Abstract
Previous studies have emphasized ethnically heterogeneous human leukocyte antigen (HLA) classical allele associations to rheumatoid arthritis (RA) risk. We fine-mapped RA risk alleles within the major histocompatibility complex (MHC) in 2782 seropositive RA cases and 4315 controls of Asian descent. We applied imputation to determine genotypes for eight class I and II HLA genes to Asian populations for the first time using a newly constructed pan-Asian reference panel. First, we empirically measured high imputation accuracy in Asian samples. Then we observed the most significant association in HLA-DRβ1 at amino acid position 13, located outside the classical shared epitope (Pomnibus = 6.9 × 10−135). The individual residues at position 13 have relative effects that are consistent with published effects in European populations (His > Phe > Arg > Tyr ≅ Gly > Ser)—but the observed effects in Asians are generally smaller. Applying stepwise conditional analysis, we identified additional independent associations at positions 57 (conditional Pomnibus = 2.2 × 10−33) and 74 (conditional Pomnibus = 1.1 × 10−8). Outside of HLA-DRβ1, we observed independent effects for amino acid polymorphisms within HLA-B (Asp9, conditional P = 3.8 × 10−6) and HLA-DPβ1 (Phe9, conditional P = 3.0 × 10−5) concordant with European populations. Our trans-ethnic HLA fine-mapping study reveals that (i) a common set of amino acid residues confer shared effects in European and Asian populations and (ii) these same effects can explain ethnically heterogeneous classical allelic associations (e.g. HLA-DRB1*09:01) due to allele frequency differences between populations. Our study illustrates the value of high-resolution imputation for fine-mapping causal variants in the MHC.
INTRODUCTION
Rheumatoid arthritis (RA) is a chronic autoimmune disease characterized by a symmetric polyarticular inflammatory arthritis, which affects up to 1% of the population worldwide (1). The majority of affected RA cases (∼70%) are seropositive for anti-citrullinated protein antibodies (ACPA), a highly specific biomarker of RA related to disease severity (2). The major histocompatibility complex (MHC) region at chromosome 6p21.3 contributes substantially to the heritability of ACPA-positive RA (3–5). Indeed, many reports have implicated consensus amino acid sequences spanning positions 70–74 in the human leukocyte antigen (HLA)-DRβ1 subunit (6) for conferring RA risk, suggesting a critical role for these so-called ‘shared epitope’ (SE) alleles in the etiology of RA (4,5,7–9). Previous studies assessing the role of the MHC in modulating RA risk demonstrated both shared and distinct features between Asians and Europeans. Even though classical SE alleles of HLA-DRB1 have been reported to confer strong risks in both continental populations (5–7), heterogeneity of effect sizes (7) and the population-specific associations of non-SE alleles in HLA-DRB1 (e.g. HLA-DRB1*09:01 risk in the Asian populations) (10,11) has made it challenging to draw definitive conclusions about the role of HLA-DRB1 in RA susceptibility.
Recently, we fine-mapped RA risk in European populations within the MHC region to three amino acid positions in HLA-DRβ1 (at positions 11 or 13, 71 and 74) and single amino acid positions in HLA-B (at position 9) and HLA-DPβ1 (at position 9) (12). All these amino acid polymorphisms are located in peptide-binding grooves of HLA molecules, suggesting a critical role for antigen binding and presentation. It is not yet known specifically whether the same alleles at position 13 or at other sites explain RA risk in non-European populations. Here, we explored in detail the possibility that our HLA amino acid variant risk model established in European populations might also explain RA risk in Asian populations.
To this end, we analyzed genetic variation in 2782 ACPA-positive RA cases and 4315 controls from China and South Korea, with each subject densely genotyped across the MHC. In order to apply imputation methods, we newly created a high-density reference panel including genotyped classical four-digit alleles of eight class I and II HLA genes in Asians. With this reference panel, we imputed sequence variation in classical HLA genes, fine-mapped the MHC association for RA risk, and compared results with previous fine-mapping findings in European populations.
RESULTS
Construction and evaluation of a pan-Asian reference panel for imputation of HLA variants
We constructed a pan-Asian reference panel with high-density SNP genotypes and four-digit classical HLA allele genotypes (n = 530; Supplementary Material, Table S1). The newly constructed Asian reference panel consists of three datasets: (i) the Singapore Chinese population (n = 91) (13); (ii) pan-Asian datasets including 111 Chinese, 119 Indian and 120 Malaysian subjects (n = 350) (13) and (iii) HapMap Phase II Japanese and Han Chinese (JPT + CHB) populations (n = 89) (14). Four-digit classical typing data for class I HLA genes (HLA-A, HLA-B and HLA-C) and class II genes (HLA-DRB1, HLA-DQA1 and HLA-DQB1) were available for all three datasets. We had access to four-digit classical typing data for HLA-DPA1 and HLA-DPB1 for datasets (i) and (ii), but not for (iii).
To evaluate the imputation accuracy of this pan-Asian reference panel, we excluded the HapMap JPT + CHB samples from the panel to avoid sample overlap, and subsequently compared imputed and genotyped classical alleles of the six HLA genes (HLA-A, B, C, DRB1, DQA1 and DQB1) in these 89 subjects (Table 1). The imputations based on this pan-Asian reference panel (n = 441; not including the HapMap JPT + CHB subjects used for validation) achieved 95.1% of genotype concordance for HLA alleles at two-digit resolution and 82.4% genotype concordance at four-digit resolution (Table 1). As reported previously, alleles with high frequencies (f ≥ 0.025) showed better correlations between imputed and genotyped dosages (average correlation coefficient = 0.85; Supplementary Material, Fig. S1A). These results are comparable with our previous assessments of HLA variant imputation (12,15,16), and we thus considered this approach to be suitable for downstream association analysis.
Table 1.
Concordance of genotyped and imputed HLA alleles in HapMap Asian populations
| Imputation reference panel | Allele | Concordance of genotyped and imputed HLA alleles in HapMap JPT+CHB (n = 89) |
||||||
|---|---|---|---|---|---|---|---|---|
| HLA-A | HLA-B | HLA-C | HLA-DRB1 | HLA-DQA1 | HLA-DQB1 | 6 HLA genes | ||
| Asian reference panel | Two-digit | 0.989 | 0.870 | 1.000 | 0.904 | 0.949 | 0.994 | 0.951 |
| (n = 441)a | Four-digit | 0.751 | 0.722 | 0.932 | 0.762 | 0.903 | 0.874 | 0.824 |
| European reference panel | Two-digit | 0.747 | 0.429 | 0.588 | 0.494 | 0.624 | 0.871 | 0.625 |
| (HapMap CEU, n = 120) | Four-digit | 0.644 | 0.352 | 0.480 | 0.280 | 0.545 | 0.366 | 0.445 |
| European reference panel | Two-digit | 0.972 | 0.894 | 0.966 | 0.826 | 0.944 | 0.916 | 0.920 |
| (T1DGC consortium, n = 5225) | Four-digit | 0.819 | 0.835 | 0.898 | 0.685 | 0.892 | 0.880 | 0.835 |
| Asian and European reference panel | Two-digit | 0.994 | 0.898 | 1.000 | 0.890 | 0.978 | 0.921 | 0.947 |
| (n = 5666; T1DGC for European)a | Four-digit | 0.864 | 0.841 | 0.932 | 0.744 | 0.972 | 0.880 | 0.872 |
The highest concordance rates for each HLA gene are indicated in bold, separately for two- and four-digit alleles.
aThe subjects used for validation (HapMap JPT + CHB) were excluded.
To compare the imputation performance of this new Asian HLA reference panel to our previous HLA panels (14,16), we also constructed three additional reference panels including European subjects (Supplementary Material, Table S1). These were (i) HapMap Europeans (CEU founders; n = 120) (14), (ii) unrelated European subjects from Type 1 Diabetes Genetics Consortium (T1DGC; n = 5,225) (16,17) and (iii) multiethnic panel combining the T1DGC European subjects and the pan-Asian panel described above (i and ii; n = 5225 + 441 = 5666).
When we used the small reference panel of HapMap CEU founders (n = 120), the imputation performance was limited (44.5% genotype concordance for four-digit alleles and average correlation coefficient = 0.49 for high-frequency alleles (f ≥ 0.025); Table 1; Supplementary Material, Figure S1B). In contrast, the large-scale reference panel from the T1DGC (n = 5225) yielded much better accuracy (83.5% genotype concordance for four-digit alleles and average correlation coefficient = 0.880 for high-frequency alleles), though slightly worse than the accuracy of the Asian-only reference panel (Table 1; Supplementary Material, Fig. S1C). The combined reference panel of Asians and Europeans (n = 5666) demonstrated comparable or better imputation accuracy than the Asian or European panel alone (87.2% genotype concordance for four-digit alleles and average correlation coefficient = 0.91 for high-frequency alleles; Table 1; Supplementary Material, Fig. S1D). The Asian reference panel showed modest imputation performance for four-digit alleles of HLA-A and HLA-B (<75.1%), whereas the combined Asian and European panel yielded improved concordance rates (>84.1%). We note that improvement in imputation accuracy was not consistent for all HLA genes; the combined Asian and European panel showed slightly lower accuracy for HLA-DRB1 alleles (76.2% genotype concordance for four-digit alleles for the Asian-only panel and 74.4% for the combined panel).
Risk of HLA-DRB1 variants in the Asian RA samples
Having demonstrated the accuracy of our imputation protocol, we imputed HLA alleles and tested them for association in two RA GWAS and one RA Immunochip datasets of Asian ancestries (a GWAS including 466 ACPA-positive RA cases and 873 controls from China (18); a GWAS including 799 ACPA-positive RA cases and 751 controls from South Korea (19); an Immunochip study including 1517 ACPA-positive RA cases and 2691 controls from South Korea (20); in total, 2782 ACPA-positive RA cases and 4315 controls). We adopted the pan-Asian imputation reference panel due to its reliable imputation accuracy on HLA-DRB1 alleles and similar genetic backgrounds to the subjects in the Asian RA GWAS, while we note that the following association analysis results did not change substantially when we adopted the large-scaled European reference panel or the combined panel of Europeans and Asians. We incorporated top five principal components (PCs) for each cohort as covariates in the logistic regression model to correct for population stratification. After adjustment with PCs, we did not observe apparent inflation of test statistics genome wide (λGC < 1.05).
We then tested two- and four-digit classical HLA alleles, amino acid polymorphisms, and SNPs within the MHC for association, and observed the top association signal at HLA-DRB1 (Fig. 1A). After conditioning on all classical HLA-DRB1 alleles, no other variants were significantly associated with RA risk with a genome-wide significance threshold (conditioned P > 5.0 × 10−8; Fig. 1B).
Figure 1.
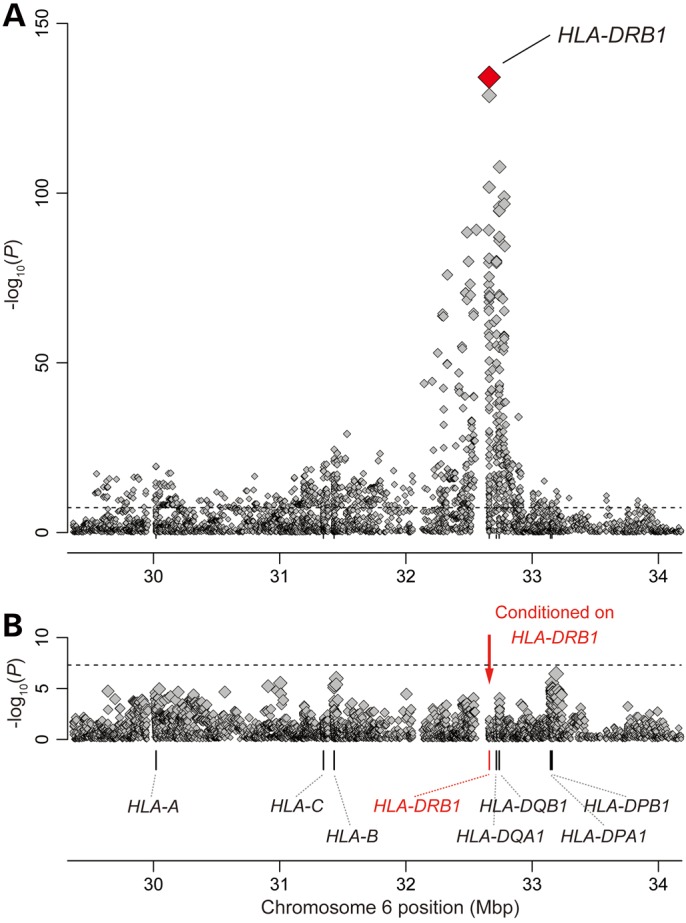
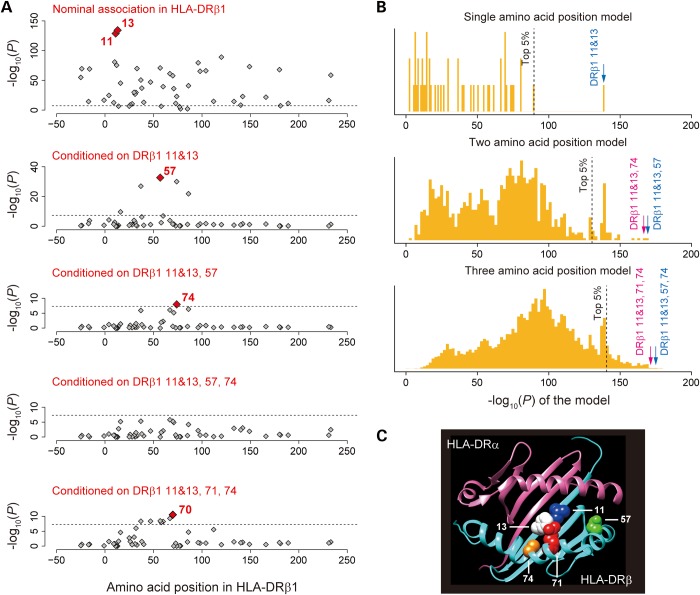
Association plots of the tested variants in the MHC region to ACPA-positive RA in Asians. Each diamond represents −log10(P) of the variants, including the SNPs, the classical HLA alleles and the amino acid polymorphisms of the HLA genes. The dotted horizontal line represents the significance threshold of P = 5.0 × 10−8. The bottom panel showed the physical positions of the HLA genes on chromosome 6 (NCBI Build 36). (A) Nominal associations in the RA GWAS of Asians, in which HLA-DRβ1 amino acid polymorphisms showed the most significant associations. (B) Conditional results on HLA-DRB1, in which no variants showed significant associations.
The most significant association across all variants tested was observed at amino acid position 13 of HLA-DRβ1 (Pomnibus = 6.9 × 10−135) followed by position 11 of HLA-DRβ1 (Pomnibus = 1.7 × 10−129), which is in tight linkage disequilibrium (LD) with position 13. Associations for amino acid positions 70–74 (which define the SE) were considerably weaker (Fig. 2A, Pomnibus > 1.6 × 10−58). Of the amino acid residues at positions 11 and 13, His13 and Val11 showed the strongest RA risk [odds ratio (OR) = 2.03 for His13 and OR = 2.16 for Val11; Supplementary Material, Table S3]. These results are consistent with our previous results in Europeans, which demonstrated the most significant RA risk at position 11 of HLA-DRβ1, notably Val11 (12).
Figure 2.
Association results for the amino acid polymorphisms of HLA-DRβ1. (A) Associations of the tested amino acid polymorphisms of HLA-DRβ1. Each diamond represents −log10(Pomnibus) of the amino acid polymorphisms. The dotted horizontal line represents the significance threshold of P = 5.0 × 10−8. The most significantly associated amino acid polymorphisms are labeled with their positions, while the positions of the amino acid polymorphisms in LD with them are labeled with parenthesis. Through the stepwise regression analysis for HLA-DRβ1, we found independent RA risks of positions 11, 13, 57 and 74 (top four panels), while amino acid polymorphisms reported in Europeans also showed better fitness of the model (positions 11, 13, 71 and 74; bottom panel). (B) Distributions of the HLA-DRβ1 amino acid position model fitness among all possible combinations of the positions. To find the best combination of HLA-DRβ1 amino acid positions to explain the RA risk in Asians, we tested the associations of all possible combination of single, two and three amino acid positions. Positions 11 and 13 were considered as a single position due to their strong LD and local vicinity. Each panel represents −log10(Pomnibus) of the model consisting of combinations of the single (top), two (middle) and three (bottom) positions of the HLA-DRβ1 amino acid polymorphisms. The dotted vertical line represents top five percentile of the −log10(P) of the model. −log10(P) of the models obtained from Asians (positions 11, 13, 57 and 74) and Europeans (positions 11, 13, 71 and 74) are highlighted with blue and red arrows, respectively. (C) 3D ribbon models for the HLA-DR protein. These structures are based on Protein Data Bank entries 3pdo (for HLA-DRβ1) (21) and prepared using UCSF Chimera version 1.7 (22). Residues at RA risk amino acid positions are highlighted as colored spheres.
Applying stepwise conditional regression analysis, we observed that independent HLA-DRβ1 amino acid positions confer independent risks on RA (Fig. 2A). When conditioning on DRβ1 positions 11 and 13, amino acid position 57 showed the most significant independent evidence of association (conditional Pomnibus = 2.2 × 10−33), with amino acid positions 37, 74 and 86 demonstrating similar levels of evidence (conditional Pomnibus = 1.2 × 10−27, 1.0 × 10−30 and 1.5 × 10−22, respectively). Conditioning on positions 11, 13 and 57 demonstrated an independent association of amino acid position 74 (conditional Pomnibus = 1.1 × 10−8). No significant associations were observed after adjusting for the effects of positions 11, 13, 57 and 74 (conditional Pomnibus > 1.6 × 10−6), suggesting that the combination of these amino acid positions explain the majority of the HLA-DRB1 risk in Asians.
Because we had previously highlighted a role for positions 11, 13, 71 and 74 in Europeans (12), we also evaluated this specific combination. We found that these amino acid positions were also able to explain the majority of the risk, while the degree of significance after conditioning was relatively less conservative (conditional Pomnibus > 2.9 × 10−11 at amino acid position 70 and others). To find the best combination of HLA-DRβ1 amino acid positions in Asians, we tested all possible combinations of single, two and three amino acid positions (Fig. 2B; positions 11 and 13 were considered as a single position due to their strong LD and local vicinity). We found that both HLA-DRβ1 amino acid models (positions at 11, 13, 57 and 74 versus positions at 11, 13, 71 and 74) demonstrated a significantly better goodness-of-fit among all possible combinations of the tested positions (permutation P<0.05, 0.001, 0.001 for single, two and three position models, respectively). Addition of the position 57 to the positions 11, 13, 71 and 74 did not demonstrate independent fitness improvement (P = 0.36), and addition of the position 71 to the positions 11, 13, 57 and 74 did not, either (P = 0.52). Genetic risk scores obtained from these two models were highly correlated (R2 = 0.96). Imputation quality scores of the residues of these amino acid positions were high (average r2 scores by SNP2HAP = 0.96), and no apparent heterogeneity among the datasets were observed (Supplementary Material, Table S3). Thus, it would be difficult to robustly distinguish these models from each other given the relatively modest sample size of the present study. We note that all the HLA-DRβ1 amino acid residues pinpointed by our association analysis, including newly suggested position 57 (23), are located in the binding groove of HLA-DR, consistent with their functional contributions to RA pathogenesis (Fig. 2C).
HLA amino acid haplotype risks on RA are shared between Asians and Europeans
We compared RA risks of amino acid polymorphism haplotypes between Asians and Europeans. For HLA-DRβ1 amino acid polymorphisms, we selected positions 11, 13, 71 and 74, based on the RA risk model from our previous European study (12). We first assessed the associations of the haplotypes defined by positions 11, 13, 71 and 74 of the HLA-DRβ1 amino acid polymorphisms, and observed significant correlation of effect sizes (expressed as the log OR) between Asians and Europeans (r = 0.944, P = 3.8 × 10−7; Fig. 3; Table 2). The Val11-His13-Lys71-Ala74 haplotype confers the greatest risk not only in Europeans (OR = 4.44, 95% confidence interval [95% CI]: 4.02–4.91) but also in Asians (OR = 3.63, 95% CI: 2.63–5.00, P = 3.4 × 10−15). When we considered the HLA-DRβ1 amino acid model based on the current Asian study (positions 11, 13, 57 and 74), we confirmed that effect sizes were significantly correlated between Asians and Europeans (r = 0.94, P = 5.0 × 10−7; Supplementary Material, Fig. S2 and Table S4).
Figure 3.
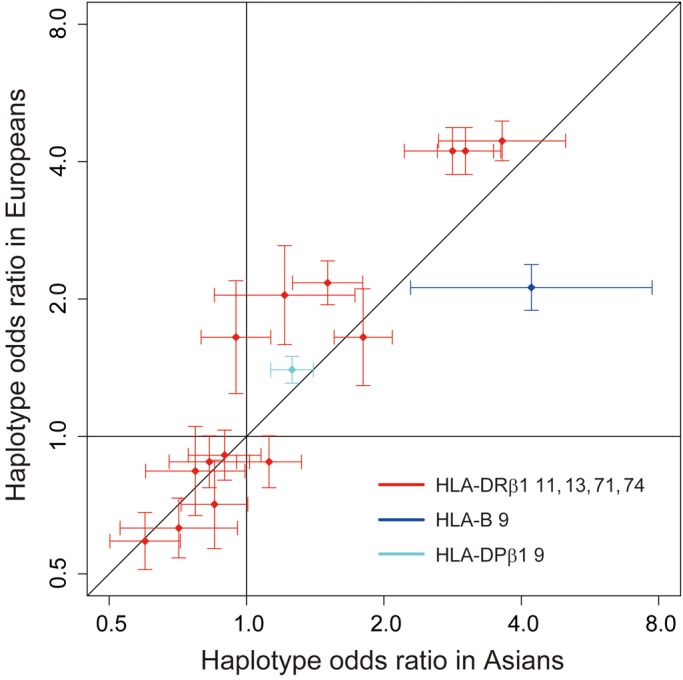
Comparison of haplotype ORs of RA risk HLA amino acid polymorphisms between Asians and Europeans. Odds ratios of the haplotypes consisting of the risk HLA amino acid polymorphisms and their 95% confidence intervals are plotted based on those in Asians (x-axis) and Europeans (y-axis).
Table 2.
Associations of HLA amino acid haplotypes with risk of RA in Asians
| HLA-DRβ1 amino acid position |
Frequencya | RA risk associationb | Classical HLA-DRB1 allelesc | |||||
|---|---|---|---|---|---|---|---|---|
| 11 | 13 | 71 | 74 | RA case | Control | OR (95% CI) | P | |
| Val | His | Lys | Ala | 0.022 | 0.009 | 3.63 (2.63–5.00) | 3.4 × 10−15 | *04:01 |
| Val | His | Arg | Ala | 0.228 | 0.092 | 3.02 (2.62–3.48) | 8.3 × 10−53 | *04:04, *04:05, *04:10 |
| Val | Phe | Arg | Ala | 0.038 | 0.017 | 2.83 (2.22–3.61) | 6.1 × 10−17 | *10:01 |
| Asp | Phe | Arg | Glu | 0.149 | 0.108 | 1.80 (1.56–2.09) | 4.0 × 10−15 | *09:01 |
| Leu | Phe | Arg | Ala | 0.068 | 0.056 | 1.51 (1.26–1.80) | 6.2 × 10−6 | *01:01 |
| Pro | Arg | Arg | Ala | 0.011 | 0.013 | 1.21 (0.85–1.73) | 0.29 | *16:02 |
| Ser | Gly | Arg | Ala | 0.080 | 0.093 | 1.12 (0.95–1.32) | 0.18 | *11:05, *12:01, *12:02, *12:03 |
| Pro | Arg | Ala | Ala | 0.095 | 0.122 | (reference) | – | *15:01, *15:02, *15:04 |
| Val | His | Arg | Glu | 0.057 | 0.074 | 0.95 (0.79–1.13) | 0.55 | *04:03, *04:06 |
| Gly | Tyr | Arg | Gln | 0.048 | 0.075 | 0.90 (0.75–1.08) | 0.24 | *07:01 |
| Ser | Gly | Arg | Leu | 0.066 | 0.092 | 0.85 (0.72–1.01) | 0.059 | *08:01, *08:02, *08:03, *08:09 |
| Ser | Ser | Arg | Ala | 0.036 | 0.057 | 0.83 (0.68–1.02) | 0.070 | *11:01, *13:12 |
| Ser | Ser | Arg | Glu | 0.021 | 0.032 | 0.77 (0.60–0.99) | 0.043 | *14:01, *14:05, *14:07 |
| Ser | Ser | Lys | Arg | 0.013 | 0.026 | 0.71 (0.53–0.96) | 0.024 | *03:01 |
| Ser | Ser | Glu | Ala | 0.049 | 0.099 | 0.60 (0.50–0.72) | 1.5 × 10−8 | *13:01, *13:02 |
| Ser | Gly | Arg | Glu | 0.014 | 0.033 | 0.56 (0.42–0.74) | 3.9 × 10−5 | *14:04 |
| HLA-B amino acid position 9 | Classical HLA-B alleles | |||||||
| Asp | 0.006 | 0.003 | 4.21 (2.29–7.74) | 3.8 × 10−6 | *08 | |||
| His, Tyr | 0.994 | 0.997 | (reference) | – |
*07, *13, *15, *18, *27, *35, *37, *38, *39, *40, *44, *46 *48, *49, *50, *51, *52, *54, *55, *56, *57, *58, *59, *67 |
|||
| HLA-DPβ1 amino acid position 9 | Classical HLA-DPB1 alleles | |||||||
| Phe | 0.862 | 0.826 | 1.26 (1.13–1.40) | 3.0 × 10−5 | *02, *04, *05, *16, *28, *31 | |||
| His, Tyr | 0.138 | 0.174 | (reference) | – | *01, *03, *09, *10, *13, *14, *15, *17, *19, *21, *26 | |||
RA: rheumatoid arthritis; OR: odds ratio.
aUnadjusted allele frequencies of the HLA amino acid residues and the haplotypes. Haplotypes with frequency ≥0.005 in controls are indicated.
bAssociations in HLA-B and HLA-DPβ1 were conditioned on the HLA-DRβ1 amino acid residues.
cClassical HLA alleles observed in the tested Asian GWAS datasets corresponding to each amino acid residue. Classical HLA-DRB1 alleles included in Shared Epitope alleles are indicated in bold. HLA gene names and HLA alleles are conventionally written in italic.
We then compared the amino acid polymorphism risks other than HLA-DRB1. We selected the HLA-B position 9 and HLA-DPβ1 position 9, which are independent risk variants in Europeans (12). We observed that the effects of these variants were significantly replicated in Asians with consistent direction (OR = 4.21, 95% CI: 2.29–7.74, P = 3.8 × 10−6 for HLA-B Asp9 and OR = 1.26, 95% CI: 1.13–1.40, P = 3.0 × 10−5 for HLA-DPβ1 Phe9; conditioned on HLA-DRβ1 polymorphisms). Collectively, the combination of amino acid polymorphisms in HLA-DRβ1, HLA-DPβ1 and HLA-B explains 5.6% of the phenotypic variance of ACPA-positive RA risk in Asians, which is <12.7% previously estimated in Europeans (12).
Trans-ethnic comparisons of RA risk HLA amino acid polymorphisms
To better understand the overlap of RA risk variants in these two populations, we compared allele frequency spectra and the LD structure around the associated amino acid polymorphisms between Asians and Europeans (Fig. 4; Supplementary Material, Table S3).
Figure 4.
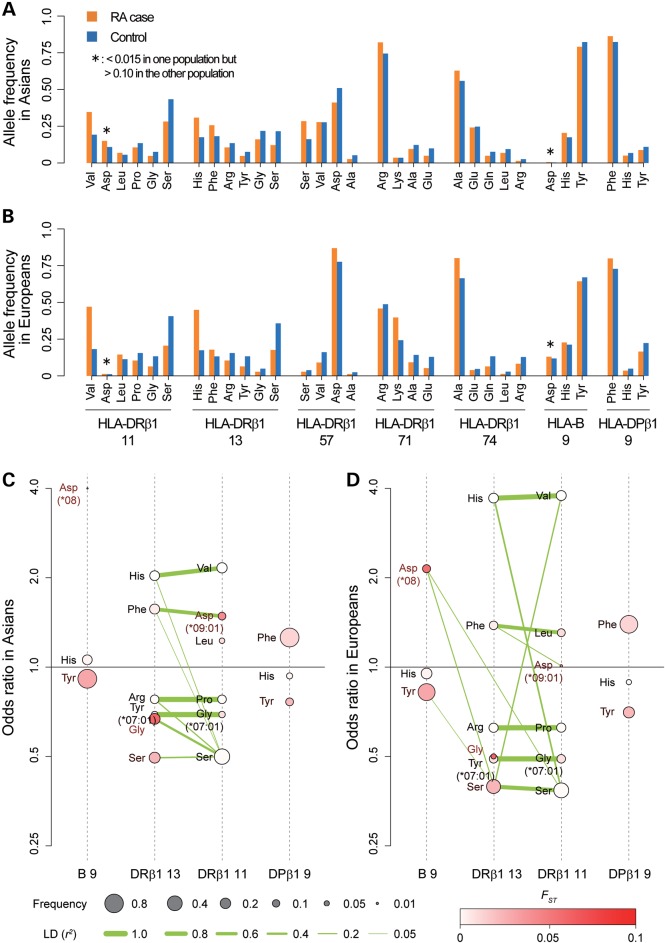
Trans-ethnic comparisons of allele frequencies and LD structure of RA risk HLA amino acid polymorphisms. Allele frequencies of amino acid polymorphisms with RA risks are indicated for RA cases and controls separately for Asians (A) and Europeans (B). Amino acid residues are ordered based on binominal OR obtained in Asians for each of the positions. Alleles of which frequencies are different between Asians and Europeans are highlighted with asterisks (<0.015 in one population but >0.10 in the other population). 2D visualization of LD structures between multiple alleles (or haplotypes) of HLA-B, HLA-DRβ1 and HLA-DPβ1 in Asians (C) and in Europeans (D). The dotted vertical axis indicates each of HLA amino acid polymorphisms. The points on the axis represent respective amino acid residues (or haplotypes), of which vertical positions represent their binominal OR of respective amino acid residues. The sizes of the points correspond to their frequencies as indicated in the legend. The density of the red color of the points corresponds to difference of the frequencies between Asians and Europeans (FST) as indicated in the legend. The segment connecting amino acid residues represents pairwise LD between them, and the thickness of the segment corresponds to the value of r2 as indicated in the legend. The segments with LD of r2< 0.05 are not indicated.
Certain alleles highlighted in our analysis had very different frequencies in European and Asian populations (Supplementary Material, Table S3). For example, HLA-DRβ1 Asp11 showed higher frequency in Asians (=0.108) but lower frequency in Europeans (=0.011; FST = 0.042), whereas HLA-B Asp9 showed lower frequency in Asians (=0.003) but higher frequency in Europeans (=0.118; FST= 0.058). We note that HLA-DRβ1 Asp11 corresponds to the classical non-SE four-digit HLA-DRB1 allele of HLA-DRB1*09:01. Investigators have previously noted that HLA-DRB1*09:01 is associated with risk of seropositive RA in Asians (10,11,24). This Asian-specific risk of HLA-DRB1*09:01 may reflect ethnically different allele frequencies of the risk amino acid residues at the same HLA-DRβ1 amino acid position (HLA-DRβ1 Asp11).
Despite these allele frequency differences between Asian and European populations, we observed that the LD structure between amino acid polymorphisms of HLA-DRβ1 positions 11 and 13 was largely consistent in both populations, with strong LD for the Val11-His13, Pro11-Arg13 and Gly11-Tyr13 haplotypes (r2 > 0.80; Fig. 4C and D). Although Val11 and His13 confer the strongest RA risk in both populations, their effect was weaker in Asians (2.16 and 2.03, respectively) than in Europeans (3.78 and 3.71, respectively; Supplementary Material, Table S3).
DISCUSSION
Association analyses of HLA genes at amino acid sites have facilitated fine-mapping efforts in immune-related diseases (12,15). In this study, we applied this approach to ACPA-positive RA in the Asian population, using a newly constructed reference panel for Asian ancestries, and demonstrated good imputation accuracy in Asian samples. Our study validated previously identified amino acid positions in HLA-DRβ1, HLA-B and HLA-DPβ1 from the European study, suggesting that genetic risk of the MHC region on ACPA-positive RA are generally shared between continental populations.
In the published European study, the most significant associations mapped to HLA-DRβ1 amino acid positions 11 and 13, both located outside of the classical SE hypothesis (HLA-DRβ1 70–74) (12). We note that this single site explains most of the variation in MHC-mediated risk for both populations, and that the residues confer similar directions and relative magnitude of risk.
We also observed that other positions in HLA-DRβ1 in European populations are generally concordant with the results presented here for Asian populations, both in terms of the specific positions identified (71 and 74 versus 57 and 74) and direction of effect of the amino acid residues at these positions. We note that amino acid position of HLA-DRβ1 57 is also located in the peptide-binding groove of HLA-DR. We cannot rule out the possibility that the observed differences could be a consequence of statistical fluctuation. Alternatively, it is possible that slight differences in the spectrum of antigens in the two populations might introduce subtle differences in which sites might play a more important role in disease susceptibility in the two populations.
In addition, we found that previously reported Asian-specific RA risk of non-SE HLA-DRB1 allele, HLA-DRB1*09:01, could be explained by ethnically different allele frequencies of the residues at same HLA-DRβ1 amino acid position. These findings illustrate the value of trans-ethnic association analysis that can exploit differences in LD structure and allele frequency among populations for the fine-mapping of causal variants. We note that the (additive) effect sizes of the ACPA-positive risk HLA variants, as well as the explained heritability estimates, were smaller in Asians than in Europeans. The different magnitudes of the effect sizes might be related to population-specific gene–gene and gene–environmental interaction that have yet to be elucidated (25).
A potential limitation of our study is the relatively modest sample size of our Asian data sets, compared in particular with the studies in European populations. As a result there is the possibility that alleles which are specific to Asian populations within HLA-DRB1 or other loci might have been missed. Such independent RA risk contributions at non-HLA genes have been frequently suggested, such as TNF, MICA and MICB genes (26–28). To investigate the role of such non-HLA gene variants as well as other classical genes such as HLA-DQ (29), future studies incorporating larger number of individuals from multiple ethnicities would be desirable.
This study also highlights the value of large-scale reference panels to achieve excellent imputation accuracy for HLA variants. Interestingly, the combined reference panel of Asians and Europeans yielded overall better accuracy than the respective panels we constructed for each single population.
In summary, through efficient genotype imputation of HLA variants and subsequent association analysis in two Asian populations, our study demonstrates significant sharing of HLA risk alleles with Europeans. Our study contributes to our understanding of HLA variants in the etiology of RA.
MATERIALS AND METHODS
Ethics statement
Our study was approved by the institutional review board at our institutions. All the enrolled subjects provided written informed consent for participation in the study.
Asian reference panel for imputation of HLA variants
For construction of the imputation reference panel of HLA variants for Asian ancestry, we enrolled 530 unrelated Asian subjects consisting of (i) HapMap Phase II Japanese and Han Chinese (JPT + CHB) populations (n = 89) (14); (ii) the Singapore Chinese population (n = 91) (13); and (iii) pan-Asian datasets including 111 Chinese, 119 Indian and 120 Malaysian subjects (n = 350). All datasets had high-density SNP genotype data and four-digit resolution of classical alleles of the class I HLA genes (HLA-A, HLA-B and HLA-C) and class II HLA genes (HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1), except that HapMap JPT + CHB subjects did not have data for HLA-DPA1 and HLA-DPB1. Part of the HLA-DRB1, HLA-DQA1 and HLA-DQB1 allele genotype data obtained from the Singapore Chinese population showed ambiguity in the resolution of four-digit alleles and provided several candidate alleles, due to similarity of the DNA sequences between these alleles and limited resolution of the genotyping methods (e.g. both HLA-DRB1*15:01 and HLA-DRB1*15:02 for candidate alleles) (13).
For each dataset, we encoded all variants including SNP, classical two- and four-digit HLA alleles, and amino acid polymorphisms of the HLA genes and combined them into a single reference panel (n = 530), using the SNP2HLA software (12,15,16). We selected the SNPs located in the region containing the entire MHC region (25–35 Mbp at chromosome 6, NCBI Build 36) which were genotyped and satisfied quality control (QC) filters in all three datasets for encoding. All variants were encoded as biallelic markers representing the presence and absence of the variants, and singletons were removed from the combined reference panel. For ambiguous four-digit alleles obtained in the Singapore Chinese dataset (13), we extracted shared amino acid sequences among the candidate four-digit alleles and encoded them into the panel, whereas non-shared amino acid sequences were encoded as missing genotypes (e.g. for the candidate four-digit allele set of HLA-DRB1*15:01 and HLA-DRB1*15:02, amino acid sequences were identical except for the amino acid position 86 of HLA-DRβ1 and were able to be encoded by using the rest of shared amino acid sequences).
Evaluation of imputation accuracy for HLA variants
To evaluate the accuracy of the imputation, we empirically compared imputed and genotyped classical HLA alleles. We adopted HapMap Phase II JPT + CHB dataset as a gold standard to assess imputation accuracy. We pruned the SNPs by selecting ones included in the GWAS array of Affymetrix Genome-wide Human SNP Array 6.0 (Santa Clara, CA, USA), to make the genotyped SNP density similar to that in the GWAS arrays. We conducted imputation of classical HLA alleles of the HapMap subjects without including genotyped HLA allele information using the rest of the Asian reference panel [Singapore Chinese and pan-Asian datasets (n = 441), which was constructed separately from the original reference panel (n = 530)] and the SNP2HLA software (12,15,16). We then compared the concordances and correlations between imputed and genotyped allele dosages. To relatively assess imputation performances among reference panels, we also performed imputation using the two previously constructed reference panels from the European populations for HLA polymorphism imputation in the same manner; HapMap CEU founder populations (n = 120) (14) and a collection from T1DGC (n = 5225) (16,17). Concordances of the alleles were calculated for overall two- alleles or four-digit alleles as described elsewhere (16,17). Correlations of the allele dosages were calculated for each of the two- and four-digit alleles separately by using Pearson's correlation test.
RA GWAS data in Asian populations
We used data from two GWAS and one Immunochip study from the Asian populations for 2782 cases and 4315 controls: one GWAS with the samples from China (466 cases and 873 controls) (18), one GWAS with the samples from South Korea (799 cases and 751 controls) (19) and one Immunochip study with the samples from South Korea (1517 cases and 2691 controls) (20). All cases met the 1987 American College of Rheumatology diagnostic criteria (30) and were confirmed to be ACPA positive.
Details of the GWAS and Immunochip data, including genotyping platforms, were described elsewhere (18–20). Each dataset was filtered with the stringent QC criteria as described elsewhere (18,19,31), including SNP and sample call rate cutoffs, exclusion of closely related relative and outliers in terms of ancestry, and SNP minor allele frequency (MAF) and Hardy–Weinberg equilibrium cutoffs. All subjects were confirmed to be of Asian ancestry using the results of principal component analysis conducted with HapMap Phase II populations (32). PCs estimated for QC-filtered GWAS subjects using whole-genome SNP data was used to correct potential population structures in the following analysis focusing on the MHC region.
Imputation of HLA variants in Asian RA GWAS data
From each GWAS, we extracted SNP genotypes located in the entire MHC region (2142 SNPs for the Chinese GWAS, 2232 SNPs for the South Korean GWAS and 5393 SNPs for the South Korean Immunochip study from 25 to 35 Mbp at chromosome 6, NCBI Build 36) to impute classical two- and four-digit HLA alleles and amino acid polymorphisms of the HLA genes along with the SNPs that were not genotyped in the GWAS. The imputation was conducted for each GWAS separately in conjunction of cases and controls together with the combined reference panel of the Asian populations (n = 530) by using the SNP2HLA software (12,15,16). We applied postimputation QC criteria of MAF > 0.5% for the association analysis.
Association analysis of HLA alleles and amino acid polymorphisms
We tested associations of the variants with the risk of ACPA-positive RA using logistic regression model assuming additive effects of the allele dosages in the log-odds scale and their fixed effects among the GWAS datasets. We defined HLA variants to include biallelic SNPs in the MHC region, two- and four-digit biallelic classical HLA alleles, biallelic HLA amino acid polymorphisms for respective residues, multiallelic HLA amino acid polymorphisms for respective positions. To account for potential population stratification, we included top five PCs estimated from each of GWAS datasets as covariates. We also included a dummy variable to represent GWAS datasets to account for study-specific confounding effects. For HLA variants with m alleles (m = 2 for biallelic variants and m > 2 for multiallelic variants), we included m − 1 alleles as independent variables in the regression model, excluding the most frequent allele as the reference. This resulted in the following logistic regression models:
where β0 is the logistic regression intercept and β1,j is the additive effects of the dosage of allele j for the variant (xj). K and L are numbers of the cohorts and PCs enrolled in the analysis, respectively (K = 3 and L = 5). yk,l is the lth PC for kth cohort, and z is the indicator dummy variable for the three cohorts. β2,k,l and β3 parameters are the effects of yk,l and z, respectively. An omnibus P-value of the variant (=Pomnibus) was obtained by a log-likelihood ratio test comparing the likelihood of null model against the likelihood of the fitted model. We assessed the significance of the improvement in fit by calculating the deviance (=−2 × the log likelihood), which follows a χ2 distribution with m − 1 degree(s) of freedom.
For the conditional analysis, we assumed the logistic regression model additionally including the variants as covariates, which we refer ‘conditional’ throughout the text. For conditional analysis on the specific HLA amino acid positions, we included the multiallelic variants of the amino acid residues as covariates. For conditional analysis on the HLA gene, we included all the amino acid positions and classical HLA alleles as covariates. Amino acid positions and the HLA genes to be included as covariates were consecutively selected in a forward stepwise fashion (33).
We compared fitness of the HLA-DRβ1 amino acid models (positions at 11, 13, 57 and 74 for the model from the current Asian study and positions at 11, 13, 71 and 74 for the model from the previous European study) by a conditional analysis. Fitness of the model was evaluated by applying ANOVA test between the model including all risk HLA-DRβ1 amino acid positions (11, 13, 57, 71 and 74) and the model not including the representative position of each model (11, 13, 71 and 74 for evaluating the model from the current Asian study and 11, 13, 57 and 74 for evaluating the model from the previous European study). Calculation of genetic risk scores of the models was described elsewhere (34).
Trans-ethnic comparisons of HLA variant risks on RA
For trans-ethnic comparisons of HLA variant risks, we obtained distributions of the amino acid polymorphism frequencies and their risks on ACPA-positive RA from the previous study in the European populations (12). Allele frequency differences of the amino acid polymorphisms between Asian and Europeans were evaluated based on frequencies of the respective residues in the controls using FST (35). Correlations of HLA-DRβ1 amino acid haplotypes between Asians and Europeans were assessed by using Pearson's correlation test for logarithm of ORs. Phenotypic variance explained by the RA risk amino acid polymorphisms were estimated using liability threshold model under the assumption of disease prevalence of 0.5% (12,36). Trans-ethnic comparison of LD between the HLA variants were assessed using the phased haplotype data in the reference panels of Asians (n = 530) and Europeans (T1DGC consortium, n = 5225).
WEB RESOURCES
The URLs for data presented herein are as follows:
SNP2HLA, http://www.broadinstitute.org/mpg/snp2hla/.
International HapMap consortium, http://hapmap.ncbi.nlm.nih.gov
T1DGC consortium, https://www.t1dgc.org/home.cfm
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institutes of Health (1R01AR062886-01, 5U01GM092691-04 and 1R01AR063759-01A1), the Arthritis Foundation, a Clinical Scientist Development Award to S.R. from the Doris Duke Foundation, the Japan Society of the Promotion of Science (JSPS), Japan Science and Technology Agency (JST), and by the Korean Health Technology R&D Project, Ministry of Health & Welfare, Korea (A121983). M.A.B. was funded by a National Health and Medical Research Council (Australia) Senior Principal Research Fellowship and Queensland Premier′s Fellowship for Science. P.I.W.D.B. is the recipient of a Vernieuwingsimpuls VIDI Award (project 016.126.354) from the Netherlands Organization for Scientific Research (NWO).
Supplementary Material
REFERENCES
- 1.Gabriel S.E. The epidemiology of rheumatoid arthritis. Rheum. Dis. Clin. North. Am. 2001;27:269–281. doi: 10.1016/s0889-857x(05)70201-5. [DOI] [PubMed] [Google Scholar]
- 2.Neogi T., Aletaha D., Silman A.J., Naden R.L., Felson D.T., Aggarwal R., Bingham C.O., III, Birnbaum N.S., Burmester G.R., Bykerk V.P., et al. The 2010 American College of Rheumatology/European League Against Rheumatism classification criteria for rheumatoid arthritis: Phase 2 methodological report. Arthritis Rheum. 2010;62:2582–2591. doi: 10.1002/art.27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl E.A., Wegmann D., Trynka G., Gutierrez-Achury J., Do R., Voight B.F., Kraft P., Chen R., Kallberg H.J., Kurreeman F.A., et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Woude D., Houwing-Duistermaat J.J., Toes R.E., Huizinga T.W., Thomson W., Worthington J., van der Helm-van Mil A.H., de Vries R.R. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009;60:916–923. doi: 10.1002/art.24385. [DOI] [PubMed] [Google Scholar]
- 5.Ding B., Padyukov L., Lundstrom E., Seielstad M., Plenge R.M., Oksenberg J.R., Gregersen P.K., Alfredsson L., Klareskog L. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–38. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregersen P.K., Silver J., Winchester R.J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 7.Okada Y., Yamada R., Suzuki A., Kochi Y., Shimane K., Myouzen K., Kubo M., Nakamura Y., Yamamoto K. Contribution of a haplotype in the HLA region to anti-cyclic citrullinated peptide antibody positivity in rheumatoid arthritis, independently of HLA-DRB1. Arthritis Rheum. 2009;60:3582–3590. doi: 10.1002/art.24939. [DOI] [PubMed] [Google Scholar]
- 8.Kazkaz L., Marotte H., Hamwi M., Angelique Cazalis M., Roy P., Mougin B., Miossec P. Rheumatoid arthritis and genetic markers in Syrian and French populations: different effect of the shared epitope. Ann. Rheum. Dis. 2007;66:195–201. doi: 10.1136/ard.2004.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes L.B., Morrison D., Kelley J.M., Padilla M.A., Vaughan L.K., Westfall A.O., Dwivedi H., Mikuls T.R., Holers V.M., Parrish L.A., et al. The HLA-DRB1 shared epitope is associated with susceptibility to rheumatoid arthritis in African Americans through European genetic admixture. Arthritis Rheum. 2008;58:349–358. doi: 10.1002/art.23166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kochi Y., Yamada R., Kobayashi K., Takahashi A., Suzuki A., Sekine A., Mabuchi A., Akiyama F., Tsunoda T., Nakamura Y., et al. Analysis of single-nucleotide polymorphisms in Japanese rheumatoid arthritis patients shows additional susceptibility markers besides the classic shared epitope susceptibility sequences. Arthritis Rheum. 2004;50:63–71. doi: 10.1002/art.11366. [DOI] [PubMed] [Google Scholar]
- 11.Lee H.S., Lee K.W., Song G.G., Kim H.A., Kim S.Y., Bae S.C. Increased susceptibility to rheumatoid arthritis in Koreans heterozygous for HLA-DRB1*0405 and *0901. Arthritis Rheum. 2004;50:3468–3475. doi: 10.1002/art.20608. [DOI] [PubMed] [Google Scholar]
- 12.Raychaudhuri S., Sandor C., Stahl E.A., Freudenberg J., Lee H.S., Jia X., Alfredsson L., Padyukov L., Klareskog L., Worthington J., et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillai N.E., Okada Y., Ong R.T., Wang X., Tantoso E., Xu W., Peterson T.A., Belawney T., Ali M., Poh W., et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum. Mol. Genet. 2014;23:4443–4451. doi: 10.1093/hmg/ddu149. [DOI] [PubMed] [Google Scholar]
- 14.de Bakker P.I.W., McVean G., Sabeti P.C., Miretti M.M., Green T., Marchini J., Ke X., Monsuur A.J., Whittaker P., Delgado M., et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The International HIV Controllers Study. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia X., Han B., Onengut-Gumuscu S., Chen W.M., Concannon P.J., Rich S.S., Raychaudhuri S., de Bakker P.I.W. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rich S.S., Concannon P., Erlich H., Julier C., Morahan G., Nerup J., Pociot F., Todd J.A. The Type 1 Diabetes Genetics Consortium. Ann. N. Y. Acad. Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 18.Jiang L., Yin J., Ye L., Yang J., Hemani G., Liu A.J., Zou H., He D., Sun L., Zeng X., et al. Novel risk loci for rheumatoid arthritis in han chinese and congruence with risk variants in europeans. Arthritis Rheum. 2014 doi: 10.1002/art.38353. doi:10.1002/art.38353. [DOI] [PubMed] [Google Scholar]
- 19.Freudenberg J., Lee H.S., Han B.G., Shin H.D., Kang Y.M., Sung Y.K., Shim S.C., Choi C.B., Lee A.T., Gregersen P.K., et al. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011;63:884–893. doi: 10.1002/art.30235. [DOI] [PubMed] [Google Scholar]
- 20.Kim K., Bang S., Lee H., Cho S., Choi C., Sung Y., Kim T., Jun J., Yoo D., Kang Y., et al. High-density genotyping of immune loci in Koreans and Europeans identifies 8 new rheumatoid arthritis risk loci. Ann. Rheum. Dis. 2014 doi: 10.1136/annrheumdis-2013-204749. doi:10.1136/annrheumdis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunther S., Schlundt A., Sticht J., Roske Y., Heinemann U., Wiesmuller K.H., Jung G., Falk K., Rotzschke O., Freund C. Bidirectional binding of invariant chain peptides to an MHC class II molecule. Proc. Natl. Acad. Sci. USA. 2010;107:22219–22224. doi: 10.1073/pnas.1014708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.Nepom B.S., Nepom G.T., Coleman M., Kwok W.W. Critical contribution of beta chain residue 57 in peptide binding ability of both HLA-DR and -DQ molecules. Proc. Natl. Acad. Sci. USA. 1996;93:7202–7206. doi: 10.1073/pnas.93.14.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada Y., Suzuki A., Yamada R., Kochi Y., Shimane K., Myouzen K., Kubo M., Nakamura Y., Yamamoto K. HLA-DRB1*0901 lowers anti-cyclic citrullinated peptide antibody levels in Japanese patients with rheumatoid arthritis. Ann. Rheum. Dis. 2010;69:1569–1570. doi: 10.1136/ard.2009.118018. [DOI] [PubMed] [Google Scholar]
- 25.Kallberg H., Padyukov L., Plenge R.M., Ronnelid J., Gregersen P.K., van der Helm-van Mil A.H., Toes R.E., Huizinga T.W., Klareskog L., Alfredsson L. Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am. J. Hum. Genet. 2007;80:867–875. doi: 10.1086/516736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguillon J.C., Cruzat A., Aravena O., Salazar L., Llanos C., Cuchacovich M. Could single-nucleotide polymorphisms (SNPs) affecting the tumour necrosis factor promoter be considered as part of rheumatoid arthritis evolution? Immunobiology. 2006;211:75–84. doi: 10.1016/j.imbio.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Kirsten H., Petit-Teixeira E., Scholz M., Hasenclever D., Hantmann H., Heider D., Wagner U., Sack U., Hugo Teixeira V., Prum B., et al. Association of MICA with rheumatoid arthritis independent of known HLA-DRB1 risk alleles in a family-based and a case control study. Arthritis. Res. Ther. 2009;11:R60. doi: 10.1186/ar2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Arbesu R., Ballina-Garcia F.J., Alperi-Lopez M., Lopez-Soto A., Rodriguez-Rodero S., Martinez-Borra J., Lopez-Vazquez A., Fernandez-Morera J.L., Riestra-Noriega J.L., Queiro-Silva R., et al. MHC class I chain-related gene B (MICB) is associated with rheumatoid arthritis susceptibility. Rheumatology (Oxford) 2007;46:426–430. doi: 10.1093/rheumatology/kel331. [DOI] [PubMed] [Google Scholar]
- 29.Vignal C., Bansal A.T., Balding D.J., Binks M.H., Dickson M.C., Montgomery D.S., Wilson A.G. Genetic association of the major histocompatibility complex with rheumatoid arthritis implicates two non-DRB1 loci. Arthritis Rheum. 2008;60:53–62. doi: 10.1002/art.24138. [DOI] [PubMed] [Google Scholar]
- 30.Arnett F.C., Edworthy S.M., Bloch D.A., McShane D.J., Fries J.F., Cooper N.S., Healey L.A., Kaplan S.R., Liang M.H., Luthra H.S., et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 31.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The International HapMap, Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 33.Okada Y., Yamazaki K., Umeno J., Takahashi A., Kumasaka N., Ashikawa K., Aoi T., Takazoe M., Matsui T., Hirano A., et al. HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn's disease. Gastroenterology. 2011;141:864–871. doi: 10.1053/j.gastro.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 34.Kurreeman F.A., Liao K., Chibnik L., Hickey B., Stahl E.A., Gainer V., Li G., Bry L., Mahan S., Ardlie K., et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am. J. Hum. Genet. 2011;88:57–69. doi: 10.1016/j.ajhg.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewontin R.C., Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viatte S., Plant D., Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nat. Rev. Rheumatol. 2013;9:141–153. doi: 10.1038/nrrheum.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.