).
Statistical Analysis
Statistical analysis was performed using a Student’s t-test. All values are expressed as the mean ± SEM of three independent experiments. Significance was set at p<0.05.
Results
sRANKL Activates Autophagy during Osteoclastogenesis
As shown in Supplemental Figure 1A, we initially treated RAW264.7 cells with sRANKL to induce osteoclast differentiation, and detected the formation of TRAP-positive multinuclear (>3) cells and F-actin rings, two significant markers of osteoclast differentiation. The up-regulated expression of several osteoclastogenesis-associated genes also confirmed the formation of osteoclasts in these cultures (Supplemental Fig. 1B).
To further determine the involvement of autophagy in sRANKL-induced osteoclastogenesis, we evaluated the expression levels of autophagy-related markers in RAW264.7 cells after treatment with sRANKL. First, the mRNA expression levels of Atg5, Beclin 1, Atg7 and Atg12 were examined, and shown to be significantly up-regulated in mature osteoclasts as compared with osteoclast precursors (Fig. 1A). Next, to further verify autophagy activation in sRANKL-induced osteoclastogenesis, we detected the LC3-II/LC3-I ratio, which is indicative of the status of autophagy, as well as changes in Atg5 protein expression. We found that the LC3-II/LC3-I ratio and Atg5 protein expression were significantly up-regulated in a time-dependent manner during osteoclastogenesis. Finally, we sought to detect the expression level of p62, a characterized adaptor protein for autophagy, during osteoclastogenesis. We found that p62 was significantly down-regulated during the initial stages of RANKL-induced osteoclastogenesis and then gradually increased over time in culture (Fig. 1B). This down-regulation of p62 protein was associated with the increase in the LC3-II/LC3-I ratio, which indicates the autophagic degradation of p62. To confirm these results, RAW264.7 cells were pretreated with the autophagy inhibitor 3-methyladenine (3-MA) and then incubated with sRANKL. As shown in Figure 1C, autophagy inhibition by 3-MA significantly rescued the degradation of p62, suggesting that the down-regulation of p62 at the initial stage of RANKL-induced osteoclastogenesis was probably due to autophagy activation. Collectively, these observations indicate that sRANKL can induce autophagy in osteoclasts.
Figure 1.

sRANKL activates autophagy in osteoclasts. (A) The mRNA expression levels of autophagy-related genes were determined by real-time quantitative PCR using total RNA isolated from RAW264.7 cells cultured to form osteoclasts. RAW264.7 cells without treatment with sRANKL were used as negative controls. mRNA expression was normalized with β-actin in the analyzed samples. (B) The expression levels of Atg5, LC3 and p62 during osteoclastogenesis were detected by western blotting. (C) RAW264.7 cells were pretreated with 3-MA (4 mM) for 4 hr, followed by incubation with sRANKL. The autophagy inhibitor 3-MA could rescue the degradation of p62 on the first day of osteoclastogenesis. The data represent the mean ± SE from three independent experiments performed in duplicate. *p<0.05; **p<0.01 versus the negative control.
Correlation between p62 Expression and F-actin Ring Formation
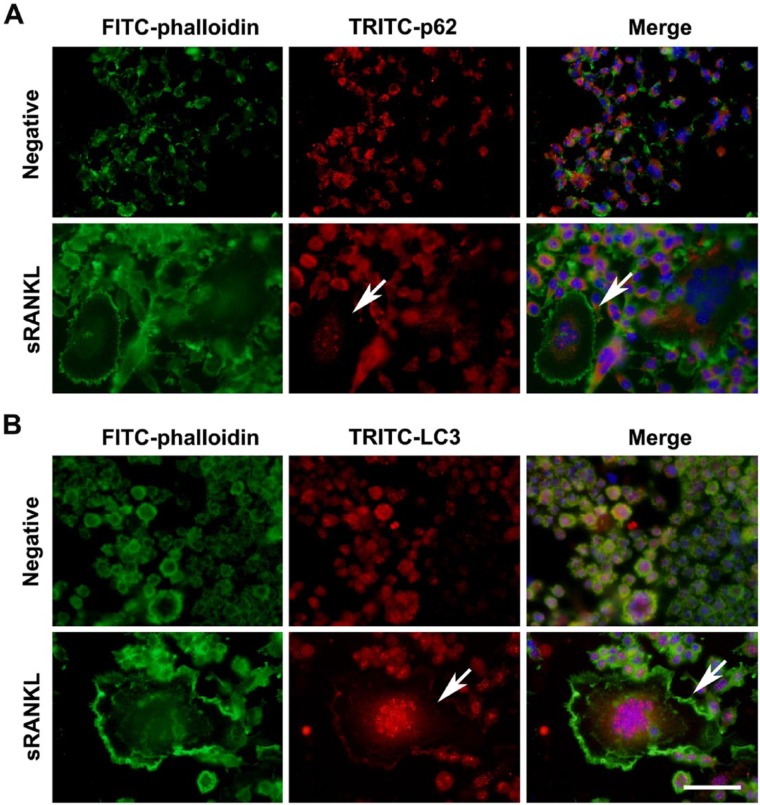
F-actin ring formation is a significant indicator of osteoclast differentiation. In order to evaluate the possible involvement of p62 in F-actin ring formation, we next investigated the localization of p62 in sRANKL-induced osteoclasts using double-labeling immunofluorescence. As shown in Figure 2A, the expression of p62 was down-regulated in the center of the F-actin ring in the sRANKL-induced osteoclasts, concomitant with an increase in LC3 accumulation (Fig. 2B). These findings suggested that there was a possible correlation between p62 expression and F-actin ring formation.
Figure 2.
Correlation between p62 expression and F-actin ring formation during osteoclastogenesis. Immunofluorescence staining was performed to locate TRITC-conjugated p62 or LC3 and FITC-labeled F-actin ring. (A) During sRANKL-induced osteoclastogenesis, p62 expression is decreased (indicated by arrows) in the center of F-actin ring. (B) During sRANKL-induced osteoclastogenesis, the accumulation of LC3 was observed (indicated by arrows) in the center of F-actin ring. Scale, 50 µm.
Knockdown of p62 Inhibits Autophagy Activation in sRANKL-induced Osteoclasts
To verify the effect of p62 on autophagy activation in sRANKL-induced osteoclasts, the expression of p62 was knocked down using a lentiviral siRNA. Both the mRNA and protein expression levels of p62 were significantly decreased in osteoclasts after treatment with p62 siRNA as compared with the negative control (Fig. 3A and 3B). As autophagy activation has been shown to degrade p62 protein and p62 is also considered to be a regulator of autophagy flux, we next studied whether p62 knockdown changes the expression of autophagy-related genes. As shown in Figure 3C, we observed that the knockdown of p62 obviously decreased the mRNA expression levels of Atg5, Atg7, LC3 and Atg12 in sRANKL-treated osteoclasts. Thus, all of the data suggest that the knockdown of p62 inhibits autophagy activation in osteoclasts induced by sRANKL.
Figure 3.

Knockdown of p62 inhibits autophagy activation in osteoclasts induced by sRANKL. (A, B) Both the mRNA and protein expression levels of p62 were decreased in osteoclasts after treatment with p62 siRNA as compared with the negative control (NC). (C) Knockdown of p62 significantly decreased the mRNA expression levels of Atg5, Atg7, LC3 and Atg12 in sRANKL-treated osteoclasts. mRNA expression was normalized against β-actin in the samples analyzed. The data represent mean ± SEM from three different experiments performed in duplicate. *p<0.05; **p<0.01 versus the negative control.
Knockdown of p62 Inhibits Osteoclastogenesis
To further investigate the role of p62 in sRANKL-induced osteoclastogenesis, the potential for RAW264.7 cells to form multinuclear osteoclasts was detected after transfection with p62 siRNA. We found that the number of TRAP-positive multinuclear cells was significantly decreased after knockdown of p62 as compared with the negative control (Fig. 4A). Also, we found that knockdown of p62 inhibited the formation of F-actin ring (Fig. 4B). We also showed a significant decrease in the mRNA expression levels of osteoclastogenesis-associated genes, TRAP, NFATc1, CatK and Fra-2 (Fig. 4C). Taken together, these results indicate that the adaptor protein p62 may play essential roles in F-actin ring formation during sRANKL-induced osteoclastogenesis.
Figure 4.
Knockdown of p62 inhibits osteoclastogenesis and F-actin ring formation. (A) In RAW264.7 cells after transfection with p62 siRNA, the number of TRAP-positive multinuclear cells was significantly decreased as compared with the negative control (NC). (B) F-actin ring formation was inhibited in osteoclasts after transfection with p62 siRNA. (C) The mRNA expression levels of osteoclastogenesis-related genes were significantly decreased in RAW264.7 cells after knockdown of p62 as compared with the negative control (NC). mRNA expression was normalized against β-actin in the samples analyzed. The data represent the mean ± SEM from three different experiments performed in duplicate. *p<0.05; **p<0.01 versus the negative control. Scale, 50 µm.
Expression of p62 during Osteoclastogenesis In Vivo
In order to investigate the role of p62 in osteoclastogenesis in vivo, we used animal models of periapicitis and periodontitis to detect the expression level of p62 in osteoclasts. For histological examination, TRAP staining was used to identify osteoclasts on the bone surface. TRAP-positive multinuclear cells were dark red to purple in color, and were observed in periapical and periodontic lesions, respectively. No osteoclasts were observed in the normal control group. We show that p62 was obviously expressed in osteoclasts in both periapicitis and periodontitis models as compared with the normal control group (Fig. 5). These results were consistent with the aforementioned in vitro findings, and again indicated that p62 was involved in osteoclastogenesis.
Figure 5.
Expression of p62 during osteoclastogenesis in vivo. The immunofluorescence results showed that p62 were obviously expressed in osteoclasts (TRAP-positive cells) in both the periapical and periodontic lesions (arrows) as compared with the normal control group. Scale, 50 µm.
Discussion
Bone is elaborately remodeled by the coordinated efforts of the bone-forming osteoblasts and the bone-resorbing osteoclasts (Lee et al. 2009; von Metzler et al. 2009). Osteoclasts originate from hematopoietic progenitors, which fuse to form multinucleated cells by an orchestrated process. RANKL and M-CSF are two main molecules required for osteoclast precursors to mature into osteoclasts (Lee et al. 2009; Kimura et al. 2014). In the presence of sRANKL, the mouse macrophage-like RAW264.7 cells have the ability to differentiate into TRAP-positive multinuclear cells (Kim et al. 2008; Asai et al. 2014). The differentiation of osteoclast precursors into mature osteoclasts occurs through a complicated process that requires a turnover of intracellular proteins and organelles (Chen and Olson 2005; Oikawa et al. 2013). Moreover, accumulating evidence demonstrates that autophagy can degrade specific proteins and organelles under physiological conditions so as to complete the cell’s self-renewal process (Orvedahl et al. 2011). In the present study, mouse RAW264.7 cells were induced to form mature osteoclasts in the presence of sRANKL. During sRANKL-induced osteoclastogenesis, we confirmed the occurrence of autophagy through the dection of autophagy-related markers.
Autophagy has been demonstrated to play pivotal roles in a wide array of physiological and pathological processes in eukaryotic cells (Mazure and Pouyssegur 2010; Ryter et al. 2014). Among approximately 30 autophagy-related genes, LC3, the first-identified mammalian protein, can bind with autophagic membranes and is generally used to monitor the process of autophagy. In addition, p62 becomes incorporated into autophagosomes and can thus also be used as an autophagy flux marker because of its characteristic accumulation accompanied by degradation (Maynard et al. 2010). Generally, autophagy is considered to be an important process in the degradation of long-lived proteins. Various studies have identified and characterized new substrates of autophagy, including p62, NBR1, NDP52 and Nix. Among these characterized substrates, p62 is the highest specific receptor protein for autophagy (Johansen and Lamark 2011). Our present study showed that p62 protein is significantly degraded at the initial stage of RANKL-induced osteoclastogenesis, and associated with the increase in autophagy-related gene expression and LC3-II/LC3-I ratio, suggesting autophagic degradation of p62. Moreover, double-labeling immunofluorescence analyses revealed a correlation between p62 expression and F-actin ring formation, and the distribution of LC3 also showed a close relationship with F-actin ring formation. Our results thus suggest a possible link between autophagic degradation of p62 and F-actin ring formation.
p62 is a scaffold protein that takes part in the regulation of autophagy, apoptosis and osteoclast formation, possessing an N-terminal PB1 domain, a C-terminal ubiquitin-associated domain (UBA) as well as the LC3-interacting region (LIR) (X Lin et al. 2013; Zhou et al. 2013). Recent studies have revealed that p62 can directly bind LC3 via its LIR motif, thereby guiding the autophagic uptake of proteins along with its own degradation. In addition, mutations in SQSTM1, the gene encoding p62, can lead to Paget’s disease, which causes various complications, including bone pain, deformity, pathological fractures and deafness; this also indicates a possible role of p62-dependent autophagy in osteoclastogenesis (Yip et al. 2006; Daroszewska et al. 2011). Correspondingly, our results showed a significant decrease in the number of TRAP-positive multinuclear cells and attenuation in the expression of autophagy-associated genes after knockdown of p62 in RAW264.7 cells. To further confirm these results, we also used siRNA assays to deplete the endogenous p62 in mouse BMMs and found that the knockdown of p62 reduced the number of TRAP-positive multinuclear cells as compared with the negative control (Supplemental Fig. 2). Importantly, double-labeling immunofluorescence results also showed that knockdown of p62 affected the F-actin ring formation and LC3 protein distribution. These results indicate that the autophagic degradation of p62 is essential for RANKL-induced osteoclastogenesis. Of note, previous studies have demonstrated that the adaptor protein p62 is required for NF-κB activation and osteoclast differentiation, and meanwhile suggested a pivotal role of atypical PKC in p62-involved autophagy and osteoclastogenesis (Duran et al. 2004; Chamoux et al. 2013). However, to the best of our knowledge, the present study provides the first direct evidence to support p62 as an important bridge protein between RANKL-induced autophagy and osteoclastogenesis. Also, we have revealed for the first time the potential involvement of autophagic degradation of p62 in the formation of F-actin rings.
In summary, the present study reveals that autophagic degradation of p62 may play an essential role in F-actin ring formation during sRANKL-induced osteoclastogenesis. Although, further studies are still needed to investigate the precise mechanism of sRANKL-induced osteoclast differentiation, our findings may shed new light onto the complexity of osteoclastogenesis-related diseases, where autophagy may develop as a new therapeutic strategy.
Supplementary Material
Footnotes
Supplementary Material: Supplementary material for this article is available on the Journal of Histochemistry & Cytochemistry Web site at http://jhc.sagepub.com/supplemental.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Fundamental Research Funds for the Central Universities (121099), National Natural Science Foundation of China (81300855), and Key Project of Science and Technology of Wuhan (201260523196).
References
- Asai K, Funaba M, Murakami M. (2014). Enhancement of RANKL-induced MITF-E expression and osteoclastogenesis by TGF-beta. Cell Biochem Funct 32:401-409. [DOI] [PubMed] [Google Scholar]
- Bloemen V, Schoenmaker T, de Vries TJ, Everts V. (2010). Direct cell-cell contact between periodontal ligament fibroblasts and osteoclast precursors synergistically increases the expression of genes related to osteoclastogenesis. J Cell Physiol 222:565-573. [DOI] [PubMed] [Google Scholar]
- Chamoux E, McManus S, Laberge G, Bisson M, Roux S. (2013). Involvement of kinase PKC-zeta in the p62/p62(P392L)-driven activation of NF-kappaB in human osteoclasts. Biochim Biophys Acta 1832:475-484. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. (2005). Unveiling the mechanisms of cell-cell fusion. Science 308:369-373. [DOI] [PubMed] [Google Scholar]
- Chung YH, Jang Y, Choi B, Song DH, Lee EJ, Kim SM, Song Y, Kang SW, Yoon SY, Chang EJ. (2014). Beclin-1 Is Required for RANKL-Induced Osteoclast Differentiation. J Cell Physiol. 229:1963-1971 [DOI] [PubMed] [Google Scholar]
- Daroszewska A, van ‘t, Hof RJ, Rojas JA, Layfield R, Landao-Basonga E, Rose L, Rose K, Ralston SH. (2011). A point mutation in the ubiquitin-associated domain of SQSMT1 is sufficient to cause a Paget’s disease-like disorder in mice. Hum Mol Genet 20:2734-2744. [DOI] [PubMed] [Google Scholar]
- Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT. (2004). The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell 6:303-309. [DOI] [PubMed] [Google Scholar]
- Holzhausen M, Rossa Junior C, Marcantonio Junior E, Nassar PO, Spolidorio DM, Spolidorio LC. (2002). Effect of selective cyclooxygenase-2 inhibition on the development of ligature-induced periodontitis in rats. J Periodontol 73:1030-1036. [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7:279-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Takahashi N. (2002). Regulatory mechanisms of osteoblast and osteoclast differentiation. Oral Dis 8:147-159. [DOI] [PubMed] [Google Scholar]
- Kim MH, Ryu SY, Bae MA, Choi JS, Min YK, Kim SH. (2008). Baicalein inhibits osteoclast differentiation and induces mature osteoclast apoptosis. Food Chem Toxicol 46:3375-3382. [DOI] [PubMed] [Google Scholar]
- Kim MH, Ryu SY, Choi JS, Min YK, Kim SH. (2009). Saurolactam inhibits osteoclast differentiation and stimulates apoptosis of mature osteoclasts. J Cell Physiol 221:618-628. [DOI] [PubMed] [Google Scholar]
- Kimura K, Kitaura H, Fujii T, Ishida M, Hakami Z, Takano-Yamamoto T. (2014). An anti-c-Fms antibody inhibits osteoclastogenesis in a mouse periodontitis model. Oral Dis 20:319-324. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. (2010). The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12:213-223. [DOI] [PubMed] [Google Scholar]
- Lee MS, Kim HS, Yeon JT, Choi SW, Chun CH, Kwak HB, Oh J. (2009). GM-CSF regulates fusion of mononuclear osteoclasts into bone-resorbing osteoclasts by activating the Ras/ERK pathway. J Immunol 183:3390-3399. [DOI] [PubMed] [Google Scholar]
- Lin NY, Beyer C, Giessl A, Kireva T, Scholtysek C, Uderhardt S, Munoz LE, Dees C, Distler A, Wirtz S, Kronke G, Spencer B, Distler O, Schett G, Distler JH. (2013). Autophagy regulates TNFalpha-mediated joint destruction in experimental arthritis. Ann Rheum Dis 72:761-768. [DOI] [PubMed] [Google Scholar]
- Lin X, Li S, Zhao Y, Ma X, Zhang K, He X, Wang Z. (2013). Interaction domains of p62: a bridge between p62 and selective autophagy. DNA and Cell Biology 32:220-227. [DOI] [PubMed] [Google Scholar]
- Liu C, Yan X, Wang HQ, Gao YY, Liu J, Hu Z, Liu D, Gao J, Lin B. (2012). Autophagy-independent enhancing effects of Beclin 1 on cytotoxicity of ovarian cancer cells mediated by proteasome inhibitors. BMC Cancer 12:622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y, Sou YS, Kageyama S, Takahashi T, Ueno T, Tanaka K, Komatsu M, Ichimura Y. (2014). LC3B is indispensable for selective autophagy of p62 but not basal autophagy. Biochem Biophys Res Commun 446:309-315. [DOI] [PubMed] [Google Scholar]
- Maynard AA, Dvorak K, Khailova L, Dobrenen H, Arganbright KM, Halpern MD, Kurundkar AR, Maheshwari A, Dvorak B. (2010). Epidermal growth factor reduces autophagy in intestinal epithelium and in the rat model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol 299:G614-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazure NM, Pouyssegur J. (2010). Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol 22:177-180. [DOI] [PubMed] [Google Scholar]
- Meng G, Xia M, Xu C, Yuan D, Schnurr M, Wei J. (2014). Multifunctional antitumor molecule 5’-triphosphate siRNA combining glutaminase silencing and RIG-I activation. Int J Cancer 134:1958-1971. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. (2010). Autophagy in mammalian development and differentiation. Nat Cell Biol 12:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. (2010). Methods in mammalian autophagy research. Cell 140:313-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberley TD, Swanlund JM, Zhang HJ, Kregel KC. (2008). Aging results in increased autophagy of mitochondria and protein nitration in rat hepatocytes following heat stress. J Histochem Cytochem 56:615-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Kuroda Y, Matsuo K. (2013). Regulation of osteoclasts by membrane-derived lipid mediators. Cell Mol Life Sci 70:3341-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Jr., Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. (2011). Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480:113-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, Kirkin V. (2014). Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 53:167-178. [DOI] [PubMed] [Google Scholar]
- Ryter SW, Mizumura K, Choi AM. (2014). The Impact of Autophagy on Cell Death Modalities. Int J Cell Biol 2014:502676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Wang L, Peng B. (2014). Kinetics of glycogen synthase kinase (GSK)3beta and phosphorylated GSK3beta (Ser 9) expression in experimentally induced periapical lesions. Int Endod J doi: 10.1111/iej.12258. [DOI] [PubMed] [Google Scholar]
- Takayanagi H. (2009). Osteoimmunology and the effects of the immune system on bone. Nat Rev Rheumatol 5:667-676. [DOI] [PubMed] [Google Scholar]
- von Metzler I, Krebbel H, Kuckelkorn U, Heider U, Jakob C, Kaiser M, Fleissner C, Terpos E, Sezer O. (2009). Curcumin diminishes human osteoclastogenesis by inhibition of the signalosome-associated I kappaB kinase. J Cancer Res Clin Oncol 135:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cao L, Kang R, Yang M, Liu L, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Tang D. (2011). Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARalpha oncoprotein. Autophagy 7:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu Y, Xu H, Zhao C, Li J, Morita Y, Yao Z, Xing L, Boyce BF. (2014). Chloroquine reduces osteoclastogenesis in murine osteoporosis by preventing TRAF3 degradation. J Clin Invest 124:297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu HF, Ang ES, Yip K, Woloszyn M, Zheng MH, Tan RX. (2009). NF-kappaB modulators in osteolytic bone diseases. Cytokine Growth Factor Rev 20:7-17. [DOI] [PubMed] [Google Scholar]
- Yip KH, Feng H, Pavlos NJ, Zheng MH, Xu J. (2006). p62 ubiquitin binding-associated domain mediated the receptor activator of nuclear factor-kappaB ligand-induced osteoclast formation: a new insight into the pathogenesis of Paget’s disease of bone. Am J Pathol 169:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Zheng M, Wang W, Rozanski GJ, Zucker IH, Gao L. (2010). Developmental changes in AT1 and AT2 receptor-protein expression in rats. J Renin Angiotensin Aldosterone Syst 11:214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen G, Ren JG, Zhao YF. (2013). Bleomycin induces endothelial mesenchymal transition through activation of mTOR pathway: a possible mechanism contributing to the sclerotherapy of venous malformations. Br J Pharmacol 170:1210-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen G, Zhang W, Xu N, Zhu JY, Jia J, Sun ZJ, Wang YN, Zhao YF. (2012). Autophagy regulates hypoxia-induced osteoclastogenesis through the HIF-1alpha/BNIP3 signaling pathway. J Cell Physiol 227:639-648. [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang HF, Ren HG, Chen D, Gao F, Hu QS, Fu C, Xu RJ, Ying Z, Wang GH. (2013). Bcl-2-dependent upregulation of autophagy by sequestosome 1/p62 in vitro. Acta Pharmacol Sin 34:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.