Abstract
The endocannabinoids, anandamide (AEA) and 2-arachidonoylglycerol (2-AG), are arachidonic acid (AA) derivatives that are known to regulate human cardiovascular functions. CYP2J2 is the primary cytochrome P450 in the human heart and is most well known for the metabolism of AA to the biologically active epoxyeicosatrienoic acids. In this study, we demonstrate that both 2-AG and AEA are substrates for metabolism by CYP2J2 epoxygenase in the model membrane bilayers of nanodiscs. Reactions of CYP2J2 with AEA formed four AEA-epoxyeicosatrienoic acids, whereas incubations with 2-AG yielded detectable levels of only two 2-AG epoxides. Notably, 2-AG was shown to undergo enzymatic oxidative cleavage to form AA through a NADPH-dependent reaction with CYP2J2 and cytochrome P450 reductase. The formation of the predominant AEA and 2-AG epoxides was confirmed using microsomes prepared from the left myocardium of porcine and bovine heart tissues. The nuances of the ligand-protein interactions were further characterized using spectral titrations, stopped-flow small-molecule ligand egress, and molecular modeling. The experimental and theoretical data were in agreement, which showed that substitution of the AA carboxylic acid with the 2-AG ester-glycerol changes the binding interaction of these lipids within the CYP2J2 active site, leading to different product distributions. In summary, we present data for the functional metabolomics of AEA and 2-AG by a membrane-bound cardiovascular epoxygenase.
Introduction
The human body contains endogenous cannabinoids (endocannabinoids) that elicit similar effects as Δ9-tetrahydrocanabinol, the principal component of cannabis. The two most well characterized endocannabinoids are anandamide (AEA) and 2-arachidonylglycerol (2-AG), which primarily exert their effects by activating the cannabinoid receptors—cannabinoid receptor 1 (CB1) and cannabinoid receptor 2 (CB2). Dysregulation of the endocannabinoids has been implicated in a wide range of pathologies, including anxiety disorders, neurodegenerative diseases, obesity, and heart disease (Kogan and Mechoulam, 2007). Therefore, a thorough understanding of all the potential metabolic pathways in the body is necessary to potentially target and alter these pathophysiological conditions.
Endocannabinoids are derivatives of arachidonic acid (AA) that are stored in the plasma membrane (Fig. 1). AA is the primary substrate for the eicosanoid-synthesizing cascade that includes three branches—the cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 epoxygenase pathways (Yang et al., 2011). Endocannabinoids provide substrate for the eicosanoid-synthesizing cascade through both direct and indirect pathways. The indirect route is through the cleavage of either the AEA ethanolamide motif by fatty acid amide hydrolase or the 2-AG glycerol motif by monoacylglycerol lipase, which produces AA that can enter one of the three branches (Yang et al., 2011). Alternatively, AEA can directly enter each of the pathways and produce a diverse range of metabolites with different pharmacologic properties when compared with the endocannabinoid parent compound. Prior to this work, 2-AG was only shown to be a direct substrate of the COX and LOX pathways, although the 2-AG epoxide metabolites had been extracted from rat kidney, spleen, and brain tissues (Chen et al., 2008). Recently, it has been shown that endocannabinoids regulate human cardiovascular functions and are important for homeostasis (Pacher and Steffens, 2009). Thus, it is of interest to evaluate the metabolism of endocannabinoids by relevant heart enzymes. In this work, we reveal the nuances of the metabolism of 2-AG and AEA by the predominant P450 in the heart, CYP2J2 epoxygenase. CYP2J2 is an extrahepatic cytochrome P450 that is highly expressed in the myocardium and surrounding aortic epithelium and has been shown to be an important regulator of cardiovascular function (Delozier et al., 2007; Alghasham et al., 2012; Panigrahy et al., 2012). CYP2J2 produces potent lipid epoxide mediators derived from AA known as epoxyeicosatrienoic acids (EETs), which are involved in pain, inflammation, kidney, and cardiovascular disease (Imig and Hammock, 2009; Rouzer and Marnett, 2011). Additionally, the epoxygenases also produce hydroxyeicosatetraeonic acids (HETE) that are hydroxylated at the 19- or 20-Ω position of AA.
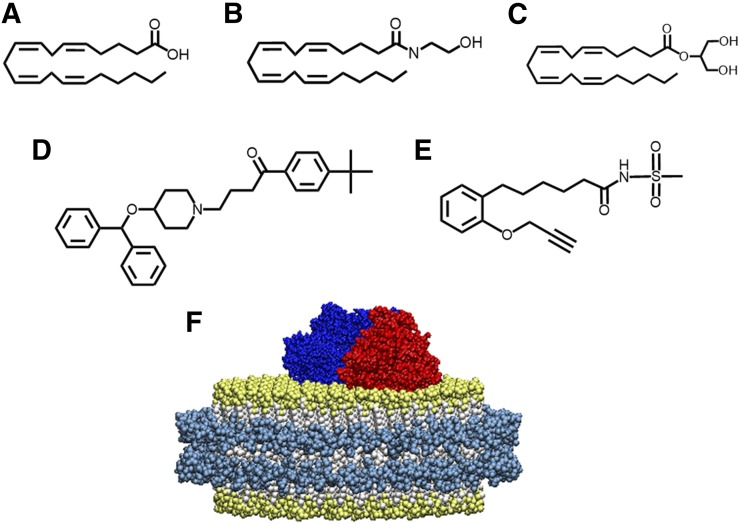
Fig. 1.
Chemical structures of substrates that bind to CYP2J2-ND. Endogenous CYP2J2 ligands include (A) arachidonic acid, (B) anandamide, and (C) 2-arachidonoyl glycerol. (D) The xenobiotic, ebastine, is a cardiotoxic CYP2J2 substrate. (E) MS-PPOH is a nonspecific P450 inhibitor. (F) Schematic of CYP2J2 and CPR incorporated into the membrane bilayers of nanodiscs. The membrane scaffold protein is represented in cyan surrounding a bilayer of phospholipids colored white with gold phosphate head groups. CYP2J2 is red, and its obligate redox partner cytochrome P450 reductase is colored blue.
Currently, there are no reports of the direct oxygenation of 2-AG by a cytochrome P450 (P450). Previous studies with several recombinant P450 epoxygenases (CYP2C8, CYP2C11, and CYP2C3) failed to produce 2-AG epoxide metabolites (Chen et al., 2008). In this work, we show that CYP2J2 epoxygenase metabolizes 2-AG to form 11,12-epoxyeicosatrienoic glycerol (EET-G) and 14,15-EET-G. Several reports have demonstrated the P450-mediated metabolism of AEA to 20-HETE-EA, 5(6)-, 8(9)-, 11(12)-, and 14(15)-epoxyeicosatrienoic ethanolamide (EET-EA) by CYP3A4, CYP4F2, CYP4X1, CYP2B6, and CYP2D6 (Snider et al., 2007, 2008; Stark et al., 2008; Sridar et al., 2011). Notably, it was demonstrated that these new metabolites provide different pharmacological profiles when compared with the parent endocannabinoid. In this study, we characterize the oxidation of 2-AG and AEA with the heart epoxygenase, CYP2J2, using both recombinantly expressed CYP2J2 and heart microsomes.
Additionally, the activities of endocannabinoids are regulated through various activation and inactivation pathways. 2-AG is primarily inactivated through hydrolysis by monoacylglycerol lipase (Rouzer and Marnett, 2011). However, inhibition of this enzyme does not completely block the 2-AG conversion to AA (Blankman et al., 2007). This indicates the presence of alternative pathways of attenuation. In this work, we demonstrate another potential pathway in which 2-AG is converted to AA by CYP2J2 and other P450s.
In summary, we elucidate the nuances of binding and metabolism of AA and its derivatives, AEA and 2-AG, by CYP2J2 using liquid chromatography–tandem mass spectrometry (LC-MS/MS), liquid chromatography-mass spectrometry (LC-MS), spectral titrations, small-molecule ligand egress studies, and molecular modeling. For all studies with recombinant protein, we used the nanoscale lipid bilayers of nanodiscs (ND) to solubilize and stabilize membrane-bound CYP2J2. These model membranes have been previously used to functionally stabilize membrane protein in solution and on surfaces (Nath et al., 2007; Bayburt and Sligar, 2010; Denisov and Sligar, 2011; Das et al., 2014; Orlando et al., 2014). Importantly, the use of these model membranes significantly reduced scattering and enabled spectroscopic characterization by increasing the solubility of the hydrophobic lipid substrates and membrane proteins.
Materials and Methods
Reagents.
NADPH and NADP were obtained from P212121 (Ann Arbor, MI). The POPC [1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine], 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, POPS [1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-l serine], and PIP2 (1-heptadecanoyl-2-arachidonoyl-sn-glycero-3-[phosphoinositol-4′,5′-bisphosphate]) were purchased from Avanti Polar Lipids (Alabaster, AL). AA, 2-AG, AEA, deuterated anandamide, MS-PPOH [N-(methylsulfonyl)-2-(2-propynyloxy)-benzenehexanamide], 14,15-EET, 11-12-EET, 8,9-EET, 5,6-EET, (±)16(17)-epoxy-4Z,7Z,10Z,13Z,19Z-docosapentaenoic acid, 2-(14,15-epoxyeicosatrienoyl glycerol), and 14,15-EET ethanolamide were obtained from Cayman Chemical (Ann Arbor, MI). Whole cow and porcine hearts were obtained fresh from the University of Illinois Meat Sciences Laboratory. Polyclonal CYP2J2 antibody (catalog ABIN1529400) and horseradish peroxidase (HRP)–conjugated donkey anti-rabbit antibody (catalog ABIN101948) were purchased from Antibodies-Online, Inc. (Atlanta, GA). Recombinant CYP2C8 supersomes were purchased from BD Biosciences Gentest (San Jose, CA). The antioxidant butylated hydroxytoluene (BHT) was obtained from Sigma-Aldrich (St. Louis, MO). All other materials and reagents used were purchased from Sigma-Aldrich and Fisher Scientific.
Recombinant Expression of CYP2J2 in Escherichia coli.
The recombinant CYP2J2 (construct D34G) was expressed and purified, as published previously (McDougle et al., 2013; Zelasko et al., 2013). Briefly, DH5α cells containing the His-tagged and N-terminally truncated CYP2J2 plasmid and pTGro7 chaperonin plasmid were cultured in 30 ml Luria Bertani media with chloroamphenicol (20 µg/ml) and ampicillin (100 µg/ml) at 37°C and 250 rpm overnight. The resulting culture was used to inoculate 500 ml Terrific Broth containing ampicillin (100 µg/ml) and chloroamphenicol (20 µg/ml). The culture was grown at 37°C and 220 rpm until reaching an optical density of 1.0, and then δ-aminolevulinic acid (500 µl 0.5 mM) was added and grown for 2 hours at 26°C and 160 rpm. A total of 1 mM isopropyl β-d-1-thiogalactopyranoside and 2 g arabinose was added to each 500 ml culture for induction and subsequently grown for an additional 44 hours. The cells were harvested via centrifugation (2800g for 10 minutes) using a JA-10 rotor (Beckman Coulter, Brea, CA) at 4°C. The cells were then sonicated in lysis buffer [dithiothreitol (DTT), 0.2 mM phenylmethanesulfonylfluoride (PMSF), 5 mg DNase, and RNase] before ultracentrifugation at 140,000g for 30 minutes with a Ti-45 rotor (Beckman Coulter) for membrane fraction isolation. Protein was extracted from the membrane fraction using 1.0% (w/v) cholate and stirred at 4°C for 4 hours. The membrane fraction was removed by ultracentrifugation at 140,000g for 30 minutes with a Ti-45 rotor (Beckman Coulter). CYP2J2 was isolated from the supernatant by running the solution through a Ni-NTA column and eluting in 0.1% (w/v) cholate, 100 mM KPi, 200 mM imidazole, and 20% glycerol with protein yield of ∼150 nmol/l.
Expression and Purification of Cytochrome P450 Reductase.
Expression of the redox partner, cytochrome P450 reductase (CPR), from Rattus norvegicus was described previously (McDougle et al., 2013). Briefly, CPR starter cultures were used to inoculate 1 l Luria Bertani supplemented with ampicillin (100 µg/ml) and riboflavin (1 mg/l) and grown for 4 hours at 37°C and 220 rpm, before induction with isopropyl β-d-1-thiogalactopyranoside (1 ml 1 mM), and then grown for 18–20 hours at 33°C and 220 rpm. Upon harvesting the cells by centrifugation (2800g for 10 minutes), the pellets were treated with 500 ml cold lysozyme buffer (75 mM Tris, pH 8.0, 0.25 M sucrose, 0.25 mM EDTA, and 0.02 mg/ml lysozyme) and allowed to stir at 4°C for 30 minutes. The resulting solution was centrifuged (2800g for 10 minutes) and resuspended in 40 ml lysis buffer (50 mM Tris, pH 8.0, and 1 mM PMSF) at 4°C before sonication on ice (5 cycles × 30 seconds) with 1-minute rests. The membrane fraction was isolated via ultracentrifugation and solubilized in column buffer [50 mM Tris, pH 7.7, 0.1 mM EDTA, 0.1 mM DTT, and 20% (v/v) glycerol] containing 0.2% (v/v) Triton X-100 for 1 hour at 4°C. The solution was subjected to ultracentrifugation (140,000g for 30 minutes), and the resulting supernatant was loaded onto 2′,5′-ADP column, followed by detergent removal using a DEAE Sepharose column.
Incorporation of CYP2J2 into Nanodiscs.
Purified CYP2J2 was incorporated into the membrane bilayers of NDs as previously described, with the only modification being the addition of POPS (McDougle et al., 2013). POPS and POPC lipid stocks were combined in chloroform in a 20:80 molar ratio and dried down under a steady stream of N2. The lipid mixture was reconstituted with 200 mM cholate in phosphate buffer (pH 7.4) and added to membrane scaffold protein MSP1D1(−) in a 65:1 molar ratio and mixed at 4°C for 1 hour. Purified CYP2J2 [100 mM KPi, pH 7.4, 20% (v/v) glycerol, and 0.1% cholate] was added to the solution in a CYP2J2:MSP molar ratio of 1:10 and mixed for several hours before detergent removal with Amberlite beads overnight. The CYP2J2-NDs were separated from empty NDs using a Ni-NTA column and eluted with 100 mM phosphate buffer containing 200 mM imidazole. Homogenous CYP2J2-ND populations were further purified using size-exclusion chromatography utilizing an Alliance 2695 analytical separation module (Waters, Milford, MA) coupled to a Waters 996 photodiode diode array detector (Waters) and a semipreparative Superdex 10/200 column (GE Healthcare) column.
Recombinant CYP2J2 Metabolism Assays Containing Arachidonic Acid, Anandamide, and 2-Arachidonoyl Glycerol.
Incubation mixtures containing CYP2J2-ND (0.2 µM), CPR (0.6 µM), and 0.1% (w/v) BHT in 100 mM phosphate buffer (pH 7.4) and either AA, AEA, or 2-AG (500 µl total volume) were equilibrated at 37°C for 5 minutes before the reaction was initiated with the addition of 1 mM NADPH. Importantly, CPR was added to the P450-nanodisc in the optimal 3:1 CPR to P450 ratio. Previous reports demonstrated the incorporation of CPR and P450 into NDs as verified by size-exclusion chromatography (Grinkova et al., 2010). Additionally, in a previous work, we prepared discs containing both CYP2J2 and CPR NDs (McDougle et al., 2013). Incubations for subsequent qualitative LC-MS analysis contained saturating concentrations of each substrate, whereas samples for quantitative kinetic analysis contained either 1, 5, 10, 20, 40, or 60 µM of each respective substrate. Incubations were reacted for 30 minutes, unless otherwise stated in the figure legends. Samples lacking NADPH were performed in parallel as a routine control. For termination of the reaction, samples were quenched with glacial acetic acid to pH of 3–4 and extracted (three times) with an equal volume of ice-cold ethyl acetate, vortexed 1 minute, and centrifuged at 1590g for 3 minutes for phase separation (Stark et al., 2008). The organic layer was removed and dried under a steady stream of N2 and then reconstituted with 100 µl methanol. AA, AEA, and 2-AG incubations were qualitatively analyzed using LC-MS, as described below. For quantitative analysis, LC-MS/MS was used to directly quantify 14,15-, 11,12-, 8,9-, and 5,6-EET turnover when AA was used as the substrates. For quantitative analysis of EET-G, extracted samples were split into two pools as described in Saponification of EET-G Regioisomers to EETs and Quantification.
Preparation of Bovine and Porcine Microsomes and Incubations.
Fresh heart microsomes were prepared from the left myocardium of female porcine and male bovine hearts using differential centrifugation methods, as described previously (Pretorius et al., 1969). Briefly, tissue from the left myocardium was dissected to remove the endo- and epicardium and minced. Tissue was weighed and diluted to 20% (w/v) using a buffer containing 250 mM sucrose, 10 mM Tris-Cl (pH 7.5), and 1 mM PMSF, and a protease inhibitor cocktail for mammalian tissue extraction (Nacalai Tesque 25955-11). Next, tissue was homogenized on ice for 1 minute at maximum speed using a metal Bio-homogenizer (Biospec, Bartlesville, OK). A low-speed centrifugation step was employed to remove large tissue debris at 3000g for 20 minutes at 4°C. The supernatant was then centrifuged at 10,000g for 20 minutes at 4°C using a Ti-45 rotor (Beckman) and then repeated once more. The resulting supernatant was centrifuged at 100,000g for 90 minutes at 4°C to pellet the sarcoplasmic reticulum containing heart P450s. The cell pellet was then resuspended in 50 mM Tris (pH 7.5), 1 mM DTT, 1 mM EDTA, and 20% glycerol with a Teflon homogenizer. The resulting microsomal solution was aliquoted and stored at −80°C until future use. Microsomal protein content was calculated using a bicinchoninic acid assay kit from Thermo Fisher Scientific (Waltham, MA). Incubations with both porcine and bovine microsomes were performed with a concentration of 1 mg/ml microsomes with saturating concentrations of AA, AEA, and 2-AG and then processed, as described, with recombinant CYP2J2-ND incubations.
Immunoblotting of CYP2J2 Located in Cow and Porcine Heart Microsomes.
Heart microsomes were prepared, as described above, and used for CYP2J2 immunoblotting following a previously described protocol (Zeldin et al., 1997). In short, samples were boiled for 10 minutes in 5× Laemmli buffer supplemented with 8 M urea and run on a 12% SDS polyacrylamide gel. Proteins were transferred to the nitrocellulose membrane and blocked with 5% low-fat milk in Tris-buffered saline with Tween 20 for 1 hour before adding the primary polyclonal antibody raised against the 250–275 segment of human CYP2J2 (Antibodies-Online, Inc.). The membrane was washed three times with Tris-buffered saline with Tween 20, and the HRP-conjugated donkey anti-rabbit antibody (Antibodies-Online, Inc.) was used for detection. Blots were visualized using a HRP development solution (Thermo Fisher Scientific) and a ChemiDoc XRS+ system (Bio-Rad, Hercules, CA).
Saponification of EET-G Regioisomers to EETs and Quantification.
For the quantification of EET-G in solution, the extracted fatty acids were cleaved from the glycerol backbone using methanolic potassium hydroxide (KOH) hydrolysis, as previously described (Capdevila et al., 1991). Extracted samples were hydrolyzed in 1 ml of methanol containing 0.4 N KOH and incubated at 50°C for 1 hour. Next, 1.5 ml Millipore water was added, and glacial acetic acid was added to achieve pH 3–4. Samples were extracted with an equal volume of ice-cold ethyl acetate (3×), vortexed 1 minute, and centrifuged at 1600g for 3 minutes for phase separation (Stark et al., 2008). The organic layer was removed and dried under a steady stream of N2 and then reconstituted with 100 µl methanol. Thus, the saponified sample contained both free EETs originally present in solution and EETs derived from EET-G hydrolysis. EET-G quantification at the time of reaction quenching was calculated using eq. 1:
| (1) |
Extracted EET-G regioisomers were converted to their corresponding EETs using methanolic KOH (0.4 N) and quantified using LC-MS/MS, as described below.
Liquid Chromatography-Electrospray Ionization-Mass Spectrometry Product Analysis.
AA, AEA, and 2-AG regioisomers were resolved using a XTerra C18 column 2.1 × 150 mm, 3.5 µM (Waters), and a Waters Alliance 2695 reversed-phase high performance liquid chromatography (HPLC) coupled to an electrospray ionization (ESI) source. For analysis of AA and AEA metabolites, the mobile phases consisted of mobile phase A (acetonitrile/H2O/formic acid, 95:5:0.1) and mobile phase B (acetonitrile/H2O/formic acid, 5:95:0.1) and were run with a linear gradient as follows: 0–1 minute, 60% A; 41 minutes, 20% A; 42 minutes, 0% A; and then returned to initial conditions. The same solvent system was used in the analysis of 2-AG metabolites with a linear gradient as follows: 0–1 minute, 70% A; 51 minutes, 20% A; 52 minutes, 0% A; and then returned to initial conditions. For ESI, a Q-TOF Ultima time-of-flight mass spectrometer was used in positive ion mode for analysis of AEA, with a spray voltage of 4.5 V and capillary temperature of 200°C. Data were collected and processed using Mass Lynx software (version 4.1), initially scanning from 200 to 800 m/z and processed for the selected AEA-EET m/z of 364. The mass and elution time of the 14,15-EET-EA moiety were confirmed using the commercially available authentic standard. The detection of AA and 2-AG metabolites was detected with the same system in negative ion mode, with a cone voltage of 35 V and a 200°C desolvation temperature, scanning from 200 to 800 m/z and processed for the selected EET m/z of 319 and EET-G m/z of 393–394. The mass and elution times of the EET regioisomers were confirmed with commercially available authentic standards. For 2-AG analysis, the mass and elution time of the commercially available 14,15-EET-G, EETs, and AA lipids were confirmed with authentic standards purchased from Cayman Chemical.
Liquid Chromatography–Tandem Mass Spectrometry for Quantitation of 14,15-, 11,12-, 8,9-, 5,6-EETs, 14,15-EET-EA, and 14,15-EET-G.
Quantification EET regioisomers derived from in vitro CYP2J2 incubations were analyzed with the 5500 QTRAP LC-MS/MS system (AB Sciex, Foster City, CA) with a 1200 series HPLC system (Agilent Technologies, Santa Clara, CA), including a degasser, an autosampler, and a binary pump. The LC separation was performed on an Agilent Zorbax Eclipse XDB C18 column (4.6 × 150 mm, 5 µm; Agilent Technologies) with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The flow rate was 0.4 ml/min. The linear gradient was as follows: 0–2 minutes, 90% A; 8 minutes, 50% A; 13–25 minutes, 25% A; 30 minutes, 20% A; 35 minutes, 15% A; and then returned to initial conditions. The autosampler was set at 5°C. The injection volume was 5 µl. Mass spectra were acquired with negative ESI, and the ion spray voltage was −4500 V. The source temperature was 450°C. The curtain gas, ion source gas 1, and ion source gas 2 were 32, 50, and 65, respectively. Multiple reaction monitoring (MRM) was used to quantify EETs, as follows: 5(6)-EET m/z 319.2→191.0; 8(9)-EET m/z 319.2→155.1; 11(12)-EET m/z 319.2→167.0; 14(15)-EET m/z 319.2→219.1; internal standard (±)16(17)-epoxy-4Z,7Z,10Z,13Z,19Z-docosapentaenoic acid m/z 343.2→274.1.
Quantification of 14,15-EET-EA samples was analyzed using the 5500 QTRAP LC/MS/MS system with a LC separation method consisting of 0–1 minute, 50% A; 5–17 minutes, 0% A; and then returned to initial conditions. The autosampler was set at 5°C. The injection volume was 2 µl. Mass spectra were acquired with positive ESI, and the ion spray voltage was 5500 V. The source temperature was 450°C. The curtain gas, ion source gas 1, and ion source gas 2 were 32, 50, and 65, respectively. MRM was used to quantify 14(15)-EET ethanolamide (m/z 364.3→346.3) with spiked anandamide-d4 (m/z 352.3→287.2) as the internal standard).
Direct quantification of the 14,15-EET-G regioisomer was analyzed using the 5500 QTRAP LC/MS/MS system with a linear gradient, as follows: 0–2 minutes, 90% A; 8 minutes, 55% A; 13–25 minutes, 40% A; 30 minutes, 30% A; 35 minutes, 25% A; 40 minutes, 2% A; 45–47 minutes, 15% A; 48–56 minutes, 0% A; and then returned to initial conditions. Mass spectra were acquired under positive (ion spray voltage at 5500 V) ESI. The source temperature was 450°C. The 14,15-EET-G (m/z 395.3→377.3) was measured in positive ESI with arachidonoyl-1-thio-glycerol (m/z 395.3→287.3) as an internal standard.
Quantification of CYP2J2-ND Rate of 2-AG Ester Cleavage.
The rate of 2-AG ester cleavage was quantified using a HPLC system consisting of an Alliance 2695 analytical separation module (Waters) coupled to a Waters 996 photodiode array and a XTerra C18 column, 1.3 Å, 2.1 mm × 50 mm, 3.5 µm pore size column (Waters). The mobile system was composed of two solutions, as follows: solvent A (H2O/ACN/AcOH 95:5:0.1) and solvent B (H2O/ACN/AcOH 5:95:0.1). The linear gradient was as follows: 0–1 minute, 60% A; 30 minutes, 20% A; 31 minutes, 0% A; then returned to initial conditions. Elution times of 12.6 minutes for 2-AG and 19.7 minutes for AA were verified with authentic standards. Quantification of free AA was calculated using AA absorbance at 204 nm. For kinetic analysis, samples containing CYP2J2-ND (50 pmol; 0.2 µM), CPR (150 pmol; 0.6 µM), and 0.1% BHT in 0.1 M phosphate buffer (pH 7.4) were incubated at 37°C for 5 minutes with 1, 5, 10, 20, 40, and 80 µM 2-AG before initiation of reaction with 1 mM NADPH at 37°C for 20 minutes. Controls lacking either NADPH or CYP2J2 were run simultaneously with 80 µM 2-AG.
CYP2J2 Molecular Operating Environment Homology Modeling.
A multiple sequence alignment was performed using the protein data bank to cross-reference the CYP2J2 sequence against 2A1, 2D6, 2E1, 2R1, 2A6, 2C8, 2B4, 2C5, 2C9, and 2A13. For construction of the CYP2J2 homology model, crystal structure Protein Data Bank coordinates were used for the isozymes with the closest homology to 2J2 and included CYP2B4 (backbone, 42% primary sequence homology), 2C8 (B region, 38% primary sequence homology), 2R1 (FG region, 40% primary sequence homology), and 2C9 (β4 region, 26% primary sequence homology). An open CYP2J2 conformation was initially used to dock AA, AEA, and 2-AG following previously described procedures (Baudry et al., 2003). Two apparent CYP2J2 active site cavities exhibited adjacent/overlapping areas near the heme in agreement with the highest propensity of ligand-binding number and were both used for docking of each respective substrate. Next, we conducted an energy minimization step for each respective ligand conformation, and the induced fitting resulted in the CYP2J2-substrate model used to examine heme and ligand distances as well as the predicted interacting active site residues. Lastly, interaction energies were calculated for each substrate to examine the energetics and feasibility of substrate binding.
Spectral Titrations of CYP2J2-ND.
Spectral binding studies were performed by titrating AA (both AA and sodium arachidonate), AEA, 2-AG, MS-PPOH, and ebastine against CYP2J2-NDs (8 µM) in 100 mM phosphate buffer (pH 7.4). Importantly, substrate stocks were prepared with dimethylsulfoxide (5 mM and 50 mM) after the addition of ethanol and acetonitrile was determined to cause type I binding. CYP2J2-ND were incrementally titrated with increasing concentrations of each putative substrate (either 5 mM or 50 mM stocks) at 22°C and allowed to equilibrate for 5 minutes before measuring the spectra. The total dilution of the original CYP2J2 concentration was ≤1.5%. Absorbance spectra were collected from 800 to 200 nm using a Cary Bio 300 UV-visible spectrophotometer (Agilent Technologies). Raw absorbance spectra were processed to the corresponding subtraction spectra using a standard MATLAB subroutine (Mathworks, Natick, MA). The absolute absorbance difference of the subtraction spectra (peak and trough) was plotted against the corresponding substrate concentration in Origin Laboratory (Northhampton, MA) and fitted using a single binding isotherm (eq. 2).
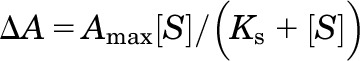 |
(2) |
where ∆A is the absorbance difference at each difference spectra peak and trough, Amax is the amplitude corresponding to maximal spin shift, Ks is the spectral dissociation constant, and [S] is the substrate concentration. None of the substrates investigated exhibited any homotropic cooperativity, and hence the titration data were fitted to a single binding isotherm.
Cyanide Binding and Equilibrium Constant (Ks) Determination for CYP2J2-ND.
The steady-state cyanide spectral equilibrium constants were determined by titrating cyanide concentrations (800 mM and 4 M stocks) against ∼3 μM CYP2J2-ND in the absence and the presence of 40 µM AA, AEA, 2-AG, and MS-PPOH until saturation was reached (Das et al., 2014). The total addition of CYP2J2 concentration was ≤3% and did not affect Soret peak height. The raw data collected were converted into the difference spectra using a MATLAB subroutine (Mathworks). The ΔA (ΔA444–414nm) was plotted against the corresponding cyanide concentration and fitted to a single binding isotherm equation using Origin Pro 8.6 (San Clemente, CA) with eq. 2, where ΔA = ΔA444–414nm.
Transient Cyanide-Binding Kinetics to CYP2J2 Active Site Measured Using Stopped-Flow Spectroscopy.
The cyanide-binding kinetics to the CYP2J2 active site in the absence and presence of varying concentrations of AA, AEA, 2-AG, and MS-PPOH was determined using stopped-flow spectroscopy. Raw data were collected using an Applied Photophysics SX.18MV (Leatherhead, UK) spectrophotometer to monitor the rate of cyanide binding. A dual syringe system was prepared, with syringe I containing CYP2J2-ND in the absence or presence of varying concentrations (10, 20, and 40 µM) of substrate and syringe 2 containing 80 mM cyanide in the absence or presence of varying concentrations (10, 20, and 40 µM) of substrate. The relatively high cyanide concentration was selected based on steady-state cyanide-binding measurements and was necessary to ensure pseudo first-order binding kinetics (Denisov et al., 2007). The parameters for rapid mixing and data collection were as follows: 1.5 ms of dead time, external triggering mechanism, photodiode array detection, and collection obtained on a logarithmic scale over 20 seconds. The raw data were converted to the difference spectra by subtracting each successive scan from the time = 0 scan using a MATLAB subroutine (Mathworks) that produced the typical 446 and 415 nm cyanide-binding peaks. The difference of absorbance was plotted against time and analyzed with a double exponential equation using Origin Pro 8.6.
| (3) |
where A1 and A2 are the amplitudes of the fast and slow phases of the reaction, respectively, and k1 and k2 are the rates of fast and slow phases of cyanide binding to the protein, determined from fitting data obtained to eq. 3.
Data Analysis.
Data were analyzed and presented as means ± S.E. For all experiments, data were collected in triplicate (n = 3) or greater. Stopped-flow data were analyzed for statistical significance using a Student’s t test where *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
CYP2J2-Mediated Metabolism of AA and AEA Determined Using Liquid Chromatography-Electrospray Ionization-Mass Spectrometry.
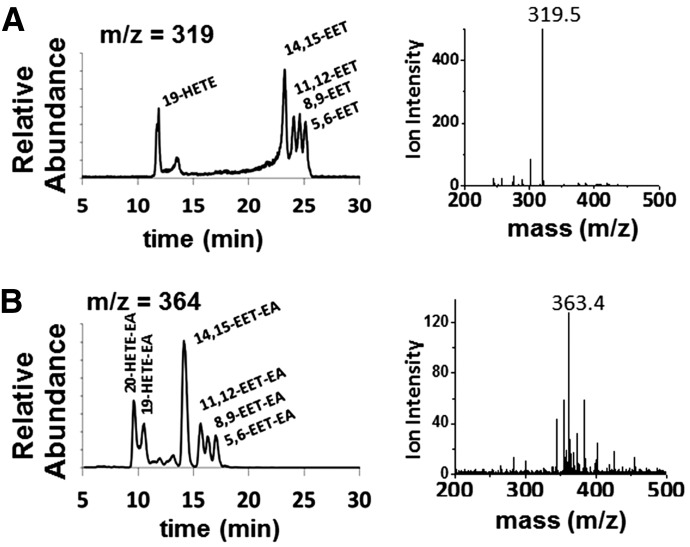
We first investigated the metabolism of AA and AEA by CYP2J2. CYP2J2 was recombinantly expressed in E. coli, purified, and assembled into NDs as described in Materials and Methods. Due to increased functional stability of the CYP2J2-CPR-nanodisc system, all of the studies utilizing recombinant CYP2J2 were first incorporated into NDs. To examine the metabolism of AA and AEA, a liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) method was developed to qualitatively assess the product formation. Importantly, the use of a slow linear gradient was necessary for separation of the regioisomers and structural identification by mass and elution time. Product analysis of CYP2J2-ND-CPR incubations with AA produced four regioisomers (m/z 319.5) corresponding to the mass and elution times of 14,15-, 11,12-, 8,9-, and 5,6-EET (Fig. 2A). The additional peaks (m/z 319), 19-HETE, was confirmed at earlier elution times (11 minutes). Similarly, analysis of CYP2J2 and AEA incubations produced the same olefin oxygenation pattern as AA, with a regioisomer corresponding to the elution times of 14,15-EET-EA, as confirmed using the authentic standard (Fig. 2B). The identity of the 11,12-, 8,9-, and 5,6-EET-EA was extrapolated based on literature describing the elution profile and peak mass confirmation. Similar to AA, the presence of additional HETE metabolites (19- and 20-HETE-EA) was observed. These results show that both AA and AEA are substrates of CYP2J2 and form four epoxide products similar to previous reports with other P450s (Snider et al., 2010).
Fig. 2.
Metabolism of AA and AEA by human CYP2J2-ND-CPR identified qualitatively using LC-ESI-MS. The different regioisomers of (A) AA (m/z 319.5 of 14,15-EET at 22.5 minutes) and (B) AEA (m/z 363.4 of 14,15-EET-EA at 14 minutes) produced six different products, including four epoxide regioisomers at the 5,6-, 8,9-, 11,12-, and 14,15-olefin positions and two HETEs.
CYP2J2-Mediated Metabolism of 2-AG Determined Using LC-ESI-MS.
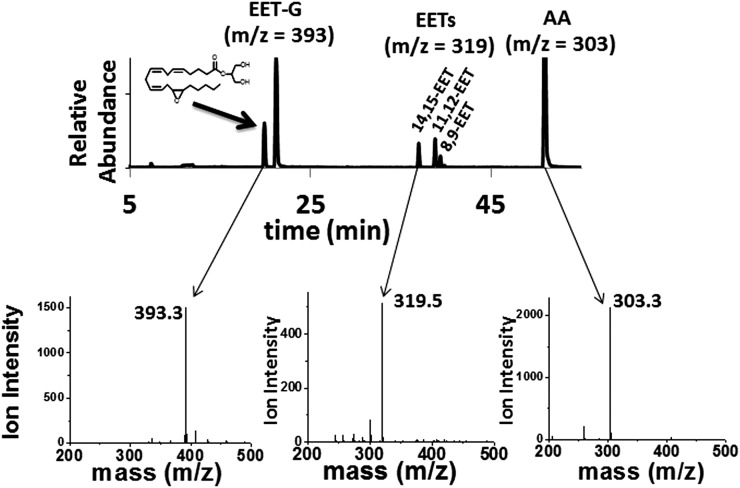
The in vitro incubations containing CYP2J2-ND-CPR and 2-AG produced the following products: two detectable 2-AG epoxide metabolites (m/z 393.3), AA (m/z 303.3) and EETs (m/z 319.5), as shown in Fig. 3. Authentic standards were used to confirm the 14,15-EET-G, EETs (14,15-, 11,12-, 8,9-, 5,6-EET), and AA metabolites. Although the 11,12-EET-G was not commercially available, the results from EET-G saponification confirmed the production of the 11,12-regioisomer.
Fig. 3.
Metabolism of 2-AG by incubations with CYP2J2-ND-CPR revealed the following metabolic profile as represented in the chromatogram that consists of an overlay of two EET-G regioisomers, 14,15-EET-G and 11,12-EET-G (m/z 393); three detectable EET regioisomers, 14,15-EET, 11,12-EET, and 8,9-EET (m/z 319); and arachidonic acid (m/z 303).
In contrast to AA and AEA, only two olefin sites of 2-AG were epoxidized to form 2-AG-EETs (m/z 393.3). Thus, the functional data suggest that the interaction of 2-AG within the CYP2J2-binding pocket is influenced by the presence of the bulky glycerol group of 2-AG (Fig. 1C). Previous incubations with other P450s have failed to produce the EET-G regioisomers (Chen et al., 2008). In this work, we report that CYP2J2 is identified as an isoform of P450 that produces epoxide 2-AG metabolites. As a control, we analyzed CYP2C8 (supersomes) with 2-AG following the same methodology and failed to observe any detectable EET-G formation (Supplemental Fig. 1). This suggests that CYP2J2 may be unique in the epoxidation of 2-AG as compared with other family 2 P450s.
Quantitative Analysis of CYP2J2-Mediated Epoxidation of 2-AG and AEA Using LC-ESI-MS/MS.
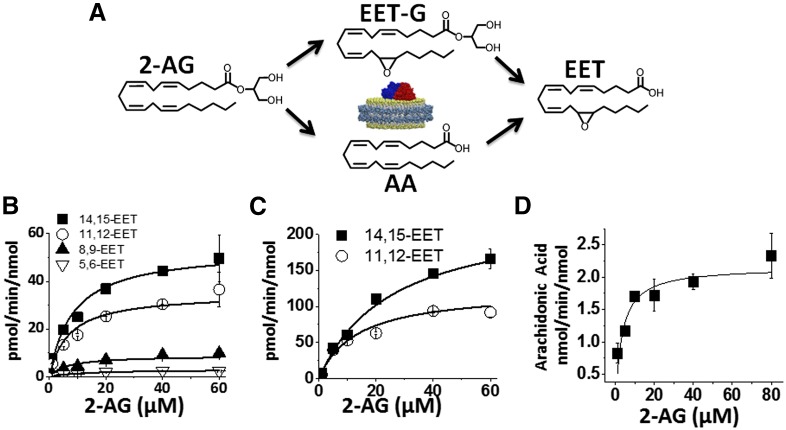
The detection and quantification of fatty acids and their metabolites have been most commonly examined using radio flow HPLC due to the high sensitivity and ease of use (Wu et al., 1996). Alternatively, detection by mass spectrometry after derivatization, coupled with gas chromatography or liquid chromatography, has also been widely used for EET analysis (Zhu et al., 1995). In this study, we successfully used HPLC coupled to a triple quadrupole detector utilizing one of the most sensitive commercially available ion traps (QTRAP 5500), which enabled a highly sensitive, accurate, and reproducible method for detection of all the EET regioisomers in MRM mode. The limit of quantitation with a 10:1 signal to noise ratio for 5(6)-EET was 5.0 ng/ml, whereas 8,9-, 11,12-, and 14,15-EET were calculated at 2.5 ng/ml. Prior to each run, a six-point standard curve was generated for each set of samples using authentic standards. Extraction efficiency was calculated for each of the EET regioisomers using the acidification and the three-step ethyl acetate extraction. The 14,15-, 11,12-, 8,9-, and 5,6-EETs were extracted from the reaction mixture with a 95% or greater efficiency. Reaction linearity was demonstrated for each of the substrates at 15, 30, and 60 minutes with R2 of 0.98 or greater. The advantages of a LC-MS/MS method are that it provides structural elucidation, quantification, and efficiency with excellent sensitivity without the need of derivatization. This method should be widely applicable for complex biologic samples with low levels of EETs. The identification of the metabolites of 2-AG and AEA was followed by quantification and measurement of the rate of formation using a LC-MS-MS method, as described in Materials and Methods (Fig. 4, B and C).
Fig. 4.
Kinetics of CYP2J2-mediated metabolism of 2-AG. (A) Schematic of the proposed routes of 2-AG metabolism by CYP2J2 that produces AA, EET-G, and EET. The kinetic rates of the three products were quantified, as described in Materials and Methods. (B) Kinetics of the formation of the four regioisomers, 14,15-, 11,12-, 8,9-, and 5,6-EET, were detected by LC-MS/MS. These correspond to the free EETs in solution that were generated either from epoxidation of AA or oxidative ester cleavage of EET-G. (C) Kinetics of the formation of the two EET-G regioisomers, 14,15-EET-G and 11,12-EET-G, analyzed as described in Materials and Methods. (D) The kinetics of CYP2J2-ND–mediated 2-AG ester cleavage and arachidonic acid production were determined by monitoring AA formation. The data were fitted to Michaelis-Menten kinetics in Origin Pro 8.6 for calculation of Vmax and Km. The kinetic rates of the epoxidation reactions and side reactions are summarized in Table 1.
The rates of formation of the two EET-G regioisomers were determined by saponification of the EET-G to the corresponding EET regioisomers. The final value of EETs arising from EET-G was obtained by subtracting the free EETs, as determined in Materials and Methods. The kinetics of EET-G formation were as follows: 14,15-EET-G Vmax = 265.4 ± 12.3 pmol/min per nanomole and Km = 32.6 ± 1.7 µM; 11,12-EET-G Vmax = 125.5 ± 5.2 pmol/min per nanomole and Km = 13.6 ± 1.3 µM (Table 1).
TABLE 1.
Summary of 2-arachidonoyl glycerol epoxidation kinetics
The product formation kinetics for CYP2J2 nanodisc incubations with 2-arachidonoyl glycerol.
| Km | Vmax | Vmax/Km | |
|---|---|---|---|
| µM | pmol/min per nmol | ||
| 14,15-EET-G | 32.6 ± 1.7 | 265.4 ± 12.3 | 8.1 ± 0.3 |
| 11,12-EET-G | 13.6 ± 1.3 | 125.5 ± 5.2 | 9.2 ± 0.2 |
| 14,15-EET | 8.3 ± 2.0 | 53.3 ± 2.9 | 6.4 ± 1.25 |
| 11,12-EET | 6.7 ± 1.3 | 34.8 ± 1.5 | 5.1 ± 0.4 |
| 8,9-EET | 6.2 ± 1.8 | 8.9 ± 1.1 | 1.4 ± 0.3 |
| 5,6-EET | 12.7 ± 3.8 | 3.1 ± 0.3 | 0.2 ± .05 |
In a separate experiment, the rate of metabolism of AA to form the four EET regioisomers was measured in the presence of saturating concentrations of AA. Catalytic rates were calculated at 103 ± 4.6, 61.5 ± 5.6, 18.4 ± 1.7, and 8.9 ± 0.1 pmol/min per nanomole CYP2J2 for 14,15-, 11,12-, 8,9-, and 5,6-EET, respectively.
Additionally, we examined the CYP2J2-mediated epoxidation of saturating amounts of AEA (60 µM) toward the production of 14,15-EET-EA, which was calculated to be 391 ± 12 pmol/min per nanomole CYP2J2. Thus, the production of the 14,15-EET-EA appears to proceed at a significantly faster rate when compared with the production of 14,15-EET from AA despite sharing a similar qualitative product profile and binding characteristics, as discussed later.
CYP2J2-Mediated 2-AG Ester Cleavage and Production of Free AA.
The LC-ESI-MS product analysis of 2-AG incubations revealed significant amounts of free AA in solution (Fig. 3). Control experiments showed that the conversion of 2-AG to AA required the presence of both NADPH and CPR for catalytic turnover, indicating that the reaction proceeded through an oxidative mechanism (controls not shown). As seen in Fig. 4D, analysis of 2-AG incubations followed Michaelis-Menten kinetics, indicating that the reaction was dependent on substrate binding to the active site. The Km of the ester cleavage reaction was calculated at 3.25 µM with a Vmax of 2.2 ± 0.2 nmol/min per nanomole CYP2J2. Samples lacking NADPH and CYP2J2 were run in parallel as controls. AA was not detected in the NADPH-free sample. Additionally, 2-AG was stable in buffer at 37°C with negligible hydrolysis. The ability of P450 to mediate ester cleavage reactions has long been reported, and rates determined in this work show similar catalytic properties of other P450 isozymes (Guengerich, 1987; Peng et al., 1995). However, this is the first account of the P450-mediated cleavage of an endogenous fatty acid ester. The same reactions were run with CYP2C8 (supersomes), which also produced AA in a NADPH-dependent reaction (Supplemental Fig. 1). However, CYP2C8 epoxygenase did not demonstrate the ability to oxygenate the 2-AG olefin bonds.
Bovine and Porcine Heart Microsome Metabolism of AA and 2-AG.
To examine whether the novel endocannabinoid epoxygenation reactions would occur within a complex environment in the presence of other proteins, we prepared heart microsomes from the tissue of bovine and porcine left ventricular myocardium. The rate of metabolism of AA, AEA, and 2-AG is summarized in Table 2. We chose bovine and porcine species because they both contain CYP2J2 with high sequence homology to humans (Supplemental Fig. 3). CYP2J2 immunoblotting of the bovine and porcine microsomes was performed using a polyclonal rabbit IgG antibody for human CYP2J2. The microsomal preparations cross-reacted with the CYP2J2 antibody demonstrating the presence of CYP2J2 in bovine and porcine hearts (Supplemental Fig. 4). First, the functionality was assessed by measuring AA metabolism by bovine and porcine microsomes, which produced 1.8 ± 0.3 and 2.08 ± 0.02 pmol min−1 mg−1 microsomal protein of EETs (all regioisomers), respectively. Next, we quantified 2-AG metabolism by directly measuring the production of the 14,15-EET-G regioisomer by bovine and porcine microsomes that was calculated at 1.17 ± 0.57 and 1.1 ± 0.55 pmol min−1 mg−1 microsomal protein, respectively. The 14,15-EET-EA isomer was produced with a higher turnover in both bovine and porcine microsomes with values of 1.96 ± 0.05 and 2.25 ± 0.07 pmol min−1 mg−1 microsomal protein, respectively.
TABLE 2.
Metabolism of arachidonic acid, 2-arachidonoyl glycerol, and anandamide by bovine and porcine heart microsomes
| Substrate | Metabolite(s) Detected | Bovine Microsomes | Porcine Microsomes |
|---|---|---|---|
| pmol min−1 mg−1 | |||
| AA | Total EET (all regioisomers) | 1.8 ± 0.3 | 2.08 ± 0.02 |
| 2-AG | 14,15-EET-G | 1.1 ± 0.57 | 1.18 ± 0.55 |
| AEA | 14,15-EET-EA | 1.96 ± 0.05 | 2.25 ± 0.07 |
CYP2J2-Mediated Lipase Activity.
CYP2J2-mediated oxidative cleavage of 2-AG into AA raised questions as to whether this enzyme could act as a lipase and use AA-containing phospholipids as direct substrates. PIP2 is present in the plasma membrane, where it is cleaved by phospholipase C to produce AA for subsequent entry into the eicosanoid cascade (Rouzer and Marnett, 2011). Thus, we incubated PIP2 with the CYP2J2-ND-CPR-NADPH system utilizing PIP2 concentrations below its critical micelle formation. In these experiments, we did not observe formation of AA or diacylglycerol (Huang et al., 2011).
Spectral Binding Studies of CYP2J2 with AA, AEA, and 2-AG.
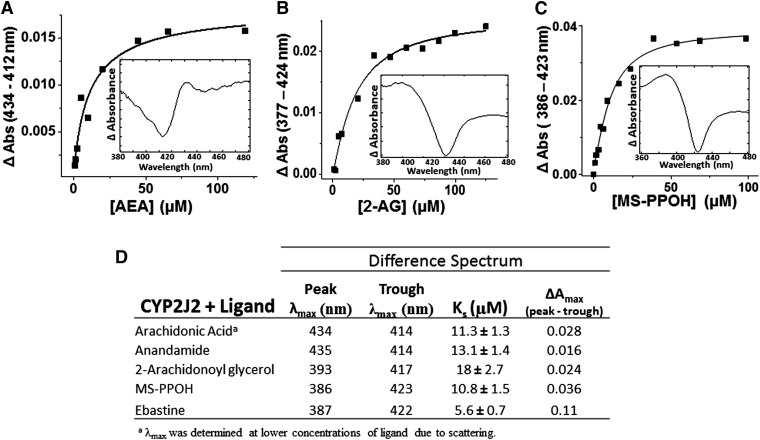
In an effort to understand the binding interactions of AA and its derivatives (AEA and 2-AG) with the active site of CYP2J2, we performed spectroscopic binding studies. First, the interactions of AA, AEA, 2-AG, ebastine, and MS-PPOH with the CYP2J2-ND active site were investigated using UV-visible spectral binding titrations (Fig. 5). As controls, the spectral binding properties of the potent epoxygenase inhibitor MS-PPOH (Falck et al., 1997) and ebastine (Fig. 5D) were investigated, and both induced type-I shifts with apparent spectral binding constants (Ks) of 10.8 ± 1.5 µM and 5.6 ± 0.7, respectively.
Fig. 5.
Equilibrium binding using absorption spectra for interactions of substrate with CYP2J2 nanodiscs. (A) AEA titration Ks was determined to be 13.1 ± 1.4 µM (inset: difference spectrum of AEA-bound CYP2J2-NDs). (B) 2-AG titration Ks was determined to be 18 ± 2.7 µM (inset: difference spectrum of 2-AG–bound CYP2J2 nanodiscs). (C) MS-PPOH titration Ks was determined to be 10.8 ± 1.5 µM (inset: difference spectrum of MS-PPOH–bound CYP2J2-NDs showing a type I binding mode). (D) Summary of the λmax (nm) of peak/trough, Ks, and ΔΑmax for binding of arachidonic acid, anandamide, 2-arachidonoyl glycerol, ebastine, and MS-PPOH to CYP2J2 nanodiscs.
The fatty acids presented with smaller changes in spin-state and minor shifts in Soret band. Specifically, the Soret band at 417 nm red-shifted by 0.5 nm with a slight decrease in amplitude for both AA and AEA. The difference spectra revealed prominent troughs for both ligands at 414 nm and broad peaks at 434 and 435 nm for AA and AEA, respectively (Fig. 5A). Both substrates were fitted to single binding isotherm with apparent Ks of 11.3 ± 1.3 µM for AA and 13.1 ± 1.4 µM for AEA (Fig. 5D). The similarities between AA and AEA spectral shifts and binding equilibrium suggest that the presence of the ethanolamide (AEA) motif does not significantly alter substrate distances and positioning in relation to the heme when compared with AA. Conversely, 2-AG spectral titrations resulted in a blue shift while slightly decreasing the Soret. The apparent Ks of binding for 2-AG was calculated at 18 ± 2.7 µM. Taken together, these data demonstrate that the modification of the carboxylic acid of the AA with the glycerol moiety affects binding in CYP2J2’s large, yet narrow binding cavity (Lafite et al., 2007). Note that the observed Soret shifts are not as prominent as xenobiotic P450 substrates, yet are consistent with the few published spectra of other fatty acids binding to membrane-bound P450s (Loughran et al., 2001). Moreover, the lack of significant Soret perturbations corresponds to the relatively low turnover rates of the AA-metabolizing epoxygenases, such as CYP2J2 (Westphal et al., 2011). Additionally, these experiments were further validated using steady-state cyanide-binding experiments in the presence of AA, AEA, and 2-AG to probe the access and binding of cyanide to the heme (Supplemental Fig. 2).
Kinetics of Cyanide Binding to CYP2J2-ND in the Presence of Substrates.
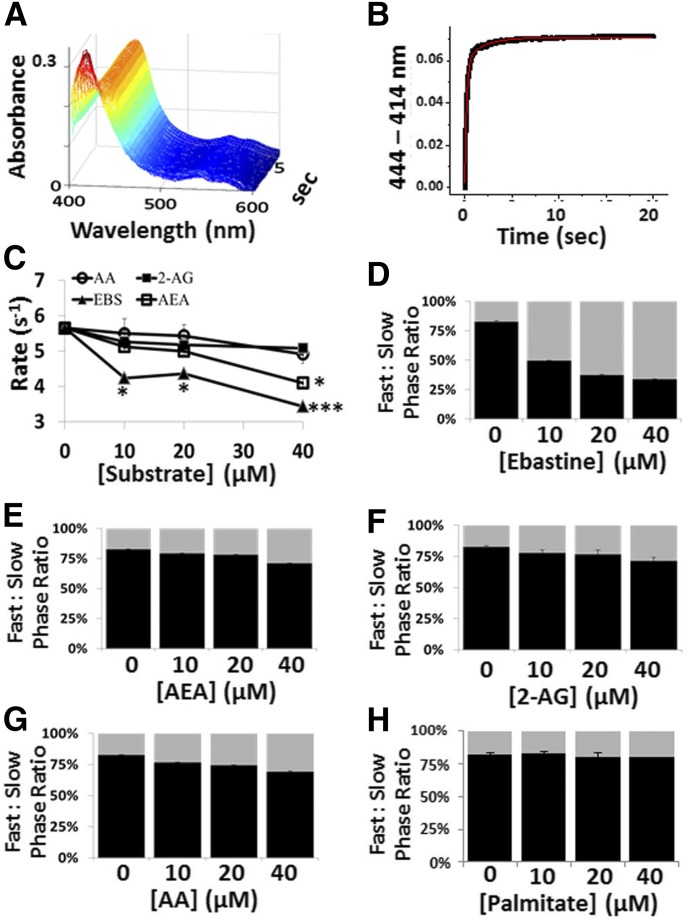
The cyanide-binding kinetics of CYP2J2 was investigated in the absence or presence of hydrophobic substrates using stopped-flow spectroscopy. For each substrate tested, rapid mixing and formation of the heme-cyanide complex red-shifted the Soret (Fig. 6A). As shown in Fig. 6B, the difference spectra Δ (A444–414nm) were analyzed as a function of time and yielded an exponential biphasic curve that was comprised of a fast and slow phase (Bidwai et al., 2013). The fast phase corresponds to the rate of cyanide binding to the ferric heme active site. Specifically, it can be used as a direct measure of substrate egress from the immediate vicinity of the heme active site to accommodate formation of the cyanide complex. In the fast phase, the presence of increasing concentrations of ebastine significantly decreased cyanide-binding rates in a concentration-dependent manner. Conversely, the fatty acid ligands only slightly decreased cyanide binding (Fig. 6C). The ratio of the fast- and slow-phase amplitudes was compared to determine how ligand concentrations affect the presence of the ratios. As seen in Fig. 6, D–G, saturating concentrations of ebastine significantly increase the ratio of the slow phase (17.4 to 66.4%), whereas the arachidonate-containing ligands only slightly increased the slow phase ∼10%. To rule out nonspecific fatty acid binding, we examined the rates of cyanide binding in the presence of saturating concentrations of palmitate as a nonbinding substrate. As shown in Fig. 6H, the presence of palmitate does not noticeably alter the ratio of the two phases. Although not dramatic, the reported results provide important characterization of the nature of fatty acid binding.
Fig. 6.
Stopped-flow cyanide-binding spectroscopy with CYP2J2-ND and AA, 2-AG, AEA, and ebastine. (A) Stopped-flow spectroscopy was used to monitor kinetics of cyanide binding to CYP2J2-ND by recording the formation of the cyanide-heme complex at 444 nm as a function of time in the presence of saturating [CN-]. (B) The difference spectra (ΔAbs 444–414 nm) was plotted against time and fitted to a double exponential equation for determination of the rate of the (C) fast phase for each substrate concentration tested and determination of statistical significance relative to the substrate-free condition when *P < 0.05 and ***P < 0.001. The ratio of the amplitudes of the fast phase (black) to slow phase (gray) was dramatically altered in the presence of (D) ebastine, whereas the presence of the polyunsaturated fatty acids such as (E) AEA, (F) 2-AG, and (G) AA was modestly changed. (H) The saturated fatty acid palmitate did not alter cyanide-binding characteristics.
CYP2J2 Homology Modeling with AA, AEA, and 2-AG.
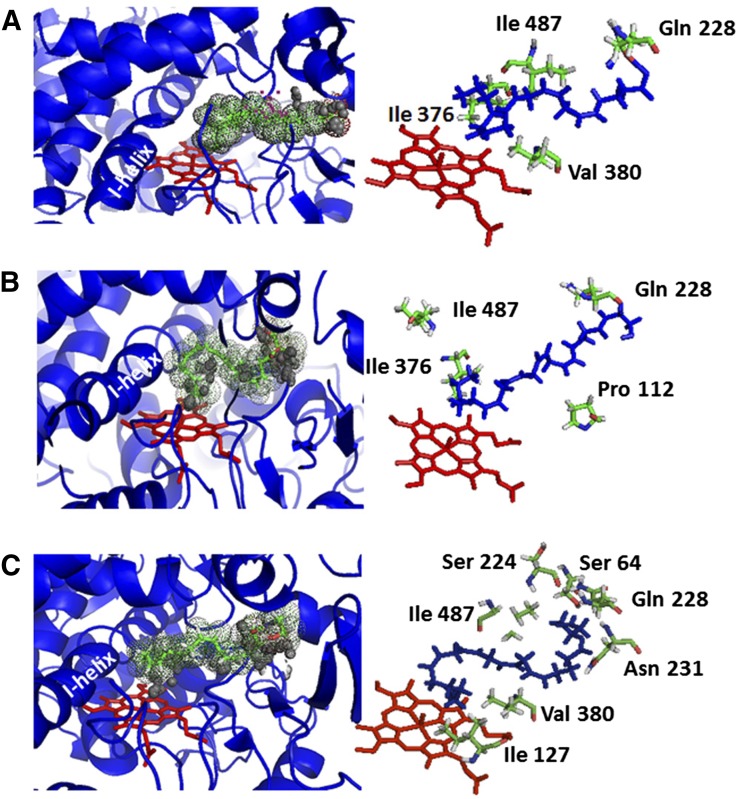
Molecular Operating Environment (Chemical Computing Group, Montreal, QC, Canada) software was used to identify substrate-binding characteristics within the active site of a CYP2J2 homology model. A hybrid structure model was constructed from the crystal Protein Data Bank coordinates of similar P450 isozymes (class-dependent sequence alignment strategy), as described in Materials and Methods (Baudry et al., 2006). This approach takes advantage of the conserved sequences that maintain the secondary and tertiary P450 folds to generate the core structures surrounding the buried catalytic sites. Given the fatty acids’ relatively low turnover, there are likely many unproductive conformations within the active site. As shown in Fig. 7A, AA was docked in the most preferred configuration for epoxidation of the 14,15-olefin when closest to the heme (5.1 Å), followed by the 11,12-, 8,9-, and 5,6-olefins with calculated heme distances of 8.6, 11.3, and 12.9 Å, respectively. As shown in Fig. 7B, the distances of the AEA 14,15-, 11,12-, 8,9-, and 5,6-olefins were calculated at 6, 8.5, 12.1, and 13.5 Å, respectively. Interestingly, 2-AG (Fig. 7C) yielded significantly different active site interactions albeit with similar heme distances of the 14,15-, 11,12-, 8,9-, and 5,6-olefin estimated at 5.9, 7.6, 9, and 11.5 Å, respectively. The interaction potential energies predict feasibility of binding and were similar for each polyunsaturated fatty acid with calculations of −68.3 kcal/mol, −69.4 kcal/mol, and −73.4 kcal/mol for AA, 2-AG, and AEA, respectively. Thus, the olefin distances of the Molecular Operating Environment model were in agreement with experimental differences observed in AA, AEA, and 2-AG metabolism. Additionally, the model predicted significantly different binding characteristics based on the presence of the carboxylate/ethanolamide or glycerol moiety. For example, a patch of hydrophobic residues appeared to mediate AA and AEA positioning within the CYP2J2 active site with Pro112, Ile487, and Ile376 common for both substrates. Interactions of polar residues were fewer with only Gln228 interacting with the polar carboxylate/ethanolamide groups. Conversely, positioning of the 2-AG used both hydrophobic residues (Ile127, Val380, and Ile487) as well as interactions of the hydrophilic glycerol moiety with polar CYP2J2 residues (Asn231, Ser224, Gln228, and Ser64). The relatively fewer polar interactions of AA and AEA might enable sampling of different conformations of these substrates in the active site leading to the epoxidation of the 8,9- and 5,6-olefin groups. Conversely, the glycerol group appears tethered and cannot sample orientations for epoxidation of the 8,9- and 5,6-olefins. In absence of a high-resolution CYP2J2 crystal structure, this model provides a useful tool to predict the interactions of AA, AEA, and 2-AG within the active site that provides CYP2J2’s regioselectivity.
Fig. 7.
Substrate-bound CYP2J2 homology model with arachidonic acid, anandamide, and 2-arachidonoyl glycerol. Proposed docking of the polyunsaturated fatty acids into the active site of CYP2J2. (Left) The substrate (A) AA, (B) AEA, and (C) 2-AG are represented as stick figures (green) surrounded by space-filling dots. The CYP2J2 ribbon structure is colored blue, and the heme-oxygen motif is in red. The interaction potential energies predict feasibility of binding and were similar for each polyunsaturated fatty acid with calculations of −69.4 kcal/mol and −73.4 kcal/mol for 2-AG and AEA, respectively. (Right) Predicted ligand-residue interactions within the CYP2J2 active site for (A) AA, (B) AEA, and (C) 2-AG.
Discussion
Extrahepatic CYP2J2 epoxygenase metabolizes AA to form EETs that are essential for regulation of blood pressure and cardiovascular function (Spector, 2009). AA-derived endocannabinoids play an important role in the regulation of the cardiovascular system. Therefore, we explored the metabolism of functionalized arachidonate-containing lipids such as the endocannabionoids with the cytochrome P450 most highly expressed in the human heart. The two most well characterized, AA-derived endocannabinoids are AEA and 2-AG, whose dysregulation has been implicated in a wide range of pathophysiological states (Pacher et al., 2006; Kogan and Mechoulam, 2007; Pacher and Steffens, 2009). In this study, we demonstrate CYP2J2-mediated metabolism of AEA and 2-AG using recombinant human CYP2J2 and heart microsomes derived from bovine and porcine tissues. The results provided in this work show the endocannanbinoids 2-AG and AEA are both substrates of human CYP2J2 and that the predominant products, 14,15-EET-EA and 14,15-EET-G, are produced in heart microsomal systems of two different mammalian species, lending credibility to the proposed pathway in tissues.
The qualitative product analysis of CYP2J2-AEA incubations by LC-MS revealed four epoxide regioisomers at the 5,6-, 8,9-, 11,12-, and 14,15-olefin positions. The quantitative analysis of 14,15-EET-EA exhibited fourfold more product when compared with the rate of formation of 14,15-EET regioisomer from AA. Previously, the oxygenation of AEA into a single hydroxy product, 20-HETE ethanolamide, as well as four epoxide regioisomers, 5,6-, 8,9-, 11,12-, and 14,15-EET-EA, was demonstrated by several hepatic P450s, including CYP2D6, CYP3A4, and CYP4F2 (Snider et al., 2010). However, this is the first report of the AEA metabolism by a P450 epoxygenase that is highly expressed in the myocardium and endocardium. Given the increasing reports of an endocannabinoid-mediated regulation of blood pressure and the cardiovascular system, the role of the oxygenated AEA metabolites may provide an interesting avenue for future studies. For instance, AEA is a partial agonist of both CB1 and CB2 receptors, whereas the 5,6-EET-EA metabolite selectively binds CB2 with 1000-fold greater affinity (Snider et al., 2009). Additionally, there is mounting evidence of CB2 cardioprotection versus CB1 cardiotoxicity, and the ability to selectively target these pathways may provide a useful therapeutic target for the treatment of heart disease (Mukhopadhyay et al., 2008; Pacher and Steffens, 2009). Moreover, less is known about the physiologic effects of the other EET-EA metabolites and whether they also exhibit differential receptor-binding profiles and in vivo effects.
Previously, the epoxide metabolites of 2-AG, 14,15-, and 11,12-EET-G were found endogenously in rat kidney and spleen tissues. These metabolites bound both CB1 and CB2 with greater affinity than 2-AG (Chen et al., 2008). Incubations with family 2 P450s, CYP2C8, CYP2C11, and CYP2C3 failed to produce these metabolites (Chen et al., 2008). Therefore, it was concluded that these epoxide metabolites do not arise from epoxidation of 2-AG through the P450 pathway. Our modeling studies show that CYP2J2 contains a relatively large active site that can accommodate 2-AG as compared with other family 2 P450s. Moreover, we show it is possible to directly oxygenate 2-AG by CYP2J2 to produce relatively high levels of both 14,15- and 11,12-EET-G. Additionally, our results show that CYP2J2 produces only two epoxide metabolites, 14,15-EET-G and 11,12-EET-G, similar to the two 2-AG epoxide metabolites identified in vivo. Thus, these results provide an alternative pathway for the biosynthesis of 2-AG epoxides through the direct enzymatic oxygenation by CYP2J2.
Due to the demonstrated beneficial functions of the endocannabinoids, there are several drugs in the market that target the enzymes that attenuate the in vivo levels of these substrates (Cravatt and Lichtman, 2003). Previous inhibition of enzymes that inactivate AEA and 2-AG has shown that the inhibition is incomplete (Blankman et al., 2007), indicating that there may be alternative pathways of attenuation of these endocannabinoids. We demonstrate in this work that 2-AG is converted to AA by CYP2J2 and CYP2C8. Subsequent control experiments confirmed that the cleavage proceeded via an oxidative NADPH-dependent CYP2J2-CPR mechanism. To our knowledge, these results provide the first evidence of a P450-mediated endocannabinoid attenuation pathway for production of free AA for subsequent metabolism in the classic eicosanoid pathways (COX, LOX, and epoxygenase). To date, there have been no reports of an oxidative ester cleavage mechanism of endogenous 2-AG. However, other P450 isozymes have long been recognized as important sources of carbonyl products through nonhydrolytic ester cleavage reactions (Guengerich, 1987; Peng et al., 1995).
The potential of CYP2J2 to produce its own substrate through the cleavage of AA from PIP2 was also explored. Several different conditions were examined, and it was concluded the PIP2 is not a CYP2J2 substrate. Considering the tissue distribution of cytochrome P450s, the role of a P450-mediated attenuation of 2-AG may constitute further examination in vivo.
The binding interactions of AA and its derivatives with the active site of CYP2J2 revealed relatively low spin state changes. Thus, the use of the nanoscale lipid bilayers of NDs was essential for the characterization of AA, AEA, and 2-AG binding in spectroscopic studies with minimal spectral scattering. In general, there are very few examples of human P450 titrations with lipid substrates in the literature, most likely due to issues with solubility, scattering, and low spin state changes. Interestingly, the few that have been published typically present with a similar spectra to what is presented in this work (Loughran et al., 2001). The ebastine, MS-PPOH, and 2-AG Soret shifts present with classic type I binding modes (Fig. 5). The binding of AA and AEA was less clear and difficult to conclusively assign the classic type I and type II shifts due to the low signal to noise ratio. However, all the binding shifts were concentration dependent and reach saturation and are present with clear peaks/troughs, lending credibility to the calculated binding constants. Although the spin-state changes of the AA and the derivatives are minor when compared with ebastine, they do correspond to the relatively low catalytic turnover of these substrates. Importantly, the relatively slow turnover rates of CYP2J2 resemble the functions of the steroid-synthesizing P450s that regulate complex homeostatic processes rather than the very fast detoxification functions of liver P450s.
The results for the CYP2J2-ND substrate-free and substrate-bound stopped-flow cyanide-binding experiments are summarized in Fig. 6 and Supplemental Table 1. For cyanide-binding studies, we employed ebastine and showed that the rates of the fast phase were significantly decreased with increasing ebastine concentrations similar to other P450-substrate systems (Denisov et al., 2007). Conversely, AA, AEA, and 2-AG only showed modest inhibition of cyanide binding as shown in Fig. 6C and indicated that they either more easily egress from the active site or are bound as to not block cyanide binding. Similarly, the ratio of the fast to slow phase was significantly decreased in the presence of increasing concentrations of ebastine and only modestly decreased in the presence of increasing concentrations of AA, AEA, and 2-AG (Fig. 6, E–G). This effect was consistent and repeatable and not due to nonspecific fatty acid binding, as evidenced by the lack of change induced by palmitate (Fig. 6H).
The enzymatic turnover of CYP2J2 is 1 to 2 orders of magnitude lower than xenobiotic metabolizing hepatic P450s, and it might be tempting to disregard the potential biologic significance of the CYP2J2 pathway based solely on these low enzymatic turnover rates. However, the extrahepatic epoxygenases, including CYP2J2, function to regulate complex homeostatic functions within the body and exhibit significantly lower turnover of the endogenous fatty acids (Westphal et al., 2011) in a highly regulated manner. Thus, the formation of these metabolites most likely represents a far more elegant mechanism for control of the many delicate homeostatic processes within organs, such as the heart and kidneys. Therefore, similar to the steroid-synthesizing P450s, the enzymatic expression levels rather than the catalytic turnover are likely the prominent mechanism for metabolic control in each respective organ.
In summary, we have demonstrated the ability of CYP2J2 to metabolize a diverse range of AA-containing substrates into a number of unique metabolites that have previously been shown to regulate important physiologic effects in vivo. Additionally, we provide important evidence that CYP2J2 can directly attenuate 2-AG levels through a NADPH-dependent oxidative cleavage mechanism, thereby producing free AA for subsequent metabolism within the classic eicosanoid cascade. We further show that the formation of the metabolites of 2-AG and AEA is in agreement with the spectral binding studies and modeling.
Supplementary Material
Acknowledgments
The authors thank Dr. Ilia Denisov and Dr. Matthew Edin for helpful discussions. The authors thank Mr. William Arnold for editing the paper. The authors greatly appreciate the contributions of Dr. Zhong Li at the Metabolomics Laboratory of Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign. The authors thank Furong Sun and Dr. Kevin Tucker for continued support at the School of Chemical Sciences mass spectrometry facility, University of Illinois at Urbana-Champaign. The authors thank Prof. Stephen Sligar for providing the gene encoding MSP1D1 and MSP1E3D1; Prof. Robert Gennis for the use of stopped-flow equipment; and Dr. Hanlin Ouyang for technical assistance. The authors thank Prof. Mary Schuler, Brendon Colón, and Eryk Radziszewski for training and assistance with the Molecular Operating Environment modeling software. The authors thank Prof. Fan and Holly Pondenis for assistance with CYP2J2 immunoblotting. The authors thank Prof. Bunick for providing the homogenizer.
Abbreviations
- AA
arachidonic acid
- AEA
anandamide
- 2-AG
2-arachidonoylglycerol
- BHT
butylated hydroxytoluene
- CB1
cannabinoid receptor 1
- CB2
cannabinoid receptor 2
- COX
cyclooxygenase
- CPR
cytochrome P450 reductase
- DTT
dithiothreitol
- EET
epoxyeicosatrienoic acid
- EET-EA
epoxyeicosatrienoic ethanolamide
- EET-G
epoxyeicosatrienoic glycerol
- ESI
electrospray ionization
- HETE
hydroxyeicosatetraeonic acid
- HPLC
reversed-phase liquid chromatography
- HRP
horseradish peroxidase
- LC-ESI-MS
liquid chromatography-electrospray ionization-mass spectrometry
- LC-MS
liquid chromatography-mass spectrometry
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- LOX
lipoxygenase
- MRM
multiple reaction monitoring
- MS-PPOH
N-(methylsulfonyl)-2-(2-propynyloxy)-benzenehexanamide
- ND
nanodisc
- P450
cytochrome P450
- PIP2
1-heptadecanoyl-2-arachidonoyl-sn-glycero-3-[phosphoinositol-4′,5′-bisphosphate]
- PMSF
phenylmethanesulfonylfluoride
- POPC
1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine
- POPS
1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-l serine
Authorship Contributions
Participated in research design: McDougle, Das.
Conducted experiments: McDougle, Kambalyal.
Performed data analysis: McDougle, Meling, Kambalyal, Das.
Wrote or contributed to the writing of the manuscript: McDougle, Das.
Footnotes
This work was supported in part through the American Heart Predoctoral Fellowship [14PRE20130015] (to D.R.M.). The Roy J. Carver Biotechnology Center’s 5500 QTrap MS was funded by the National Institutes of Health National Center for Research Resources [Grant S10-RR024516]. The School of Chemical Sciences Q-TOF Ultima mass spectrometer was purchased in part with a grant from the National Science Foundation, Division of Biological Infrastructure [DBI-0100085].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Alghasham A, Ali A, Ismail H, Dowaidar M, Settin AA. (2012) CYP2J2 -50 G/T and ADRB2 G46A gene polymorphisms in Saudi subjects with hypertension. Genet Test Mol Biomarkers 16:1027–1031. [DOI] [PubMed] [Google Scholar]
- Baudry J, Li W, Pan L, Berenbaum MR, Schuler MA. (2003) Molecular docking of substrates and inhibitors in the catalytic site of CYP6B1, an insect cytochrome p450 monooxygenase. Protein Eng 16:577–587. [DOI] [PubMed] [Google Scholar]
- Baudry J, Rupasinghe S, Schuler MA. (2006) Class-dependent sequence alignment strategy improves the structural and functional modeling of P450s. Protein Eng Des Sel 19:345–353. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Sligar SG. (2010) Membrane protein assembly into nanodiscs. FEBS Lett 584:1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwai AK, Meyen C, Kilheeney H, Wroblewski D, Vitello LB, Erman JE. (2013) Apolar distal pocket mutants of yeast cytochrome c peroxidase: hydrogen peroxide reactivity and cyanide binding of the TriAla, TriVal, and TriLeu variants. Biochim Biophys Acta 1834:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankman JL, Simon GM, Cravatt BF. (2007) A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol 14:1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila JH, Dishman E, Karara A, Falck JR. (1991) Cytochrome P450 arachidonic acid epoxygenase: stereochemical characterization of epoxyeicosatrienoic acids. Methods Enzymol 206:441–453. [DOI] [PubMed] [Google Scholar]
- Chen JK, Chen J, Imig JD, Wei S, Hachey DL, Guthi JS, Falck JR, Capdevila JH, Harris RC. (2008) Identification of novel endogenous cytochrome p450 arachidonate metabolites with high affinity for cannabinoid receptors. J Biol Chem 283:24514–24524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Lichtman AH. (2003) Fatty acid amide hydrolase: an emerging therapeutic target in the endocannabinoid system. Curr Opin Chem Biol 7:469–475. [DOI] [PubMed] [Google Scholar]
- Das A, Varma SS, Mularczyk C, Meling DD. (2014) Functional investigations of thromboxane synthase (CYP5A1) in lipid bilayers of nanodiscs. ChemBioChem 15:892–899. [DOI] [PubMed] [Google Scholar]
- Delozier TC, Kissling GE, Coulter SJ, Dai D, Foley JF, Bradbury JA, Murphy E, Steenbergen C, Zeldin DC, Goldstein JA. (2007) Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos 35:682–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, McLean MA, Sligar SG. (2007) The one-electron autoxidation of human cytochrome P450 3A4. J Biol Chem 282:26865–26873. [DOI] [PubMed] [Google Scholar]
- Denisov IG, Sligar SG. (2011) Cytochromes P450 in nanodiscs. Biochim Biophys Acta 1814:223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck JR, Belosludtsev YY, Reddy KK, Reddy KM, Shortt MF, Chauhan K, Capdevila JH, Wei SZ. (1997) Eicosanoid biosynthesis: differential inhibition of cytochrome P450 epoxygenase and omega-hydroxylase. Bioorg Med Chem Lett 7:3053–3056. [Google Scholar]
- Grinkova YV, Denisov IG, Sligar SG. (2010) Functional reconstitution of monomeric CYP3A4 with multiple cytochrome P450 reductase molecules in nanodiscs. Biochem Biophys Res Commun 398:194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. (1987) Oxidative cleavage of carboxylic esters by cytochrome P-450. J Biol Chem 262:8459–8462. [PubMed] [Google Scholar]
- Huang W, Jiang D, Wang X, Wang K, Sims CE, Allbritton NL, Zhang Q. (2011) Kinetic analysis of PI3K reactions with fluorescent PIP2 derivatives. Anal Bioanal Chem 401:1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imig JD, Hammock BD. (2009) Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat Rev Drug Discov 8:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan NM, Mechoulam R. (2007) Cannabinoids in health and disease. Dialogues Clin Neurosci 9:413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafite P, André F, Zeldin DC, Dansette PM, Mansuy D. (2007) Unusual regioselectivity and active site topology of human cytochrome P450 2J2. Biochemistry 46:10237–10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran PA, Roman LJ, Miller RT, Masters BS. (2001) The kinetic and spectral characterization of the E. coli-expressed mammalian CYP4A7: cytochrome b5 effects vary with substrate. Arch Biochem Biophys 385:311–321. [DOI] [PubMed] [Google Scholar]
- McDougle DR, Palaria A, Magnetta E, Meling DD, Das A. (2013) Functional studies of N-terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Sci 22:964–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P, Mohanraj R, Bátkai S, Pacher P. (2008) CB1 cannabinoid receptor inhibition: promising approach for heart failure? Congest Heart Fail 14:330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, Grinkova YV, Sligar SG, Atkins WM. (2007) Ligand binding to cytochrome P450 3A4 in phospholipid bilayer nanodiscs: the effect of model membranes. J Biol Chem 282:28309–28320. [DOI] [PubMed] [Google Scholar]
- Orlando BJ, McDougle DR, Lucido MJ, Eng ET, Graham LA, Schneider C, Stokes DL, Das A, Malkowski MG. (2014) Cyclooxygenase-2 catalysis and inhibition in lipid bilayer nanodiscs. Arch Biochem Biophys 546:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Bátkai S, Kunos G. (2006) The endocannabinoid system as an emerging target of pharmacotherapy. Pharmacol Rev 58:389–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Steffens S. (2009) The emerging role of the endocannabinoid system in cardiovascular disease. Semin Immunopathol 31:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahy D, Edin ML, Lee CR, Huang S, Bielenberg DR, Butterfield CE, Barnés CM, Mammoto A, Mammoto T, Luria A, et al. (2012) Epoxyeicosanoids stimulate multiorgan metastasis and tumor dormancy escape in mice. J Clin Invest 122:178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HM, Raner GM, Vaz AD, Coon MJ. (1995) Oxidative cleavage of esters and amides to carbonyl products by cytochrome P450. Arch Biochem Biophys 318:333–339. [DOI] [PubMed] [Google Scholar]
- Pretorius PJ, Pohl WG, Smithen CS, Inesi G. (1969) Structural and functional characterization of dog heart microsomes. Circ Res 25:487–499. [DOI] [PubMed] [Google Scholar]
- Rouzer CA, Marnett LJ. (2011) Endocannabinoid oxygenation by cyclooxygenases, lipoxygenases, and cytochromes P450: cross-talk between the eicosanoid and endocannabinoid signaling pathways. Chem Rev 111:5899–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Kornilov AM, Kent UM, Hollenberg PF. (2007) Anandamide metabolism by human liver and kidney microsomal cytochrome p450 enzymes to form hydroxyeicosatetraenoic and epoxyeicosatrienoic acid ethanolamides. J Pharmacol Exp Ther 321:590–597. [DOI] [PubMed] [Google Scholar]
- Snider NT, Nast JA, Tesmer LA, Hollenberg PF. (2009) A cytochrome P450-derived epoxygenated metabolite of anandamide is a potent cannabinoid receptor 2-selective agonist. Mol Pharmacol 75:965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Sikora MJ, Sridar C, Feuerstein TJ, Rae JM, Hollenberg PF. (2008) The endocannabinoid anandamide is a substrate for the human polymorphic cytochrome P450 2D6. J Pharmacol Exp Ther 327:538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Walker VJ, Hollenberg PF. (2010) Oxidation of the endogenous cannabinoid arachidonoyl ethanolamide by the cytochrome P450 monooxygenases: physiological and pharmacological implications. Pharmacol Rev 62:136–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector AA. (2009) Arachidonic acid cytochrome P450 epoxygenase pathway. J Lipid Res 50:S52–S56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridar C, Snider NT, Hollenberg PF. (2011) Anandamide oxidation by wild-type and polymorphically expressed CYP2B6 and CYP2D6. Drug Metab Dispos 39:782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Dostalek M, Guengerich FP. (2008) Expression and purification of orphan cytochrome P450 4X1 and oxidation of anandamide. FEBS J 275:3706–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal C, Konkel A, Schunck WH. (2011) CYP-eicosanoids—a new link between omega-3 fatty acids and cardiac disease? Prostaglandins Other Lipid Mediat 96:99–108. [DOI] [PubMed] [Google Scholar]
- Wu S, Moomaw CR, Tomer KB, Falck JR, Zeldin DC. (1996) Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J Biol Chem 271:3460–3468. [DOI] [PubMed] [Google Scholar]
- Yang J, Dong H, Hammock BD. (2011) Profiling the regulatory lipids: another systemic way to unveil the biological mystery. Curr Opin Lipidol 22:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelasko S, Palaria A, Das A. (2013) Optimizations to achieve high-level expression of cytochrome P450 proteins using Escherichia coli expression systems. Protein Expr Purif 92:77–87. [DOI] [PubMed] [Google Scholar]
- Zeldin DC, Foley J, Goldsworthy SM, Cook ME, Boyle JE, Ma J, Moomaw CR, Tomer KB, Steenbergen C, Wu S. (1997) CYP2J subfamily cytochrome P450s in the gastrointestinal tract: expression, localization, and potential functional significance. Mol Pharmacol 51:931–943. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Schieber EB, McGiff JC, Balazy M. (1995) Identification of arachidonate P-450 metabolites in human platelet phospholipids. Hypertension 25:854–859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.