Abstract
One of the most crucial concerns of cancer research pertains to the differences between the neoplastic cells in tumor specimens in vivo and their counterparts in cell lines. The huge amount of results deposited in cancer genetic databases allows to address this issue from a wider perspective. Our analysis of the Sanger Institute Catalog Of Somatic Mutations In Cancer (COSMIC) database v61 showed a lower percentage of homozygous mutations in a group of tumor suppressor genes in surgical samples (in vivo) in comparison to their frequency in cell lines (in vitro). Similarly, the mutations resulting in the lack of protein (e.g., nonsense mutations or whole gene deletions) of several tumor suppressor genes (TSGs) were more frequently observed in vitro than in vivo. In this article, we suggest two potential explanations of these data. Firstly, TSG heterozygous mutations resulting in the modified protein (e.g., missense mutations) may be gradually (when the specific molecular context is achieved) changed to homozygous mutations resulting in the lack of protein during carcinogenesis. Secondly, among different independent pathways of tumorigenesis, those leading to homozygous nonsense mutations are characteristic for cells which are more efficiently stabilized in vitro. To conclude, these observations may be interesting for researchers working with cell line in vitro models illustrating the extent to which they reflect the tumors in vivo.
Electronic supplementary material
The online version of this article (doi:10.1007/s13277-014-2444-5) contains supplementary material, which is available to authorized users.
Keywords: Tumor suppressor genes, Quasi-sufficiency, Three hits hypothesis, Two hits hypothesis
Introduction
It is an obvious fact that cancer cell lines bear only a partial resemblance to their origin. This issue has been addressed many times [1–6]; however, thus far, no practical guidelines on how to address this incongruity have been proposed, despite the fact that the basic and preclinical research is to a high extent based on cell line analysis (mostly due to the lack of a better model for a large scale use). We have already described the discrepancies in the TP53 mutational profile between surgical samples and cell lines [7]. Moreover, we have recently reported differences in the status of several tumor suppressor genes (TSGs) between glioma cancer cell lines and surgical specimens [8]. Since a better model is not available, recognizing such differences might facilitate the adequate result interpretation, and thus, may be useful in various research projects from comparative biology studies to in vitro drug testing. Therefore, we decided to perform a comparison of in vivo and in vitro mutational profiles for other tumor suppressor genes. This would be the next step in understanding the discrepancies between the surgical samples and cell lines both in the frequency of homo- and heterozygous mutations and in their effect on protein sequence and structure.
Currently, the foundations of oncology are based on Knudson hypothesis (two-hit hypothesis), according to which tumorigenesis requires the elimination of both alleles of given tumor suppressor gene [9]. However, this model does not recognize the influence of the first allele elimination on cell biology. The exceptions to this pattern, such as the single-heterozygous mutations of TP53, APC, or PTEN in tumor samples, are usually explained as a consequence of dominant-negative effect (DNE), gain of function (GOF), and haploinsufficiency. Recently, Berger et al. proposed an extension of the two hit model, the continuum/quasi-sufficiency model, suggesting that one allele elimination/mutation is sufficient to importantly influence cell biology and certain genes, such as PTEN, can be entirely eliminated only at the latest stages of carcinogenesis, since their action temporarily protects preneoplastic cells against senescence/apoptosis [10]. This extended model inspired our analyses, since we assumed that cancer cell lines represent the most advanced stages of carcinogenesis. Furthermore, various neoplastic cells require special conditions to survive in vitro, for example, those with IDH1 mutation or EGFRvIII [6, 11]. All these premises encouraged us to inquire if (and why) the status of tumor suppressor genes differs between surgical tumor specimens and cell lines.
Materials and methods
The mutation frequency and microsatellite instability (MSI) data was extracted from the Sanger Institute Catalog Of Somatic Mutations In Cancer (COSMIC) database (v61 release) which gathers information on somatic mutations taken from the literature and in-house sequencing in human tumor samples and tumor cell lines [12–15]. For each gene in the database, the frequency of normal and mutated samples was extracted. Moreover, the mutated samples were divided into groups according to mutation zygosity (homozygosity, heterozygosity) and type (Table 1). Two main mutation groups were distinguished: mutations modifying or partially abolishing the protein function (mostly missense mutations) and mutations completely abolishing the protein function (mostly nonsense mutations and whole gene deletions) (Table 1). Splicing site mutations were not classified into either group because of their usually unpredictable effect on protein structure and function. The analyses were performed for each gene with Fisher’s exact test for 2 × 2 contingency tables calculated with Matlab 2010b (Mathworks) and R 2.15.1 package. The analysis comprised the comparison of homo- versus heterozygous mutations and those partially versus completely abolishing the protein function mutations in all their possible combinations.
Table 1.
Classification of mutation type on the basis of the effect on protein structure and function
| Modified protein (mutations modifying or partially abolishing the protein function) | Complex, deletion in frame |
| Complex, insertion in frame | |
| Deletion, in frame | |
| Insertion, in frame | |
| Substitution, missense | |
| No protein (mutations completely abolishing the protein function) | Complex, frame shift |
| Deletion, frame shift | |
| Insertion, frame shift | |
| No detectable mRNA/protein | |
| Substitution, nonsense | |
| Whole gene deletion |
Splicing site mutations were not classified into any of group, because of unpredictable effect on protein structure and sequence
Results
Six hundred seventy cancer-associated genes were analyzed in 142,961 samples (137,708 tumor samples and 5,253 cell lines). The analysis of the summarized data (Supplementary tables) revealed the significant discrepancies in the frequencies of different mutation types between the surgical samples and cell lines. In oncogenes, as expected, there are almost no mutations resulting in the lack of protein and the proportions of missense heterozygous and missense homozygous mutations are retained in cell lines (exemplary data in Table 2: oncogene). In TSG, we observed two distinct recurrent patterns: a significant increase in the rate of homozygous mutations with the retention of the missense/nonsense proportion in cell lines or a simultaneous increase of the proportion of both homozygous and nonsense mutations in cell lines. TP53 is an exemplary gene following the first pattern. In tumor samples, the frequencies of heterozygous and homozygous mutations are comparable (both for missense and nonsense), while the homozygous mutations are dominant in cell lines (the missense/nonsense proportion was retained; data and results of the statistical analyses in Table 2: classical two hits). Moreover, RB1, NF1, NOTCH1, and PTEN presented similar changes. The second pattern may be exemplified by the SMAD4 gene, for which the proportion of nonsense homozygous mutations significantly increases in cell lines, while that of other mutation types decreases. For CDKN2A and APC, the mutation proportions were initially (in tumor samples) strongly shifted towards nonsense homozygous; however, the increase of this mutational type with the subsequent decrease of the other types of mutations was statistically significant. BRCA2, SOCS1, STK11, MSH6, and SMARCA4 followed such a pattern as well (Table 2: three hits). Furthermore, we observed a group of genes for which the proportion of nonsense homozygous mutations in tumor samples was similar or higher than in the previously mentioned genes in cell lines. No further increase of this proportion was observed in cell lines; however, this may imply that such genes follow a similar pattern, but the process is already advanced in tumor samples (Table 2: three hits/final stage). Finally, we observed several less commonly analyzed genes which may potentially be classified as following the second pattern; however, the number of described samples was not large enough for a reliable analysis (Table 2: three hits/small group).
Table 2.
A comparison of subgroups of homo/heterozygous mutation occurrence (HO/HE) and occurrence of mutations resulting in modified protein or lack of protein (MS/NS) in human cancer cell lines and surgical samples
| Group | Gene | Number of samples | %MS-HE | %MS-HO | %NS-HE | %NS-HO | MS-HE//NS-HO | NS-HE//NS-HO | MS-HE//MS-HO | MS-HE//NS-HE | MS-HO//NS-HO | MS-HO//NS-HE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classical 2 hits | TP53 | TS | 401 | 43.9 | 36.2 | 10.2 | 9.7 | p < 0.001 | p < 0.001 | p < 0.001 | p > 0.1 | p > 0.1 | p < 0.001 |
| CL | 566 | 9.2 | 68.9 | 2.5 | 19.4 | ||||||||
| RB1 | TS | 47 | 10.6 | 21.3 | 34.0 | 34.0 | p = 0.003 | p < 0.001 | p = 0.007 | p > 0.1 | p > 0.1 | p < 0.001 | |
| CL | 102 | 1.0 | 33.3 | 2.0 | 63.7 | ||||||||
| NF1 | TS | 96 | 22.9 | 14.6 | 32.3 | 30.2 | p > 0.1 | p = 0.013 | p = 0.007 | p > 0.1 | p = 0.098 | p < 0.001 | |
| CL | 40 | 12.5 | 42.5 | 7.5 | 37.5 | ||||||||
| NOTCH1 | TS | 326 | 46.9 | 2.5 | 50.0 | 0.6 | p < 0.001 | p < 0.001 | p < 0.001 | p > 0.1 | p = 0.141 | p < 0.001 | |
| CL | 67 | 34.3 | 2.5 | 29.9 | 17.9 | ||||||||
| PTEN | TS | 270 | 23.0 | 20.7 | 24.8 | 31.5 | p < 0.001 | p < 0.001 | p < 0.001 | p > 0.1 | p > 0.1 | p < 0.001 | |
| CL | 184 | 4.3 | 37.0 | 3.3 | 55.4 | ||||||||
| 3 hits | BRCA2 | TS | 17 | 35.3 | 17.6 | 11.8 | 35.3 | p = 0.042 | p < 0.001 | p = 0.028 | p > 0.1 | p = 0.047 | p > 0.1 |
| CL | 17 | 0.0 | 35.3 | 17.6 | 47.1 | ||||||||
| SMAD4 | TS | 173 | 12.7 | 52.6 | 9.8 | 24.9 | p < 0.001 | p = 0.091 | p = 0.033 | p > 0.1 | p = 0.003 | p > 0.1 | |
| CL | 105 | 2.9 | 43.8 | 4.8 | 48.6 | ||||||||
| SOCS1 | TS | 18 | 55.6 | 22.2 | 22.2 | 0.0 | p = 0.003 | p = 0.061 | p = 0.074 | p > 0.1 | p > 0.1 | p > 0.1 | |
| CL | 15 | 13.3 | 40.0 | 13.3 | 33.3 | ||||||||
| STK11 | TS | 87 | 36.8 | 24.1 | 20.7 | 18.4 | p < 0.001 | p < 0.001 | p = 0.002 | p > 0.1 | p = 0.005 | p = 0.005 | |
| CL | 61 | 4.9 | 24.6 | 1.6 | 68.9 | ||||||||
| MSH6 | TS | 26 | 53.8 | 11.5 | 30.8 | 3.8 | p < 0.001 | p < 0.001 | p = 0.002 | p > 0.1 | p > 0.1 | p = 0.004 | |
| CL | 20 | 5.0 | 35.0 | 0.0 | 60.0 | ||||||||
| SMARCA4 | TS | 13 | 38.5 | 46.2 | 7.7 | 7.7 | p = 0.063 | p > 0.1 | p > 0.1 | p > 0.1 | p = 0.040 | p > 0.1 | |
| CL | 44 | 22.7 | 29.5 | 2.3 | 45.5 | ||||||||
| APC | TS | 199 | 8.5 | 9.5 | 27.6 | 54.3 | p = 0.126 | p < 0.001 | p > 0.1 | p > 0.1 | p = 0.025 | p > 0.1 | |
| CL | 65 | 4.6 | 3.1 | 9.2 | 83.1 | ||||||||
| CDKN2A | TS | 1,813 | 3.6 | 61.7 | 1.8 | 32.8 | p < 0.001 | p < 0.001 | p < 0.001 | p > 0.1 | p < 0.001 | p < 0.001 | |
| CL | 9,900 | 0.3 | 46.6 | 0.1 | 53.0 | ||||||||
| 3 hits/final stage | BAP1 | TS | 32 | 3.1 | 40.6 | 6.3 | 50.0 | p > 0.1 | p = 0.019 | p > 0.1 | p > 0.1 | p > 0.1 | p = 0.054 |
| CL | 9 | 0.0 | 33.3 | 44.4 | 22.2 | ||||||||
| FAM123B | TS | 97 | 2.1 | 37.1 | 14.4 | 46.4 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | |
| CL | 7 | 14.3 | 14.3 | 0.0 | 71.4 | ||||||||
| NF2 | TS | 247 | 2.8 | 17.0 | 0.8 | 79.4 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p = 0.001 | p > 0.1 | |
| CL | 25 | 4.0 | 32.0 | 0.0 | 64.0 | ||||||||
| SMARCB1 | TS | 179 | 0.6 | 16.2 | 0.6 | 82.7 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | |
| CL | 20 | 5.0 | 15.0 | 0.0 | 80.0 | ||||||||
| VHL | TS | 415 | 4.6 | 39.0 | 2.9 | 53.5 | p = 0.091 | p = 0.036 | p > 0.1 | p > 0.1 | p > 0.1 | 0.073 | |
| CL | 26 | 11.5 | 38.5 | 11.5 | 38.5 | ||||||||
| 3 hits/small group | CDH10 | TS | 6 | 83.3 | 16.7 | 0.0 | 0.0 | p > 0.1 | N/A | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 |
| CL | 7 | 28.6 | 28.6 | 14.3 | 28.6 | ||||||||
| MLH1 | TS | 9 | 44.4 | 22.2 | 0.0 | 33.3 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | |
| CL | 13 | 15.4 | 23.1 | 0.0 | 61.5 | ||||||||
| MSH2 | TS | 9 | 44.4 | 0.0 | 33.3 | 22.2 | p > 0.1 | p > 0.1 | p = 0.076 | p > 0.1 | p > 0.1 | p = 0.004 | |
| CL | 15 | 13.3 | 26.7 | 13.3 | 46.7 | ||||||||
| SETD2 | TS | 5 | 40.0 | 40.0 | 0.0 | 20.0 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | p > 0.1 | |
| CL | 10 | 20.0 | 30.0 | 10.0 | 40.0 | ||||||||
| Oncogene | EGFR | TS | 768 | 81.6 | 17.8 | 0.3 | 0.3 | p > 0.1 | N/A | p > 0.1 | N/A | N/A | N/A |
| CL | 26 | 84.6 | 15.4 | 0.0 | 0.0 | ||||||||
| KIT | TS | 846 | 77.3 | 21.9 | 0.8 | 0.0 | p > 0.1 | N/A | p > 0.1 | N/A | N/A | N/A | |
| CL | 13 | 61.5 | 38.5 | 0.0 | 0.0 | ||||||||
| KRAS | TS | 1,601 | 58.5 | 41.3 | 0.1 | 0.0 | p > 0.1 | N/A | p > 0.1 | N/A | N/A | N/A | |
| CL | 177 | 58.8 | 41.2 | 0.0 | 0.0 | ||||||||
Classical 2 hits, the genes which followed the 2-hit pattern; 3 hits, the genes which followed the 3-hit pattern; 3 hits/final stage, the genes which may have followed the 3-hit pattern, but the proportion of homozygous nonsense mutations is very high in tumors; 3 hits/small group, the genes which may have followed the 3-hit pattern, but the groups were not numerous enough to verify that
TS tumor sample, CL cell line, MS mutation resulting in modified protein, NS mutation resulting in lack of protein, HO homozygous mutation, HE heterozygous mutation
Since the number of cases with the reported status of zygosity was relatively low, we performed an additional analysis of the proportion of homo-/heterozygous and missense/nonsense mutations separately to verify the observed discrepancies in more numerous groups. In most genes, the results of both analyses were concordant (data in Table 3). Two of the genes initially classified as following the first pattern (PTEN and RB1) showed a statistically significant increase in the proportion of nonsense mutations in cell lines in comparison to tumor samples. On the other hand, for two genes classified as following the second pattern, (SOCS1 and MSH6) the increase of the proportion of nonsense mutations in cell lines was not observed. In the genes potentially following the second pattern, the analysis of wider groups implies the increase in the proportion of homozygous and nonsense mutations; however, the statistical analyses do not confirm its significance.
Table 3.
A comparison of homo/heterozygous mutation occurrence (HO/HE) and occurrence of mutations resulting in modified protein or lack of protein (MS/NS) in human cancer cell lines and surgical samples
| Group | Gene | M tumor | % M tumor | M line | % M line | p value M | HO + HE tumor | % HO tumor | HO + HE line | % HO line | p value HO/HE | MS + NS tumor | % MS tumor | MS + NS line | % MS line | p value MS/NS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classical 2 hits | TP53 | 14,015 | 75.03 | 681 | 65.36 | p < 0.001 | 419 | 46.30 | 567 | 88.71 | p < 0.001 | 13,071 | 81.99 | 677 | 79.32 | p = 0.082 |
| RB1 | 214 | 7.00 | 151 | 14.80 | p < 0.001 | 56 | 50.00 | 122 | 98.36 | p < 0.001 | 167 | 19.16 | 121 | 9.09 | p = 0.019 | |
| NF1 | 320 | 14.95 | 39 | 4.56 | p < 0.001 | 104 | 56.73 | 39 | 84.62 | p = 0.002 | 283 | 22.97 | 40 | 22.50 | p > 0.1 | |
| NOTCH1 | 926 | 13.85 | 90 | 8.47 | p < 0.001 | 295 | 15.93 | 55 | 34.55 | p = 0.002 | 894 | 59.51 | 91 | 57.14 | p > 0.1 | |
| PTEN | 1,793 | 11.50 | 395 | 18.68 | p < 0.001 | 281 | 54.09 | 186 | 94.09 | p < 0.001 | 1,626 | 42.19 | 369 | 30.08 | p < 0.001 | |
| 3 hits | BRCA2 | 50 | 2.55 | 16 | 1.85 | p > 0.1 | 17 | 52.94 | 16 | 68.75 | p > 0.1 | 46 | 52.17 | 18 | 33.33 | p > 0.1 |
| SMAD4 | 251 | 8.11 | 124 | 10.39 | p = 0.022 | 182 | 80.22 | 110 | 92.73 | p = 0.004 | 243 | 44.03 | 115 | 20.00 | p < 0.001 | |
| SOCS1 | 38 | 3.95 | 17 | 1.96 | p = 0.014 | 18 | 27.78 | 17 | 76.47 | p = 0.007 | 40 | 55 | 16 | 56.25 | p > 0.1 | |
| STK11 | 200 | 5.41 | 69 | 6.51 | p > 0.1 | 96 | 41.67 | 60 | 93.33 | p < 0.001 | 181 | 51.38 | 64 | 18.75 | p < 0.001 | |
| MSH6 | 122 | 7.11 | 19 | 1.99 | p < 0.001 | 26 | 26.92 | 28 | 77.78 | p = 0.002 | 109 | 27.52 | 23 | 39.13 | p > 0.1 | |
| SMARCA4 | 36 | 7.86 | 50 | 4.66 | p = 0.015 | 13 | 61.54 | 50 | 76.00 | p > 0.1 | 35 | 91.43 | 44 | 36.36 | p < 0.001 | |
| APC | 2,121 | 20.20 | 139 | 11.69 | p < 0.001 | 205 | 65.85 | 75 | 88.00 | p < 0.001 | 2,074 | 8.00 | 94 | 5.32 | p > 0.1 | |
| CDKN2A | 2,513 | 12.29 | 1,172 | 38.45 | p < 0.001 | 1,848 | 94.59 | 1,014 | 99.61 | p < 0.001 | 2,369 | 17.56 | 966 | 10.14 | p < 0.001 | |
| 3 hits/final stage | BAP1 | 99 | 10.44 | 10 | 13.89 | p > 0.1 | 33 | 90.91 | 9 | 55.56 | p = 0.028 | 91 | 26.37 | 10 | 30.00 | p > 0.1 |
| FAM123B | 114 | 8.76 | 8 | 0.99 | p < 0.001 | 98 | 83.67 | 8 | 75.00 | p > 0.1 | 112 | 7.14 | 7 | 28.57 | p > 0.1 | |
| NF2 | 506 | 23.94 | 35 | 3.56 | p < 0.001 | 277 | 96.75 | 33 | 93.94 | p > 0.1 | 447 | 9.62 | 24 | 4.17 | p > 0.1 | |
| SMARCB1 | 240 | 19.67 | 44 | 16.18 | p > 0.1 | 184 | 98.91 | 23 | 95.65 | p > 0.1 | 230 | 13.04 | 35 | 17.14 | p > 0.1 | |
| VHL | 1,123 | 22.44 | 90 | 8.65 | p < 0.001 | 441 | 92.52 | 30 | 76.67 | p = 0.009 | 1,018 | 40.57 | 85 | 45.88 | p > 0.1 | |
| 3 hits/small group | CDH10 | 13 | 6.16 | 5 | 10.2 | p > 0.1 | 6 | 33.33 | 5 | 40.00 | p > 0.1 | 10 | 100 | 7 | 71.43 | p > 0.1 |
| MLH1 | 52 | 3.35 | 19 | 2.17 | p > 0.1 | 13 | 61.54 | 19 | 89.47 | p = 0.091 | 43 | 60.47 | 14 | 35.71 | p > 0.1 | |
| MSH2 | 37 | 3.26 | 15 | 1.84 | p = 0.064 | 11 | 36.36 | 15 | 80 | p = 0.043 | 29 | 34.48 | 15 | 13.33 | p > 0.1 | |
| SETD2 | 16 | 9.94 | 14 | 1.75 | p < 0.001 | 6 | 50.00 | 14 | 57.14 | p > 0.1 | 13 | 53.85 | 10 | 40.00 | p > 0.1 | |
| Oncogene | EGFR | 11,257 | 22.41 | 113 | 6.43 | p < 0.001 | 793 | 18.16 | 29 | 17.24 | p > 0.1 | 11,219 | 95.73 | 84 | 85.71 | p < 0.001 |
| KIT | 5,986 | 27.81 | 50 | 4.14 | p < 0.001 | 1,053 | 18.33 | 15 | 33.33 | p > 0.1 | 5,368 | 99.61 | 29 | 100 | p > 0.1 | |
| KRAS | 21,938 | 22.89 | 631 | 18.34 | p < 0.001 | 1,604 | 41.40 | 205 | 41.46 | p > 0.1 | 21,806 | 99.99 | 394 | 100 | p > 0.1 |
Classical 2 hits, the genes which followed the 2-hit pattern; 3 hits, the genes which followed the 3-hit pattern; 3 hits/final stage, the genes which may have followed the 3-hit pattern, but the proportion of homozygous nonsense mutations is very high in tumors; 3 hits/small group, the genes which may have followed the 3-hit pattern, but the groups were not numerous enough to verify that
M mutated sample, MS mutation resulting in modified protein, NS mutation resulting in lack of protein, HO homozygous mutation, HE heterozygous mutation
The analysis of microsatellite data from 810 cell lines and 720 primary samples revealed that the frequency of MSI was moderately higher in the former. High-frequency MSI (MSI-H) was detected in 66 cell lines and in 36 primary samples (8 vs. 5 %, p = 0,014). Any MSI (including high- and low-frequency MSI-L) was detected in 106 cell lines and in 69 primary samples (13 vs. 8.5 %, p = 0.036) (Table 4).
Table 4.
A comparison of microsatellite instability frequency in human cancer cell lines and surgical samples. Fisher’s exact test results
| MSS | MSI | |||
|---|---|---|---|---|
| MSI-H | MSI-L | Total | ||
| Tumor | 651 | 36 | 33 | 69 |
| Line | 704 | 66 | 40 | 106 |
| MSI-H vs. others | p = 0.01384 | Any MSI vs. MSS | p = 0.03624 | |
MSS microsatellite stability, MSI microsatellite instability, MSI-H high-frequency microsatellite instability (detected in at least 2 markers), MSI-L low-frequency microsatellite instability (detected in 1 marker)
Discussion
In general, it is well known that cancer cell lines cannot be seen as a direct representation of the tumors; however, the extent of the differences between them may be underestimated. A deep analysis of mutation databases offers insight into this issue from another perspective. The presented study showed that homozygous mutations of many tumor suppressor genes are significantly more frequent in cell lines than in tumor samples. Similarly, nonsense mutations of such genes occur more frequently in vitro than in vivo. The quasi-sufficiency hypothesis proposed by Berger et al. offers a partial explanation for these discrepancies [9] (justifying the higher incidence of single heterozygous mutations in tumor samples, but not the preference of cell lines towards nonsense mutations). PTEN is an exemplary gene with such characteristics. Preneoplastic cells require a heterozygous mutation of this gene (with its function partially retained) during the early stages of carcinogenesis, as the cells without PTEN activity become senescent or die. Apparently, the hyperactivation of the PI3K pathway may be unfavorable at the early stages of carcinogenesis [16]. Most authors suggest that the complete PTEN elimination requires a prior neutralization of the genes required for oncogene-induced senescence (e.g., TP53, CDKN2A) [17–19].
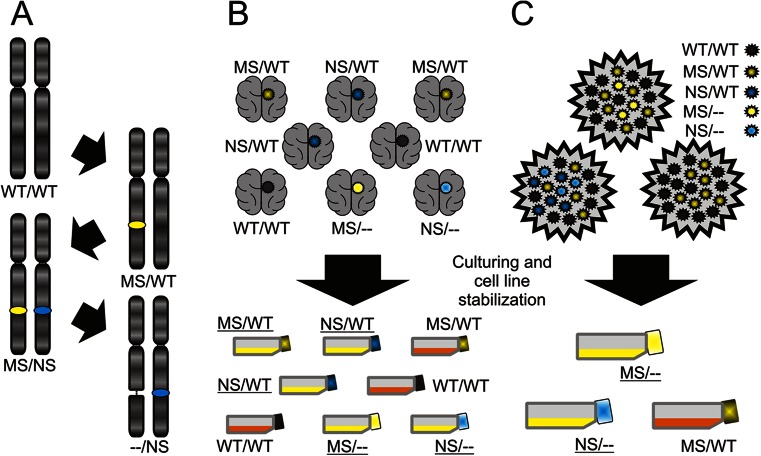
Both missense and nonsense mutations of the PTEN gene almost equally eliminate the phosphatase activity of the protein; thus, the gradual change from missense heterozygous to nonsense homozygous would not be expected in this case [20]. On the other hand, genes such as BRCA2 and SOCS1 show the complex differences between surgery samples and cell lines—a significantly higher frequency of homozygous and nonsense (i.e., resulting in the complete lack of protein) mutations in the latter (Table 2). For such genes, we propose the “three hit” model (Fig. 1a). At the initial stages of carcinogenesis, a missense TSG mutation is sufficient/optimal through altering (yet not completely eliminating) the protein function. Next, a nonsense mutation is generated within the other allele. Finally, the allele with the missense mutation is deleted, causing the lack of protein. Other forms of in vivo changes are also possible, e.g., the missense mutation may be directly changed into a nonsense one etc.
Fig. 1.
The hypotheses potentially explaining the differences in mutational profiles. a In vivo transformation of missense into nonsense mutations; this process may occur through various mechanisms, for example, a missense mutation of one allele is followed by a nonsense mutation of the other alleles and next by a deletion of the allele with the missense mutation. Missense mutations are marked as yellow spots, nonsense mutations as blue ones. b In vitro selection of tumor specimens which may give rise to stabilized cell lines. Obviously, the mutational status of a single gene may not determine the stabilization efficiency; however, the cumulative influence of all mutations may affect the probability of the successful cell line stabilization. The color of the tumor represents the mutation type of given gene (legend in the top right-hand corner), which is further reflected by the color of the cap of the respective culture flask. Stabilized cell lines are marked as yellow bottles with underlined labels; the others are marked as orange bottles with normal labels. c In vitro selection of cells within a tumor specimen which may give rise to a stabilized cell line. Again, within a heterozygous tumor the cells with certain molecular profiles may be more predisposed to the stabilization as a cell line. The color of the cells within tumors represents the mutation type of given gene (legend in the top right-hand corner), which is further reflected by the color of the cap of the respective culture flask. Stabilized cell lines are marked as yellow bottles with underlined labels; the others are marked as orange bottles with normal labels
These data may also offer an alternative explanation, assuming that among the multiple independent carcinogenic pathways, these with nonsense mutations are more adaptable in vitro and allow for more efficient cell line stabilization. This may refer both to intratumoral heterogeneity and to differences between cases (Fig. 1b, c). Therefore, cell lines would represent only a subgroup of cells/cases observed in vivo. Here, the transformation into the more advanced tumor stages is not accompanied by the missense to nonsense change. Nonetheless, still, the nonsense mutations may hypothetically cause some biological changes associated with the ease and effectiveness of cell line stabilization, e.g., the cells isolated from tumors with nonsense mutations may be more proliferative. CDKN2A is an exemplary gene whose molecular characteristic supports the in vitro adaptation hypothesis.
These hypotheses may seem mutually exclusive; however, both may be partially responsible for the observed differences. Still, irrespective of the underlying mechanism, such discrepancies are an incentive to consider the respective genes as following the quasi-sufficiency hypothesis.
Finally, we inquired whether the defective DNA damage response and repair mechanisms might be responsible for the observed differences in mutational profiles. For that purpose, we compared the MSI detection rates in cell lines and primary samples. Although the differences were statistically significant, microsatellite instability may not be the sole explanation of the observed results, due to its generally low detection rates (Table 4). Nonetheless, it may be one of the aspects affecting the cell line stabilization effectiveness.
The database analysis provides an additional argument that average cells from a cell line are more advanced in tumorigenesis than average cells from the corresponding surgical sample, e.g., the frequencies of detected TSG mutations are much higher in cell lines; however, the more thorough analysis of cell lines has to be emphasized.
In conclusion, we report the preferential character of nonsense mutations of TSGs in the most advanced cancer cells. Apparently, during the early stages of tumorigenic transformation, the complete elimination of many TSG may be lethal or otherwise unfavorable and becomes possible only when the required cellular/molecular context is achieved. Our hypothesis of “three hits” is a modification of the “continuum” model by Berger et al. [10]. Clearly, the potential transformation of missense to nonsense mutations during carcinogenesis requires more data. Alternatively, the presented data may be explained by the hypothesis that cell lines originating from tumors/cells with nonsense mutations are more easily stabilized in comparison to those with missense mutations. The differences in the mutational characteristics of TSG between tumor samples and cell lines may indicate the lack of the appropriate in vitro representation of tumors in vivo, which is particularly important from the drug testing perspective.
This study was supported by the National Science Center Grant No. 2011/01/B/NZ4/07832. Calculations in this paper have been carried out within Upper Silesian Center for Scientific Computational Science and Engineering (POIG.02.03.01-24-099/13).
Electronic supplementary material
Below is the link to the electronic supplementary material.
(XLS 3430 KB)
(XLS 453 KB)
(XLS 245 KB)
(XLS 216 KB)
(XLS 199 KB)
(XLS 58 KB)
(XLS 47 KB)
(XLS 35 KB)
Acknowledgments
Conflicts of interest
None.
Footnotes
Ewelina Stoczynska-Fidelus and Michal Bienkowski equally contributed.
References
- 1.Ertel A, Verghese A, Byers SW, Ochs M, Tozeren A. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol Cancer. 2006;5(1):55. doi: 10.1186/1476-4598-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6(12):909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 4.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wistuba II, Bryant D, Behrens C, Milchgrub S, Virmani AK, et al. Comparison of features of human lung cancer cell lines and their corresponding tumors. Clin Cancer Res. 1999;5(5):991–1000. [PubMed] [Google Scholar]
- 6.Piaskowski S, Bienkowski M, Stoczynska-Fidelus E, Stawski R, Sieruta M, et al. Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br J Cancer. 2011;104(6):968–970. doi: 10.1038/bjc.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoczynska-Fidelus E, Szybka M, Piaskowski S, Bienkowski M, Hulas-Bigoszewska K, et al. Limited importance of the dominant-negative effect of TP53 missense mutations. BMC Cancer. 2011;11(1):243. doi: 10.1186/1471-2407-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoczynska-Fidelus E, Piaskowski S, Bienkowski M, Banaszczyk M, Hulas-Bigoszewska K, et al. The failure in the stabilization of glioblastoma-derived cell lines: spontaneous in vitro senescence as the main culprit. PLoS One. 2014;9(1):e87136. doi: 10.1371/journal.pone.0087136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68(4):820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476(7359):163–9. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoczynska-Fidelus E, Och W, Rieske P, Bienkowski M, Banaszczyk M, et al. Spontaneous in vitro senescence of glioma cells confirmed by an antibody against IDH1R132H. Anticancer Res. 2014;34(6):2859–67. [PubMed] [Google Scholar]
- 12.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010 (Database issue): D652–D657. [DOI] [PMC free article] [PubMed]
- 13.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet. 2008; CHAPTER: Unit–10.11. [DOI] [PMC free article] [PubMed]
- 14.Forbes S, Clements J, Dawson E, Bamford S, Webb T, et al. COSMIC 2005. Br J Cancer. 2006;94(2):318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Astle MV, Hannan KM, Ng PY, Lee RS, George AJ, et al. AKT induces senescence in human cells via mTORC1 and p53 in the absence of DNA damage: implications for targeting mTOR during malignancy. Oncogene. 2012;31(15):1949–1962. doi: 10.1038/onc.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumour suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Trotman LC, Shaffer D, et al. Crucial role of p53- dependent cellular senescence in suppression of Pten-deficient tumourigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumour xenografts to suppress prostate tumourigenesis. J Clin Invest. 2010;120:681–693. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han SY, Kato H, Kato S, Suzuki T, Shibata H, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000;60(12):3147–51. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 3430 KB)
(XLS 453 KB)
(XLS 245 KB)
(XLS 216 KB)
(XLS 199 KB)
(XLS 58 KB)
(XLS 47 KB)
(XLS 35 KB)