Abstract
Background
The study of intestinal microbiota has been revolutionized by the use of molecular methods, including terminal restriction fragment length polymorphism (T-RFLP) analysis. A number of microbiota studies of Crohn’s disease patients have examined samples from stool or from the neoterminal ileum with a standard biopsy forceps, which could be contaminated by colonic bacteria when the forceps passes through the colonoscope channel.
Objective
To determine whether sheathed biopsy forceps are able to obtain terminal ileal microbiota samples with less colonic bacterial contamination compared to unsheathed (standard) biopsy forceps.
Design
Prospective randomized single center-study.
Patients and Methods
We obtained four (paired) biopsy specimens from adjacent locations in the terminal ileum using the sheathed and standard forceps of 27 consecutive subjects undergoing colonoscopy and characterized the microbiota using T-RFLP. We calculated the Bray Curtis similarity index (BCI) between samples (sheathed vs. unsheathed forceps) within patients and tested for significant differences across all patients.
Results
There was not a significant difference in the microbial diversity of samples obtained using sheathed vs. unsheathed forceps. The difference in microbial diversity between patients was much greater than the variability within patients by proximal vs. distal site or by forceps type.
Limitations
T-RFLP is based on PCR amplification, so it is not always sensitive to rare bacterial species.
Conclusion
Standard unsheathed forceps appear to be sufficient for microbiota sample collection from the terminal ileum.
Keywords: Terminal Ileum, Terminal restriction fragment length polymorphism, Microbiota, Microbial diversity, Crohn’s disease
Introduction
The gastrointestinal tract of humans is inhabited by a complex microbial community that is made up of 100 trillion cells, ten times the number of human cells in the body (1). There is wide variation in the specific composition of the microbiota of the GI tract between individuals, but the microbial community appears to be relatively stable within an individual (2). The composition of the intestinal microbiota has been implicated in the recurrence of Crohn’s disease, since antibiotics (metronidazole and ornidazole) delay the recurrence in the neo-terminal ileum after surgical resection (3, 4). The study of the intestinal microbiota has been revolutionized by the use of molecular-based methods, many of which target the 16S rRNA-encoding gene. These methods circumvent the main disadvantage of traditional culture-based approaches, which fail to grow and identify more than half of the species in the gut (5–7). Terminal restriction fragment length polymorphism (T-RFLP) is a molecular method that utilizes restriction enzyme digestion to identify 16S sequence heterogeneity. Sequence heterogeneity is then used as a proxy for different bacteria. T-RFLP is suitable for rapid, low-cost analysis of samples from a multiple subjects (8). This approach has been used to study the intestinal microbiota in many published studies (9–16).
Microbiota studies have traditionally been conducted on human biopsy samples from subjects using a standard unsheathed biopsy forceps. Contamination with bacteria that are not from the biopsy site (2, 17–20) can occur when the forceps passes through the colonoscope channel, particularly if colonic contents have been suctioned through the channel during the procedure. Eckburg, et al. showed that the abundance of constituent bacterial species of the microbiota is patchy and that the distribution of these species is heterogeneous from the small bowel to the colon (2). It is clear that human cecal microbiota differs quantitatively and qualitatively from the fecal microbiota (2, 21). Furthermore, the local gut associated microbiota is more likely to be involved in the pathogenesis of IBD than the fecal microbiota (19, 20). Thus, in order to study the role of the microbiota in particular locations (i.e. the neo-terminal ileum), it is necessary to obtain biopsies from the site itself, rather than assuming that the fecal microbiota is representative of proximal locations in the GI tract. The aim of this study was to determine whether sterile sheathed forceps are needed to obtain ileal microbiota samples that are not contaminated with colonic bacteria.
Methods
We recruited 40 consecutive subjects from University of Michigan Medical Procedures Center scheduled for a clinically-indicated colonoscopy. Inclusion criteria were ≥18 years of age and successful performance of a full colonoscopy with terminal ileal intubation after ≤ 3 attempts. Exclusion criteria were an inability or unwillingness to consent to the study, use of antibiotics in the last 3 months, inability to intubate the terminal ileum within 3 minutes, and any medical contraindication for obtaining biopsies.
Making the Sheathed forceps from Radial Jaw 4 Forceps
We used the Radial Jaw 4 (RJ4) single use biopsy forceps (Boston Scientific, Natick, United States) and modified them by covering the entire length with FEP (Fluorinated Ethylene Propylene) tubing to form a sheath (Zeus #0000048562). To seal this tubing we incorporated a wax plug at the tip of the forceps. The wax plug was composed of 4 parts turnip wax (International group# Microsere 5981A, which meets the FDA requirements set forth in 21 CFR 172.886 for use in food articles) and 1 part petrolatum jelly (Vaseline) to obtain a plug compatible with sterilization that was easily extruded when the terminal ileum was reached. After forceps assembly in our laboratory, these were sterilized in ethylene oxide at the University of Michigan Hospital sterile supply services facility. Successful sterilization was indicated by change of color on a gas sterilization test strip. The University of Michigan Biomedical Engineering Unit certified the sheathed forceps for use in human subjects.
Assessment of Sheath Resistance to Contamination
To determine whether sterility of the sheathed forceps was maintained during passage through the colonoscope, a colonoscope contamination ex vivo experiment was performed without a human subject. A colonoscope channel was deliberately contaminated by suctioning a slurry of human stool through the colonoscope prior to insertion of either unsheathed or sheathed forceps. Each type of forceps was passed through the colonoscope, and extended out of the colonoscope (and sheath, if present). The forceps were opened and closed, then withdrawn into the colonoscopy (and sheath, if present). The forceps were withdrawn from the colonoscope, extended and opened so that the jaws could be shaken in 1 mL of sterile saline. Bacterial contamination of the saline solution was assayed by testing for bacterial 16S rRNA by PCR using a broad-range forward primer 8F (Integrated DNA Technologies 5-AGAGTTTGATCCTGGCTCAG-3) and a broad-range reverse primer 1525R (Integrated DNA Technologies 5-AGA AAG GAG GTG ATC CAG CC-3). PCR products were visualized on a 0.8% agarose gel stained with ethidium bromide and digitally photographed.
Biopsy Sample collection from Human Terminal Ileum
Before collecting each biopsy sample with a given forceps, the cecum was irrigated with water to produce a soup of stool, and one quarter of the liquid contents were suctioned through the scope to simulate the worst-case scenario of significant stool contamination of the colonoscope channel. Subjects were randomized to which forceps (sheathed vs. unsheathed) were used first in a 1:1 allocation ratio. The allocation sequence was generated with the uniform function in Stata 10.1 by PDRH, and allocations were written on paper and sealed in opaque security envelopes until the patient was consented and sedated for colonoscopy. The participants were consented, and envelopes opened and randomization order assigned by MNC and JF. We obtained 2 pairs of biopsy specimens from adjacent locations (proximal vs. distal) in the terminal ileum using the sheathed and unsheathed forceps. The biopsy specimens were collected from a single pass with a new sterile (sheathed or unsheathed) forceps used for each biopsy specimen. Each biopsy specimen was transferred from the spike of the forceps using a sterile 20 gauge blunt needle to a sterile 2 mL screw-cap cryovial kept on dry ice. The tissue samples were immediately snap frozen in liquid nitrogen (−70° C) and transferred to the lab. The participants and the analysts of T-RFLP results (LAJ, SW, VAY) were blinded to the type of forceps used for each sample. The study coordinators (MNC, JF) and the endoscopist (PDRH) could not be blinded to the forceps type.
Terminal Restriction Fragment Length Polymorphism (T-RFLP)
The microbiota of the terminal ileum was characterized for each sample using terminal restriction fragment length polymorphism (T-RFLP). This technique generates a community fingerprint based on the composition of the constituent bacterial members (12, 22). Total DNA was extracted from biopsy samples as follows: The entire biopsy sample (20–50mg) was transferred to a Mo Bio Ultra Clean Fecal DNA tube (Mo Bio Laboratories Inc, Carlsbad, CA) containing 350 microL of ATL buffer (Qiagen DNAeasy extraction kit, Qiagen). Tissue was disrupted in ATL buffer by homogenization for 1 min using a BioSpec mini- beadbeater (BioSpec Products, Bartlesville, OK). After disruption, samples were digested with 40 microL of 20 mg/ml proteinase K (Qiagen) at 55° C for 1 hour. Total DNA was extracted per manufacturer’s instructions using the Qiagen DNAeasy extraction kit (Qiagen) and eluted with 30 microL AE buffer. DNAs were stored at −80° prior to PCR analysis.
The 16S rRNA encoding gene was amplified by PCR using a 6-FAM-5’-labeled, broad-range forward primer 6-FAM-8F (Integrated DNA Technologies 5-AGAGTTTGATCCTGGCTCAG-3)(23) and a conventional, broad-range reverse primer 1525R (24)(Integrated DNA Technologies 5-AGA AAG GAG GTG ATC CAG CC-3). PCR was performed with 70 ng of DNA in illustra PuReTaq™ Ready-To-Go™ PCR beads (GE Healthcare, Piscataway, NJ). Cycling conditions consisted of an initial denaturing step at 94°C for 2 minute followed by 30 cycles of 94°C 30 seconds/ 58°C 45 seconds/ 72°C 90 seconds and a final 4 minute extension at 72°C. With every PCR run, a blank (no DNA template control) was included. Amplification was confirmed by visualization of a single 1.4kb PCR product on a 0.8% agarose gel. Biopsy specimens were considered adequate if 16S rRNA PCR products could be amplified from all four biopsy specimens from the same patient.
Amplicons were column purified (QIAquick PCR Purification Kit, Qiagen, Inc.) per manufacturer’s instructions. 200–300 ng of amplicon DNA (as determined by Nanodrop spectrophotometer measurement (Thermo Scientific, Wilmington, DE)) was digested with 20 units of the restriction enzyme MspI (New England Biolabs) for 2 hours at 37°C.(25) Restriction fragments were column purified with the QIAquick PCR kit according to manufacturer’s instructions and eluted with 30 microL of EB buffer. DNA concentration was determined using the Nanodrop spectrophotometer.
Fragments were separated on an automated capillary sequencer (Applied Biosystems 3730XL DNA Analyzer). 100 ng of Msp I-digested amplicons were loaded in duplicate onto a CE plate containing ROX1000 size standard. As a control, 100 ng of undigested amplicons were run in duplicate on the same plate. Fluorescently labeled terminal fragments generated chromatogram peaks and were identified using Peak Scanner Software (Applied Biosystems). Peaks corresponding to fragments between 50 and 1000 base pairs (bp) in length were used in the analysis (reproducibility of peaks inside this range is high) and the height of each peak was obtained and used as a proxy for fragment abundance.
Visualization and statistical analysis of community similarity
For community comparisons, a count matrix was generated, where terminal fragment lengths (50–1000 bp) were rows, biopsy samples were columns, and each element in the matrix was the height of the given peak in a biopsy sample. The matrix was loaded into the EstimateS program (26) and the similarity between all communities was calculated using the Bray-Curtis similarity index (BCI). BCI is an ecological diversity index that ranges in value between 1 (communities are identical) and 0 (communities are completely different) and takes into account the presence/absence of a species as well as its relative abundance (27). Because the results of T-RFLP identify distinct terminal restriction fragments (TRFs), rather than species, ecological diversity measures were adapted to use TRFs. To visualize the similarity between all communities, a neighbor-joining dendrogram was constructed based on all pairwise BCI values using the MEGA4 program (28).
Since four samples (2 sheathed and 2 unsheathed) were collected from each patient, we tested for differences in the average BCI between samples within patients and between patients by forceps type. Analysis of variance (ANOVA) was used to test for statistical differences between these groups (SAS statistical software, SAS Institute, Cary, NC). If significant sample contamination occurred with the standard forceps, a sizable increase in microbial diversity should be detected in the BCI of the unsheathed samples.
TRF richness (S) was calculated as the total number of individual T-RFLP peaks present in each biopsy sample. We also calculated the Shannon-Wiener diversity index (H’), which is maximized when there are many species (TRFs) present in equal proportions. This was calculated based on the number of TRFs (S), and the relative abundance of each TRF (pi), as follows:
where pi is the proportion of the ith peak relative to the sum of all peak heights.
Sample size, Ethical Approval, and Funding
The sample size determination was based on prior analyses of intestinal microbial data from our microbiome core. The standard deviation in the Shannon Diversity Index was expected to be less than 0.5 for paired samples. We pre-specified a significant difference in the SDI as a change of at least 0.5. With a power of 95%, and a two-sided alpha of 0.05, this would require a minimum of 13 subjects per group. Given that these were estimates, we increased the sample size to 16 for a margin of safety. We also allowed for 4 cases of dropouts or sample processing problems, increasing our planned sample size to 20 per group, or 40 overall.
This study was approved by the University of Michigan Institutional Review Board (IRB-MED) on January 17, 2008. The protocol for this study is posted for public review at: http://www.med.umich.edu/higginslab/protocols.html. This study was funded by an Endoscopic Research Award from the American Society of Gastrointestinal Endoscopy. The funder (ASGE) had no role in the study analysis or writing of the manuscript.
Results and Discussion
Sheath sterility
Two pairs of unsheathed or sheathed forceps were passed through a deliberately contaminated colonoscope and analyzed by PCR for the bacterial 16S rRNA gene. Both unsheathed samples were contaminated with bacteria, while the sheathed samples did not have detectable bacterial DNA (data not shown).
Patient Samples
We obtained complete T-RFLP profiles from all four biopsy sites for 108 samples from 27 subjects. Of the 40 consented subjects, the first three subjects served as a pilot sample in which the biopsy specimen collection protocol was refined, but no microbiota analyses were done on these samples as poor quality DNA was obtained. In another six subjects, biopsies were not attempted due to endoscopy scheduling pressures and/or because ileal intubation could not be achieved in 3 minutes or less, as agreed to with the Institutional Review Board. In another four subjects, at least one of the 4 biopsy specimens did not yield any T-RFLP peaks, so they were excluded from the analysis. Despite these dropouts, randomization by forceps type was successful and samples were collected using sheathed forceps first in 15 patients and unsheathed first in 12 patients. The baseline demographics of these subjects and their indications for colonoscopy are presented in Supplementary Table 1.
T-RFLP Analysis
It is important to note that T-RFLP uses broad-range nucleotide primers to amplify the same genetic locus (the 16S rRNA encoding gene) from all bacterial chromosomes represented in bulk extracted (community) DNA. Therefore, PCR amplicons represent all members of the bacterial community of a sample. Since the forward primer in the T-RFLP PCR is fluorescently labeled and an endonuclease is used to digest amplicons at unique restriction sites, the length of terminal restriction fragments represents unique bacterial members of the community (i.e. different alleles of the 16S locus vary in restriction sites) and can be quantified using a capillary nucleotide sequencer (25). Amplicons of the same size increase the fluorescent signal (chromatogram peak height) so that the area within a chromatogram peak is a proxy for the abundance of a particular bacterium. Therefore, data generated by T-RFLP is both the length of a terminal fragment (presence of a particular bacterium) and the abundance of a terminal fragment (the abundance of a particular bacterium). One community ecology metric (a standard of measurement) that incorporates both the presence/absence of a species and its relative abundance is called the Bray-Curtis similarity index. This metric was used to evaluate the similarity between the communities (samples) in this study. To visualize the similarity among the communities, a dendrogram was constructed based on all pairwise (sample-to-sample) Bray-Curtis values (Figure 3). To appreciate the similarity of two particular communities, one simply needs to note the branch length that connects each community. Shorter branch lengths represent similar communities, while longer branch lengths represent dissimilar communities.
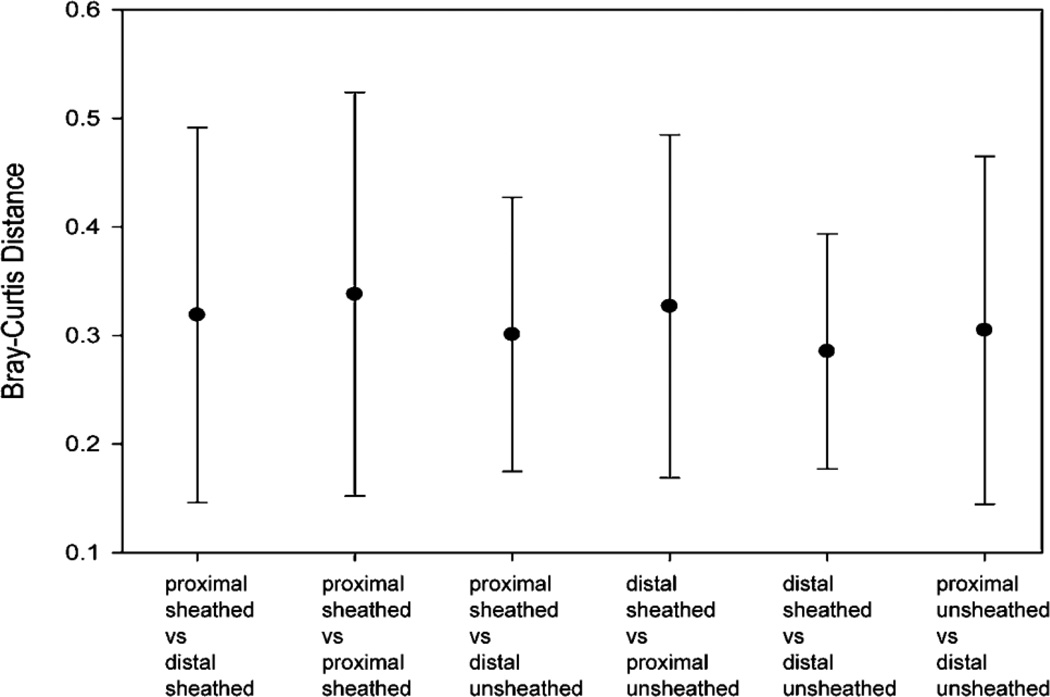
Figure 3.
Microbial diversity measured by Bray-Curtis index (BCI). The mean difference in BCI between samples (pairwise analysis within individual patients) is presented. Error bars represent the SE of the mean. No significant differences by location or forceps type were found.
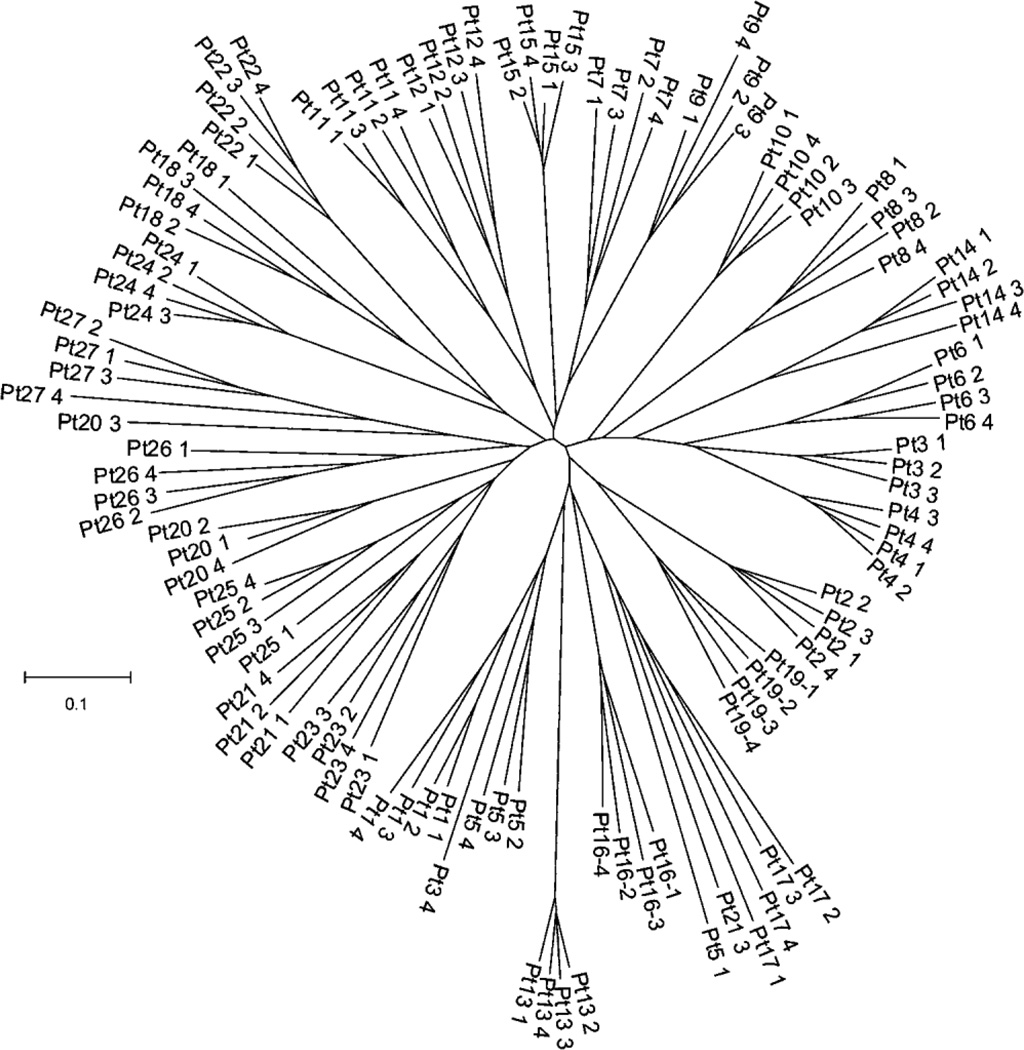
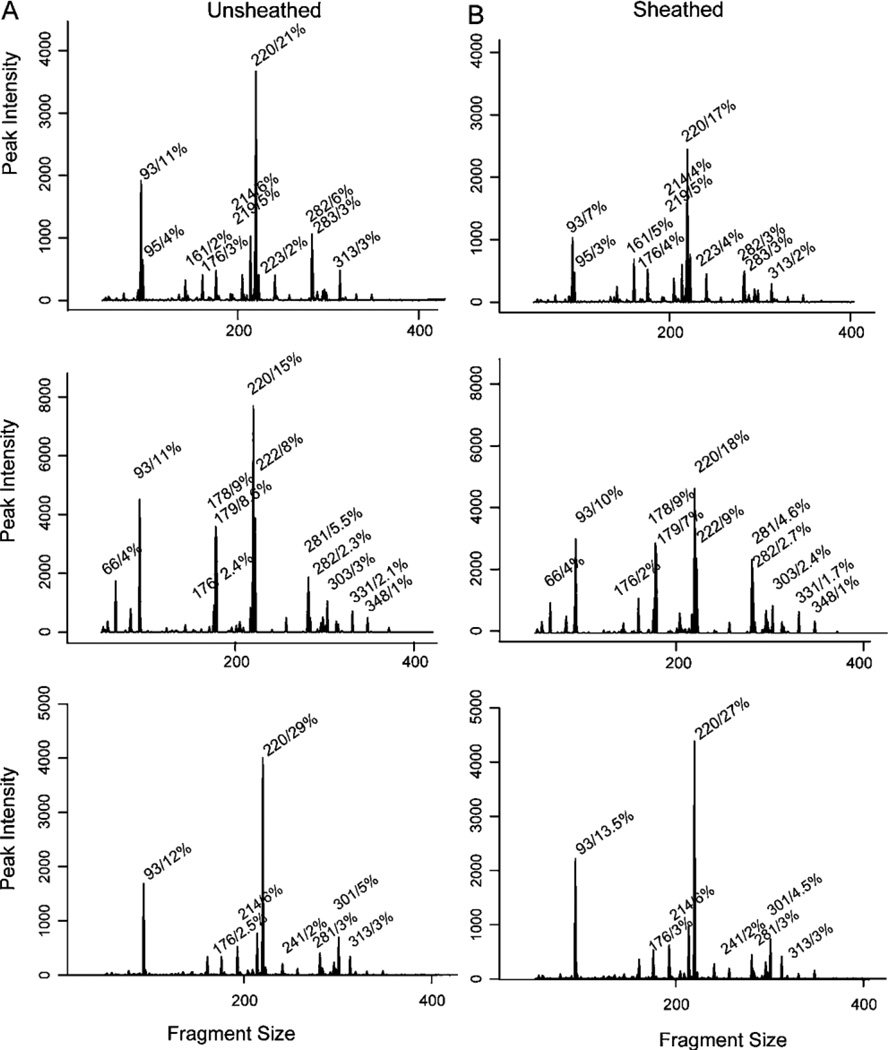
For the majority of patients, the tissue-associated microbiota obtained by both types of forceps (sheathed vs. unsheathed) in a given patient were similar, as illustrated by the T-RFLP chromatograms (Figure 1A and 1B), yet distinct from the microbiota of other patients, as illustrated by the dendrogram (Figure 2A).With few exceptions, samples obtained from an individual patient clustered together, indicating that samples from a single patient were more similar to each other than to samples from other patients. Also, no obvious clustering of sheathed and unsheathed samples was observed, indicating that the same bacterial community was sampled using the sheathed and unsheathed forceps.
Figure 1.
Bacterial Community Diversity Dendrogram. This neighbour-joining dendrogram was based on all pairwise comparisons between samples taken with sheathed (n=2) and unsheathed (n=2) forceps from 27 patients. Each sample is represented by a branch in the tree and branch lengths (the length of the line that connects two tips) represent the similarity among bacterial communities in these samples (the shorter the length, the more similar the communities).
Figure 2.
Representative terminal restriction fragment length polymorphism (T-RFLP) chromatograms from unsheathed versus sheathed forceps. Representative T-RFLP profiles of unsheathed (A) compared to sheathed (B) biopsy samples from three patients. T-RFLP fragments (in base pairs) are represented as individual peaks. The peak height (relative fluorescence) corresponds to the relative abundance of each T-RFLP fragment. The most abundant individual fragments are labeled with respect to fragment size (in base pair) and relative percentage (decimal).
The difference in the microbiota between patients was much greater than the variability within patients, as has been observed in previously published studies (2). To determine whether sheathed and unsheathed forceps sample the same microbiota within patients, we tested for significant differences in diversity, as measured by the Bray-Curtis Index. There was no significant difference in the microbial diversity for sheathed samples vs. unsheathed samples (Figure 2B). We did multiple comparisons of the diversity of the microbiota, however in all groups the BCI measures overlapped. On average, the same microbiota was obtained from biopsies taken with sheathed and unsheathed forceps.
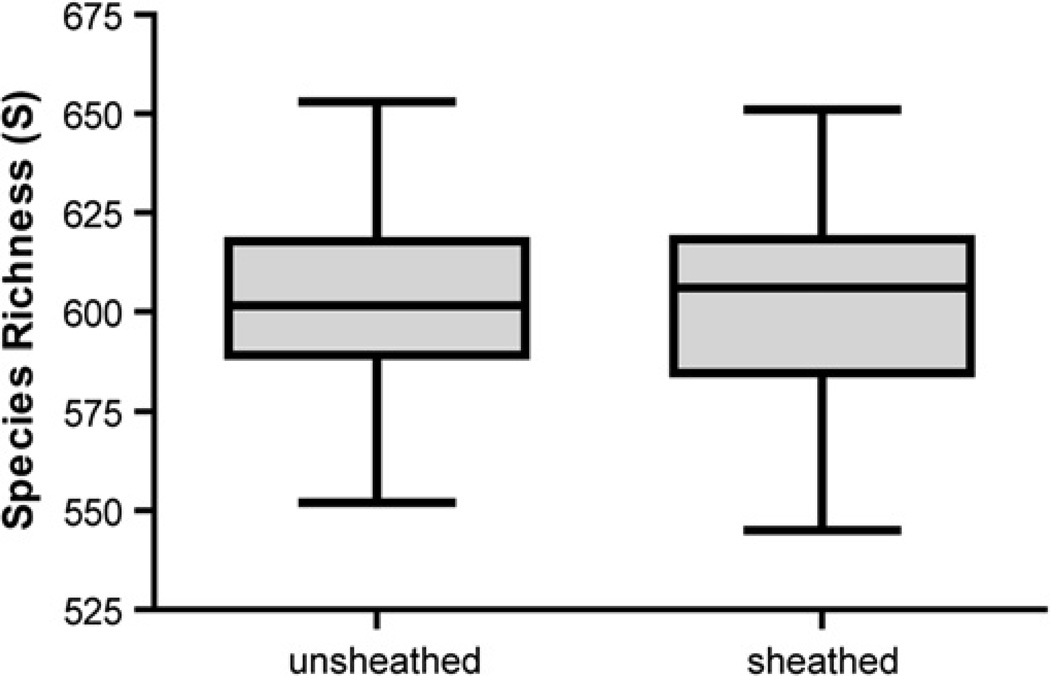
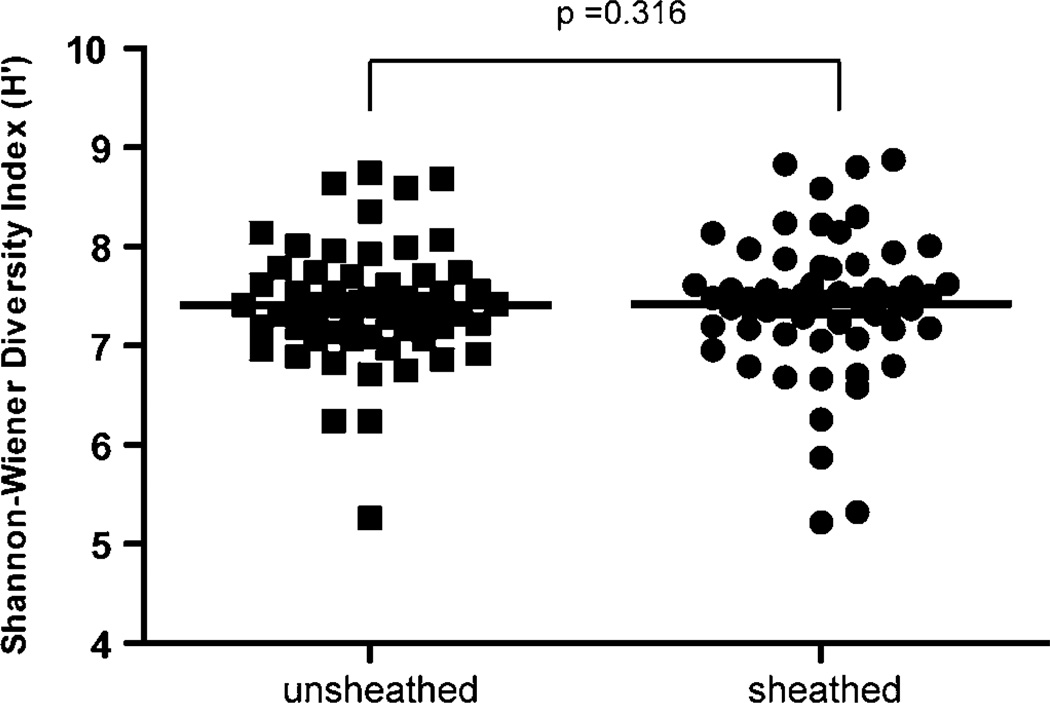
The mean TRF richness was virtually identical in the unsheathed vs. sheathed samples, (602.57 vs. 601.50), (t-test p=0.803) (Figure 3A). A similar result was obtained using the Shannon-Wiener index (H'). On average, H' was similar between forceps samples (7.51 for unsheathed vs. 7.38 for sheathed) and was not statistically different (t-test p=0.316) (Figure 3B). Thus, the overall microbial diversity was not significantly different between samples obtained with the sheathed and unsheathed forceps. The range of H' values calculated for sheathed samples was wider than that obtained for unsheathed forceps. This observation suggests that samples obtained using unsheathed forceps have less variability in TRF richness (TRF presence) or evenness (relative abundance of organisms). It is difficult to tell from these data the source of the variation in sheathed samples (richness or evenness), but it is clear that sheathed forceps do not consistently sample less or more bacterial diversity than unsheathed forceps. It is important to point out that these results are only as relevant as the individuals from which they were obtained. Given the uniqueness of the human GI tract microbiota, it is possible that the mean and range of microbial diversity reported here (H' values) may change as more individuals are sampled, but on average we find little evidence that these changes will be influenced by whether the samples are taken with sheathed vs. unsheathed forceps.
The main limitations to this study are those related to the sensitivity of the PCR amplification and those related to the detection of PCR fragments. T-RFLP relies on PCR amplification with broad-range primers, which do not amplify the 16S sequences of some rare bacteria. In addition, two distinct species of bacteria can potentially produce the same T-RFLP peak in their profile, potentially obscuring some species diversity. Also, the ability of automated capillary sequencers to size fractionate fluorescently labeled PCR amplicons is dependent on the abundance of the amplicons, so organisms at low abundance are not represented in the analysis. More sensitive techniques, like next-generation DNA sequencing, can identify low abundance species in the gut microbiota (29). The most important difference between T-RFLP and these newer techniques is one of scale. Where T-RFLP data represent the most abundant microbial TRFs, next-generation sequencing can identify rarer members of the microbial community. So it is possible that characterizations using newer techniques would show real differences between samples collected with sheathed vs. unsheathed forceps, but results obtained from T-RFLP suggest that the diversity among the dominant gut microbes will be similar.
In this worst-case scenario test, with deliberate and substantial contamination of the colonoscope channel with cecal contents, no significant differences were found in TRF diversity or TRF richness. This may be due to the use of forceps with a biopsy spike which is covered by an outer cup. Contamination of forceps in the colonoscope channel may only occur on the outside of the forceps cup, and it is possible that biopsies are adequately protected from bacterial contamination by the contents of the colonoscope channel once they are enclosed in the forceps cup.
Conclusion
Based on T-RFLP data, sheathed forceps do not appear to be required for microbiota sample collection from the terminal ileum. There are many factors that affect the composition of gut microbiota like diet, recent use of antibiotics, concomitant intestinal disease, and host genotype, which make it difficult to compare the microbiota between different individuals (30, 31). The main strength of our study is that comparisons were made between paired biopsies in individual patients, thus eliminating the need to account for the above factors. While more sensitivity for rare species could be obtained with other techniques, this worst-case contamination study did not find significant differences. Based on this data, we would recommend the following for obtaining biopsies for microbial analysis. First, taking usual care to minimize contamination of the colonoscope channel, or flushing with sterile saline before passing the forceps; and use of standard forceps in which the biopsy is retained with a biopsy spike and enclosed by a forceps cup during withdrawal of the forceps. We hope that this approach will accelerate the analysis of the microbiota of the neo-terminal ileum in Crohn’s disease to elucidate which bacteria contribute to post-operative recurrence.
Supplementary Material
Figure 4.
Terminal restriction fragment (TRF) richness (S). The middle line in the box represents the mean TRF richness. There is overlap between the TRF richness of unsheathed and sheathed forceps with the mean TRF richness of unsheathed forceps being only slightly greater than that of sheathed forceps (t-test, p=0.803).
Figure 5.
Terminal restriction fragment (TRF) diversity as measured by the Shannon-Wiener diversity index (H’). The middle line represents the mean Shannon-Wiener diversity index for each group. The Shannon-Wiener diversity index for each individual patient is represented by a filled square (sheathed) or filled circle (unsheathed). The H’ overlaps between unsheathed and sheathed forceps with the mean H’ of unsheathed forceps being only slightly more than that of sheathed forceps. This is not statistically significant (t-test, p=0.316).
Take-Home Message.
For study of the microbiota of the mucosal surface of the intestine (e.g. to study the microbiota that cause Crohn’s disease recurrence in the of the neo-terminal ileum after surgical resection), standard biopsy forceps can be considered sufficient for microbiota sampling.
Acknowledgments
Grant Support:
This study was funded by and Endoscopic Research Award from the American Society of Gastrointestinal Endoscopy.
Institutions participating in the study: University of Michigan
Abbreviations
- T-RFLP
Terminal restriction fragment length polymorphism
- TRF
Terminal Restriction Fragment
- BCI
Bray-Curtis similarity Index
- IBD
Inflammatory Bowel Disease
- PCR
Polymerase Chain Reaction
- rRNA
ribosomal RiboNucleic Acid
Footnotes
Disclosure:
The authors have no potential conflicts of interest or competing interests to disclose.
Authors’ Contributions:
Maneesh Dave was involved in the conception and design; laboratory work, analysis and interpretation of the data; drafting of the article; critical revision of the article for important intellectual content and the final approval of the article.
Laura Johnson was involved in the laboratory work, analysis, interpretation of data and drafting of the article, and final approval of the manuscript.
Seth Walk was involved in the analysis and interpretation of data and critical revision of the manuscript, and final approval of the manuscript.
Vincent B Young was involved in the analysis and interpretation of data and critical revision of the manuscript, and final approval of the manuscript.
Ryan Stidham was involved in the laboratory work, and final approval of the manuscript.
Meghana N. Chaudhary was involved in gathering data, maintaining regulatory documents, and final approval of the manuscript.
Jessica FunNell was involved in gathering data, maintaining regulatory documents, and final approval of the manuscript.
Peter D.R. Higgins is the corresponding senior author who was involved in conception and design, analysis and interpretation of the data; critical revision of the article for important intellectual content, and the final approval of the article.
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Gut editions and any other BMJPGL products to exploit all subsidiary rights, as set out in our licence (http://group.bmj.com/products/journals/instructions-for-authors/licence-forms).
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006 Feb 24;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005 Jun 10;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutgeerts P, Van Assche G, Vermeire S, D'Haens G, Baert F, Noman M, et al. Ornidazole for prophylaxis of postoperative Crohn's disease recurrence: a randomized, double-blind, placebo-controlled trial. Gastroenterology. 2005 Apr;128(4):856–861. doi: 10.1053/j.gastro.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Rutgeerts P, Hiele M, Geboes K, Peeters M, Penninckx F, Aerts R, et al. Controlled trial of metronidazole treatment for prevention of Crohn's recurrence after ileal resection. Gastroenterology. 1995 Jun;108(6):1617–1621. doi: 10.1016/0016-5085(95)90121-3. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46(8):535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 6.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999 Nov;65(11):4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furrie E. A molecular revolution in the study of intestinal microflora. Gut. 2006 Feb;55(2):141–143. doi: 10.1136/gut.2005.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004 Mar;42(3):1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uronis JM, Arthur JC, Keku T, Fodor A, Carroll IM, Cruz ML, et al. Gut microbial diversity is reduced by the probiotic VSL#3 and correlates with decreased TNBS-induced colitis. Inflamm Bowel Dis. Jun 17; doi: 10.1002/ibd.21366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zella GC, Hait EJ, Glavan T, Gevers D, Ward DV, Kitts CL, et al. Distinct microbiome in pouchitis compared to healthy pouches in ulcerative colitis and familial adenomatous polyposis. Inflamm Bowel Dis. Sep 15; doi: 10.1002/ibd.21460. [DOI] [PubMed] [Google Scholar]
- 11.Kohyama A, Ogawa H, Funayama Y, Takahashi K, Benno Y, Nagasawa K, et al. Bacterial population moves toward a colon-like community in the pouch after total proctocolectomy. Surgery. 2009 Apr;145(4):435–447. doi: 10.1016/j.surg.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Kuehl CJ, Wood HD, Marsh TL, Schmidt TM, Young VB. Colonization of the cecal mucosa by Helicobacter hepaticus impacts the diversity of the indigenous microbiota. Infect Immun. 2005 Oct;73(10):6952–6961. doi: 10.1128/IAI.73.10.6852-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishikawa J, Kudo T, Sakata S, Benno Y, Sugiyama T. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand J Gastroenterol. 2009;44(2):180–186. doi: 10.1080/00365520802433231. [DOI] [PubMed] [Google Scholar]
- 14.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 5(3):e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. May;1(3):138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. May-Jun;44(5):354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006 Dec;12(12):1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 18.Bibiloni R, Mangold M, Madsen KL, Fedorak RN, Tannock GW. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J Med Microbiol. 2006 Aug;55(Pt 8):1141–1149. doi: 10.1099/jmm.0.46498-0. [DOI] [PubMed] [Google Scholar]
- 19.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002 Jan;122(1):44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 20.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005 Jul;43(7):3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marteau P, Pochart P, Dore J, Bera-Maillet C, Bernalier A, Corthier G. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl Environ Microbiol. 2001 Oct;67(10):4939–4942. doi: 10.1128/AEM.67.10.4939-4942.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackwood CB, Marsh T, Kim SH, Paul EA. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl Environ Microbiol. 2003 Feb;69(2):926–932. doi: 10.1128/AEM.69.2.926-932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt TM, Relman DA. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol. 1994;235:205–222. doi: 10.1016/0076-6879(94)35142-2. [DOI] [PubMed] [Google Scholar]
- 24.Osborne CA, Galic M, Sangwan P, Janssen PH. PCR-generated artefact from 16S rRNA gene-specific primers. FEMS Microbiol Lett. 2005 Jul 15;248(2):183–187. doi: 10.1016/j.femsle.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 25.Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997 Nov;63(11):4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colwell RK. EstimateS: Statistical estimation of species richness and shared species from samples. 2006 Available from: purl.oclc.org/estimates. [Google Scholar]
- 27.Beals EW. In: Bray-Curtis Ordination: An effective strategy for analysis of multivariate ecological data. MacFadyen A, Ford ED, editors. Orlando, FL: Advances in Ecological Research; 1984. [Google Scholar]
- 28.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007 Aug;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 29.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009 Nov 11;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen Y-Y, et al. High-Fat Diet Determines the Composition of the Murine Gut Microbiome Independently of Obesity. Gastroenterology. 2009;137(5):1716.e2–1724.e2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008 Nov 18;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.