Abstract
Multidrug resistances and the failure of chemotherapies are often caused by the expression or overexpression of ATP-binding cassette transporter proteins such as the multidrug resistance protein, P-glycoprotein (P-gp). P-gp is expressed in the plasma membrane of many cell types and protects cells from accumulation of toxins. P-gp uses ATP hydrolysis to catalyze the transport of a broad range of mostly hydrophobic compounds across the plasma membrane and out of the cell. During cancer chemotherapy, the administration of therapeutics often selects for cells which overexpress P-gp, thereby creating populations of cancer cells resistant to a variety of chemically unrelated chemotherapeutics. The present study describes extremely high-throughput, massively parallel in silico ligand docking studies aimed at identifying reversible inhibitors of ATP hydrolysis that target the nucleotide-binding domains of P-gp. We used a structural model of human P-gp that we obtained from molecular dynamics experiments as the protein target for ligand docking. We employed a novel approach of subtractive docking experiments that identified ligands that bound predominantly to the nucleotide-binding domains but not the drug-binding domains of P-gp. Four compounds were found that inhibit ATP hydrolysis by P-gp. Using electron spin resonance spectroscopy, we showed that at least three of these compounds affected nucleotide binding to the transporter. These studies represent a successful proof of principle demonstrating the potential of targeted approaches for identifying specific inhibitors of P-gp.
Introduction
One long-standing problem of significant medical consequence in the chemotherapeutic treatment of cancer is multidrug resistance (MDR), which appears as either an acquired or inherent resistance to chemically diverse pharmaceuticals (Kartner and Ling, 1989; Ford and Hait, 1990; Harris and Hochhauser, 1992). Although MDR can be caused by a number of different mechanisms (Binkhathlan and Lavasanifar, 2013), it is often linked to the overexpression of P-glycoprotein (P-gp) (Gros et al., 1986; Roninson et al., 1986; Gottesman and Pastan, 1993).
P-gp in humans is expressed from the multidrug resistance 1 gene (MDR1). As a member of the ATP-binding cassette (ABC) transporter family (subclass ABCB1), it exists as a plasma membrane protein of 1280 residues (Gottesman and Pastan, 1993; Persidis, 1999; Binkhathlan and Lavasanifar, 2013). Three-dimensional structures of homologs of human P-gp from several bacterial species are known (Dawson and Locher, 2006, 2007; Ward et al., 2007), as well as eukaryotic transporters from Caenorhabditis elegans (Jin et al., 2012) and Mus musculus (Aller et al., 2009; Li et al., 2014). Most eukaryotic P-gps are monomeric and composed of two relatively symmetrical halves connected by a linker polypeptide. Each half is homologous to the homodimeric bacterial ABCB1 transporters (Chen et al., 1986) and possesses an N-terminal domain containing six transmembrane (TM) helices followed by a C-terminal nucleotide-binding domain (NBD). Transport substrates appear to bind to multiple binding sites within the TM domains (Dey et al., 1997; Shapiro and Ling, 1997; Loo et al., 2003, 2009; Lugo and Sharom, 2005). The nucleotide-binding and the TM domains of P-gp and other ABC transporters appear to undergo very large conformational changes during the catalytic cycle (Hollenstein et al., 2007; Lee et al., 2008; Aller et al., 2009; Verhalen and Wilkens, 2011; Verhalen et al., 2012; Zoghbi and Altenberg, 2013, 2014; Li et al., 2014). Some of the structural models show tightly integrated, closed NBD dimers with nucleotides bound at each site while the TM domains are opened to the exterior of the cell (Dawson and Locher, 2006, 2007; Ward et al., 2007). In nucleotide-free structures, the TM domains are opened to the interior of the cell with the NBDs disengaged and sometimes widely separated (Aller et al., 2009; Ward et al., 2007; Jin et al., 2012). Molecular dynamics simulations of the MalK maltose transporter indicate that opening of the NBD dimer is a direct result of ATP hydrolysis at either nucleotide-binding site (Wen and Tajkhorshid, 2008). Targeted molecular dynamics studies of human P-gp based on crystal structures of homologs in various conformations visualized the large, concerted conformational changes required for a catalytic transport cycle (Wise, 2012).
P-gp has been actively investigated as a pharmacologic target in MDR cancers for several decades. P-gp inhibitors have been classified into three generations of compounds (Binkhathlan and Lavasanifar, 2013). Generation 1 included compounds already approved as therapeutics for other indications, including verapamil, quinine, quinidine, and cyclosporine A. These agents failed at clinically reversing MDR because the high concentrations required to inhibit P-gp resulted in unacceptable side effects (Palmeira et al., 2012). The second-generation compounds were more effective, but many of these compounds, like the first-generation compounds, were transport substrates for P-gp, requiring relatively high concentrations. Some also affected cytochrome P450 CYP3A isozymes, altering the pharmacokinetics of other drugs (Binkhathlan and Lavasanifar, 2013). The best of the third-generation P-gp inhibitors appears to be tariquidar (Stewart et al., 2000; Walker et al., 2004). However, ongoing clinical trials of tariquidar and other inhibitors have reported only limited successes in reversing MDR (Binkhathlan and Lavasanifar, 2013).
Here, we report efforts aimed at identifying novel inhibitors of P-gp that might be useful as lead compounds for reversing MDR in cancers. We employed very high-throughput, massively parallel computational screens of a very large database of drug-like structures (Irwin and Shoichet, 2005; Irwin et al., 2012) to one hypothetically critical conformation of P-gp, a partially opened outward structure described previously (Wise, 2012). Our present study identifies potential inhibitors that specifically target the NBDs of P-gp with limited interactions at the drug-binding sites. We hypothesized that compounds that strongly bind to the power-transducing structures of P-gp but not well to the drug-pumping structures would potentially inhibit P-gp catalyzed transport without being transported out of the cell. Drug-like compounds with these attributes may make good cotherapeutic leads to be developed against MDR in cancer chemotherapies.
Materials and Methods
In Silico Screening.
A database of commercially available compounds that includes molecules with “clean” drug-like characteristics was obtained from the ZINC Web site (http://zinc.docking.org/) (Irwin and Shoichet, 2005; Irwin et al., 2012), and about half these compounds (5,277,160) were used for in silico docking experiments with a human P-gp structural conformation as described earlier (Wise, 2012). The modeled protein conformation represents a conformation of P-gp where the NBDs are fully engaged similar to those of the Sav1866 crystal structure (Dawson and Locher, 2006). Nucleotides were removed from the protein before docking studies were performed. Ligand docking used the Autodock 4.2 program (http://autodock.scripps.edu/) (Goodsell et al., 1996; Morris et al., 1996; Osterberg et al., 2002) and the high-performance computational facilities of the Center for Scientific Computing at Southern Methodist University. Ligands were converted to pdbqt file format using the AutoDockTools Python scripts (Morris et al., 2009). Autodock parameters included 100 Lamarckian genetic algorithm runs per ligand with mutation rates of 0.02, crossover set at 0.8, and a population size of 300, with 3,000,000 evaluations and 27,000 generations per tested ligand. Roughly 5 million of these docking studies against the P-gp NBDs were analyzed in the first round of the in silico screening experiments.
As a second part to the experiment, docking against the drug-binding domains (DBD) of the same P-gp structure was performed. This experiment used two overlapping grid maps that encompassed the entire P-gp molecule minus the NBDs. Two grids were required because of the size of the desired zone. A subset of ligands that was found to dock to P-gp with estimated binding affinities of less than 200 nM at the NBD was used in the secondary docking experiments to the DBDs. Only ligands predicted to bind the NBD at least 100-fold more tightly than the DBD were considered for further analysis.
ATP Hydrolysis Assays.
A well-established activity assay for ATP hydrolyzing enzymes (Delannoy et al., 2005; Hoffman et al., 2010) was used that couples the hydrolysis of ATP by P-gp to the oxidation of NADH to NAD+ through the reactions of pyruvate kinase and lactate dehydrogenase (Vogel and Steinhart, 1976). These reactions were adapted to medium-throughput conditions on 96-well plates. To test the effects of the in silico identified potential inhibitors, a total volume of 5 μl of various concentrations of experimental compounds dissolved in dimethylsulfoxide (DMSO) was added to the wells in a 96-well plate containing 45μl of equilibration buffer (20% glycerol, 50 mM Tris-Cl, pH 7.5). To each of these wells, 50 μl of a solution containing 80–100 μg/ml lipid-activated, concentrated P-gp (see below) in equilibration buffer in the absence or presence of 300 μM verapamil (dissolved in water) was added.
After incubating the plate for 30 minutes at 37°C, we added to each well 100 µl coupled enzyme assay cocktail (50 mM Tris-Cl, pH 7.5, 24 mM MgSO4, 20 mM KCl, 2 mM PEP, 0.014 mg/ml pyruvate kinase, 0.0288 mg/ml lactate dehydrogenase, 1.13 mM NADH, and 4 mM ATP), prewarmed to 37°C. Immediately after this addition, ATPase activity was assayed for 20–30 minutes. ATP hydrolysis was recorded as the absorbance decrease at 340 nm using a BioTek Eon plate reader (BioTek, Winooski, VT). An extinction coefficient for NADH of 6300 M−1cm−1 was used.
All assays were performed in duplicate or triplicate and were repeated at least two times using different P-gp preparations. Control experiments were performed that showed that none of the identified inhibitors affected the other enzymes in the coupling assays. Dose response curves and inhibitor concentration values (IC20, IC50, and IC80 values) were calculated from the data as described by Copeland (Copeland, 2000, 2013) using a four-parameter logistic fit.
Lipid Activation.
To stabilize the protein for the ATP hydrolysis assays, lipid activation of the purified P-gp was performed. Lipid stocks were prepared from l-α-phosphatidyl-choline from soybean (∼40%) (Sigma-Aldrich, St. Louis, MO) by adding lipid stock buffer (50 mM Tris-Cl, pH 7.5, 0.1 mM EGTA) to solid lipid in glass test tubes. Lipid stock concentrations were 25 mg/ml or 100 mg lipid/ml buffer. The suspension was flushed with argon and subsequently sonicated in a bath sonicator at room temperature for 15–30 minutes until the mixture clarified. The lipid suspension was supplemented with 2 mM dithiothreitol and stored at 4°C under argon until use. To activate the P-gp, we prewarmed lipid stock to 37°C, mixed with concentrated protein (see below) at a ratio of 2:1 (w/w) and then diluted with equilibration buffer at 37°C to a final concentration of typically 100 µg P-gp per ml activated P-gp mix. This mixture was incubated at room temperature for 30 minutes to 1 hour before use in the coupled enzyme assays.
Preparation of Isolated P-gp.
The P-gp used in these assays was a close relative of the human P-gp that is found in Mus musculus (MDR3, 87% identical to human P-gp). A mutant form of the mouse P-gp where all intrinsic cysteine amino acids had been replaced by alanine residues was recombinantly expressed in the yeast Pichia pastoris (Lerner-Marmarosh et al., 1999; Tombline et al., 2006). This cysteine-less mouse P-gp was used for assaying the potential inhibitor compounds. Purification of the protein was performed essentially as described by Delannoy et al. (2005) with small modifications resulting in highly enriched P-gp in mixed micelles containing dodecyl maltoside and lysophosphatidyl choline. To obtain concentrated P-gp, the elution fraction of the Ni-NTA column was concentrated 70- to 100-fold using Amicon Ultra centrifugal filters (Amicon/Millipore Corp., Billerica, MA) with a molecular mass cutoff of 100,000 Da. This procedure enriches both protein and micelles and ensures retained solubilization of the membrane protein.
Electron Spin Resonance Measurements.
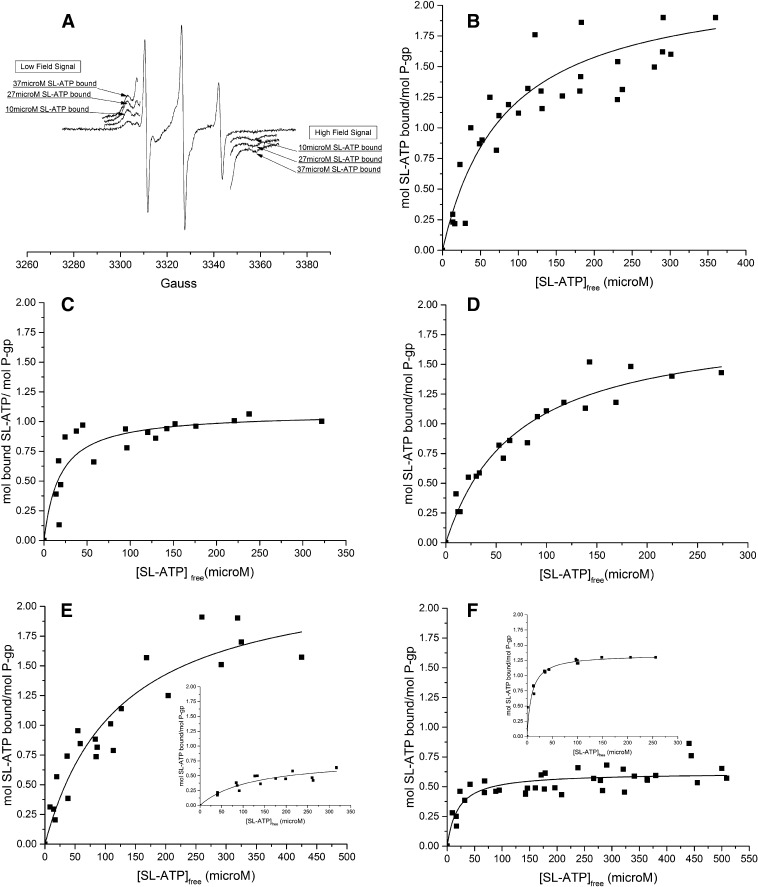
All electron spin resonance (ESR) spectroscopy measurements were performed using a Bruker EMX 6/1 ESR spectrophotometer (Bruker, Karlsruhe, Germany) operating in X-band mode and equipped with a high-sensitivity cavity, as described previously (Delannoy et al., 2005). Briefly, a total of 30–40 μl of sample was pipetted into 50-μl glass capillaries. The top of the capillary was closed and inserted into a quartz cuvette. ESR spectra were acquired at a microwave frequency of 9.33 GHz, microwave power of 12.63 mW, 100 kHz modulation frequency, and a resolution of 1024 points. The center field of the scan was at 3325 G, and an area of 100 G was scanned with a peak-to-peak modulation amplitude of 1 G. The time constant was set to 10.240 milliseconds, and the conversion time to 163.84 milliseconds, resulting in a total time sweep of 167.772 seconds. The signal gain was adjusted for the spin-labeled (SL)–ATP concentrations in the different experiments.
All samples in the ESR experiments were dissolved in buffer containing 50 mM Tris-HCl, pH 7.4, 50 mM NaCl, 20% glycerol, 0.1% dodecyl maltoside, 0.01% lysophosphatidyl choline, and 300 mM imidazole with or without the experimental inhibitor compounds present. All ESR measurements were at room temperature. Titration experiments were performed using increasing concentrations of 2′3′-SL-ATP, essentially as described in Delannoy et al. (2005). SL-ATP is a substrate of P-gp and is hydrolyzed to the corresponding SL-ADP by the enzyme (Delannoy et al., 2005). Due to this fact, the phosphorylation state of the spin-labeled nucleotide is not known throughout the course of an experiment. The spin-labeled nucleotide is therefore referred to as SL-ANP. The amount of protein-bound SL-ANP was determined as the difference between the known total concentration of SL-ATP added and the free spin-labeled nucleotide observed in the experiment. The latter was measured by comparing the amplitude of the high field ESR signal of SL-ANP at each point to a standard curve of SL-ATP generated in the absence of protein.
The amplitudes of the high-field signals were determined using WINEPR (Bruker) software for each concentration of SL-ATP used. Moles of SL-ANP bound per mole of P-gp were plotted over the free SL-ANP concentration, and nonlinear, hyperbolic curve fitting of the results was performed using Origin 8.5.1 (OriginLab, Northampton, MA) to determine the maximum binding and apparent affinity for the spin-labeled nucleotide. The equation used for fitting the curves was y = P1 * x/(P2 + x), where P1 corresponds to the maximum binding of SL-ANP (moles of SL-ANP bound per mole P-gp), and P2 equals the apparent dissociation constant for SL-ANP. The P-gp concentrations in titration experiments ranged from 20 to 100 μM in the presence of 2 mM MgSO4, 150 µM verapamil, and 0, 25, 50, or 100 µM of experimental compound dissolved in DMSO. For the experiments shown in Fig. 4, the starting concentration of P-gp was 50 μM and the concentration of inhibitor was as given in the figure legend. The final volume of the assay was 40 μl. Between three and five individual titration experiments were performed for each compound and concentration using at least two different enzyme preparations. The concentration of SL-ATP ranged from 25 to 500 µM.
Fig. 4.
Spin-labeled nucleotide binding (SL-ANP) in the presence of inhibitors to P-gp. Between 30 and 50 µM Pgp was incubated at no inhibitor or at 25, 50, or 100 µM inhibitor compound with increasing concentrations of SL-ATP. The ESR signal of the free, not protein bound SL-ATP in the presence of P-gp with or without inhibitor was compared with a standard curve obtained in the absence of P-gp. (A) Freely tumbling, not-enzyme–bound SL-ANP results in ESR spectra consisting of three relatively sharp, equidistant signals. The signal amplitude of the high-field signal of the free SL-ANP was used to determine the amount of free spin-labeled nucleotide. For comparison, the ESR signals of increasing amounts of protein-bound SL-ANP are pointed out at the left (low field) and the right (high field) of the signal of the free SL-nucleotide. The parts of the ESR spectra shown have been rerecorded at increased signal gain and a modulation amplitude of 2 for better visualization. (B–F) The moles of SL-ANP bound per moles of P-gp were determined as described in Materials and Methods and were plotted against free spin–labeled nucleotide. (B) Binding of SL-ANP to P-gp in the absence of P-gp inhibitor but in the presence of 150 µM verapamil. Nonlinear regression as described in Materials and Methods extrapolated to maximal binding of 2.2 ± 0.2 mol/mol and an apparent Kd of 88 ± 22 µM. (C–F), SL-ANP binding to P-gp as in (B), except for the inclusion of 50 µM of inhibitor compounds 19, 29, 34 and 45, respectively. Maximum binding and apparent Kd were determined to be (C) 1.1 ± 0.1 mol/mol and 18± 6 µM in the presence of 50 µM compound 19; (D) 1.9 ± 0.1 mol/mol and 71 ± 12 µM in the presence of 50 µM compound 29; (E) 2.1 ± 0.2 mol/mol and 101 ± 31 µM in the presence of 50 µM compound 34; and (F) 0.6 ± 0.02 mol/mol and 20 ± 5 µM in the presence of 50 µM compound 45. (E, insert) SL-ANP binding to P-gp in the presence of 100 µM of inhibitor compound 34 results in maximum binding of 0.80 ± 0.09 mol/mol and an apparent dissociation constant of 121± 33 µM. (F, insert) In the presence of 25 µM of compound 45, maximum binding of SL-ANP to P-gp extrapolates to 1.3 ± 0.04 with an apparent Kd of 9.1 ± 1.5 µM.
Chemicals.
Potential inhibitors of P-gp identified through in silico screening were purchased in small quantities through SIA MolPort (Riga, Latvia). Dodecyl maltoside was purchased from Inalco Pharmaceuticals (San Luis Obispo, CA). Lysophosphatidyl choline was a generous gift from Lipoid GmbH (Ludwigshafen, Germany).
Results
Targeted In Silico Screening for Inhibitors of Human P-gp Using a Large Database of Drug-like Small Molecules.
The goal of this study was to identify inhibitors of P-gp that interrupt ATP-binding to or hydrolysis by P-gp by interacting at the NBDs of P-gp. Using targeted molecular dynamics techniques, we previously visualized structural changes in human P-gp as the transporter transitions from one conformational state to another during a catalytic transport cycle (Wise, 2012). In the present study, we chose one conformation of P-gp for docking analysis that corresponded to a structural conformation of P-gp where the NBDs are fully engaged and the DBDs are partially opened outward. This P-gp conformation, which is similar to that observed in the crystal structural model of the bacterial Sav1866 ABC transporter (Dawson and Locher, 2006), was used as the protein target because it possesses fully formed adenine nucleotide-binding sites. We hypothesized that compounds that interfere with nucleotide binding to P-gp or that disrupt the nucleotide binding sites might make good inhibitors of ATP binding or hydrolysis and thus be inhibitors of drug transport.
Irwin and Shoichet have made publicly available a computational resource containing the three-dimensional coordinates of over 20 million commercially obtainable compounds, including subsets that possess the chemical characteristics often found in approved medicinal drugs (see the ZINC database at http://zinc.docking.org/) (Irwin and Shoichet, 2005; Irwin et al., 2012). In the first round of docking, about 5.3 million out of about 11 million of these drug-like molecules (subset 13 of the ZINC database) were individually tested for binding interactions at the NBDs of human P-gp using the AutoDock4.2 program (Morris et al., 2009), as described in Materials and Methods. The results of these calculations were subsequently analyzed for the estimated binding affinity of the ligands to P-gp. Compounds were identified that were predicted to have potentially strong binding interactions at the P-gp NBD structures. We disregarded any compounds for further study that had predicted estimated Kd values that were more than 200 nM.
This first set of 180,142 putative P-gp ligands that possessed predicted affinities to the NBDs of P-gp of at least 200 nM (subset 1) was further analyzed for possible interaction at the DBDs of P-gp in a second round of docking experiments that targeted the transport drug binding sites of P-gp. Ultimately, compounds that interacted strongly at NBDs but weakly at the DBDs of P-gp were sought. The docking results for subset 1 compounds at either of two target grids at the DBD of P-gp (see Materials and Methods) were therefore analyzed for predicted binding affinities at the drug-binding sites. The calculated affinities at the NBDs and the DBDs of P-gp were then compared for each compound in subset 1.
Those molecules with estimated Kd values at the DBD of P-gp that were ≥100 times that of the projected Kd at the NBDs (subset 2) were considered to be potential inhibitor candidates for P-gp that specifically target the NBDs of P-gp. Subset 2 consisted of approximately 250 molecules. Of these initial computational leads, 35 compounds were purchased from commercial sources (numbered 11–45) and used in in vitro biochemical assays to determine their effectiveness in inhibiting ATP-hydrolysis by isolated P-gp. These 35 compounds were representative of the various binding clusters observed in silico both within and outside the nucleotide-binding sites on P-gp.
Screening for ATP Hydrolysis Inhibition by In Silico Identified Putative P-gp Inhibitors.
The ability of the molecules identified using in silico methods to function as potential inhibitors of P-gp was tested by assessing their capability to inhibit ATP hydrolysis using purified P-gp. ATP hydrolysis by P-gp is stimulated by transport substrates such as the calcium channel blocker verapamil. Verapamil has been found to inhibit P-gp catalyzed transport by competing for transport cycles (reviewed in Palmeira et al., 2012). Depending on whether a transport substrate such as verapamil is present in the assay, purified P-gp preparations exhibit ATP hydrolysis rates that vary over a range from about 50 nmol min−1 mg−1 (basal activity) to about 750–1000 nmol min−1 mg−1 (stimulated activity).
In our initial screening of the in silico–generated hits of subset 2, we tested the effect on basal ATP hydrolysis activity (inhibition and or stimulation) of the commercially available compounds chosen from subset 2 in assays not containing verapamil. Compounds that stimulated the basal activity of P-gp in the absence of verapamil were assumed to achieve the stimulation by binding to the drug-binding sites of P-gp similar to transport substrates such as verapamil and would therefore not support our premise of finding compounds that specifically interact with the NBDs and not the DBDs. Several of the compounds tested did indeed stimulate basal rates of hydrolysis by P-gp a factor of 1.5 or more (data not shown) and were not further pursued in this work. Only compounds that in addition to stimulating basal activity also showed strong inhibition of stimulated activity were further pursued in this study.
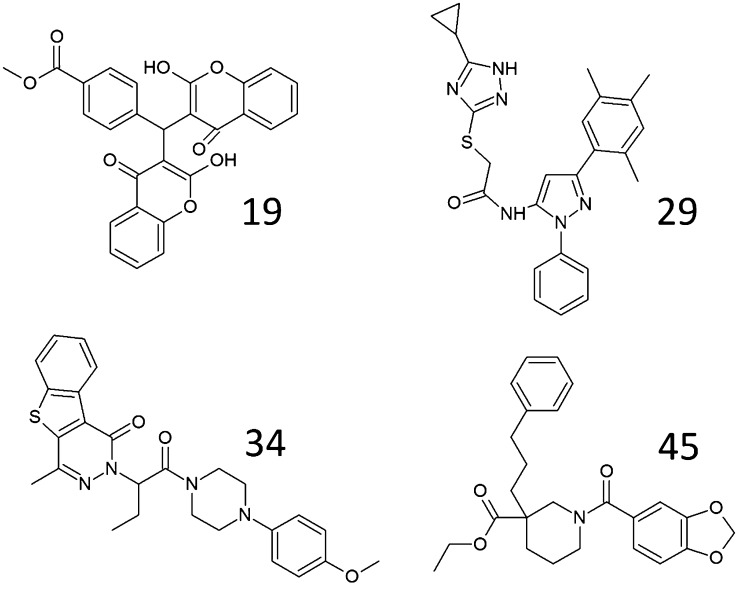
To test the ability of the selected subset 2 compounds to inhibit verapamil-stimulated ATP hydrolysis activities of P-gp, verapamil was added at 150 µM final concentration to assays containing purified P-gp. ATP hydrolysis was determined at near physiologic ATP concentrations (2 mM ATP). Figure 1A shows the relative ATP-hydrolysis activities of P-gp in the presence of 25 µM of a subset of compounds tested. Compounds that in separate experiments induced strong stimulation of basal activity are not shown. Four of the 35 compounds tested were observed to inhibit verapamil-stimulated ATPase activities by P-gp: compound 19 [methyl 4-[bis(2-hydroxy-4-oxochromen-3-yl)methyl]benzoate (ZINC 09973259, CID 4694077)], compound 29 [2-[(5-cyclopropyl-1H-1,2,4-triazol-3-yl)sulfanyl]-N-[2-phenyl-5-(2,4,5-trimethylphenyl)pyrazol-3-yl]acetamide (ZINC 08767731, CID 17555821)], compound 34 [2-[1-[4-(4-methoxyphenyl)piperazin-1-yl]-1-oxobutan-2-yl]-4-methyl-[1]benzothiolo[2,3-d]pyridazin-1-one (ZINC 09252021, CID 22514118)], and compound 45 [ethyl 1-(1,3-benzodioxole-5-carbonyl)-3-(3-phenylpropyl)piperidine-3-carboxylate (ZINC 15078148, ZINC 15078146, CID 26410703, CID 45252040)] (Fig. 1A). Compounds that did not show significant inhibition of ATP hydrolysis in these assays were not further considered. A side-by-side comparison of verapamil-stimulated and basal ATP hydrolysis assays in the presence of the four confirmed inhibitors of P-gp is presented in Fig. 1B. The figure shows that under the conditions used (i.e., in the presence of 2 mM ATP) all compounds inhibited both basal (Fig. 1B, gray bars) and stimulated (Fig. 1B, black bars) ATPase activities. In all cases, the stimulated ATP hydrolysis rate was more strongly affected by the compounds.
Fig. 1.
The effects of putative inhibitors of P-gp on ATP hydrolysis activities. (A) ATP hydrolysis activities of highly enriched P-gp preparations were assayed at final concentrations of 2 mM Mg ATP, 150 µM verapamil, and 25 µM of the indicated compound. The figure presents only the results of those compounds that did not significantly stimulate basal ATPase activity by P-gp. Results were normalized to uninhibited control samples that exhibited specific verapamil-stimulated activities ranging between 750 and 1000 nmol min−1 mg−1. (B) A side-by-side comparison of basal (gray bars) and verapamil-stimulated (black bars) ATPase activities of P-gp in the presence of 25 µM of the indicated inhibitor. The data were normalized to uninhibited control samples that exhibited specific basal ATP hydrolysis activities between 50 and 75 nmol min−1 mg−1 and specific verapamil-stimulated activities ranging between 750 and 1000 nmol min−1 mg−1. All data represent averages of at least three experiments in duplicate using at least two different enzyme preparations. The error bars given represent standard error of the mean.
The structures of the four targeted inhibitors of P-gp ATP hydrolysis activity are shown in Fig. 2. It should be noted that both compounds 34 and 45 were used as racemic mixtures of the respective enantiomeric compounds.
Fig. 2.
Chemical structures of four inhibitors of P-gp catalyzed ATP hydrolysis identified through in silico screening methods. Top row: Chemical structures of compounds 19 and 29. Bottom row: Chemical structures of compounds 34 and 45.
Dose Response Testing of Identified P-gp Inhibitors.
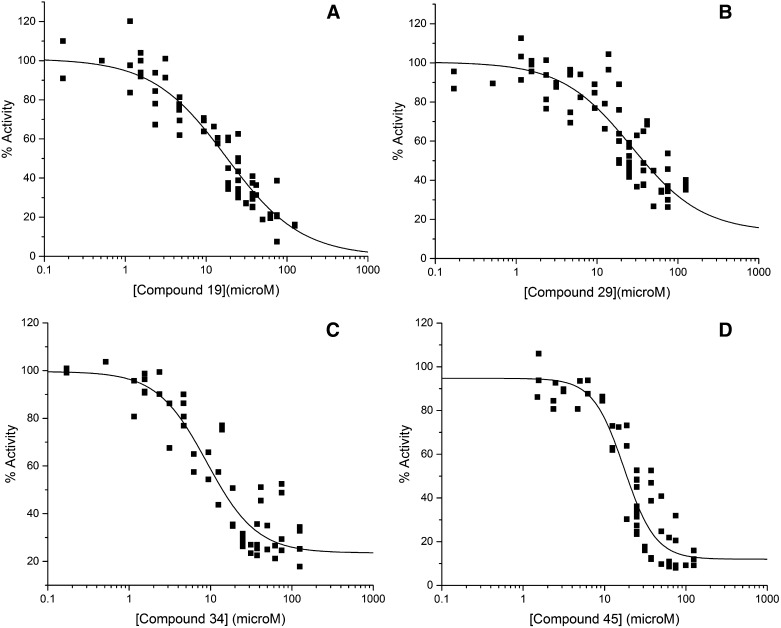
The four identified P-gp inhibitors were investigated further in ATP hydrolysis dose-response inhibition curves. Figure 3 shows the verapamil-stimulated ATP hydrolysis activities of P-gp preparations in the presence of varying concentrations of inhibitor compounds after normalizing to verapamil-stimulated P-gp activity measured in the absence of inhibitor. The IC20, IC50, and IC80 inhibition values were calculated as described in Materials and Methods for the inhibitors in these assays, and the results of these calculations are presented in Table 1. IC50 values in the range of 10−5 M were observed for all four P-gp inhibitors in these experiments.
Fig. 3.
Dose-response curves for the identified inhibitors of P-gp. Concentration dependence of inhibition of the four identified inhibitor compounds (abbreviated 19, 29, 34, and 45). (A) Compound 19, (B) compound 29, (C) compound 34, and (D) compound 45. Each plot represents the composite results of three to five individual experiments performed on at least three independently isolated P-gp preparations. ATPase activity was determined at 2 mM MgATP as described in Materials and Methods. The curves shown were calculated as described in Materials and Methods using a four-parameter logistic fit as described in Copeland (2000, 2013).
TABLE 1.
Inhibition constant values calculated from dose-response curves for the inhibition of verapamil-stimulated ATPase activities of P-gp
ATP assays were performed as described in Materials and Methods in the presence of 150 µM verapamil with the indicated concentrations of inhibitors. The DMSO vehicle did not exceed 5% (v/v). MgATP concentration was at 2 mM. Values are given with S.E.M.
| Inhibitor | IC20 | IC50 | IC80 |
|---|---|---|---|
| µM | |||
| Compound 19 | 4 ± 0.7 | 18 ± 1.5 | 77 ± 8.1 |
| Compound 29 | 7 ± 1.6 | 27 ± 6.8 | 108 ± 47.0 |
| Compound 34 | 3 ± 0.7 | 9 ± 1.4 | 24 ± 6.0 |
| Compound 45 | 10 ± 1.5 | 18 ± 1.5 | 32 ± 4.6 |
Probing Inhibition Mechanisms of Identified P-gp Inhibitors Using ESR Spectroscopy and Spin-Labeled ATP.
The possible mechanism of inhibition of ATP hydrolysis by the identified P-gp inhibitors was investigated as in Delannoy et al. (2005) by measuring the maximal binding stoichiometry and binding affinity of an analog of ATP, SL-ATP, to P-gp both in the presence and absence of the identified inhibitors. These methods allowed us to directly follow binding of SL-nucleotide to P-gp under equilibrium binding conditions. Because SL-ATP is a substrate for P-gp catalyzed ATP hydrolysis (Delannoy et al., 2005), both SL-ATP and SL-ADP can potentially bind to P-gp. We therefore refer to the spin-labeled nucleotide in these experiments as SL-ANP.
ESR spectra were acquired for known concentrations of SL-ATP and compared with spectra acquired in the presence of known concentrations of P-gp, verapamil, and the different inhibitor molecules. Free, not-enzyme–bound, SL-ANP results in ESR spectra with three relatively sharp, equidistant signals (see Fig. 4A). The high-field signal of the free SL-ANP can be seen as the sharp signal on the right. The relatively broad signals of the enzyme-bound spin-labeled nucleotide can be seen to the left (low field) and right (high field) of the three signals resulting from the free SL-ANP. The signal gain was increased for better visualization of SL-ANP bound to P-gp. The high-field signal of the freely mobile, unbound SL-ANP was used to calculate the amount of free SL-ANP–analog in the binding assays with P-gp (Materials and Methods) (Fig. 4A). The amount of P-gp–bound SL-nucleotide was then calculated as the difference between the known amount of total SL-ATP added and the measured free SL-ANP, using a standard curve of increasing amounts of SL-ATP as a reference (see Materials and Methods) (Delannoy et al., 2005). The results of SL-ANP binding experiments to P-gp in the absence or presence of P-gp inhibitors are presented in Fig. 4, B–F.
In the absence of drug, but in the presence of 5% (v/v) drug vehicle DMSO, maximum binding to P-gp was found to extrapolate to 2.2 ± 0.2 mol SL-ANP bound per mol of P-gp with an apparent Kd of 88 ± 21.6 µM (Fig. 4B). SL-ATP titrations in the presence of experimental drug-like compounds were all compared with standard curves generated in the presence of 5% DMSO and the experimental compound to control for potential compound–SL-ATP interactions. In an initial set of titrations, stoichiometric amounts of about 50 µM P-gp and 50 µM experimental compounds were mixed, and increasing amounts of SL-ATP were added (Fig. 4, C–F). Compounds 19 and 45 significantly decreased the stoichiometry of SL-nucleotide binding under these conditions to approximately 1 or 0.6 mol/mol, respectively (see Fig. 4, C and F). These compounds also caused a decrease of the apparent Kd of P-gp for SL-ANP binding to about 20 µM. Compounds 29 and 34 did not show an obvious change in maximal nucleotide binding or the apparent dissociation constants for SL-ANP when incubated with P-gp at 50 µM (Fig. 4, D and E).
When titrations were performed in the presence of 100 µM of compound 34, however, maximum binding of SL-ANP to P-gp was decreased to 0.8 mol/mol, while the dissociation constant for SL-ANP binding did not change significantly (Fig. 4E, insert). Titrations in the presence of 100 µM of compound 45 were not significantly different from those in the presence of 50 µM of compound 45 (not shown). However, when the concentration of compound 45 was decreased to 25 µM while P-gp remained at 50 µM, maximal binding of about 1.3 moles SL-ANP/mol of P-gp was observed (Fig. 4F, insert), again with decreased apparent Kd. Compound 29 did not significantly affect SL-nucleotide binding at either 50 µM or 100 µM concentration.
Comparing Success Rates to Other Methods.
Schmidt et al. (2007, 2008) reported in two studies that a series of dioxolane-derivatized piperazines and several phenothiazines and related compounds resensitized drug-resistant colon cancer or porcine kidney epithelial cells to vincristine. The cell lines used in these studies were shown to stably express P-gp at moderately high-expression levels. We took 10 of the best compounds identified in (Schmidt et al., 2007, 2008) and assayed them for their effect on ATP hydrolysis by P-gp as if they had passed our docking criteria, although none of them actually did pass these computational criteria (not shown). One of the 10 compounds was observed to moderately inhibit ATPase activity of P-gp, but ESR titrations as described earlier did not indicate any interaction of the compound with the NBD (not shown), indicating that inhibition of P-gp ATP hydrolysis was not caused by binding to or close to the nucleotide binding sites. The hit rate for this quantitative structure activity relationships cell culture resensitization selection method (1 in 10) compares favorably with our computational hit rate (4 in 35) of ATP hydrolysis inhibitors, where three of the four identified inhibitors adversely affected SL-nucleotide binding to P-gp.
This comparison shows that the computational selection method used in this work was at least as good as the quantitative structure activity relationships cell culturing system in identifying inhibitors of ATPase by P-gp and was arguably better at targeting these inhibitors to the NBDs of P-gp.
Discussion
Screening for Inhibitors Targeted to the Nucleotide-Binding Domains of P-gp.
Our goal was to identify inhibitors of P-gp that interrupt ATP-binding or hydrolysis by interacting at the NBDs of P-gp. We employed a structural homology model of a conformation of human P-gp that possesses fully formed nucleotide-binding sites that are similar to those observed in the crystal structure of the bacterial Sav1866 ABC transporter (Dawson and Locher, 2006). In this conformation, the Sav1866 transporter bound both ADP and AMPPNP [5′-adenylyl-β,γ-imidodiphosphate] (Dawson and Locher, 2006, 2007). To identify drug-like molecules that interact strongly at the NBDs of P-gp, we targeted the entire composite of the N-terminal and C-terminal NBDs of P-gp in massively parallel in silico docking experiments. A large set of drug-like molecules (∼11 million compounds) was obtained from the ZINC chemical database, and about half of these were employed in docking calculations. A subset of ∼180,000 molecules was identified that was predicted to interact with the NDBs of P-gp with estimated affinities less than 200 nM. It should be noted that the ATP-binding sites in the protein structure were left unoccupied and freely available for docking in these experiments. In a secondary screen, this subset 1 of identified molecules was docked to the DBDs of P-gp using the same programs. A much smaller subset of about 250 compounds was identified that was predicted to bind weakly at the DBDs but have high-affinity interactions at the NBDs. These criteria were deemed prognostic of putative P-gp inhibitor molecules that may interfere with ATP use by P-gp while not being transport substrates of the pump.
Screening In Silico Hits Using ATP Hydrolysis Assays.
From the approximately 250 compounds that were predicted to bind tightly at the NBDs and significantly less strongly at the DBDs of P-gp, 35 were analyzed in biochemical assays using isolated P-gp for effects on basal or transport substrate stimulated rates of ATP hydrolysis. We expected that compounds that interacted with the DBDs of P-gp despite AutoDock predictions to the contrary might stimulate the basal ATP hydrolysis rates similar to other transport substrates. Compounds that did stimulate basal rates over 1.5-fold were therefore not considered further.
Four compounds, 19, 29, 34, and 45, showed significant inhibition of verapamil-stimulated ATP hydrolysis activities when assayed at 25 µM both at high ATP (10 mM, not shown) and near physiologic ATP concentration (2 mM ATP, Fig. 1, A and B). The structural formulas for these four compounds are shown in Fig. 2. Compounds 29 and 34 cause slight activations of basal ATPase activity rates of P-gp when assayed at 10 mM ATP (not shown), indicating that these compounds may interact with the DBDs of P-gp to some degree. Compounds 19 and 45 did not stimulate basal hydrolysis rates at either ATP concentration, suggesting that these compounds do not productively interact with the DBDs or that the net effect of interaction at these or other sites is to inhibit P-gp catalyzed ATP hydrolysis.
It is interesting to note that transport-drug–stimulated ATP hydrolysis was much more affected by the four inhibitor compounds than basal activity (see Fig. 1B). One possible explanation for this observation may be that the compounds tested were selected for binding to NBDs in their closed conformation, which corresponds to the formation of complete nucleotide-binding sites (Dawson and Locher, 2006, 2007). It seems feasible that in a transport-drug–stimulated state, more of the enzyme adopts the closed NBD conformation and is thus more susceptible to binding of and being inhibited by the inhibitor candidates than enzymes performing only basal level hydrolysis. In addition, previous results from our group showed that binding of transport substrate indeed altered the conformation of the nucleotide-binding sites (Delannoy, 2005; Hoffman, Wise and Vogel, unpublished data). This again may cause better binding of the inhibitor molecules to the NBDs.
Dose-Response Curves.
In further tests using isolated P-gp preparations, the inhibitor concentrations in verapamil-stimulated ATP hydrolysis assays were varied, and the residual activities were monitored. Figure 3 shows the results of these assays for the four inhibitor compounds. Affinities as expressed through IC50 values ranged from 9 to 27 µM for the four inhibitors (Table 1). These values are high but were deemed appropriate for initial screening hits. Lead optimization studies are in progress to lower these IC50 values to a more useful pharmacologic range.
Mechanisms of Inhibition by the Four Identified Inhibitors.
In a series of experiments designed to elucidate whether the four inhibitors indeed interacted with the NBDs of P-gp, spin-labeled analogs of ATP were used in equilibrium binding studies to determine whether the compounds affected ATP binding to P-gp. These studies take advantage of the fact that protein-bound SL-ANP and free SL-ANP have different, well-resolved ESR spectra that enable the assessment of binding of the analog under equilibrium conditions. The measured maximum binding was in good agreement with values previously reported by us (Delannoy et al., 2005) and others (Qu et al., 2003). The difference between the previously reported Kd of P-gp for SL-ANP in the presence of verapamil of 180 µM (Delannoy et al., 2005) and the Kd reported in this article may be due to the presence of DMSO and/or the somewhat improved purity of the protein.
Inclusion of compounds 19 or 45 at concentrations equimolar to protein (about 50 µM) resulted in maximum binding of about 1 mol/mol SL-ANP in the presence of compound 19 and of about 0.5 mol/mol in the presence of the same concentration of compound 45. The apparent Kd was decreased to about 20 µM in the presence of either compound. Lowering of the number of accessible nucleotide-binding sites in addition to lowering of the dissociation constant for nucleotide binding strongly suggests that these two inhibitors are directly affecting the ATP binding sites of P-gp. Earlier studies have shown that it is possible to inhibit ATP hydrolysis by P-gp by abolishing function at a single NBD (e.g., Urbatsch et al., 1998, 2003) . Such “half-site reactivity” might be expected from agents that interact strongly at one of the nucleotide-binding sites of P-gp. In earlier experiments, we observed that SL-ANP binding to vanadate-inhibited P-gp was reduced to about 1 mol/mol and also exhibited a decreased Kd for SL-ANP binding (Delannoy, unpublished data). The fact that binding of SL-ANP was reduced to less than 1 mol/mol in the presence of equimolar concentration of compound 45 may be explained by a strong interaction of compound 45 with P-gp that affects binding to both nucleotide-binding sites. Because the Kd for SL-ANP and compound 45 are within the same overall range, we speculate that in equilibrium a subset of the protein molecules has no inhibitor bound and is therefore able to bind the nucleotide. The number of accessible nucleotide binding sites did not further decrease when the concentration of compound 45 was increased to 100 µM, but about doubled when the compound 45 concentration was reduced to 25 µM. Because compound 45 was obtained as a racemic mixture of the two enantiomers, these results suggest that both enantiomers actively interact with the P-gp NBDs
At equimolar inhibitor and transporter concentrations, no effect on the stoichiometry of nucleotide binding was observed with inhibitor 34. When the concentration of 34 was doubled to 100 µM, however, reduced binding of about one mol/mol SL-ANP was observed. This may indicate that two molecules of compound 34 are needed to interact with each molecule of P-gp to exert its inhibitory effect. Alternatively, because compound 34 was also obtained as a racemic mixture, this result may be explained with only one of the stereoisomers binding to the protein and affecting nucleotide accessibility of the nucleotide-binding sites.
Compound 29 did not affect the binding stoichiometry in these tests nor did it significantly affect the Kd for SL-ANP at any concentration tested, suggesting that compound 29 did not directly interact with the P-gp nucleotide binding sites but may cause inhibition of ATP-hydrolysis by blocking crucial conformational transitions of the enzyme.
Correlation of the Best Docking Pose of the Inhibitors with Effects on Nucleotide Binding.
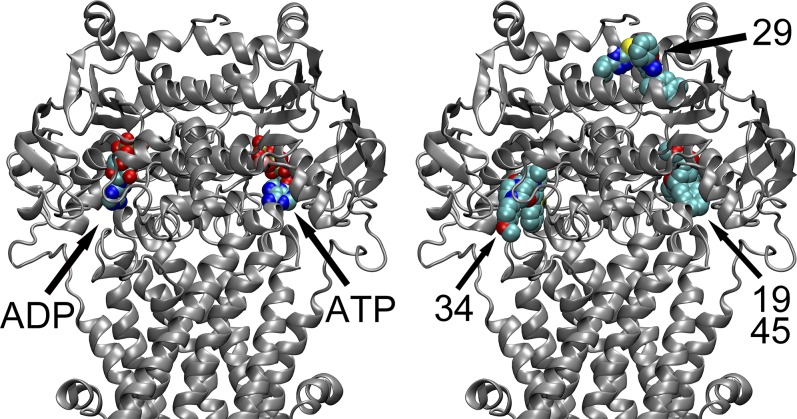
It is remarkable that the highest affinity docking pose shown for each of the four identified P-gp inhibitors correlates very well with the effect these inhibitors have on SL-ANP binding to P-gp. Compounds 19, 34, and 45 reduced SL-ANP binding by at least 1 mol/mol, and the best docks for these inhibitors are directly at the nucleotide-binding sites (Fig. 5). The fact that compound 34 needed to be in 2-fold excess over protein to affect nucleotide binding may be due to the fact that a racemic mixture of the compound was used. Conversely, compound 29 neither affected the stoichiometry nor the affinity of P-gp for SL-ANP, and its best dock is found between the two NBDs of P-gp and not directly within the nucleotide-binding sites (Fig. 5). This direct correlation of the best docking modes of the inhibitors to the possible mechanism of inhibition by the inhibitors was unanticipated.
Fig. 5.
The location of docked inhibitors in the human P-gp model. The protein is shown in cartoon representation in silver-gray. Only the cytoplasmic portion of P-gp has been shown with the two NBDs oriented horizontally. P-gp inhibitors and nucleotides (ADP or ATP) are shown in van der Waal spheres color coded to atom types (O in red, N in blue, P in orange, C in turquoise, Mg in pink, S in yellow). Nonpolar hydrogens have been left out for clarity. Ligands have been labeled in the figure with arrows. Left: The location of nucleotide bound to the human P-gp model used for docking is shown. Note that nucleotides were removed from the model before docking was performed. For orientation, the catalytic glutamate residues E556 and E1201 are located on the right and left sides of the representation, respectively. Right: The approximate locations of the highest affinity interaction predicted by docking studies for the four identified inhibitors of P-gp are shown relative to the protein structure. Both compounds 19 and 45 bound to the E556-binding site are superposed in the representation. The figure shows that three of the four identified inhibitors (19, 34, and 45) are bound in the nucleotide-binding sites. These are the same compounds that altered ATP binding in the ESR titration experiments described. Compound 29 was predicted to bind outside the nucleotide-binding sites in the docking experiments. This compound did not alter the binding characteristics of SL-ATP at the concentrations used for the experiments reported here. Residues within 5 Å of the putative docking clusters of the inhibitor molecules are presented in Table 2.
TABLE 2.
Identification of protein residues in P-gp that were within 5.0 Å of docked inhibitors
| Compound | Compound 19 (ATP-binding site) | Compound 29 (Allosteric binding site) | Compound 34 (ADP-binding site) | Compound 45 (ATP-binding site) |
|---|---|---|---|---|
| Residues within 5.0 Å of docked inhibitors | ILE 160 | THR 422 | ARG 262 | ILE 160 |
| GLY 161 | VAL 423 | THR 483 | GLY 161 | |
| ASP 164 | ALA 424 | LEU 516 | ASP 164 | |
| PHE 399 | LEU 552 | PRO 517 | PHE 399 | |
| TYR 401 | LEU 554 | HIS 518 | TYR 401 | |
| PRO 402 | THR 563 | ASP 521 | PRO 402 | |
| ILE 409 | GLU 566 | THR 522 | ILE 409 | |
| ASN 428 | ALA 567 | LEU 523 | LEU 410 | |
| SER 429 | VAL 569 | VAL 524 | ASN 428 | |
| GLY 430 | GLN 570 | GLY 525 | SER 429 | |
| CYS 431 | VAL 571 | GLU 526 | GLY 430 | |
| GLY 432 | LEU 573 | ALA 529 | CYS 431 | |
| LYS 433 | ASP 574 | GLN 530 | GLY 432 | |
| SER 434 | ARG 577 | LEU 531 | LYS 433 | |
| THR 435 | THR 582 | VAL 801 | SER 434 | |
| GLN 438 | ILE 583 | SER 802 | THR 435 | |
| LEU 443 | VAL 584 | TRP 803 | THR 436 | |
| TYR 444 | ARG 588 | PHE 804 | GLN 438 | |
| GLN 475 | SER 590 | ASP 805 | TYR 444 | |
| GLU 556 | THR 591 | ASN 809 | GLN 475 | |
| ILE 585 | VAL 592 | ASN 1043 | GLU 556 | |
| HIS 587 | ARG 593 | TYR 1044 | ILE 585 | |
| PHE 601 | ASN 594 | PRO 1045 | HIS 587 | |
| ARG 905 | ALA 595 | ARG 1047 | ARG 905 | |
| ILE 1127 | ASP 596 | VAL 1052 | ILE 1127 | |
| PHE 1157 | VAL 1273 | SER 1077 | PHE 1157 | |
| ILE 1158 | GLN 1274 | THR 1078 | ILE 1158 | |
| LEU 1161 | GLY 1276 | GLN 1081 | LEU 1161 | |
| PRO 1162 | THR 1277 | TYR 1087 | PRO 1162 | |
| ASN 1163 | LYS 1278 | GLN 1118 | ASN 1163 | |
| THR 1167 | ARG 1279 | LYS 1164 | ||
| VAL 1169 | GLN 1280 | THR 1167 | ||
| THR 1174 | THR 1174 | |||
| GLN 1175 | GLN 1175 | |||
| LEU 1176 | LEU 1176 | |||
| SER 1177 | SER 1177 | |||
| GLY 1178 | GLY 1178 | |||
| GLY 1179 | GLY 1179 | |||
| GLN 1180 | GLN 1180 |
Other groups have shown that flavonoids are strong modulators of P-gp activity and that they interact with the NBDs of the enzyme (Di Pietro et al., 2002). It is of interest to point out that one of the inhibitors identified here, compound 19, contains both the A and C rings of flavinols with a modification of the B ring (Fig. 3). Compound 19 may therefore be considered a flavonol analog. Comparison of the binding modality of 19 with those of flavonoids reported in Badhan and Penny (2006) shows that the modified B ring of 19 is in close contact with the tyrosine of the A-loop (Y401 in our study; Y1044 in the Badhan and Penny study), very similar to the B-rings of flavonoids studied by Badhan and Penny (2006).
Conclusion
The present study demonstrates that high-throughput in silico docking studies can be used to generate inhibitor leads that target specific areas within ABC transporters responsible for MDR in cancer chemotherapies. The novel “subtractive” screening methods employed here that target one section of the protein and deemphasize binding at a different section of the protein may be generalizable. Although exploratory in nature, the presented study yielded productive leads for both catalytic sites and an allosteric site inhibitors. These inhibitor-binding sites are now being investigated more thoroughly in current experiments. We are also expanding the docking selections to other conformations of P-gp from the targeted molecular dynamics trajectories. This approach, pioneered by McCammon and colleagues (Amaro et al., 2008), adds a protein dynamic component to docking studies such as presented here.
It is easy to foresee the use of similar subtractive screening methods applied to other difficult drug discovery projects that may generate sets of drug leads that discriminate between desired drug targets and other proteins that should be excluded from interactions with the leads. Optimization studies to increase the affinities of the P-gp inhibitors identified in these studies are currently in progress.
Acknowledgments
The authors thank Isolina Rossi, Stephen Christopher Wheelis, Mindy McClean, and Collette Marchesseault for assistance in the growth of Pichia pastoris and the preparation of P-gp, and the Hamilton family of Dallas for their sustained support of Southern Methodist University (SMU) undergraduate research through the Hamilton Undergraduate Research Scholars Program and the SMU Undergraduate Research Assistantship Program for support of the undergraduate researchers. Computational resources were provided by the SMU Center for Scientific Computing. A special thanks to Joe Gargiula and Amit Kumar of the SMU Office of Information Technology for their assistance with this work and to Dr. Jürgen Zirkel, Lipoid GmbH, Germany, for the generous gift of lysophosphatidyl choline. The authors also thank Dr. Ina Urbatsch, Texas Tech University, Lubbock, for initially providing cultured recombinant Pichia pastoris cells for some of the experiments described, and Matthias Schmidt, Martin-Luther-University, Halle, Germany, for generous gifts of modulator compounds.
Abbreviations
- ABC
ATP-binding cassette
- AMPPNP
5′-adenylyl-β,γ-imidodiphosphate
- compound 19
methyl 4-[bis(2-hydroxy-4-oxochromen-3-yl)methyl]benzoate (ZINC 09973259, CID 4694077)
- compound 29
2-[(5-cyclopropyl-1H-1,2,4-triazol-3-yl)sulfanyl]-N-[2-phenyl-5-(2,4,5-trimethylphenyl)pyrazol-3-yl]acetamide (ZINC 08767731, CID 17555821)
- compound 34
2-[1-[4-(4-methoxyphenyl)piperazin-1-yl]-1-oxobutan-2-yl]-4-methyl-[1]benzothiolo[2,3-d]pyridazin-1-one (ZINC 09252021, CID 22514118)
- compound 45
ethyl 1-(1,3-benzodioxole-5-carbonyl)-3-(3-phenylpropyl)piperidine-3-carboxylate (ZINC 15078148, ZINC 15078146, CID 26410703, CID 45252040)
- DBD
drug-binding domain
- DMSO
dimethylsulfoxide
- ESR
electron spin resonance
- MDR
multidrug resistance
- NBD
nucleotide-binding domain
- P-gp
P-glycoprotein
- SL
spin-labeled
- SL-ATP
2′,3′-(2,2,5,5,-tetramethyl-3-pyrroline-1-oxyl-3-carboxylic acid ester) ATP
- SL-ANP
2′,3′-(2,2,5,5,-tetramethyl-3-pyrroline-1-oxyl-3-carboxylic acid ester) -ATP or -ADP “2′,3′-” indicates a rapid equilibrium of the ester bond between the 2′- and 3′-hydroxyl groups of the adenosine
- TM
transmembrane
Authorship Contributions
Participated in research design: Brewer, Follit, Vogel, Wise.
Conducted experiments: Brewer, Follit, Wise.
Performed data analysis: Brewer, Follit, Vogel, Wise.
Wrote or contributed to the writing of the manuscript: Brewer, Follit, Vogel, Wise.
Footnotes
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant 1R15-GM094771-01A1] (to P.D.V. and J.G.W.), and the Southern Methodist University (SMU) Research Council, the SMU Dedman College Dean’s Research Council, the Dedman College Center for Drug Discovery, Design and Delivery, and the Communities Foundations of Texas, Dallas. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
References
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, et al. (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323:1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. (2008) Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci USA 105:17278–17283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badhan R, Penny J. (2006) In silico modelling of the interaction of flavonoids with human P-glycoprotein nucleotide-binding domain. Eur J Med Chem 41:285–295. [DOI] [PubMed] [Google Scholar]
- Binkhathlan Z, Lavasanifar A. (2013) P-glycoprotein inhibition as a therapeutic approach for overcoming multidrug resistance in cancer: current status and future perspectives. Curr Cancer Drug Targets 13:326–346. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB. (1986) Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 47:381–389. [DOI] [PubMed] [Google Scholar]
- Copeland RA. (2000) Enzymes: a Practical Introduction to Structure, Mechanism, and Data Analysis, 2nd ed. Wiley, New York. [Google Scholar]
- Copeland RA. (2013) Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists, 2nd ed Wiley, Hoboken, NJ. [PubMed] [Google Scholar]
- Dawson RJP, Locher KP. (2006) Structure of a bacterial multidrug ABC transporter. Nature 443:180–185. [DOI] [PubMed] [Google Scholar]
- Dawson RJP, Locher KP. (2007) Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett 581:935–938. [DOI] [PubMed] [Google Scholar]
- Delannoy S, Urbatsch IL, Tombline G, Senior AE, Vogel PD. (2005) Nucleotide binding to the multidrug resistance P-glycoprotein as studied by ESR spectroscopy. Biochemistry 44:14010–14019. [DOI] [PubMed] [Google Scholar]
- Dey S, Ramachandra M, Pastan I, Gottesman MM, Ambudkar SV. (1997) Evidence for two nonidentical drug-interaction sites in the human P-glycoprotein. Proc Natl Acad Sci USA 94:10594–10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A, Conseil G, Pérez-Victoria JM, Dayan G, Baubichon-Cortay H, Trompier D, Steinfels E, Jault JM, de Wet H, Maitrejean M, et al. (2002) Modulation by flavonoids of cell multidrug resistance mediated by P-glycoprotein and related ABC transporters. Cell Mol Life Sci 59:307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JM, Hait WN. (1990) Pharmacology of drugs that alter multidrug resistance in cancer. Pharmacol Rev 42:155–199. [PubMed] [Google Scholar]
- Goodsell DS, Morris GM, Olson AJ. (1996) Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit 9:1–5. [DOI] [PubMed] [Google Scholar]
- Gottesman MM, Pastan I. (1993) Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem 62:385–427. [DOI] [PubMed] [Google Scholar]
- Gros P, Ben Neriah YB, Croop JM, Housman DE. (1986) Isolation and expression of a complementary DNA that confers multidrug resistance. Nature 323:728–731. [DOI] [PubMed] [Google Scholar]
- Harris AL, Hochhauser D. (1992) Mechanisms of multidrug resistance in cancer treatment. Acta Oncol 31:205–213. [DOI] [PubMed] [Google Scholar]
- Hoffman AD, Urbatsch IL, Vogel PD. (2010) Nucleotide binding to the human multidrug resistance protein 3, MRP3. Protein J 29:373–379. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJP, Locher KP. (2007) Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol 17:412–418. [DOI] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. (2012) ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MS, Oldham ML, Zhang Q, Chen J. (2012) Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490:566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartner N, Ling V. (1989) Multidrug resistance in cancer. Sci Am 260:44–51. [DOI] [PubMed] [Google Scholar]
- Lee J-Y, Urbatsch IL, Senior AE, Wilkens S. (2008) Nucleotide-induced structural changes in P-glycoprotein observed by electron microscopy. J Biol Chem 283:5769–5779. [DOI] [PubMed] [Google Scholar]
- Lerner-Marmarosh N, Gimi K, Urbatsch IL, Gros P, Senior AE. (1999) Large scale purification of detergent-soluble P-glycoprotein from Pichia pastoris cells and characterization of nucleotide binding properties of wild-type, Walker A, and Walker B mutant proteins. J Biol Chem 274:34711–34718. [DOI] [PubMed] [Google Scholar]
- Li J, Jaimes KF, Aller SG. (2014) Refined structures of mouse P-glycoprotein. Protein Sci 23:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. (2003) Simultaneous binding of two different drugs in the binding pocket of the human multidrug resistance P-glycoprotein. J Biol Chem 278:39706–39710. [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. (2009) Identification of residues in the drug translocation pathway of the human multidrug resistance P-glycoprotein by arginine mutagenesis. J Biol Chem 284:24074–24087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo MR, Sharom FJ. (2005) Interaction of LDS-751 and rhodamine 123 with P-glycoprotein: evidence for simultaneous binding of both drugs. Biochemistry 44:14020–14029. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Huey R, Olson AJ. (1996) Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des 10:293–304. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS. (2002) Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins 46:34–40. [DOI] [PubMed] [Google Scholar]
- Palmeira A, Sousa E, Vasconcelos MH, Pinto MM. (2012) Three decades of P-gp inhibitors: skimming through several generations and scaffolds. Curr Med Chem 19:1946–2025. [DOI] [PubMed] [Google Scholar]
- Persidis A. (1999) Cancer multidrug resistance. Nat Biotechnol 17:94–95. [DOI] [PubMed] [Google Scholar]
- Qu Q, Russell PL, Sharom FJ. (2003) Stoichiometry and affinity of nucleotide binding to P-glycoprotein during the catalytic cycle. Biochemistry 42:1170–1177. [DOI] [PubMed] [Google Scholar]
- Roninson IB, Chin JE, Choi KG, Gros P, Housman DE, Fojo A, Shen DW, Gottesman MM, Pastan I. (1986) Isolation of human mdr DNA sequences amplified in multidrug-resistant KB carcinoma cells. Proc Natl Acad Sci USA 83:4538–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Teitge M, Castillo ME, Brandt T, Dobner B, Langner A. (2008) Synthesis and biochemical characterization of new phenothiazines and related drugs as MDR reversal agents. Arch Pharm (Weinheim) 341:624–638. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Ungvári J, Glöde J, Dobner B, Langner A. (2007) New 1,3-dioxolane and 1,3-dioxane derivatives as effective modulators to overcome multidrug resistance. Bioorg Med Chem 15:2283–2297. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Ling V. (1997) Positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. Eur J Biochem 250:130–137. [DOI] [PubMed] [Google Scholar]
- Stewart A, Steiner J, Mellows G, Laguda B, Norris D, Bevan P. (2000) Phase I trial of XR9576 in healthy volunteers demonstrates modulation of P-glycoprotein in CD56+ lymphocytes after oral and intravenous administration. Clin Cancer Res 6:4186–4191. [PubMed] [Google Scholar]
- Tombline G, Urbatsch IL, Virk N, Muharemagic A, White LB, Senior AE. (2006) Expression, purification, and characterization of cysteine-free mouse P-glycoprotein. Arch Biochem Biophys 445:124–128. [DOI] [PubMed] [Google Scholar]
- Urbatsch IL, Beaudet L, Carrier I, Gros P. (1998) Mutations in either nucleotide-binding site of P-glycoprotein (Mdr3) prevent vanadate trapping of nucleotide at both sites. Biochemistry 37:4592–4602. [DOI] [PubMed] [Google Scholar]
- Urbatsch IL, Tyndall GA, Tombline G, Senior AE. (2003) P-glycoprotein catalytic mechanism: studies of the ADP-vanadate inhibited state. J Biol Chem 278:23171–23179. [DOI] [PubMed] [Google Scholar]
- Verhalen B, Ernst S, Börsch M, Wilkens S. (2012) Dynamic ligand-induced conformational rearrangements in P-glycoprotein as probed by fluorescence resonance energy transfer spectroscopy. J Biol Chem 287:1112–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhalen B, Wilkens S. (2011) P-glycoprotein retains drug-stimulated ATPase activity upon covalent linkage of the two nucleotide binding domains at their C-terminal ends. J Biol Chem 286:10476–10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G, Steinhart R. (1976) ATPase of Escherichia coli: purification, dissociation, and reconstitution of the active complex from the isolated subunits. Biochemistry 15:208–216. [DOI] [PubMed] [Google Scholar]
- Walker J, Martin C, Callaghan R. (2004) Inhibition of P-glycoprotein function by XR9576 in a solid tumour model can restore anticancer drug efficacy. Eur J Cancer 40:594–605. [DOI] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G. (2007) Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci USA 104:19005–19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen PC, Tajkhorshid E. (2008) Dimer opening of the nucleotide binding domains of ABC transporters after ATP hydrolysis. Biophys J 95:5100–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise JG. (2012) Catalytic transitions in the human MDR1 P-glycoprotein drug binding sites. Biochemistry 51:5125–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi ME, Altenberg GA. (2013) Hydrolysis at one of the two nucleotide-binding sites drives the dissociation of ATP-binding cassette nucleotide-binding domain dimers. J Biol Chem 288:34259–34265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi ME, Altenberg GA. (2014) ATP binding to two sites is necessary for dimerization of nucleotide-binding domains of ABC proteins. Biochem Biophys Res Commun 443:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]