Abstract
Genetic variants in UBQLN1 gene have been linked to neurodegeneration and mutations in UBQLN2 have recently been identified as a rare cause of amyotrophic lateral sclerosis (ALS).
Objective
To test if genetic variants in UBQLN1 are involved in ALS.
Methods
102 and 94 unrelated patients with familial and sporadic forms of ALS were screened for UBQLN1 gene mutations. Single nucleotide variants were further screened in a larger set of sporadic ALS (SALS) patients and unrelated control subjects using high-throughput Taqman genotyping; variants were further assessed for novelty using the 1000Genomes and NHLBI databases. In vitro studies tested the effect of UBQLN1 variants on the ubiquitin-proteasome system (UPS).
Results
Only two UBQLN1 coding variants were detected in the familial and sporadic ALS DNA set; one, the missense mutation p.E54D, was identified in a single patient with atypical motor neuron disease consistent with Brown-Vialetto-Van Laere syndrome (BVVLS), for whom c20orf54 mutations had been excluded. Functional studies revealed that UBQLN1E54D protein forms cytosolic aggregates that contain mislocalized TDP-43 and impairs degradation of ubiquitinated proteins through the proteasome.
Conclusions
Genetic variants in UBQLN1 are not commonly associated with ALS. A novel UBQLN1 mutation (E45D) detected in a patient with BVVLS altered nuclear TDP-43 localization in vitro, suggesting that UPS dysfunction may also underlie the pathogenesis of this condition.
Keywords: Amyotrophic lateral sclerosis, Drosophila, motor neuron disease, TDP-43, ubiquilins
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset degenerative motor neuron disease (MND). The incidence of ALS is 1-2 per 100,000, the prevalence is 4-6 per 100,000 and the lifetime ALS risk is approximately 1/600 (McGuire et al. 1996; Mitsumoto et al. 1998). While most ALS cases are sporadic (SALS), approximately 10% are familial (FALS). The most common genetic cause in FALS is the recently defined expansion of a hexanucleotide repeat in the non-coding region of C9orf72 and mutations in the gene encoding cytosolic superoxide dismutase 1 (SOD1), which together may account for at least 50% of FALS cases (Dejesus-Hernandez et al. 2011; Renton et al. 2011; Rosen et al. 1993). Mutations in TAR DNA binding protein (TARDBP) and fused-in-sarcoma/translated-in-liposarcoma (FUS/TLS) genes each account for about 5% of FALS cases (Kabashi et al. 2008; Kwiatkowski et al. 2009; Sreedharan et al. 2008; Vance et al. 2009). Less frequently, mutations in genes encoding vesicle-associated membrane protein-associated protein B (VAPB), senataxin (SETX), dynactin (DCTN1), alsin (ALS2), angionenin (ANG), optineurin (OPTN), valosin-containing protein (VCP) are predicted to cause FALS (Chen et al. 2004; Greenway et al. 2006; Johnson et al 2010a; Maruyama et al. 2010; Münch et al. 2005; Nishimura et al. 2004; Yang et al. 2001). Recently, some cases of X-linked ALS have been ascribed to mutations in UBQLN2 (Deng et al. 2011).
Genetic variants in ubiquilin 1 (UBQLN1) have been linked to neurodegeneration. Two family-based cohorts showed a positive association between an intronic variant (UBQ-8i) and Alzheimer disease, although this association was not consistently confirmed (Bensemain et al 2006; Bertram et al. 2005; Bertram et al. 2007; Brouwers et al. 2006; Slifer et al. 2005; Smemo et al. 2006). The plausibility of this association was reinforced by the observation that UBQLN1 interacts with polyubiquitinated TDP-43 protein through its ubiquitin-associated (UBA) domain and regulates the transport of TDP-43 to both the proteasome and the autophagy systems through an ubiquitin-like (UBL) domain (Heir et al. 2006; Kim et al. 2009; Ko et al. 2004). During our study, mutations in UBQLN2 were identified in 2% of dominant X-linked ALS families and ubiquilin pathology has been observed on autopsy-confirmed ALS without mutations in this gene (Brettschneider et al. 2012; Deng et al. 2011), which reinforces our interest in UBQLN1 as a candidate ALS gene. Here, we report our mutational analysis of the UBQLN1 gene in ALS.
Subjects and Methods
The study was approved by the local ethics committee; participants provided written informed consent. The diagnosis of ALS was based on the revised El Escorial criteria including patients affected with possible, probable or definite ALS (Brooks et al. 2000). SOD1, TARDBP and FUS/TLS mutations were ruled out in all cases; hexanucleotide expansion in the c9orf72 gene was ruled out in ALS patients identified as carriers of UBQLN1 variants.
Genotyping analysis
Three single nucleotide polymorphisms (SNPs) in the UBQLN1 gene (rs2780995, rs 2781002, rs12344615 [UBQ-8i]) were tested for association with risk for ALS in more than 900 SALS cases and healthy controls.
Sequence analysis
102 and 94 unrelated patients with FALS and SALS, respectively, were screened for UBQLN1 gene mutations. Whole genome amplification was performed on blood DNA (Illustra Genomiphi V2 DNA Amplification kit; GE HealthCare cat. #25-6600-31). UBQLN1 exons 1 through 11, and exon-intron junctions, were amplified by PCR (primers available on request). AmpliTaq Gold PCR Master Mix 2500U (Applied Biosystems cat. #4327059) was used for touchdown PCR. The high GC content of exon1 (70%) required addition of 3 μl of a 10X Combinatorial Enhancer Solution (10X CES) with betaine 0.54 M betaine, 1.34 mM DTT, 1.34% DMSO, and 11 μg/ml BSA in 30 μl PCR reaction volume.
DNA samples were resolved by capillary electrophoresis on an ABI 3730XL DNA Analyzer.
High-throughput SNP genotyping was performed using TaqMan assay for each novel variant in a larger set of SALS patients and unrelated control subjects.
Analysis of alternative splicing
Total lymphoblast RNA was extracted and reverse transcribed to complementary DNA (cDNA) using oligodT primers, according to the manufacturer’s instructions (Invitrogen cat#74104, 18080-051). cDNA amplification was carried out with two previously reported specific PCR primers that annealed to exon 7 and exon 9 (Bertram et al. 2005). Two transcripts were extracted from agarose gel (Qiagen cat# 28706) and directly sequenced to confirm their specificity for UBQLN1 sequence.
Cell culture and transfection
HEK293T, Hela-CCL2 and FUS-GFP stable cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Sigma, Poole, UK) supplemented with 10% fetal calf serum (Sigma) at 5% CO2 , 37°C (Bosco et al. 2010). The cDNA of human UBQLN1 was cloned into the PCR-Blunt vector (Invitrogen K2700-20). Site-directed mutagenesis was performed to create the mutant constructs (Stratagene cat#200522-5). Subsequently the wild-type (wt) and mutant UBQLN1 cDNAs were cloned into MoPrP.Xho inserted into the pBluescript vector (Stratagene cat# 212205) at the Xho site (gift from Dr. David Borchelt). Flag-tagged UBQLN1 mutant constructs were derived from pCS2-Flag-UBQLN1 (Addgene, Plasmid 8663). Various plasmids were transiently transfected into HEK293T or Hela-CCL2 cells using Lipofectamin2000 (Invitrogen) following manufacturer’s instructions.
Fly strains
Flies were raised on standard food medium and kept at 25°C. To generate UAS-hUBQLN1, UAS-hUBQLN1-S503C, UAS-hUBQLN1-E54D, UAS-HA-hUBQLN1, UAS-HA-hUBQLN1-S503C and UAS-HA-hUBQLN1-E54D constructs, corresponding DNA fragments were synthesized and cloned into the pUAST vector (GenScript, Inc). These constructs were confirmed by DNA sequencing and microinjected into wildtype (w1118) Drosophila embryos to generate transgenic lines (Rainbow Transgenic Flies, Inc.). Individual transgenic lines were crossed with GMR-Gal4 or other Gal4 drivers for expression and phenotypic analyses.
Western blots and immunoprecipitation
Levels of wildtype (wt) and mutant UBQLN1 and TDP-43 proteins in transfected HEK293T or Hela-CCL2 cells were determined using western immunoblotting according to standard methods (Lu et al. 2009a). For fly protein expression analysis, adult flies were frozen with dry ice and vortexed to remove their heads. Approximately 20 heads from each genotype were homogenized in cold lysis buffer (50 mM Tris-HCl, pH 7.5, 150 Mm sodium chloride, 1% Nondidet P40, 0.5% sodium deoxycholate, 1 tablet complete Mini protein inhibitor cocktail/10 mL). Homogenates were centrifuged at 4°C for 20 min at 13,000 rpm. Protein concentrations were determined using Bradford Assay (Bio-Rad).
Supernatants containing 5 μg of protein were mixed with 4X SDS sample buffer (1 M Tris-HCl, pH 6.8, 8% SDS, 40% glycerol, 0.1% bromophenol blue) and boiled for 5 min. These samples were then separated on a 10% polyacrylamide SDS gel and transferred to a PVDF membrane (Bio-Rad) in a wet transfer system at 4°C for 60 min at 100V. The membrane was incubated in the blocking solution TBST (25 mM Tris-HCl, 137 mM NaCl, 3 mM KCl, pH 7.4, and 0.1% Tween-20) containing 5% milk at room temperature for 1 hr. The membrane was then incubated with anti-dTDP-43 antibody (1:1000 in blocking solution) at room temperature for 3 hr or at 4°C overnight, and finally with anti-rabbit HRP-conjugated secondary antibody (Jackson ImmunoResearch; 1:10,000) for 1 hr (Lu et al 2009b). The signal was visualized with chemiluminescent substrate (Supersignal West Pico, Pierce). For other western blot analyses, the primary antibodies were anti-hTDP-43 antibody (1:2000; Proteintech, 10782-2-AP), anti-ubiquilin antibody (1:2000; Abcam, ab3341), mouse monoclonal anti-actin antibody (1:8,000, Sigma, A2228) and β-actin antibody (1:2000; Cell Signaling, 4967).
Immunostaining
Anti-UBQLN1 antibody (Abcam, ab3341 and ab96870), anti-EGFP antibody (Clontech, 632375) and anti-TDP-43 antibody (Proteintech, 10782-2-AP and 60019-2-Ig) were used as primary antibodies. Goat anti-rabbit Alexa Fluor 568 and goat anti-mouse Alexa Fluor 488 (Invitrogen) were used as secondary antibodies. Cells were washed three times in PBS and mounted with Vectashield (Vector Laboratories Ltd, Peterborough, UK). Immunostaining signals were examined with the IX-70 microscope or a Leica TCS-SP inverted scanning confocal microscope.
UPS reporter assay
HEK293T cell lines were grown in 6-well plates and co-transfected with a UPS-reporter vector encoding UbG76V-GFP (Addgene, 11941), and an expression vector encoding either human UBQLN1wt or UBQLN1E54D/S503C. Cells were harvested 36 hr after transfection and resuspended in PBS. The fluorescence intensities were measured by flow cytometry. All flow-cytometry data were collected and analysed using a MoFlo cell sorter and BD FACSCaliburs. Argon-ion (488 nm) laser were used for excitation. The GFP signals were collected using 530/540-nm band pass filter. In all experiments, data were gated on GFP labelled cells. At least 50,000 such events were recorded in each experiment. Data were collected from three independent experiments.
Statistic analyses
Pearson’s chi-squared test, Fisher’s exact test and two-tailed unpaired Student’s t-test were performed considering p < 0.05 as significant value. Association analyses were based on allele-based comparisons of ALS cases vs. healthy controls using a chi-square statistic and computing allelic odds ratios and 95% confidence intervals.
Results
Genetic studies
No significant association was detected for any of the three UBQLN1 SNPs previously associated with risk for Alzheimer disease (Supplementary Table 1). To further evaluate a role for UBQLN1 in ALS, we sequenced all coding exons of UBQLN1 in 102 FALS and 92 SALS cases. We identified four variants by direct sequencing: two missense SNPs in exon 1 (c.162 G>T, p.E54D) and exon 10 (c.1508 C>G, p.S503C), a silent SNP in exon 8 (c.1685 C>T, p.L442L), and a non-coding change located 24bp upstream of exon 10 (i10:-24 A>T).
For three of these variants (E54D, S503C, L442L), we next performed a TaqMan assay in 1,474 SALS patients and 1,378 unrelated control subjects. The non-coding change (i10:-24 A>T) was detected by TaqMan analysis in one of the first 380 controls and was thus considered to be a benign polymorphism. Neither of the two missense variants was detected in our set of 1,378 controls. The p.S503C variant was identified in an additional patient with definite SALS; it was not list in the 1000Genomes database (http://www.1000genomes.org/home) but was present in dbSNP (rs151070080) and in the NHLBI exome database (http://evs.gs.washington.edu/EVS/); we therefore conclude that this is a rare polymorphism. The p.E54D variant was not detected in the dbSNP, 1000Genomes or NHLBI databases and thus is potentially disease-specific (Table 1).
Table 1.
Results of UBQLN1 mutational screening
| Variant | Sequencing | TaqMan Assay | Total Cases | Total Alleles | Control Allele | Chi-square Test ((2)) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon | SNP | AA | FALS | SALS | SALS | control | FALS/SALS | FALS/SALS | Frequency ((1)) | P | OR | 95% CI |
| 1 | c.162 G>T | E54D | 1/102 | 0/92 | 0/1474 | 0/1378 | 1/102 | 1/204 | 0/2,756a | 0.0002 | 40.64 | 1.649-1001 |
| 8 | c.1685 C>T | L442L | 0/102 | 1/92 | 0/1474 | 0/1378 | 1/1566 | 1/3,132 | 21/4,940a,b | 0.001 | 0.07 | 0.010-0.557 |
| 10 | c.1508 C>G | S503C | 0/102 | 1/92 | 1/1474 | 0/1378 | 2/1566 | 2/3,132 | 6/8398a,c | 0.8905 | 0.89 | 0.180-4.432 |
| intron | ilO:-24 A>T | no | 0/102 | 1/92 | no | 1/380 | 1/92 | 1/184 | 1/760a | 0.2756 | 4.15 | 0.258-66.67 |
Control frequency includes data from
Taqman assay
1,000 Genome database
NHLBI exome server, European-American population
Total alleles in FALS or SALS cases compared to control allele frequency
The E54D mutation was present in a heterozygous form in an Italian female patient diagnosed with an early-onset motor neuron disease characterized by bulbar palsy. There were no affected relatives or consanguinity in the family. After ruling out other disease processes that might explain the signs of motor neuron degeneration, this patient was diagnosed with clinically possible ALS according to El Escorial criteria (Brooks et al. 2000). Subsequent clinical examinations revealed the presence of sensorineuronal hearing loss that, together with the early onset of the motor neuron symptoms, suggested the diagnosis of Brown-Vialetto-Van Laere syndrome (BVVLS) (Mégarbané et al. 2000). We therefore excluded mutations in all five exons of the c20orf54 gene, which reportedly cause some (but not all) cases of BVVLS (Green et al. 2010; Johnson et al. 2010b). These data suggest that the UBQLN1 E54D mutation is causal. Because the glutamate residue at position 54 is highly conserved, one suspects that its mutation to aspartate is functionally significant, although PMut (http://mmb2.pcb.ub.es:8080/PMut/), PolyPhen (http://genetics.bwh.harvard.edu/pph/) and SIFT ( http://sift.jcvi.org) predicted that suggest this change is not likely to be functionally deleterious.
Lastly, L442L was identified in a male patient diagnosed with definite SALS. This C-to-T nucleotide change, which is located 7 bp upstream of the 3′ end of exon 8, was well preserved across species. Statistically, this variant is under-represented in ALS cases (Table 1B); however, its biological significance is unclear. We note that there normally are two spliced variants of UBQLN1: TV1 (entire coding region) and TV2 (excludes exon 8). We did not detect an impact of this variant on the alternative splicing of exon 8 (see Methods, data not shown).
Functional studies
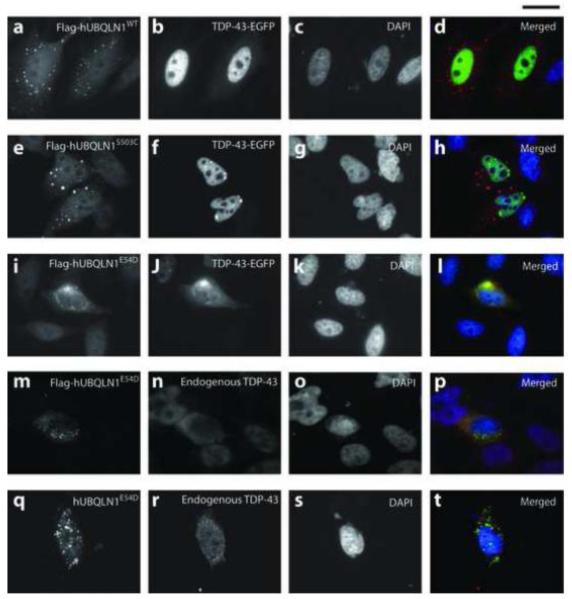
We next studied the functional significance of E54D variant since it was novel, evolutionarily conserved, and located within UBL domain, and interestingly was identified in a BVVLS patient of unknown genetic cause. S503C was also tested in parallel although its presence in controls predicted a wild type response. We first investigated the effect of wt and mutant hUBQLN1 on the TDP-43 distribution (Fig 1). When co-expressed with Flag-hUBQLN1E54D, hTDP-43-EGFP was mislocalized from the nucleus to the cytoplasm in 30% of the co-transfected cells, whereas this phenotype was observed in only 9.5% and 12% of cells when co-expressing Flag-hUBQLN1WT and Flag-hUBQLN1S503C, respectively. Furthermore, TDP-43 and UBQLN1 protein were clearly identified as co-stained cytoplasmic aggregates when co-expressed with Flag-hUBQLN1E54D but not when co-expressed with UBQLN1WT (Fig 1-l). Similar cellular phenotypes were obtained when testing the effect of tagged and untagged UBQLN1WT, UBQLN1E54D, and UBQLN1S503C constructs on endogenous TDP-43 (Fig 1 m-t, Supplementary Figure 1 and 2). Although some cytosolic distribution of endogenous TDP-43 was observed when expressing UBQLN1WT and UBQLN1S503C, no aggregates were identified in any of these cases. It thus might be argued that the finding of cytosolic TDP-43 aggregation induced by UBQLN1E54D is a consequence of high levels of UBQLN1 expression rather than the missense mutation. However, we note that western immunoblotting showed approximately equivalent expression of hUBQLN1 or Flag-hUBQLN1 in wt and mutant forms (Fig 2). These data strongly support the view that the mislocalization and aggregation effects of TDP-43 in cells transfected with UBQLN1E54D is a consequence of the E54D mutation and not due to high levels of UBQLN1 expression.
Fig.1. hUBQLN1E54D perturbs the distribution of TDP-43 protein in HEK293 cells.
Co-expression of Flag-hUBQLN1E54D and TDP-43-EGFP redistributes TDP-43 to the cytoplasm of HEK293 cells, where TDP-43 co-localizes with mutant UBQLN1E54D, primarily in aggregate forms (i-l). This phenotype was not observed with UBQLN1WT (a-d) or UBQLN1S503C (e-h), although a loss of nuclear TDP-43 was observed in the latter case. Flag-hUBQLN1E54D and hUBQLN1E54D reproduced the results seen with UBQLN1E54D (m-t).
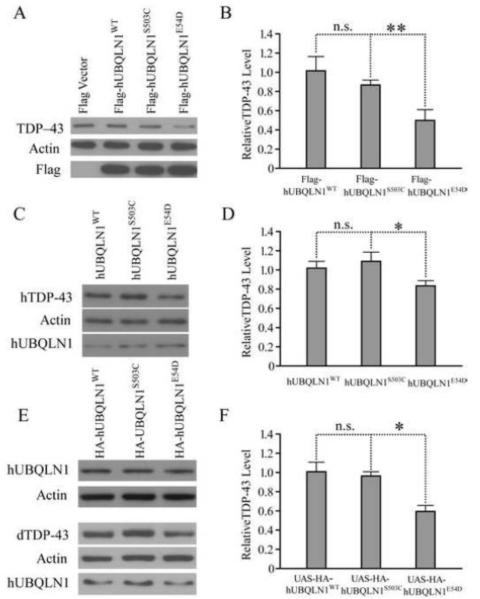
Fig. 2. hUBQLN1E54D reduces the level of endogenous TDP-43 in both HEK293 cells and D. melanogaster.
HEK293T cells were transfected with tagged (Fig 2A and B) and untagged (Fig 2C and D) human UBQLN1WT, UBQLN1S503C and UBQLNE54D constructs. Endogenous TDP-43 levels were significantly reduced in cells transfected with PrP-UBQLN1E54D and Flag-UBQLN1E54D when compared to hUBQLN1WT; transfection with hUBQLN1S503C did not produce this effect. The same effect was also evident after over-expression of UAS-HA-hUBQLN1WT, UAS-HA-hUBQLN1S503C and UAS-HA-hUBQLN1E54D in the eye of D. melanogaster (Fig.2 E and F).
As a further approach, we quantified the endogenous levels of the soluble fraction of TDP 43 protein when over-expressing untagged and tagged UBQLN1 constructs in both HEK293T cells and D. melanogaster (Fig 2). In these experiments, forced expression of Flag-hUBQLN1E54D in HEK293T cells significantly reduces soluble TDP-43 levels (Fig 2A and B). To exclude the possibility that this effect is due to over-expression of the mutant protein or adverse effects of the Flag tag, we also expressed non-tagged hUBQLN1WT, hUBQLN1S503C, or hUBQLN1E54D whose expression is driven by the weaker PrP promoter. A reduction of endogenous TDP-43 was also observed in HEK293T cells expressing hUBQLN1E54D (Fig 2C and D). To determine if this in vitro effect of hUBQLN1E54D is evident in vivo, we generated multiple UAS-transgenic fly lines that express hUBQLN1WT, hUBQLN1S503C, or hUBQLN1E54D under specific Gal4 drivers, such as eye-specific GMR-Gal4. We first selected transgenic lines that express these proteins at comparable levels, such that their biological effects can be compared (Fig 2E). We found that expression of hUBQLN1E54D in the eye of D. melanogaster also led to a reduction in the level of endogenous dTDP-43 (Fig 2E and F) although no obvious morphological phenotype was observed. By contrast, no significant differences were found when hUBQLN1S503C was over-expressed.
In a related functional study, we also assessed the impact of UBQLN1 mutants on the degradation of ubiquitinated proteins through the proteasome using a GFP-tagged, mutant ubiquitin (UbG76V-GFP) as a reporter, as previously reported (Dantuma et al. 2000). Expression of UBQLN1E54D but not UBQLN1S503C slightly enhanced accumulation of UbG76V-GFP, as compared to UBQLN1wt (Supplementary Fig 3). This change was statistically significant but modest and thus is of uncertain biological significance.
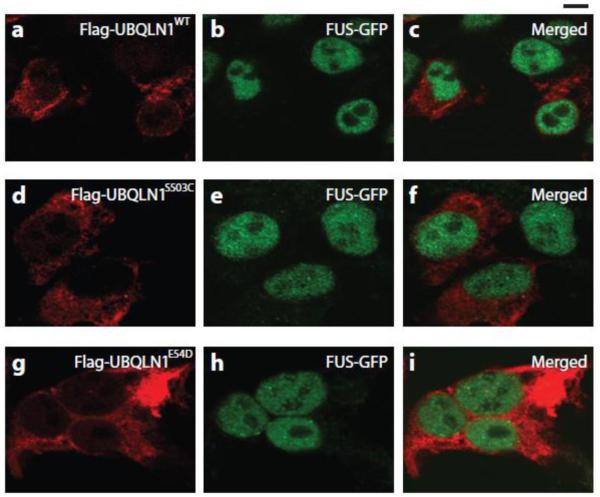
Finally, because TDP-43 and FUS/TLS have similar domain structures, are involved in RNA binding and transport, and are both implicated in FALS and SALS, we tested the possibility that co-expression of UBQLN1wt, UBQLN1S503C and UBQLN1E54D with GFP-WT-FUS would cause cytosolic retention of FUS/TLS, as it did with TDP-43 (Ito et al. 2011). This was not observed when UBQLN1E54D was co-expressed with GFP-WT-FUS (Fig 3).
Fig. 3. UBQLN1 mutants did not influence FUS distribution in HEK293 cells.
Nuclear distribution of FUS protein was not affected when co-expressing Flag-hUBQLN1E54D/S503C/WT and FUS-GFP.
Discussion
Despite the absence of association between any of the three UBQLN1 SNPs and ALS, and despite the fact that UBQLN1 lacks the PXX domain that is mutated in UBQLN2 in ALS, we screened for UBQLN1 mutations in an ALS cohort for two reasons. UBQLN1 interacts directly with polyubiquitinated TDP-43 (Kim et al. 2009; Ko et al. 2004), the main component of cytoplasmic inclusions in ALS (Neumann et al. 2006). And, there is distinct ubiquilin pathology in autopsy-confirmed ALS cases without mutations in UBQLN2 (Deng et al. 2011), raising the possibility that there may be mutations in other ubiquilin genes.
By direct sequencing of the UBQLN1 gene, we have not detected novel UBQLN1 gene variants in ALS. However, we have identified one novel coding variant (E54D) in a juvenile-onset form of MND (BVVLS). This variant was not detected in 1,378 unrelated control subjects or in three large databases of DNA variants. We conclude that polymorphisms in UBQLN1 (a PXX domain-lacking ubiquilin) do not commonly contribute to risk for ALS, while rare variants may contribute to the pathogenesis of atypical motor neuron disease.
We recognize that the E54D variant may be a rare polymorphism. Indeed, it is striking that apparently normal individuals typically harbour ~200 novel SNPs per exome (30 Mb) (http://useast.ensembl.org/index.html). Given that the coding sequence for the major UBQLN1 protein is 589 amino acids, encoded by a cDNA domain of 1,767 bp (Ensemble entry ENST00000376395), one would estimate that the number of novel variants in UBQLN1 in each individual is 200 × 1,767/30×106, or 1.2×10−2. Since we performed full mutational screening on 196 individuals, we would therefore expect 196 × 1.2×10−2 = 2.3 (i.e. 2 or 3) novel SNPs from the analysis.
These considerations prompt caution in interpreting the E54D variant as causal in BVVL. We therefore undertook studies to define potential adverse functional effects of this variant. The ubiquilin proteins (1-4) function as adaptor proteins that chaperone cargos to proteasomes and autophagosomes. Chaperone partners include TDP-43, presenilin-1, huntingtin and amyloid precursor protein (Hiltunen et al. 2006; Kim et al. 2009; Stieren et al. 2011; Thomas et al. 2006; Wang et al. 2007). Essential for the linking of adaptor protein–partner complexes to the proteosome is the ubiquitin-like domain (UBL) in the N-terminus of UBQLN1. It is therefore interesting that the predicted glutamate-to-aspartate amino acid change at position 54 involves a residue that is highly conserved across species within this UBL domain. We were intrigued to find that UBQLN1E45D induces cytosolic mislocalization of TDP-43, primarily in aggregate forms that co-localize with the UBQLN1 protein. While it is possible that cytosolic retention of TDP-43 is a non-specific reaction to stress induced by UBQLN1E45D, we note that there is clear documentation that UBQLN1 can directly interact with TDP-43 both in yeast and in vitro (Kim et al. 2009).
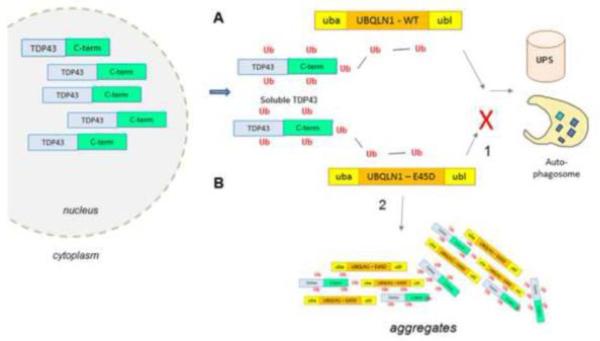
Our data suggest that the UBQLN1E45D variant may weakly, but significantly, inhibit ubiquitin degradation, as assayed using a UbG76V-GFP reporter. The weakness of effect could explain the absence of phenotype in transgenic flies when hUBQLN1E54D is expressed in the eye and neurons of D. Melanogaster, whereas the ubiquitous overexpression of both mutant and wt forms prevented flies from developing into the adult stage (data not shown). We hypothesize that one of two pathogenic events arises: (1) this UBQLN1E54D mutant in the UBL domain prevents normal interactions with proteasome and autophagosomes, leading to secondary aggregation with binding partners such as TDP-43. (2) Alternatively, the mutations may enhance aggregation of UBQLN1 with TDP-43, secondarily impairing trafficking to proteasomes and autophagosomes (Fig 4).
Fig. 4. Speculative model of the effect of UBQLN1E54D on TDP-43.
The E54D mutation in UBQL-1 is presumed to prevent the UBL domain from interacting with the proteasome or autophagosomes. Consequently, polyubiquitinated proteins that bind UBQL1, like TDP-43, are not degraded and accumulate in cytoplasm.
The presence of ubiquilin protein in aggregates in autopsies from ALS patients is consistent with the view that perturbations in protein turnover are important elements in ALS pathogenesis and with the finding that UBQLN2 mutations are infrequently detected in ALS cases (Deng et al. 2011; Millecamps et al. 2012), even though, as we report here, genetic variations in UBQLN1 are not commonly associated with ALS. The concept that protein aggregation and distressed protein turnover is critical in neurodegeneration is firmly supported by an immense body of experimental data both in humans and in animal and cellular models. Potentially of particular importance is the interplay between the ubiquilins and RNA-binding proteins like TDP-43, given recent discoveries of causative ALS genes that implicate disruptions of RNA metabolism as the fundamental agent in ALS pathogenesis, at least in a subset of cases.
Supplementary Material
Highlights.
Genetic variants in UBQLN1 are not associated with amyotrophic lateral sclerosis.
Novel UBQLN1 mutation (E54D) in a patient with atypical motor neuron disease.
Dysfunction in the UPS may contribute to the pathogenesis of BVVLS.
Acknowledgments
Study funding: This study was supported by the National Institutes of Health (5RO1NS050557 to R.H.B. and 5RO1NS057553 to F.-B.G.), the Angel Fund, the ALS Association, P2ALS, Pierre L. de Bourgknecht ALS Research Foundation, the ALS Therapy Alliance, and the Alfonso Martin Escudero Foundation (Madrid) (R.H.B.), and UMMS Startup Fund (F.-B.G.). PS was supported through the auspices of Dr. H. Robert Horvitz, an Investigator at the Howard Hughes Medical Institute in the Department of Biology at the Massachusetts Institute of Technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors and their individual contribution to the manuscript: Paloma González-Pérez: genotyping, mutational analysis, analysis and interpretation of the data, drafting manuscript.
Yubing Lu: functional studies of wildtype and mutant UBQLN1, analysis and interpretation of the data, manuscript revision.
Ru-Ju Chian: cloning of WT and mutant UBQLN1, manuscript revision.
Peter Sapp: genotyping, manuscript revision.
Diane Mckenna-Yasek: collection of clinical information, manuscript revision.
Rudolph E. Tanzi: conceptualization of the study, manuscript revision.
Lars Bertram: conceptualization of the study, statistical analyses, manuscript revision.
Fen-Biao Gao: design of experiments in Drosophila and cell cultures, interpretation of the data, drafting and revision of the manuscript.
Robert H. Brown: design, conceptualization of the study, interpretation of the data, drafting and manuscript revision.
Disclosure of all authors’ financial relationships R.H.B. is a co-founder of AviTx, which targets development of therapies.
REFERENCES
- Bensemain F, Chapuis J, Tian J, Shi J, Thaker U, Lendon C, et al. Association study of the Ubiquilin gene with Alzheimer’s disease. Neurobiol Dis. 2006;22(3):691–3. doi: 10.1016/j.nbd.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, Ramasamy K, et al. Family-based association between Alzheimer’s disease and variants in UBQLN1. N Engl J Med. 2005;352(9):884–94. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19(21):4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J, Van Deerlin VM, Robinson JL, Kwong L, Lee EB, Ali YO, et al. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol. 2012 Mar 18; doi: 10.1007/s00401-012-0970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Miller RG, Swash M, Munsat TL, World Federation of Neurology Research Group on Motor Neuron Diseases El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(5):293–9. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- Brouwers N, Sleegers K, Engelborghs S, Bogaerts V, Serneels S, Kamalik, et al. The UBQLN1 polymorphism, UBQ-8i, at 9q22 is not associated with Alzheimer’s disease with onset before 70 years. Neurosci Lett. 2006;392(1-2):72–4. doi: 10.1016/j.neulet.2005.08.064. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74(6):1128–35. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quatifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18(5):538–43. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- Dejesús-Hernández M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of c9orf72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–15. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P, Wiseman M, Crow YJ, et al. Brown-Vialetto-Van Laere syndrome, a ponto-bulbar palsy with deafness, is caused by mutations in c20orf54. Am J Hum Genet. 2010;86(3):485–9. doi: 10.1016/j.ajhg.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38(4):411–13. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- Heir R, Ablasou C, Dumontier E, Elliot M, Fagotto-Kaufmann C, Bedford FK. The UBL domain of PLIC-I regulates aggregosome formation. EMBO Rep. 2006;7(12):1252–8. doi: 10.1038/sj.embor.7400823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen M, Lu A, Thomas AV, Romano DM, Kim M, Jones PB, et al. Ubiquilin 1 modulates amyloid precursor protein trafficking and A-Beta secretion. J Biol Chem. 2006;281(43):32240–53. doi: 10.1074/jbc.M603106200. [DOI] [PubMed] [Google Scholar]
- Ito D, Suzuki N. Conjoint pathologic cascades mediated by ALS/FTD-U linked RNA-binding proteins TDP-43 and FUS. Neurology. 2011;77(17):1636–43. doi: 10.1212/WNL.0b013e3182343365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, et al. Exome Sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010a;68(5):857–64. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Gibbs JR, Van Maldergem L, Houlden H, Singleton AB. Exome sequencing in Brown-Vialetto-van Laere syndrome. Am J Hum Genet. 2010b;87(4):567–9. doi: 10.1016/j.ajhg.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PM, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40(5):572–4. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kim SH, Shi Y, Hanson KA, Williams LM, Sakasai R, Bowler MJ, et al. Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, Ubiquilin 1. J Biol Chem. 2009;284(12):8083–92. doi: 10.1074/jbc.M808064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566(1-3):110–4. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lu Y, Wang F, Li Y, Ferris J, Lee JA, Gao FB. The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum Mol Genet. 2009a;18(3):454–62. doi: 10.1093/hmg/ddn373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ferris J, Gao FB. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol Brain. 2009b;2:30. doi: 10.1186/1756-6606-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465(7295):223–6. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- McGuire V, Longstreth WT, Jr, Koepsell TD, van Belle G. Incidence of amyotrophic lateral sclerosis in three countries in western Washington state. Neurology. 1996;47(2):571–3. doi: 10.1212/wnl.47.2.571. [DOI] [PubMed] [Google Scholar]
- Mégarbané A, Desguerres I, Rizkallah E, Delague V, Nabbout R, Barois A, et al. Brown-Vialetto-van Laere syndrome in a large inbred Lebanese family: Confirmation of autosomal recessive inheritance? Am J Med Genet. 2000;92(2):117–21. doi: 10.1002/(sici)1096-8628(20000515)92:2<117::aid-ajmg7>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Millecamps S, Corcia P, Cazeneuve C, Boillee S, Seilhean D, Danel-Brunaud V, et al. Mutations in UBQLN2 are rare in French amyotrophic lateral sclerosis. Neurobiol Aging. 2012 Apr 33;(4):839, e1–3. doi: 10.1016/j.neurobiolaging.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Mitsumoto H, Chad DA, Pioro EP. Amyotrophic Lateral sclerosis. Oxford Univ. Press; New York: 1998. [Google Scholar]
- Münch C, Rosenbohm A, Sperfeld AD, Uttner I, Reske S, Krause BJ, et al. Heterozygous R1101K mutation of the DCTN1 gene in family with ALS and FTD. Ann Neurol. 2005;58(5):777–80. doi: 10.1002/ana.20631. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75(5):822–31. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in c9orf72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentah A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Slifer MA, Martin ER, Haines JL, Pericak-Vance MA. The ubiquilin 1 gene and Alzheimer’s disease. N Engl J Med. 2005;352(26):2752–3. doi: 10.1056/NEJM200506303522618. [DOI] [PubMed] [Google Scholar]
- Smemo S, Nowotny P, Hinrichs AL, Kauwe JS, Cherny S, Erickson K, et al. Ubiquilin 1 polymorphisms are not associated with late-onset Alzheimer’s disease. Ann Neurol. 2006;59(1):21–6. doi: 10.1002/ana.20673. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, et al. TDP-43 mutations in familial ad sporadic Amyotrophic lateral Sclerosis. Science. 2008;319(5870):1668–72. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieren ES, El Ayadi A, Xiao Y, Siller E, Landsverk ML, Oberhauser AF, et al. Ubiquilin 1 is a molecular chaperone for the amyloid precursor protein. J Biol Chem. 2011;286(41):35689–98. doi: 10.1074/jbc.M111.243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AV, Herl L, Spoelgen R, Hiltunen M, Jones PB, Tanzi RE, et al. Interaction between presenilin1 and ubiquilin 1 as detected by fluorescence lifetime imaging microscopy and a high-throughput fluorescent plate reader. J Biol Chem. 2006;281(36):26400–7. doi: 10.1074/jbc.M601085200. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360(2):423–7. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29(2):160–5. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.