Abstract
Genetic diversity was investigated in 60 individuals of Primula obconica from four natural populations (from Hubei, central China, and Sichuan, south‐west China) and from cultivated material. Inter‐simple sequence repeat (ISSR) techniques produced 249 polymorphic bands and identified 60 ISSR genotypes. Based on Shannon’s index and Nei’s genetic diversity, the genetic diversity detected in all natural populations of P. obconica was much higher than that in the cultivated plants, and that in the three Dalaoling (Hubei) populations was higher than that in the Wawushan (Sichuan) population. UPGMA cluster analysis showed that there was no distinct genetic differentiation between populations from the Mt Dalaoling area and the Mt Wawushan area. This study provides a population‐level genetic profile of P. obconica, which was previously poorly known but which is important for Primula breeding and cultivation.
Key words: Primula obconica, genetic diversity, ISSR, genotype, cluster analysis, China
INTRODUCTION
Primula obconica Hance (Primulaceae), the poison primrose, is mainly distributed in central and south‐west China where it grows in calcareous soils at altitudes of 800–2000 m (Zheng, 1993; Hu and Kelso, 1996). It was first introduced to Britain from Yichang (Ichang) County, Hubei (Hupeh) Province, China, in 1880 and rapidly became established as a popular ornamental house‐plant worldwide (Hausen, 1978; Richards, 1993). However, P. obconica is reported to be a significant cause of allergic contact dermatitis, particularly in Europe and the United States (Rook and Wilson, 1965; Aplin and Lovell, 2001) owing to the allergic compounds primin (2‐methoxy‐6‐pentyl‐1,4‐benzoquinone) and miconidin (2‐methoxy‐6‐pentyl‐1,4‐dihydroxybenzene) that can be isolated from leaves, stems and flowers (including pedicel and calyx) of many cultivars (Horper and Marner, 1995, 1996; Krebs and Christensen, 1995; Christensen and Larsen, 2000a). Breeding of primin‐free cultivars of P. obconica has therefore become a focus of attention within the horticultural industry (Christensen and Larsen, 2000b). Recently, the volatile oil was extracted and analysed from wild P. obconica from Yichang, Hubei (central China) but the two allergic compounds were not found (Nan et al., 2002a; L. P. Christensen, pers. comm.). This research suggested that wild P. obconica in China is a potential genetic resource for horticultural use since the allergic compounds do not appear to be present. Thus, further comparative studies of the genetic and chemical diversity among natural populations and cultivars of P. obconica are needed.
In recent years a number of molecular markers such as RAPD (random amplified polymorphic DNA; Hu and Quiros, 1991; Munthali et al., 1992), AFLP (amplified fragment length polymorphism; Vos et al., 1995), SSR (simple sequence repeats; Zietkiewicz et al., 1994) and ISSR (inter‐simple sequence repeats; Salimath et al., 1995; Wolfe and Randle, 2001) have been widely used to detect genetic diversity in plants (Karp et al., 1996; Wolfe and Liston, 1998). In particular, ISSR markers can be highly variable within a species and have the advantage over RAPDs in utilizing longer primers that allow more stringent annealing temperatures (Tsumura et al., 1996; Wolfe and Liston, 1998; Camacho and Liston, 2001) and revealing many more polymorphic fragments (Fang and Roose, 1997). In the present study the ISSR technique was used to examine natural populations of P. obconica from central and south‐west China as well as some glasshouse‐grown plants to evaluate the population‐level genetic diversity of P. obconica.
MATERIALS AND METHODS
Sampling
During July and August 2001, four natural populations of P. obconica in central (Hubei) and south‐west (Sichuan) China were sampled: three natural populations (D1–D3) were from the Mt Dalaoling area of Yichang, Hubei, and one (W) was from the Mt Wawushan area, Sichuan. A few living plants of P. obconica (G) were introduced from Yunnan and had been cultivated in a glasshouse at the Wuhan Botanical Garden, Academia Sinica for over 10 years. They were also sampled in September 2001 for comparison with the natural populations (Table 1).
Table 1.
Sampling for ISSR survey and genetic diversity of P. obconica
| Population | No. of individuals | Latitude | Longitude | Altitude (m) | PPB | I | I* | h | h* |
| D1 | 20 | 31°04′ | 110°52′ | 850 | 84·5 | 0·309 | 0·209 | 0·202 | 0·141 |
| D2 | 13 | 31°06′ | 110°57′ | 930 | 82·3 | 0·260 | 0·192 | 0·167 | 0·129 |
| D3 | 18 | 31°03′ | 110°55′ | 800 | 84·8 | 0·294 | 0·197 | 0·193 | 0·133 |
| W | 6 | 29°41′ | 102°52′ | 1360 | 68·9 | 0·196 | 0·117 | 0·131 | 0·079 |
| G | 3 | 30°33′ | 114°23′ | 35 | 18·2 | 0·045 | 0·045 | 0·031 | 0·031 |
Populations D1–D3 sampled from Mt Dalaoling area, Hubei, central China; W from Mt Wawushan area, Sichuan, south‐west China. G, plants grown in a glasshouse at the Wuhan Botanical Garden, Hubei.
PPB, Percentage of polymorphic bands; I, Shannon index (all individuals); I*, Shannon index calculated using mean values for three randomly selected individuals; h, Nei’s genetic diversity (all individuals); h*, Nei’s genetic diversity calculated using mean values for three randomly selected individuals.
Total DNA extraction
Fresh leaves of individual plants were collected and preserved in silica gel until required for DNA isolation. Total DNA was extracted using a modified CTAB protocol (Doyle and Dolye, 1987). Dried leaf material was ground in liquid nitrogen, transferred to a 1·5 ml Eppendorf tube holding 800 µl preheated 2 × CTAB extraction buffer containing 0·2 % mercaptoethanol and incubated at 64 °C for 60 min. Subsequently, 600 µl cold chloroform : isoamylalcohol (24 : 1, v/v) was added, and the mixture was shaken gently for 2 min and spun down at 10 000 r.p.m. for 10 min at room temperature. The supernatant was reserved and mixed with 2/3 volume ice‐cold isopropanol. DNA was then recovered as a pellet by centrifugation at 12 000 r.p.m. for 8 min, washed in 70 % ethanol, dried, and resuspended in 100 µl TE buffer. DNA quality and quantity were checked on 1·0 % agarose gels.
ISSR PCR amplification
Ninety primers from the UBC set 9 of ISSR primers (http://www.biotech.ubc.ca/) were tested for PCR. Reaction volumes were 10 µl, and consisted of 1 µl 10 × reaction buffer, 3 mm MgCl2, 300 µm dNTPs, 0·25 µm primer, 2 % formamide, 10 ng DNA template and 1 U Taq DNA polymerase (Genda Tech. Corp., Toronto, Canada). The thermocycler programme for PCR was set for 1·5 min at 94 °C, followed by 45 cycles of 45 s at 94 °C, 45 s annealing at 51 or 52 °C (Table 2) and 1·5 min extension at 72 °C, and a final extension cycle of 7 min at 72 °C.
Table 2.
Primers used for ISSR amplification
| Primer | Sequence | Annealing temperature (°C) |
| 807 | (AG)8T | 51 |
| 811 | (GA)8C | 52 |
| 814 | (CT)8A | 52 |
| 827 | (AC)8G | 52 |
| 834 | (AG)8CTT | 52 |
| 845 | (CT)8AGG | 52 |
| 846 | (CA)8AGT | 52 |
| 848 | (CA)8AGC | 52 |
| 852 | (TC)8AGA | 51 |
| 856 | (AC)8CTA | 52 |
| 858 | (TG)8AGT | 52 |
| 868 | (GAA)5GAA | 52 |
| 888 | CGTAGTCGT(CA)7 | 52 |
| 890 | ACGACTACG(GT)7 | 51 |
Amplification products were resolved electrophoretically on 1·5 % agarose gels in 1 × TAE buffer by loading the entire reaction volumes into prepared wells. Gels were run until a bromophenol blue indicator dye ran 10 cm from the well. Gels were stained with ethidium bromide and bands were visualized and photographed under UV light. Molecular weights were estimated using a 100 bp DNA ladder (Shengong Inc., Shanghai, China).
Data analysis
All ISSR bands were scored as present (1) or absent (0). The number of bands, the percentage of polymorphic bands (PPB) and the number of ISSR genotypes were calculated manually, and the Shannon index of diversity (I) and Nei (1973) genetic diversity (h) were calculated using POPGENE version 1.31 (Yeh et al., 1999) (available free from http://www.ualberta.ca/∼fyeh/). Two separate analyses of genetic diversity were conducted, one including all the individuals and the other using average scores of three individuals for each population (randomly selected about ten times).
The Nei and Li (1979) coefficient for measuring pairwise band similarities between individuals was calculated using NTSYSpc version 2.02 (Rohlf, 1998). A clustering analysis of all individuals was performed using the unweighted pair‐group method with an arithmetic average (UPGMA) using NTSYSpc 2.02.
RESULTS AND DISCUSSION
By comparing the effects of magnesium concentration and annealing temperature during amplification, 14 of the 90 primers were chosen for further analysis (Table 2). For the 60 P. obconica samples the total number of bands scored for the 14 primers was 308, with the PPB being 68–84 % among natural populations and 18·2 % for the cultivated population (Table 1). The size range of PCR fragments was 300–2200 bp. The average number of bands per population per primer ranged from 9·2 to 17·1, and the average number of bands per primer for the species was 22·0. The gene diversity (h) in all populations of P. obconica ranged from 0·031 to 0·202 (Table 1). Among the four natural populations, the gene diversity of the three Dalaoling populations (h = 0·193–0·202) was higher than that of the Wawushan population (h = 0·131), and the values of Shannon’s index (I) also showed the same trend (Table 1). Since the genetic diversity results may correlate with sample size of a population, an equal number of individuals (three) randomly selected from each population was analysed. Results showed that the values of gene diversity (h*) and Shannon’s index (I*) obtained from three random individuals of each population were lower than the original values of h and I, but the trend was same for each index (Table 1). In particular, among the three Dalaoling populations the smallest number of individuals was sampled from population D2, which also had the lowest genetic diversity. Human activity in the Mt Wawushan and Mt Dalaoling areas is believed to be a major factor in the dramatic decrease in population size of P. obconica (Shen et al., 2000; Yuan, pers. comm.).
The number of bands and polymorphic bands produced by each primer varied. The highest number of bands (24) was produced by primers 834, 846 and 888, and the highest number of polymorphic bands (20) by primers 845, 852, 888 and 890 (Table 3). The number of ISSR genotypes within each population distinguished by each primer is also shown in Table 3. Since the maximum number of genotypes within each population is equal to the number of individuals in the same population, the genotypic diversity (number of genotypes/number of individuals; Wolfe and Randle, 2001) for each population is 1·0.
Table 3.
Number of bands per primer and number of ISSR genotypes determined for each population per primer
| No. of polymorphic | ISSR genotypes | ||||||
| Primer | No. of bands | bands | D1(20) | D2(13) | D3(18) | W(6) | G(3) |
| 807 | 20 | 14 | 19 | 13 | 18 | 6 | 2 |
| 811 | 22 | 18 | 18 | 13 | 18 | 6 | 2 |
| 814 | 19 | 14 | 20 | 13 | 15 | 6 | 2 |
| 827 | 21 | 17 | 20 | 10 | 18 | 6 | 2 |
| 834 | 24 | 16 | 20 | 11 | 17 | 6 | 2 |
| 845 | 23 | 20 | 19 | 12 | 17 | 5 | 2 |
| 846 | 24 | 17 | 19 | 13 | 18 | 6 | 3 |
| 848 | 22 | 18 | 20 | 13 | 16 | 6 | 2 |
| 852 | 21 | 20 | 12 | 13 | 15 | 6 | 3 |
| 856 | 22 | 19 | 20 | 13 | 18 | 6 | 2 |
| 858 | 21 | 19 | 20 | 13 | 11 | 4 | 2 |
| 868 | 23 | 17 | 20 | 13 | 18 | 6 | 3 |
| 888 | 24 | 20 | 20 | 13 | 16 | 6 | 2 |
| 890 | 22 | 20 | 20 | 13 | 18 | 6 | 1 |
| Total | 308 | 249 | 20 | 13 | 18 | 6 | 3 |
Sample sizes in parentheses. Bold type indicates that each individual in a population was found to have a unique banding pattern.
A dendrogram of all 60 individuals of P. obconica based on the ISSR markers was generated using the Nei–Li similarity coefficient matrix and the UPGMA cluster method (Fig. 1). In this dendrogram all individuals of each population form a distinct cluster suggesting that there is genetic differentiation between populations. However, the Wawushan population cluster is shown to be more closely related to the cluster consisting of two Dalaoling populations (D1 and D3) than the other Dalaoling population (D2), indicating that there is no distinct genetic differentiation between populations from the Mt Daloling area and the Mt Wawushan area. In contrast, the cluster of cultivated plants is distinct from that of the four natural populations. The genetic diversity of P. ovalifolia, which grows alongside P. obconica, has also been investigated recently using ISSR markers (Nan et al., 2002b). As for P. obconica, results showed that there is no distinct genetic differentiation between populations of P. ovalifolia from the Mt Dalaoling area (two populations) and those from the Mt Wawushan area (three populations).
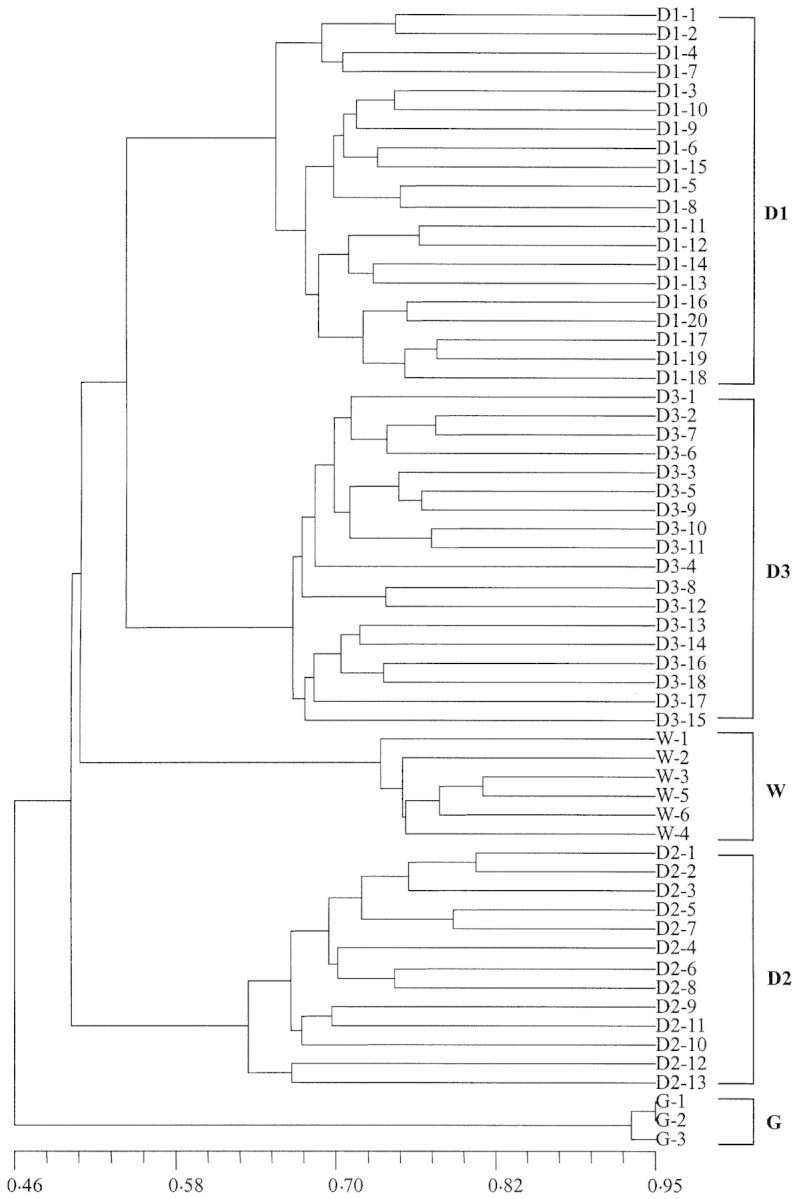
Fig. 1. Dendrogram of P. obconica individuals based on ISSR markers generated using the Nei and Li (1979) similarity coefficient and the UPGMA clustering method. Populations D1–D3 sampled from Mt Dalaoling area, Hubei, central China; W from Mt Wawushan area, Sichuan, south‐west China; G, plants grown in a glasshouse at the Wuhan Botanical Garden, Hubei.
This study has increased the understanding of population‐level genetic diversity of P. obconica, which was previously poorly known but which is important for Primula breed ing and cultivation. Volatile oils from populations of P. obconica in central and south‐west China have been analysed. Further comparative studies are being undertaken including an assessment of phytochemical variation among and within natural populations and of the relationship between genetic and chemical diversity of cultivars of this species.
ACKNOWLEDGEMENTS
We thank Professors Chi‐Ming Hu and Gang Hao for their help in specimen identification, Zien Zhao, Yafu Yuan and Yinying Shi for fieldwork, and Shuguang Jian, Fengxiao Tan, Yuguo Wang, Yalin Peng, Yaqing Du and Tian Tang for technical assistance. This work was supported by the National Science Foundation of China (39899370), Academia Sinica (STZ‐01–36) and the High‐Tech Research and Development Programme (863) of China (2002 AA231041).
Supplementary Material
Received: 7 March 2002; Returned for revision: 24 August 2002; Accepted: 26 October 2002 Published electronically: 12 December 2002
References
- AplinCG, Lovell CR.2001. Contact dermatitis due to hardy primula specices and their cultivars. Contact Dermatitis 44: 23–29. [DOI] [PubMed] [Google Scholar]
- CamachoFJ, Liston A.2001. Population structure and genetic diversity of Botrychium pumicola (Ophioglossaceae) based on inter‐simple sequence repeats (ISSR). American Journal of Botany 88: 1065–1070. [PubMed] [Google Scholar]
- ChristensenLP, Larsen E.2000a Direct emission of three allergen primin from intact Primula obconica plants. Contact Dermatitis 42: 149–153. [DOI] [PubMed] [Google Scholar]
- ChristensenLP, Larsen E.2000b Primin‐free Primula obconica plants available. Contact Dermatitis 43: 45–46. [PubMed] [Google Scholar]
- DoyleJJ, Doyle JL.1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- FangDQ, Roose ML.1997. Identification of closely related citrus cultivars with inter‐simple sequence repeat genetics. Theoretical and Applied Genetics 95: 408–417. [Google Scholar]
- HausenBM.1978. On the occurrence of the contact allergen primin and other quinoid compounds in species of the family of Primulaceae. Archive of Dermatitis Research 261: 311–321. [DOI] [PubMed] [Google Scholar]
- HorperW, Marener FJ.1995. Phenols and quinones from leaves of Primula obconica Natural Product Letters 6: 163–170. [Google Scholar]
- HorperW, Marener FJ.1996. Biosynthesis of primin and miconidin and its derivatives. Phytochemistry 41: 451–456. [Google Scholar]
- HuCM, Kelso S.1996. Primulaceae. In: Wu CY, Raven PH, eds. Flora of China, Myrsinaceae through Loganiaceae, Vol. 15. Beijing: Science Press and St Louis: Missouri Botanical Garden, 118–119. [Google Scholar]
- HuJ, Quiros CF.1991. Identification of broccoli and cauliflower cultivars with RAPD markers. Plant Cell Reporter 10: 505–511. [DOI] [PubMed] [Google Scholar]
- KarpA, Seberg O, Buiatti M.1996. Molecular techniques in the assessment of botanical diversity. Annals of Botany 78: 143–149. [Google Scholar]
- KrebsM, Christensen LP.1995. 2‐Methoxy‐6‐pentyl‐1,4‐dihydroxy benzene (miconidin) from Primula obconica‐a possible allergen. Contact Dermatitis 33: 90–93. [DOI] [PubMed] [Google Scholar]
- MunthaliM, Ford‐Lloyd BV, Newbury HJ.1992. The random amplification of polymorphic DNA for fingerprinting plants. PCR Methods and Applications 1: 274–276. [DOI] [PubMed] [Google Scholar]
- NanP, Peng S, Zhang Y, Zhong Y.2002a Composition of volatile oil of Primula obconica in Central China. N atural Product Letters 16: 249–253. [DOI] [PubMed] [Google Scholar]
- NanP, Peng S, Ren H, Shi S, Tian C, Zhong Y.2002b Genetic diversity of Primula ovalifolia from central and southwest China based on ISSR markers. J ournal of Genetics and Molecular Biology 13: 119–123. [Google Scholar]
- NeiM.1973. Analysis of gene diversity in subdivided population. Proceedings of National Academy of Sciences USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NeiM, Li WH.1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of National Academy of Sciences USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RichardsAJ.1993. Primula. London: Batsford Ltd. [Google Scholar]
- RohlfFJ.1998. NTSYSpc: numerical taxonomy and multivariate analysis system, version 2.02. Exeter Software, Setauket, New York. [Google Scholar]
- RookA, Wilson HTH.1965. Primula dermatitis. British Medical Journal I: 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SalimathSS, De Oliverira AC, Godwin ID, Bennetzen JL.1995. Assessment of genomic origins and genetic diversity in the genus Eleusine with DNA markers. Genome 38: 757–763. [DOI] [PubMed] [Google Scholar]
- ShenZH, Zhang XS, Jin YX.2000. An analysis of the topographical patterns of the chief woody species at Dalaoling mountain in the Three Gorges region. Acta Phyoecologica 24: 581–589. [Google Scholar]
- TsumuraY, Ohba K, Strauss SH.1996. Diversity and inheritance of inter‐simple sequence repeat polymorphisms in douglas‐fir (Pseudotsuga menziesii) and sugi (Cryptomeria japonica). Theoretical and Applied Genetics 92: 40–45. [DOI] [PubMed] [Google Scholar]
- VosP, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M.1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WolfeAD, Liston A.1998. Contributions of PCR‐based methods to plant systematics and evolutionary biology. In: Soltis PS, Soltis DE, Doyle JJ, eds. Molecular systematics of plants: DNA sequencing. New York: Kluwer, 43–86. [Google Scholar]
- WolfeAD, Randle CP.2001. Relationships within and among species of the holoparasitic genus Hyobanche (Orobanchaceae) inferred from ISSR banding patterns and nucleotide sequences. Systematic Botany 26: 120–130. [Google Scholar]
- YehFC, Boyle T, Yang RC, Ye Z, Xiyan JM.1999. POPGENE, the user friendly shareware for population genetic analysis, version 1.31. University of Alberta and Centre for International Forestry Research. [Google Scholar]
- ZhengZ.1993. Plants in Hubei. Wuhan: Wuhan University Press. [Google Scholar]
- ZietkiewiczE, Rafalski A, Labuda D.1994. Genome fingerprinting by simple sequence repeats (SSR)‐anchored PCR amplification. Genomics 20: 176–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


