Abstract
Proteome analysis, and more recently DNA chip technology, has led to the identification of a large number of genes that are implicated in the anaerobic response of plants. As a result, an increasingly complex picture of the response in terms of biochemical and regulatory processes is emerging. A challenge is to find out more about the function of these newly identified gene products, and how they contribute to flooding tolerance. Our approach has been to manipulate levels of candidate gene products (using sense and antisense constructs) in the model system Arabidopsis thaliana, combined with biochemical and phenotypic analysis of the resulting transgenic plants.
Keywords: Key words: Review, anaerobiosis, gene expression, Arabidopsis thaliana, microarrays, gene manipulation.
INTRODUCTION
The anaerobic response of plant cells was first studied in maize roots. Using two‐dimensional electrophoresis, it was shown that a set of about 20 anaerobic proteins was synthesized during low oxygen treatment, while synthesis of the normal aerobic proteins was drastically repressed (Sachs et al., 1980). Many of these induced proteins were subsequently identified as enzymes of the glycolytic and fermentation pathways (Kelley and Freeling 1984a, b; Kelley, 1989; Dolferus et al., 2000). Proteomics identified a further 46 proteins (Chang et al., 2000). Associated with the increased number of gene products shown to be involved in response to low oxygen stress is a steady increase in the number of gene products that are implicated in processes other than sugar breakdown, glycolysis and fermentation, suggesting that many other biochemical and metabolic processes are modulated during low oxygen stress. For instance, xyloglucan endotransglycosylase, a cell wall loosening enzyme (Sachs et al., 1996), is induced, as is the ethylene biosynthetic enzyme ACC synthase (1‐aminocyclopropane‐1‐carboxylate synthase; Olson et al., 1995).
Proteome analysis has advanced our understanding of the anaerobic response, but the more sensitive DNA microarray technology (Schena, 1996; Jordan, 1998; Ramsay, 1998; Cheung et al., 1999; Duggan et al., 1999; Henn, 1999; Richmond and Somerville, 2000) has the potential to generate more information about the different biochemical and regulatory processes that take place in the plant during low oxygen conditions. Microarrays offer the possibility of identifying virtually all genes that are up‐ or down‐regulated by low oxygen treatment. The high sensitivity of microarrays makes it possible to compare the timing of induction of a large set of genes during low oxygen treatment and to identify those genes that are potentially involved in the regulation of different aspects of the response. A limitation is that microarray experiments only detect changes in steady state mRNA levels and cannot distinguish between transcriptional and post‐transcriptional aspects of gene regulation. Post‐transcriptional control and selective translation of mRNA have been shown to occur during low oxygen stress (Bailey‐Serres and Dawe, 1996; Bailey‐Serres et al., 1999).
Gene products identified by proteomics and genomics will require analysis by approaches that determine the function of each gene in the anaerobic response and how each gene or gene product contributes to survival. This can be achieved by a reverse genetics approach, by expressing the gene in sense or antisense orientation. The phenotype of the resulting transgenic plants will require analysis at the biochemical/physiological level, and in terms of flooding tolerance. The use of a model system like Arabidopsis thaliana facilitates this approach. First, the arabidopsis genome sequence is completely known (http://www.arabidopsis.org). Secondly, arabidopsis is readily transformed (Clough and Bent, 1998), making it relatively easy to screen for the function of genes using over‐expressing and knock‐out transgenic lines. A large number of arabidopsis knock‐out lines using T‐DNA insertions and transposons are available (http://www.jic.bbsrc.ac.uk/staff/michael‐bevan/atis/Resources1.html) for analysis. Thirdly, web sites such as the arabidopsis functional genomics consortium (http://afgc.stanford.edu/) and the Stanford microarray database (http://genome‐www4.stanford.edu/MicroArray/SMD/) are important resources assisting in the elucidation of the function of arabidopsis genes.
CAN FLOODING TOLERANCE BE MANIPULATED IN ARABIDOPSIS?
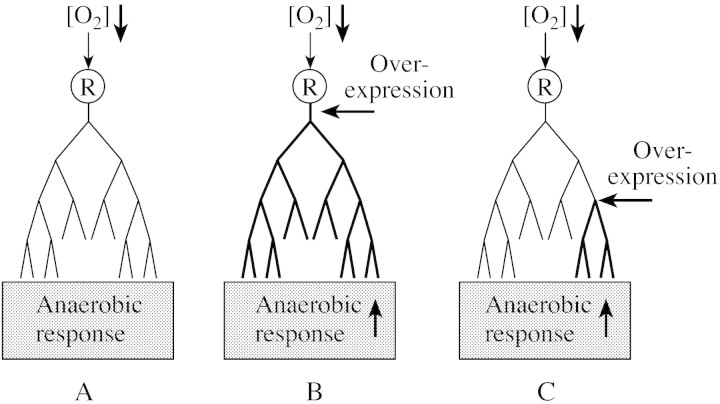
The anaerobic response pathway starts with the perception of a single signal (e.g. oxygen levels decreasing below a threshold level) leading to the activation of a first set of genes. This may then be followed by activation of a secondary set of signal transduction events and/or a cascade of biochemical pathways which are part of the defence/adaptation mechanisms put in place by the stress (Fig. 1A). Cross‐talk and interactions between different aspects of the response may take place at any stage of the response. At different stages during the response new signals can be generated and signal transduction components induced/activated, leading to activation of new aspects of the response.
Fig. 1. Schematic representation of the anaerobic response. The anaerobic response consists of a complex tree‐like cascade of gene expression events, starting with a single initial stimulus (decreased oxygen levels) perceived by a receptor (‘R’), and a signal transduction event leading to the induction of several biochemical reactions as part of the response (A). Manipulation of the initial regulatory and signal transduction event may lead to amplification of the entire response pathway (B). Alternatively, gene manipulation using genes that are induced later in the response may lead to a partial amplification of the response pathway. Depending on the importance of this gene, this may still lead to amplification of the response and improved survival of low oxygen conditions (C).
Successful manipulation of the anaerobic response will require detailed knowledge of how the response is activated and which genes are critical in mounting an efficient metabolic reply to the deficiencies caused by the stress. Knowledge of the early signal transduction and regulatory events that lead to the establishment of the entire response pathway and the genes involved may enable us to manipulate the entire response cascade (Fig. 1B). To date, only one transcription factor with a role in the anaerobic response has been identified (AtMYB2; Hoeren et al., 1998). Gene manipulation using regulatory components has been successful in the manipulation of freezing tolerance using the AP2 domain transcription factor CBF1/DREB1B (Stockinger et al., 1997; Jaglo‐Ottosen et al., 1998; Thomashow, 2001), improving the drought and salinity response using another AP2 domain transcription factor DREB1A (Yamaguchi‐Shinozaki and Shinozaki, 2001), enhancing NaCl tolerance using AtGSK1 (Piao et al., 2001), and increasing freezing and low temperature tolerance using the transcription factor ABI3 (Tamminen et al., 2001).
An alternative approach to manipulating the anaerobic response is to use genes that are known to play a role in the metabolic response (Fig. 1C). Metabolic engineering is expected to be successful only if the genes that are used contribute enough to the response to affect plant survival. That this approach may be successful is demonstrated by the addition of genes to give increased production of the metabolite glycine betaine, resulting in improved drought tolerance (Hanson et al., 1994; Huang et al., 2000; Sakamoto and Murata, 2000). More recently, it was shown that manipulation of ABA biosynthesis using the 9‐cis‐epoxycarotenoid dioxygenase gene (AtNCED3) resulted in improved drought tolerance in arabidopsis (Thompson et al., 2000; Iuchi et al., 2001).
MANIPULATION USING REGULATORY FACTORS: ATMYB2
The GT‐ and GC‐motifs located in the promoter of the arabidopsis ADH1 gene are critical for the expression of the gene under a variety of abiotic stress conditions (Dolferus et al., 1994). The GT‐motif, which is found in the promoter of many anaerobically induced genes, binds AtMYB2, a transcription factor of the Myb family (Hoeren et al., 1998). AtMYB2, which was first identified as a drought‐inducible transcription factor (Urao et al., 1993, 1996), is induced by all stresses known to induce ADH1 (low oxygen, cold, drought, wounding). Transient assays using protoplasts as well as particle bombardment experiments demonstrated that over‐expression of AtMYB2 leads to enhanced ADH1‐promoter‐driven GUS expression (Hoeren et al., 1998). AtMYB2 mRNA induction is synchronized with accumulation of ADH1 mRNA under low oxygen conditions, with AtMYB2 peaking slightly before ADH1 (AtMYB2: 2–4 h; ADH1: 4–6 h).
Arabidopsis AtMYB2 is a good target for the manipulation of the anaerobic response. When AtMYB2 was introduced into arabidopsis under the control of a strong constitutive promoter (35S‐AtMYB2‐3′NOS), none of the resulting transgenic plants over‐expressed AtMYB2 mRNA, and Southern blot hybridizations showed that in some of the transgenic plants the construct was truncated or reorganized. These results may indicate that strong constitutive over‐expression of AtMYB2 in arabidopsis is lethal. To avoid this problem, we used a dexamethasone‐inducible promoter system to drive AtMYB2 expression (Aoyama and Chua, 1997). We obtained transgenic arabidopsis plants with high inducible levels of AtMYB2 mRNA. But dexamethasone showed a strong negative effect on ADH1 expression, making it impossible to establish the effect of AtMYB2 on the expression of ADH1 and other anaerobically induced genes. Dexamethasone treatment was shown to have toxic effects and cause induction of defence‐related genes in arabidopsis (Kang et al., 1999). When we attempted to use the classic antisense approach we were unable to find transformants with significantly down‐regulated AtMYB2 mRNA levels, possibly due to the low expression levels of the gene. We are currently using a more efficient method (Waterhouse et al., 1998; Smith et al., 2000) to down‐regulate AtMYB2 expression in arabidopsis.
Curiously, treatment with the protein synthesis inhibitor cycloheximide resulted in a strong up‐regulation of AtMYB2 mRNA levels, both under stressed and unstressed conditions, while induction of ADH1 was inhibited (Hoeren et al., 1998). Cycloheximide has been shown to induce mRNA of many genes in plants and other organisms, especially in the case of signal transduction components (Otani et al., 1990; Wisdom and Lee, 1991; Shen et al., 1993; Berberich and Kusano, 1997; Horvath et al., 1998). Numerous reports indicate that cycloheximide stabilizes mRNAs that are turning over quickly, and that this could involve sequences in the 5′ and 3′ untranslated regions (UTR) of the mRNA, as well as in the coding region of the message (Fen and Daniel, 1991; Peppel et al., 1991; Gil and Green, 1996; Memon et al., 1996; Chan and Yu, 1998; Dickey et al., 1998; Marchant and Bennett, 1998; Yeilding et al., 1998; Johnson et al., 2000). We expect that post‐transcriptional regulation mechanisms that increase transcript levels via mRNA stabilization may be important in AtMYB2 regulation. Post‐transcriptional regulation mechanisms have been shown to play an important role in the regulation of other anaerobically induced genes (Gallie and Bailey‐Serres, 1997; Fennoy et al., 1998).
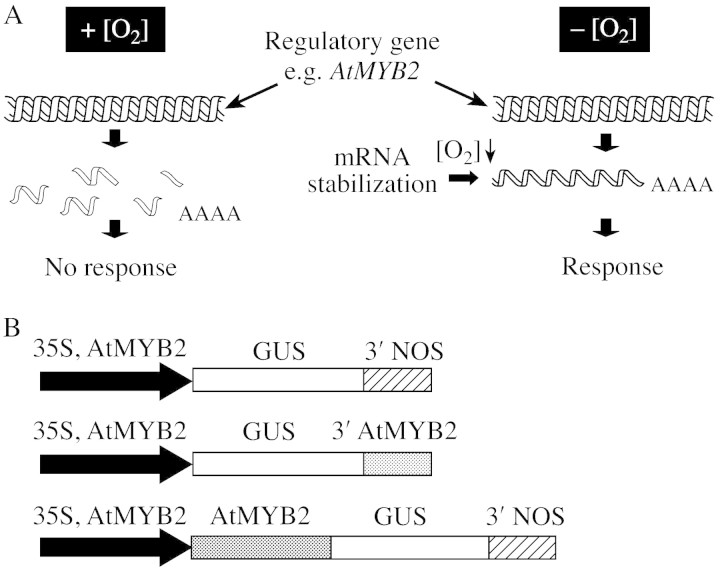
These observations indicate that the genes encoding the regulatory factors required to establish the anaerobic response are transcribed under normal conditions, but their transcripts are degraded very rapidly. Under low oxygen conditions, when energy supply becomes critical, a fast response could be obtained through stabilization of the transcripts and subsequent translation (Fig. 2A). We are currently studying AtMYB2 expression in more detail using reporter gene constructs including the 5′ and 3′ UTR, as well as various parts of the coding region in transgenic arabidopsis plants (Fig. 2B). Detailed knowledge of AtMYB2 regulation is essential to understand how the factor works and how it can be used to manipulate the anaerobic response.
Fig. 2. Model and approaches used to study the regulation of the anaerobic response using AtMYB2 as a model. A, Model for the regulation of the anaerobic response, based on post‐transcriptional regulation and mRNA stability. Regulatory factors such as AtMYB2 are expressed under normal aerobic conditions, but mRNA instability prevents the mRNA from being translated into a functional transcription factor. Low oxygen conditions lead to stabilization of the message and activation of the anaerobic response. This mode of gene activation may conserve energy when it is most needed, and it may lead to quicker responses than via transcriptional regulation mechanisms. B, Constructs used to study post‐transcriptional regulation of AtMYB2 expression in transgenic arabidopsis plants. One set of constructs aims to compare the effect of the AtMYB2 3′ UTR on GUS mRNA stability, by comparing expression levels using the AtMYB2 3′ UTR and the 3′ UTR of the nopaline synthase gene (NOS). A similar approach is used to study the effect of the AtMYB2 coding region, by fusing different parts of AtMYB2 (5′, 3′ and full‐length) to the GUS reporter gene. Both the 35S promoter and the AtMYB2 promoter were used to drive these constructs.
MANIPULATION OF THE FERMENTATION PATHWAYS IN ARABIDOPSIS
Most genes induced by low oxygen stress encode enzymes involved in sugar metabolism, or the glycolytic and fermentation pathways (Dennis et al., 2000). In arabidopsis and many other plants, the fermentation pathway enzymes are not normally present and are induced de novo under low oxygen conditions. In flooding‐tolerant plants, alcohol fermentation is more important than the lactic acid or alanine fermentation pathways, and ADH null mutants are more sensitive to low oxygen conditions than wild‐type plants (Roberts et al., 1984a, b, 1985; Kennedy et al., 1992; Ellis et al., 1999). Many studies have compared the importance of these pathways between different plant species, but it remains to be established how amplification or knock‐out of these pathways really affects flooding tolerance within one species. The fermentation pathways are relatively easy targets for gene manipulation; they consist of only one or two genes. We have cloned the genes involved in these pathways in arabidopsis and produced sense and antisense lines to increase or decrease the production of fermentation pathway enzymes (Table 1). To test the effect of these changes a survival assay was used (Ellis et al., 1999; Ellis and Setter, 1999) involving three screening conditions. Three‐week‐old arabidopsis plants were transferred directly to a 0·1 % O2 environment for either 24 h (no hypoxic pre‐treatment, NHPT 24 h) or 48 h (NHPT 48 h), or plants were first incubated in a 5 % O2 environment before transfer to a 0·1 % O2 environment for 48 h (hypoxic pre‐treatment, HPT; Fig. 3). We found that over‐expression of the arabidopsis pyruvate decarboxylase genes PDC1 and PDC2 resulted in improved survival especially under the 48 h NHPT conditions. Strong over‐expression of arabidopsis lactate dehydrogenase (LDH1) did not cause increased sensitivity to low oxygen stress, nor did over‐expression of ADH1 improve survival. These results show that by changing plant metabolism by manipulating one gene of the alcohol fermentation pathway, it is possible to increase plant survival under low oxygen conditions (K. P. Ismond et al., unpubl. res.). Our findings confirm previous reports on the importance of the alcohol fermentation pathway under low oxygen conditions and the critical role of the PDC enzyme as a rate‐limiting regulatory step in this pathway (Roberts et al., 1984a, b, 1985; Ricard et al., 1994).
Table 1.
Overview of transgenic arabidopsis lines over‐expressing fermentation pathway genes in sense or antisesense orientation
| Plant line | Gene used (GenBank ID) | Fold mRNA expression compared with C24 |
| C24 | Wild‐type line | 1× |
| 35S‐ADH1 | Arabidopsis ADH1 (M12196) | 12·1× |
| EMS4/7 | EMS‐induced ADH1 null mutant (allyl alcohol selection) | 0·03× |
| 35S‐PDC1 | Arabidopsis PDC1 (U71121) | 9× |
| 35S‐1CDP | Arabidopsis PDC1, antisense (U71121) | Undetectable |
| 35S‐PDC2 | Arabidopsis PDC2 (U71122) | 23× |
| 35S‐2CDP | Arabidopsis PDC2, antisense (U71122) | 0·4× |
| 35S‐LDH1 | Arabidopsis LDH1 (AF043130) | 602× |
| 35S‐1HDL | Arabidopsis LDH1, antisense (AF043130) | 0·8× |
| 35S‐AlaAT1 | Barley AlaAT1 (Z26322) | N/A |
| 35S‐1TAalA | Arabidopsis AlaAT1, antisense (AF275372) | 0·6× |
Only the PDC1 and PDC2 over‐expressing lines performed better than the C24 wild type in survival assays.
mRNA levels were quantified using Northern blot hybridizations and phosphor imager analysis (Dolferus et al., 1994).
ADH1, Alcohol dehydrogenase; PDC1, PDC2, pyruvate decarboxylase; LDH1, lactate dehydrogenase; AlaAT1, alanine aminotransferase.
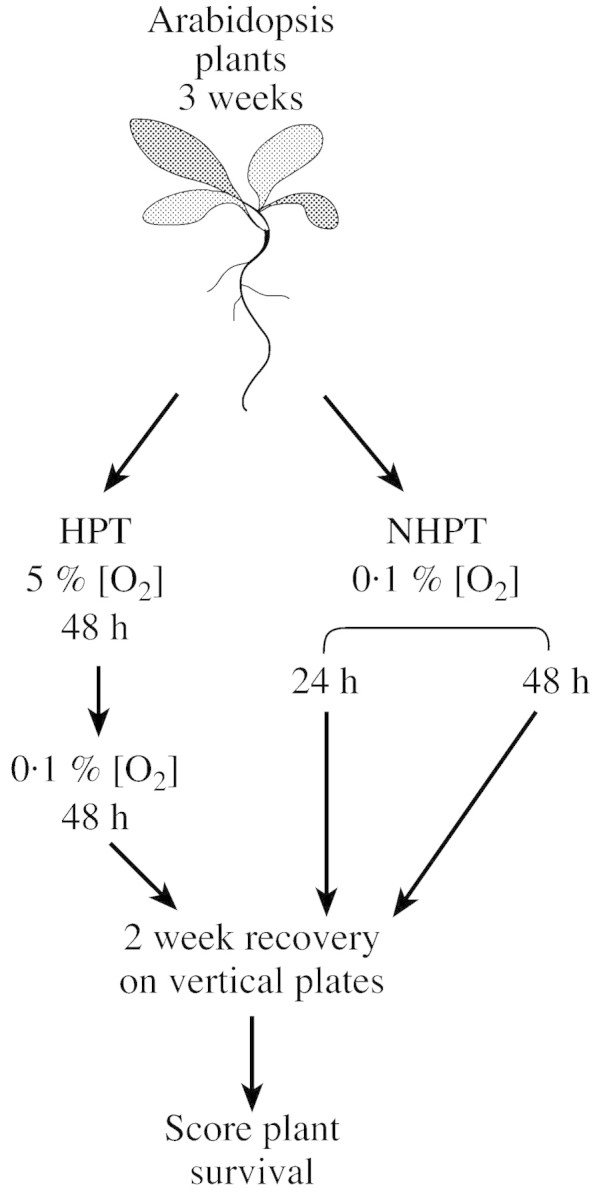
Fig. 3. Selection scheme used for screening transgenic arabidopsis lines for improved survival under low oxygen conditions. Three‐week‐old arabidopsis plants were subjected to three selective treatments, followed by a recovery period on vertical plates to score root growth and shoot survival. HPT, Hypoxic pre‐treatment; NHPT, no hypoxic pre‐treatment.
Although it still remains to be established whether PDC‐over‐expressing arabidopsis plants are better able to survive flooding conditions in the field, it is possible that the PDC gene can be used to improve flooding tolerance in plants where levels of the PDC enzyme are limiting. Experiments are in progress to test whether LDH1 knock‐outs or transgenic plants over‐expressing the complete alcohol fermentation pathway (PDC + ADH) can further improve plant survival.
ADH1 IS INDUCED BY WOUNDING: DO STRESS RESPONSES OVERLAP?
The arabidopsis ADH1 gene is not only induced by low oxygen stress, but also by cold, drought and the hormone ABA. These abiotic stresses only induce the gene in roots and not in leaves. The GT‐ and GC‐motifs in ADH1 are critical for all these stress responses, and induction by dehydration, cold and ABA also requires the G‐Box‐1 element (Dolferus et al., 1994; de Bruxelles et al., 1996). The transcription factor AtMYB2 is induced in roots by the same stresses that activate ADH1 (Hoeren et al., 1998).
We found that ADH1 is also induced in leaves by mechanical wounding (G. L. de Bruxelles et al., unpubl. res.). Expression is limited to about ten cell layers in the immediate vicinity of the wound site, and ABA measurements as well as ABA mutants indicate that ABA is involved in the process. The wounding response in leaves requires the same promoter elements as the response to abiotic stresses in roots.
The role of ADH1 under stress conditions other than low oxygen stress remains unknown. Is ADH1 part of a set of genes in plants, with common regulatory features and functional properties, that are recruited under a variety of stress conditions? The low oxygen stress response may consist of different sets of genes, with different regulatory and functional properties in the response, and some of these genes could be shared by other stress responses.
MICROARRAYS: THE ANSWER TO MANY QUESTIONS?
The possibilities of manipulating the anaerobic response using the genes we know so far are limited. Using microarray technology, we expect to identify many more genes involved in this response. We are currently applying this approach with the aim of identifying genes that are affected in the earlier stages of the low oxygen response. We also aim to identify those genes which are induced specifically by anaerobic conditions, and those genes which are also activated by a wider range of stresses (wounding, drought, cold, salinity). cDNA libraries were constructed from anaerobically induced arabidopsis roots that were also treated with cycloheximide, as an enrichment step for rapidly turning‐over transcripts that would normally be under‐represented. Two sets of microarrays were prepared: a 3·5 K array, containing 1000 cDNA clones from our cDNA library together with 2500 arabidopsis ESTs from genes involved in plant development and metabolism. A 10 K array, containing 10 000 random clones from our library was also prepared.
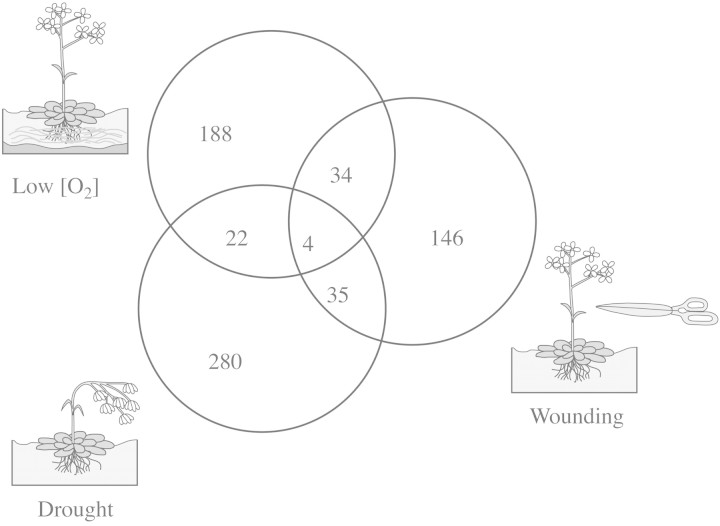
We have completed the screening of the 3·5 K array using RNA probes from plant roots that were treated for 0·5, 2, 4 and 20 h in a 0·5 % oxygen environment. We found 249 genes that were significantly affected by the treatment during this time course. The spectrum of genes that were changed during the first 0·5 h of treatment was significantly different from that of the genes induced later (2–20 h). About 20 % of the differentially expressed genes were down‐regulated at the 0·5 h time point, but not all of these genes were repressed throughout the 20‐h treatment period, and other genes were found to be down‐regulated at other stages of the anaerobic response. Transcription factors and signal transduction components were found to change mainly during the early part of the response, whereas most metabolic genes were induced during later stages (E. J. Klok et al., unpubl. res.). The time‐course experiment also allowed us to discriminate between different sets of genes according to their kinetics of induction or repression (E. J. Klok et al., unpubl. res.). This grouping of genes allowed us to identify different biological processes and regulatory events involved in the anaerobic response. Screening of the same array with RNA from wounded leaves gave a similar number of genes (220), but only 34 of these genes were also present in the gene list from anaerobically treated roots. Screening for genes affected by drought stress resulted in the identification of 342 genes. Only four genes were affected by all three stress treatments, while the overlap with low oxygen and wounding treatment was larger (Fig. 4). We plan to screen the larger 10 K microarray for genes induced by anaerobiosis and other stresses (cold, drought, salinity), and the hormone ABA, to further dissect the functional overlap between different stresses.
Fig. 4. Overlap in gene expression in arabidopsis plants treated by different stress conditions. Microarrays were used to identify the overlap in gene expression between the low oxygen response (248), drought (341) and wounding (219).
Microarrays have generated a wealth of new information about the anaerobic response. The real challenge has become the functional analysis of these genes and elucidation of their role in the anaerobic response. This would require production of large numbers of transgenic plants and tedious examination of this material. However, by focusing on certain aspects of the response and carrying out additional microarray screening using carefully chosen conditions, it may be possible to reduce the number of target genes significantly. For instance, if the focus is on transcription factors involved in establishing the response, time‐course experiments revealing gene expression profiles will greatly reduce the number of clones to be considered for further analysis. Screening for genes that are induced by other stress treatments will eliminate genes that are not specific to the anaerobic response, but could also lead to the identification of functional units that are required for different stress responses. Further progress in the field of bioinformatics (e.g. http://www.gene‐regulation.de/; Hehl and Wingender, 2001) will allow us to identify common regulatory elements in the promoter region of gene clusters identified in microarray experiments. This will ultimately lead to a better understanding of the anaerobic response and how it is regulated.
Supplementary Material
Received: 4 October 2001; Returned for revision: 9 November 2001; Accepted: 21 November 2001
References
- AoyamaT, Chua NH.1997. A glucocorticoid‐mediated transcriptional induction system in transgenic plants. Plant Journal 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Bailey‐SerresJ, Dawe RK.1996. Both 5′ and 3′ sequences of maize adh1 mRNA are required for enhanced translation under low‐oxygen conditions. Plant Physiology 112: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey‐SerresJ, Rochaix JD, Wassenegger M, Filipowicz W.1999. Plants, their organelles, viruses and transgenes reveal the mechanisms and relevance of post‐transcriptional processes. EMBO Journal 18: 5153–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BerberichT, Kusano T.1997. Cycloheximide induces a subset of low temperature‐inducible genes in maize. Molecular and General Genetics 254: 257–283. [DOI] [PubMed] [Google Scholar]
- ChanMT, Yu SM.1998. The 3′ untranslated region of a rice α‐amylase gene functions as a sugar‐dependent mRNA stability determinant. Proceedings of the National Academy of Sciences of the USA 95: 6543–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChangWW, Huang L, Shen M, Webster C, Burlingame AL, Roberts JKM.2000. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low‐oxygen environment, and identification of proteins by mass spectrometry. Plant Physiology 122: 295–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CheungVG, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G.1999. Making and reading microarrays. Nature Genetics 21: 15–19. [DOI] [PubMed] [Google Scholar]
- CloughSJ, Bent AF.1998. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de BruxellesGL, Peacock WJ, Dennis ES, Dolferus R.1996. Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiology 111: 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DennisES, Dolferus R, Ellis M, Rahman M, Wu Y, Hoeren FU, Grover A, Ismond KP, Good AG, Peacock WJ.2000. Molecular strategies for improving waterlogging tolerance in plants. Journal of Experimental Botany 51: 89–97. [PubMed] [Google Scholar]
- DickeyLF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF.1998. Light regulation of Fed‐1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell 10: 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DolferusR, Jacobs M, Peacock WJ, Dennis ES.1994. Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiology 105: 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DolferusR,Klok EJ, Ismond KP, Delessert C, Wilson S, Good AG, Peacock WJ, Dennis ES.2000. Molecular basis of the anaerobic response in plants. IUBMB Life 51: 79–82. [DOI] [PubMed] [Google Scholar]
- DugganDJ, Bittner M, Chen Y, Meltzer P, Trent JM.1999. Expression profiling using cDNA microarrays. Nature Genetics 21: 10–14. [DOI] [PubMed] [Google Scholar]
- EllisMH, Setter TL.1999. Hypoxia induces anoxia tolerance in completely submerged rice seedlings. Journal of Plant Physiology 154: 219–230. [Google Scholar]
- EllisMH, Dennis ES, Peacock WJ.1999. Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiology 119: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FenZ, Daniel TO.1991. 5′ untranslated sequences determine degradative pathway for alternate PDGF B/c‐sis mRNAs. Oncogene 6: 953–959. [PubMed] [Google Scholar]
- FennoySL, Nong T, Bailey‐Serres J.1998. Transcriptional and post‐transcriptional processes regulate expression in oxygen‐deprived roots of maize. Plant Journal 15: 727–735. [DOI] [PubMed] [Google Scholar]
- GallieDR, Bailey‐Serres J.1997. Eyes off transcription! The wonderful world of post‐transcriptional regulation. Plant Cell 9: 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GilP, Green PJ.1996. Multiple regions of the Arabidopsis SAUR‐AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO Journal 15: 1678–1686. [PMC free article] [PubMed] [Google Scholar]
- HansonAD, Rathinasabapathi B, Rivoal J, Burnet M, Dillon MO, Gage DA.1994. Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proceedings of the National Academy of Sciences of the USA 91: 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HehlR, Wingender E.2001. Database‐assisted promoter analysis. Trends in Plant Science 6: 251–255. [DOI] [PubMed] [Google Scholar]
- HennW.1999. Genetic screening with the DNA chip: a new Pandora’s box? Journal of Medical Ethics 25: 200–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HoerenF, Dolferus R, Wu Y, Peacock WJ, Dennis ES.1998. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase (ADH1) gene by low oxygen. Genetics 149: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HorvathDM, Huang DJ, NH, Chua NH.1998. Four classes of salicylate‐induced tobacco genes. Molecular Plant‐Microbe Interactions 11: 895–905. [DOI] [PubMed] [Google Scholar]
- HuangJ, Hirji R, Adam L, Rozwadowski KL, Hammerlindl JK, Keller WA, Selvaraj G.2000. Genetic engineering of glycinebetaine production toward enhancing stress tolerance in plants: metabolic limitations. Plant Physiology 122: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IuchiS, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi‐Shinozaki K, Shinozaki K.2001. Regulation of drought tolerance by gene manipulation of 9‐cis‐epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant Journal 27: 325–333. [DOI] [PubMed] [Google Scholar]
- Jaglo‐OttosenKR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF.1998. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106. [DOI] [PubMed] [Google Scholar]
- JohnsonMA, Perez‐Amador MA, Lidder P, Green PJ.2000. Mutants of Arabidopsis defective in a sequence‐specific mRNA degradation pathway. Proceedings of the National Academy of Sciences of the USA 97: 13991–13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JordanBR.1998. Large‐scale expression measurement by hybridization methods: from high‐density membranes to ‘DNA chips’. Journal of Biochemistry 124: 251–258. [DOI] [PubMed] [Google Scholar]
- KangHG, Fang Y, Singh KB.1999. A glucocorticoid‐inducible transcription system causes severe growth defects in Arabidopsis and induces defence‐related genes. Plant Journal 20: 127–133. [DOI] [PubMed] [Google Scholar]
- KelleyPM.1989. Maize pyruvate decarboxylase mRNA is induced anaerobically. Plant Molecular Biology 13: 213–222. [DOI] [PubMed] [Google Scholar]
- KelleyPM, Freeling M.1984a Anaerobic expression of maize fructose‐1,6‐diphosphate aldolase. Journal of Biological Chemistry 259: 14180–14183. [PubMed] [Google Scholar]
- KelleyPM, Freeling M.1984b Anaerobic expression of maize glucose phosphate isomerase. Journal of Biological Chemistry 259: 673–677. [PubMed] [Google Scholar]
- KennedyRA, Rumpho ME, Fox TC.1992. Anaerobic metabolism in plants. Plant Physiology 84: 1204–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MarchantA, Bennett MJ.1998. The Arabidopsis AUX1 gene: a model system to study mRNA processing in plants. Plant Molecular Biology 36: 463–471. [DOI] [PubMed] [Google Scholar]
- MemonAR, Meng B, Mullet JE.1996. RNA‐binding proteins of 37/38 kDa bind specifically to the barley chloroplast psbA 3′‐end untranslated RNA. Plant Molecular Biology 30: 1195–1205. [DOI] [PubMed] [Google Scholar]
- OlsonDC, Oetiker JH, Yang SF.1995. Analysis of LE‐ACS3, a 1‐aminocyclopropane‐1‐carboxylic acid synthase gene expressed during flooding in the roots of tomato plants. Journal of Biological Chemistry 270: 14056–14061. [DOI] [PubMed] [Google Scholar]
- OtaniY, Quinones S, Saus J, Kurkinen M, Harris ED.1990. Cycloheximide induces stromelysin mRNA in cultured human fibroblasts. European Journal of Biochemistry 192: 75–79. [DOI] [PubMed] [Google Scholar]
- PeppelK, Vinci JM, Baglioni C.1991. The AU‐rich sequences in the 3′ untranslated region mediate the increased turnover of interferon mRNA induced by glucocorticoids. Journal of Experimental Medicine 173: 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PiaoHL, Lim JH, Kim SJ, Cheong G‐W, Hwang I.2001. Constitutive over‐expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced tolerance in Arabidopsis. Plant Journal 27: 305–314. [DOI] [PubMed] [Google Scholar]
- RamsayG.1998. DNA chips: state‐of‐the art. Nature Biotechnology 16: 40–44. [DOI] [PubMed] [Google Scholar]
- RicardB, Couée I, Raymond P, Saglio PH, Saint‐Ges V, Pradet A.1994. Plant metabolism under hypoxia and anoxia. Plant Physiology and Biochemistry 32: 1–10. [Google Scholar]
- RichmondT, Somerville S.2000. Chasing the dream: plant EST microarrays. Current Opinion in Plant Biology 3: 108–116. [DOI] [PubMed] [Google Scholar]
- RobertsJKM, Callis J, Wemmer D, Walbot V, Jardetzky O.1984a Mechanisms of cytoplasmic pH regulation in hypoxic maize root tips and its role in survival under anoxia. Proceedings of the National Academy of Sciences of the USA 81: 3368–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RobertsJKM, Callis J, Jardetzky O, Walbot V, Freeling M.1984b Cytoplasmic acidosis as a determinant of flooding intolerance in plants. Proceedings of the National Academy of Sciences of the USA 81: 6029–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RobertsJKM, Andrade FH, Anderson IC.1985. Further evidence that cytoplasmic acidosis is a determinant of flooding intolerance in plants. Plant Physiology 77: 492–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SachsMM, Freeling M, Okimoto R.1980. The anaerobic proteins of maize. Cell 20: 761–767. [DOI] [PubMed] [Google Scholar]
- SachsMM, Subbaiah CC, Saab IN.1996. Anaerobic gene expression and flooding tolerance in maize. Journal of Experimental Botany 47: 1–15. [Google Scholar]
- SakamotoA, Murata N.2000. Genetic engineering of glycinebetaine synthesis in plants: current status and implications for enhancement of stress tolerance. Journal of Experimental Botany 51: 81–88. [PubMed] [Google Scholar]
- SchenaM.1996. Genome analysis with gene expression microarrays. Bioessays 18: 427–431. [DOI] [PubMed] [Google Scholar]
- ShenQ, Uknes SJ, Ho TH.1993. Hormone response complex in a novel abscisic acid and cycloheximide‐ inducible barley gene. Journal of Biological Chemistry 268: 23652–23660. [PubMed] [Google Scholar]
- SmithNA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM.2000. Total silencing by intron‐spliced hairpin RNAs. Nature 407: 319–320. [DOI] [PubMed] [Google Scholar]
- StockingerEJ, Gilmour SJ, Thomashow MF.1997. Arabidopsis thalianaCBF1 encodes an AP2 domain‐containing transcriptional activator that binds to the C‐repeat/DRE, a cis‐acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proceedings of the National Academy of Sciences of the USA 94: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TamminenI, Makela P, Heino P, Palva ET.2001. Ectopic expression of ABI3 gene enhances freezing tolerance in response to abscisic acid and low temperature in Arabidopsis thaliana Plant Journal 25: 1–8. [DOI] [PubMed] [Google Scholar]
- ThomashowMF.2001. So what’s new in the field of plant cold acclimation? Lots! Plant Physiology 125: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ThompsonAJ, Jackson AC, Symonds RC, Mulholland BJ, Dadswell AR, Blake PS, Burbidge A, Taylor IB.2000. Ectopic expression of a tomato 9‐cis‐epoxycarotenoid dioxygenase gene causes over‐production of abscisic acid. Plant Journal 23: 363–374. [DOI] [PubMed] [Google Scholar]
- UraoT, Noji M, Yamaguchi‐Shinozaki K, Shinozaki K.1996. A transcriptional activation domain of ATMYB2, a drought‐inducible Arabidopsis Myb‐related protein. Plant Journal 10: 1145–1148. [DOI] [PubMed] [Google Scholar]
- UraoT, Yamaguchi‐Shinozaki K, Urao S, Shinozaki K.1993. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WaterhousePM, Graham MW, Wang MB.1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proceedings of the National Academy of Sciences of the USA 95: 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WisdomR, Lee W.1991. The protein‐coding region of c‐myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes and Development 5: 232–243. [DOI] [PubMed] [Google Scholar]
- Yamaguchi‐ShinozakiK, Shinozaki K.2001. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress‐inducible transcription factor. Novartis Foundation Symposium 236: 176–186. [PubMed] [Google Scholar]
- YeildingNM, Procopio WN, Rehman MT, Lee WMF.1998. c‐myc mRNA is down‐regulated during myogenic differentiation by accelerated decay that depends on translation of regulatory coding elements. Journal of Biological Chemistry 273: 15749–15757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.