Abstract
Transposon tagging with modified maize Ds–GUS constructs was used to isolate genes induced by oxygen deprivation in Arabidopsis thaliana. Seedlings of 800 gene‐trap (DsG) and 600 enhancer‐trap (DsE) lines were grown on vertically positioned plates for 1 week, oxygen deprived for up to 24 h and stained for GUS activity. Oxygen deprivation induced intricate patterns of gene expression in seedlings of 65 lines. The insertion site and phenotypes of 15 lines were examined. Surprisingly, none of the insertions were into genes that encode known anaerobic polypeptides. Insertions were identified within or adjacent to genes encoding proteins of regulatory, enzymatic, mitochondrial protein import and unknown function, as well as adjacent to genes encoding a putative receptor‐like kinase and putative sensor‐histidine kinase. Four lines had significantly lower ADH activity after 24 h of oxygen deprivation and three of these showed reduced stress tolerance. Two lines with wild‐type levels of ADH were low‐oxygen intolerant. Paradoxically, several lines had significantly higher ADH activity after 12 h of oxygen deprivation but reduced stress tolerance. Caffeine treatment, which increased ADH specific activity in wild‐type seedlings under aerobic conditions, was sufficient to increase GUS staining in seven of the 15 lines, providing evidence that these genes may be regulated by cytosolic calcium levels. These results demonstrate the effectiveness of the Ds–GUS tagging system in the identification of genes that are regulated in response to oxygen deprivation and a calcium second messenger.
Keywords: Key words: Transposon tagging, gene expression pattern, anoxia, hypoxia, alcohol dehydrogenase, caffeine, cytosolic calcium, second messenger.
INTRODUCTION
Oxygen deprivation causes rapid and dramatic alterations in gene expression in many plant species (Sachs et al., 1996; Drew, 1997). The ability to produce ATP through glycolysis and regenerate NAD+ through ethanolic fermentation is required for short‐term survival of flooding. In certain species, moderate oxygen deprivation (hypoxia) promotes the development of root aerenchyma, which further increases flood tolerance (Drew, 1997). The initial report on the response of maize (Zea mays L.) seedling roots to anoxia described the synthesis of 20 soluble proteins synthesized at high levels (Sachs et al., 1980). By use of genetic, biochemical and immunological methods, alcohol dehydrogenase (ADH) isozymes were identified as major ‘anaerobic polypeptides’ (Sachs and Freeling, 1978; Sachs et al.,1980). Subsequent studies revealed that many species induce the synthesis of enzymes involved in sucrose consumption, glycolysis, ethanolic fermentation and aerenchyma development in response to oxygen deprivation (Drew, 1997;Vartapetian and Jackson, 1997; Dennis et al., 2000). The identification of genes that encode these enzymes has been facilitated by the high abundance of their mRNAs in stressed cells, whereas the identification of genes encoded by rare mRNAs that are up‐regulated by stress has been difficult.
The cellular mechanisms that underlie the perception of oxygen deprivation and the signal transduction processes responsible for the metabolic and developmental alterations are poorly understood. Ethylene has also been implicated in gene regulation after prolonged oxygen deprivation in Arabidopsis thaliana (Peng et al., 2001), whereas several studies have shown an increase in cytosolic calcium as an early second messenger in the low‐oxygen response. Reports on maize, arabidopsis and rice (Oryza sativa L.) have shown that an increase in cytosolic calcium is required for the elevated expression of ADH (Subbaiah et al., 1994a, b; Sedbrook et al., 1996; Chung and Ferl, 1999; Tsuji et al., 2000). The induction of ADH under low‐oxygen stress may require an influx of calcium from the apoplastic space in arabidopsis, since the presence of the calcium channel blockers lanthium or gadolinium chloride dampened the increase in accumulation of the mRNA of the single‐copy ADH gene (Sedbrook et al., 1996) and expression of an ADH–GUS reporter gene construct in arabidopsis (Chung and Ferl, 1999), but not the increase in ADH activity in maize seedlings (Subbaiah et al., 1994b). Calcium release from mitochondria and possibly other intracellular stores is also involved, since Ruthenium Red, which blocks endomembrane calcium channels, reduced cytosolic calcium levels and ADH expression under anoxia in cultured maize cells (Subbaiah et al., 1994a, 1998). A hypoxia‐induced calcium signal was linked to increases in cellulase activity and promotion of cell death leading to cortical aerenchyma formation in maize seedling roots (He et al., 1996). Increases in maize root ADH and cellulase specific activities were observed following exposure to caffeine (Subbaiah et al., 1994a; He et al., 1996), which promoted a rapid increase in cytosolic calcium levels in cultured cells (Subbaiah et al., 1994a). Exposure of cells to 2·5 mm caffeine resulted in an immediate increase in cytosolic calcium levels and a three‐fold increase in ADH specific activity after 24 h. Interestingly, the steady‐state accumulation of alternative oxidase (AOX1) mRNA was reduced in response to anoxia in a calcium‐dependent manner in rice seedlings (Tsuji et al., 2000), indicating that a rise in cytosolic calcium levels can also negatively affect gene expression.
Arabidopsis mutants with reduced ADH expression were recently obtained by negative selection with allyl‐alcohol (Conley et al., 1999). We reasoned that the implementation of a mutagenesis system that allows for the identification of genes, without bias for gene expression level or redundancy, could yield genes that are important in controlling ADH expression and other aspects of the response to oxygen deprivation. Towards this end we employed two genetically engineered transposable DNA elements as mutagens in arabidopsis. The DsG (gene trap) element possesses the uidA (β‐glucuronidase, GUS) reporter gene preceded by a short intron and three splice acceptor site motifs and the kanamycin resistance gene (NPTII) positioned between Ds terminal inverted repeats (Sundaresan et al., 1995). The insertion of the DsG element into exonic or coding regions, with the GUS open reading frame in the same orientation as the resident gene, can result in transcriptional and translational fusion of the gene with GUS. Thus, GUS expression is modulated by the 5′ cis‐acting transcription and translational elements at the site of insertion. The DsE (enhancer trap) element differs from the DsG element in that the –46 region of the CaMV 35S promoter is positioned upstream of the GUS open reading frame (Sundaresan et al., 1995). The insertion of this element into a chromosomal region results in GUS expression only if an enhancer is in the vicinity. The X‐Gluc histochemical assay for GUS activity allows for the assessment of expression patterns of the DsG or DsE ‘transposant’ lines. The transposon‐tagged genes can be identified by amplification of genomic DNA at the site of insertion by use of thermal asymmetric interlaced polymerase chain reaction (TAIL‐PCR), DNA sequence analysis and BLASTN search of arabidopsis genomic sequences. This transposon‐tagging system has been successfully used to identify genes involved in plant development (reviewed in Springer, 2000) and provides a promising method for identification of genes regulated by abiotic stress.
We present the results of a screen of 1400 GTR (DsG: gene trap UC Riverside) and ETR (DsE: enhancer trap UC Riverside) lines that yielded 15 transposants with increased expression in seedlings in response to oxygen deprivation. A subset of these lines showed increased GUS staining in response to caffeine treatment. The screen identified genes not previously implicated in response to oxygen deprivation.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana ecotype Landsberg erecta seed lines included wild‐type, and adh‐null CS8095 obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH, USA). Seeds were surface sterilized, cold treated at 4 °C for 2 days, and grown on solid medium [0·43 % MS salts (Sigma, St Louis, MO, USA), 1 % (w/v) sucrose, 0·3 % (w/v) phytagel (Sigma), adjusted to pH 5·7 with NaOH] under 16 h illumination at 22 °C in Petri dishes held vertically in racks to avoid growth of roots into the solid medium. Transposant lines were selected for use by growth on plates that additionally contained 50 µg/ml kanamycin (Sigma) and 0·45 µg/ml napthalene acetamide (NAM) (Sigma), added to the media after cooling to 50 °C. The Ds gene trap (DsG) and enhancer trap (DsE) lines were generated from the stable Ac, DsG and DsE starter lines as described previously (Sundaresan et al., 1995). Briefly, females with DsG or DsE were crossed with males containing a stable Ac element, F1 progeny were self‐pollinated, and NAM insensitive (lacking a stable Ac element) and kanamycin resistant (possessing a transposed Ds element) seedlings were propagated and self‐pollinated to obtain the F3 generation. Seeds were grown on moistened compost for bulking and genetic analysis.
Oxygen deprivation stress and caffeine treatments
Ten to twenty F3 seeds were grown vertically on solid MS medium plates (90 mm) for 7 d and the plates were transferred to an airtight chamber (35 cm length, 19 cm width and 19 cm height) under dim light for oxygen deprivation treatment. The chamber was plumbed to allow entry and exit of gas mixtures. Oxygen deprivation treatment was carried out by gassing with 99·998 % argon for the designated times (6, 12 and 24 h). The control treatment was performed in a similar setting, except the chamber was left uncovered, open to air. Chamber humidity was maintained by bubbling the gas mixture into water. For caffeine treatment, 7‐d‐old seedlings were gently transferred to solid 0·5 × MS medium plates additionally containing 0, 2·5, 5 or 10 mm caffeine and returned to the growth environment for the time indicated. Seedlings were harvested for in situ staining for GUS or ADH activity or frozen directly in liquid nitrogen and stored at –80 °C.
GUS and ADH activity stains
For GUS staining, seedlings were placed into each well of a 24‐well microtitre plate and submerged in 500 µl staining solution [100 mm NaPO4, pH 7·0, 10 mm EDTA, 0·1 % (v/v) Triton X‐100, 1 mg/ml 5‐bromo‐4‐chloro‐3‐indoxyl‐beta‐d‐glucronic acid, 100 µg/ml chloramphenicol, 2 mm potassium ferricyanide and 2 mm potassium ferrocyanide] and processed as described previously (Sundaresan et al., 1995). The dish was placed under vacuum in a desiccator for 10 min, removed, wrapped in aluminium foil and incubated at 37 °C for 48 h. The staining solution was replaced with 70 % ethanol to remove chlorophyll. For ADH staining, seedlings were placed in a 24‐well microtitre dish well containing 1·5 ml of 10 mm Tris (pH 7·3), frozen at –20 °C and thawed at room temperature three times, with the Tris‐buffer replaced after the first two cycles. For staining, seedlings were submerged in 1·5 ml of freshly prepared ADH stain (1 m Tris–HCl, pH 8·0, 10 mg/ml NAD+, 5 mg/ml Nitro blue tetrazolium chloride, 1 mg/ml phenazine methosulfate, 95 % ethanol) and transferred to the dark for 1 h. A control was performed using ADH stain without ethanol to ensure that the endogenous substrate was removed from the seedlings during the freeze–thaw treatment. Staining patterns were recorded with a LEICA DMR microscope with a RT colour SPOT camera (Diagnostic Inc., Sterling Heights, MI, USA).
ADH activity assay
ADH enzyme activity was measured as described previously (Bailey‐Serres and Dawe, 1996). Seedlings were homogenized in extraction buffer (100 mm Tris–HCl, pH 8·0, 4 mm DTT) and a clarified extract was obtained by centrifugation for 5 min at 14 000 g at 4 °C. ADH activity was assayed in 1 ml activity buffer (150 mm Tris–HCl, pH 8, 300 µm NAD+, 0·66 mm ethanol) by the addition of 160 µl cell extract and measurement of absorbance at 340 nm for 1 min. One unit of ADH activity was defined as 1 µmol of reduced NAD+ per minute per milligram of protein. Protein concentration was determined by using a protein assay kit (Bio Rad Laboratories, Hercules, CA, USA) with bovine serum albumin as the standard.
Identification of transposant insertion sites
Genomic DNA was isolated from inflorescence tissue by pulverization under liquid nitrogen and hydration in 600 µl of DNA extraction buffer [5 m urea, 0·3 m NaCl, 50 mm Tris–HCl, pH 8·0, 20 mm EDTA and 5 % (v/v) N‐lauryl sarkosine], incubation at room temperature for 10 min, extraction with one volume of phenol–chloroform–isoamyl alcohol (25 : 24 : 1), concentration by isopropanol precipitation, and resuspension in 50 µl TE (10 mm Tris–HCl, pH 8·0, 1 mm EDTA). Thermal asymmetric interlaced PCR (TAIL‐PCR) was carried out using arbitrary degenerate primers and Ds specific primers essentially as described (Liu et al., 1995; Tsugeki et al., 1996) with the modification of Martienssen and Springer (1998) to selectively amplify genomic sequence flanking the Ds element. TAIL‐PCR products were cloned using the pGEM‐T vector system as described by the manufacturer (Promega, Madison, WI, USA). Plasmid DNA was purified using the QIAprep spin mini‐plasmid prep kit (Qiagen, Chatsworth, CA, USA) and the insert was sequenced by cycle sequencing at the DNA sequencing facility (University of Maine). The site and orientation of the Ds transposant insertion was identified by use of the BLAST alignment search tool (Altschul et al., 1990) and the GenBank database (www.ncbi.nlm.nih.gov). The copy number of Ds element insertions in each transposant line was determined by Southern blot analysis of 10 µg of EcoRI digested genomic DNA probed with a Ds fragment as described previously (Sundaresan et al., 1995).
RESULTS
Establishment of an oxygen deprivation stress system for arabidopsis seedlings
Our goal was to identify transposant lines with Ds insertions into genes that are induced in seedlings in response to low‐oxygen. To determine an appropriate duration of oxygen deprivation stress, we examined 7‐d‐old wild‐type and adh‐null (Landsberg erecta ecotype) seedlings grown on vertically orientated plates over a 48 h time course of oxygen deprivation. Table 1 compares the ADH specific activity, primary root tip viability, lateral root development and appearance 48 h after treatment of these lines. ADH specific activity increased dramatically in wild‐type seedlings after 6 h of oxygen deprivation, reaching a maximum of six‐fold over the aerobic level after 36 h. As expected, the adh‐null line had extremely low ADH enzymatic activity. Both wild‐type and adh‐null plants showed increased lateral root development when deprived of oxygen for 6 h. The proliferation of lateral roots was observed in wild‐type but not in adh‐null seedlings after 12 and 24 h of oxygen deprivation. After 24 h of stress, the aerial organs of wild‐type plants were green, erect and 95 % of the seedlings had continued growth of the primary root tip as well as lateral roots (Table 1). Primary root tip survival decreased dramatically after 36 h of oxygen deprivation in the wild‐type. adh‐null seedlings stressed for 12 h were clearly distinguishable from the wild‐type; cotyledons were dark green and wilted, primary root tip viability was 50 % and lateral roots failed to develop. Thus, severe ADH deficiency resulted in oxygen deprivation sensitivity that was observable within 12 h. ADH activity was monitored in wild‐type seedlings by histochemical staining in situ (data not shown). Staining was detected at low levels in vasculature and ground tissue of the root, hypocotyl and cotyledons under aerobic conditions. ADH activity staining was high throughout the root after 6 h of oxygen deprivation and in the cotyledons after 12 h. ADH activity staining was especially concentrated in the hydathode at the tip of the cotyledon after 12 and 24 h of oxygen deprivation.
Table 1.
ADH specific activity, root survival and seedling appearance after oxygen deprivation and caffeine treatment of 7‐d‐old seedlings of arabidopsis
| Duration of | ADH specific activity (U/mg protein ± SE) | Primary root tip survival 48 h after treatment (% seedlings) | Seedling appearance 48 h after treatment | |||
| oxygen deprivation | ||||||
| (h) | Wild‐type | adh‐null | Wild‐type | adh‐null | Wild‐type | adh‐null |
| 0 | 0·30 ± 0·002 | 0·001 ± 0·0002 | 100 | 100 | Green, erect | Green, erect |
| 6 | 0·40 ± 0·01 | 0·001 ± 0·0002 | 100 | 100 | Green, erect lateral root initiation and elongation | Green, erect lateral root initiation and elongation |
| 12 | 0·80 ± 0·01 | 0·002 ± 0·0003 | 100 | 50 | Green, erect increased lateral roots | Dark green, slightly wilted no lateral roots |
| 20 | 1·00 ± 0·02 | 0·001 ± 0·0001 | 100 | 20 | Green, erect increased lateral roots | Dark green, slightly wilted |
| 24 | 1·07 ± 0·02 | 0·001 ± 0·0002 | 95 | 10 | Green, erect increased lateral roots | Dark green,wilted |
| 36 | 2·03 ± 0·01 | 0·001 ± 0·0001 | 0 | 0 | Green, erect | Very dark green, wilted |
| 48 | 1·40 ± 0·03 | 0·001 ± 0·0003 | 0 | 0 | Dark green, wilted | Very dark green, wilted |
| Caffeine treatment of wild‐type (mm) | ADH specific activity after 24 h treatment (U/mg protein ± SE) | Primary root tip survival 48 h after treatment (% seedlings) | Root elongation 48 h after treatment (cm) | Seedling appearance 48 h after treatment | ||
| 0 | 0·25 ± 0·08 | 100 | 1·32 ± 0·01 | Green, erect | ||
| 2·5 | 2·2 ± 0·31 | 100 | 0·52 ± 0·01 | Green, erect | ||
| 5 | 2·9 ± 0·31 | 100 | 0·40 ± 0·01 | Light green, erect | ||
| 10 | 4·0 ± 0·93 | 0 | 0·00 ± 0·00 | Light brown, wilted | ||
ADH specific activity data are the mean ± s.e. from three independent experiments.
To determine if caffeine treatment affects ADH expression in arabidopsis, 7‐d‐old wild‐type seedlings were transferred to MS plates that additionally contained caffeine (0, 2·5, 5 or 10 mm) and returned to the growth chamber for 24 h. Exposure to caffeine efficiently promoted an increase in ADH specific activity in the absence of oxygen deprivation (Table 1). Exposure of wild‐type seedlings to 2·5 mm caffeine resulted in a seven‐fold increase in ADH specific activity, above the maximum level observed over the time course of oxygen deprivation. Treatment with caffeine also resulted in decreased root elongation rates in a dosage dependent manner, with the 10 mM caffeine resulting in primary root tip death within 48 h. These results support the hypothesis that ADH regulation in arabidopsis is mediated by cytosolic calcium.
Screening of the gene and enhancer trap lines
To identify genes induced by oxygen deprivation, F3 seedlings of 800 GTR and 600 ETR transposants were grown on vertically positioned plates for 7 d and subjected to oxygen deprivation (6, 12 or 24 h). GUS activity in intact seedlings was detected by histochemical staining with X‐Gluc. For comparison, seedlings grown in air were harvested at the same time as the stressed seedlings and stained for GUS activity. Similar to the frequencies previously reported (Sundaresan et al., 1995), 28 % of the GTR and 35 % of the ETR lines expressed GUS at the seedling stage (Table 2). GUS staining decreased in 10 % of the GTR and 12·5 % of the ETR lines, and increased in 6 % of the GTR and 2·5 % of the ETR lines. The GTR and ETR lines in which no GUS activity was detected in seedlings may express the GUS gene at another developmental stage or not at all. The patterns of GUS staining in the lines showing increased GUS expression under oxygen deprivation were diverse and cell‐type specific. GUS activity was observed at elevated levels throughout the root, hypocotyl and cotyledons in five lines. In other lines, GUS staining increased throughout an organ (ubiquitous), in specific tissues such as the vasculature, or specialized cell types such as the primary root tip, lateral root meristems, root hairs, trichomes or shoot apex (Table 3). Several lines showed induced expression in specific regions of the cotyledon such as the lateral margin or hydathode. Fifteen lines with a reproducible increase in GUS staining under oxygen deprivation were selected for further characterization (described below).
Table 2.
GUS histochemical staining of 7‐d‐old vertically grown seedlings following control and oxygen deprivation treatments
| GUS | Gene trap (% total) | Enhancer trap (% total) |
| Undetected before and after oxygen deprivation | 576 (72) | 390 (65) |
| Unaffected by oxygen deprivation | 95 (12) | 121 (20) |
| Decreased by oxygen deprivation | 79 (10) | 74 (12·5) |
| Increased by oxygen deprivation | 50 (6) | 15 (2·5) |
| Total F3 lines screened | 800 | 600 |
Table 3.
Location of GUS activity in seedlings after oxygen deprivation
| Organ | Region | No. GTR lines | No. ETR lines |
| Root | Ubiquitous | 9 | 3 |
| Primary tip | 7 | 2 | |
| Lateral root | 9 | 1 | |
| Vasculature | 6 | 3 | |
| Hair | 6 | 0 | |
| Elongation zone | 1 | 1 | |
| Hypocotyl | Ubiquitous | 6 | 2 |
| Vasculature | 6 | 0 | |
| Colet | 4 | 3 | |
| Cotyledon | Ubiquitous | 15 | 4 |
| Vasculature | 8 | 1 | |
| Margin | 5 | 2 | |
| Hydathode | 3 | 0 | |
| Trichome | 1 | 0 | |
| Leaf | Petiole | 1 | 0 |
| Shoot apex | Apical | 1 | 3 |
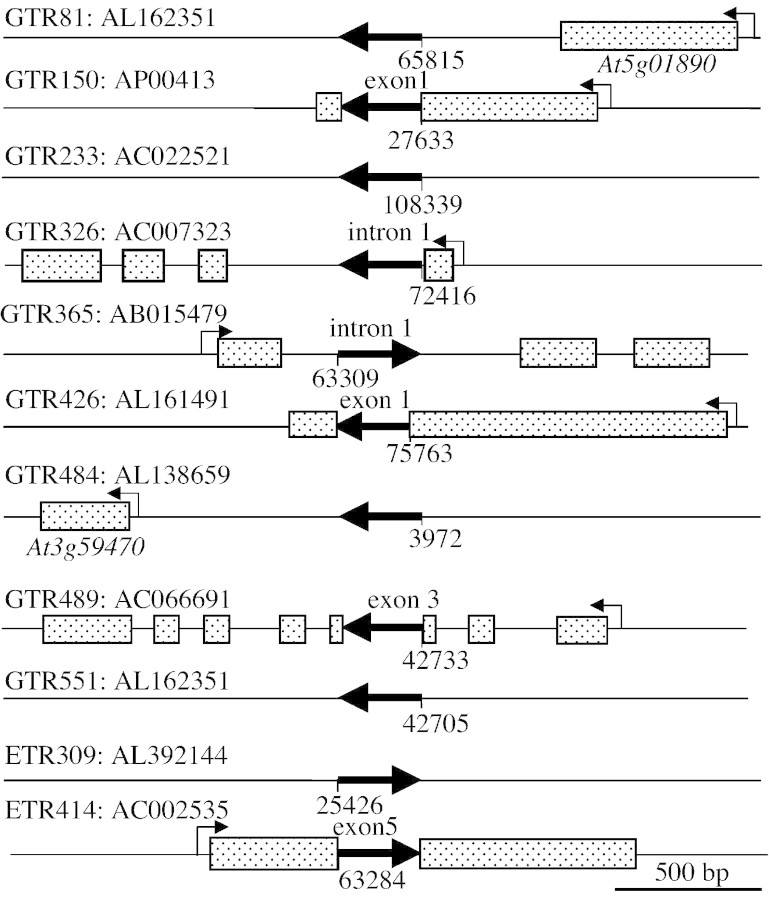
The site of transposon insertion into these lines was determined by TAIL‐PCR amplification and sequencing of the genomic DNA flanking the Ds element. The insert was localized in the genome by BLAST search of arabidopsis genomic sequences (Fig. 1). Southern blot hybridization on genomic DNA from each line with a Ds probe confirmed that each transposant possessed a single Ds insertion (data not shown). The insertion site in five GTR lines and one ETR line was within an exon or intron of an annotated protein‐coding gene (Fig. 1). In all five lines the orientation of the GUS coding sequence was the same as the transcription unit. These insertions are predicted to result in a translational fusion of GUS with the protein coding sequence and may cause gene disruption. The insertion site of four GTR lines and one ETR line was into a region of the arabidopsis genome where no genes have been annotated. In each of these cases we found no corresponding expressed sequence tag (EST) or evidence that the insertion was into a protein‐coding gene. The site of insertion has not yet been determined successfully in four lines. Transposant insertions were identified on all five haploid chromosomes of arabidopsis: four on chromosome 5, three on chromosome 1, two on chromosome 3 and one on chromosomes 2 and 4. Interestingly, GTR81 and GTR551 were situated 23 kb apart on chromosome 5 (BAC T20L15) but arose from transposition of two different parental DsG elements (DsG6, GTR81; DsG7, GTR551).
Fig. 1.Ds insertion site in GTR and ETR lines. The arrow indicates the transposon insertion and the direction of transcription of uidA (GUS). Shaded boxes indicate exonic sequence at the site of insertion. Ds element not shown to scale. Orientation shown is consistent with the GenBank accession.
Screening of the GTR and ETR lines uncovered genes that are up‐regulated in response to low‐oxygen but were not previously implicated in the response to oxygen deprivation. Since our motive was to identify genes involved in signal transduction mechanisms that regulate responses to oxygen deprivation, we examined the induction of ADH specific activity and ADH in situ staining of these lines (Table 4). We also assessed root tip survival following 24 h of oxygen deprivation and the effect of caffeine treatment on expression of the reporter gene. Analyses were performed on lines that were homozygous or segregating (expected genotypic ratios of 1/4 wild‐type, 1/2 hemizygous Ds, 1/4 homozygous Ds) as indicated in Table 4. Two GTR (GTR150, GTR426) and two ETR (ETR414, ETR528) lines had significantly elevated ADH activity after 12 h of oxygen deprivation; three of these showed reduced primary root tip survival after 24 h of oxygen deprivation (GTR150, GTR426, ETR528). Four lines (GTR81, GTR233, GTR326, GTR551) had significantly reduced ADH activity after 24 h of oxygen deprivation and three of these (GTR233, GTR326, GTR551) showed reduced primary root tip survival after 24 h of oxygen deprivation. Three lines (GTR365, GTR404, GTR489) had wild‐type ADH activity but were low‐oxygen intolerant. Seven lines (GTR81, GTR150, GTR404, GTR424, GTR426, ETR414, ETR528) showed inducible GUS expression after transfer to plates with solid medium containing 5 mm caffeine for 24 h, suggesting that their regulation is modulated by cytosolic calcium (Fig. 3).
Table 4.
Characteristics of GTR and ETR lines with increased GUS expression in response to oxygen deprivation
| ADH enzymatic activity (U/mg protein ± s.e.) | Primary root | |||||||||||||||||||||
| GUS histochemical staining | ADH histochemical activity | survival of | GenBank | Gen/protein | ||||||||||||||||||
| (h oxygen deprivation) | (h oxygen deprivation) | Location of GUS | 24 h oxygen | Accesional/BAC | encoded at | |||||||||||||||||
| activity after | deprivation | Insertion site | transposon insertion | |||||||||||||||||||
| Line | 0 | 6 | 12 | 24 | 0 | 6 | 12 | 24 | oxygen deprivation | (% seedlings) | Chromosome | site; position | ||||||||||
| GTR81 | R + | + | ++ | + | 0·001 ± 0·001a | 0·35 ± 0·02 | 0·98 ± 0·005 | 0·001 ± 0·000a | Vasculature | 80 | AL162351/T20L15 | No known gene at site; | ||||||||||
| H + | + | +++ | + | + | + | +++ | – | 65815 | 352 bp 3′ of LRR RLK gene | |||||||||||||
| C + | + | ++ | + | 5 | ||||||||||||||||||
| GTR150 | R + | + | ++ | +++ | 0·05 ± 0·01 | 0·71 ± 0·03 | 1·67 ± 0·16a | 0·01 ± 0·001a | Root vasculature, | 10 | AP000413/K7L4. 10 | Unknown protein | ||||||||||
| + | ++ | +++ | – | lateral meristems | 27633 | Ser/Pro‐rich; exonic | ||||||||||||||||
| 3 | ||||||||||||||||||||||
| GTR233 | C + | + | +++ | + | 0·02 ± 0·002a | 0·40 ± 0·0001 | 0·62 ± 0·006 | 0·02 ± 0·005a | Cotyledon | 20 | AC022521/T14P4 | No known gene at site; | ||||||||||
| + | ++ | ++ | – | hydathode | 108339 1 | 2·4 kb 5′ to polygalacturonase gene | ||||||||||||||||
| GTR326 | R + | + | + | +++ | 0·01 ± 0·002a | 0·38 ± 0·006 | 0·98 ± 0·03 | 0·13 ± 0·01a | Primary root tip, | 30 | AC007323/T25K16.15 | Unknown protein | ||||||||||
| H + | + | + | + | + | ++ | +++ | + | cotyledon | (segregating) | 72416 | SYT‐motif containing; | |||||||||||
| C + | + | + | ++ | 1 | intron 1 | |||||||||||||||||
| GTR365 | R + | + | + | +++ | 0·09 ± 0·02 | 0·50 ± 0·03 | 0·99 ± 0·003 | 1·40 ± 0·15 | Root tips, ubiquitous, | 25 | AB015479/MTE17.23 | Unknown protein, | ||||||||||
| C + | + | + | +++ | + | ++ | ++ | +++ | esp. vascular | (segregating) | 63309 | similar to yeast mit. | |||||||||||
| 5 | Tim17; intron 1 | |||||||||||||||||||||
| GTR404 | R + | ++ | ++ | ++ | 0·08 ± 0·005 | 0·29 ± 0·008 | 0·72 ± 0·005 | 0·99 ± 0·01 | Root hairs | 10 | Not determined | |||||||||||
| + | ++ | +++ | +++ | |||||||||||||||||||
| GTR424 | R + | + | ++ | + | 0·13 ± 0·01 | 0·29 ± 0·005 | 0·66 ± 0·013 | 0·87 ± 0·005 | Seed coat, hypocotyl, | 80 | Not determined | |||||||||||
| H + | ++ | +++ | + | + | ++ | +++ | +++ | colet, vasculature | ||||||||||||||
| C + | + | ++ | – | |||||||||||||||||||
| GTR426 | C + | + | ++ | + | 0·22 ± 0·003 | 0·32 ± 0·01a | 1·60 ± 0·13a | 0·08 ± 0·01a | Cotyledon | 25 | AL161491/At4g01070 | Flavonoid | ||||||||||
| + | ++ | +++ | + | vasculature | (segregating) | 75763 | 3‐O‐glucosyl‐transferase; | |||||||||||||||
| 4 | exonic | |||||||||||||||||||||
| GTR484 | R + | + | ++ | + | 0·06 ± 0·005 | 0·36 ± 0·13 | 0·60 ± 0·007 | 0·78 ± 0·04 | Root vasculature | 80 | AL138659/T16L24 | No known gene at site; | ||||||||||
| + | ++ | +++ | +++ | (segregating) | 3972 | 1 kb upstream of gene of | ||||||||||||||||
| 3 | unknown function | |||||||||||||||||||||
| GTR489 | R + | + | ++ | + | 0·10 ± 0·01 | 0·39 ± 0·02 | 0·73 ± 0·01 | 1·99 ± 0·02a | Vasculature | 20 | AC066691/T6J19.2 | Beta‐glucosidase; | ||||||||||
| C + | + | ++ | ++ | + | ++ | +++ | ++++ | (segregating) | 42733 | exon 3 | ||||||||||||
| 1 | ||||||||||||||||||||||
| GTR550 | R – | + | ++ | + | 0·15 ± 0·01 | 0·32 ± 0·03 | 0·78 ± 0·02 | 0·98 ± 0·05 | Seed coat, hypocotyl, | 85 | Not determined | |||||||||||
| H + | ++ | ++ | + | + | ++ | +++ | +++ | colet, vasculature | ||||||||||||||
| C – | + | ++ | + | |||||||||||||||||||
| GTR551 | R + | + | + | ++ | 0·008 ± 0·001a | 0·34 ± 0·05 | 0·99 ± 0·04 | 0·002 ± 0·000a | Mesophyll, | 10 | AL162351/T20L15 | No known gene at site; | ||||||||||
| H + | + | + | +++ | + | ++ | +++ | – | ubiquitous | 42705 | 3·3 kb 5′ of a | ||||||||||||
| C + | ++ | +++ | +++ | 5 | protein kinase gene | |||||||||||||||||
| ETR309 | R + | + | ++ | +++ | 0·45 ± 0·07 | 0·60 ± 0·002 | 0·97 ± 0·01 | 1·40 ± 0·06 | Root ground tissue, | 80 | AL392144/MAJ23 | No known gene at site; | ||||||||||
| ++ | +++ | +++ | ++++ | vasculature, hairs, | 25426 | 1·5 kb 5′ of a | ||||||||||||||||
| not meristem | 5 | putative sensorhistidine kinase gene | ||||||||||||||||||||
| ETR414 | C + | + | +++ | ++ | 0·41 ± 0·06 | 0·97 ± 0·01a | 1·99 ± 0·09a | 0·66 ± 0·05 | Cotyledon | 90 | AC002535 T30B22·15 | Pectin methylesterase; | ||||||||||
| ++ | +++ | ++++ | ++ | mesophyl | 63284 | exon 5 | ||||||||||||||||
| 2 | ||||||||||||||||||||||
| ETR528 | H – | + | + | ++ | 0·05 ± 0·006 | 0·53 ± 0·01 | 1·70 ± 0·08a | 0·008 ± 0·003a | Cotyledon margins, | 25 | Not determined | |||||||||||
| C – | + | ++ | +++ | + | ++ | ++++ | – | stipules | ||||||||||||||
R, root; H, hypocotyls; C, cotyledon and leaflets; +, ++, +++, increasing enzymatic staining intensity; –, no GUS activity detected.
Oxygen deprived samples were compared with aerobic control samples at same time point.
Lines were homozygous unless indicated as segregating. Segregating lines were expected to be 1/4 homozygous for the Ds insertion.
The superscript ‘a’ indicates a statistically significant difference in ADH specific activity from the wild‐type (Student’s t‐test, P < 0·01).
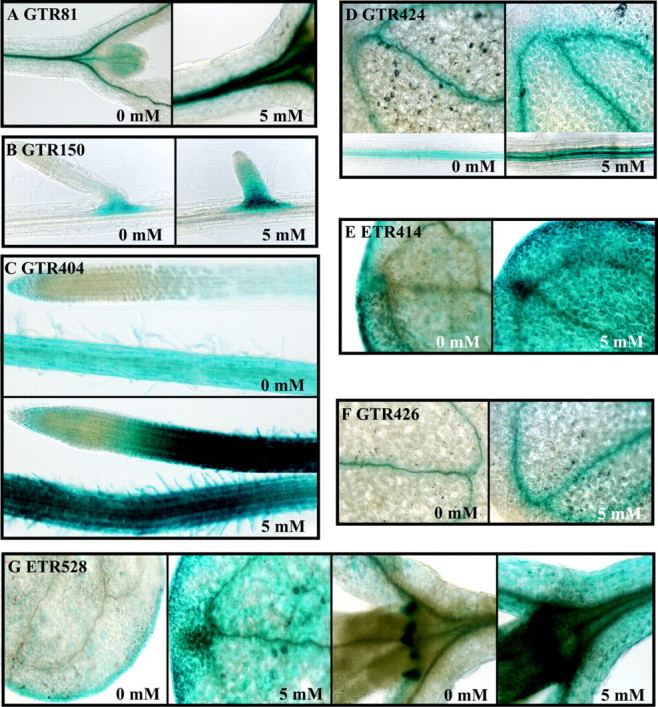
Fig. 3. Histochemical staining of GUS enzymatic activity in 7‐d‐old seedlings following transfer to vertically positioned plates with solid medium containing 5 mm caffeine for 24 h. GUS activity was found in: A, GTR81, shoot apex, leaf primordia and hypocotyl vasculature; B, GTR150, lateral root primordia; C, GTR404, induced throughout root except the tip; D, GTR424, cotyledon and root vasculature; E, ETR414, induced throughout cotyledons; F, GTR426, cotyledon vasculature; G, ETR528, cotyledons and stipules, induced ubiquitously by caffeine treatment.
Characterization of gene and enhancer trap lines with induced expression under low‐oxygen
GTR81 possesses a DsG insertion into a region of chromosome 5 that does not appear to encode a protein (Table 4; Fig. 1). The flanking genes identified by gene prediction algorithms encode a ring H2‐finger protein (At5g01880) in the opposite orientation and a leucine‐rich receptor‐like protein kinase (LRR RLK) (At5g01890), similar to CLAVATA1, in the same orientation as the GUS reporter. The insertion site is 352 bp downstream of the most 3′ predicted polyadenylation site of the LRR RLK gene (EST AV545971). GUS expression in GTR81 was detected in the vascular tissue of the root, hypocotyl and cotyledon (Fig. 2A). In GTR81 homozygotes, ADH specific activity was significantly lower than wild‐type after 24 h of oxygen deprivation, but primary root tip survival was not clearly affected. The effect on ADH specific activity indicates that the insertion affects the expression of a gene that plays a role in stress tolerance. GUS activity was up‐regulated in seedlings that had been transferred to plates containing 5 mm caffeine for 24 h (Fig. 3A), suggesting that GUS expression in GTR81 was mediated by an increase in cytosolic calcium.
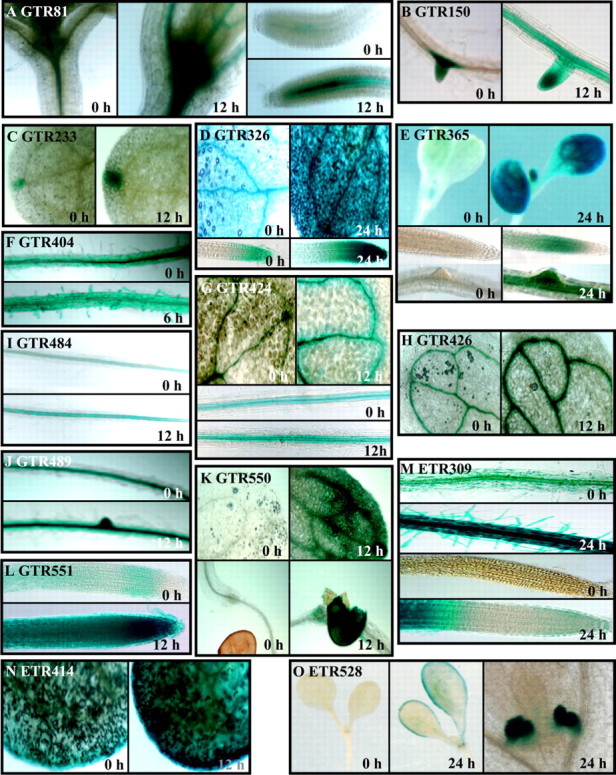
Fig. 2. Histochemical detection of GUS enzymatic activity in gene trap (GTR) and enhancer trap (ETR) lines. Seedlings were grown on vertically positioned plates for 7 days and then deprived of oxygen for the time indicated or not stressed (0 h, harvested from control plates at the same time as treated). GUS enzymatic activity was detected by staining with X‐Gluc for 48 h. GUS staining in: A, GTR81, shoot apex and root vasculature; B, GTR150, lateral root primordia; induced in root vasculature in response to oxygen deprivation; C, GTR233, hydathode; D, GTR326, cotyledons and root tip; E, GTR365, induced in cotyledons, leaf primordia, root vasculature, lateral root primordia and root elongation zone; F, GTR404, root; G, GTR424, cotyledon and root vasculature; H, GTR426, cotyledon vasculature; I, GTR484, root vasculature; J, GTR489, root vasculature and ground tissue; K, GTR550, cotyledon vasculature and mesophyll, and seed coat; L, GTR551, induced throughout root; M, ETR309, root hairs, vasculature, ground tissue and root differentiation zone; N, ETR414, cotyledons; O, ETR528, cotyledon margins and stipules.
GTR150 possesses a DsG insertion into the exon of a gene predicted to encode a serine and proline‐rich protein of unknown function on chromosome 3 (At3g15300, EST: AI995951, 23·7 kDa, 9·44 pI) (Table 4; Fig. 1). GUS reporter activity was localized to lateral root primordia under aerobic conditions and spread to the root vasculature in response to 12 and 24 h of oxygen deprivation (Table 4; Fig. 2B). Root tips of homozygous GTR150 seedlings died after 24 h of oxygen deprivation (Table 4). GTR150 homozygotes showed significantly higher levels of ADH specific activity as compared with wild‐type after 12 h of oxygen deprivation. However, ADH activity was extremely low after 24 h of oxygen deprivation. We also observed that GUS activity was stimulated by caffeine treatment (Fig. 3B), suggesting that regulation of this gene may be mediated by changes in cytosolic calcium. A stimulation of lateral root development in response to oxygen deprivation was observed in this line as in wild‐type.
GTR233 possesses a DsG insertion into a region of chromosome 1 that is not known to encode a polypeptide (Table 4; Fig. 1). A polygalacturonase gene (At1g02460) is 2·4 kb downstream and in the same orientation as the GUS reporter. GUS activity was only detectable in the hydathode of the cotyledon under control conditions, and was up‐regulated after low‐oxygen treatment. The hydathode is a specialized structure involved in secretion of water under high humidity (Fig. 2C). Interestingly, ADH activity detected by in situ staining was high in the hydathode after 12 h of oxygen deprivation (data not shown). ADH specific activity in GTR233 homozygotes was similar to wild‐type after 6 and 12 h of oxygen deprivation but was significantly lower after 24 h. Primary root tip survival in homozygous GTR233 seedlings was significantly lower than wild‐type after 24 h of stress. This result raises the interesting possibility that the hydathode plays a role in low‐oxygen tolerance. However, the possibility that this gene may be expressed at levels below the limit of detection in other cell types cannot be excluded.
GTR326 displayed GUS activity in the vascular and ground tissue in the primary root tip, hypocotyl and cotyledons of seedlings under aerobic conditions and after 6 and 12 h of oxygen deprivation; GUS activity was greatly enhanced after 24 h of oxygen deprivation, especially in the root tip (Fig. 2D). This line possesses a DsG insertion into the first intron of a gene encoding an unknown protein (not annotated by MIPS; EST: AV553796, 21 kDa, 7·06 pI) (Table 4; Fig. 1). The amino‐terminal region of this protein has high identity to the SNH domain characterized in the SYT transcriptional activator of humans. This domain of SYT is involved in protein–protein interactions that regulate chromatin remodelling (Thaete et al., 1999; Nagai et al., 2001). GTR326 showed wild‐type levels of ADH induction. However, only 30 % of seedlings segregating for the insertion (1/4 homozygous for insertion expected) had viable primary root tips after 24 h of oxygen deprivation. GUS staining in this line was not inducible by treatment with caffeine. These results indicate that disruption of a gene that is up‐regulated by prolonged oxygen deprivation and encoding an SNH domain results in sensitivity without affecting ADH expression.
GTR365 displayed very low levels of GUS activity in aerobic seedlings and extensive activity following 24 h of oxygen deprivation (Fig. 2E). GUS staining was localized in the root elongation zone and cotyledon vascular and mesophyll cells following stress. This line possesses a DsG insertion into intron 1 of a gene encoding a protein of unknown function (At5g55510; no EST, 22·3 kDa, 9·08 pI) (Table 4; Fig. 1). The predicted protein has 45 % sequence similarity to a yeast mitochondria protein, Tim17, which is involved in import of matrix proteins (Kurz et al., 1999; Moro et al., 1999). An arabidopsis EST corresponding to a closely related gene (AV442618) was found. In seedlings segregating for this insertion, ADH production was unaffected even after 24 h of anoxia. However, the observed reduction in primary root tip viability after 24 h oxygen deprivation suggests that this gene may be required for endurance to low‐oxygen stress. The induction of this gene in response to oxygen deprivation raises the intriguing possibility that import of mitochondrial proteins may be differentially regulated when oxidative phosphorylation is impaired. GUS expression in this transposant was not stimulated by caffeine.
GTR426 displayed very low levels of GUS activity in the cotyledon vasculature in air‐grown seedlings; GUS staining was intensified after 12 h of oxygen deprivation (Fig. 2H). The DsG was inserted into the exonic region of a gene encoding a flavonoid 3‐O‐glucosyl transferase (FGT) (At4g01070; EST: AF360262, 52·9 kDa, 6·13 pI). GTR426 seedlings segregating for the insertion had significantly higher ADH specific activity after 12 h of oxygen deprivation but significantly reduced ADH specific activity at 24 h compared with wild‐type. Consistent with the observation that maintenance of ADH activity is required for primary root tip viability, GTR426 showed reduced root recovery after 24 h of oxygen deprivation. Caffeine treatment induced GUS activity in this line (Fig. 3F). FGT catalyses the last step in the anthocyanin biosynthesis pathway. Anthocyanins are a class of flavonoids derived from phenylalanine and acetyl CoA that perform an important function involved in many plant processes (reviewed by Harborne and Williams, 2000). Specific flavonoids including anthocyanin act as antioxidants, which could reduce damage by reactive oxygen species produced in cells experiencing oxygen deprivation or upon re‐oxygenation.
GTR484 possesses a DsG insertion into a region of chromosome 3 that is not known to encode a protein (Table 4; Fig. 1). The nearest annotated gene encodes a protein of unknown function (At3g59470) and is 1 kb from the insertion site, with the transcription unit in the same orientation as the GUS reporter. A fructokinase gene (At3g59480) is situated 2 kb downstream in the opposite orientation. GUS activity was reproducibly elevated in the root vasculature after 12 h of oxygen deprivation (Fig. 2I) but not in response to caffeine treatment. ADH specific activity in seedlings segregating for this mutation was not significantly different from wild‐type after 12 and 24 h of oxygen deprivation.
GTR489 possesses a DsG element in exon 3 of a gene encoding a β‐glucosidase (At1g66270; EST AV551893, 59·6 kDa, pI 7·01) on chromosome 1 (Table 4; Fig. 1). Seedlings grown in air displayed GUS activity in the root vasculature; GUS activity was detected throughout the root and cotyledon after 12 h of oxygen deprivation (Table 4; Fig. 2J). The expression of this GUS reporter was not affected by caffeine treatment. Seedlings segregating for this insertion showed normal induction of ADH specific activity after 6 and 12 h but had elevated levels of ADH after 24 h of oxygen deprivation. Nevertheless, primary root tips of only 20 % of GTR489 seedlings survived 24 h of oxygen deprivation, indicating that expression of this enzyme may be required for low‐oxygen tolerance. This enzyme was also shown to be synthesized in maize root tips under hypoxia (Chang et al., 2000). This enzyme may be required to mobilize stored carbohydrate reserves.
GTR551 possesses a DsG insertion into a region on chromosome 5 where there is no known gene or EST (Table 4; Fig. 1). The DsG insertion was located 3·3 kb 5′ of a protein kinase gene (At5g01820), in the same orientation as the GUS reporter. This line showed a low level of GUS activity in mesophyll cells under aerobic conditions and ubiquitous expression in mesophyll cells following 12–24 h of oxygen deprivation (Table 4; Fig. 2L). Expression was not affected by caffeine treatment. In seedlings homozygous for the GTR551 insertion, ADH specific activity was similar to wild‐type levels for the first 12 h of oxygen deprivation, but dramatically lower than wild‐type after 24 h. Not surprisingly, given the dramatic reduction in ADH activity at 24 h, this line showed extremely reduced survival of the primary root tip. These results indicate that an unidentified genetic element disrupted by this insertion is required for survival of extended low‐oxygen stress.
The site of transposant insertion in GTR404, GTR424 and GTR550 has not been successfully confirmed. Nevertheless, the pattern of GUS expression and alterations in induction of ADH specific activity suggest that these lines merit further investigation. GUS expression in GTR404 was detected in the root vascular and ground tissue under aerobic conditions and was specifically elevated in root hairs in response to oxygen deprivation (Fig. 2F) and exposure to 5 mm caffeine (Fig. 3C). GTR404 homozygotes were low‐oxygen sensitive, even though they had wild‐type levels of ADH specific activity. GUS staining was primarily in the vasculature of all seedling organs and the seed coat in GTR424 and GTR550 (Fig. 2G and K). Diffuse GUS activity was apparent in the mesophyll of GTR550. GTR424 showed induced GUS activity in seedlings in response to 5 mm caffeine (Fig. 3D), whereas GTR550 did not.
The DsE insertion into ETR309 was located on chromosome 5, 2·8 kb downstream of a gene encoding a protein of unknown function (At5g10710) and 1·4 kb upstream of a putative sensor histidine kinase (At5g10720) (Table 4; Fig. 1). The two resident genes and the GUS ORF are in the same orientation. The putative sensor histidine kinase gene, which has no representative EST, is a member of the large family of histidine kinases of plants that includes the ethylene response sensor, cytokinin receptor (CRE1) and photoreceptors (reviewed in Suzuki et al., 2000). GUS activity in ETR309 is detected at low levels under control conditions in the root mesophyll and vasculature. Oxygen deprivation caused a progressive intensification in GUS staining in cells throughout the root, with the exception of cells in the tip meristem and zone of elongation (Table 4; Fig. 2M). ETR309 homozygotes showed no alteration in ADH specific activity or low‐oxygen sensitivity. DsE elements do not need to disrupt gene function for reporter gene expression; however, as in this example, the observation indicates that a nearby gene, such as the putative histidine kinase gene, may have a role in the response to oxygen deprivation.
ETR414 possess a single Ds insertion in exon 5 of a putative pectin methylesterase gene (At2g47550) on chromosome 2 (Table 4; Fig. 1). GUS activity was detected at low levels in the cotyledon mesophyll under control conditions and at increased levels following 12 h of oxygen deprivation (Table 4; Fig. 2N). After 12 h of stress, ETR414 homozygotes also showed significantly higher levels of ADH specific activity than wild‐type seedlings (Table 4). Although ETR414 seedlings survived 24 h of oxygen deprivation, ADH specific activity declined between 12 and 24 h, suggesting that tolerance may be altered in this line. ETR414 seedlings showed increased GUS staining in response to 5 mm caffeine (Fig. 3E), indicating that regulation of this gene may be dependent upon cytosolic free calcium. Pectins are secreted into cell walls in a highly methlyesterfied form and are demethylated by pectin methylesterase. This results in the release of acidic pectins and methanol as well as increased cell wall rigidity and calcium chelation (Micheli, 2001). The regulation of expression of this enzyme could play a role in modulation of apoplastic free calcium and/or cell wall rigidity in response to oxygen deprivation.
The site of DsE insertion in ETR528 is under investigation due to the intriguing GUS staining patterns observed in these lines. ETR528 displayed a unique staining pattern in the cotyledon margin, lateral root meristem and stipules in response to 12 h of oxygen deprivation (Table 4; Fig. 2O). The gene tagged by the DsE in ETR528 appears to be calcium‐regulated and required for low‐oxygen tolerance.
ADH specific activity in GTR150 (unknown Ser/Pro rich protein), ETR414 (pectin methylesterase) and ETR528 (unknown) homozygotes was abnormally high after 12 h of oxygen deprivation and low after 24 h (Table 4), providing evidence that ADH expression may be both positively and negatively regulated in the wild‐type. An effect on primary root tip survival was observed for GTR150 and ETR528 but not ETR414, perhaps reflecting the less severe reduction in ADH specific activity in ETR414 at 24 h. Significantly elevated ADH specific activity was observed after 12 h in the low‐oxygen sensitive line GTR426 (FGT). These observations suggest that these lines may possess mutations in genes that directly or indirectly influence ADH expression.
CONCLUSIONS
The results demonstrate the effectiveness of gene‐ and enhancer‐trap mutagenesis to discover genes that are regulated in response to oxygen deprivation in arabidopsis seedlings. The collection of transposant lines we identified showed many distinct and intriguing patterns of reporter gene expression in response to oxygen deprivation. In seven cases GUS expression was also stimulated by treatment with caffeine, an effective inducer of ADH specific activity in arabidopsis. Surprisingly, none of the transposon insertions were into genes that encode known anaerobic polypeptides. Gene‐trap insertions were identified in genes encoding FGT required for anthocyanin biosynthesis, β‐glucosidase, pectin methylesterase and proteins of unknown function including a serine/proline‐rich protein and an SNH domain‐containing protein. A gene trap insertion adjacent to an LRR RLK and an enhancer trap insertion adjacent to a putative histidine kinase gene were identified. Several of the insertions were observed in regions of the arabidopsis genome in which no genes have been identified by gene annotation algorithms. Nevertheless, a subset of these insertion events appears to effect ADH activity and/or low‐oxygen tolerance. DsG insertions that do not appear to disrupt a protein‐encoding gene but that show GUS expression has been identified on several occasions and could perhaps result in an insertion near to a sequence that can act as a minimal promoter or from the capture of the GUS open reading frame by an antisense transcript or a non‐coding RNA (P. Springer, unpublished data). Another intriguing observation was that several lines showed abnormally high induction of ADH specific activity but were low‐oxygen intolerant. We anticipate that further implementation of this screening procedure will allow for the identification of genes that are involved in the response to low‐oxygen stress including components of the calcium‐mediated signal transduction pathway.
Acknowledgements
We thank Marcela Pierce, David Holding for generating transposant lines, Barbara Jablonska for maintaining the gene/enhancer trap collection and Kathy Miranda‐Szick for continuing encouragement and advice. This research was funded by the USDA/NRI Competitive Grants Program (97‐35100‐4191 to J.B.S.) and the Southwest Consortium (99‐N02 to P.S. and J.B.S.).
Supplementary Material
Received: 23 August 2001; Returned for revision: 20 November 2001; Accepted: 16 January 2001
References
- AltschulSF, Gish W, Miller W, Myers EW, Lipman DJ.1990. Basic local alignment search tool. Journal Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bailey‐SerresJ, Dawe RK.1996. Both 5′ and 3′ sequences of maize adh1 mRNA are required for enhanced translation under low oxygen conditions. Plant Physiology 112: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChangWWP, Huang L, Shen M, Webster C, Burlingame AL, Roberts JKM.2000. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to low‐oxygen environment and identification of proteins by mass spectrometry. Plant Physiology 122: 295–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChungHJ, Ferl RJ.1999. Arabidopsis alcohol dehydrogenase expression in both shoots and roots is conditioned by root growth environment. Plant Physiology 121: 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ConleyTR, Peng HP, Shih MC.1999. Mutations affecting induction of glycolytic and fermentative genes during germination and environ mental stresses in Arabidopsis. Plant Physiology 119: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DennisES, Dolferus R, Ellis M, Rahman M, Wu Y, Hoeren FU, Graover A, Ismond KP, Good AG, Peacock WJ.2000. Molecular strategies for improving waterlogging tolerance in plants. Journal of Experimental Botany 51: 89–97. [PubMed] [Google Scholar]
- DrewMC.1997. Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annual Review of Plant Physiology and Plant Molecular Biology 48: 223–250. [DOI] [PubMed] [Google Scholar]
- HarborneJB, Williams CA.2000. Advances in flavonoid research since 1992. Phytochemistry 55: 481–504. [DOI] [PubMed] [Google Scholar]
- HeCJ, Morgan PW, Drew MC.1996. Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiology 112: 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KurzM, Martin H, Rasow J, Pfanner N, Ryan MT.1999. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Molecular Biology of the Cell 10: 2461–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiuYG, Whittier, RF.1995. Efficient isolation and mapping of Arabidopsis thaliana T‐DNA insert junctions by thermal asymmetric interlaced PCR. Plant Journal 8: 457–463. [DOI] [PubMed] [Google Scholar]
- MartienssenRA, Springer PS.1998. Enhancer and gene trap transposon mutagenesis in Arabidopsis. www.arabidopsis.org/springer.html
- MicheliF.2001. Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends in Plant Science 6: 414–419. [DOI] [PubMed] [Google Scholar]
- MoroF, Sirrenberg C, Schneider HC, Neupert W, Brunner M.1999. The Tim17‐23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO Journal 18: 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NagaiM, Tanaka S, Tsuda M, Endo S, Kato H, Minami A, Hiraga H, Nishihara H, Sawa H, Nagashima K.2001. Analysis of transforming activity of human synovial sarcoma‐associated chimeric protein SYT‐SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proceedings of the National Academy of Sciences of the USA 27: 3843–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PengHP, Chan CS, Shih MC, Yang SF.2001. Signaling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiology 126: 742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SachsMM, Freeling M.1978. Selective synthesis of alcohol dehydrogenase during anaerobic treatment of maize. Molecular and General Genetics 161: 111–115. [Google Scholar]
- SachsMM, Freeling M, Okimoto R.1980. The anaerobic proteins of maize. Cell 20: 761–767. [DOI] [PubMed] [Google Scholar]
- SachsMM, Subbaiah CC, Saab IN.1996. Anaerobic gene expression and flooding tolerance in maize. Journal of Experimental Botany 47: 1–15. [Google Scholar]
- SedbrookJC, Kronebusch PJ, Borisy GG, Trewavas AJ, Masson PH.1996. Transgenic AEQUORIN reveals organ specific cytosolic Ca2+ responses to anoxia in Arabidopsis thaliana seedlings. Plant Physiology 111: 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SpringerPS.2000. Gene traps: tools for plant development and genomics. Plant Cell 12: 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SubbaiahCC, Bush DS, Sachs MM.1994a Elevation of cytosolic calcium precedes anoxia gene expression in maize suspension‐cultured cells. Plant Cell 6: 1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SubbaiahCC, Zhang J, Sachs MM.1994b Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiology 105: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SubbaiahCC, Bush DS, Sachs MM.1998. Mitochondrial contribution to the anoxic Ca2+ signal suspension cultured cells. Plant Physiology 118: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SundaresanV, Springer P, Volpe T, Jones JDG, Dean C, Ma H, Martienssen R.1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes and Development 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- SuzukiT, Sakurai K, Imamura A, Nakamura A, Ueguchi C, Mizuno T.2000. Compilation and characterization of histidine‐containing phosphotransmitters implicated in His‐to‐Asp phosphorelay in plants: AHP signal transducers of Arabidopsis thaliana Bioscience Biotechnology and Biochemistry 64: 2486–2489. [DOI] [PubMed] [Google Scholar]
- ThaeteC, Brett D, Monaghan P, Whitehouse S, Rennie G, Rayner E, Cooper CS, Goodwin G.1999. Functional domains of the SYT and SYT‐SSX synovial sarcoma translocation proteins and co‐localization with the SNF protein BRM in the nucleus. Human Molecular Genetics 8: 585–591. [DOI] [PubMed] [Google Scholar]
- TsugekiR, Kochieva EZ, Fedoroff NV.1996. A transposon insertion in the Arabidopsis SSR16 gene causes an embryo‐defective lethal mutation. Plant Journal 10: 479–489. [DOI] [PubMed] [Google Scholar]
- TsujiH, Nakazono M, Saisho D, Tsutsumi N, Hirai A.2000. Transcript levels of the nuclear‐encoded respiratory genes in rice decrease by oxygen deprivation: evidence for involvement of calcium in expression of the alternative oxidase 1a gene. FEBS Letters 471: 201–204. [DOI] [PubMed]
- VartapetianBB, Jackson MB.1997. Plant adaptations to anaerobic stress. Annals of Botany 79 (Suppl. A): 3–20.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.