Abstract
Tolerance to low oxygen availability is likely to be due to the interaction of several factors. Sugar availability is one of the elements required to support anaerobic metabolism. In cereal grains the availability of soluble sugars is limited, while starch is stored in large amounts. Degradation of starch under anoxia is therefore needed to avoid sugar starvation leading to rapid cell death. The striking difference in the ability to produce α‐amylase when comparing the anoxia‐tolerant rice (Oryza sativa L.) grains with grains of other cereals is not easily explained. Rice is able to respond to gibberellins under anoxia, but the response is too slow to explain the rapid production of α‐amylase enzyme. In the present work we demonstrated that α‐amylase production during the first 2 d after imbibition is mostly due to the activity of the Ramy3D gene, encoding for the G and H isoforms of α‐amylase. The induction of Ramy3D transcription is likely to result from a low sugar content in the grains incubated under anoxia. The ability of rice embryos to sense sugars under anoxia is reported.
Keywords: Key words: α‐amylase, anaerobiosis, anoxia, cereal, Oryza sativa, rice, sugar sensing
INTRODUCTION
Sugar availability plays an important role in plant tolerance to anoxia (Perata et al., 1997a, 1998; Vartapetian and Jackson, 1997). Sugars such as glucose and sucrose are rapidly channelled to fermentative metabolism as soon as oxygen availability decreases below a threshold value (usually below 1 % O2), but the amount of hexoses and disaccharides stored in plant cells is usually limited: the ability to degrade starchy reserves becomes crucial for survival under prolonged anoxia (Perata et al., 1998).
A set of enzymes is needed for starch degradation, namely α‐amylase, β‐amylase, α‐glucosidase and debranching enzyme, but only α‐amylase is considered to play a major role in starch degradation (Dunn, 1974; Sun and Henson, 1991). α‐Amylase is produced in rice seeds under anoxia (Perata et al., 1992), while it is not in the anoxia‐intolerant cereals (wheat, barley) (Guglieminetti et al., 1995a). The successful production of α‐amylase in rice is very likely to be responsible for the successful degradation of starch taking place in the endosperm since other starch‐degrading enzymes are unlikely to be able to initiate the process of starch degradation. Indeed, in the absence of α‐amylase starch is not degraded, and anoxia‐intolerant cereals such as wheat and barley suffer soon from sugar starvation, and eventually die (Perata et al., 1996). Remarkably, in a recent report Arpagaus and Braendle (2000) demonstrated that α‐amylase also plays an important role for carbohydrate metabolism in the anoxia‐stress‐tolerant rhizomes of Acorus calamus L. The authors compared α‐amylase activities in Acorus calamus rhizomes with the enzyme activity in the non‐tolerant tubers of Solanum tuberosum L., revealing the ability of the tolerant plant to maintain a high level of α‐amylase under anoxia, together with a higher‐than‐aerobic amount of soluble carbohydrates. On the other hand, potato tubers suffer from sugar starvation, a likely consequence of the low α‐amylase activity found in the tubers of these species (Arpagaus and Braendle, 2000).
The mechanisms allowing the successful production of α‐amylase under anoxia in rice seeds is largely unknown. Anoxic rice embryoless half‐grains respond to exogenous gibberellic acid (GA), but with great delay when compared with the rapid induction of α‐amylase mRNA accumulation triggered by GA under aerobic conditions (Perata et al., 1993). Intriguingly, the appearance of the α‐amylase protein is unaffected by anoxia as demonstrated by immunoblot analysis (Guglielminetti et al., 1995a). We have subsequently shown that anoxic treatment results in the production of α‐amylase isoforms not observed in aerobic rice seedlings (Perata et al., 1997a). Comparison of the isoelectric point of these anaerobically induced α‐amylase isoforms with that of characterized rice α‐amylase isoforms (Yamaguchi et al., 1995; Mitsui et al., 1996) allows us to tentatively identify the anoxic isoforms as isoforms G and H, while isoforms A and B are observed in extracts from both aerobic and anaerobic seedlings (Perata et al., 1997a). This observation is of interest, since isoforms A and B are encoded by the Ramy1A gene, while isoforms G and H are encoded by the Ramy3D gene. In a recent report, Hwang et al. (1999) characterized the effects of anoxia on the pattern of expression of α‐amylase genes. While Ramy1A appears to be repressed by anoxia, Ramy3D is anoxia‐induced (Hwang et al., 1999). Furthermore Ramy1A and Ramy3D differ in their respective mechanisms of regulation, the former being hormonally modulated and the latter sugar modulated. Remarkably, the sugar content in germinating rice grains drops under the concentration of 100 mm during aerobic germination only transiently, between 0·5 and 1 d, reaching 152 mm after 2 d and up to 548 mm after 5 d of germination (Yu et al., 1996). Under anoxia, the sugar content of rice grains drops from nearly 100 mm (dry grain content) to 30 mm after 2 d of anoxic germination (our unpublished observation). The threshold for complete repression of RAmy3D transcription is between 30 and 100 mm glucose (Morita et al., 1998).
The complexity of the rice α‐amylase multigene family, its modulation by hormones, sugars, anoxia, and the cross‐talk between hormonal and sugar signals (Perata et al., 1997b) suggest that anoxia modulations of α‐amylase genes such as RAmy3D could be mediated by the anoxia‐induced sugar depletion. In this paper this hypothesis will be investigated.
MATERIALS AND METHODS
Plant material
Rice grains (Oryza sativa L. cv. Nipponbare) were used. Grains were separated into two parts, one‐third of the intact grain containing the embryo, and two‐thirds of the grain lacking the embryo (embryoless). When isolated embryos were used, embryos were dissected from sterilized grains (shaken in 5 % sodium hypochlorite for 1 h; washed in sterile water with shaking for 2 h) using a scalpel. Only intact embryos with no starch or aleurone tissue adhering to the scutellar tissue were used. Incubation of embryoless half‐grains or embryos was carried out in test tubes, each containing four embryoless half‐grains/embryos and 500 µl of 5 mm CaCl2 containing 5 µg of chloramphenicol. Incubation was carried out at 27 °C with vigorous shaking. When used, 1 µm GA or 100 mm glucose was added. Treatments were carried out in a growth chamber in air or in an anaerobic incubator (Anaerobic System Model 1025; Forma Scientific, Marietta, OH, USA).
Chemicals
The commercially available compounds were purchased from Sigma (St Louis, MO, USA).
Gene specific probes
The gene‐specific probes for the detection on Ramy1A and Ramy3D probes were prepared by PCR labelling as described by Hwang et al. (1999). The primers used were as described by Hwang et al. (1999), designed to amplify the 3′ untranslated region of the genes. The probe for rRNA was a rice rRNA probe.
RNA isolation and gel blots
RNA extraction was performed by using the aurintricarboxylic acid method as previously described (Perata et al., 1997a). The amount of total RNA loaded in electrophoresis was 20 µg. RNA was electrophoresed on 1 % agarose‐formaldehyde gels, and blotted on nylon membrane (BrightStar‐Plus®; Ambion, Woodward Austin, TX, USA) by using the procedure suggested by the manufacturer. Membranes were prehybridized and hybridized using the NorthernMax® kit (Ambion). Equal loading was checked by reprobing with a rRNA and ubiquitin cDNA probe (not shown).
Chimeric gene constructs
Using the polymerase chain reaction technology, HindIII and XhoI restriction endonuclease sites were created at the 5′ flanking region (–422 to –65) of the RAmy3D gene from the rice genomic clone (pOSg1·5S). The nucleotide sequence and other characteristics of the gene have been reported before (Huang et al., 1990; Mitsunaga et al., 1994). The amplified promoter was attached, using the HindIII and XhoI restriction endonuclease sites of a truncated minimal (–46) cauliflower mosaic virus (CaMV) 35S promoter (Benfey et al., 1989), to the sequence coding for the Escherichia coli β‐glucuronidase (GUS) gene with a modified ATG initiation codon. The first intron from the mung bean catalase gene was inserted into the 5′ untranslated sequence (Tanaka et al., 1990); this construct (RAmy3D promoter/–46 of CaMV 35S promoter/intron of catalase gene/gusA/pUC19) is identified as RAmy3D‐GUS. As an internal standard, we used the 35S‐LUC clone (pREXΦLUC), a construct of the 35S promoter fused with the luciferase gene (LUC) (Mitsuhara et al., 1996) gifted from Dr Hirochika (National Institute of Agrobiological Resources, Tsukuba). The 35S‐LUC construct expression in rice embryo was unaffected by the sugars and other chemicals used in our experiments.
Transient expression system
Experiments were performed with particle bombardment co‐delivery of Ramy3D‐GUS and 35S‐LUC for data normalization, as described by Umemura et al. (1998). Bombardment was performed according to the instruction provided by the manufacturer (BioRad, Hercules, CA, USA) by using a 1100 p.s.i. He pressure and the sample holder closed to the gun (5 cm from the stopping screen). The bombardment was repeated twice on each plate containing 30–40 embryos. After bombardment, embryos were transferred in Petri dishes containing liquid Murashige–Skoog salt mixture and 2 mg/l 2,4‐D, eventually supplemented with filtered‐sterilized glucose. Each experiment was repeated three times on different days and with freshly prepared batches of reagents and rice embryos. Each independent experiment consisted of three replicates of five embryos each. All repeated experiments gave consistent results. The reported data are means of the obtained results from a representative experiment.
RESULTS AND DISCUSSION
Effects of anoxia on the expression of RAmy1A and RAmy3D genes in embryoless half‐grains
We tested the GA‐responsiveness of Ramy1A in rice embryoless half‐grains (including the aleurone) under aerobic or anaerobic conditions (Fig. 1A). While Ramy1A is readily induced by GA under aerobic conditions, its transcript does not accumulate under anoxia (Fig. 1A). Addition of glucose does not alter the effects of anoxia and only slightly represses the aerobic induction of the gene (Fig. 1A). Anoxia thus exerts drastically negative effects on the expression of RAmy1A, hampering (or, most likely, delaying) the action of GA (Fig. 1A). Indeed, α‐amylase induction is also GA‐dependent under anoxia, but the process takes at least 6 d to produce as much α‐amylase mRNA as that produced in 3 d under aerobic conditions (Perata et al., 1993). Hwang et al. (1999) have demonstrated that RAmy1A is expressed during anaerobic germination in anoxic rice aleurones, but expression is lower, with appearance of a clear signal in Northern blots delayed (6‐d anoxic comparable with 2‐d aerobic).
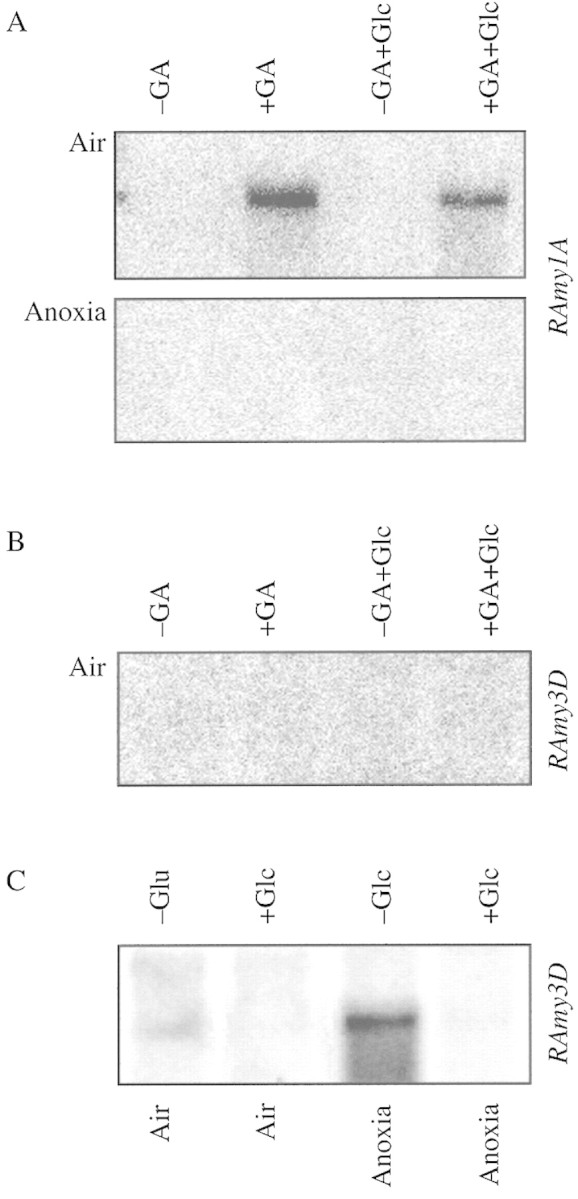
Fig. 1. Effects of anoxia, gibberellic acid and glucose on Ramy1A and Ramy3D mRNA accumulation in embryoless half‐grains. A, Northern blot analysis of Ramy1A mRNA accumulation in embryoless half‐grains incubated for 2 d in presence/absence of 1 µm GA and, when used, in presence of 100 mm glucose under aerobic/anaerobic conditions. B, Northern blot analysis of Ramy3D mRNA accumulation in embryoless half‐grains incubated for 2 d in presence/absence of GA in aerobic conditions and, when used, in presence of 100 mm glucose. C, Northern blot analysis of Ramy3D mRNA accumulation in embryoless half‐grains incubated for 2 d in air or anoxia in presence/absence of 100 mm glucose.
A different result was obtained when studying the pattern of expression of RAmy3D (Fig. 1B and C). RAmy3D is not induced by GA (Fig. 1B), which is not surprising since the GA‐response cis‐acting element (GARE) is not found in the RAmy3D promoter (Mitsui and Itoh, 1997). Under aerobic conditions RAmy3D is not expressed regardless of glucose presence/absence (Fig. 1B). Hwang et al. (1999) have shown that RAmy3D transcript is not detectable in the aerobic rice aleurone, even after a 6‐d‐long incubation. RAmy3D mRNA is easily detectable in the anoxic embryoless half grain, but this anoxia‐triggered expression is repressed in the presence of glucose (Fig. 1C). This result is of interest for at least three reasons: (1) this result is the only available evidence for the competence of the aleurone tissue for sugar repression of α‐amylase genes; (2) it explains why RAmy3D cannot be expressed under aerobic conditions, since the strong and rapid expression of RAmy1A (Fig. 1A, AIR) results in a high sugar concentration near the aleurone itself, repressing Ramy3D expression; (3) sugar repression of RAmy3D gives a possible explanation for its transient expression under anaerobic conditions (Hwang et al., 1999), since its own expression in the aleurone results in the production of α‐amylase which degrades starch with the production of glucose units whose presence down‐regulates RAmy3D expression.
Effects of anoxia on the expression of RAmy1A and RAmy3D genes in embryos
RAmy1A expression in the embryo is not affected by exogenous GA, a consequence of endogenous gibberellin synthesis in the embryo (data not shown; Panabieres et al., 1989). The expression of RAmy1A in the aerobic embryo is down‐regulated by glucose (Fig. 2A; see also Morita et al., 1998), while under anoxia glucose alleviates the negative effects due to oxygen absence (Fig. 2A, Air versus Anoxia). This apparent discrepancy is explained by the strong negative effect of anoxia on GA‐signalling at the level of RAmy1A transcription (see Fig. 2A). Glucose feeding (Fig. 2A), boosting anaerobic metabolism and the energy status of the embryo, appears to restore, at least partly, the ability to produce the RAmy1A transcript under anoxia.
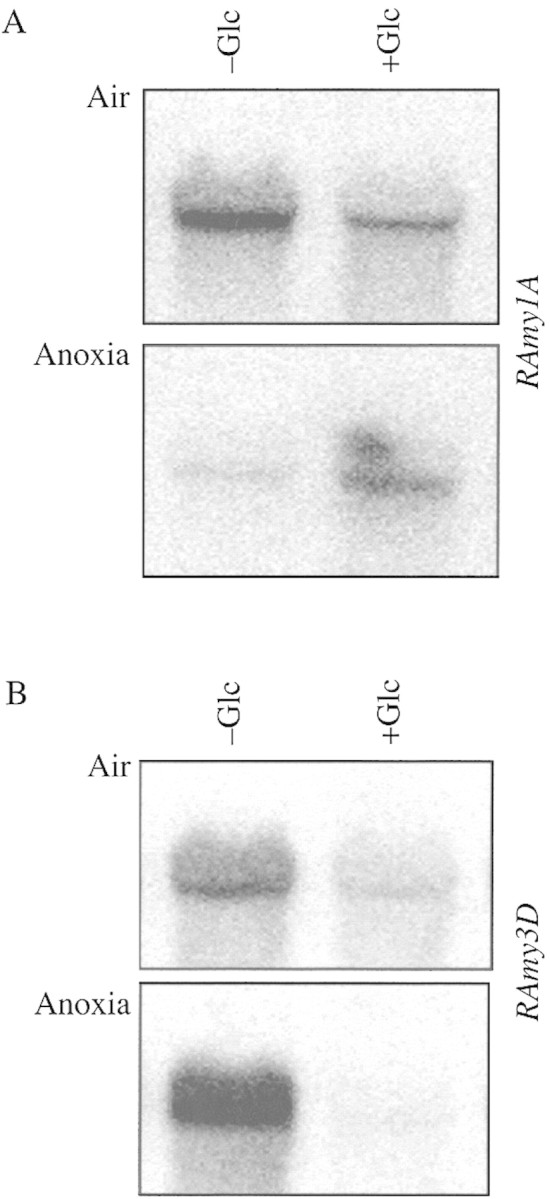
Fig. 2. Effects of anoxia and glucose on Ramy1A and Ramy3D mRNA accumulation in rice embryos. A, Northern blot analysis of Ramy1A mRNA accumulation in embryos incubated for 2 d in presence/absence of 100 mm glucose under aerobic/anaerobic conditions. B, Northern blot analysis of Ramy3D mRNA accumulation in embryos incubated for 2 d in presence/absence of 100 mm glucose under aerobic/anaerobic conditions.
RAmy3D, whose expression is solely modulated by sugars, is strongly induced under anoxia, but glucose counteracts this inductive effect (Fig. 2B).
We propose that the effects of anoxia on RAmy3D are likely to be mediated by sugar availability. Under anoxia the level of soluble sugars drops (Guglielminetti et al., 1995b; Perata et al., 1996), a prerequisite for the expression of RAmy3D (Fig. 2B). Furthermore, the inductive effects of anoxia on the expression of RAmy3D are easily counteracted by exogenous glucose, which indicates that a direct anaerobic induction of RAmy3D expression is not likely.
Relative importance of RAmy1A and RAmy3D under anoxia
Assuming that the ability to degrade starch is of importance for rice survival under anoxia, and that α‐amylase is a prerequisite for starch degradation, it is likely that production of this starch‐degrading enzyme during the first days after imbibition under anoxia is crucial for survival. In our experiments we used isolated embryos and aleurones (embryoless half‐grains) to avoid interferences in the expression of α‐amylase genes in each tissue from metabolites coming from nearby tissues. The amount of expression of RAmy1A and RAmy3D in Figs 1 and 2 was quantified and compared with the maximal expression level detected (Fig. 3). After 2 d of incubation under aerobic conditions, both RAmy1A and RAmy3D are expressed at a higher level in the embryo compared with expression in the aleurones. In the embryos, the aerobic expression of RAmy1A and RAmy3D is comparable (Fig. 3A) while, under anoxia, RAmy3D expression is strongly enhanced and that of RAmy1A repressed (Fig. 3A). In the embryoless half‐grains (Fig. 3B), RAmy1A expression predominates under aerobic conditions, while its transcript is not detected under anoxia, where RAmy3D is expressed instead. The RAmy3D‐encoded isoenzymes (isoforms G and H) are likely to be responsible for starch degradation in anaerobic rice embryos during the first 2 d of germination.
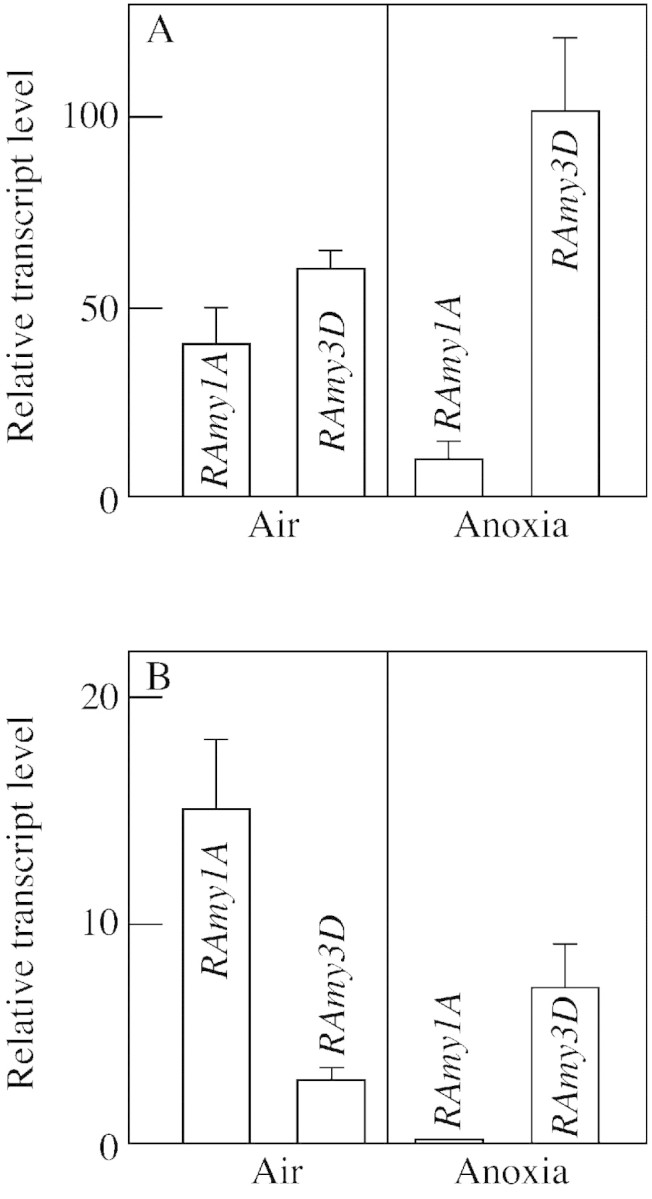
Fig. 3. Relative importance of expression of Ramy1A versus Ramy3D under aerobic and anaerobic conditions. Quantitative data (± s.e.) from replicated (n = 3) Northern blots performed as described in Figs 1 and 2 were normalized for electrophoresis loading differences on the basis of rRNA hybridization of the same blots. A relative value of 100 is assigned to the higher level of expression detected. A, Relative accumulation of Ramy1A and Ramy3D mRNAs in aerobic and anaerobic isolated embryos incubated in the presence of 1 µm GA. B, Relative accumulation of Ramy1A and Ramy3D mRNAs in aerobic and anaerobic isolated embryoless half‐grains incubated in the presence of 1 µm GA.
Anoxia tolerance correlates with the ability to produce α‐amylase under anoxia, and RAmy3D thus appears to play an important role in rice tolerance to oxygen deprivation, at least during the first days of anaerobiosis, since RAmy1A expression takes place at a level comparable with the aerobic one after 6 d of anoxia (Perata et al. 1993). There is little evidence concerning other Amy3 genes in other cereals. As discussed by Hwang et al. (1999) further knowledge about the existence and pattern of expression of Amy3 genes in other cereals may contribute to our understanding of the importance of this α‐amylase gene subfamily in anoxia tolerance.
Sugar sensing under anoxia: glucose‐induced repression of RAmy3D transcription
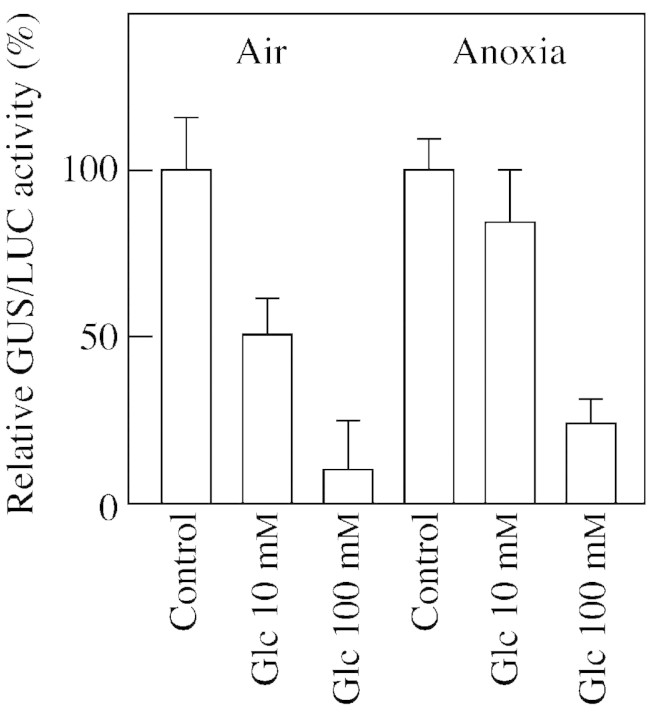
We tested whether sugar repression of RAmy3D under anoxia occurs at the level of transcription, as demonstrated previously for aerobic rice embryos (Morita et al., 1998). Transient expression experiments were performed using a RAmy3D‐GUS construct. The results indicate that 100 mm glucose, a concentration commonly found in rice grains germinating under aerobic conditions (Yu et al., 1996), is able to repress by 60–70 % transcription of RAmy3D under anoxia (Fig. 4), a repression only slightly less effective than under aerobic conditions (Fig. 4). The effects of glucose are not glucose‐specific, since sucrose and fructose are also able to elicit the same effects, while an osmotic effect can be ruled out, since 100 mm mannitol is unable to trigger any repression (data not shown). To our knowledge, this is the first available evidence of the ability of plants to sense sugars under anoxia. This evidence is of importance, since some genes modulated by anoxia are also sugar modulated (Koch et al., 2000). For instance, the sucrose synthase Sh1 is a well‐known anoxia‐induced gene (Springer et al., 1986), but is also a sugar‐starvation‐induced gene (Koch et al., 1992). Since anoxia is known to trigger sugar starvation, it is tempting to speculate that the induction of Sh1 under anoxia could be mediated by sugar starvation. However, sugar and oxygen signal do not appear to overlap for all sugar‐modulated genes: an invertase gene which is starvation‐induced is not induced by anoxia (Koch et al., 2000). Furthermore, the alcohol dehydrogenase Adh1 gene is sugar induced (Koch et al., 2000), which is counter‐intuitive assuming that anoxia triggers sugar starvation. Overall, it is tempting to speculate that some anoxia‐modulated genes are in fact sugar‐modulated genes whose expression under anoxia is the consequence of the anoxia‐induced altered sugar status (RAmy3D, Sh1), while other genes are independently modulated by sugars or anoxia (invertases, alcohol dehydrogenase).
Fig. 4. Effect of glucose on the repression of RAmy3D promoter activity under aerobic and anaerobic conditions. Transformation was performed by bombardment with Ramy3D‐GUS co‐delivered with 35S‐LUC. After trasformation the embryos were subsequently incubated for 2 d on a glucose‐free medium (Control) or a medium containing glucose (10–100 mm). Data were normalized by using the 35S‐LUC construct as internal standard. Relative GUS/LUC activity is expressed as control = 100. Data are means ± s.e. (n = 3).
CONCLUSION
The ability of rice grains to sense sugars under anoxia is suggestive of a complex control of gene regulation under low oxygen availability. Sugar starvation is likely to be a common phenomenon occurring in plant tissues experiencing hypoxic or anoxic conditions and it is tempting to speculate that sugar signalling cross‐talking with hormone and oxygen perception can mediate some of the responses to anaerobiosis.
ACKNOWLEDGEMENTS
This work was supported in part by CNR Target Project on Biotechnology.
Supplementary Material
Received: 29 August 2001; Returned for revision: 16 November 2001; Accepted: 18 January 2002
References
- ArpagausS, Braendle R.2000. The significance of α‐amylase under anoxia stress in tolerant rhizomes (Acorus calamus L.) and non‐tolerant tubers (Solanum tuberosum L. var. Désirée). Journal of Experimental Botany 51: 1475–1477. [PubMed] [Google Scholar]
- BenfeyPN, Ren L, Chua N‐H.1989. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue‐specific expression patterns. EMBO Journal 8: 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DunnG.1974. A model for starch breakdown in higher plants. Phytochemistry 13: 1341–1346. [Google Scholar]
- GuglielminettiL, Yamaguchi J, Perata P, Alpi A.1995a Amylolytic activities in cereal seeds under aerobic and anaerobic conditions. Plant Physiology 109: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GuglielminettiL, Perata P, Alpi A.1995b Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiology 108: 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HuangJ, Yamaguchi J, Akita S.1999. Changes in α‐amylase isoforms during emergence of rice in submerged soil. Plant Production Science 2: 12–13. [Google Scholar]
- HuangN, Koizumi N, Reinel S, Rodriguez RL.1990. Structural organization and differential expression of rice α‐amylase genes. Nucleic Acid Research 18: 7007–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HwangYS, Thomas BR, Rodriguez RL.1999. Differential expression of rice α‐amylase genes during seedling development under anoxia. Plant Molecular Biology 40: 911–920. [DOI] [PubMed] [Google Scholar]
- KochKE, Nolte KD, Duke ER, McCarty DR, Avigne WT.1992. Sugar levels modulate differential expression of maize sucrose synthase genes. The Plant Cell 4: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KochKE, Ying Z, Wu Y, Avigne WT.2000. Multiple paths of sugar‐sensing and a sugar/oxygen overlap for genes of sucrose and ethanol metabolism. Journal of Experimental Botany 51: 417–427. [DOI] [PubMed] [Google Scholar]
- MitsuharaI, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, Ueno K, Mochizuki A, Tanimoto H, Tsugawa H.1996. Efficient promoter cassettes for enhanced expression of foreign genes in dycotyledonous and monocotyledonous plants. Plant Cell Physiology 37: 49‐59. [DOI] [PubMed]
- 29.MitsuiT, Itoh K.1997. The α‐amylase multigene family. Trends in Plant Science 2: 255–261. [Google Scholar]
- MitsuiT, Yamaguchi J, Akazawa T.1996. Physicochemical and serological characterization of rice α‐amylase isoforms and identification of their corresponding genes. Plant Physiology 110: 1395–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MitsunagaS, Rodriguez RL, Yamaguchi J.1994. Sequence‐specific interactions of a nuclear protein factor with the promoter region of a rice gene for α‐amylase, RAmy3D Nucleic Acid Research 22: 1948–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MoritaA, Umemura T, Kuroyanagi M, Futsuhara Y, Perata P, Yamaguchi J.1998. Functional dissection of a sugar‐repressed α‐amylase gene (Ramy1A) promoter in rice embryos. FEBS Letters 423: 81–85. [DOI] [PubMed] [Google Scholar]
- PanabieresF, Kerhardy F, Montembault A, Daussant J, Delseny M.1989. Induction of α‐amylase isoenzymes by gibberellic acid in imbibed rice half‐seeds. Plant Science 64: 15–23. [Google Scholar]
- PerataP, Pozueta‐Romero J, Akazawa T, Yamaguchi J.1992. Effect of anoxia on starch breakdown in rice and wheat seeds. Planta 188: 611–618. [DOI] [PubMed] [Google Scholar]
- PerataP, Geshi N, Yamaguchi J, Akazawa T.1993. Effect of anoxia on the induction of α‐amylase in cereal seeds. Planta 191: 402–408. [Google Scholar]
- PerataP, Guglielminetti L, Alpi A.1996. Anaerobic carbohydrate metabolism in wheat and barley, two anoxia‐intolerant cereal seeds. Journal of Experimental Botany 47: 999–1006. [Google Scholar]
- PerataP, Guglielminetti L, Alpi A.1997a Mobilization of endoperm reserves in cereal seeds under anoxia. Annals of Botany 79 (Suppl. A): 49–56. [Google Scholar]
- PerataP, Matsukura C, Vernieri P, Yamaguchi J.1997b Sugar repression of a gibberellin‐dependent signaling pathway in barley embryos. The Plant Cell 9: 2197–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PerataP, Loreti E, Guglielminetti L, Alpi A.1998. Carbohydrate metabolism and anoxia tolerance in cereal grains. Acta Botanica Neerlandica 47: 269–283. [Google Scholar]
- SpringerB, Werr W, Starlinger P, Bennett DC, Zokolica M, Freeling M.1986. The Shrunken gene on chromosome 9 of Zea mays L. is expressed in various plant tissues and encodes an anaerobic protein. Molecular and General Genetics 205: 461–468. [DOI] [PubMed] [Google Scholar]
- SunZ, Henson CA.1991. A quantitative assessment of the importance of barley seed α‐amylase, debranching enzyme, and α‐glucosidase in starch degradation. Archives of Biochemistry and Biophysics 284: 298–305. [DOI] [PubMed] [Google Scholar]
- TanakaA, Mita S, Otha S, Kyozuka J, Shimamoto K, Nakamura K.1990. Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficent splicing of the intron. Nucleic Acid Research 18: 6767–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UmemuraT‐A, Perata P, Futsuhara Y, Yamaguchi J.1998. Sugar sensing and α‐amylase gene repression in rice embryos. Planta 204: 420–428. [DOI] [PubMed] [Google Scholar]
- VartapetianBB, Jackson MB.1997. Plant adaptation to anaerobic stress. Annals of Botany 79: 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YamaguchiJ, Geshi N, Mitsunaga S, Itoh S, Umemura T, Masui H, Mitsui T.1995. Expression of RAmy3D‐protein (isoform H) in rice seedlings. In: Noda K and Mares DJ, eds. Seventh International Symposium on Pre‐harvest Sprouting in Cereals 1995, Center for Academic Societies Japan, Osaka, 405–410.
- YuSM, Lee YC, Fang SC, Hwa SF, Liu LF.1996. Sugars act as signal molecules and osmotica to regulate the expression of α‐amylase genes and metabolic activities in germinating cereal grains. Plant Molecular Biology 30: 1277–1289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.