Abstract
Technologies for genome-wide sequence interrogation have dramatically improved our ability to identify loci associated with complex human disease. However, a chasm remains between correlations and causality that stems, in part, from a limiting theoretical framework derived from Mendelian genetics, and an incomplete understanding of disease physiology. Here we propose a set of criteria, akin to Koch’s postulates for infectious disease, for assigning causality between genetic variants and human disease phenotypes.
“…Thus it is easy to prove that the wearing of tall hats and the carrying of umbrellas enlarges the chest, prolongs life, and confers comparative immunity from disease; for the statistics show that the classes which use these articles are bigger, healthier, and live longer than the class which never dreams of possessing such things. It does not take much perspicacity to see that what really makes this difference is not the tall hat and the umbrella, but the wealth and nourishment of which they are evidence, and that a gold watch or membership of a club in Pall Mall might be proved in the same way to have the like sovereign virtues….”
George Bernard Shaw, The Doctor’s Dilemma (Preface), 1909.
Distinguishing correlation from causality is the essence of experimental science. Nowhere is the need for this distinction greater today than in complex disease genetics, where proof that specific genes have causal effects on human disease phenotypes remains an enormous burden and challenge. Given the potential scientific and medical payoffs of disease gene discovery (Chakravarti, 2001), we argue in this essay of the need for a rigorous examination of the assumptions under which we connect genes to phenotypes. This is particularly so in this age of routine -omic surveys which can produce more false positive than true positive findings (Kohane et al., 2006). Moreover, genomic mapping and sequencing approaches that are invaluable for producing a list of unbiased candidates are, by themselves, insufficient for implicating specific gene(s) in a disease or biological process. Consequently, we suggest that specific genetic criteria, analogous to Koch’s postulates in microbiology, need to be satisfied in order to promote the role of one or more genes as being ‘causal,’ rather than just ‘associated,’ in a disease process (Brown and Goldstein, 1992; Falkow, 1988; Falkow, 2004) (Box 1).
Box 1: Koch’s Postulates for Complex Human Diseases & Traits.
Candidate gene variants are enriched in patients;
Disruption of the gene in a model system gives rise to a phenotype that is accepted as relevant and “equivalent” to the human phenotype;
Phenotype in model system can be rescued with wild-type human alleles;
Phenotype in model system cannot be rescued with mutant human alleles.
Below we discuss the nature of ‘proof’ we desire in order to make fundamental discoveries in human pathophysiology. We admit at the outset that the answers are not straightforward, and that there are serious technical and intellectual impediments to demonstrating causality for the common complex disorders of man where multiple interacting genes are involved. We acknowledge that even unproven candidate genes may lead to significant insight into disease pathophysiology. Nevertheless, the casual conflation of ‘mapped locus’ to ‘proven gene’ is a constant source of confusion and obfuscation in biology and medicine that requires remedy. We hope to offer some concrete suggestions, however difficult they may be to satisfy, since incorrect knowledge is worse than no knowledge at all (Brown and Goldstein, 1992).
Consider that two types of genomic surveys, one horizontal and the other vertical, are now routine for attempting to understand human biology and disease. In horizontal or broad surveys, we can obtain the full genome sequence in tens to hundreds of thousands of individuals to sort out which genomic segments are important, and which innocent bystanders, to a particular comparison between individuals, such as those with versus without coronary artery disease, or cases with early versus late onset of dementia. In contrast, in vertical or deep surveys, we examine the effects of the genome as the DNA information gets processed, and its encoded functions get executed through its transcriptome, proteome and effectors such as the metabolome. Both of these classes of studies are relevant to analysis of a disease of unknown etiology and have re-emphasized the long-held suspicion that studying genes one-at-a-time may not be meaningful since a genes’ effect is usually pleiotropic, context-dependent, and contingent upon the state of many other genetic and non-genetic factors (Chin et al., 2012). In turn, this implies that proving a genes’ specific role in a biological process, either in wild-type or mutant form, may not be straight-forward since its role may only be evident when examined in relation to its biochemical partners, and in particular contexts of diet, pathogen exposure, etc. (Zerba et al., 1996). This is a particular problem in genetic studies of any outbred non-experimental organism, such as the human, and studies of human disease where investigations are observational not experimental. It is the strong belief of contemporary human geneticists that uncovering the genetic underpinnings of any disease, however complex, is the surest unbiased route to understanding its pathophysiology, and, thus, enabling its future rational therapies (Brooke et al., 2008). Consequently, for this view to prevail, we should require experimental evidence, be they in cells, tissues, experimental models or the rare patient, for the role of a specific gene in a disease process. We discuss here the types of evidence that we consider incontrovertible.
Success in this difficult task requires us to solve a logical conundrum: how can we understand the genes underlying a phenotype if some of these component factors, in isolation, do not have recognizable phenotypes on their own? We know that even in a simple model organism, budding yeast that synthetic lethality—where death or some other phenotype occurs only through the conspiracy of mutations at two different genes—is widely prevalent (Costanzo et al., 2010). Interactions of greater complexity and involving more than two genes are also known in yeast (Hartman et al., 2001) and must be true for humans as well. A human genome will typically harbor 20 genes that are fully inactivated, without any overt disease phenotype, presumably due to the buffering by other genes (MacArthur et al., 2012). Acknowledging this complexity, there are two general ways forward. First, at this stage of our knowledge, perhaps we should not worry about ‘all’ of the genes in a disease, in many ways an undefinable goal, but rather those whose effects are demonstrable, i.e., through a mutation that, irrespective of its interactions, can by itself affect a critical pathway. Second, as we unravel the effects of multiple genes on a phenotype we should advance the same criterion, namely, that a set of mutations affects that same critical process. Both of these goals are approachable, particularly with recent advances in genome editing technologies that allow the creation of multiple mutations within a single experimental organism (Wang et al., 2013). The question then is how ‘complex’ are complex traits and diseases?
The new genetics: understanding the function of variation
With the rediscovery of Mendel’s rules of transmission more than 100 years ago, there was a vicious debate on the relative importance of single gene versus multifactorial inheritance (Provine, 1971). Geneticists quickly, and successfully, focused on deciphering the specific mechanisms of gene inheritance and understanding the physiology of the gene in lieu of answering why some phenotypes had complex etiology and transmission. Nevertheless, the rare examples of deciphering the genetic basis of complex phenotypes, such as for truncate (wing) in Drosophila (Altenburg and Muller, 1920), clearly emphasized that traits were more than the additive properties of multiple genes. Today, it is quite clear that Mendelian inheritance of traits, including diseases, is the exception not the rule. Nevertheless, the entire language of genetics is in terms of individual genes for individual phenotypes, with one function, rather than the ensemble and emergent property of genomes. This absence of a specific genetics language for the proper description of the multigenic architecture of traits (the ensemble), remains as an impediment to our understanding of the nature and degree of genetic complexity of the phenotype.
The case of amyotrophic lateral sclerosis (ALS), a devastating, progressive motor neuron disease, illustrates this point (Ludolph et al., 2012). Despite the lack of evidence, we largely describe ALS as being ‘heterogeneous’ and comprised of single gene mutations that can individually lead to disease. In 1993, mutations in superoxide dismutase 1 (SOD1) were identified in an autosomal dominant form of the disease; subsequently, the disorder has become synonymous with aberrant clearance of free radicals as its central pathology. What is often not appreciated, however, is that fewer than 10% of all cases of ALS are familial and even fewer follow an apparent Mendelian pattern. Even within this subset of cases, more than 20 distinct genes, spanning other pathways including RNA homeostasis, have been identified, and SOD1 represents a minority of cases. The molecular etiology for the majority of the sporadic forms of the disease remains unclear, and the scientific problem in understanding ALS is more than simply identification of additional genes. We may ask: can SOD1 and the other described gene mutations lead to ALS by themselves? Are these the key rate-limiting steps to ALS or simply one of several required in concert? Is the aberrant clearance of free radicals the fundamental defect or one of many such pathologies or a common downstream consequence? Given the diversity and number of deleterious, even loss-of-function, genetic variants in all of our genomes (1000 Genomes Project Consortium et al., 2012; MacArthur et al., 2012), and, in the absence of stronger evidence bearing on these questions, it is fair to assume that ALS patients harbor multiple mutations with a plurality of molecular defects and that free radical metabolism is only one of a set of canonical pathophysiologies that define the disease. No doubt, this plurality is the case for cancer (Vogelstein et al., 2013), Crohn’s disease (Jostins et al., 2012) and even rare developmental disorders such as Hirschsprung disease (McCallion et al., 2003). In all of these cases, a richer genetics vocabulary may improve our understanding of the phenotypes through recognizing what we know and what we don’t; our current language limits us to describing genes not phenotypes.
Molecular biology, genetics’ twin, on the other hand, appears to have been far more successful in deciphering and describing not only its individual components (e.g., DNA, RNA, protein) but also their mutual relationships (e.g., DNA-binding protein), and ensembles (e.g., transcriptional complex), although this is also far from complete (Watson et al., 2007). We not only understand the structure of individual genes, how their molecular functions get executed, but we are also starting to learn how functions get regulated through a diversity of cis- and trans-acting functions. The consequences of the primary and interaction effects are often well understood, even though not completely described, at both the molecular and cellular levels (Alberts et al., 2007). There are also improving technologies and understanding of the structures and functions of ensembles of proteins and cells, and how these interact and communicate with one another to create complexity (Ilsley et al., 2013). Although the use of genetic tools and genetic perspectives are fundamental to this progress, these advances have not as yet led to a major revision of our understanding of trait or disease variation. The major reason for this discrepancy is that, with few exceptions (Raj et al., 2010), molecular and cell biology has focused on the impact of deleting or overexpressing genes, and not grappled with the consequences of allelic variation.
Classical Mendelian genetics has been a boon to uncovering biology from yeast to humans whenever a mutation with a simple inheritance pattern can be isolated. This approach has been revolutionary in the unicellular yeast, particularly because genetics (and gene manipulation), biochemistry and cell biology was melded to understand function at a variety of levels. This kind of multi-level approach has been less straightforward, but still largely successful, for a metazoan such as Drosophila where more genes and multiple specialized cells often rescue the effects of a mutation or enhance its minor effect. These lessons suggest to us that the current approach, based strictly on genetic variation, to understanding complex human disease is also grossly insufficient, and, as in yeast and flies, will require the contemporaneous analysis of the molecular biology, biochemistry and physiology of the genes within a mapped locus to even identify the disease gene, let alone understand its functions. Success in this endeavor will require a synthesis of many biological disciplines that includes the role of genetic variation as intrinsic to the biological process, not an aspect to be ignored.
Consequently, melding variation-based genetic and molecular biological thinking is of critical importance for both fields, and is central to our understanding of mechanisms of trait variation, including inter-individual variation in disease risk. If most disease, in most humans, is the consequence of the effects of variation at many genes, then knowledge of their functional relationships, rather than merely their identities, is central to understanding the phenotype. This is clearly a problem of ‘Systems Biology’ but one that incorporates genetic variation directly. The ability to integrate the realities of such widespread genetic variation, which are ultimately at the causal root of disease mechanisms, with systems biology approaches to understand functional contingencies, is central to the challenge of deciphering complex human disease. Importantly, it is likely to spur new thinking in both fields.
Genetic dissection of complex phenotypes
Genetic transmission rules imply that, even in an intractable species such as us, one can map genomic segments that must contain a disease or trait gene. The lure and success of this method is that we can map a disease locus in the absence of any knowledge of the underlying biology of the phenotype. Such mapping requires identification of the segregation of common sites of variation across the genome, now easy to identify through sequencing, and recognition of a genomic segment identical-by-descent in affected individuals, both within and between families. This task has become easier and more powerful as sequencing technology has improved to provide a nearly complete catalog of variants above 1% frequency in the population; further improvements to sample rarer variants are ongoing (1000 Genomes Project Consortium et al., 2012). Consequently, genetic mapping, once the province of rare Mendelian disorders, is now applicable to any human trait or disease. In fact, more than 2,000 confirmed loci, each containing multiple genes, affecting susceptibility to more than 100 medically relevant traits (e.g., blood pressure) and disease (e.g., hypertension) are now known (Hindorff et al., 2009). For most complex traits examined, many such loci have been mapped, but the vast majority of the specific genes remain unidentified. We can sometimes guess at a candidate gene within the locus (Jostins et al., 2012), sometimes implicate a gene by virtue of an abundance of rare variants among affected individuals (Jostins et al., 2012), in rare circumstances use therapeutic modulation of a pathway to pinpoint the gene (Moon et al., 2004), and sometimes identify one by painstaking experimental dissection (Musunuru et al., 2010), but, generally, identification of the underlying gene has not become easier. In fact, most of the mapped loci underlying complex traits remain unresolved at the gene or mechanistic level.
Despite the beginning clues to human disease pathophysiology that complex disease mapping is providing, and the slow identification of individual genes, it appears highly unlikely that we can understand traits and diseases this way. There is indeed evidence for scenarios in which variation in complex traits, including risk of complex disease, is mediated by a myriad of variants of minute effect, spread evenly across the genome (Yang et al., 2011). Therefore, we need other approaches to override this bottleneck.
For Mendelian disorders, gene identification within a locus is made possible by each mutation being necessary and sufficient for the phenotype, being functionally deleterious and rare, and, having an inheritance pattern consistent with the phenotype. It’s the mutation that eventually reveals the biology and explains the phenotype. Any component locus for a complex disease has no such restriction, since the causal variants are neither necessary nor sufficient, nor coding (in fact, they are frequently non-coding and regulatory) nor rare (Emison et al., 2010; Jostins et al., 2012). Currently, the major attempts to overcome this impediment involve reliance on single severe mutations at the very same component genes and demonstrating Mendelian inheritance of the same or similar phenotype, and/or identifying single genes with a demonstrable excess of rare coding variants. The first of these two strategies is a strong unproven hypothesis, and probably not universally true, while the second relies on very large sample sizes of patients and suffers from the unknown functional effect of the majority of rare coding variants. Consequently, these strategies themselves depend on the hidden biology we seek, and are applicable only to the most common human diseases. It appears to us that ignorance of biology has become rate-limiting for understanding disease pathophysiology, except perhaps for the Mendelian disorders. There are two ways to get out of this vicious cycle.
One approach may be to use a set of model traits and diseases and use their existing mapped loci to identify a small set of the component genes by brute-force (or, luck) and use the uncovered biology to infer which other genes in their ‘pathways’ can explain the disease. This approach has been highly profitable in Crohn’s disease – a common, inflammatory disorder whose root causes have remained cryptic until genome-wide association studies identified a large number of loci, with fundamental defects in mucosal immunity (Graham and Xavier, 2013), but not in Type 2 Diabetes where the pathophysiology awaits clarification (Groop and Pociot, 2013). Although we suspect that the numbers of pathways involved are fewer than the numbers of genes involved, this is merely suspicion. Nevertheless, can we reduce the complexity of the problem by identifying all of the relevant pathways? Despite uncertainty, this approach has the advantage of leading to specific testable hypotheses. The second approach is to focus research on why the disease is complex in the first place. Although the genome is linear, its expression and biology are highly non-linear and hierarchical, being sequestered in specific cells and organelles (Ilsley et al., 2013). Understanding this hierarchy, the province of Systems Biology, is critical to the solution of the complex inheritance problem (Yosef et al., 2013). Even more importantly, this approach might, through the effect of mutations, allow us to decipher cell circuitry, understand which pathways are limiting and which are redundant. This last aspect is critical: as we argue below, with our current state of knowledge, we are likely to have our greatest success with understanding how genes map onto pathways, and how pathways map onto disease, before a true quantitative understanding of disease biology emerges. One might counter that existing gene ontologies do precisely that, but, even in yeast, this appears to be highly incomplete (Dutkowski et al., 2013).
Proving causality: Molecular Koch’s postulates
The evidence that a specific gene is involved in a particular human disease has historically been non-statistical and based on our experience with identifying mutations in Mendelian diseases. The chief criteria have been to demonstrate co-segregation with the phenotype in families, exclusivity of the mutation to affected individuals (rare alleles absent in controls) and the nature of the mutation (a plausibly deleterious allele at a conserved site within a protein). Unfortunately, as already mentioned, all of these rules break down in complex phenotypes where neither co-segregation nor exclusivity to affecteds nor obviously deleterious alleles are likely; moreover, many mutations are suspected to be non-coding and in a diversity of regulatory RNA molecules. Consequently, statistical evidence of enrichment has been the mainstay, but this has two negative consequences: first, scanning across the genome or multiple loci covering tens to hundreds of megabases requires very large sample sizes and very strict levels of significance to guard against the many expected false positive findings; second, genetic effects that are small or genes with only a few causal alleles are notoriously difficult to detect although they may be very important to understanding pathogenesis. This difficulty translates into a low power of detection, since common disease alleles cannot be distinguished from bystander associated alleles, whereas rare alleles are observed too infrequently to provide statistical significance. Consequently, although many genes are ‘named’ as being responsible in a complex disease or disease process, proof of their involvement is either absent or circumstantial and not direct.
We need to move beyond lists of plausible genes, to provide rigorous proof for their role in disease. In the late 19th century when bacteria were first shown to cause human disease, they were indiscriminately implicated in all manner of disease with little proof (Brown and Goldstein, 1992). One particularly embarrassing example was alcaptonuria, which Sir Archibald Garrod subsequently showed, by genetic inheritance, to be an inborn error of metabolism. We are likely to repeat this “witch-hunt” unless we are careful to note that mapping a locus is not equivalent to identifying the gene, and that identifying a gene and its mutations at a locus depends on numerous untested assumptions (mutational type, mutational frequency in cases and controls, coding or regulatory, cell autonomy). So what might be rigorous proof of an attractive candidate? In microbiology, Robert Koch set out three postulates that had to be satisfied to connect a specific bacterium (amongst the multitudes encountered, not unlike current genome analysis) to a disease: the agent had to be isolated from an affected subject, the agent had to produce disease when transmitted to an animal, and the agent had to be recoverable from an animal’s lesion (Falkow, 1988). For any human disease where many loci have been mapped we can propose analogous postulates: (1) a specific candidate gene identified by mapping with variants is enriched in patients, (2) demonstration of a mutant phenotype among bearers of mutations in the same gene in a ‘model system,’ (3) rescue of the mutant phenotype using wild-type human alleles, and, (4) failure to rescue the mutant phenotype using human mutant alleles. In principle, this is applicable to both single genes and collections of a few genes (Wang et al., 2013).
The key to these analyses is the equivalence of the laboratory model phenotype; this cannot be arbitrary but one carefully chosen to be analogous to the human phenotype. In other words, we require success at two levels: demonstration that a mutant allele leads to a specific mutant phenotype in a model system, and, demonstration and acceptance that the model and human phenotypes are equivalent. The keys to these analyses are delineation of the terms ‘model system’ and ‘phenotype.’ First, many types of reagents could comprise the ‘model system’ including human cells and tissues, animal models, and human volunteers in rare circumstances (e.g., therapeutic interventions against a pathway). Eventually, even computational models of tissue physiology, such as for the cardiac system (Guyton et al., 1972), might be helpful. Second, many types of phenotypes could be considered from a biochemical or cellular correlate of the disease to an analogous pathology in animal model systems. Simply because we cannot follow Koch to the letter in human patients does not absolve us from the responsibility of demonstrating a rigorous level of proof. This is particularly true if we are to pursue therapeutic targets for these diseases.
It is clear that the majority of complex diseases do not harbor this level of proof today; neither do most monogenic disorders. As the case for Marfan syndrome demonstrates, the identification of fibrillin 1 mutations were insufficient to identify therapies without the concomitant understanding of its pathophysiology (Brooke et al., 2008). Animal models are attractive because of the ability to do experimental manipulations that test predictions of gene function, but these experiments test the function of a gene in a context that is decidedly different from a human patient. However imperfect animal models are, progress in the direction of understanding causality have been very beneficial when gene disruptions alone, perhaps at more than one gene, have taught us fundamental lessons in pathophysiology (Farago et al., 2012). In many cases, investigators have also demonstrated that disease results only when combined with a potent environmental insult. When known, such as the effect of dietary cholesterol vis-à-vis genes involved in cholesterol metabolism in atherosclerosis, such environmental exposures to gene-deficient mouse models have provided a tight circle of proof (Plump et al., 1992). A recent example of gestational hypoxia modulating the effect of Notch signaling and leading to scoliosis in mice and in human families shows how environmental factors beyond diet can be examined even for congenital disorders (Sparrow et al., 2012). Despite these successes, pursuit of Koch’s postulates faces other challenges. For example, mutations in the same gene might not reveal an identical phenotype in humans and in an animal model even if molecular pathways are conserved. This is a particular problem for behavioral phenotypes where brain circuitry may have evolved quite differently in humans and other mammals, challenging our ability to model behavior accurately. Nevertheless, such an analysis might reveal an underlying neural phenotype or a molecular or cellular correlate that is in common and subject to testing of the postulates.
Ultimately, a lack of understanding of fundamental physiology is the biggest impediment to our understanding of genetically complex human disease. A unique aspect of genetics research seldom appreciated is that genetic effects are chronic biological exposures and as such can pinpoint the earliest stages of disease not readily studied otherwise. To fulfill this potential contribution of genetics to physiology, genetic studies that can inform disease pathogenesis should be intrinsic to additional mapping. In reality, we still do not fully understand the pathogenesis stemming from some of the earliest identified human disease genes. With better understanding of disease mechanism, it seems likely that many disorders that we think of as “genetic” may have ameliorative diet, exercise, or other benign environmental “treatments”. But this goal is unlikely to be achieved in the absence of a superior understanding of the biology of hierarchical function within genomes, how variation alters these functions and how these altered functions lead to human disease. Koch’s postulates can be a guiding light for these discoveries.
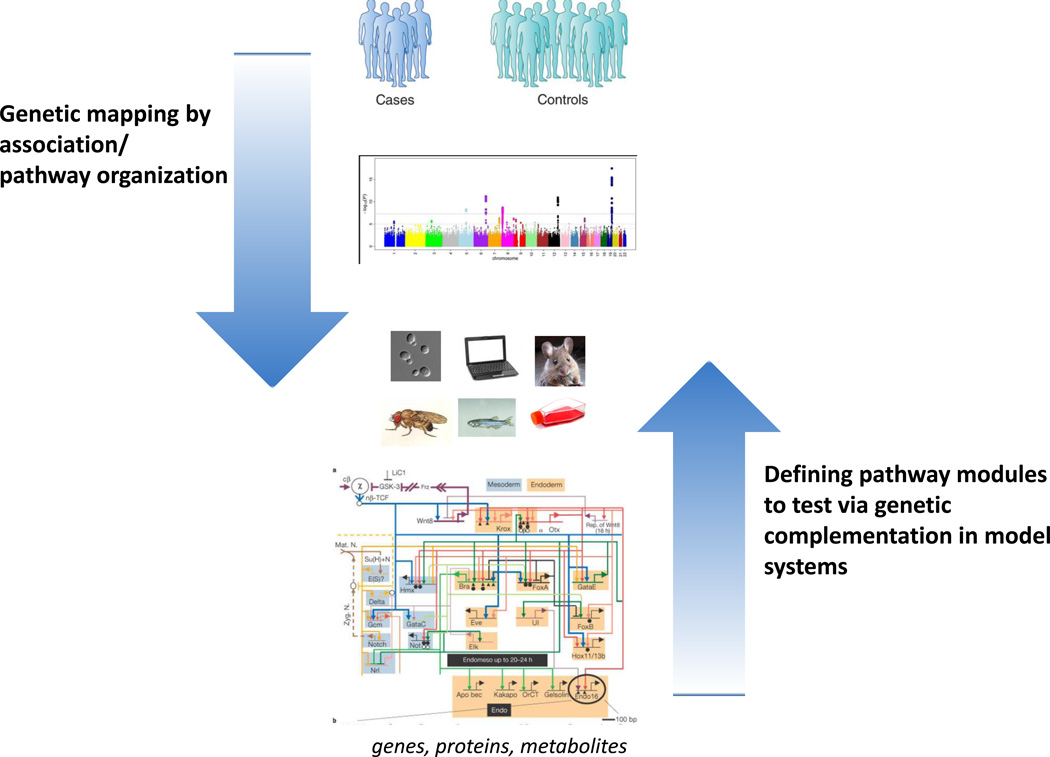
Figure 1.
Complementary approaches for proving genetic causality and understanding pathophysiology of complex disease. Genetic association studies in humans combined with prior knowledge about biological pathways generate testable hypotheses. Prior knowledge and systems-level quantitative analysis can help predict where (anatomically) and when (developmentally) disruptions to a particular pathway will lead a to a specific morphological or biochemical phenotype, which can then be tested in an appropriate model system while adhering to the postulates outlined in Box 1.
Acknowledgement
We thank Donna Krasnewich of the NIGMS for early discussions and hosting a meeting that was the genesis of many of the ideas discussed here.
Reference List
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Robert K, Walter P. Molecular Biology of the Cell. 5th edition. New York: Garland Science; 2007. [Google Scholar]
- Altenburg E, Muller HJ. The Genetic Basis of Truncate Wing,-an Inconstant and Modifiable Character in Drosophila. Genetics. 1920;5:1–59. doi: 10.1093/genetics/5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke BS, Habashi JP, Judge DP, Patel N, Loeys B, Dietz HC., 3rd Angiotensin II blockade and aortic-root dilation in Marfan's syndrome. N. Engl. J. Med. 2008;358:2787–2795. doi: 10.1056/NEJMoa0706585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Koch's postulates for cholesterol. Cell. 1992;71:187–188. doi: 10.1016/0092-8674(92)90346-e. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. To a future of genetic medicine. Nature. 2001;409:822–823. doi: 10.1038/35057281. [DOI] [PubMed] [Google Scholar]
- Chin BL, Ryan O, Lewitter F, Boone C, Fink GR. Genetic variation in Saccharomyces cerevisiae: circuit diversification in a signal transduction network. Genetics. 2012;192:1523–1532. doi: 10.1534/genetics.112.145573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkowski J, Kramer M, Surma MA, Balakrishnan R, Cherry JM, Krogan NJ, Ideker T. A gene ontology inferred from molecular networks. Nat. Biotechnol. 2013;31:38–45. doi: 10.1038/nbt.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emison ES, Garcia-Barcelo M, Grice EA, Lantieri F, Amiel J, Burzynski G, Fernandez RM, Hao L, Kashuk C, West K, et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am. J. Hum. Genet. 2010;87:60–74. doi: 10.1016/j.ajhg.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkow S. Molecular Koch's postulates applied to bacterial pathogenicity--a personal recollection 15 years later. Nat. Rev. Microbiol. 2004;2:67–72. doi: 10.1038/nrmicro799. [DOI] [PubMed] [Google Scholar]
- Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 1988;10(Suppl 2):S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- Farago AF, Snyder EL, Jacks T. SnapShot: Lung cancer models. Cell. 2012;149:246–246. e1. doi: 10.1016/j.cell.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Graham DB, Xavier RJ. From genetics of inflammatory bowel disease towards mechanistic insights. Trends Immunol. 2013;34:371–378. doi: 10.1016/j.it.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groop L, Pociot F. Genetics of diabetes - Are we missing the genes or the disease? Mol. Cell. Endocrinol. 2013 doi: 10.1016/j.mce.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Guyton AC, Coleman TG, Granger HJ. Circulation: overall regulation. Annu. Rev. Physiol. 1972;34:13–46. doi: 10.1146/annurev.ph.34.030172.000305. [DOI] [PubMed] [Google Scholar]
- Hartman JL, 4th, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilsley GR, Fisher J, Apweiler R, Depace AH, Luscombe NM. Cellular resolution models for even skipped regulation in the entire Drosophila embryo. Elife. 2013;2:e00522. doi: 10.7554/eLife.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohane IS, Masys DR, Altman RB. The incidentalome: a threat to genomic medicine. JAMA. 2006;296:212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- Ludolph AC, Brettschneider J, Weishaupt JH. Amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2012;25:530–535. doi: 10.1097/WCO.0b013e328356d328. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallion AS, Emison ES, Kashuk CS, Bush RT, Kenton M, Carrasquillo MM, Jones KW, Kennedy GC, Portnoy ME, Green ED, Chakravarti A. Genomic variation in multigenic traits: Hirschsprung disease. Cold Spring Harb. Symp. Quant. Biol. 2003;68:373–381. doi: 10.1101/sqb.2003.68.373. [DOI] [PubMed] [Google Scholar]
- Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plump AS, Smith JD, Hayek T, Aalto-Setala K, Walsh A, Verstuyft JG, Rubin EM, Breslow JL. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 1992;71:343–353. doi: 10.1016/0092-8674(92)90362-g. [DOI] [PubMed] [Google Scholar]
- Provine WB. The Origins of Theoretical Population Genetics. Chicago & London: The University of Chicago Press; 1971. [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow DB, Chapman G, Smith AJ, Mattar MZ, Major JA, O'Reilly VC, Saga Y, Zackai EH, Dormans JP, Alman BA, et al. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell. 2012;149:295–306. doi: 10.1016/j.cell.2012.02.054. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R. Molecular Biology of the Gene. 7th Edition. San Francisco: CSHL Press & Benjamin Cummings; 2007. [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerba KE, Ferrell RE, Sing CF. Genotype-environment interaction: apolipoprotein E (ApoE) gene effects and age as an index of time and spatial context in the human. Genetics. 1996;143:463–478. doi: 10.1093/genetics/143.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]