Abstract
Cytochrome P450 2D6 (CYP2D6), a major drug-metabolizing enzyme, is responsible for metabolism of approximately 25% of marketed drugs. Clinical evidence indicates that metabolism of CYP2D6 substrates is increased during pregnancy, but the underlying mechanisms remain unclear. To identify transcription factors potentially responsible for CYP2D6 induction during pregnancy, a panel of genes differentially expressed in the livers of pregnant versus nonpregnant CYP2D6-humanized (tg-CYP2D6) mice was compiled via microarray experiments followed by real-time quantitative reverse-transcription polymerase chain reaction(qRT-PCR) verification. As a result, seven transcription factors—activating transcription factor 5 (ATF5), early growth response 1 (EGR1), forkhead box protein A3 (FOXA3), JUNB, Krüppel-like factor 9 (KLF9), KLF10, and REV-ERBα—were found to be up-regulated in liver during pregnancy. Results from transient transfection and promoter reporter gene assays indicate that KLF9 itself is a weak transactivator of CYP2D6 promoter but significantly enhances CYP2D6 promoter transactivation by hepatocyte nuclear factor 4 (HNF4α), a known transcriptional activator of CYP2D6 expression. The results from deletion and mutation analysis of CYP2D6 promoter activity identified a KLF9 putative binding motif at -22/-14 region to be critical in the potentiation of HNF4α-induced transactivation of CYP2D6. Electrophoretic mobility shift assays revealed a direct binding of KLF9 to the putative KLF binding motif. Results from chromatin immunoprecipitation assay showed increased recruitment of KLF9 to CYP2D6 promoter in the livers of tg-CYP2D6 mice during pregnancy. Taken together, our data suggest that increased KLF9 expression is in part responsible for CYP2D6 induction during pregnancy via the potentiation of HNF4α transactivation of CYP2D6.
Introduction
Cytochrome P450 2D6 (CYP2D6), a major drug-metabolizing enzyme expressed in the liver and extrahepatic organs (such as the brain, kidney, and intestine), mediates the hepatic metabolism of approximately 25% of marketed drugs including antidepressants and antipsychotics (Yu et al., 2004; Zanger et al., 2004). In addition to metabolizing drugs, CYP2D6 is implicated in the development of Parkinson disease in Caucasian populations with decreased CYP2D6 activity (Lu et al., 2014). Also, decreased CYP2D6 activity levels in the brain are associated with higher perfusion levels in the regions linked to alertness or serotonergic function, suggesting a functional role of CYP2D6 in the brain (Kirchheiner et al., 2011). However, the factors governing the regulation of CYP2D6 expression have been studied to only a limited extent. For example, pregnancy is known to induce hepatic elimination of CYP2D6 substrates (Hogstedt et al., 1985; Wadelius et al., 1997; Tracy et al., 2005). The underlying molecular mechanisms remain unclear, in part due to our lack of understanding of the transcriptional regulation of CYP2D6.
Hepatocyte nuclear factor 4α (HNF4α) is a transcription regulator of CYP2D6 expression (Cairns et al., 1996; Lemberger et al., 1996; Kharitonenkov et al., 2005). In HepG2 cells, ectopically expressed HNF4α enhances promoter activity of CYP2D6 by its binding to a promoter region (i.e., -53/-41 of CYP2D6) (Cairns et al., 1996). Knockdown of HNF4α expression significantly decreases the transcription of CYP2D6 (Corchero et al., 2001; Kharitonenkov et al., 2005). HNF4α harboring the G60D polymorphism is unable to transactivate CYP2D6 promoter activity in HEK293 cells, and it is associated with decreased metabolism of a CYP2D6 substrate in humans (Lee et al., 2008). These studies indicate key roles of HNF4α in CYP2D6 regulation. Of note, HNF4α controls constitutive expression of many genes involved in basic hepatic functions (e.g., nutrient metabolism and blood clotting), being a master regulator of hepatic genes (Gonzalez, 2008). This suggests that a mechanism that fine-tunes HNF4α action on different HNF4α-target genes probably exists. The identities of such putative mechanisms remain unknown.
Krüppel-like factor (KLF) 9 is a member in the KLF transcription factor family, and it is capable of either activating or repressing target gene expression in a promoter-specific context. KLF9 is involved in various physiologic functions. KLF9 modulates signaling pathways involving progesterone receptor (PR) (Zhang et al., 2003) such that Klf9-null female mice exhibit defects in the reproductive system (e.g., uterine hypoplasia and decreased litter size) (Simmen et al., 2004). In epidermis, KLF9 mediates the proliferation of keratinocytes by cortisol (Sporl et al., 2012). Biologic actions of KLF9 are mediated either by its direct binding to the promoters of its target genes such as CYP1A1 (Imataka et al., 1992) or by coactivation of other transcription factors. In endometrial cells, KLF9 binds to PR and enhances transcriptional activation of PR target genes (Zhang et al., 2002, 2003).
Previously, we recapitulated CYP2D6 induction during pregnancy in an animal model (Koh et al., 2014). In tg-CYP2D6 mice whose genome harbors the human CYP2D6 gene plus 2.5-kb of its upstream regulatory region, CYP2D6 expression was significantly enhanced at term pregnancy. This was accompanied by increased recruitment of HNF4α to the promoter of CYP2D6 (Koh et al., 2014). The enhanced HNF4α activity during pregnancy was attributed in part to the decreased expression of small heterodimer partner (SHP) (Koh et al., 2014), a corepressor that inhibits HNF4α activity via physical interaction (Zhou et al., 2010). Interestingly, our results showed that other target genes of HNF4α (such as Hes6 and ApoC2) did not exhibit similar increases in expression to CYP2D6 during pregnancy in tg-CYP2D6 mice (Koh et al., 2014). These results suggest a potential role of promoter contexts in modulating HNF4α transactivation of its target genes and the presence of additional regulatory factors potentially involved in CYP2D6 regulation during pregnancy.
In this study, we show that KLF9 expression is up-regulated during pregnancy in the livers of tg-CYP2D6 mice. The roles of KLF9 in HNF4α transactivation of the CYP2D6 promoter as well as in CYP2D6 induction during pregnancy were investigated. Our results illustrate the interplay among hepatic transcription factors KLF9, HNF4α, and SHP contributing to the regulation of CYP2D6 expression during pregnancy.
Materials and Methods
Animals.
Elsewhere, tg-CYP2D6 mice were previously described (Corchero et al., 2001). Adult female (8-week-old) mice were mated with male mice of a similar age. Mating between adult mice was confirmed by the presence of vaginal plugs (day 0). All procedures were approved by the Institutional Animal Care and Use Committee at the University of Illinois at Chicago.
Plasmids.
Luciferase vectors harboring upstream regulatory regions of CYP2D6 were previously described elsewhere (Koh et al., 2014). The mutation pGL3-CYP2D6 constructs [i.e., mutations at K1 to K5 sites (see Fig. 3B for their locations),] were made using a QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer’s protocol using pGL3-CYP2d6(−205/+90) as a template. The primer sequences are listed in Supplemental Table 1. Expression vectors for human REV-ERBα, KLF10, and forkhead box protein A3 (FOXA3) were purchased from Thermo Scientific (Hanover Park, IL). An expression vector for KLF9 was purchased from Thermo Scientific and subcloned into pCMV-Sport6 vector. The pcDNA3.1-ATF5 and pcDNA-JUNB were gifts from Drs. Nathalie Wong (Chinese University of Hong Kong) and Jeremy J.W. Chen (National Chung-Hsing University), respectively. The pcDNA3.1-his-EGR1 and pcDNA3-HNF4α (the longest isoform b) were received from Drs. Masahiko Negishi (U.S. National Institute of Environmental Health Sciences) and Frances M. Sladek (University of California Riverside), respectively. The CYP2D6 promoter vector harboring mutated HNF4α response element was provided by Dr. H. Hara (Gifu Pharmaceutical University, Gifu, Japan).
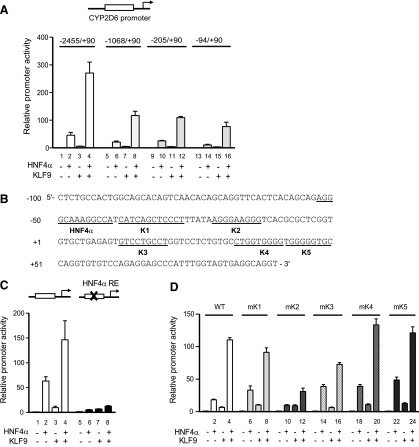
Fig. 3.
KLF9 action on CYP2D6 promoter requires cis-element(s) located in −94/+90 of CYP2D6. (A) HEK293T cells were transfected with the expression vectors (HNF4α and/or KLF9) and pCMV-Renilla vector, along with a 5′-deletion promoter construct harboring −2455/+90, −1068/+90, −205/+90, or −94/+90 of the CYP2D6 gene. After 48 hours, dual luciferase assays were performed (triplicate experiment, mean ± S.D.). Data shown are relative promoter activities in cells transfected with HNF4α and/or KLF9 versus cells transfected with empty vector. (B) Putative KLF9- (K1 to K5) and HNF4α-binding sites are underlined. (C) HEK293T cells were transfected with expression vectors (HNF4α and/or KLF9) and pCMV-Renilla vector, along with the pGL3-CYP2D6 vector carrying the wild-type (WT; lanes 1–4) or mutated HNF4α response element (lanes 5–8). After 48 hours, dual luciferase assays were performed (triplicate experiment, mean ± S.D.). (D) HEK293T cells were transfected with expression vectors (HNF4α and/or KLF9) and pCMV-Renilla vector, along with the pGL3-CYP2D6 vector carrying the WT or mutated KLF9-binding sequences (i.e., mK1 to mK5). After 48 hours, dual luciferase assays were performed (triplicate experiment, mean ± S.D.).
Western Blot.
KLF9 protein expression levels were determined by using an antibody from Santa Cruz Biotechnology (Santa Cruz, CA).
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction.
Total RNA was isolated from mouse liver tissues using TRIzol (Life Technologies, Carlsbad, CA) and converted to cDNA using High Capacity cDNA Archive Kit (Life Technologies). Resulting cDNA samples were subject to real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis using StepOnePlus Real-Time PCR System and the primers listed in Supplemental Table 1. The fold increase in mRNA levels during pregnancy was determined after normalizing the gene expression levels to those of mouse β-actin (2−ΔΔCt method).
Microarray and Promoter Analysis.
Total RNA was isolated from mouse liver tissues at pre-pregnancy (virgin), at term (21 days of pregnancy), and 7 days postpartum (n = 2/time point) using TRIzol. The cDNA synthesis, modification, hybridization, and labeling on Affymetrix GeneChip MG430 2.0 arrays were performed using kits from Affymetrix (Santa Clara, CA) as described in the manufacturer's instructions. The raw microarray data were quantile normalized and summarized using Affymetrix Power Tools (GEO database: GSE50166). The local pooled error method (Jain et al., 2003), implemented in the R Statistical Packages (http://www.r-project.org/), was used to identify differentially expressed genes, considering its efficiency with a small number of replicated arrays. The false-discovery rate was controlled at 1% using rank-invariant resampling based estimation (Jain et al., 2005). The relative change in expression (fold change > 2.0 or 1.5) was used to obtain a list of differentially expressed genes at term pregnancy and postpartum, compared with the control group (virgin). The prediction of putative transcription factor binding sites in the CYP2D6 promoter was performed using the MatInspector software (Genomatix, Munich, Germany).
Luciferase Reporter Assay.
HepG2 or HEK293T cells were seeded in 12-well plates at a density of 4.5 × 105 cells/ml, and on the next day, transfected with 0.3 μg of luciferase construct, 0.1 μg of expression plasmid (or empty vector as a control), and 0.002 μg of Renilla expression vector (Promega, Madison, WI) using Fugene HD transfection reagent (Promega) according to the manufacturer’s protocol. The transfected cells were grown for 48 hours and harvested for determination of luciferase activities using a luciferase assay kit (Promega). At least two independent experiments were performed in triplicate.
Coimmunoprecipitation Assay.
HEK293T cells were cotransfected with expression vectors for 3xFLAG-tagged KLF9 and HNF4α, and lysed in lysis buffer (0.5% Nonidet P-40, 50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.25% Triton × 100, and protease inhibitor cocktail) and centrifuged at 13,000 rpm for 30 minutes. The resulting supernatant was incubated overnight with magnetic beads (Dynabeads protein-A; Life Technologies) coated with HNF4α (Santa Cruz Biotechnology) or FLAG-M2 antibody (Sigma-Aldrich, St. Louis, MO) at 4°C. The precipitated proteins were eluted in SDS-PAGE sample buffer and subject to SDS-PAGE and Western blot.
Chromatin Immunoprecipitation Assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described elsewhere with minor modifications (Fang et al., 2007). Briefly, livers were finely minced and incubated in phosphate-buffered saline containing 1% formaldehyde at room temperature for 15 minutes, and glycine was added to stop the crosslinking reaction. Cell pellets were resuspended in hypotonic buffer (15 mM HEPES, pH 7.9, 60 mM KCl, 2 mM EDTA, 0.5% bovine serum albumin, 0.15 mM spermine, 0.5 mM spermidine, 0.32 M sucrose) and lysed by homogenization. Nuclei were pelleted and resuspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 2 mM EDTA, 1% SDS). The samples were sonicated to shear DNA to the length ranging from 100 to 500 bp. After centrifugation, the chromatin sample was immunoprecipitated overnight with magnetic beads (Life Technologies) coated with KLF9 antibody (Santa Cruz Biotechnology) or immunoglobulin G (IgG) at 4°C. The immune complexes were collected, and the magnetic beads were extensively washed, followed by elution of bound chromatin. Genomic DNA was purified by PCR Clean-up kit (Promega) and used as a template for qPCR. Primer sequences are listed in Supplemental Table 1.
Electrophoretic Mobility Shift Assay.
Electrophoretic mobility shift assays (EMSAs) were performed following the manufacturer’s protocol (Gel Shift Assay Systems; Promega). Briefly, nuclear extracts were prepared from human embryonic kidney 293T (HEK293T) cells transiently transfected with KLF9 or empty vector (as a negative control) using the CelLytic Nuclear Extraction Kit (Sigma-Aldrich). A [32P] dATP-labeled double stranded oligonucleotide (Supplemental Table 1) was incubated with nuclear extract (20 μg) at room temperature in the presence or absence of nonradioactive DNA duplex probe (for competition) or KLF9 antibody (Santa Cruz Biotechnology). The reaction mixture was resolved on a native 4% (w/v) polyacrylamide gel. The signal was visualized by using PhosphorImager (Typhoon Trio, GE Healthcare).
Statistical Analysis.
For comparison of two groups, statistical differences were determined by Student’s t test. For statistical testing of more than two groups, a one-way analysis of variance test followed by post hoc Dunnett’s test was performed.
Results
KLF9 Expression Is Enhanced during Pregnancy.
To identify potential transcription factors involved in CYP2D6 regulation during pregnancy, microarray gene expression profiling was performed using RNA samples extracted from the livers of tg-CYP2D6 mice at various gestational time points (n = 2/group): virgin, 21 days of pregnancy (G21), and 7 days postpartum (PP7). The results showed that 555 genes were upregulated at G21 (versus virgin) and/or downregulated at PP7 (versus G21), and 239 genes showed the opposite pattern (GSE50166; Supplemental Tables 2 and 3). Subsequent qRT-PCR analyses (additional time points including virgin, G7/14/21, and PP7, n = 4/group) led to the identification of seven up-regulated genes [i.e., activating transcription factor 5 (ATF5), early growth response 1 [EGR1], forkhead box protein A3 [FOXA3], JUNB, KLF9, KLF10, and REV-ERBα)] (Fig. 1). SHP was found to be down-regulated during pregnancy as we reported in a recent study (Koh et al., 2014).
Fig. 1.
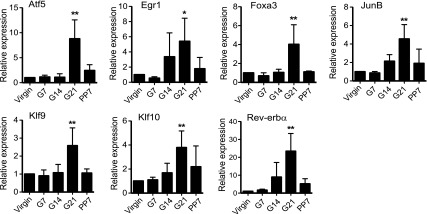
Transcription factors are differentially expressed during pregnancy in the livers of tg-CYP2D6 mice. Liver tissues were collected from tg-CYP2D6 mice at pre-pregnancy (virgin), 7, 14, and 21 days of pregnancy (G7, G14, and G21, respectively), or 7 days postpartum (PP7), and total RNA was isolated from the tissues. The mRNA expression levels of the respective transcription factors were determined by qRT-PCR (n = 4, mean ± S.D.; *P < 0.05, **P < 0.01 versus virgin).
KLF9 Potentiates HNF4α-Mediated Transactivation of CYP2D6.
To examine the functional impact of differentially regulated transcription factors on CYP2D6 transactivation, reporter gene assays were performed using a luciferase vector where luc expression is driven by 2.5-kb CYP2D6 promoter (i.e., pGL3-CYP2D6). HNF4α, a positive regulator of basal CYP2D6 expression (Cairns et al., 1996; Corchero et al., 2001; Lee et al., 2008), was included as a positive control and led to a marked increase in CYP2D6 promoter as expected (Fig. 2A). None of the transcription factors upregulated during pregnancy was found to directly modulate CYP2D6 promoter activity in a statistically significant manner. Although statistically insignificant, KLF9 appeared to enhance the CYP2D6 promoter activity (Fig. 2A).
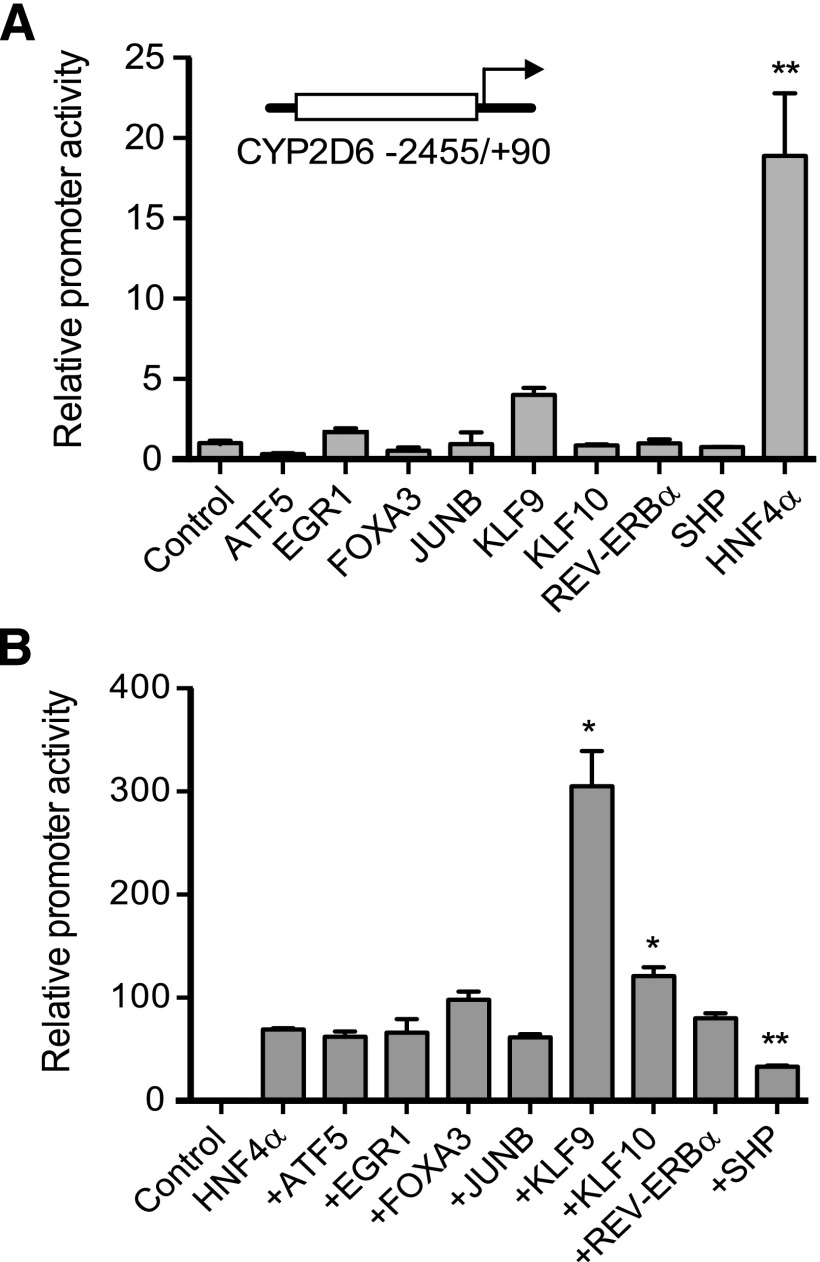
Fig. 2.
KLF9 enhances HNF4α-mediated transactivation of CYP2D6 promoter. (A) HEK293T cells were transfected with the pGL3-CYP2D6 vector (−2455/+90), an expression vector for each of the transcription factors (ATF5, EGR1, FOXA3, JUNB, KLF9, KLF10, REV-ERBα, or empty vector), and the pCMV-Renilla vector. After 48 hours, dual luciferase assays were performed (n = 3, mean ± S.D.; **P < 0.01 versus control). (B) HEK293T cells were transfected with the pGL3-CYP2D6 vector (−2455/+90), HNF4α expression vector plus an expression vector for each of the transcription factors (ATF5, EGR1, FOXA3, JUNB, KLF9, KLF10, REV-ERBα, or empty vector), and the pCMV-Renilla vector. After 48 hours, dual luciferase assays were performed (n = 3, mean ± S.D.; *P < 0.05, **P < 0.01 versus HNF4α alone).
Given the key role that HNF4α plays in CYP2D6 induction during pregnancy (Koh et al., 2014) and the evidence that HNF4α activity is functionally modulated by other transcription factors (Gonzalez, 2008), we examined whether the differentially expressed transcription factors affect HNF4α-mediated transactivation of CYP2D6 promoter. Our group has recently reported that SHP functions as a negative modulator of HNF4α action on CYP2D6 promoter (Koh et al., 2014). Indeed, SHP repressed the HNF4α-induced transactivation of CYP2D6 promoter as previously shown (Fig. 2B). Among the seven transcription factors tested, KLF9 exhibited the most prominent effect in enhancing HNF4α-induced transactivation of the CYP2D6 promoter (Fig. 2B); the coexpression of KLF9 and HNF4α led to a 304-fold increase in CYP2D6 promoter activity while 7- and 69-fold increases in the promoter activity were shown in cells transfected with KLF9 or HNF4α alone, respectively. The rest of the transcription factors exhibited insignificant or minor effects on HNF4α activity.
Proximal Regulatory Region of CYP2D6 Mediates the KLF9 Action on CYP2D6 Promoter.
To map the cis-elements responsible for the potentiating effect of KLF9 on HNF4α-mediated CYP2D6 transactivation, promoter reporter gene assays were performed using 5′-deletion constructs of CYP2D6 promoter. The four deletion constructs of CYP2D6 promoter retained the potentiating effect of KLF9 on HNF4α-induced CYP2D6 promoter transactivation although the extent of potentiation slightly varied among the constructs (Fig. 3A). Given that 5′-deletion of the CYP2D6 regulatory region up to -94 did not abolish the potentiating effect of KLF9 on HNF4α-induced transactivation of CYP2D6 promoter, we further explored potential cis-element(s) responsible for KLF9 action within the proximal 94-bp region of the CYP2D6 promoter. In silico analysis of the proximal region (−100/+90) of CYP2D6 using MatInspector (Genomatix, Munich, Germany) revealed multiple putative binding sites (K1 to K5) for KLF9 (Fig. 3B) and the previously reported HNF4α response element (Cairns et al., 1996).
Before examining the functional importance of putative KLF9-binding sites, we confirmed the role of the HNF4α response element in mediating the KLF9 effect on CYP2D6 promoter activity. Reporter gene assays were performed using CYP2D6 promoter vectors harboring wild-type or mutated HNF4α response element sequences. The mutation of the HNF4α response element abrogated not only HNF4α-induced transactivation of CYP2D6 promoter activity (Fig. 3C, lane 2 versus 6), but also the potentiating effects of KLF9 (Fig. 3C, lane 4 versus 8), indicating that KLF9 action on CYP2D6 promoter activity requires HNF4α binding to the DNA.
Next, reporter assays were performed using promoter constructs harboring mutated sequences at the putative KLF9-binding sites. Overall, mutations at putative KLF9-binding sites other than the K2 position did not have a major impact on the potentiating effect of KLF9 on the transactivation of CYP2D6 promoter activity (Fig. 3D). Mutations at the K2 position (mK2) substantially attenuated HNF4α transactivation of CYP2D6 promoter activity in KLF9-cotransfected cells (Fig. 3D, lane 4 versus 12).
KLF9 Binds to CYP2D6 Promoter.
The importance of the K2 site was further validated by examining its interaction with KLF9 via electrophoretic mobility shift assay. Nuclear extracts were prepared from HEK293T cells transfected with a KLF9 expression vector (or empty vector as control) and incubated with radiolabeled K2 probe. Results from the KLF9 consensus sequence (a positive control) identified a band shift likely arising from KLF9 binding to the probe (Fig. 4A). The signal of this band was successfully competed away by the nonradiolabeled probe and blocked by KLF9 antibody (Fig. 4A, lanes 4 and 5). Similarly, the K2 probe exhibited a shifted band that can be competed away by the nonradiolabeled probe with intact K2 sequence (Fig. 4A, lanes 8 and 9). Signal of the band did not decrease when the nonradiolabeled probe contained the mutated K2 sequence (Fig. 4A, lane 10). Addition of KLF9 antibody to the mixture decreased the signal of shifted band (Fig. 4A, lane 11). Similar results were obtained when nuclear extracts were prepared from HEK293T cells transfected with 3xFLAG-tagged KLF9 (Fig. 4B). Together, these data support that KLF9 binds to the K2 site of CYP2D6 promoter.
Fig. 4.
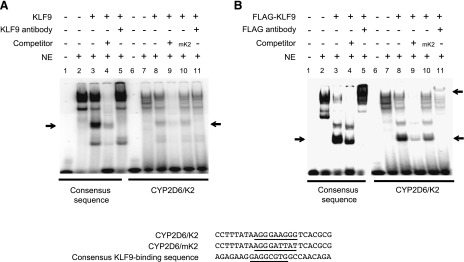
KLF9 binds to CYP2D6 promoter sequences. Cell lysates from HEK293T cells transiently transfected with KLF9 (A) or 3xFLAG-tagged KLF9 vector (B) (or empty vector as control) were incubated with 32P-labeled probes harboring a consensus KLF9-binding sequence (left) or CYP2D6/K2 (right). The mixture was resolved on nondenaturing polyacrylamide gel. The lower and upper arrows indicate locations of shifted and super-shifted bands by apparent KLF9 binding to the DNA, respectively. The heavy band at the top likely represents nonspecific binding. The sequences of probes with the putative binding sequences underlined are shown in the bottom panel.
KLF9 Does Not Physically Interact with HNF4α.
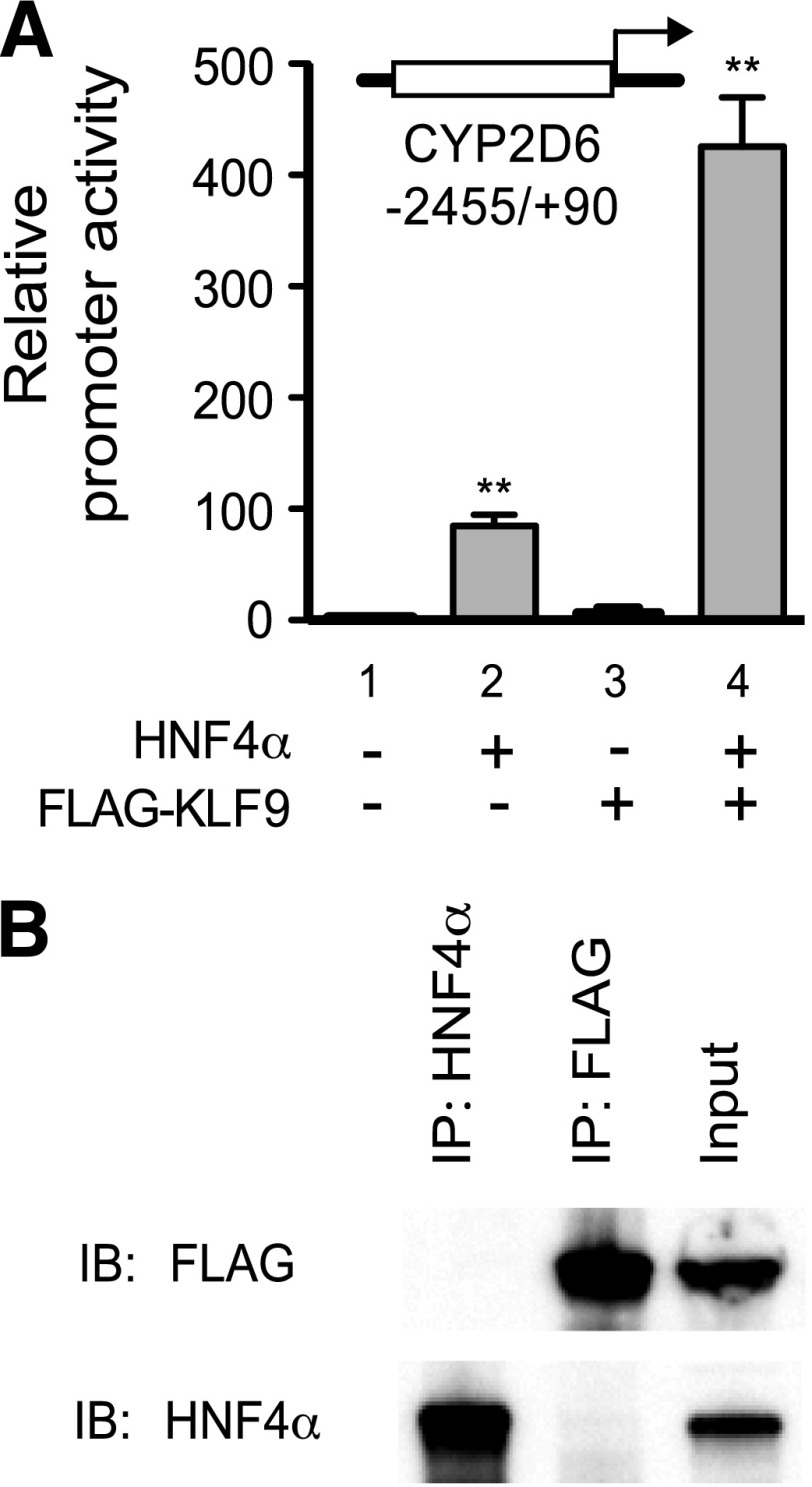
To examine whether KLF9 and HNF4α physically interact with each other for the synergistic activation of transcription from the CYP2D6 promoter, coimmunoprecipitation assays were performed using 3xFLAG-tagged KLF9. First, we verified that the FLAG-tagging of KLF9 did not interfere with the ability of KLF9 to potentiate the HNF4α transactivation of the CYP2D6 promoter activity using promoter reporter gene assays (Fig. 5A). To examine physical interaction between HNF4α and KLF9, HNF4α and 3xFLAG-KLF9 were ectopically expressed in HEK293T cells, and then immunoprecipitation was performed using antibodies against HNF4α and FLAG, respectively. The pulled-down proteins were detected using Western blot. The results showed a lack of KLF9 in proteins pulled down by HNF4α (or vice versa) (Fig. 5B), indicating that KLF9 and HNF4α do not directly bind to each other.
Fig. 5.
KLF9 does not bind to HNF4α in vitro. (A) HEK293T cells were transfected with the pGL3-CYP2D6 vector (−2455/+90), the expression vectors (HNF4α and/or 3xFLAG-tagged KLF9), and pCMV-Renilla vector. After 48 hours, dual luciferase assays were performed (triplicate experiment, mean ± S.D.; **P < 0.01 versus lane 1). (B) HEK293T cells were cotransfected with the 3xFLAG-tagged KLF9 and HNF4α expression vector. Cell lysates were prepared, and immunoprecipitation was performed using magnetic beads coated with antibodies against HNF4α (lane 1) or FLAG (lane 2). Immunoprecipitates were analyzed by Western blot. One percent of the input (lane 3) was loaded onto the gel as a control.
KLF9 Functionally Interacts with SHP.
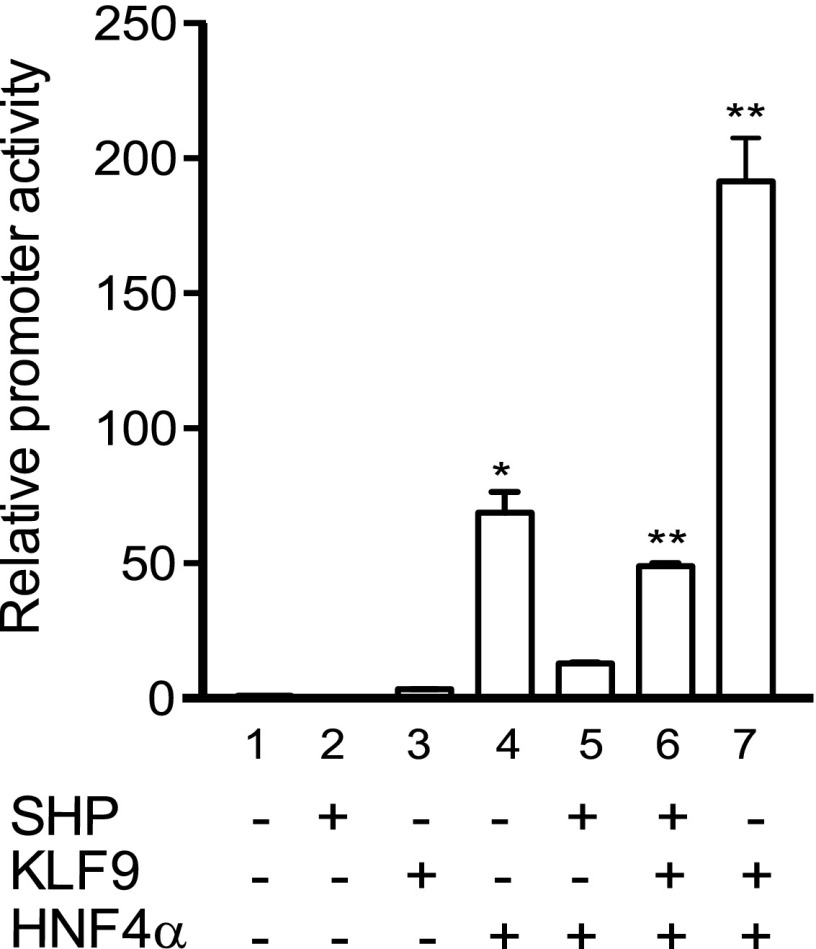
We have previously shown that SHP represses CYP2D6 promoter activity by inhibiting HNF4α-mediated transactivation and that SHP expression decreases during pregnancy (Koh et al., 2014). The current study, on the other hand, shows that KLF9 expression increases in mouse livers during pregnancy. Given that both KLF9 and SHP exert their actions on the CYP2D6 promoter via modulating HNF4α activity, we examined whether KLF9 and SHP interact functionally using cotransfection and promoter reporter gene assays. Additional expression of KLF9 enhanced CYP2D6 promoter activity as compared with the cells transfected with SHP and HNF4α (Fig. 6, lane 5 versus 6). The potentiating effect of KLF9 was greater when cells were not transfected with SHP (Fig. 6, lane 6 versus 7). This suggests the possibility that KLF9 may efficiently potentiate the HNF4α transactivation of CYP2D6 promoter activity during pregnancy due to the concomitant decline in SHP expression. The functional interplay among SHP, KLF9, and HNF4α was abrogated by the mutation of the HNF4α-binding response element in the CYP2D6 promoter (data not shown), indicating the key role of HNF4α in KLF9- and SHP-mediated regulation of CYP2D6 expression.
Fig. 6.
Functional interplay between SHP and KLF9 in regulating HNF4α-induced transactivation of CYP2D6 promoter. HEK293T cells were transfected with pGL3-CYP2D6 (−870/+65), expression vectors (HNF4α, KLF9, or SHP) along with pCMV-Renilla. After 48 hours, luciferase assays were performed (triplicate experiment, mean ± S.D.; * P < 0.05, **P < 0.01 versus lane 1).
KLF9 Recruitment to CYP2D6 Promoter Increases during Pregnancy.
Previously, we showed that HNF4α recruitment to CYP2D6 promoter increases the in liver at term pregnancy as compared with the pre-pregnancy level (Koh et al., 2014). To determine whether increased KLF9 expression contributes to the enhanced CYP2D6 expression during pregnancy, KLF9 recruitment to the CYP2D6 promoter was examined in tg-CYP2D6 mouse liver tissues collected at different gestational time points. The increased KLF9 expression was first verified by using Western blot (Fig. 7A). The ChIP results showed that KLF9 recruitment to the CYP2D6 promoter encompassing the K2 site increased during pregnancy and then returned to the pre-pregnancy levels after delivery, while KLF9 was not recruited to distal CYP2D6 genomic region without KLF9 binding site (Fig. 7B). We further examined whether the increased KLF9 expression during pregnancy can also influence other target genes. KLF9 is known to transrepress Cyp1a1 promoter via direct KLF9 binding to the promoter (Imataka et al., 1992). As expected, our results showed that KLF9 recruitment to Cyp1a1 promoter was significantly increased at term pregnancy (Fig. 7C), and this led to decreased Cyp1a1 expression (Fig. 7D). We also examined the recruitment of KLF9 recruitment to the promoters of previously known HNF4α target genes, Hes6 and ApoC2. KLF9 recruitment to ApoC2 promoter was minimal (data not shown), but KLF9 was unexpectedly found to be enriched on the promoter of Hes6 (Fig. 7E; KLF9/IgG signal ratio >1), indicating the presence of sequences for direct or indirect KLF9 binding in Hes6 promoter. Interestingly, however, the extent of KLF9 recruitment to the promoter did not differ among different time points (Fig. 7E).
Fig. 7.
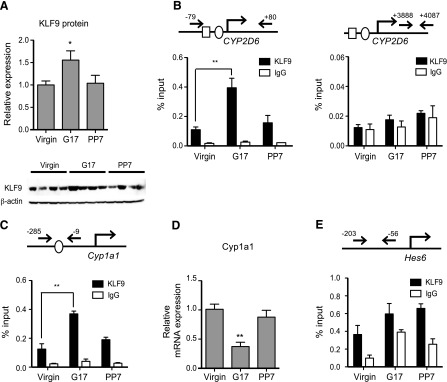
KLF9 recruitment to CYP2D6 promoter increases during pregnancy. Liver tissues were collected from tg-CYP2D6 mice at pre-pregnancy (virgin), 17 days of pregnancy (G17), or 7 days postpartum (PP7). (A) Protein expression levels of the KLF9 and β-actin were determined by Western blot analysis (n = 4, mean ± S.D.; *P < 0.05 versus virgin). (B, C, and E) ChIP assays were performed using antibodies against KLF9. KLF9 recruitment to CYP2D6 (B), Cyp1a1 (C), and Hes 6 (E) was examined (n = 4/time point, mean ± S.D.; * P < 0.05, **P < 0.01 versus virgin). The circle and rectangle in the gene promoters represent KLF9 and HNF4α binding sites, respectively. (D) mRNA expression level of Cyp1a1 was determined by qRT-PCR (n = 4, mean ± S.D.; **P < 0.01 versus virgin).
Discussion
CYP2D6-mediated drug metabolism is increased during pregnancy, but the underlying mechanisms remain to be fully elucidated. In our previous study, using tg-CYP2D6 mice as an in vivo model, we recapitulated CYP2D6 induction during pregnancy and established the key role of HNF4α in CYP2D6 induction during pregnancy (Koh et al., 2014). In this study, we report that KLF9 is a novel regulator of CYP2D6 expression, enhancing HNF4α-mediated transcription from the CYP2D6 promoter and potentially contributing to CYP2D6 induction during pregnancy.
Results from our previous study suggest that pregnancy enhances HNF4α transactivation in a gene-specific manner (Koh et al., 2014). For example, while the mRNA level of CYP2D6 and HNF4α recruitment to CYP2D6 promoter were increased during pregnancy, this did not occur with other HNF4α target genes (e.g., Hes6). In our current study, we searched for transcription factors that may be involved in CYP2D6 regulation by fine-tuning HNF4α activity. In tg-CYP2D6 mice, we found that expression of seven hepatic transcription factors (including KLF9) increases during pregnancy as compared with pre-pregnancy or after delivery. The identified transcription factors modulated CYP2D6 promoter activity to a minor extent when tested individually. However, when their effects on HNF4α-mediated transactivation of CYP2D6 promoter were examined, KLF9 was found to significantly enhance the HNF4α action on CYP2D6 promoter activity, indicating functional synergy between KLF9 and HNF4α. Results from deletion and mutation assays indicate that direct binding of KLF9 and HNF4α to the proximal region of the CYP2D6 promoter likely contributes to the functional interactions between two transcription factors. Importantly, the HNF4α response element located at -53/-41 and the KLF9-binding site at -22/-14 (i.e., K2) were found to play a key role. However, direct physical interaction between HNF4α and KLF9 appears unnecessary for this functional synergy based on the results from ChIP assays. Intermediary coregulators may be involved in the functional interaction between KLF9 and HNF4α. For example, a recent study showed that GATA4 forms a complex with KLF9 and HNF4α, and this interaction is important for activating genes involved in thyroid hormone homeostasis (Ohguchi et al., 2008). Taken together, our study revealed a novel role for KLF9 that it regulates CYP2D6 expression by enhancing HNF4α transactivation of CYP2D6 promoter activity.
What causes the up-regulation of KLF9 during pregnancy remains unclear. Previous studies have shown that cortisol induces KLF9 mRNA expression in keratinocytes and the hippocampus (Bagamasbad et al., 2012; Sporl et al., 2012). Additionally, in developing rat brain, thyroid hormone up-regulates KLF9, increasing the number and length of neurites (Denver et al., 1999). During pregnancy, it has been shown that the plasma concentrations of free cortisol and thyroid hormones increase 1.5- to 2-fold by the third trimester as compared with the pre-pregnancy levels (Soldin et al., 2004; Jung et al., 2011). Our initial investigations using primary human hepatocytes showed that cortisol or thyroid hormone did not alter CYP2D6 expression (data not shown). In line with these findings, there is no clinical report on drug-drug interactions between corticosteroids (or thyroid hormones) and CYP2D6 substrates to date. Currently, we cannot rule out a possibility that these results are affected by as-yet-unknown, inherent limitations of primary hepatocytes as a model to study CYP2D6 regulation. Further investigations would be necessary to determine the role of rising concentrations of cortisol and thyroid hormone in CYP2D6 induction during pregnancy.
Previous studies have shown that growth factors functionally modulate KLFs by triggering posttranslational modification or cytoplasm-to-nucleus shuttling of KLFs (Lomberk and Urrutia, 2005; Daftary et al., 2012). For example, the phosphorylation of KLF16 (one of the KLF family members) modulates its transcriptional activity assessed by reporter gene expression, and certain serum factors have been shown to significantly decrease the nuclear translocation of KLF16 and its transcriptional activity (Daftary et al., 2012). Also, mutations of serines to alanines in the KLF9 protein have been shown to significantly increase KLF9 transactivation of a target gene promoter (Pei et al., 2011), suggesting that phosphorylation of KLF9 may govern functionality of KLF9. During pregnancy, multiple physiologic changes occur along with alterations in plasma concentrations of various hormones and growth factors. For example, placenta growth factor is a vascular endothelial growth factor uniquely produced by placenta, and its levels increase gradually over the course of pregnancy (Tjoa et al., 2001). Whether any of these pregnancy-specific growth factors could modulate KLF9 activity and subsequently CYP2D6 expression during pregnancy remains to be determined.
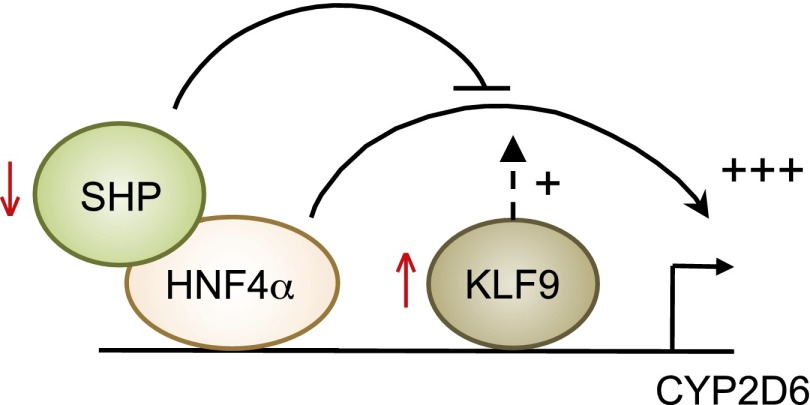
We previously showed that SHP is a repressor of HNF4α transactivation of CYP2D6 promoter activity and that pregnancy is accompanied by decreased expression of SHP (Koh et al., 2014). Considering that both SHP and KLF9 modulate HNF4α activity, we examined whether any crosstalk exists between SHP and KLF9 for HNF4α-induced transactivation of CYP2D6. Results from our study indicate that the changes in expression of SHP and KLF9 (i.e., decreased and increased expression, respectively) lead to synergistic HNF4α-induced transactivation of CYP2D6 promoter (Fig. 8). This interplay may potentially account for the differential regulation of HNF4α target genes during pregnancy. Possibly, CYP2D6 expression is activated by synergistic functional interaction among SHP, KLF9, and HNF4α. This functional interaction appears to occur in a gene-specific manner during pregnancy. For example, we found Hes6 to be another gene where both KLF9 and HNF4α are enriched at its promoter; however, despite the increased expression of KLF9 at term pregnancy, KLF9 recruitment at the Hes6 promoter did not change during pregnancy. The underlying mechanisms for gene-specific interplay among KLF9, SHP, and HNF4α warrant further investigation.
Fig. 8.
Working model for CYP2D6 induction during pregnancy. The working model illustrates the interplay of SHP and KLF9 in regulating CYP2D6 expression.
In conclusion, using CYP2D6-humanized mice, we identified KLF9 as a novel regulator of CYP2D6 expression during pregnancy, providing mechanistic insight into CYP2D6 induction during pregnancy. These results may provide a basis to improve drug therapy during pregnancy. Also, considering the roles of CYP2D6 in the pathophysiology of neurologic diseases (in addition to drug metabolism), better understanding of the regulation of CYP2D6 expression may allow for the identification of new drug targets. In this regard, whether the regulatory pathways involving KLF9, SHP, and HNF4α play a role in governing CYP2D6 expression in nonpregnant subjects is currently being investigated.
Supplementary Material
Acknowledgments
The authors thank Dr. Wooin Lee for critical reading of the manuscript.
Abbreviations
- ChIP
chromatin immunoprecipitation
- HEK293T
human embryonic kidney 293T cells
- HNF4α
hepatocyte nuclear factor 4α
- KLF9
Krüppel-like factor 9
- PP
postpartum
- PR
progesterone receptor
- qRT-PCR
real-time quantitative reverse-transcription polymerase chain reaction
- SHP
small heterodimer partner
Authorship Contributions
Participated in research design: Koh, Pan, McLachlan, Jeong.
Conducted experiments: Koh, Pan.
Contributed new reagents or analytic tools: Urrutia.
Performed data analysis: Koh, Pan, Zhang, McLachlan, Jeong.
Wrote or contributed to the writing of the manuscript: Koh, Pan, Zhang, McLachlan, Urrutia, Jeong.
Footnotes
This work was supported by the National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant HD065532], the National Institutes of Health (Grant DK52913) to R.U., and the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567)
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Bagamasbad P, Ziera T, Borden SA, Bonett RM, Rozeboom AM, Seasholtz A, Denver RJ. (2012) Molecular basis for glucocorticoid induction of the Krüppel-like factor 9 gene in hippocampal neurons. Endocrinology 153:5334–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns W, Smith CA, McLaren AW, Wolf CR. (1996) Characterization of the human cytochrome P4502D6 promoter. A potential role for antagonistic interactions between members of the nuclear receptor family. J Biol Chem 271:25269–25276. [DOI] [PubMed] [Google Scholar]
- Corchero J, Granvil CP, Akiyama TE, Hayhurst GP, Pimprale S, Feigenbaum L, Idle JR, Gonzalez FJ. (2001) The CYP2D6 humanized mouse: effect of the human CYP2D6 transgene and HNF4α on the disposition of debrisoquine in the mouse. Mol Pharmacol 60:1260–1267. [DOI] [PubMed] [Google Scholar]
- Daftary GS, Lomberk GA, Buttar NS, Allen TW, Grzenda A, Zhang J, Zheng Y, Mathison AJ, Gada RP, Calvo E, et al. (2012) Detailed structural-functional analysis of the Krüppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J Biol Chem 287:7010–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver RJ, Ouellet L, Furling D, Kobayashi A, Fujii-Kuriyama Y, Puymirat J. (1999) Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. Evidence for a role in neurite outgrowth. J Biol Chem 274:23128–23134. [DOI] [PubMed] [Google Scholar]
- Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. (2007) Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol 27:1407–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FJ. (2008) Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet 23:2–7. [DOI] [PubMed] [Google Scholar]
- Högstedt S, Lindberg B, Peng DR, Regårdh CG, Rane A. (1985) Pregnancy-induced increase in metoprolol metabolism. Clin Pharmacol Ther 37:688–692. [DOI] [PubMed] [Google Scholar]
- Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. (1992) Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J 11:3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Cho H, O’Connell M, Lee JK. (2005) Rank-invariant resampling based estimation of false discovery rate for analysis of small sample microarray data. BMC Bioinformatics 6:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Thatte J, Braciale T, Ley K, O’Connell M, Lee JK. (2003) Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics 19:1945–1951. [DOI] [PubMed] [Google Scholar]
- Jung C, Ho JT, Torpy DJ, Rogers A, Doogue M, Lewis JG, Czajko RJ, Inder WJ. (2011) A longitudinal study of plasma and urinary cortisol in pregnancy and postpartum. J Clin Endocrinol Metab 96:1533–1540. [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA, et al. (2005) FGF-21 as a novel metabolic regulator. J Clin Invest 115:1627–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchheiner J, Seeringer A, Godoy AL, Ohmle B, Maier C, Beschoner P, Sim EJ, Viviani R. (2011) CYP2D6 in the brain: genotype effects on resting brain perfusion. Mol Psychiatry 16:237,333–241. [DOI] [PubMed] [Google Scholar]
- Koh KH, Pan X, Shen HW, Arnold SL, Yu AM, Gonzalez FJ, Isoherranen N, Jeong H. (2014) Altered expression of small heterodimer partner governs cytochrome P450 (CYP) 2D6 induction during pregnancy in CYP2D6-humanized mice. J Biol Chem 289:3105–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Cha EY, Jung HJ, Shon JH, Kim EY, Yeo CW, Shin JG. (2008) Genetic polymorphism of hepatocyte nuclear factor-4α influences human cytochrome P450 2D6 activity. Hepatology 48:635–645. [DOI] [PubMed] [Google Scholar]
- Lemberger T, Saladin R, Vázquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J. (1996) Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem 271:1764–1769. [DOI] [PubMed] [Google Scholar]
- Lomberk G, Urrutia R. (2005) The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem J 392:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Peng Q, Zeng Z, Wang J, Deng Y, Xie L, Mo C, Zeng J, Qin X, Li S. (2014) CYP2D6 phenotypes and Parkinson’s disease risk: a meta-analysis. J Neurol Sci 336:161–168. [DOI] [PubMed] [Google Scholar]
- Ohguchi H, Tanaka T, Uchida A, Magoori K, Kudo H, Kim I, Daigo K, Sakakibara I, Okamura M, Harigae H, et al. (2008) Hepatocyte nuclear factor 4α contributes to thyroid hormone homeostasis by cooperatively regulating the type 1 iodothyronine deiodinase gene with GATA4 and Kruppel-like transcription factor 9. Mol Cell Biol 28:3917–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Yao Y, Yang Y, Liao K, Wu JR. (2011) Krüppel-like factor KLF9 regulates PPARγ transactivation at the middle stage of adipogenesis. Cell Death Differ 18:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen RC, Eason RR, McQuown JR, Linz AL, Kang TJ, Chatman L, Jr, Till SR, Fujii-Kuriyama Y, Simmen FA, Oh SP. (2004) Subfertility, uterine hypoplasia, and partial progesterone resistance in mice lacking the Krüppel-like factor 9/basic transcription element-binding protein-1 (Bteb1) gene. J Biol Chem 279:29286–29294. [DOI] [PubMed] [Google Scholar]
- Soldin OP, Tractenberg RE, Hollowell JG, Jonklaas J, Janicic N, Soldin SJ. (2004) Trimester-specific changes in maternal thyroid hormone, thyrotropin, and thyroglobulin concentrations during gestation: trends and associations across trimesters in iodine sufficiency. Thyroid 14:1084–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spörl F, Korge S, Jürchott K, Wunderskirchner M, Schellenberg K, Heins S, Specht A, Stoll C, Klemz R, Maier B, et al. (2012) Krüppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc Natl Acad Sci USA 109:10903–10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjoa ML, van Vugt JM, Mulders MA, Schutgens RB, Oudejans CB, van Wijk IJ. (2001) Plasma placenta growth factor levels in midtrimester pregnancies. Obstet Gynecol 98:600–607. [DOI] [PubMed] [Google Scholar]
- Tracy TS, Venkataramanan R, Glover DD, Caritis SN, National Institute for Child Health and Human Development Network of Maternal-Fetal-Medicine Units (2005) Temporal changes in drug metabolism (CYP1A2, CYP2D6 and CYP3A Activity) during pregnancy. Am J Obstet Gynecol 192:633–639. [DOI] [PubMed] [Google Scholar]
- Wadelius M, Darj E, Frenne G, Rane A. (1997) Induction of CYP2D6 in pregnancy. Clin Pharmacol Ther 62:400–407. [DOI] [PubMed] [Google Scholar]
- Yu AM, Idle JR, Gonzalez FJ. (2004) Polymorphic cytochrome P450 2D6: humanized mouse model and endogenous substrates. Drug Metab Rev 36:243–277. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Raimundo S, Eichelbaum M. (2004) Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol 369:23–37. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhang XL, Michel FJ, Blum JL, Simmen FA, Simmen RC. (2002) Direct interaction of the Krüppel-like family (KLF) member, BTEB1, and PR mediates progesterone-responsive gene expression in endometrial epithelial cells. Endocrinology 143:62–73. [DOI] [PubMed] [Google Scholar]
- Zhang XL, Zhang D, Michel FJ, Blum JL, Simmen FA, Simmen RC. (2003) Selective interactions of Kruppel-like factor 9/basic transcription element-binding protein with progesterone receptor isoforms A and B determine transcriptional activity of progesterone-responsive genes in endometrial epithelial cells. J Biol Chem 278:21474–21482. [DOI] [PubMed] [Google Scholar]
- Zhou T, Zhang Y, Macchiarulo A, Yang Z, Cellanetti M, Coto E, Xu P, Pellicciari R, Wang L. (2010) Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J Biol Chem 285:24871–24881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.