Abstract
Submergence tolerance is an important agronomic trait for rice grown in South‐East Asia, where flash flooding occurs frequently and unpredictably during the monsoons. Although mapping locations of one major and several minor quantitative trait loci (QTL) were known previously, improving submergence tolerance in agronomically desirable types of rice has not been achieved. KDML105 is jasmine rice widely grown in rain‐fed lowland regions of Thailand. This cultivar is very intolerant of submergence stress. To improve submergence tolerance in this cultivar, three submergence‐tolerant cultivars, FR13A, IR67819F2‐CA‐61 and IR49830‐7‐1‐2‐2, were cross‐pollinated with KDML105. Transferring the major QTL for submergence tolerance was facilitated by four back‐crossings to the recipient KDML105. Molecular markers tightly linked to the gene(s) involved were developed to facilitate molecular genotyping. We demonstrated that individuals of a BC4F3 line that retained a critical region on chromosome 9 transferred from tolerant lines were also tolerant of complete submergence while retaining all the agronomically desirable traits of KDML105. In addition, effects of secondary QTLch2 were detected statistically in back‐cross progenies. Effects of secondary QTLch7 were not statistically significant. The close association between tightly linked markers of the tolerance locus on chromosome 9 and submergence tolerance in the field demonstrates the considerable promise of using these markers in lowland rice breeding programmes for selecting increased submergence tolerance.
Keywords: Key words: Submergence tolerance, QTL analysis, plant breeding, back‐crossing, rice (Oryza sativa L.), marker‐assisted selection, graphical genotype.
INTRODUCTION
Flash flooding greatly depresses grain yield in lowland rain‐fed rice. Genetic determinants of traits responsive to submergence have been studied in three mapping populations taking a quantitative trait locus (QTL) approach. These populations comprised (1) recombinant inbred lines derived from a cross between FR13A and CT6241‐17‐1‐5‐1, (2) doubled haploid lines derived from a cross between IR49830‐7‐1‐2‐2 and CT62417‐1‐5–1 and (3) F2 lines derived from KDML105 and Jao Hom Nin (Toojinda et al., 2002). A major QTL for submergence tolerance contributed by FR13A, a submergence‐tolerant landrace from India, has been identified on chromosome 9 in all mapping studies (Xu et al., 1996; Nandi et al., 1997; Xu et al., 2000; Toojinda et al., 2002). In addition, secondary QTL that influence submergence tolerance have been located in chromosomes 1, 2, 5, 7, 10 and 11 (Siangliw et al., 2002; Toojinda et al., 2002). Fine mapping of the QTL on chromosome 9 has been carried out using BAC‐end sequences (Kamolsukyunyong et al., 2001; Vanavichit et al., 2001) and chromosome landing (Xu et al., 2000), and useful molecular markers for the major QTL for submergence tolerance are now defined and well developed. A further step for breeding submergence‐tolerant rice is to transfer the relevant QTL from submergence‐tolerant donors into a variety of high culinary quality such as Khao Dok Mali 105 (KDML105), a well‐known traditional Thai jasmine rice variety that is aromatic and soft‐cooked.
Several donors of submergence tolerance are available in addition to FR13A itself. These include DH206 (IR67819F2‐CA‐61), a doubled haploid line derived from a cross between FR13A and CT6241‐7‐1‐2‐2. They also include IR49830‐7‐1‐2‐2, an improved variety developed at IRRI (Mackill et al., 1993). Here, we describe back‐crossing to transfer submergence tolerance simultaneously from three donors into the susceptible but otherwise highly desirable fragrant Thai cultivar KDML 105. The experiment used PCR‐based markers tightly linked to the major QTL for submergence tolerance located on chromosome 9. The back‐cross progenies were genotyped and phenotyped to validate the effects of submergence QTL. The introgressed QTL on chromosome 9 was displayed using tightly linked markers. Graphical genotyping of the tolerance locus demonstrated the potential value of these markers for breeding programmes that adopt marker‐aided selection as a substitute for time consuming and labour‐intensive phenotypic screening.
MATERIALS AND METHODS
Plant material
Three submergence‐tolerant parents, FR13A (FR), IR49830‐7‐1‐2‐2 (IR) and DH206 (IR67819F2‐CA‐61) (DH) were used as submergence‐tolerant donors. While FR, a photoperiod‐sensitive landrace from India, is highly submergence tolerant, it is agronomically undesirable, being low yielding and possessing long awns, poor cooking quality and lacking aroma. IR, an agronomically improved cultivar possesses submergence‐tolerance genes derived from FR and can produce heavy yields (Mackill et al., 1993). DH is a doubled haploid line derived from a cross between FR and CT6241‐17‐1‐5‐1. CT6241 was developed by Centro International de Agriculura Tropical (CIAT). It is agronomically poor but non‐photoperiod sensitive and highly intolerant to submergence. To improve submergence tolerance, the well‐known Thai jasmine rice KDML105 (KD) was crossed with all three submergence‐tolerant donors. KD is adaptable to a medium‐to‐deep water regime but not to complete submergence caused by flash flooding. More than 100 F1 seeds were produced from each cross. F1 seeds of each cross were used to back‐cross to KD to produce 50–100 BC seeds. To maintain the acceptable agronomic characteristics of KD, back‐crossing was implemented in a crossing scheme. Only 20–25 BC plants were randomly selected to back‐cross to KD in each of three further generations. The back‐crossing method was applied repeatedly from June 1995 to May 1999 until the BC4F2 generation was produced. Four hundred and sixty‐seven seeds of BC4F2 were obtained. DNA was extracted from 3‐week‐old leaf tissue of the BC4F2 population using the CTAB–NaCl method (Rogers and Bendich, 1994) and used for marker genotyping at the targeted QTL regions. BC4F3 lines were produced by selfing all the BC4F2 individuals. These were evaluated for submergence tolerance in September 2000 in outdoor conditions. Based on the phenotypic result of the test, the nine most tolerant BC4F3 lines were re‐evaluated for their tolerance to submergence in February 2001. The photoperiod sensitivity of the nine most submergence‐tolerant lines was established under controlled‐environment conditions 100 d after sowing.
Evaluation of submergence tolerance in BC4F3
The BC4F3 population was evaluated for its ability to survive under totally submerged conditions using an outdoor lagoon at Agronomy Field, Kasetsart University, Kamphangsaen Campus, Thailand in September 2000. The BC4 population was direct‐seeded in one‐row plots, 75 cm in length, with 25 cm between rows, on a puddled seedbed constructed in the base of a 60 m2 pond. FR and IR were used as submergence‐tolerant checks, and CT6241 and KD as susceptible checks. These same checks were included in every tenth plot. Four weeks after seeding, all plants were completely submerged for 8 d. The water level was maintained 60–100 cm above the tallest plants to prevent leaf tips from emerging into the air. Each line was replicated twice in a randomized complete block design. The experiment was repeated by re‐testing the nine most tolerant BC families under almost identical conditions in February 2001. In both tests the following traits were assessed.
Percentage plant survival (PPS).
The ability of the plant to survive after experiencing submergence stress for 8 d was estimated 10 d after desubmergence. Plants were judged to have been killed if no new growth was visible at this time. Percentage survival of BC4 progenies ranged from 0 to 100. No plants of the susceptible checks (KD and CT6241) survived, while 100% of the tolerant check line (FR) survived. In 2001, plant survival of BC4 progenies was calculated as a percentage by comparing the number of plants surviving 10 d after desubmergence and the number of plants present before submergence.
Plant elongation (PE).
Plant elongation was calculated as the difference in height (cm) between plants before and after 8 d of submergence. Plant heights were taken from ten randomly selected plants of each line. The measurement was taken as the distance from the soil surface to the tip of the longest leaf.
Relative plant elongation under submergence (RPE).
The term ‘relative plant elongation’ expresses the difference in elongation of submerged plants compared with non‐submerged controls as a percentage (Toojinda et al., 2002). Plants showing RPE of less than 100 had leaf elongation suppressed by submergence. RPE values over 100 % indicate that leaf elongation was stimulated by submergence.
Leaf senescence (LS).
Leaf senescence was measured by a hand‐held SPAD‐502 chlorophyll meter (Minolta Camera Co., Ltd, Japan) that assesses leaf greenness based on the amount of chlorophyll. Ten randomly selected youngest leaves for each individual line were used. The measurements were made at the base, middle and tip of each leaf, and average LS–SPAD values of 30 readings were used (Toojinda et al., 2002). Plants showing high SPAD values indicate non‐senescent leaves with a high content of chlorophyll.
Development of tightly linked markers to submergence tolerance QTL
The QTL on chromosome 9 (SubQTL) is a primary determinant of submergence tolerance (Xu et al., 1996; Nandi et al., 1997; Toojinda et al., 2002). Physical mapping (Kamolsukyunyong et al., 2001) and sequencing methods (Vanavichit et al., 2001) were performed on this region of chromosome 9. The restriction fragment length polymorphism (RFLP) marker R1164, that is linked particularly closely to the QTL, was used to select bacterial artificial chromosomes (BAC) and P1 artificial chromosome (PAC) clones. BAC and PAC contigs were assembled. The tightly linked markers were then developed from the BAC end as RFLP probes and were designated as 126G1R and 180D1R from previous work (Kamolsukyunyong et al., 2001). To use the marker efficiently, primers were developed based on the sequence of the RFLP marker generated from the BAC ends, thus converting the markers into sequence tag site (STS), a PCR‐based method. RB0783, an expressed sequence tag (EST) marker from the Rice Genome Project (RGP), Japan was also found linked in the region of interest. The markers mentioned were also used in graphical genotyping (Young and Tanksley, 1989) which is useful for viewing the chromosomal segment of BC lines.
Molecular genotyping
Markers flanking one major and two minor QTL for submergence tolerance were used for genotyping the 467 BC4F2 progenies. Seven markers, RM285, R1164, 126G1R, 180D1R, RB0783, RM219 and RM105, were used for genotyping the major QTL on chromosome 9, two markers, RM221 and RM240, were used for genotyping a minor QTL on chromosome 2, and two markers, OSR4 and RM11, were used for genotyping a minor QTL on chromosome 7. R1164 and 180D1R were RFLP markers. The non‐radioisotopic detection method based on DIG (digoxygenin) of RFLP was done following recommended procedures (Boehringer Mannheim, Biochemica, Mannheim, Germany). The STS marker from 126G1R was tested by PCR with the forward primer (5′‐CGTTGAAAAGGTGAGGAAAG‐3′) and reverse primer (5′‐TAAATTACGACCTGAAACC‐3′). PCR‐amplification was performed in a 10 µl volume containing 40 ng of DNA template, 10 mm Tris–HCl (pH 8·3), 50 mm KCl, 1·5 mm MgCl2, 0·25 mm dNTPs, 1·25 pmol each of forward and reverse primers and 0·2 units of Taq DNA polymerase (Promega, Madison, WI, USA). PCR was as follows. Initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturing at 94 °C for 1 min, annealing at 50 °C for 1 min and extension at 72 °C for 1 min. A final incubation at 72 °C for 5 min was allowed to complete primer extension followed by a hold at 4 °C. The primer sequence, PCR components and PCR profile of SSLP markers (with RM and OSR prefixes) were performed according to Akagi et al. (1996), Chen et al. (1997) and Temnykh et al. (2000). The primer sequence of RB0783 was as follows: forward 5′‐CTGCTCCGACGACCTGATGG‐3′ and reverse 5′‐TCT CTGTGCGTCCTAATTGC‐3′. For the RB0783 marker, the PCR amplication was performed using the following PCR conditions: 94 °C for 5 min followed by 35 cycles of denaturing at 94 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 1·30 min. A final incubation at 72 °C for 7 min was allowed to complete primer extension and hold at 4 °C. After amplification, reactions were stopped with 5 µl of loading buffer (95 % formamide, 20 mm EDTA, 0·025 % bromophenol blue, 0·025 % xylene cyanol). The PCR products were denatured at 94 °C for 5 min and were quenched on ice before loading in the sequencing gel. For STS and SSLP markers, 2 µl of PCR product was separated by electrophoresis on gels containing 4·5 % denaturing polyacrylamide gel with 8 m urea in 0·5 × TBE (89 mm Tris–borate, 2 mm EDTA, pH 8·0) buffer. Electrophoresis was done at 60 W constant power using a 100‐well comb of a standard DNA sequencing unit (Bio‐Rad, Hercules, CA, USA). On the other hand, the SSCP marker RB0783 was separated on 3·5 % non‐denaturing polyacrylamide/bisacrylamide 29 : 1 gels for 10 h at 27 °C, 5 W constant power in 1 × TBE (178 mm Tris–borate, 4 mm EDTA, pH 8·0) buffer. Amylose content and aroma were examined using SSLP markers RM00 (Garland et al., 2000) and RM342 (Lanceras et al., 2000) linked to the relevant genes.
Whole‐genome scanning for favourable genetic background
The top nine most submergence‐tolerant BC4F2 lines were fingerprinted with 47 SSLP markers (approx. four markers spanning each chromosome) according to Chen et al. (1997). The result was entered into SupergeneTM software (Boutin et al., 1995) showing graphically the genotype of each line as well as the percentage similarity to KD of the selected lines. Genetic similarity among KD, heterozygotes and donor parents was assessed by fingerprinting analysis using Jaccard coefficient in the Numerical Taxonomy and Multivariate Analysis System (NTSYS) software, version 2·01e.
Statistical and QTL analyses
The correlation and QTL analyses based on single‐marker analysis using ANOVA and regression‐based software (STATGRAPHICS 2·1) were used for detecting significant correlations between the responsive traits and submergence stress, and significant associations between markers and traits.
RESULTS
Phenotypic distribution of submergence tolerance in BC4 population
Tightly linked markers at three target QTL were used to determine the presence of introgressed QTL against the genetic background of KD. FR and DH showed the highest tolerance score while IR was moderately tolerant. On the other hand, KD and CT6241 were killed after 8 d of complete submergence.
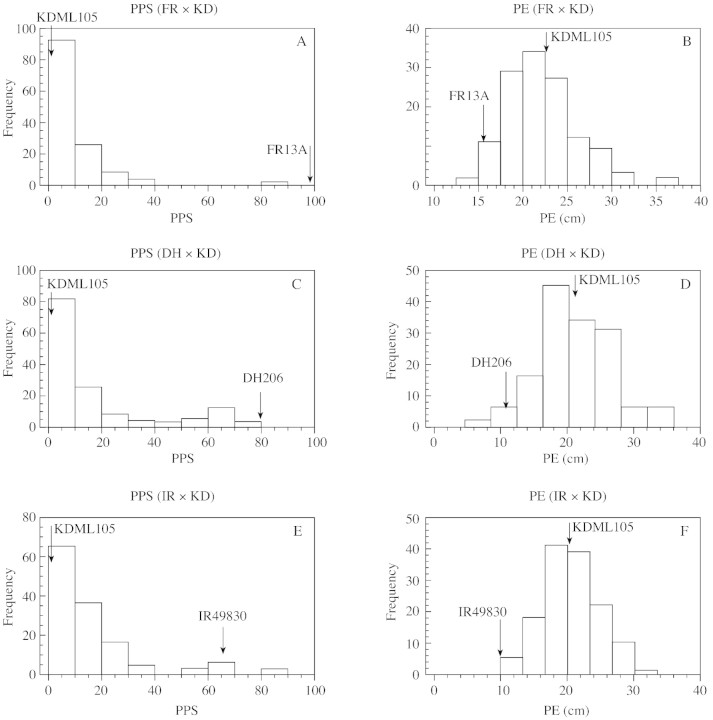
PE and PPS both showed continuous distributions. The distribution of PPS across the BC4 population, however, was skewed toward the KD (intolerant) phenotype (Fig. 1). Susceptible plants elongated rapidly underwater at a rate of 2·5 cm d–1, compared with 1·0 cm d–1 for tolerant plants. Transgressive segregation was observed in some BC4 progenies that elongated even faster than susceptible KDML105 or slower than the tolerant parent. In February 2001, the second experiment tested the nine BC4 lines showing greatest tolerance. This was done to confirm the dominating impact of the major QTL region on submergence tolerance. The 95 percentile was used as a parameter for comparing two replications. There was no significant difference between replications in this experiment and one made in 2000 (data not shown). The nine selected BC lines retained good tolerance, giving a high percentage of plant survival after 10 d underwater.
Fig. 1. Distribution in BC4 progeny of plant elongation during submergence (cm) and plant survival (% of each line) after experiencing 10 d submergence stress. Tests were made in the year 2000 on each back‐cross (FR × KD, DH × KD and IR × KD). The frequency axes refer to the number of lines falling in each elongation or survival class.
A high correlation of the PPS was found between the two test years for the top nine BC lines (Table 1). PE and RPE also showed high correlations and the two traits had a negative correlation with PPS. LS measured by the SPAD machine showed high positive correlation with percentage plant survival, while leaf senescence had low correlation with both PE and RPE.
Table 1.
Correlation coefficients among traits related to submergence tolerance in the whole BC population in 2000 (top part) and between nine BC lines showing most submergence tolerance during 2000 compared with a similar test conducted in 2001 (bottom part)
| PE | PPE | ||||
| PPS | –0·03 | 0·01 | |||
| PE | 0·87** | ||||
| PPS 2001 | RPE 2001 | PE 2000 | PE 2001 | LS 2001 | |
| PPS 2000 | 0·67** | –0·46* | 0·18 | –0·49* | 0·64** |
| PPS 2001 | –0·14 | 0·14 | –0·06 | 0·64** | |
| RPE 2001 | 0·07 | 0·87** | 0·02 | ||
| PE 2000 | –0·03 | 0·18 | |||
| PE 2001 | –0·09 |
For descriptions of traits, see Materials and Methods.
PPS, Percentage plant survival; PE, plant elongation; RPE, relative plant elongation under submergence; LS, leaf senescence.
* and **, significant effect at P = 0·05 and 0·01, respectively.
Genetic contribution of the introgressed QTL
Regression analysis detected associations between single marker and trait performance. Significant associations were found with several markers on chromosomes 2, 7 and 9 as shown in Table 2. QTL for PPS and PE on chromosome 9 were significant in the BC4 progenies. The PPS QTL contributed 61 % of the phenotypic variation and was tightly linked to marker 126G1R. This association was contributed by each of the three donor lines. Low phenotypic variance explained (PVE) obtained for elongation ability, as controlled by the chromosome 9 QTL, was contributed by KD. The parents contributing the traits for slow elongation and higher survival rates were actually reversed in the QTL controlled by chromosome 2, i.e. they came from the intolerant parents. PVE of 3·8 % and 2·3 % were found for PPS and PE, respectively. This finding was not observed on chromosome 7. The major QTL on chromosome 9 and the minor QTLs located on chromosomes 2 and 7 were analysed in the FR13A × CT6241 RI population (Siangliw, 2002; Toojinda et al., 2002). It is probable that the location of minor QTLs varied with different genetic backgrounds. The incidence of finding the PPS and PE QTL in the same region suggests either tight linkage or strong pleiotropy. This could be further understood by refining the traits in a substitution population.
Table 2.
DNA markers with significant effects in simple regression models on elongation ability and plant survival
| Elongation ability (PE) | Percentage plant survival (PPS) | |||||
| Marker | R 2 | Effect | Contributor | R 2 | Effect | Contributor |
| R1164 (Ch. 9) | 1·11 %* | +1·7 | KDML105 | 26 %** | –20·9 | FR13A, IR49830, DH206 |
| 126G1R (Ch. 9) | 1·8 %** | +2·8 | KDML105 | 61 %** | –41·72 | FR13A, IR49830, DH206 |
| 180D1R (Ch. 9) | 1·4 %** | +2·26 | KDML105 | 49 % ** | –33·99 | FR13A, IR49830, DH206 |
| RM240 (Ch. 2) | 0·6 % | – | – | 0 % | – | – |
| RM221 (Ch. 2) | 2·3 %** | –3·8 | FR13A, IR49830, DH206 | 3·8 %** | +4·1 | KDML105 |
| OSR4 (Ch. 7) | 0·4 %* | +9·0 | KDML105 | 0 % | – | – |
| RM11 (Ch. 7) | 0 % | – | – | 0 % | – | – |
The percentage values indicate the percentage of the variation in each trait that was linked to the markers.
Negative and positive effects indicate donors and recurrent parents contributing favourable QTL alleles, respectively.
The markers used are designated 126G1R and 180G1R which were BAC end markers.
* and **, significant effect at P = 0·05 and 0·01, respectively.
Recovery rate of the desirable parent
The top nine most tolerant BC4F2 lines were selected from three crosses (FR × KD, DH × KD and IR × KD). Four SSR markers per chromosome were used to scan the whole genome. The nine tolerant individuals showed 90, 87, 90, 83, 90, 93, 90, 93, 93 % similarity to KD (Fig. 2).
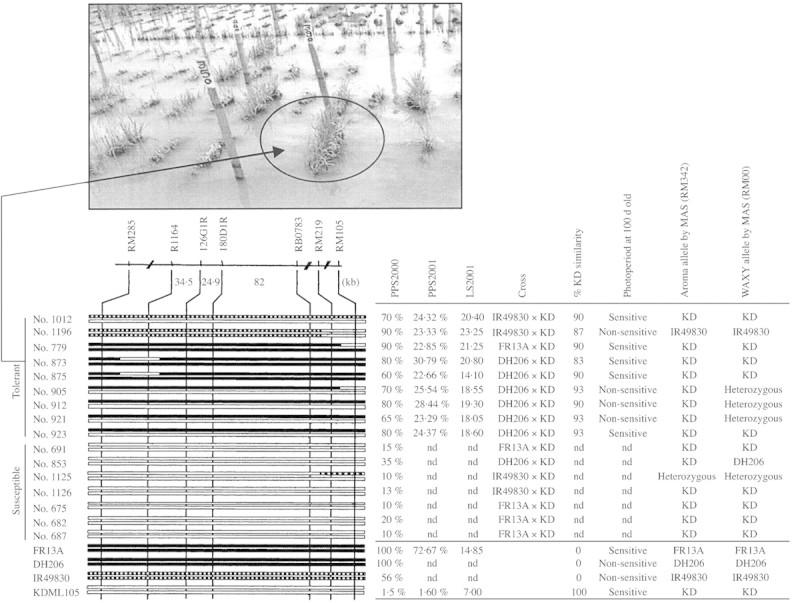
Fig. 2. Graphical genotypes of the most tolerant and susceptible BC4F2 progenies in the SubQTL region on the short arm of rice chromosome 9. Distances are in kilobases (kb). Trait descriptions were specified in Materials and Methods. ‘nd’ refers to no data. The picture at the top shows the appearance of a promising submergence‐tolerant BC line (no. 873) after 10 d submergence.
Graphical genotype
The extent of the introgressed region at the chromosome 9 QTL in BC4F2 is graphically displayed in Fig. 2. Nine tolerant and seven susceptible lines were compared based on the allelic compositions at two marker loci from R1164 to RB0783, where the QTL for submergence tolerance was mapped. The PPS for the test made in the year 2000 obtained from the nine most tolerant and seven most susceptible BC lines ranged from 60 to 90 % and 10 to 35 %, respectively. In 2001, the PPS were much lower, ranging from 22·9 to 30·8 % in the BC4 progenies, 45 % in FR but only 1·6 % in KD. The difference in the PPS for the two years was due to the method of scoring. In 2000, PPS was visually scored, while in 2001, the PPS was obtained by actual counting. LS measured by SPAD after desubmergence showed that the tolerant group contained more chlorophyll than KD after submergence. The most important region for submergence tolerance is flanked by markers R1164 and RB0783, where the genotypes at the interval correspond with PPS and LS. The graphical genotypes also revealed that the SubQTL is dominant since BC lines containing heterozygotes at markers R1164 and RB0783 were as tolerant as those containing homozygous tolerant alleles. Among these tolerant BC lines, the percentage similarity to KD background ranged from 83 to 93 %, implying that chromosome segments outside the introgressed sub region were very much the same as in KD itself. This is an indication that a variety of KD with submergence tolerance had been produced. To confirm further that other traits contributed by KD were still maintained, photoperiod sensitivity (chromosome 6), aroma (chromosome 8) and amylose content (chromosome 6) were also evaluated. Photoperiod sensitivity was obtained phenotypically while aroma and amylose content were examined using SSLP markers linked to the relevant genes (Garland et al., 2000; Lanceras et al., 2000). The result showed that BC lines 1012, 779, 873, 875, and 923 were photoperiod sensitive, aromatic and retained the same high cooking quality as KD itself. Some BC lines such as 905, 912, and 921 were photoperiod insensitive but still retained submergence tolerance, good aroma and the soft‐cooking character. In the near future, further purification could achieve various improved forms of KD tailored even closer to the needs of rice farmers.
DISCUSSION
Chromosomal regions that influence submergence tolerance have been located by QTL analysis. In the present study, we tested the effects of such QTL by back‐crossing into a single susceptible genotype. Phenotypic variation among BC lines in terms of plant survival (PPS) and leaf senescence (LS) was used to indicate the minimum segments acquired from the donor in order to attain a satisfactory level of tolerance. In all cases, the region located within the region flanked by markers RM285–RM105 from the tolerance donors was the prerequisite for submergence tolerance. The distribution of PPS and PE showed that only a small proportion of the BC lines attained a level of submergence tolerance that matched their tolerance donors. Particularly in the FR cross, no BC lines were as tolerant as the donor. For example, in 2000, the best BC line (no. 779) attained only 90 % of the PPS of FR. Especially in 2001, when screening was more stressful, all progeny performed poorly. In the two other crosses, derived from DH and IR, small numbers of BC lines were as good or better than their tolerance donors. We hypothesized that because FR contains all genes necessary for top performance under submergence stress, it is difficult to breed for even better tolerance in its progeny. The distribution of PPS and PE supported this idea. In the FR cross, almost all BC lines displayed below 40 % PPS compared with 70 % for FR. Therefore, there is a smaller chance of obtaining the most tolerant genotypes from this cross. As both KD and FR were strongly photoperiod sensitive, selection was more constrained and limited during back‐crossing. Selection was more favourable in the other crosses as more BC lines were distributed above 40 % PE, whereas few transgressive segregants were identified.
An alternative hypothesis is that these BC lines did not carry all necessary genes from FR to sustain the complete tolerance. This idea was supported by the fact that no tolerant BC lines carried, from their tolerance donors, a complete homozygous dominant chromosomal segment lying between RM285 and RM105. The two best BC lines lacked a fragment near RM105 while others missed other segments. Although the QTLch9 region between RM285 and RM105 contributed up to 61 % PVE of PPS, there were other regions on QTLch2 and QTLch7 that might complete QTLch9 functionality. This idea was supported by the fact that the BC line no. 779, of which the PPS in the year 2000 screening approached 90 %, was missing alleles of QTLch2 and QTLch7 donated by FR. The effect was more pronounced in 2001 where screening conditions were more rigid. When BC lines were missing all three QTL (QTLch9, QTLch2 and QTLch7), they were completely susceptible. However, this interaction was not the same when new generations of FR, DH and IR, were used as donors. On the other hand, BC lines derived from the other two donors, DH and IR, were as tolerant or even more tolerant than their tolerance donors. This result supported a role for QTLch2 in tolerance control. In conjunction with QTLch9, QTLch2 significantly affected elongation ability. Both QTLs consisted of alleles with opposite genetic effects that may cause the transgressive segregation in both BC populations.
ACKNOWLEDGEMENTS
This research was supported financially by the European Commission INCO‐DC Rice‐for‐Life project (IC18‐CT96‐00078) and by the National Center for Genetic Engineering and Biotechnology at Kasetsart University, Thailand.
Supplementary Material
Received: 6 August 2001; Returned for revision: 21 November 2001; Accepted: 21 February 2002
References
- AdkinSW, Shiraishi T, McComb JA.1990. Submergence tolerance of rice – a new glasshouse method for the experimental submergence of plants. Physiologia Plantarum 80: 642–646. [Google Scholar]
- AkagiH, Yokozeki Y, Inagaki A, Fujimura T.1996. Microsatellite DNA markers for rice chromosomes. Theoretical and Applied Genetics 93: 1071–1077. [DOI] [PubMed] [Google Scholar]
- BoutinSR, Young ND, Lorenzen LL, Shoemaker RC.1995. Marker based pedigrees and graphic genotypes generated by supergene software. Crop Science 35: 1703–1707. [Google Scholar]
- ChenX, Temnykh S, Xu Y, Cho YG, McCouch SR.1997. Development of a microsatellite framework map providing genome‐wide coverage in rice (Oryza sativa L.). Theoretical and Applied Genetics 95: 553–567. [Google Scholar]
- GarlandS, Lewin L, Blakeney A, Reinke R.2000. PCR‐based molecular markers for the fragrance gene in rice (Oryza sativa L.). Theoretical and Applied Genetics 101: 364–371. [Google Scholar]
- HaqueQA, Hille Ris Lambers, D, Tepora NM, dela Cruz QD.1989. Inheritance of submergence tolerance in rice. Euphytica 41: 247–251. [Google Scholar]
- KamolsukyunyongW, Ruanjaichon V, Siangliw M, Kawasaki S, Sasaki T, Vanavichit A, Tragoonrung S.2001. Mapping of quantitative trait locus related to submergence tolerance in rice with aid of chromosome walking. DNA Research8 (4): 163–171. [DOI] [PubMed]
- LancerasJC, Huang Z‐L, Naivikul O, Vanavichit A, Ruanjaichon V, Tragoonrung S.2000. Mapping of genes for cooking and eating qualities in Thai jasmine rice (KDML105). DNA Research 7: 93–101. [DOI] [PubMed] [Google Scholar]
- MackillDJ, Amante MM, Vergara BS, Sarkarung S.1993. Improved semidwarf rice lines with tolerance to submergence of seedings. Crop Science 33: 749–753. [Google Scholar]
- MohantyHK, Khush GS.1985. Diallel analysis of submergence tolerance in rice, Oryza sativa L. Theoretical and Applied Genetics 70: 467–473. [DOI] [PubMed] [Google Scholar]
- NandiS, Subudhi PK, Senadhira D, Manigbas NL, Sen‐Mandi S, Huang N.1997. Mapping QTLs for submergence tolerance in rice by AFLP and selective genotyping. Molecular and General Genetics 255: 1–8. [DOI] [PubMed] [Google Scholar]
- PaladaM, Vergara BS.1972. Environmental effects on the resistance of rice seedlings to complete submergence. Crop Science 12: 209–212. [Google Scholar]
- RogersS.O. and Bendich AJ.1994. Extraction of total cellular DNA from plants, algae and fungi. Plant Molecular Biology Manual DI, 1–8.
- SetterTL, Laureles EV.1996. The beneficial effect of reduced elongation growth on submergence tolerance of rice. Journal of Experimental Botany 47: 1551–1559. [Google Scholar]
- SiangliwM, Toojinda T, Tragoonrung S and Vanavichit A.2002. Mapping genes controlling traits related to submergence tolerance in rice recombinant inbred lines. Rice Genetic IV. Proceedings of the Fourth International Rice Genetics Symposium, 22–27 October 2000, Los Banos, Philippines New Delhi (India): Science Publishers, Inc. and Los Banos (Philippines): International Rice Research Institute (in press).
- SinhaMM, Saran S.1988. Inheritance of submergence tolerance in lowland rice. Oryza 25: 351–354. [Google Scholar]
- SuprihatnoB, Coffman WR.1981. Inheritance of submergence tolerance in rice (Oryza sativa L.). SABRAO Journal 13: 98–108. [Google Scholar]
- TemnykhS, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR.2000. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theoretical and Applied Genetics 100: 697–712. [Google Scholar]
- ToojindaT, Siangliw M, Tragoonrung S, Vanavichit A.2003. Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Annals of Botany 91: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanavichitA, Ruanjaichon V, Techayingpaiboon D, Kamol sukyunyong W, Jamparuang M, Jaturapahu T, Nimlek A, Wachana S, Toojinda T, Tragoonrung S.2001. Sequence rice chromosome 9: platform for understanding structural and functional genomics. Plant and Animal Genome IX. Abstract available from http://www.intl‐pag.org/pag/9/abstracts/P01_08.html.
- XuK, Mackill DJ.1996. A major locus for submergence tolerance mapped on rice chromosome 9. Molecular Breeding 2: 219–224. [Google Scholar]
- XuK, Xu X, Ronald PC, Mackill DJ.2000. A high‐resolution linkage map of the vicinity of the rice submergence tolerance locus Sub1 Molecular and General Genetics 263: 681–689. [DOI] [PubMed] [Google Scholar]
- YanJ, Zhu J, He C, Benmoussa M, Wu P.1999. Molecular marker‐assisted dissection of genotype × environment interaction for plant type traits in rice (Oryza sativa L.). Crop Science 39: 538–544. [Google Scholar]
- YoungND and Tanksley SD.1989. Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theoretical and Applied Genetics 77: 95–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.