Abstract
MicroRNAs (miRNAs) provide insight into both the biology and clinical behavior of many human cancers, including nasopharyngeal carcinoma (NPC). The dysregulation of miRNAs in NPC results in a variety of tumor-promoting effects. Furthermore, several miRNAs are prognostic markers for NPC. In addition to cellular miRNAs, NPC samples also often contain miRNAs encoded by Epstein-Barr virus, and these miRNAs may impact NPC biology by targeting both cellular and viral genes. Given their numerous putative roles in NPC development and progression, a thorough understanding of the impact of miRNA dysregulation in NPC is expected to shed light on useful biomarkers and therapeutic targets for the clinical management of this disease. In this review, we describe the efforts to date to identify and characterize such miRNAs in the context of NPC.
Keywords: Nasopharyngeal carcinoma, microRNA, gene regulation, small RNAs, Epstein-Barr virus
MicroRNAs (miRNAs) have been shown to provide insight into both the biology and clinical behavior of numerous human cancers, including nasopharyngeal carcinoma (NPC). miRNAs are known to function as both tumor suppressor genes and oncogenes, and their dysregulation has been found to be related to disease prognosis and clinical outcome. Hence, the examination of miRNA dysregulation in NPC can (1) provide useful insight into the biological workings of this disease, aiding in the development of novel targeted therapies and (2) provide clinical prognostic and predictive biomarkers to aid physicians in treatment decision making, and thus improve outcome for future NPC patients.
miRNAs
miRNAs are endogenous, small (18–25 nt), non-protein-coding RNA molecules[1]. Originally discovered in C. elegans, miRNAs have now been identified in over 200 different species[2]. In general, miRNAs bind to transcripts of target protein-coding genes in a sequence-specific manner, functioning primarily to decrease the transcript and/or protein levels of their targets. However, miRNA-target interactions resulting in increased protein levels have also been noted[3],[4]. In recent decades, miRNAs have been increasingly recognized as important genetic regulators in the mammalian system[1]. Moreover, numerous miRNAs have been reported to function as both oncogenes and tumor suppressors, regulating tumor initiation and progression at all levels[5].
miRNA Biogenesis and Function
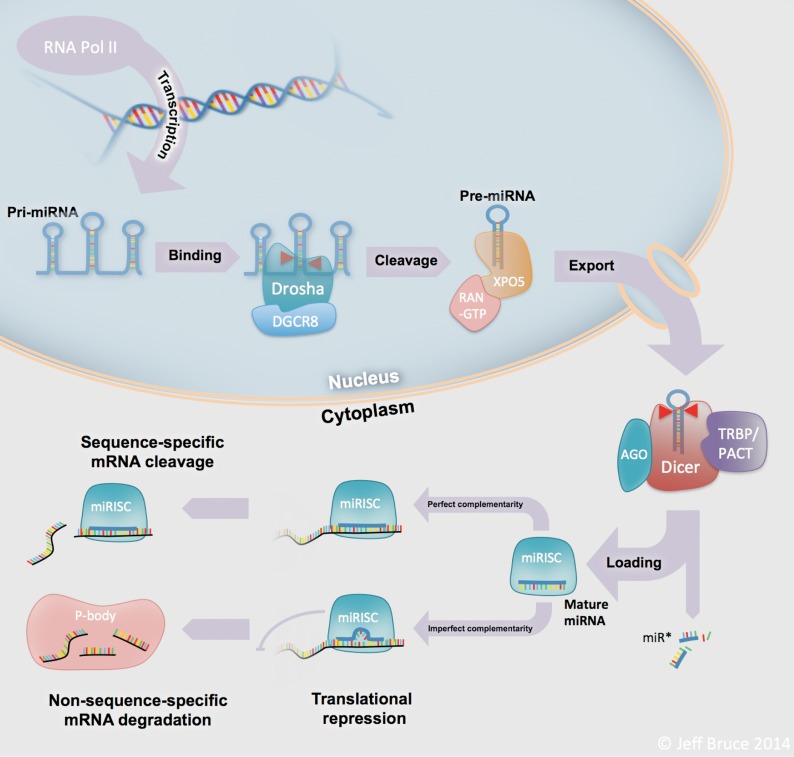
The biogenesis of miRNAs is a multistep process that is tightly regulated within the cell. Figure 1 depicts the steps in canonical miRNA processing—from transcription in the nucleus to the interaction with mRNAs in the cytoplasm. Genes that encode miRNAs can be located in intergenic regions or within the exons or introns of other genes. Transcription of these miR-genes is performed predominantly by RNA polymerase II (Pol II), though RNA Pol III performs transcription in some cases[6],[7]. A variety of Pol II–associated transcription factors direct miRNA transcription, thereby regulating miRNA expression at the level of transcription[8]. The RNA product that is transcribed is called a primary miRNA (pri-miRNA). These pri-miRNAs vary in length from hundreds to thousands of base pairs and exist in diverse stem-loop structures[6]. Following transcription, the pri-miRNA is cleaved within the nucleus by the type III RNase Drosha, in association with DiGeorge syndrome critical region gene 8 (DGCR8), at the stem of the loop structure to release a ∼70 nt precursor miRNA (pre-miRNA)[6]. This pre-miRNA is then exported out of the nucleus by the dsRNA-specific transporting complex composed of Exportin-5 and GTP-bound ras-related nuclear protein (RAN-GTP)[9].
Figure 1. MicroRNA (miRNA) biogenesis.
miRNAs are processed through a complex series of highly regulated steps in the nucleus and the cytoplasm, from transcription through to their functional roles as transcript and protein level regulators. Abbreviations: RNA Pol II, RNA polymerase type II; pri-miRNA, primary microRNA; pre-miRNA, precursor microRNA; DGCR8, Drosha-DiGeorge syndrome critical region gene 8; XPO5, nuclear export factor exportin 5; RAN, ras-related nuclear protein; GTP, guanosine tri-phosphate; AGO, Argonaute; TRBP, TAR (HIV-1) RNA-binding protein 2; PACT, protein kinase, interferon-inducible double stranded RNA; Dicer, Dicer 1 ribonuclease Type III; miR*, passenger strand from mature miRNA duplex; miRISC, microRNA RNA-induced silencing complex; P-body, processing body.
In the cytoplasm, the pre-miRNA is further processed by Dicer and its cofactor trans-activation-responsive RNA-binding protein (TRBP) to release the ∼22 nt, mature miRNA duplex[6]. One strand of this duplex, termed the “guide strand,” is then preferentially incorporated into the miRNA-inducible silencing complex (mi-RISC)[6]. Although this process is not completely understood, the guide strand is almost always the strand with the 5′ terminus that is least thermodynamically stable[10]. The miRISC is a multipart entity with several potential members. The key functional element of miRISC is the Argonaute protein (in mammals, one of AGO1-4)[6]. In addition to Argonaute, several other proteins may be involved in miRISC, including the P-body marker fragile X mental retardation 1 protein (FMRP)[11] and the de-capping activator RCK/p54[12],[13].
Once a mature miRNA is incorporated, miRISC can then bind to a target mRNA at a sequence-specific binding site. A particular miRNA may bind to hundreds of different genes, sometimes even onto multiple sites for a given target mRNA. Binding occurs with either perfect complementarity or, more often, imperfect complementarity[5]. In these instances of imperfect complementarity, there is often a short, ∼6–8 nt “seed” region, located near the 5′ end of the miRNA, which appears to be of paramount importance in terms of dictating binding to specific target mRNAs[1]. These seed regions are often conserved among species and form the basis for many of the current in silico target prediction algorithms[6],[14]–[16].
miRNAs can regulate expression of their targets through either mRNA degradation or translational inhibition. In instances of perfect or near-perfect miRNA-mRNA complementarity, degradation of target mRNAs can be mediated by AGO2, the only Argonaute protein with “slicer” activity[6]. In cases of imperfect complementarity, all 4 Argonaute proteins are capable of inhibiting protein translation as part of miRISC[6]. In addition, miRNA-target relationships with imperfect binding can also result in mRNA degradation through a non-sequence-specific mechanism within cytoplasmic processing bodies (P-bodies)[17]. In most cases, the net effect of miRISC binding to a target mRNA is a decrease in its protein levels. However, recent reports have demonstrated a few instances wherein protein levels is actually increased[6]. For example, miR-10A can bind the 5′ untranslated region (UTR) of the mRNA transcript for several ribosomal genes, increasing the expression of these genes[3].
miRNAs in Cancer
As the list of miRNAs has grown, so has our knowledge regarding their biological functions. Indeed, miRNAs have been found to play a role in most, if not all, cellular processes, including many pathways related to cancer development and progression. miRNAs function as tumor suppressor genes or oncogenes, with some miRNAs mediating contradictory roles in different diseases[5]. Aberrant miRNA expression and function have been described in a wide variety of human malignancies, with chromosomal amplifications/deletions, point mutations, or epigenetic alterations as potential causes[5].
Comprehensive miRNA expression profiling has been performed in a variety of human cancers, yielding a number of interesting observations. These include expression signatures that are capable of distinguishing cancer cells from normal cells[5] or one cancer from another[18]; predicting response to a particular drug[19]; or predicting patient outcome[20],[21]. Indeed, miRNA signatures capable of predicting patient outcome have been developed for a number of human cancers including lung cancer[22],[23], breast cancer[24], brain caner[25], and chronic lymphocytic leukemia[26]. Prognostic, predictive, and biological roles have also been described for miRNAs in NPC, as discussed below.
Human miRNA expression in NPC
The first study on the global profiling of miRNAs in NPC was published in 2008 by Paul Ahlquist's group at the National Cancer Institute (NCI)[27]. Using a microarray-based approach to profile 31 laser-capture microdissected NPC samples and 10 normal nasopharyngeal epithelial samples, they discovered several miRNAs to be dysregulated in NPC[27]. In particular, miR-29c was significantly down-regulated in NPC, and several miR-29c targets involved in extracellular matrix synthesis and function were identified and validated[27]. Subsequently, other groups have identified a number of dysregulated miRNAs in both nasopharyngeal tumor and blood samples from patients with NPC (Table 1).
Table 1. Summary of microRNA (miRNA) expression in studies using primary nasopharyngeal carcinoma (NPC) samples.
| Patients with NPC vs. healthy controls |
Prognostic association |
||
| Up-regulated | Down-regulated | Positive | Negative |
| *miR-16[57] | let-7g[58] | miR-18a[59] | *miR-9[31] |
| ŦmiR-17[60] | *miR-9[31] | ŦmiR-22[61] | miR-26a[62] |
| miR-18a[59] | miR-26a[11],[36],[58],[62] | miR-93[62] | miR-29c[29],[62] |
| ŦmiR-20a[60] | miR-26b[36] | ŦmiR-572[61] | miR-30e[62] |
| *miR-21[57] | miR-29c[27],[62] | ŦmiR-638[61] | miR-451[63] |
| *miR-24[57] | ŦmiR-29c[60] | miR-98[36] | |
| miR-93[62] | miR-30e[62] | miR-142-3p[62] | |
| miR-141[64] | miR-34b[58] | ŦmiR-1234[61] | |
| miR-144[65] | miR-101[36] | ||
| miR-146a[66],[67] | miR-138[68] | ||
| miR-155[38] | miR-142-3p[62] | ||
| *miR-155[37],[69] | miR-200b[41] | ||
| *miR-214-3p[31] | miR-216b[44] | ||
| miR-214-3p[70] | miR-218[71] | ||
| miR-663[72] | ŦmiR-223[60] | ||
| *miR-3135a[31] | miR-375[30] | ||
| *miR-378[57] | |||
| miR-451[63] | |||
The miRNAs included in this table Satisfy each of the following criteria: (1) a statistically significant alteration in their expression was identified in specimens from patients with NPC; and (2) some degree of validation (either in additional samples, using an alternate analytical method, or functional validation) was reported. Patients with NPC vs. healthy controls: for these miRNAs, levels were significantly increased (“up-regulated”) or decreased (“down-regulated”) in tumors and plasma from patients with NPC compared with nasopharyngeal tissues and plasma from healthy controls. Prognostic association: expression higher (“positive”) or lower (“negative”) in tumor and plasma samples from patients with poor prognosis. Superscripts: expression measured in *plasma and Ŧserum. All others were discovered using tumor samples.
In addition to alterations in miRNA expression in NPC, we and others have demonstrated phenotypic roles for miRNAs in nasopharyngeal tumorigenesis using in vitro and in vivo models. The first miRNA characterized in the context of NPC was miR-29c, which was the main focus of the first miRNA profiling reported by Sengupta et al.[27]. In this initial study, the authors demonstrated that miR-29c plays a potential tumor suppressive role, targeting mRNAs that encode extracellular matrix proteins (collagens 3A1, 4A1, and 15A1, and laminin γ1)[27]. They postulated that suppression of miR-29c will subsequently increase the migration and invasion of NPC cells through up-regulation of these components of the extracellular matrix. However, they stopped short of testing this hypothesis in the initial study, and further reports supporting these claims have not been published to date. Subsequent studies by two other groups corroborated the role of miR-29c as a tumor suppressor in NPC, but these reports differed in their causal mechanisms and putative targets. While Liu et al.[28] demonstrated that down-regulation of miR-29c resulted in the promotion of NPC cell migration and invasion through increased expression of T-cell lymphoma invasion and metastasis 1 (TMP1), Zhang et al.[29] showed that miR-29c knock-down resulted in increased resistance to radiotherapy and cisplatin through up-regulation of the anti-apoptotic regulators Mcl-1 and Bcl-2. Thus, much like other miRNAs, miR-29c can act through a number of pathways to suppress the proliferation, survival, and motility of NPC cells.
In 2011, we reported that miR-375 is a potential tumor suppressor in head and neck cancers, including NPC[30]. We discovered that metadherin (MTDH), a newly emerging oncogene, was an important target for down-regulation by miR-375 and that MTDH overexpression was particularly detrimental to NPC patients, resulting in an increased risk of distant relapse[30].
Another moderately well studied miRNA in the context of NPC is miR-9. Not only has miR-9 been identified as a potential circulating biomarker of advanced NPC[31], several potential targets and functions of miR-9 have also been reported. Recently, two studies describing putative tumor suppressive mechanisms for miR-9 in NPC have been published[32],[33]. Lu et al.[32] reported that hypermethylation and subsequent underexpression of miR-9 led to up-regulation of its putative target C-X-C chemokine receptor type 4 (CXCR4), resulting in increased cell growth, migration, and invasion through activation of the Mitogen-activated protein kinase (MAPK) pathway. In contrast, Gao et al.[33] postulated a role for miR-9 in modulating the immune response to NPC by targeting several interferon (IFN)-induced genes, multiple members of the major histocompatibility complex (MHC) class I molecule, and a number of interleukins and related genes.
Other potential, functionally active miRNAs in NPC include miR-26a[34],[35], miR-98[36], miR-155[37],[38], miR-200a/b[39]–[41], miR-205[42],[43], and miR-216b[44]. Proposed targets of these miRNAs represent regulators of important processes such as the epithelial-to-mesenchymal transition (EMT)[39], as well as signaling pathways including Notch[41], phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)-Akt[42], and MAPK[44]. Intriguingly, the function, pathways, and targets affected by many of the miRNAs involved in nasopharyngeal tumorigenesis overlap. Two such miRNAs are miR-26a and miR-218, which both target enhancer of zeste homolog 2 (EZH2), resulting in decreased oncogenic properties of migration, invasion, and cell survival[35],[36]. This overlapping functional impact of multiple miRNAs adds an increased layer of complexity when attempting to elucidate their biological role; however, this property could also increase our confidence in the biological importance of affected genes and pathways, given that there are multiple mechanisms that malignant cells exploit to activate or inhibit the same gene or pathway.
miRNAs encoded by Epstein-Barr virus in NPC
Epstein-Barr virus (EBV), which is present in the vast majority of NPC, also encodes a number of its own miRNAs. Twenty-five EBV-encoded pre-miRNAs, which are processed into 44 mature miRNA sequences, have been verified to date[2]. These miRNA emanate from two major regions of the EBV genome: (1) the BamHI-A rightward transcripts (BART) and (2) the open reading frame of the BHRF1 gene. Originally cloned from EBV-infected Burkitt's lymphoma cells[45], BHRF1-encoded miRNAs do not appear to be expressed in EBV-positive NPC primary tissues[46],[47]. Indeed, in the first report of comprehensive EBV miRNA profiling in primary NPC, the majority of samples showed substantial expression of all 35 interrogated BART-encoded miRNAs but no expression of the 4 interrogated BHRF1-encoded miRNAs. In contrast, NPC-derived cell lines C666-1 and HONE-Akata expressed all 4 BHRF1-derived miRNAs in addition to the 35 interrogated BART-encoded miRNAs[46]. Subsequent studies have identified several targets of EBV miRNAs, both host and viral. Viral targets include latent membrane protein 1 (LMP1), putatively targeted by several miRNAs from the BART region (ebv-miR-BART1, 9, 16, 17)[46],[48],[49], and LMP2A, a target of ebv-miR-BART22[50]. Subsequent functional experiments suggest that modulation of these viral proteins by BART-encoded miRNA can influence multiple cellular properties including cell proliferation, survival, and evasion of host immune response[48]–[50]. Host targets of BART-encoded miRNAs include the pro-apoptotic effectors p53 up-regulated modulator of apoptosis (PUMA)[51], Bcl-2 interacting mediator of cell death (Bim)[52], and translocase of outer mitochondrial membrane 22 homolog (TOMM22)[53], as well as several genes thought to influence host immune response, including MHC class I-related chain B (MICB)[54], importin 7 (IPO7)[53],[55], and Dicer[56]. Overall, EBV-encoded miRNAs play a complementary role to the viral proteins expressed in NPC, contributing to evasion of the host immune response and promoting the survival and proliferation of NPC cells.
Conclusions
In summary, the interrogation of miRNAs in the context of NPC has provided both clinical and biological insight into the behavior of this disease. Further development of strategies to measure and manipulate miRNAs and their targets in a clinical setting would be required before such findings can be translated into improvements in the management of this disease.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. 2009;42:1316–1329. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 8.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz DS, Hutvagner G, Du T, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 11.Jin P, Zarnescu DC, Ceman S, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 12.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eulalio A, Rehwinkel J, Stricker M, et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.John B, Enright AJ, Aravin A, et al. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Valencia-Sanchez MA, Hannon GJ, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian S, Lui WO, Lee CH, et al. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–2026. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 19.Osaki M, Takeshita F, Ochiya T. MicroRNAs as biomarkers and therapeutic drugs in human cancer. Biomarkers. 2008;13:658–670. doi: 10.1080/13547500802646572. [DOI] [PubMed] [Google Scholar]
- 20.Jay C, Nemunaitis J, Chen P, et al. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- 21.Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst. 2012;104:528–540. doi: 10.1093/jnci/djs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 23.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 25.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 27.Sengupta S, den Boon JA, Chen IH, et al. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci U S A. 2008;105:5874–5878. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N, Tang LL, Sun Y, et al. miR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett. 2013;329:181–188. doi: 10.1016/j.canlet.2012.10.032. [DOI] [PubMed] [Google Scholar]
- 29.Zhang JX, Qian D, Wang FW, et al. MicroRNA-29c enhances the sensitivities of human nasopharyngeal carcinoma to cisplatin-based chemotherapy and radiotherapy. Cancer Lett. 2013;329:91–98. doi: 10.1016/j.canlet.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 30.Hui AB, Bruce JP, Alajez NM, et al. Significance of dysregulated metad-herin and microRNA-375 in head and neck cancer. Clin Cancer Res. 2011;17:7539–7550. doi: 10.1158/1078-0432.CCR-11-2102. [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Xu X, Liu X, et al. Predictive value of miR-9 as a potential biomarker for nasopharyngeal carcinoma metastasis. Br J Cancer. 2014;110:392–398. doi: 10.1038/bjc.2013.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Luo H, Liu X, et al. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis. 2014;35:554–563. doi: 10.1093/carcin/bgt354. [DOI] [PubMed] [Google Scholar]
- 33.Gao F, Zhao ZL, Zhao WT, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2013;431:610–616. doi: 10.1016/j.bbrc.2012.12.097. [DOI] [PubMed] [Google Scholar]
- 34.Lu J, He ML, Wang L, et al. miR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 35.Yu L, Lu J, Zhang B, et al. miR-26a inhibits invasion and metastasis of nasopharyngeal cancer by targeting EZH2. Oncol Lett. 2013;5:1223–1228. doi: 10.3892/ol.2013.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alajez NM, Shi W, Hui AB, et al. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 2010;1:e85. doi: 10.1038/cddis.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X, Wang Y, Sun Y, et al. miR-155 up-regulation by LMP1 DNA contributes to increased nasopharyngeal carcinoma cell proliferation and migration. Eur Arch Otorhinolaryngol. 2014;271:1939–1945. doi: 10.1007/s00405-013-2818-0. [DOI] [PubMed] [Google Scholar]
- 38.Du ZM, Hu LF, Wang HY, et al. Upregulation of miR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One. 2011;6:e19137. doi: 10.1371/journal.pone.0019137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia H, Cheung WK, Sze J, et al. miR-200a regulates epithelial-mesenchymal to stem-like transition via ZEB2 and beta-catenin signaling. J Biol Chem. 2010;285:36995–37004. doi: 10.1074/jbc.M110.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia H, Ng SS, Jiang S, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 2010;391:535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 41.Yang X, Ni W, Lei K. miR-200b suppresses cell growth, migration and invasion by targeting Notch1 in nasopharyngeal carcinoma. Cell Physiol Biochem. 2013;32:1288–1298. doi: 10.1159/000354527. [DOI] [PubMed] [Google Scholar]
- 42.Qu C, Liang Z, Huang J, et al. miR-205 determines the radioresis-tance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle. 2012;11:785–796. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D, Wang S, Liu Q, et al. SZ-685C exhibits potent anticancer activity in both radiosensitive and radioresistant NPC cells through the miR-205-PTEN-Akt pathway. Oncol Rep. 2013;29:2341–2347. doi: 10.3892/or.2013.2376. [DOI] [PubMed] [Google Scholar]
- 44.Deng M, Tang H, Zhou Y, et al. miR-216b suppresses tumor growth and invasion by targeting kras in nasopharyngeal carcinoma. J Cell Sci. 2011;124:2997–3005. doi: 10.1242/jcs.085050. [DOI] [PubMed] [Google Scholar]
- 45.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 46.Cosmopoulos K, Pegtel M, Hawkins J, et al. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carci-noma. J Virol. 2009;83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu JY, Pfuhl T, Motsch N, et al. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo AK, To KF, Lo KW, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramakrishnan R, Donahue H, Garcia D, et al. Epstein-Barr virus BART9 miRNA modulates LMP1 levels and affects growth rate of nasal NK T cell lymphomas. PLoS One. 2011;6:e27271. doi: 10.1371/journal.pone.0027271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lung RW, Tong JH, Sung YM, et al. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. 2009;11:1174–1184. doi: 10.1593/neo.09888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choy EY, Siu KL, Kok KH, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marquitz AR, Mathur A, Nam CS, et al. The Epstein-Barr virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolken L, Malterer G, Erhard F, et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. 2010;7:324–334. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Nachmani D, Stern-Ginossar N, Sarid R, et al. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 55.Yang IV, Wade CM, Kang HM, et al. Identification of novel genes that mediate innate immunity using inbred mice. Genetics. 2009;183:1535–1544. doi: 10.1534/genetics.109.107540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iizasa H, Wulff BE, Alla NR, et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 2010;285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Luo HN, Tian WD, et al. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol Ther. 2013;14 doi: 10.4161/cbt.26170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li T, Chen JX, Fu XP, et al. MicroRNA expression profiling of nasopharyngeal carcinoma. Oncol Rep. 2011;25:1353–1363. doi: 10.3892/or.2011.1204. [DOI] [PubMed] [Google Scholar]
- 59.Luo Z, Dai Y, Zhang L, et al. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis. 2013;34:415–425. doi: 10.1093/carcin/bgs329. [DOI] [PubMed] [Google Scholar]
- 60.Zeng X, Xiang J, Wu M, et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS One. 2012;7:e46367. doi: 10.1371/journal.pone.0046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu N, Cui RX, Sun Y, et al. A four-miRNA signature identified from genome-wide serum miRNA profiling predicts survival in patients with nasopharyngeal carcinoma. Int J Cancer. 2014;134:1359–1368. doi: 10.1002/ijc.28468. [DOI] [PubMed] [Google Scholar]
- 62.Liu N, Chen NY, Cui RX, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13:633–641. doi: 10.1016/S1470-2045(12)70102-X. [DOI] [PubMed] [Google Scholar]
- 63.Liu N, Jiang N, Guo R, et al. miR-451 inhibits cell growth and invasion by targeting MIF and is associated with survival in nasopharyngeal carcinoma. Mol Cancer. 2013;12:123. doi: 10.1186/1476-4598-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L, Deng T, Li X, et al. MicroRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis. 2010;31:559–566. doi: 10.1093/carcin/bgp335. [DOI] [PubMed] [Google Scholar]
- 65.Zhang LY, Ho-Fun Lee V, Wong AM, et al. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis. 2013;34:454–463. doi: 10.1093/carcin/bgs346. [DOI] [PubMed] [Google Scholar]
- 66.Lung RW, Wang X, Tong JH, et al. A single nucleotide polymorphism in microRNA-146a is associated with the risk for nasopharyngeal carcinoma. Mol Carcinog. 2013;52(Suppl. 1):E28–E38. doi: 10.1002/mc.21937. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Y, Chen X, Jing M, et al. Expression of miRNA-146a in nasopharyngeal carcinoma is upregulated by Epstein-Barr virus latent membrane protein 1. Oncol Rep. 2012;28:1237–1242. doi: 10.3892/or.2012.1933. [DOI] [PubMed] [Google Scholar]
- 68.Liu X, Lv XB, Wang XP, et al. miR-138 suppressed nasopharyngeal carcinoma growth and tumorigenesis by targeting the CCND1 oncogene. Cell Cycle. 2012;11:2495–2506. doi: 10.4161/cc.20898. [DOI] [PubMed] [Google Scholar]
- 69.Liu X, Luo HN, Tian WD, et al. Diagnostic and prognostic value of plasma microrna deregulation in nasopharyngeal carcinoma. Cancer Biol Ther. 2013;14:1133–1142. doi: 10.4161/cbt.26170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng M, Ye Q, Qin Z, et al. miR-214 promotes tumorigenesis by targeting lactotransferrin in nasopharyngeal carcinoma. Tumour Biol. 2013;34:1793–1800. doi: 10.1007/s13277-013-0718-y. [DOI] [PubMed] [Google Scholar]
- 71.Alajez NM, Lenarduzzi M, Ito E, et al. miR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71:2381–2391. doi: 10.1158/0008-5472.CAN-10-2754. [DOI] [PubMed] [Google Scholar]
- 72.Yi C, Wang Q, Wang L, et al. miR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012;31:4421–4433. doi: 10.1038/onc.2011.629. [DOI] [PubMed] [Google Scholar]