Abstract
Background
Alterations in stress-related gene-expression may play a role in stress-related drinking and the risk for alcohol dependence.
Methods
Microarrays were used to measure changes in gene-expression in peripheral blood in non-smoking, social drinking subjects exposed to three types of personalized imagery: neutral, stressful (but not alcohol-related), and alcohol-related cues. Gene-expression was measured at baseline, immediately after, and 1 hour after stimulus presentation. Subjects were allowed to drink up to 750cc of beer in a “taste-test” following stimulus presentation in each imagery condition, and the amount of beer consumed was recorded. Gene-expression levels were compared in 2 groups of non-smoking subjects (n=11/group): heavy drinkers (HD, defined as regular alcohol use over the past year of at least 8 standard drinks/week for women and at least 15 standard drinks/week for men), and moderate drinkers (MD, defined as up to 7 standard drinks/week for women and 14 standard drinks/week for men). Expression of microRNA10 (miR-10a) and microRNA 21 (miR-21) was assessed by quantitative real-time PCR (qRT-PCR).
Results
After correction for multiple testing (FDR<0.05), 79 genes were identified that changed by > 1.3 fold in the HD group, but not the MD group, following exposure to stress. No changes were observed for any of these genes in either group following exposure to neutral or alcohol-related imagery. Pathway analysis suggested that many of these genes, form part of the TAR-RNA binding protein (TRBP)-associated complex and are positively regulated by miR-10a and MiR-21. Expression of both miR-10a and miR-21 was up-regulated following psychological stress in HD, but not MD subjects, however the differences between groups were not statistically significant. Expression levels of both microRNAs was correlated (miR-10a, R2= 0.59, miR-21 R2= 0.57) with amount drunk in HD, but not MD subjects.
Conclusions
Expression of miR-10a, miR-21 and several of their target genes is regulated by acute psychological stress, and is correlated with stress-induced drinking in a laboratory setting. Alterations in miRNA expression may be one mechanism linking psychological stress with changes in gene-expression and increased alcohol intake in binge/heavy drinkers.
Keywords: alcohol dependence, microRNA, stress-related drinking behavior, gene-expression, microarray, TAR-RNA binding protein (TRBP)-associated complex
Introduction
A variety of clinical and pre-clinical studies have implicated alterations in the neural mechanisms mediating the response to stress and the hypothalamic-pituitary-adrenal (HPA) axis in both the development of alcohol dependence and the risk for relapse in recovered alcoholics (Sinha et al., 2009, Sinha et al., 2011). Abnormalities in HPA function have been identified in alcohol-dependent subjects in a variety of states including active drinking, acute withdrawal, and 4 weeks post-withdrawal (Adinoff et al., 2005). Alcohol dependent subjects show higher basal levels of cortisol, lower basal levels of adrenocorticotropic hormone (ACTH), and a blunted cortisol/ACTH response to stress (Sinha et al., 2011). Alcohol-dependent subjects exhibit altered responses to both serotonergic and noradrenergic agonists (Krystal et al., 1996) and altered cardiovascular responses to physical and psychological stress (Bernardy et al., 2003). Animal studies show that alcohol treated rats have elevated basal levels of epinephrine and norepinephrine, and a blunted response to mild stress (Rasmussen et al., 2006). Stress induced anxiety and stress- induced alcohol craving as well as dysregulated HPA response have been shown to predict subsequent treatment outcomes including days in aftercare alcohol treatment and time to alcohol relapse (Sinha et al., 2011).
In animal models, alcohol has been shown to modulate the activity of a number of important stress-related signaling pathways including those involving cyclic adenosine 3′, 5′-monophosphate (cAMP)-dependent protein kinase A (PKA), protein kinase C (PKC), the tyrosine kinase Fyn, and phospholipase D (PLD) (reviewed in (Newton and Messing, 2006)). Alterations in these molecular pathways have been implicated in the molecular pathogenesis of addiction in general (Chao and Nestler, 2004), and addiction to alcohol in particular (Rodd et al., 2007). However, with current technology there are no methods to examine changes in gene-expression in the brains of living subjects. Thus, the extent to which such changes have occurred in the brain or may serve to regulate drinking behavior cannot be assessed while the patient is alive.
Peripheral cells, including lymphocytes, express many of the same signaling molecules implicated as possible mediators of the long-term physiological effects of alcohol. These include the CREB, phosphatidyl inositol, ERK, and EGFR pathways. Lymphocytes also express a large number of neurotransmitter receptors including dopamine receptors, cannabinoid receptors, β-adrenergic receptors, mineralocorticoid and glucocorticoid receptors, muscarinic acetylcholine receptors, substance P receptors, GABA A receptors, serotonin receptors and transporters (reviewed in (Gladkevich et al., 2004)). A microarray study evaluating the comparability of gene expression in blood and brain (Sullivan et al., 2006) found that whole blood shares significant gene expression similarities with multiple CNS tissues. These similarities in gene-expression between neural tissue and peripheral lymphoid tissue, combined with the inability to directly sample gene-expression in the living brain, have led to the suggestion that gene-expression in blood could provide useful biomarkers related to alcoholism and addiction (Mayfield and Harris, 2009).
The development of alcohol dependence has been postulated to occur along a continuum from voluntary consumption motivated by rewarding or hedonic effects to habitual and ultimately compulsive use (Everitt and Robbins, 2005). A key factor, hypothesized to play a role in mediating the transition from voluntary to compulsive use is stress-induced alterations in gene-expression (Sinha, 2009b). However, little, if any, information is available regarding how such changes may influence drinking behavior in human subjects. In a previous study, we demonstrated alterations in the expression of multiple stress-related genes in the peripheral blood of subjects with alcohol dependence compared to either heavy or moderate drinkers (Beech et al., 2012). However, since gene-expression was analyzed at only a single time-point, we were unable to determine the relationship of these changes to drinking behavior.
In the present study, we used a well-characterized laboratory model for stress induced drinking (Fox et al., 2012, Sinha et al., 2009) in combination with real-time monitoring of stress-induced changes in peripheral blood gene-expression to assess the relationships among stress, gene-expression and drinking behavior in two groups of subjects: heavy drinkers (HD, defined as regular alcohol use over the past year of at least 8 standard drinks/week for women and at least 15 standard drinks/week for men), and moderate drinkers (MD, defined as up to 7 standard drinks/week for women and 14 standard drinks/week for men). In this paradigm, subjects were exposed to three types of personalized imagery: neutral, stressful (but not alcohol-related), and alcohol-related cues. Gene-expression was measured at baseline, immediately after, and 1 hour after stimulus presentation. Subjects were allowed to drink up to 750cc of beer in an “alcohol taste-test” following stimulus presentation in each imagery condition, and the amount of beer consumed was recorded. Pathway analysis using DAVID software (Huang et al., 2009) and GeneGO Metacore® software was used to identify functional pathways and potential regulatory factors among those genes showing altered expression in the stress condition. Expression of potential regulatory microRNAs related to these genes was assessed using qRT-PCR.
Materials and Methods
Subjects
All procedures involving human subjects were approved by the Yale Human Investigation Committee and are in accordance with the Helsinki declaration of 1975. All subjects provided written informed consent at the time of enrollment in the study. Subjects were non-smoking social drinkers who reported “liking beer”, between the ages of 21 to 50 years, and were recruited from the community through local advertisements. Subjects were admitted for a 3- day hospital stay to the Yale Hospital Research Unit at Yale-New Haven Hospital for participation in the study. During this period, they were required to stay on the unit, within a controlled environment. All subjects reported no lifetime or current history of any drug dependence (including alcohol dependence), did not meet criteria for any Axis I DSM-IV psychiatric diagnoses, and provided negative urine toxicology screens during initial appointments and at inpatient admission for laboratory sessions. To define the relationship between gene-expression and drinking behavior, the subjects were divided into 2 subgroups: HD and MD, as defined by the NIAAA (NIAAA, 2005). Subjects with current or prior history of binge drinking or alcohol abuse, but not dependence, could be included in the HD group. Drinking behavior was further characterized using the Alcohol Use Disorders Identification Test (AUDIT) (Babor et al., 2001). The AUDIT is a 10-item screening instrument designed to identify drinking behaviors and distinguish between low- risk drinkers and individuals with hazardous and harmful patterns of alcohol consumption to provide the appropriate intervention. Subjects were also asked how many days they had consumed alcohol in the past 30 days.
Exclusion criteria included current nicotine users/smokers, current dependence on another psychoactive substance, current use of opiates or past history of opiate abuse/dependence; current use of any psycho- active drugs, including anxiolytics, antidepressants, naltrexone, or antabuse; any psychotic disorder or current psychiatric symptoms requiring specific attention, including need for psychiatric medications for current major depression and anxiety disorders; any significant current medical condition such as neurological, cardiovascular, endocrine, renal, liver, and thyroid pathology; subjects on medications for any medical condition; women on oral contraceptives, peri- and postmenopausal women, and those with hysterectomies; and pregnant and lactating women.
Imagery script development procedures
Before the laboratory sessions, guided imagery scripts for stress, alcohol cue and neutral relaxing states were developed for each individual subject. The stress imagery script was based on that subject’s description of a recent personal stressful event that was experienced as ‘most stressful’. Most stressful was determined by having the subjects rate their individual level of distress on a 10-point Likert scale where ‘1 = not at all stressful’ and ‘10 = the most stress they felt recently in their life’. Only situations rated as eight or above were accepted as appropriate for script development. Examples include a breakup with a significant other or unemployment-related stress. Trauma-related situations and those with explicit alcohol cues were not allowed. The alcohol-related cue script was based on individual situations that included alcohol-related stimuli and resulted in subsequent alcohol use (e.g. buying alcohol and being at a bar, watching others drink alcohol). Alcohol-related situations that occurred in the context of negative affect or psychological distress were not allowed. A neutral script was developed from the participants’ individual experiences of commonly experienced neutral- relaxing situations, such as a summer day relaxing at the beach or a fall day reading at the park. Details of each elicited situation was described using the scene construction questionnaire, based on methods developed by Lang et al (Lang et al., 1980) and further adapted in our previous studies (Sinha, 2009a). Scripts were developed using a standardized format, based on specific stimulus and response details of each situation and then audiotaped for presentation in the laboratory sessions. Order of imagery condition and script type was randomly assigned across the 3 experimental days and counterbalanced across subjects.
Habituation and imagery training session
One day before the laboratory sessions subjects were brought into the testing room where they were acclimatized to specific aspects of the study procedures, such as the subjective rating forms and then trained in relaxation and imagery procedures (Sinha, 2001).
Laboratory sessions
On each of the three testing days, subjects had a standard healthy lunch at noon, and were brought into the testing room at 1400 hours by the research nurse. A blood pressure cuff was placed on the subject’s preferred arm to monitor blood pressure and a pulse sensor was placed on the subject’s forefinger to obtain a measure of pulse. This was followed by a 1-h adaptation period. At 1500 hours, subjects were provided headphones and the audiotape presented the instructions for the imagery procedure and the script for guided imagery. The length of each script was approximately 5min. After imagery, subjects remained in the testing room for an additional 75min to examine recovery from the imagery exposure and for repeated assessments of mood and alcohol craving. In addition, immediately following imagery and assessments, subjects were also presented with the “alcohol taste test” on each day, a widely used and validated procedure to experimentally assess motivation for alcohol consumption and amount of alcohol consumed following a variety of challenges (de Wit et al., 2003).
After the imagery period and ratings of mood and craving, subjects were presented with a tray of two 12-Oz beer mugs with chilled beer and a glass of water and ice. Subjects then participated in a 10 minute “taste test” in which they were asked to taste the beer in each mug to identify whether the brand and type of beer was the same or different in each mug. They were told that they could drink as much as they wished to make this determination. The amount of beer consumed on each experimental day was recorded. Blood draws for RNA collection were performed at baseline (1 hour prior to presentation of the guided imagery stimuli), immediately after and 1 hour after stimulus presentation on each of the test days. Blood samples for measurement of cortisol and ACTH were also collected at these times.
Cardiovascular Measures
A Critikon Dinamap 120 Patient Monitor (GE Medical Systems, Tampa, FL) was used to assess the BP, and a pulse sensor was attached to the subject’s finger to provide a continuous measure of pulse.
HPA axis measures
ACTH and cortisol samples were obtained in heparinized tubes, and blood samples for basal norepinephrine determination were collected in tubes containing EGTA and reduced glutathione. All tubes were placed on ice immediately after drawing and then aliquoted after being centrifuged at 4°C within 30 minutes of collection. Blood samples for HPA axis measures were stored at −70°C and processed at the Yale Center for Clinical Investigation Core Laboratories using standard radioimmunoassay procedures, as described previously (Fox et al., 2012)
Sample Preparation and Microarray Analysis
Blood samples were collected directly into PAXgene blood RNA tubes (QIAGEN, Valencia, CA) and stored frozen at −80° C. until processing to reduce variation due to differences in sample processing. The choice to use whole blood rather than PBMCs or some other cell fraction for analysis of gene expression is based on several factors. Primary among these is the desire to capture gene expression profiles that are as close as possible to those that exist in vivo. Collecting blood samples directly into PAXgene blood RNA tubes, which lyse the cells and prevents degradation of the RNA present, prevents changes in gene expression associated with differences in storage or handling of the samples prior to RNA extraction. Total RNA and microRNA were isolated from 10 cc whole blood using the PAXgene Blood RNA Isolation kit (QIAGEN, Valencia, CA) using a modified protocol (Kruhoffer et al., 2007) that allows total RNA, microRNA, and DNA from a single PAXgene blood RNA tube. Total RNA samples were depleted of globin mRNA message using GLOBINclear hybridization capture technology (Ambion, Austin, TX). Globin-reduced total RNA underwent cDNA synthesis and overnight in vitro transcription utilizing the Illumina TotalPrep RNA Amplification Kit (Ambion). Biotinylated cRNA (1.5 μg) was hybridized onto an Illumina Sentrix Beadchip (Human-6v2) then scanned on a BeadArray Reader. Microarray hybridization and scanning were carried out at the Yale Center for Genome Analysis. All data have been deposited into the NCBI-GEO repository, accession number GSE59206.
qRT-PCR analysis
qRT-PCR for microRNA10 (miR-10a) and microRNA 21 (miR-21) was carried out using the TaqMan® “Universal PCR Master Mix” Protocol (Applied Biosystems) and Real-Time PCR probes listed on the NCBI Probe Database (miR-10a: TaqMan assay name: has-miR-10a, Assay ID: 000387; miR-21: TaqMan assay name: has-miR-21: Assay ID: 000397). Expression level of each miRNA immediately after and 1 hour after stimulus presentation was normalized to the baseline level of expression for that subject on the same test day.
Normalization and Data Analysis
Statistical analysis of microarray data was carried out at the Keck Foundation Biotechnology Biostatistics Resource (http://keck.med.yale.edu/biostats). Illumina BeadStudio software was used to generate probe and gene expression profiles of each sample. Quantile normalization was carried out using the package incorporated in the Illumina BeadStudio software package. Further statistical analysis was carried out on all genes with a detection p-value <0.01 as determined using the Illumina BeadStudio software (i.e. a 99% probability that expression was above background) in > 90% of samples. To identify genes showing differential expression, multivariate analysis of variance (MANOVA) for repeated measures (Hand, 1987) was carried out for each group of subjects (MD or HD) under each of the test conditions (Stress, Alcohol-Cue or Neutral) using Partek® Genomic Suite (Partek Inc., St. Louis, MO, USA.) Results were co-varied for the effects of age, race, sex, and batch. P-values were adjusted to control the group-wise false discovery rate (FDR) at <0.05 (Reiner et al., 2003). Network analysis was carried out using the DAVID Functional Classification tool (Huang et al., 2009) and GeneGO Metacore® software (GeneGO, Inc., Encinitas, CA, USA).
Results
Demographics and Drinking Behavior During Laboratory Sessions
Demographic information on the two groups of subjects is summarized in Table 1. MD (n=11) and HD (n=11) subjects did not differ significantly by sex, race, or age. As expected, the HD subjects had higher AUDIT (Babor et al., 2001) scores (HD=13.1, MD=4.5, p<.0001) and number of drinks/week (HD=16.7, MD=3.7, p=2.45E-05) than MD subjects. There was also a trend for higher number of drinking days during the past 30 days (HD=13.3, MD=7.2, p=0.06). In the laboratory sessions, the HD subjects drank more than the MD subjects during the taste test under each of the test conditions, however the amounts consumed by each group did not differ significantly by test condition (MD-neutral cue: 271.6 ± 62.8 ml, MD-alcohol cue: 267.9 ± 62.1 ml, MD-stress cue: 273.4 ± 55.8; HD-neutral cue: 483.8 ± 71.0, HD-alcohol cue: 518.4 ± 68.8, HD-stress cue: 543.2 ± 72.9).
Table 1.
Subjects included in between group comparisons- Summary of demographic information.
| MD subjects (n=11) | HD subjects (n=11) | P-value | |
|---|---|---|---|
| Male | 8 (73%) | 8 (73%) | N.S. |
| Female | 3 (27%) | 3 (27%) | N.S. |
| Caucasian | 8 (73%) | 10 (91%) | N.S. |
| African American | 3 (27%) | 1 (9%) | N.S. |
| Age (avg.) | 28.5 | 29.9 | N.S. |
| AUDIT score | 4.5 | 13.1 | <.0001 |
| # Drinking days in past month | 7.2 | 13.3 | 0.06 |
| # of Drinks/ week (avg.) | 3.7 | 16.7 | 2.45E-05 |
Categorical variables (race, sex) were compared between groups using the chi-squared statistic, while numerical variables (age, AUDIT scores, number of drinking days/month, and number of drinks/week) were compared using Student’s t-test.
Differential Gene Expression in Heavy vs. moderate Drinkers During Stress, Alcohol Cue and Neutral Conditions
To identify genes showing differential expression in each group under the each of the test conditions (Stress, Alcohol-Cue or Neutral), a repeated measures ANOVA was carried out for each gene on the array. Table 2 shows the number of genes showing differential regulation with a nominal p value <0.05 and Fold change > 1.3 in each group immediately after cue presentation and 1 hour after cue presentation. As shown in Table 2, there were 92 genes with altered expression (68 up-regulated and 24 down-regulated) in heavy drinkers one hour after the stress cue presentation, while none of the other conditions showed more than 2 genes with altered expression. To compare the proportion of genes with differential expression levels between conditions, we performed a statistical test between the rates based on Poisson approximations. The results showed a highly significant difference between the HD-group/stress condition and all of the other conditions tested (p= 1.7E-20).
Table 2.
Number of Differentially expressed genes (fold-change > 1.3, p < 0.05) seen in HD and MD subjects immediately after and 1 hour after cue presentation under each test condition (Neutral, Alcohol-cue, or Stress).
|
|
|||||
|---|---|---|---|---|---|
| Drinking Group | Cue | # of differentially expressed genes: Immediately after cue | # of differentially expressed genes: 1 hr. after cue | ||
|
| |||||
| Up-regulated FC > 1.3 | Down-regulated FC > 1.3 | Up-regulated FC > 1.3 | Down-regulated FC > 1.3 | ||
|
|
|||||
| Moderate | Neutral | 0 | 0 | 0 | 0 |
| Moderate | Alcohol | 0 | 0 | 2 | 0 |
| Moderate | Stress | 0 | 0 | 0 | 0 |
| Heavy | Neutral | 1 | 0 | 0 | 0 |
| Heavy | Alcohol | 0 | 0 | 1 | 0 |
| Heavy | Stress | 2 | 0 | **68 | **24 |
The proportion of differentially expressed genes differed significantly between the HD-Stress condition and all other conditions testes (P=1.7E-20)
Of the 92 genes showing stressed-induced changes in expression in the HD group, 79 (55 up-regulated and 24 down-regulated) remained significant after correction for multiple testing (FDR< 0.05). These genes, and the associated fold-change and FDR-corrected p-values are listed in Table 3. To identify functional relationships among the differentially expressed genes we carried out pathway analysis using DAVID (Huang et al., 2009) and GeneGO Metacore® software. Analysis using the GeneGO software package identified three process networks that were significantly associated with the list of differentially expressed genes: Translation: Elongation-Termination (P=7.02E-09); Translation: Initiation (P=1.28E-05); and Proteolysis in cell cycle and apoptosis (P=1.93E-03). Analysis with DAVID identified 2 biological processes associated with our gene list: Translational elongation (GO:0006414; FDR corrected P=1.60E-04); and Translation (GO:0006412; FDR corrected P=1.36E-02). Examination of the “REACTOME” list in DAVID associated with our gene list showed a group of 6 genes (as well as 2 transcripts with predicted similarity) associated with “3′ UTR-mediated translational regulation” (the only process significantly associated with this list).
Table 3.
Genes showing altered expression in heavy drinkers 1 hour after exposure to stress cue.
| Table 3a: Genes showing selective up-regulation 1 hour after psychological stress in heavy drinkers. | ||||
|---|---|---|---|---|
| GENE SYMBOL | Level of Induction in Heavy drinkers: (1 hr post-stress/ baseline) | FDR corrected p-value | REFSEQ_ID | DEFINITION |
| LOC653773 | 1.65 | 0.044 | XM_938755.2 | PREDICTED: Homo sapiens similar to ribosomal protein L31 (LOC653773), mRNA. |
| RPL9 | 1.64 | 0.012 | NM_001024921.2 | Homo sapiens ribosomal protein L9 (RPL9), transcript variant 2, mRNA. |
| LOC389404 | 1.63 | 0.016 | XR_018133.2 | PREDICTED: Homo sapiens misc_RNA (LOC389404), miscRNA. |
| LOC100134504 | 1.58 | 0.046 | XM_001725687.1 | PREDICTED: Homo sapiens hypothetical protein LOC100134504 (LOC100134504), mRNA. |
| LOC727865 | 1.57 | 0.029 | XR_038568.1 | PREDICTED: Homo sapiens misc_RNA (LOC727865), miscRNA. |
| LOC727865 | 1.53 | 0.023 | XR_038568.1 | PREDICTED: Homo sapiens misc_RNA (LOC727865), miscRNA. |
| LOC644315 | 1.53 | 0.024 | XR_017529.2 | PREDICTED: Homo sapiens misc_RNA (LOC644315), miscRNA. |
| LOC651436 | 1.52 | 0.027 | XM_940587.1 | PREDICTED: Homo sapiens similar to ribosomal protein L9 (LOC651436), mRNA. |
| LOC100129742 | 1.51 | 0.023 | XR_037977.1 | PREDICTED: Homo sapiens misc_RNA (LOC100129742), miscRNA. |
| RPS3A | 1.51 | 0.023 | NM_001006.3 | Homo sapiens ribosomal protein S3A (RPS3A), mRNA. |
| RSL24D1 | 1.50 | 0.023 | NM_016304.2 | Homo sapiens ribosomal L24 domain containing 1 (RSL24D1), mRNA. |
| RPS3A | 1.50 | 0.027 | NM_001006.3 | Homo sapiens ribosomal protein S3A (RPS3A), mRNA. |
| LOC100132291 | 1.49 | 0.038 | XM_001726077.1 | PREDICTED: Homo sapiens similar to hCG2027326 (LOC100132291), mRNA. |
| RPL9 | 1.49 | 0.044 | NM_001024921.2 | Homo sapiens ribosomal protein L9 (RPL9), transcript variant 2, mRNA. |
| EVI2A | 1.46 | 0.023 | NM_014210.2 | Homo sapiens ecotropic viral integration site 2A (EVI2A), transcript variant 2, mRNA. |
| LOC100131387 | 1.44 | 0.040 | XM_001725100.1 | PREDICTED: Homo sapiens similar to mCG146274 (LOC100131387), mRNA. |
| LOC285741 | 1.44 | 0.016 | XR_017420.2 | PREDICTED: Homo sapiens misc_RNA (LOC285741), miscRNA. |
| LOC647673 | 1.43 | 0.024 | XM_936731.1 | PREDICTED: Homo sapiens similar to Translationally-controlled tumor protein (TCTP) (p23) (Histamine-releasing factor) (HRF) (Fortilin) (LOC647673), mRNA. |
| LOC100128060 | 1.42 | 0.024 | XM_001723512.1 | PREDICTED: Homo sapiens similar to mCG19129, transcript variant 2 (LOC100128060), mRNA. |
| RPS17 | 1.41 | 0.047 | NM_001021.3 | Homo sapiens ribosomal protein S17 (RPS17), mRNA. |
| LOC391833 | 1.41 | 0.040 | XR_038761.1 | PREDICTED: Homo sapiens misc_RNA (LOC391833), miscRNA. |
| LOC646819 | 1.41 | 0.023 | XR_017610.2 | PREDICTED: Homo sapiens misc_RNA (LOC646819), miscRNA. |
| RPS3A | 1.40 | 0.023 | NM_001006.3 | Homo sapiens ribosomal protein S3A (RPS3A), mRNA. |
| LOC647030 | 1.40 | 0.038 | XR_041205.1 | PREDICTED: Homo sapiens misc_RNA (LOC647030), miscRNA. |
| HINT1 | 1.40 | 0.028 | NM_005340.3 | Homo sapiens histidine triad nucleotide binding protein 1 (HINT1), mRNA. |
| LOC646527 | 1.39 | 0.018 | XR_016632.2 | PREDICTED: Homo sapiens misc_RNA (LOC646527), miscRNA. |
| LOC100132499 | 1.38 | 0.020 | XM_001725693.1 | PREDICTED: Homo sapiens similar to mCG7602 (LOC100132499), mRNA. |
| LOC644790 | 1.38 | 0.024 | XM_927887.3 | PREDICTED: Homo sapiens hypothetical LOC644790 (LOC644790), mRNA. |
| HOPX | 1.38 | 0.004 | NM_139212.2 | Homo sapiens HOP homeobox (HOPX), transcript variant 3, mRNA. |
| LOC389156 | 1.37 | 0.020 | XR_019429.2 | PREDICTED: Homo sapiens misc_RNA (LOC389156), miscRNA. |
| LOC100130775 | 1.37 | 0.039 | XR_042325.1 | PREDICTED: Homo sapiens misc_RNA (LOC100130775), miscRNA. |
| CD52 | 1.36 | 0.036 | NM_001803.2 | Homo sapiens CD52 molecule (CD52), mRNA. |
| LOC729646 | 1.36 | 0.030 | XR_015611.2 | PREDICTED: Homo sapiens misc_RNA (LOC729646), miscRNA. |
| LOC730029 | 1.36 | 0.027 | XM_001724847.1 | PREDICTED: Homo sapiens similar to hCG1997137, transcript variant 2 (LOC730029), mRNA. |
| LOC727821 | 1.36 | 0.026 | XR_037166.1 | PREDICTED: Homo sapiens misc_RNA (LOC727821), miscRNA. |
| RPL31 | 1.35 | 0.049 | NM_000993.4 | Homo sapiens ribosomal protein L31 (RPL31), transcript variant 1, mRNA. |
| LY96 | 1.35 | 0.026 | NM_015364.2 | Homo sapiens lymphocyte antigen 96 (LY96), mRNA. |
| CDKN1B | 1.34 | 0.004 | NM_004064.2 | Homo sapiens cyclin-dependent kinase inhibitor 1B (p27, Kip1) (CDKN1B), mRNA. |
| CD69 | 1.34 | 0.012 | NM_001781.1 | Homo sapiens CD69 molecule (CD69), mRNA. |
| LOC641848 | 1.34 | 0.016 | XM_935588.1 | PREDICTED: Homo sapiens similar to ribosomal protein S3a (LOC641848), mRNA. |
| LOC100134273 | 1.34 | 0.023 | XM_001724343.1 | PREDICTED: Homo sapiens similar to mCG7602 (LOC100134273), mRNA. |
| RPS15A | 1.33 | 0.049 | NM_001019.4 | Homo sapiens ribosomal protein S15a (RPS15A), transcript variant 2, mRNA. |
| LOC388076 | 1.33 | 0.030 | XM_001722259.1 | PREDICTED: Homo sapiens hypothetical LOC388076 (LOC388076), mRNA. |
| LOC100132528 | 1.33 | 0.004 | XR_038720.1 | PREDICTED: Homo sapiens misc_RNA (LOC100132528), miscRNA. |
| GNG10 | 1.33 | 0.020 | NM_001017998.2 | Homo sapiens guanine nucleotide binding protein (G protein), gamma 10 (GNG10), mRNA. |
| LOC730029 | 1.32 | 0.026 | XM_001724848.1 | PREDICTED: Homo sapiens similar to hCG1997137, transcript variant 3 (LOC730029), mRNA. |
| GZMK | 1.32 | 0.019 | NM_002104.2 | Homo sapiens granzyme K (granzyme 3; tryptase II) (GZMK), mRNA. |
| LOC441506 | 1.31 | 0.016 | XR_017565.2 | PREDICTED: Homo sapiens misc_RNA (LOC441506), miscRNA. |
| LOC286512 | 1.31 | 0.012 | XR_038196.1 | PREDICTED: Homo sapiens misc_RNA (LOC286512), miscRNA. |
| SRP9 | 1.31 | 0.012 | NM_003133.2 | Homo sapiens signal recognition particle 9kDa (SRP9), mRNA. |
| LOC389156 | 1.31 | 0.030 | XR_017589.2 | PREDICTED: Homo sapiens misc_RNA (LOC389156), miscRNA. |
| LOC440737 | 1.30 | 0.019 | XM_496446.3 | PREDICTED: Homo sapiens similar to 60S ribosomal protein L35 (LOC440737), mRNA. |
| EEF1B2 | 1.30 | 0.019 | NM_001037663.1 | Homo sapiens eukaryotic translation elongation factor 1 beta 2 (EEF1B2), transcript variant 3, mRNA. |
| EFHA1 | 1.30 | 0.015 | NM_152726.1 | Homo sapiens EF-hand domain family, member A1 (EFHA1), mRNA. |
| GZMA | 1.30 | 0.024 | NM_006144.2 | Homo sapiens granzyme A (granzyme 1, cytotoxic T-lymphocyte- associated serine esterase 3) (GZMA), mRNA. |
| Table 3 b) Genes showing selective down-regulation in heavy drinkers 1 hour after psychological stress. | ||||
|---|---|---|---|---|
| GENE SYMBOL | Level of down- regulation in Heavy drinkers: (−1 *baseline /1 hr post-stress) | FDR corrected p-value | REFSEQ_ID | DEFINITION |
| ADIPOR1 | −1.30 | 0.012 | NM_015999.2 | Homo sapiens adiponectin receptor 1 (ADIPOR1), mRNA. |
| TNS1 | −1.30 | 0.027 | NM_022648.3 | Homo sapiens tensin 1 (TNS1), mRNA. |
| SELENBP1 | −1.30 | 0.043 | NM_003944.2 | Homo sapiens selenium binding protein 1 (SELENBP1), mRNA. |
| VWCE | −1.30 | 0.023 | NM_152718.2 | Homo sapiens von Willebrand factor C and EGF domains (VWCE), mRNA. |
| BAT3 | −1.31 | 0.004 | NM_080702.2 | Homo sapiens HLA-B associated transcript 3 (BAT3), transcript variant 2, mRNA. |
| BSG | −1.31 | 0.012 | NM_198590.1 | Homo sapiens basigin (Ok blood group) (BSG), transcript variant 3, mRNA. |
| ASCC2 | −1.31 | 0.023 | NM_032204.3 | Homo sapiens activating signal cointegrator 1 complex subunit 2 (ASCC2), mRNA. |
| IFIT1L | −1.31 | 0.016 | NM_001010987.1 | Homo sapiens interferon-induced protein with tetratricopeptide repeats 1-like (IFIT1L), mRNA. |
| RBM38 | −1.31 | 0.004 | NM_017495.4 | Homo sapiens RNA binding motif protein 38 (RBM38), transcript variant 1, mRNA. |
| FKBP8 | −1.31 | 0.012 | NM_012181.3 | Homo sapiens FK506 binding protein 8, 38kDa (FKBP8), mRNA. |
| EPB42 | −1.32 | 0.043 | NM_000119.1 | Homo sapiens erythrocyte membrane protein band 4.2 (EPB42), mRNA. |
| TMOD1 | −1.32 | 0.038 | NM_003275.2 | Homo sapiens tropomodulin 1 (TMOD1), mRNA. |
| E2F2 | −1.32 | 0.024 | NM_004091.2 | Homo sapiens E2F transcription factor 2 (E2F2), mRNA. |
| RUNDC3A | −1.32 | 0.016 | NM_006695.3 | Homo sapiens RUN domain containing 3A (RUNDC3A), mRNA. |
| ALAS2 | −1.33 | 0.038 | NM_001037967.1 | Homo sapiens aminolevulinate, delta-, synthase 2 (ALAS2), nuclear gene encoding mitochondrial protein, transcript variant 2, mRNA. |
| C16orf35 | −1.33 | 0.020 | NM_001039476.1 | Homo sapiens chromosome 16 open reading frame 35 (C16orf35), transcript variant 2, mRNA. |
| LOC389599 | −1.33 | 0.024 | XM_001131588.1 | PREDICTED: Homo sapiens similar to amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 2 (LOC389599), mRNA. |
| SLC6A8 | −1.34 | 0.012 | NM_005629.1 | Homo sapiens solute carrier family 6 (neurotransmitter transporter, creatine), member 8 (SLC6A8), mRNA. |
| KRT1 | −1.35 | 0.023 | NM_006121.3 | Homo sapiens keratin 1 (KRT1), mRNA. |
| SLC6A10P | −1.35 | 0.024 | NR_003083.2 | Homo sapiens solute carrier family 6 (neurotransmitter transporter, creatine), member 10 (pseudogene) (SLC6A10P) on chromosome 16. |
| ALAS2 | −1.36 | 0.024 | NM_001037968.1 | Homo sapiens aminolevulinate, delta-, synthase 2 (ALAS2), nuclear gene encoding mitochondrial protein, transcript variant 3, mRNA. |
| C5orf4 | −1.37 | 0.019 | NM_032385.3 | Homo sapiens chromosome 5 open reading frame 4 (C5orf4), mRNA. |
| HBA1 | −1.37 | 0.026 | NM_000558.3 | Homo sapiens hemoglobin, alpha 1 (HBA1), mRNA. |
| ATP6V0C | −1.37 | 0.019 | XM_001130742.1 | PREDICTED: Homo sapiens ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c (ATP6V0C), mRNA. |
Level of induction is the ratio of expression for a given gene at one hour after stress exposure/ its expression at baseline (i.e. 1-fold induction=no change). For down-regulated genes this number is expressed as the negative inverse of the fold induction (−1*baseline/ expression at 1 hour after stress). Thus, a 1.3 fold induction corresponds to 30% increase in expression while a −1.3 fold down- regulation corresponds to a 30% decrease.
a) Genes showing selective up-regulation 1 hour after psychological stress in heavy drinkers.
b) Genes showing selective down-regulation 1 hour after psychological stress in heavy drinkers.
Closer examination of the list of “3′ UTR-mediated translational-regulation”-related genes revealed that four of these genes (RPL9, RPL31, RPS3A, and RPS17) are part of the TAR-RNA binding protein (TRBP)-associated complex (Chi et al., 2011). Three of these ((RPL9, RPS3A, and RPS17) have been shown to be positively regulated by miR-10a (Orom et al., 2008), while the fourth (RPL31) has been reported to be positively regulated by a different microRNA, miR-21 (Li et al., 2009). Thus, changes in expression of these genes might be related to changes in specific regulatory microRNAs. To explore this hypothesis, we carried out qRT-PCR for both miR-10a and miR-21 using microRNA isolated from both MD and HD subjects at all three time-points on the stress days. Both miR-10a and MiR-21 showed increased expression 1 hour after the stress cue presentation in HD, but not MD subjects (miR-21: HD, fold induction=1.59 ± 0.32; MD, fold induction=1.01 ± 0.24; miR-10a: HD fold induction=1.24 ± 0.16, MD, fold induction=1.05 ± 0.15), however the differences between groups were not statistically significant.
Relationship Between Stress-induced MicroRNA (miR-10a and miR-21) Expression and Alcohol Consumption
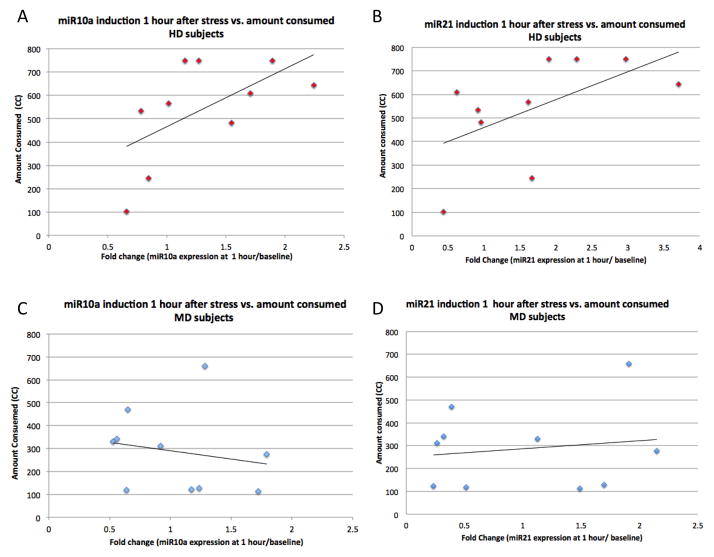
To better understand the relationship between these changes and individual differences in drinking behavior in the two groups, we plotted the fold-induction of miR-10a and miR-21 for each subject 1 hour after the stress cue presentation against the amount of beer consumed by that subject during the 10 minute taste test. As shown in Figure 1, in HD subjects, induction of both miRNA10a (Figure 1A) and miRNA21 (Figure 1B) 1 hour after exposure to the stress cue was correlated with the amount of alcohol consumed (HD subjects: induction of miRNA10a vs. amount consumed, R2= 0.59; induction of miRNA21 vs. amount consumed, R2= 0.57), while in MD subjects, there was no association between inductions of these miRNAs and the amount of alcohol consumed (Figure 1C, 1D) (MD subjects: induction of miRNA10a vs. amount consumed, R2= −0.19; induction of miRNA21 vs. amount consumed, R2= 0.15).
Figure 1. Correlation between induction of miR-10a and miR-21 1 hour after stress-exposure and amount of beer consumed by HD and MD subjects.
Induction of both miR-10a and miR-21 was correlated with the amount of beer consumed in HD, but not MD subjects. Level of induction is measured as the expression at 1 hour after stress/baseline expression of that miRNA for each subject. A: miR-10a induction 1 hour after stress vs. amount of beer consumed in HD subjects; B: miR-21 induction 1 hour after stress vs. amount of beer consumed in HD subjects; C: miR-10a induction 1 hour after stress vs. amount of beer consumed in MD subjects; D: miR-21 induction 1 hour after stress vs. amount of beer consumed in MD subjects.
Relationship Between MicroRNA (miR-10a and miR-21) Expression and Peripheral Measures of Stress Reactivity
To explore the relationships among MiRNA expression changes, drinking behavior and established measures of stress reactivity we assessed a number of physiologic measures of stress previously shown to be related to stress-induced craving for alcohol (Sinha et al., 2009, Sinha et al., 2011). These included baseline measurements of heart rate, systolic and diastolic blood pressure, cortisol and corticotropin (ACTH) levels as well as changes in each of these measurements following exposure to stress. None of these measures differed significantly between HD and MD subjects in the current group of subjects. Of the measures tested, induction of ACTH was most closely correlated with drinking behavior in both MD and HD subjects. ACTH induction was also closely correlated with induction of miR21, but not miR10 in both groups. Correlations of each of these measures to induction of miR-10a and miR-21, as well as the amount of beer consumed in the taste test is shown in Table 4.
Table 4.
Correlation of peripheral measures of stress with microRNA induction and amt. of beer consumed after stress exposure.
| Peripheral measure | Amt drunk (MD) |
Amt drunk (HD) |
Amt drunk (combined) |
miR21 induction (MD) |
miR21 induction (HD) |
miR21 induction (combined) |
miR10 induction (MD) |
miR10 induction (HD) |
miR10 induction (combined) |
|---|---|---|---|---|---|---|---|---|---|
| ACTH | *0.59 | 0.53 | *0.56 | *0.73 | 0.29 | *0.49 | 0.46 | 0.13 | 0.32 |
| ACTH_diff | *0.57 | *0.73 | *0.65 | *0.83 | *0.69 | *0.70 | 0.37 | 0.12 | 0.32 |
| ACTH/CORT | 0.42 | 0.53 | *0.53 | *0.88 | 0.22 | *0.49 | *0.68 | 0.40 | *0.54 |
| CORT | 0.05 | −0.26 | −0.10 | −0.16 | −0.03 | −0.09 | −0.26 | −0.40 | −0.32 |
| CORT_diff | −0.08 | 0.35 | 0.02 | 0.34 | 0.38 | 0.26 | 0.22 | −0.17 | 0.01 |
| DBP | *0.62 | *0.61 | 0.28 | −0.19 | 0.04 | −0.18 | −0.42 | 0.27 | −0.18 |
| DBP_diff | −0.08 | 0.11 | 0.11 | −0.14 | 0.13 | 0.09 | −0.28 | 0.04 | −0.04 |
| HR | 0.08 | 0.3 | 0.11 | −0.23 | −0.15 | −0.19 | −0.23 | 0.49 | 0.1 |
| HR_diff | −0.06 | −0.37 | −0.30 | *0.65 | *−0.63 | −0.29 | 0.30 | −0.17 | −0.05 |
| SBP | *0.59 | 0.36 | 0.21 | −0.32 | 0.37 | −0.03 | −0.49 | 0.49 | −0.05 |
| SBP_diff | 0.21 | −0.25 | −0.09 | −0.01 | −0.36 | −0.21 | −0.2 | −0.23 | −0.24 |
Peripheral measures of stress/ HPA axis activation were correlated with the amount of beer consumed during the taste test and with the induction of miR21 and miR10 during the 1 hour after exposure to the stress cue. Significant correlations are marked with an asterisk.
Abbreviations: ACTH: Adrenocorticotropin; ACTH_diff change in ACTH from baseline; ACTH/CORT: ratio of ACTH/Cortisol; CORT: Cortisol level, CORT_diff: change in CORT from baseline; DBP: diastolic blood pressure; DBP_diff: change in DBP from baseline; HR: heart rate; HR_diff: change in HR from baseline; SBP: Systolic blood pressure; SBP_diff: change in SBP from baseline;
All values are shown as Area Under the Curve (auc) for the period of time from stress exposure to the last measurement.
Discussion
In this study we used whole genome microarrays to identify genes that were differentially expressed in peripheral blood of non-smoking moderate/non-binge (MD) versus heavy-binge (HD) socially drinking subjects, in response to three different personalized imagery conditions: neutral-relaxing, alcohol-cue, and stress-cue. Imagery scripts were standardized for script style and structure but were based on real events from each subject’s personal history. After each condition, subjects were presented with a try of two chilled mugs of beer, which served as a discrete alcohol cue in each condition, prior to the alcohol taste task. We found that in MD subjects, there were few, if any, significant changes in any of the genes expressed in peripheral blood under any of the test conditions. However, in HD subjects there were a number of genes whose expression was specifically altered following exposure to the stress cue. Peripheral measures of stress (heart rate, systolic and diastolic blood pressure, cortisol and ACTH levels as well as changes in each of these measurements following exposure to stress) did not differ significantly between HD and MD subjects, thus the differences in gene-expression are not due solely to differences in HPA axis reactivity.
Pathway analysis showed that some of the genes showing altered expression encode components of the TAR-RNA binding protein (TRBP)-associated complex (Chi et al., 2011). Three of these genes ((RPL9, RPS3A, and RPS17) have been shown to be positively regulated by miR-10a (Orom et al., 2008), while a fourth (RPL31) has been reported to be positively regulated by a different microRNA, miR-21 (Li et al., 2009). We found that expression of both miR-10a and miR-21 was increased following exposure to stress and was positively correlated with the amount of alcohol consumed in HD, but not MD subjects.
TRBP was first identified and cloned based on its high affinity binding to the small hairpin trans-activation responsive (TAR) RNA of HIV-1 (Gatignol et al., 1991). More recently, TRBP has been shown be a constituent of the RNA-induced silencing complex (RISC) serving as a Dicer co- factor in the processing of the ~70 nucleotide pre-microRNAs (miRNAs) to 21–25 nucleotide mature miRNAs (Chendrimada et al., 2005). Ethanol-induced alterations in the processing of pre-microRNAs has been proposed as a mechanism for ethanol-induced teratogenicity (Miranda, 2012). TRBP also interacts with a large number of proteins involved in regulation of diverse cellular functions including protein synthesis, RNA post-translational modifications, cellular growth and proliferation, gene expression and cell death, and may play a role in dampening the cellular response to stress (Daniels and Gatignol, 2012).
miRNAs are a class of short approximately 22 nucleotide long regulatory molecules that are involved in regulation of a large number of cellular functions. Recent evidence suggests that these molecules play key roles in the response to stress at both the cellular and organismic levels (Leung and Sharp, 2010). Intriguingly, some miRNAs, which normally suppress expression of target transcripts, may become activators of expression during stress (Leung and Sharp, 2007). Changes in miRNA expression have been proposed to play a role in gene-environment interactions related to the risk for various human diseases (Hudder and Novak, 2008). Changes in specific miRNAs have also been linked to long-lasting changes in stress-responsiveness induced by early life stress in animal models (Uchida et al., 2010), and regulation of stress induced anxiety (Haramati et al., 2011).
Given the well-known association between stress and risk for addiction, it is perhaps not surprising that changes in miRNA expression have also been proposed to play a role in risk for addiction to various substances (Hollander et al., 2010), including alcohol (Nunez and Mayfield, 2012). miR9 has been reported to play a key role in the development of alcohol tolerance by down- regulating the alpha subunit of BK channel, a high conductance calcium- and voltage-dependent potassium channel (Pietrzykowski et al., 2008). Alterations in miRNA expression profiles have also been linked to ethanol-mediated gut epithelial dysfunction, inflammatory GI disease, and ethanol- induced GI cancer (Miranda et al., 2010). Altered miRNA expression profiles have been demonstrated in post-mortem tissue from the brains of human alcoholics (Lewohl et al., 2011), and increased plasma level of miR122 has been proposed as a marker for hepatic damage from a variety of causes including alcoholic hepatitis (Zhang et al., 2010).
In the present study, we examined the expression of two specific miRNAs, miR-10a and miR- 21, previously shown to regulate the expression of some of the genes that were up-regulated by stress in the HD subjects in this sutdy. miR-10a has been shown to bind to the 5′ UTR of ribosomal protein mRNAs (including 3 on our list) and enhance their translation (Orom et al., 2008). miR-10a is also an important regulator of differentiation in neuroblastoma cells (Foley et al., 2011), and up-regulation of miR-10a is linked to ethanol-induced teratogenesis, through its effects on the Hoxa1 gene (Wang et al., 2009). In the peripheral circulation, expression of miR-10a has been reported to be a specific marker of regulatory T cells, and the level of miR-10a expression is inversely correlated with susceptibility to autoimmune disease (Jeker et al., 2012).
miR-21 is overexpressed in many cancers including glioblastoma and contributes to tumor resistance to chemotherapy (Li et al., 2009), while down-regulation of miR-21 inhibits the EGFR pathway and suppresses growth of glioblastoma cells (Zhou et al., 2010). miR-21 is also overexpressed in human gastric cancer tissues and cell lines (Zhang et al., 2008). These data indicate that miR-21 is an important negative regulator of apoptosis. Previous studies have shown that miR-21 can be down-regulated in vitro by alcohol in neural stem cell/ neural progenitor cells (NSC/NPCs) (Miranda et al., 2010), while chronic ethanol feeding enhances miR-21 induction during liver regeneration (Dippold et al., 2012).
Intriguingly, previous reports have shown that expression of miR-21 is regulated by ethanol in a GABAAR dependent manner (Sathyan et al., 2007). Ethanol has long been known to exert many of its intoxicating effects by potentiating GABA mediated inhibitory synaptic transmission in the brain (Crews et al., 1996). Thus, our results, in which the stress-induced increase in expression of miR-10a and miR-21 was correlated with increased consumption of alcohol in heavy drinkers, support a heuristic feed-forward model in which psychological stress in vulnerable individuals promotes increased alcohol consumption, increased alcohol consumption leads to altered GABAAR signaling, which leads to alterations in miRNA expression, that then lead to changes in stress-responsive gene-expression, and increased vulnerability to stress-induced drinking. This model provides a number of testable hypotheses that can be explored in future studies using both animal and human laboratory models of stress-induced drinking behavior.
Limitations of this study include a relatively small sample-size and the fact that stress activated genes were assayed at the level of gene-expression rather than protein or functional assays, since it is well known that many post-transcriptional and post-translational modifications can affect the level and function of these proteins independently of changes in gene-expression. Also, since whole blood was used as the source of total RNA, it is possible that changes in RNA expression involving specific cell-types or blood compartments may have occurred, but were not detected in the current experiments due to dilution by RNA from other blood compartments. In addition, the direct effects of alcohol on the expression of these genes were not controlled for, since the final time-point for gene-expression was after the test taste was completed. However, the fact that no changes were seen in either group following the neutral-relaxing or alcohol-related cues, and the amount of alcohol consumed by subjects in each group did not vary significantly by test condition, suggests that these changes are not solely due to direct effects of alcohol on the genes in question. In addition, we surveyed only two of several hundred miRNAs. It is likely that other miRNA species may be involved in mediating the effects observed here.
Conclusions
In the present study, we found that stress-related expression of both miR-10a and miR-21 was positively correlated with the amount of alcohol consumed following exposure to the stress cue in HD, but not MD subjects. HD subjects also exhibited selective changes in the expression of a number of genes following exposure to the stress cue that was not seen following exposure to either neutral-relaxing or alcohol-related cues and was not seen in MD subjects under any of the test conditions. These results suggest that the mechanisms governing stress-related expression of both miRNAs and their down-stream targets are altered in subjects with a history of heavy drinking. Alterations in miRNA expression may be one mechanism linking psychological stress with changes in both gene-expression and drinking behavior.
Acknowledgments
This work was supported by the State of Connecticut, Department of Mental Health and Addiction Services, through its support for the Connecticut Mental Health Center and grants from the NIH: R21-AA018388 (Beech), R01-AA013892, (Sinha), Yale CTSA (UL1-RR024139) and the NIH Common Fund Support for Interdisciplinary Research via grants UL1-DE019586 (Sinha) and PL1-DA024859 (Sinha).
References
- Adinoff B, Junghanns K, Kiefer F, Krishnan-Sarin S. Suppression of the HPA axis stress-response: implications for relapse. Alcohol Clin Exp Res. 2005;29:1351–1355. doi: 10.1097/01.ALC.0000176356.97620.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babor T, Higgins-Biddle J, Saunders J, Monteiro M. World Health Organization, editor. Guidelines for use in primary care. 2. Department of Mental Heath and Substance Dependence; 2001. AUDIT. The Alcohol Use Disorders Identification Test. [Google Scholar]
- Beech RD, Qu J, Leffert JJ, Lin A, Hong KA, Hansen J, Umlauf S, Mane S, Zhao H, Sinha R. Altered expression of cytokine signaling pathway genes in peripheral blood cells of alcohol dependent subjects: preliminary findings. Alcohol Clin Exp Res. 2012;36:1487–1496. doi: 10.1111/j.1530-0277.2012.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardy NC, King AC, Lovallo WR. Cardiovascular responses to physical and psychological stress in female alcoholics with transitory hypertension after early abstinence. Alcohol Clin Exp Res. 2003;27:1489–1498. doi: 10.1097/01.ALC.0000085587.00498.38. [DOI] [PubMed] [Google Scholar]
- Chao J, Nestler EJ. Molecular neurobiology of drug addiction. Annu Rev Med. 2004;55:113–132. doi: 10.1146/annurev.med.55.091902.103730. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YH, Semmes OJ, Jeang KT. A proteomic study of TAR-RNA binding protein (TRBP)-associated factors. Cell & bioscience. 2011;1:9. doi: 10.1186/2045-3701-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Morrow AL, Criswell H, Breese G. Effects of ethanol on ion channels. Int Rev Neurobiol. 1996;39:283–367. doi: 10.1016/s0074-7742(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Daniels SM, Gatignol A. The multiple functions of TRBP, at the hub of cell responses to viruses, stress, and cancer. Microbiology and molecular biology reviews: MMBR. 2012;76:652–666. doi: 10.1128/MMBR.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit H, Soderpalm AH, Nikolayev L, Young E. Effects of acute social stress on alcohol consumption in healthy subjects. Alcohol Clin Exp Res. 2003;27:1270–1277. doi: 10.1097/01.ALC.0000081617.37539.D6. [DOI] [PubMed] [Google Scholar]
- Dippold RP, Vadigepalli R, Gonye GE, Hoek JB. Chronic ethanol feeding enhances miR-21 induction during liver regeneration while inhibiting proliferation in rats. Am J Physiol Gastrointest Liver Physiol. 2012;303:G733–743. doi: 10.1152/ajpgi.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat Neurosci. 2005;8:1481–1489. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- Foley NH, Bray I, Watters KM, Das S, Bryan K, Bernas T, Prehn JH, Stallings RL. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011;18:1089–1098. doi: 10.1038/cdd.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox HC, Anderson GM, Tuit K, Hansen J, Kimmerling A, Siedlarz KM, Morgan PT, Sinha R. Prazosin effects on stress- and cue-induced craving and stress response in alcohol-dependent individuals: preliminary findings. Alcohol Clin Exp Res. 2012;36:351–360. doi: 10.1111/j.1530-0277.2011.01628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- Gladkevich A, Kauffman HF, Korf J. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2004;28:559–576. doi: 10.1016/j.pnpbp.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Hand DJaTCC. Multivariate Analysis of Variance and Repeated Measures: a practical approach for behavioural scientists. Chapman and Hall; London: 1987. [Google Scholar]
- Haramati S, Navon I, Issler O, Ezra-Nevo G, Gil S, Zwang R, Hornstein E, Chen A. MicroRNA as repressors of stress-induced anxiety: the case of amygdalar miR-34. J Neurosci. 2011;31:14191–14203. doi: 10.1523/JNEUROSCI.1673-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hudder A, Novak RF. miRNAs: effectors of environmental influences on gene expression and disease. Toxicol Sci. 2008;103:228–240. doi: 10.1093/toxsci/kfn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G, Lund AH, Bluestone JA. MicroRNA 10a marks regulatory T cells. PLoS ONE. 2012;7:e36684. doi: 10.1371/journal.pone.0036684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhoffer M, Dyrskjot L, Voss T, Lindberg RL, Wyrich R, Thykjaer T, Orntoft TF. Isolation of microarray-grade total RNA, microRNA, and DNA from a single PAXgene blood RNA tube. J Mol Diagn. 2007;9:452–458. doi: 10.2353/jmoldx.2007.060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, Webb E, Cooney NL, Kranzler HR, Southwick SW, Heninger GR, Charney DS. Serotonergic and noradrenergic dysregulation in alcoholism: m-chlorophenylpiperazine and yohimbine effects in recently detoxified alcoholics and healthy comparison subjects. Am J Psychiatry. 1996;153:83–92. doi: 10.1176/ajp.153.1.83. [DOI] [PubMed] [Google Scholar]
- Lang PJ, Kozak MJ, Miller GA, Levin DN, McLean AJ. Emotional Imagery: Conceptual Structure and Pattern of Somato-Visceral Response. Psychophysiology. 1980;17:179–192. doi: 10.1111/j.1469-8986.1980.tb00133.x. [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD. Up-regulation of microRNAs in brain of human alcoholics. Alcohol Clin Exp Res. 2011;35:1928–1937. doi: 10.1111/j.1530-0277.2011.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Harris RA. Gene expression profiling in blood: new diagnostics in alcoholism and addiction? Neuropsychopharmacology. 2009;34:250–251. doi: 10.1038/npp.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC. MicroRNAs and Fetal Brain Development: Implications for Ethanol Teratology during the Second Trimester Period of Neurogenesis. Frontiers in genetics. 2012;3:77. doi: 10.3389/fgene.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, Sampson W, Hereld D. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34:575–587. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton PM, Messing RO. Intracellular signaling pathways that regulate behavioral responses to ethanol. Pharmacol Ther. 2006;109:227–237. doi: 10.1016/j.pharmthera.2005.07.004. [DOI] [PubMed] [Google Scholar]
- NIAAA. Series Helping Patients Who Drink Too Much: A Clinician’s Guide. National Institute on Alcohol Abuse and Alcoholism; Rockvile, MD: 2005. Helping Patients Who Drink Too Much: A Clinician’s Guide. Vol. NIH Publication No. 07–3769. [Google Scholar]
- Nunez YO, Mayfield RD. Understanding Alcoholism Through microRNA Signatures in Brains of Human Alcoholics. Frontiers in genetics. 2012;3:43. doi: 10.3389/fgene.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen DD, Wilkinson CW, Raskind MA. Chronic daily ethanol and withdrawal: 6. Effects on rat sympathoadrenal activity during “abstinence”. Alcohol. 2006;38:173–177. doi: 10.1016/j.alcohol.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, Jerome RE, Lumeng L, Nurnberger JI, Jr, Edenberg HJ, McBride WJ, Niculescu AB. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2007;7:222–256. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27:8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R. Imagery Development Procedures Manual. Yale University; 2001. [Google Scholar]
- Sinha R. Modeling stress and drug craving in the laboratory: implications for addiction treatment development. Addict Biol. 2009a;14:84–98. doi: 10.1111/j.1369-1600.2008.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R. Stress and addiction: a dynamic interplay of genes, environment, and drug intake. Biol Psychiatry. 2009b;66:100–101. doi: 10.1016/j.biopsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Fox HC, Hong KA, Bergquist K, Bhagwagar Z, Siedlarz KM. Enhanced Negative Emotion and Alcohol Craving, and Altered Physiological Responses Following Stress and Cue Exposure in Alcohol Dependent Individuals. Neuropsychopharmacology. 2009;34:1198–1208. doi: 10.1038/npp.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Fox HC, Hong KI, Hansen J, Tuit K, Kreek MJ. Effects of Adrenal Sensitivity, Stress- and Cue-Induced Craving, and Anxiety on Subsequent Alcohol Relapse and Treatment Outcomes. Arch Gen Psychiatry. 2011 doi: 10.1001/archgenpsychiatry.2011.49. (May 2nd Epub online) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Funato H, Hobara T, Otsuki K, Yamagata H, McEwen BS, Watanabe Y. Early life stress enhances behavioral vulnerability to stress through the activation of REST4-mediated gene transcription in the medial prefrontal cortex of rodents. J Neurosci. 2010;30:15007–15018. doi: 10.1523/JNEUROSCI.1436-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, Wang J, Zhou SF, Li Y. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Human reproduction (Oxford, England) 2009;24:562–579. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, Sun S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Laboratory investigation; a journal of technical methods and pathology. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, Zhang W, Kang C. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Laboratory investigation; a journal of technical methods and pathology. 2010;90:144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]