Abstract
Background
Long non-coding RNAs (lncRNAs) recently have been implicated in many biological processes and diseases. Atherosclerosis is a major risk factor for cardiovascular disease. However, the functional role of lncRNAs in atherosclerosis is largely unknown.
Methods and Results
We identified lincRNA-p21 as a key regulator of cell proliferation and apoptosis during atherosclerosis. The expression of lincRNA-p21 was dramatically down-regulated in atherosclerotic plaques of ApoE−/− mice, an animal model for atherosclerosis. Through loss- and gain-of function approaches, we showed that lincRNA-p21 represses cell proliferation and induces apoptosis in vascular smooth muscle cells (VSMCs) and mouse mononuclear macrophage cells in vitro. Moreover, we found that inhibition of lincRNA-p21 results in neointimal hyperplasia in vivo in a carotid artery injury model. Genome-wide analysis revealed that lincRNA-p21 inhibition dysregulated many p53 targets. Furthermore, lincRNA-p21, a transcriptional target of p53, feeds back to enhance p53 transcriptional activity, at least in part, via binding to mouse double minute 2 (MDM2), an E3 ubiquitin-protein ligase. The association of lincRNA-p21 and MDM2 releases MDM2 repression of p53, enabling p53 to interact with p300 and bind to the promoters/enhancers of its target genes. Finally, we show that lincRNA-p21 expression is decreased in coronary artery disease patients.
Conclusions
Our studies identify lincRNA-p21 as a novel regulator of cell proliferation and apoptosis and suggest that this lncRNA could serve as a therapeutic target to treat atherosclerosis and related cardiovascular disorders.
Keywords: lncRNA, lincRNA-p21, p53, MDM2, atherosclerosis, cell proliferation, apoptosis
Introduction
Atherosclerosis is one of the most common vascular disorders. Proliferation of vascular smooth muscle cells (VSMCs) and the formation of neointima dominate atherosclerosis lesion development. p53, an essential molecule in cell cycle and apoptosis control, also plays a central role in atherosclerosis 1-3. Inactivation of p53 stimulates the development of atherosclerosis 4-6. A complex network that includes p300 and Mouse Double Minute 2 (MDM2) regulates p53. p300 is an acetyltransferase which acetylates p53 to enhance its activity 7-9. MDM2, an E3 ubiquitin-protein ligase, degrades p53 via the ubiquitin-proteasome pathway 10, 11. MDM2 also blocks P300 interaction with p53, thereby inhibiting p53 acetylation and decreasing its activity 12-14. Intriguingly, whereas MDM2 is a key regulator of the fate and activity of p53, the transcription of MDM2 itself is under the control of p53, establishing a p53/MDM2 negative feedback loop.
More than 90% of the human genome is transcribed 15-17. Whereas protein-coding genes account for less than 2% of the human genome, non-coding RNAs (ncRNAs) are important components of the mammalian transcriptome 18, 19. There are several classes of ncRNAs, including the well-known microRNAs (miRNAs), which are ~21-23nt long and have been proven to play a key role in the regulation of gene expression and function in a variety of biological and pathological processes 20-23. Long non-coding RNAs (lncRNAs), also known as long intergenic non-coding RNAs (lincRNAs), constitute another class of ncRNAs. Defined as non-coding transcripts longer than 200 nucleotides, at least a subset of lncRNAs are likely to have biological activity 24-26. Numerous studies have already shown the involvement of lncRNAs in cancer development 27-29. However, the role of lncRNAs in cardiovascular system is less understood 30. A recent study discovered Braveheart (Bvht), a heart-associated murine lncRNA, and demonstrated that Bvht is essential for the maintenance of the fate of cardiomyocytes 31.
Previous studies linked ANRIL (CDKN2B-AS), a lncRNA located at human chromosome 9p21.3, with increased coronary artery disease risk 32. However, the molecular nature of how this lncRNA regulates atherosclerosis process is unclear.
LincRNA-p21 was initially identified as a direct transcriptional target of p53 33. LincRNA-p21 appears to function as a component of the p53 pathway, at least in part, by physically interacting with a p53 repressive complex to downregulate many p53 target genes 33. lincRNA-p21 also acts as a suppressor of translation by directly associating with target mRNAs 34. Despite these studies, the biological function of lincRNA-p21 remains elusive.
In this study, we examined the functional role of lincRNA-p21 in the pathogenesis of atherosclerosis. We showed that the expression level of lincRNA-p21 was lower in atherosclerotic plaques of the ApoE−/− mice. Importantly, we found that inhibition of lincRNA-p21 increased cell proliferation and neointima formation in injured carotid arteries. Among the genes dysregulated by lincRNA-p21 inhibition was MDM2. We show that MDM2 interacts with lincRNA-p21 to relieve its repression of p53. Our study therefore uncovered a critical function of lincRNA-p21 in atherosclerosis.
Materials and Methods
Cell culture, transfection, Cellular proliferation and apoptosis analysis
Human vascular smooth muscle cell line HA-VSMC and mouse mononuclear macrophage cell line RAW264.7 were purchased from ATCC and cultured following the instructions of manufacturers. Cells were transfected using Lipofectamine 2000.
Cells were seeded in 96-well plates, and cell proliferation was tested using the Cell Counting Kit-8 (CCK-8) assay Kit. Proliferating HA-VSMCs were quantified by Ki67 staining. For cellular apoptosis assay, HA-VSMCs were seeded in 48-well plates and TUNEL assay was performed using the ApopTag® Plus In Situ Apoptosis Fluorescein Detection Kit.
RNA isolation, qRT-PCR analysis and Unbiased gene expression profiling
Total RNAs were isolated using TRIzol reagent. Reverse transcription (RT) and real time quantitative PCR (qRCR) were performed, following manufacturer's instructions. PCR primers are listed in Supplemental Table 1. Unbiased genome-wide transcriptome profiling was performed using the HUGENE 2.0 ST array (Affymetrix), which interrogates a total of 40,716 Refseq transcripts. Arrays hybridization, signal detection and data analysis were performed as described 35,36.
RNA immunoprecipitation, RIP assay
RNA immunoprecipitation (RIP) experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit following the manufacturer's instructions. Two independent MDM2 antibodies were used.
Biotin RNA pull-down assay and deletion mapping and bioinformatics
RNA pull-down assay and deletion mapping were performed as described previously 37. Briefly, the pcDNA3.1-lincRNA-p21 plasmid was used as template to synthesize biotinylated lincRNA-p21 transcripts. For biotinylated RNA generation, PCR products were used for in vitro transcription with the Biotin RNA Labeling Mix and T7 RNA polymerase. Nuclear proteins were extracted using Nuclear and Cytoplasmic Protein Extraction Kit. After incubation, binding and washing, beads were boiled in SDS buffer, and retrieved protein was detected by standard western blotting.
CatRAPID 38 and RPIseq 39, online protein-RNA binding predictors, were used to test the potential binding of MDM2 to lincRNA-p21.
Chromatin immunoprecipitation and ChIP-Seq assays
For immunoprecipitation (IP), cells were lysed in cell lysis buffer and the whole cell extracts were incubated with protein A Sepharose beads combined with antibodies against p53, or with control IgG for 6 hours at 4°C. The Chromatin Immunoprecipitation (ChIP) Assay Kit was used for ChIP-qPCR assays, according to the manufacturer's instructions.
ChIP-Seq was performed from HA-VSMC following NimbleGen protocols for chromatin immunoprecipitation and amplification with minor modifications. The ChIP DNA was converted into Illumina sequencing libraries following the NEBNext ChIP-Seq Sample Prep Master Mix Set1 protocol. Multiplex adaptor and TrueSeq indexes were used. Reads were aligned using Bowtie2 40, and regions with enriched signal compared to input were identified using MACS, using the default and suggested threshold of 1E-5 41. The peaks for selected loci were visualized using IGV (Broad Institute, MA)
Injury-induced mouse model of carotid artery neointimal hyperplasia
Local lentivirus-mediated gene transfer into injured carotid arteries was performed as described previously 42, 43. Briefly, C57BL/6J mice underwent metal wire injury of the common carotid artery (N=5 for each group). After local injury and heparin injection, 20ul of recombinant lentivirus Si-mlincRNA-p21 (1×108 UT/ml) and cntl-SiRNA(1×108 UT/ml) were instilled into the common carotid artery and allowed to dwell for 30 minutes. Uninjured arteries were used as sham control.
For cell proliferation and apoptosis, immunofluorescence and TUNEL assays, respectively, were performed. Sections were incubated with antibodies against Ki67. Terminal deoxynucleotidyl transferase-mediated nick-end labeling (TUNEL) assays were performed on paraffin sections using the ApopTag® Plus In Situ Apoptosis Fluorescein Detection Kit according to the manufacturer's procedure.
Clinical inclusion criteria
The coronary artery disease (CAD) group contained patients with greater than 80% coronary artery stenosis and the control group contained patients without clinically significant coronary artery occlusion were recruited. All procedures were conducted in compliance protocols approved by the Third Military Medical University Ethics Committee, and written informed consent was received from all participants. Details of all probands are presented in Supplemental Tables 2 and 3.
Statistical analysis
All data are expressed as mean ± SD unless otherwise stated. Data from each experiment were individually analyzed and statistics applied. We used Student's unpaired t-test to compare 2 independent groups. In experiments comparing multiple time points, separate t-tests were used for each time point. For a comparison of ≥3 groups, 1-way ANOVA was used. GraphPad-Prism 5.0 and SPSS 17.0 were used to perform the statistical analyses. Values of p< 0.05 were considered significant.
An expanded Methods section is included in the online-only Data Supplement.
Results
LincRNA-p21 regulates cell proliferation and apoptosis
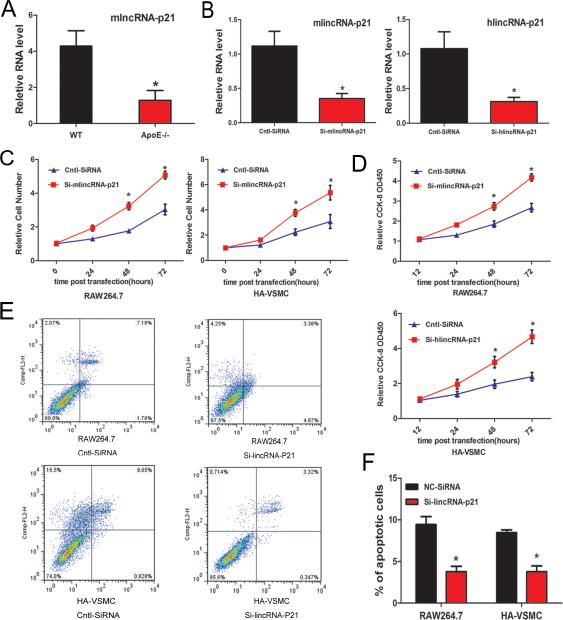
Given the vital role of p53 in the pathogenesis of atherosclerosis and the recent report that p53 regulates the expression of lincRNA-p21 33, we hypothesized that lincRNA-p21 is also involved in the development of atherosclerosis. We first examined the expression of lincRNA-p21 in aortic atherosclerotic plaques of ApoE−/− mice fed a high-fat diet, a widely used animal model of atherosclerosis. Indeed, we found that the expression of lincRNA-p21 was substantially lower in the aortic plaques of ApoE−/− mice when compared with that of wild type control mice, suggesting that lincRNA-p21 may play a role in atherosclerosis (Figure 1A).
Figure 1.
LincRNA-p21 regulates cell proliferation and apoptosis. (A) lincRNA-p21 transcript expression in atherosclerotic plaques of ApoE knockout mice (ApoE−/−) and wild-type C57 control mice (WT) was measured by qRT-PCR. N=5. (B) Si-RNAs were designed to knockdown mouse lincRNA-p21 (mlincRNA-p21) or human lincRNA-p21 (hlincRNA-p21) in RAW264.7 or HA-VSMCs, respectively. The expression of lincRNA-p21 was quantified by qRT-PCR. (C) Knockdown of lincRNA-p21 increased cell proliferation. Relative number of RAW264.7 and HA-VSMC cells at different time points was calculated after lincRNA-p21 knockdown. (D) Knockdown of lincRNA-p21 increased cell viability. The proliferation and viability of RAW264.7 and HA-VSMC cells were measured using the Cell Counting Kit-8 (CCK-8) colorimetric assay after lincRNA-p21 knockdown. (E) Knockdown of lincRNA-p21 inhibits apoptosis. Apoptosis of RAW264.7 and HA-VSMC cells was measured and quantified via Annexin-V conjugated FACS analysis. (F) Quantification of apoptosis. All values are the average of at least 3 biological replicates and data shown are the mean±standard deviation (SD). * P<0.05 relative to control.
Next, we investigated the function of lincRNA-p21 in cell proliferation and apoptosis. We used the mouse macrophage cell line RAW264.7 and the human vascular smooth muscle cell line HA-VSMC, both of which have been widely used to study atherosclerosis in vitro. We designed small interfering RNA (siRNA) to inhibit mouse lincRNA-p21 (mlincRNA-p21) and human lincRNA-p21 (hlincRNA-p21) expression. The efficiency of siRNA transfection and the inhibition of endogenous lincRNA-p21 were tested and confirmed by qRT-PCR (Figure 1B). Inhibition of lincRNA-p21 substantially increased total cell numbers in both RAW264.7 and HA-VSMC cells (Figure 1C). Increased cell proliferation and viability in these cells were further confirmed using an independent sensitive colorimetric assay (Figure 1D). Furthermore, lincRNA-p21 knockdown decreased apoptosis in both cell lines (Figures 1E and 1F). Together, these data indicate that lincRNA-p21 suppresses cell proliferation and induces apoptosis.
LincRNA-p21 regulates the expression of p53 target genes
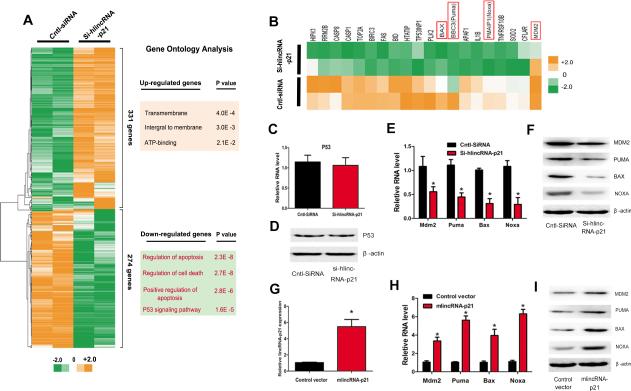
In order to understand the molecular mechanism by which linRNA-p21 regulates cell proliferation and apoptosis, we performed unbiased gene array analysis to measure gene expression changes in lincRNA-p21 knockdown cells. We found that 331 and 274 genes were up- and down-regulated more than two folds, respectively, when linRNA-p21 was knocked down (Figure 2A). Gene ontology (GO) analysis indicated that the down-regulated genes are over-represented for functional terms related to apoptosis, cell death and the p53 signaling pathway (Figure 2A). Further examination revealed that many of the down-regulated genes were previously reported p53 transcriptional targets (Figure 2B). We asked whether the expression of p53 itself was affected by lincRNA-p21 depletion. To our surprise, we detected no change in the expression of either p53 transcript or protein when lincRNA-p21 was inhibited (Figures 2C and 2D). We confirmed that siRNA lincRNA-p21 depletion in HA-VSMCs decreased the mRNA and protein levels of p53 downstream target genes Puma 44, 45, Bax 46, Noxa 47, and MDM2 (Figures 2E and 2F). Conversely, we overexpressed lincRNA-p21 in RAW264.7 cells (Figures 2G). Indeed, overexpression of lincRNA-p21 induced the expression of these p53 target genes, consistent with their role in the regulation of cell proliferation and apoptosis (Figures 2H and 2I).
Figure 2.
LincRNA-p21 is required for the expression of p53-downstream genes. (A) Hierarchical clustering analyses of 331 up-regulated and down-regulated 274 genes in lincRNA-p21 knockdown samples, relative to control-siRNA (Cntl-siRNA). The results of gene ontology (GO) analysis of differentially expressed genes are presented (right panels). (B) Heat map of twenty p53 down-stream genes that are down-regulated in lincRNA-p21 knockdown samples. (C-D) p53 mRNA (C) and protein (D) expression levels were determined by qRT-PCR and Western blotting, respectively. (E) The mRNA levels of Mdm2, Puma, Bax, and Noxa were determined by qRT-PCR. (F) The protein levels of MDM2, PUMA, BAX, and NOXA were determined by Western blotting. (G) The expression of lincRNA-p21 was determined by qRT-PCR in RAW264.7 cells. (H) The mRNA levels of Mdm2, Puma, Bax, and Noxa were determined by qRT-PCR. (I) The protein levels of MDM2, PUMA, BAX, and NOXA were determined by Western blotting. All values are the average of at least 3 biological replicates and data shown are the mean±SD. * P<0.05 relative to control.
LincRNA-p21 interacts with MDM2
The above results indicated that lincRNA-p21 participates in the regulation of p53-dependent target gene expression without altering the expression level of p53 itself, suggesting that lincRNA-p21 might instead modulate the transcriptional activity of p53. Previous work showed that p300 and MDM2 play important roles in the regulation of p53 activity. P300 is an acetyltransferase which acetylates p53 and thereby enhance its activity. MDM2 antagonizes p53 by enhancing its degradation via the ubiquitin-proteasome pathway and by blocking its binding and acetylation by p300 7-9.
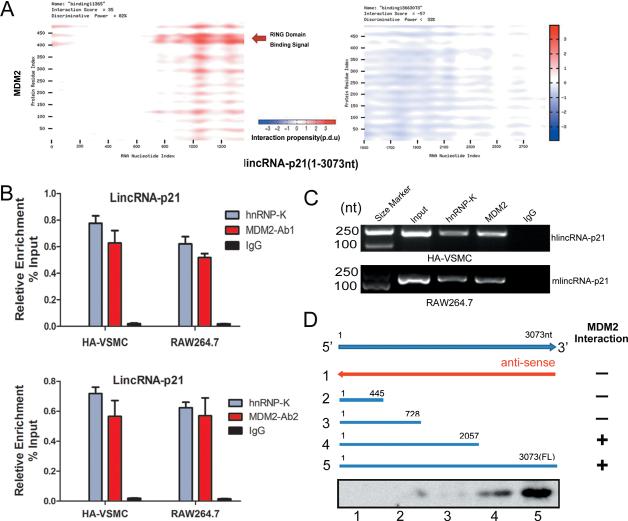
The MDM2 protein contains several conserved structural domains including an N-terminal p53 interaction domain and a C-terminal RING domain (amino acid residues 430-480), which confers its E3 ubiquitin ligase activity. Intriguingly, the C-terminal Ring domain also binds to RNA, indicating MDM2 could function as a RNA binding protein (RBP). To explore possible MDM2 binding to lincRNA-p21, we used computational approaches to assess the likelihood of protein-RNA interaction. catRAPID, a predictor of protein-RNA binding38 (http://big.crg.cat/gene_function_and_evolution/services/catrapid), predicted nt 700-1500 of the lincRNA-p21 binding to the RING domain (amino acid residues 400-480) of MDM2 with the discriminative power of 82%. In contrast, the nt 1501-3000 region of the lincRNA-p21 presented no interaction signal (Figure 3A).
Figure 3.
LincRNA-p21 physically interacts with MDM2. (A) Predicted interaction of lincRNA-p21 (nucleotide positions 1000-1500) and MDM2 protein (amino acid residues 400-450) were predicted to interact. (B) RNA Immunoprecipitation (RIP) experiments were performed using MDM2 antibody 1 (upper) or antibody 2 (lower) in both HA-VSMC and RAW264.7 cells. qRT-PCR was performed to detect pulled-down lincRNA-p21. hnRNP-K antibody and IgG were used as positive and negative controls, respectively. (C) MDM2 associated lincRNA-p21 was detected by regular RT-PCR assays. (D) Mapping the MDM2 interaction region of lincRNA-p21. Biotinylated RNAs corresponding to different fragments of lincRNA-p21 or its antisense sequences (red line) were co-incubated with cell lysates and associated MDM2 proteins were detected by Western blotting.
Next, we examined the direct binding of lincRNA-p21 and MDM2 through RNAImmunoprecipitation (RIP) in human HA-VSMC and mouse RAW264.7 cells. We used two independent anti-MDM2 antibodies in order to demonstrate the specificity of the interaction. RIP results showed that lincRNA-p21 interacts with MDM2 protein in both cell lines (Figures 3B and 3C). Deletion-mapping experiments showed that the nt728-2057 region of the lincRNA-p21 mediates the interaction with the RING domain of the MDM2 protein in mouse RAW264.7 cells (Figure 3D).
LincRNA-p21 regulates the transcriptional activity of p53
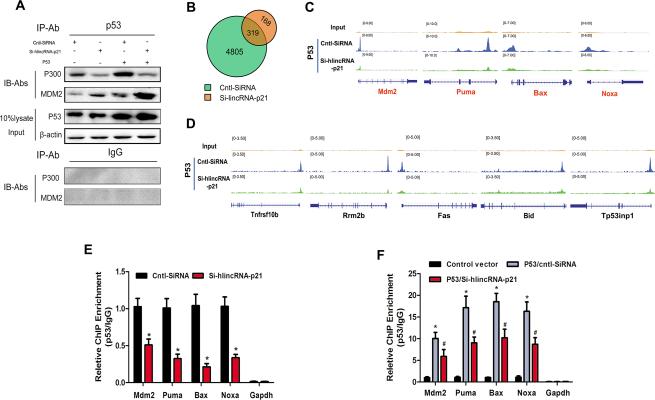
To understand how the lincRNA-p21/MDM2 interaction affects the formation of the p53/p300/MDM2 complex, co-immunoprecipitation (Co-IP) experiments were performed. We confirmed the interaction of p300 and p53, and MDM2 and p53, consistent with prior reports 12-14. As expected, the interaction of p300/p53 and MDM2/p53 increased in p53 overexpressing cells. LincRNA-p21 knockdown decreased p300/p53 interaction and increased MDM2/p53 interaction. In contrast, no interaction was detected when IgG was used for IP, demonstrating the specificity of the co-immunoprecipitation interaction assays (Figure 4A).
Figure 4.
LincRNA-p21 modulates p53 activity by augmenting p53-p300 interaction. (A) Co-Immunoprecipitation (Co-IP) assays to detect the interaction of p53, p300, and MDM2. HA-VSMCs were transfected with Si-hlincRNA-p21, or control SiRNA (Cntl-SiRNA), in the presence or absence of p53 overexpression. Cell lysates were immunoprecipitated with p53 antibodies (or IgG to serve as a negative control) and associated proteins were detected using p300 and MDM2 antibodies, respectively. 10% of the input cell lysates were loaded as controls. (B-E) ChIP-Seq assays were carried out using p53 antibodies in HA-VSMCs after lincRNA-p21 knockdown. (B) Venn diagram showing p53 ChIP-Seq peaks in control or lincRNA-p21 knockdown cells. (C) Distribution of reads obtained by ChIP-Seq in lincRNA-p21 knockdown or control HA-VSMCs at four loci highly associated with the p53–apoptosis pathway (D) Distribution of reads in lincRNA-p21 knockdown or control HA-VSMCs at five additional loci associated with the p53–pathway (E) ChIP-qPCR assays. Mdm2, Puma, Bax and Noxa promoter/enhancer regions containing p53 binding sites were quantified by PCR in control or lincRNA-p21 knockdown cells. (F) ChIP-qPCR assays. Mdm2, Puma, Bax and Noxa promoter/enhancer regions containing p53 binding sites were quantified by PCR in control or lincRNA-p21 knockdown cells, with or without p53 overexpression. Fold change of enrichment was determined relative to IgG controls. All values are the average of at least 3 biological replicates and data shown are the mean±SD. * P<0.01 relative to control, # p<0.05 relative to p53/Cntl-SiRNA.
We next investigated if inhibition of lincRNA-p21, which affected interactions between p300, MDM2, and p53 proteins without altering p53 protein level, influenced p53 binding to the promoters/enhancers of its target genes. We performed p53 chromatin immunoprecipitation (ChIP) followed by high-throughput DNA sequencing (ChIP-Seq) experiments in HA-VSMCs treated with control or lincRNA-p21 siRNA. We obtained 36-53 million sequence reads from each experimental sample, and greater than 95% of them uniquely aligned to the human genome (Supplemental Figure 1). In control siRNA-treated samples, we identified more than 4,800 p53-bound regions. Intriguingly, lincRNA-p21 knockdown diminished p53 binding at many of these regions (Figure 4B), suggesting that lincRNA-p21 is required for p53 to bind to many of its targets. We compared the peak distributions of Mdm2, Puma, Bax and Noxa, four known p53-regulated genes, in control and lincRNA-p21 knockdown cells and found that knockdown of lincRNA-p21 dramatically reduced the association of p53 to the promoters/enhancers of these genes (Figure 4C). Similarly, we observed that lincRNA-P21 knockdown diminished the binding of p53 to promoters/enhancers of many additional targets (Figure 4D). ChIP-PCR assays further confirmed the above observation (Figure 4E). To verify that lincRNA-p21 functionally regulates p53 recruitment to target regulatory regions, we overexpressed p53 with or without knocking down endogenous lincRNA-p21 and measured the binding of p53 to the regulatory regions of target genes. ChIP results showed that p53 recruitment to its target promoters/enhancers was diminished when lincRNA-p21 was knocked down (Figure 4F). These observations suggest that lincRNA-p21, a transcriptional target of p53, can feed back and regulate the activity of p53, at least in part, by modulating the interaction of p53, p300 and MDM2.
LincRNA-p21 modulates the function of p53 in regulating cell proliferation and apoptosis
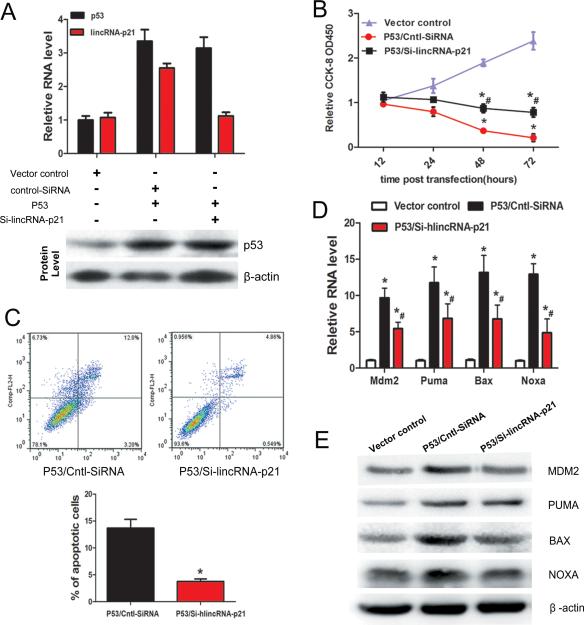
To investigate the interplay of p53 and lincRNA-p21 on cell proliferation, apoptosis and atherosclerosis, we overexpressed p53 in human VSMC cells, with or without lincRNA-p21 knockdown. Whereas overexpression of p53 induced the expression of endogenous lincRNA-p21, lincRNA-p21 knockdown had no effect on p53 expression, consistent with the prior observations 33 (Figure 5A). Overexpression of p53 inhibited VSMC proliferation and viability, evidenced by a dramatic decrease in direct cell counting (Figure 5B). Inhibition of lincRNA-p21 suppressed p53-mediated inhibition of VSMC proliferation, indicating that lincRNA-p21 modulates p53-dependent cell proliferation (Figure 5B). Similarly, p53-induced apoptosis was markedly repressed in lincRNA-p21 knockdown cells (Figure 5C). Moreover, lincRNA-p21 inhibited the stimulatory effect of p53 overexpression on levels of Mdm2, Puma, Bax and Noxa (Figures 5D and 5E). Together, these results demonstrate that lincRNA-p21 functions through the p53 pathway to regulate cell proliferation and apoptosis.
Figure 5.
LincRNA-p21 regulates p53-dependent cell proliferation and apoptosis. (A) HA-VSMCs were transfected with pCMV vector (vector control), p53 overexpression vector (p53), together with Si-lincRNA-p21 (or cntl-SiRNA). The mRNA and protein expression levels of lincRNA-p21 and p53 were determined by qRT-PCR and Western blotting, respectively. (B) The proliferation and viability of HA-VSMCs were measured using the Cell Counting Kit-8 (CCK-8) assay after p53 overexpression and lincRNA-p21 knockdown. *P<0.01 relative to control; #p<0.01 relative to p53/Cntl-SiRNA. (C) LincRNA-p21 suppressed p53-induced apoptosis. Cellular apoptosis of HA-VSMCs was measured and quantified via Annexin-V conjugated FACS analysis. The percentage of apoptotic cells was calculated (lower panel). (D) Depletion of lincRNA-p21 inhibited p53-mediated activation of transcription of down-stream target genes. The mRNA levels of Mdm2, Puma, Bax, and Noxa were determined by qRT-PCR. (E) The protein levels of MDM2, PUMA, BAX, and NOXA were determined by Western blotting. All values are the average of at least 3 biological replicates and data shown are the mean±SD. * P<0.01 relative to control; #p<0.01 relative to p53/Cntl-SiRNA.
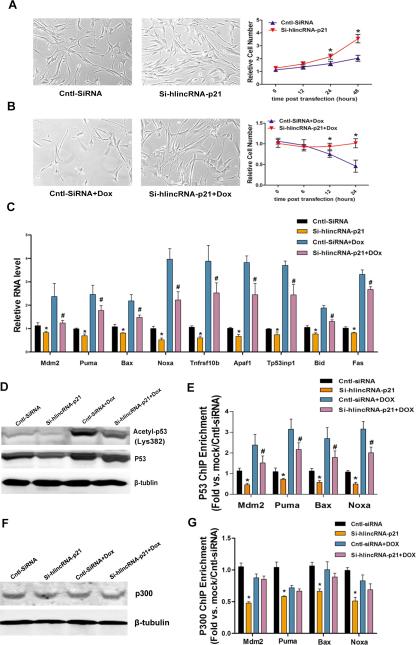
It is well established that p53 regulates cell proliferation and apoptosis in response to stress 48. We tested whether the functional involvement of lincRNA-p21 and p53 in cell proliferation is regulated by treatment of doxorubicin (Dox), an anti-cancer chemotherapy drug which also causes cardiotoxicity 49. Knockdown lincRNA-p21 increased whereas Dox treatment reduced cell number in culture (Figures 6A and 6B), consistent with previous observation. Si-lincRNA-p21 partially suppressed the Dox-induced reduction of cell number (Figure 6B). We further examined apoptosis and cell proliferation under such conditions. We found that Dox treatment significantly induces apoptosis, marked by increased TUNEL staining, which is partially suppressed when lincRAN-p21 was knocked down (Supplemental Figure 2A). Conversely, Dox treatment reduced cell proliferation, evidenced by decreased Ki67 labeling. Knockdown of lincRNA-p21 restored Dox-inhibited cell proliferation (Supplemental Figure 2B). Next, we examined the expression of p53 target genes in cells treated with Dox and si-lincRNA-p21. We found that whereas Dox treatment increased the expression of p53 target genes, knocking down endogenous lincRNA-p21 partially suppresses such increase of gene expression (Figure 6C).
Figure 6.
LincRNA-p21 regulates p53-dependent cell proliferation and apoptosis in response to stresses. (A-B) Relative cell number of HA-VSMCs at different time points was calculated after lincRNA-p21 knockdown without (A) or with doxorubicin treatment (B). Cntl-SiRNA treatment serves as controls. (C) The mRNA levels of indicated p53 target genes were determined by qRT-PCR. (D) The protein levels of total and acetylated p53 were determined by Western blotting. (E) p53 ChIP-qPCR assays. Mdm2, Puma, Bax and Noxa promoter/enhancer regions were quantified by PCR in control or lincRNA-p21 knockdown cells with or without doxorubicin treatment. (F) The protein levels of p300 were determined by Western blotting. (G) P300 ChIP-qPCR assays. Mdm2, Puma, Bax and Noxa promoter/enhancer regions were quantified by PCR in control or lincRNA-p21 knockdown cells with or without doxorubicin treatment. All values are the average of at least 3 biological replicates and data shown are the mean±SD. * P<0.01.
We asked whether knockdown of lincRNA-p21 might affect p53 protein level and its acetylation status under stress condition. We found that Dox treatment induced the level of acetylated p53 significantly, which was reduced when endogenous lincRNA-p21 was inhibited (Figure 6D). Next, we investigated whether Dox treatment and lincRNA-p21 knockdown could alter the binding of p53 to the promoters/enhancers of its target genes. ChIP-PCR showed that Dox enhances the binding of p53 to its targets, which was reduced when lincRNA-p21 was knocked down (Figure 6E). In contrast, we found that Dox treatment and lincRNA-p21 knockdown did not affect the expression level of p300 proteins (Figure 6F), neither did such treatment alter p300 binding to p53 target genes (Figure 6G). Together, these studies suggest that linRNA-p21 modulates the activities and functions of p53 in regulating its target gene expression in response to stresses.
LincRNA-p21 inhibits neointima formation in carotid arteries
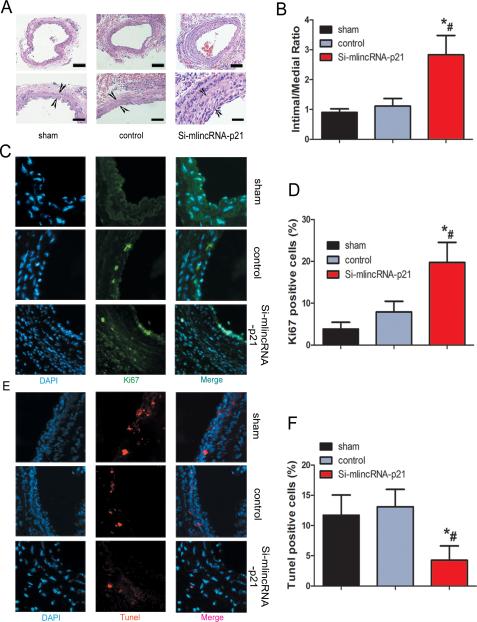
Next, we investigated the involvement of lincRNA-p21 in the formation of neointima in vivo, using the classic murine carotid artery injury model 50. Recombinant lentivirus vector expressing lincRNA-p21 siRNA or control siRNA was injected into the injured area of mouse carotid arteries. These mice were then fed with high fat diet for one month, and neointima formation was examined. We verified the reduction of lincRNA-p21 expression in local-injury carotid tissues after lentivirus-si-lincRNA-p21 injection (Supplemental Figure 3). Knockdown of lincRNA-p21 resulted in dramatic neointimal hypertplasia when compared with controls (Figure 7A). Quantification of intima-media thickness confirmed a significant increase after siRNA-lincRNA-p21 injection (Figure 7B). We asked whether inhibition of lincRNA-p21 affected cell proliferation and apoptosis in vivo. We performed immunostaining on vessel sections to detect the proliferation marker Ki67. The fraction of Ki67+ cells increased in siRNA-lincRNA-p21 treated vessels (Figures 7C and 7D). LincRNA-p21 siRNA-treated vessels also showed decreased apoptosis, as assessed using the terminal deoxynucleotidyl transferase-mediated nick-end labeling (TUNEL) assay (Figures 7E and 7F).
Figure 7.
Inhibition of lincRNA-p21 results in increased neointima formation. (A) Lentivirus vectors for lincRNA-p21 knockdown (si-mlincRNA-p21), or control si-RNA (control) were in-site injected into the injured area of mouse carotid arteries. Sham operation serves as controls. Carotid arteries were harvested 30 days later after feeding mice with high-fat diet. H&E staining was performed to show the thickness of neointima. (B) Quantification of the intima-media thickness of sham, control si-RNA and si-lincRNA-p21 treated samples. (C) Representative immunofluorescence images of Ki67 in mouse carotid arteries. DAPI staining marks cell nuclei. (D) Quantification of the Ki67 positive signals of sham, control si-RNA and si-lincRNA-p21 treated samples. (E) Representative immunofluorescence images of TUNEL staining in mouse carotid arteries. DAPI staining marks cell nuclei. (F) Quantification of the TUNEL positive signals of sham, control si-RNA and si-lincRNA-p21 treated samples. All values are the average of at least 3 biological replicates and data shown are the mean±SD. * P<0.05 relative to control.
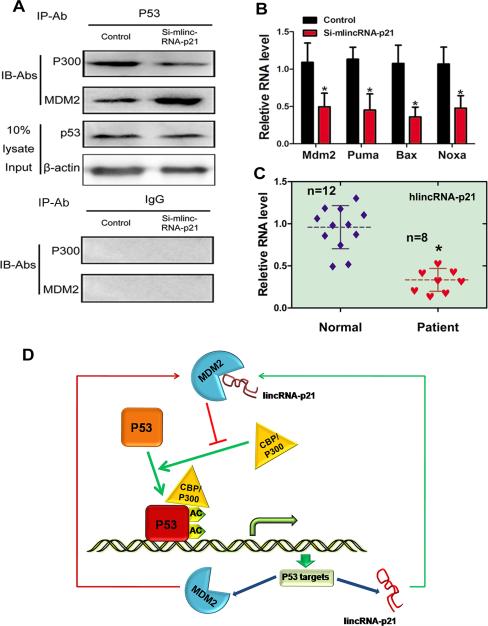
To further uncover the role of lincRNA-p21 on the interaction of p53-p300-MDM2 proteins in vivo, we examined the effect of lincRNA-p21 knockdown on p300/p53 and MDM2/p53 interactions in carotid tissues. Consistent with in vitro studies, there was a reduced binding of p300 and p53 in si-lincRNA-p21 samples. Conversely, the association of MDM2 and p53 was increased in si-lincRNA-p21 samples (Figure 8A). As a result, the expression levels of p53 targets Mdm2, Puma, Bax and Noxa were repressed in si-lincRNA-p21 treated vessels in vivo (Figure 8B).
Figure 8.
Role of lincRNA-p21 in p53-mediated atherosclerosis. (A) Cell lysates from si-lincRNA-p21 (or cntl-siRNA) treated carotid arteries were immunoprecipitated with p53 antibodies (or IgG to serve as a negative control) and associated proteins were detected using anti- p300 and MDM2 antibodies, respectively. 10% cell lysate were loaded to serve as controls. (B) The mRNA levels of Mdm2, Puma, Bax, and Noxa were determined by qRT-PCR from si-lincRNA-p21 (or cntl-siRNA) treated carotid artery samples. (C) RNAs isolated from coronary artery tissues of coronary artery disease patiens and aorta tissues of control patients were subjected to qRT-PCR assays to detect lincRNA-p21 expresson levels. The solid and dashed horizontal lines represent standard deviation (SD) and mean respectively. n=8 for each experimental groups. (D) Schematic representation of a working model by which lincRNA-p21 feeds back on the function of p53 via binding to MDM2.
Decreased lincRNA-p21 expression in patients with coronary heart disease
Lastly, to determine whether dysregulated lincRNA-p21 expression is associated with coronary artery disease, we examined the expression of human lincRNA-p21, by quantitative RT-PCR assays, using total RNAs isolated from coronary artery tissues of coronary artery disease patients and artery tissues of control patients. Indeed, expression of lincRNA-p21 was more than 50% lower in coronary artery disease patients compared to control patients (Figure 8C). Similarly, we tested lincRNA-p21 level in another set of coronatry artery disease and control patients using total RNAs isloated from peripheral blood mononuclear cells and we found that lincRNA-p21 level was also decreased in coronatry artery disease patients (Supplemental Figure 4). These results implicate lincRNA-p21 in the development of atherosclosis and coronary artery disease.
Discussion
In this study, we indentified lincRNA-p21 as a key regulator of cell proliferation and apoptosis. We showed that lincRNA-p21 represses cell proliferation and induces apoptosis in vitro and in vivo. Knockdown of endogenerous lincRNA-p21 accelerated neointima formation in injured carotid arteries. Mechanistically, we found that lincRNA-p21 directly binds to MDM2, leading to p53 release from MDM2 and binding to p300, which thereby enhances p53 activity. This finding is significant because it implicates non-coding RNAs in cardiovascular diseases such as atherosclerosis, and suggests that modulation of the activity of non-coding RNAs such as lincRNA-p21 may be a novel therapeutic approach to treat human cardiovascular disease. It is well-known that p53 plays an important role on the pathogenesis of atherosclerosis 1-6. The expression and transcriptional activity of p53 is tightly regulated at multiple levels. Especially, post-transcriptional regulation by ubiquitination and acetylation are known to be essential for the function of p53 proteins. MDM2, a direct transcriptional target gene of p53, appears to take part in both pathways. On one hand, p53 proteins can be degraded by MDM2 via ubiquitination pathway 10, 11. On the other hand, p53 is well-known to be acetylated by the acetyltransferase p300, resulting in dramatic induction of p53 activity 7-9. Interestingly, this effect is antagonized by p53 interaction with MDM2, which inhibits the formation of the p300-p53 complex. Clearly, this dynamic interaction between p300, MDM2, and p53 critically regulates the activity and function of p53, and positions MDM2 as a core regulator of p53 activity 21, 51, 52. A prior study showed that lincRNA-p21, which was identified as a transcriptional target of p53, feeds back to stimulate p53 function through its interaction with the RNA-binding protein hnRNP-K 33. Our study defines a second, MDM2-dependent mechanism by which lincRNA-p21 regulates p53 activity: lincRNA-p21 directly binds to MDM2, reducing p53-MDM2 interaction and increasing p53-p300 interaction. Consequently, the transcriptional activity of p53 is increased. Therefore, our investigation uncovered a novel mechanism by which p53 and lincRNA-p21 regulate each other's expression and activity in a feedback manner (Figure 8D).
Of note, we observed that lincRNA-p21 utilizes its middle region (nt 728-2053) for the binding of MDM2. Hurte et al. localized the hnRNP-k-binding region to the 5’ region (nt 1-778) of lincRNA-p21 33. Apparently, lincRNA-p21 uses two distinct regions to bind to two different proteins. It will be interesting to determine whether lincRNA-p21 binds to MDM2 and hnRNP-k simultaneously and whether binding to one protein modulates the binding to the other. Furthermore, it will be important to determine whether such association is regulated in pathophysiological conditions, in particular the cardiovascular diseases. Are there functional correlations of lincRNA-p21 binding to both MDM2 and hnRNP-k? Our present study uncovered the involvement of lincRNA-p21 in atherosclerosis, at least in part, by interplaying with p53 in a feedback mechanism. It will be interesting to explore whether lincRNA-p21 can interact with epigenetic factors, like PRC1 and PRC2 to regulate the expression of atherosclerosis-related genes. Furthermore, we speculate that variants of lincRNA-p21 may be genetically linked to atherosclerosis. Given the existing of large numbers of lncRNAs and their important biological function, it is not unreasonable to predict that additional novel lncRNAs will be identified and linked to atherosclerosis and cardiovascular disease in the near future.
Supplementary Material
Acknowledgements
We thank members of our laboratories for advice and support. We thank Dr. William Pu for stimulating discussion and careful reading of the manuscript.
Funding Sources: This work is supported by grants from National Natural Science Foundation of China (30925018, 31130029), National Basic Research Program of China (2013CB531104, and 2012CB517801). DZ Wang is supported by the March of Dimes Foundation and the National Institutes of Health (HL085635, HL116919). ZP Huang is supported by the NIH training grant T32 HL007572. DZ Wang was an Established Investigator of the American Heart Association.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Guevara NV, Kim HS, Antonova EI, Chan L. The absence of p53 accelerates atherosclerosis by increasing cell proliferation in vivo. Nat Med. 1999;5:335–9. doi: 10.1038/6585. [DOI] [PubMed] [Google Scholar]
- 2.Merched AJ, Williams E, Chan L. Macrophage-specific p53 expression plays a crucial role in atherosclerosis development and plaque remodeling. Arterioscler Thromb Vasc Biol. 2003;23:1608–14. doi: 10.1161/01.ATV.0000084825.88022.53. [DOI] [PubMed] [Google Scholar]
- 3.van Vlijmen BJ, Gerritsen G, Franken AL, Boesten LS, Kockx MM, Gijbels MJ, Vierboom MP, van Eck M, van De Water B, van Berkel TJ, Havekes LM. Macrophage p53 deficiency leads to enhanced atherosclerosis in APOE*3-Leiden transgenic mice. Circ Res. 2001;88:780–6. doi: 10.1161/hh0801.089261. [DOI] [PubMed] [Google Scholar]
- 4.Mercer J, Bennett M. The role of p53 in atherosclerosis. Cell Cycle. 2006;5:1907–9. doi: 10.4161/cc.5.17.3166. [DOI] [PubMed] [Google Scholar]
- 5.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer J, Figg N, Stoneman V, Braganza D, Bennett MR. Endogenous p53 protects vascular smooth muscle cells from apoptosis and reduces atherosclerosis in ApoE knockout mice. Circ Res. 2005;96:667–74. doi: 10.1161/01.RES.0000161069.15577.ca. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 8.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–41. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–9. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 11.Kubbutat MH, Jones SN. Vousden KH. Regulation of p53 stability by Mdm2. Nature. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 12.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 13.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–60. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 14.Thut CJ, Goodrich JA, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–86. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–23. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 16.Gerstein M. Genomics: ENCODE leads the way on big data. Nature. 2012;489:208. doi: 10.1038/489208b. [DOI] [PubMed] [Google Scholar]
- 17.Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, Segal E. Genomics: ENCODE explained. Nature. 2012;489:52–5. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- 18.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 21.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 23.Espinoza-Lewis RA, Wang DZ. MicroRNAs in heart development. Curr Top Dev Biol. 2012;100:279–317. doi: 10.1016/B978-0-12-387786-4.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 26.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–6. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–89. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 30.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 36.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 37.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellucci M, Agostini F, Masin M, Tartaglia GG. Predicting protein associations with long noncoding RNAs. Nat Methods. 2011;8:444–5. doi: 10.1038/nmeth.1611. [DOI] [PubMed] [Google Scholar]
- 39.Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics. 2011;12:489. doi: 10.1186/1471-2105-12-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, Day DS, Ho JW, Song L, Cao J, Christodoulou D, Seidman JG, Crawford GE, Park PJ, Pu WT. A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome Res. 2013;23:917–27. doi: 10.1101/gr.149674.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature protocols. 2012;7:1728–40. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohno T, Gordon D, San H, Pompili VJ, Imperiale MJ, Nabel GJ, Nabel EG. Gene therapy for vascular smooth muscle cell proliferation after arterial injury. Science. 1994;265:781–4. doi: 10.1126/science.8047883. [DOI] [PubMed] [Google Scholar]
- 43.Varenne O, Pislaru S, Gillijns H, Van Pelt N, Gerard RD, Zoldhelyi P, Van de Werf F, Collen D, Janssens SP. Local adenovirus-mediated transfer of human endothelial nitric oxide synthase reduces luminal narrowing after coronary angioplasty in pigs. Circulation. 1998;98:919–26. doi: 10.1161/01.cir.98.9.919. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–82. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 45.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–94. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 46.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–9. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 47.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–8. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 48.Perry ME. The regulation of the p53-mediated stress response by MDM2 and MDM4. Cold Spring Harbor perspectives in biology. 2010;2:a000968. doi: 10.1101/cshperspect.a000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takemura G, Fujiwara H. Doxorubicin-induced cardiomyopathy from the cardiotoxic mechanisms to management. Progress in cardiovascular diseases. 2007;49:330–52. doi: 10.1016/j.pcad.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 50.Hui DY. Intimal hyperplasia in murine models. Curr Drug Targets. 2008;9:251–60. doi: 10.2174/138945008783755601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–40. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, Kumar S, Howley PM, Livingston DM. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–15. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.