Abstract
Mutations in dystrophin lead to Duchenne muscular dystrophy, which is among the most common human genetic disorders. Dystrophin nucleates assembly of the dystrophin–glycoprotein complex (DGC), and a defective DGC disrupts an essential link between the intracellular cytoskeleton and the basal lamina, leading to progressive muscle wasting. In vitro studies have suggested that dystrophin phosphorylation may affect interactions with actin or syntrophin, yet whether this occurs in vivo or affects protein function remains unknown. Utilizing nanoflow liquid chromatography mass spectrometry, we identified 18 phosphorylated residues within endogenous dystrophin. Mutagenesis revealed that phosphorylation at S3059 enhances the dystrophin–dystroglycan interaction and 3D modeling utilizing the Rosetta software program provided a structural model for how phosphorylation enhances this interaction. These findings demonstrate that phosphorylation is a key mechanism regulating the interaction between dystrophin and the DGC and reveal that posttranslational modification of a single amino acid directly modulates the function of dystrophin.
INTRODUCTION
Dystrophin is one of the key proteins involved in the maintenance of skeletal muscle structure and for force transmission. It forms a sarcomeric glycoprotein complex termed the dystrophin–glycoprotein complex (DGC)—a multiprotein structure comprising dystrophin, dystroglycans, sarcoglycans, syntrophins, nNOS and dystrobrevins (1–3). The DGC plays a critical role in linking the subsarcolemmal actin cytoskeleton to the extracellular matrix (ECM) and confers membrane protection during the transmission of contractile forces. The loss of dystrophin protein, which is responsible for Duchenne muscular dystrophy (DMD), results in destabilization of this link such that the DGC fails to assemble on the sarcolemma (4). Individual myofibers are thus not able to properly transmit the forces of contraction to the ECM, and membrane instability leads to exhaustive cycles of degeneration and regeneration, an influx of inflammatory mediators with progressive muscle wasting, and extreme weakness contributed by the replacement of muscle tissue with fibrotic and adipose tissue (5).
The 427 kDa dystrophin protein comprises an N-terminal actin-binding domain, a central rod domain consisting of 24 spectrin-like repeats and 4 hinge regions, a cysteine-rich (CR) domain comprises a WW domain, two EF-hand like domains and a ZZ domain, and finally a C-terminal (CT) domain containing protein-binding sites for the syntrophins and dystrobrevins (6). Through the generation of transgenic mdx mice, we have previously demonstrated that the CR and CT domains alone are sufficient to restore the DGC at the sarcolemma as demonstrated by the overexpression of Dp71 (7). Subsequently, we showed that overexpression of the Dp116 isoform (which also comprises mostly the CR and CT domains) was not only sufficient to restore the DGC to the sarcolemma but also could maintain muscle mass and extend the lifespan of mdx/utrn−/− mice without altering the dystrophic phenotype (8). These and related studies also showed that the CR domain is the only portion of dystrophin indispensible for functional activity (6,8).
Mutagenesis studies have revealed that the CR domain is essential for dystrophin function by forming a binding site for the transmembrane protein, β-dystroglycan (9). However, analysis of patients with the milder Becker muscular dystrophy revealed large deletions within the rod domain still form highly (but not fully) functional proteins, indicating that parts of the dystrophin protein are not required for physiological function (10,11). In light of this finding, research turned to developing miniaturized dystrophin proteins for treating DMD. Deletion analysis revealed that microdystrophin constructs lacking either spectrin repeats 4–23 and the CT domain (µDys ΔR4-R23/ΔCT) or spectrin repeats 2–21 and the CT domain (µDys ΔR2-R21/ΔCT) were highly functional and could reverse the dystrophic pathology when delivered muscles of mdx mice (12). Subsequent systemic delivery of the µDys ΔR4-R23/ΔCT using adeno-associated viral (AAV) vectors demonstrated that this truncated protein could protect the dystrophic muscles and prolong the lifespan of the severely affected mdx/utrn−/− mouse (13). However, while µDys ΔR4-R23/ΔCT protein significantly improved force generation and protection from contraction-induced injury of dystrophic muscles, these were not restored to control levels (13). Therefore, research continues into improving the function of miniaturized dystrophin proteins with replacement of hinge 2 with hinge 3 in the µDys ΔR4-R23/ΔCT protein, the inclusion of the nNOS-binding site, or the inclusion of helix-1 from the CT-domain (14–16).
Phosphorylation is a key posttranslational mechanism that is well known to modulate protein stability and/or function. In vitro and in vivo studies have shown that dystrophin can be phosphorylated, particularly within the CT region (7). Furthermore, other DGC members including the syntrophins, the α-dystrobrevins and β-dystroglycan have been shown to be phosphorylated in this complex (17–23). In vitro studies have demonstrated that dystrophin can be phosphorylated by a large number of kinases including cAMP-dependent/cGMP-dependent protein kinase (DGC-PK), CaM kinase, casein kinase II, protein kinase C, p44 MAP kinase and p34cdc2 protein kinase (22,24–30). However, little is known about whether these posttranslational modifications occur in vivo. Furthermore, although some evidence suggests phosphorylation within the dystrophin rod domain affects F-actin binding, and phosphorylation within the dystrophin CT domain inhibits syntrophin-binding in vitro (26,31), it remains unclear whether phosphorylation affects the functions of dystrophin and the assembly of the DGC within striated muscle.
Understanding the biological implications of dystrophin posttranslational modification may prove valuable for developing therapeutics to combat muscle wasting in DMD and in other muscle diseases where dystrophin protein levels have been observed to decline with disease progression, such as cancer cachexia (32). We hypothesized that multiple amino acids within the dystrophin protein would be phosphorylated in the endogenous state in vivo and that this would modulate dystrophin protein function. Utilizing LC–MS/MS we identified 18 novel sites of phosphorylation within the endogenous dystrophin protein that occur in healthy and/or dystrophic skeletal muscle. Furthermore, we demonstrated that phosphorylation of a specific serine residue within the dystrophin WW domain increases the association between dystrophin and β-dystroglycan. Together, these findings reveal for the first time that posttranslational modification of a single amino acid can alter the function of the dystrophin protein. This discovery could advance novel therapeutics for muscular dystrophy and other muscle wasting conditions.
RESULTS
Dystrophin is phosphorylated on multiple serine, threonine and tyrosine residues in healthy and dystrophic skeletal muscle
Although the dystrophin protein can be phosphorylated by different kinases when incubated in vitro, it remains unclear to what extent dystrophin is phosphorylated in vivo. To examine whether dystrophin is phosphorylated in its endogenous state we performed a series of mass spectrometric analyses to identify phosphorylated amino acids within the dystrophin protein isolated from skeletal muscles of either healthy C57BL/6 mice or from transgenic mice overexpressing the FLAG-tagged Dp116 transgene on a dystrophin/utrophin-deficient background. Dp116 is a naturally occurring short dystrophin isoform that is normally expressed in the peripheral nervous system. Overexpression of this isoform on the dystrophin/utrophin-deficient background can prolong the lifespan of affected mice without preventing the dystrophic muscle pathology (8,33). Analysis of the Dp116 protein from these mice therefore allows for an in-depth analysis of dystrophin phosphorylation in a shorter protein in diseased muscle.
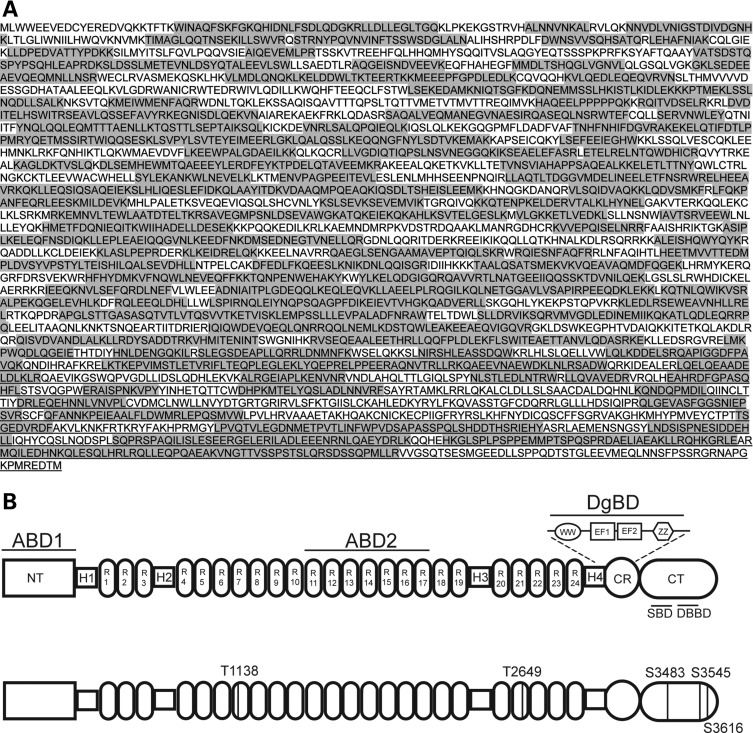
Initial analyses from the muscles of C57Bl/6 mice revealed 26% sequence coverage of the full-length dystrophin protein (Dp427) and identified one phosphorylated amino acid in the CT region (S3483). By using multiple digestion enzymes (i.e. trypsin, chymotryspin and GluC), and utilizing both the full-length dystrophin protein isolated from either C57BL/6 mouse muscle or differentiated C2C12 cells (an immortalized myogenic cell line), we could increase the overall sequence coverage to 63% (Fig. 1A) and identify a total of five phosphorylated amino acids (Fig. 1B). These included two threonine residues (T1138 and T2649) in the rod domain and three serine residues (S3483, S3545 and S3616) in the CT domain (Fig. 1B).
Figure 1.
Mass spectrometric analysis of the dystrophin protein resulted in identification of multiple phosphorylation sites with good sequence coverage. (A) The domain structures of the full-length dystrophin protein (Dp427; top) and the location of the phosphorylated amino acids identified in Dp427 (bottom). Phosphorylated threonine (T) residues were identified in R7 and R21 of Dp427, and three phosphorylated serine (S) residues were identified in the CT. (B) Peptide identification by mass spectrometric analysis resulted in 63% sequence coverage, with the identified regions shaded in gray. ABD, actin-binding domain; NT, N-terminal; H, hinge region; R, spectrin repeat; WW, WW domain; EF, EF-like region; ZZ, ZZ domain; CR, cysteine-rich region, CT, C-terminal domain.
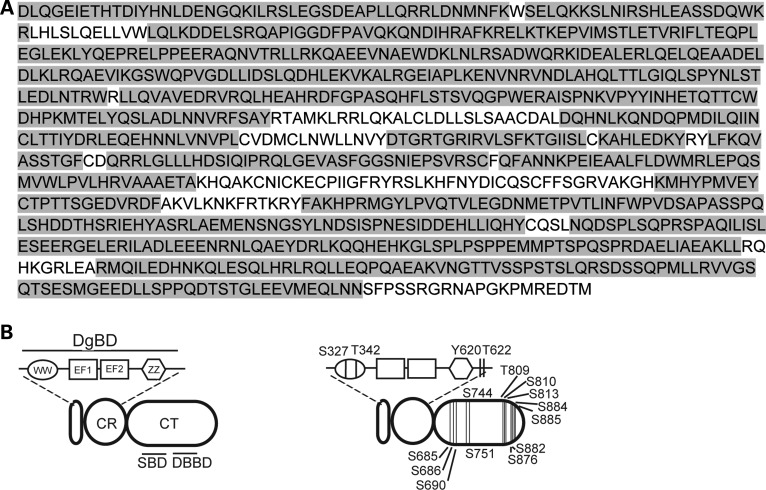
To increase protein sequence coverage, particularly near the CR and CT regions (which are the major sites of protein–protein interactions); the short dystrophin isoform Dp116 isolated from transgenic mice overexpressing the FLAG-tagged Dp116 transgene on a dystrophin/utrophin-deficient background was also analyzed. Mass spectrometric analysis of Dp116 by chymotrypsin digestion resulted in 84% sequence coverage of the Dp116 protein (Fig. 2A) and identified 16 phosphorylated amino acids within the CR and CT domains (Fig. 2B; S327, T342, Y620, T622, S690, S744, T809, S810, S813, S876, S882, S884 and S885). Furthermore, two of these phosphorylated amino acids (S813, analogous to S3545; and S884, analogous to S3616) were previously identified in the full-length dystrophin protein (Fig. 1B). Comprehensive lists of all of the peptides identified from Dp427 (Supplementary Material, Table S1) or Dp116 (Supplementary Material, Table S2) demonstrated both phosphorylated and non-phosphorylated forms of most peptides identified with a phosphorylated residue. The peptides containing phosphorylated amino acids identified from either Dp427 or Dp116 are summarized in Table 1. The consensus sequence and kinase responsible for these phosphorylation events were predicted based on previously published consensus sequences (34,35). Together, these results confirm that the dystrophin protein is subject to phosphorylation on multiple sites in healthy and dystrophic skeletal muscle.
Figure 2.
Mass spectrometric analysis of the Dp116 protein resulted in identification of multiple phosphorylation sites with good sequence coverage. (A) The domain structures of the Dp116 dystrophin isoform (Dp116; left) and the location of the phosphorylated amino acids identified Dp116 (right). Four phosphorylated amino acids were identified in the CR and another 12 were identified in the CT. (B) Peptide identification by mass spectrometric analysis resulted in 84% sequence coverage, with the identified regions shaded in gray. WW, WW domain; EF, EF-like region; ZZ, ZZ domain, CR, cysteine-rich region; CT, C-terminal domain.
Table 1.
All of the dystrophin peptides containing a phosphorylated amino acid that were identified from either Dp427 or Dp116 are shown
| Dystrophin isoform | Peptide location (relative to Dp427 sequence) | Mass | Peptide sequence | Phosphorylated residue number | Likely consensus sequence | Predicted kinase |
|---|---|---|---|---|---|---|
| Dp427 | 1135–1145 | 1413.7 | ELNT(P)QWDHICR | T1138 | [S/T]pXXX, where X may be asp or glu; [S/T]pQ | CK2; ATM kinase |
| Dp427 | 2640–2661 | 2604.3 | LLRDYSADDT(P)RKVHMITENINT | T2649 | XXX[S/T]p, where X may be asp or glu; [S/T]pXK/R | CK 1; CDK2,5 |
| Dp116 | 3055–3065 | 1272.7 | ERAIS(P)PNKVPY | S3059 (S327 in Dp116) | S(p)P; S(p)X[K/R] | Erk1, ERK2; CDK2, CDK5 |
| Dp116 | 3067–3085 | 2346 | INHETQTT(P)CWDHPKMTELY | T3074 (T342 in Dp116) | Unknown | ? |
| Dp116 | 3066–3085 | 2605.1 | YINHETQTT(P)CWDHPKMTELY | T3074 (T342 in Dp116) | Unknown | ? |
| Dp116 | 3406–3433 | 3285.5 | INFWPVDSAPAS(P)SPQLSHDDTHSRIEHY | S3417 (S685 in Dp116) | XXX[S/T]p, where X may be asp or glu; PXS(p)P | CK1 |
| Dp116 | 3344–3365 | 2701.1 | KMHYPMVEY(P)CTPTTSGEDVRDF | Y3352 (Y620 in Dp116) | XXXY(p) | Receptor tyrosine kinase, where any X is glu |
| Dp116 | 3344–3363 | 2701.1 | KMHYPMVEYCT(P)PTTSGEDVRDF | T3354 (T622 in Dp116) | XT(p)P | ERK1, 2 |
| Dp116 | 3406–3420 | 1614.8 | INFWPVDSAPASS(P)PQ | S3418 (S686 in Dp116) | XXX[S/T]p, where X may be asp or glu; PXS(p)P | CK1; GSK-3 |
| Dp116 | 3406–3433 | 3285.5 | INFWPVDSAPASS(P)PQLSHDDTHSRIEHY | S3418 (S686 in Dp116) | XXX[S/T]p, where X may be asp or glu; PXS(p)P | CK1; GSK-3 |
| Dp116 | 3410–3433 | 2725.2 | PVDSAPASS(P)PQLSHDDTHSRIEHY | S3418 (S686 in Dp116) | XXX[S/T]p, where X may be asp or glu; PXS(p)P | CK1; GSK-3 |
| Dp116 | 3406–3433 | 3285.5 | INFWPVDSAPASSPQLS(P)HDDTHSRIEHY | S3422 (S690 in Dp116) | S(p)XXX | CK2 |
| Dp116 | 3473–3488 | 1829.9 | NQDS(P)PLSQPRSPAQIL | S3476 (S744 in Dp116) | DS(p)P | ERK1, ERK2, CK1 |
| Dp427 | 3483–3496 | 1570.8 | S(P)PAQILISLESEER | S3483 (S751 in Dp116) | PXS(p)P | ERK2, GSK3 |
| Dp116 | 3527–3556 | 3511.7 | KGLSPLPSPPEMMPT(P)SPQSPRDAELIAEAKLL | T3541 (T809 in Dp116) | PT(p)SP | ERK1 |
| Dp116 | 3527–3558 | 3511.7 | KGLSPLPSPPEMMPTS(P)PQSPRDAELIAEAKLL | S3542 (S810 in Dp116) | PXS(p)P | ERK2, GSK3 |
| Dp427 | 3526–3550 | 2685.3 | HKGLSPLPSPPEMMPTSPQS(P)PRDAE | S3545 (S813 in Dp116) | PXS(p)P | ERK2, GSK3 |
| Dp116 | 3527–3558 | 3511.7 | KGLSPLPSPPEMMPTSPQS(P)PRDAELIAEAKLL | S3545 (S813 in Dp116) | PXS(p)P | ERK2, GSK3 |
| Dp116 | 3591–3620 | 3226.4 | EQPQAEAKVNGTTVSSPS(P)TSLQRSDSSQPM | S3608 (S876 in Dp116) | XPS(p)X | ERK1 |
| Dp116 | 3591–3620 | 3226.4 | EQPQAEAKVNGTTVSSPSTSLQRS(P)DSSQPM | S3614 (S882 in Dp116) | S(p)XXX | CK2 |
| Dp116 | 3591–3622 | 3468.6 | EQPQAEAKVNGTTVSSPSTSLQRS(P)DSSQPMLL | S3614 (S882 in Dp116) | S(p)XXX | CK2 |
| Dp116 | 3591–3622 | 3468.6 | EQPQAEAKVNGTTVSSPSTSLQRS(P)DSSQPMLL | S3614 (S882 in Dp116) | S(p)XXX | CK2 |
| Dp116 | 3591–3620 | 3226.4 | EQPQAEAKVNGTTVSSPSTSLQRSDS(P)SQPM | S3616 (S884 in Dp116) | RXXS(p) | CaMKII |
| Dp116 | 3591–3622 | 3468.6 | EQPQAEAKVNGTTVSSPSTSLQRSDS(P)SQPMLL | S3616 (S884 in Dp116) | RXXS(p) | CaMKII |
| Dp116 | 3601–3622 | 2374.1 | GTTVSSPSTSLQRSDS(P)SQPMLL | S3616 (S884 in Dp116) | RXXS(p) | CaMKII |
| Dp116 | 3590–3622 | 3581.7 | LEQPQAEAKVNGTTVSSPSTSLQRSDS(P)SQPMLL | S3616 (S884 in Dp116) | RXXS(p) | CaMKII |
| Dp116 | 3590–3622 | 3581.7 | LEQPQAEAKVNGTTVSSPSTSLQRSDS(P)SQPMLL | S3616 (S884 in Dp116) | RXXS(p) | CaMKII |
| Dp427 | 3614–3623 | 1212.5 | SDS(P)SQPMLLR | S3616 (S884 in Dp116) | RXXS(p) | CaMKII |
| Dp116 | 3614–3623 | 1132.6 | SDS(P)SQPMLLR | S3616 (S884 in Dp116) | RXXS(p) | CaMKII |
| Dp116 | 3601–3622 | 2374.1 | GTTVSSPSTSLQRSDSS(P)QPMLL | S3617 (S885 in Dp116) | S(p)XP | Erk1 |
The phosphorylated reside is indicated in parentheses. The likely kinase consensus sequence and possible enzymes are predicted for each phosphorylation.
Site-directed mutagenesis of identified amino acids reveals requirements for DGC binding
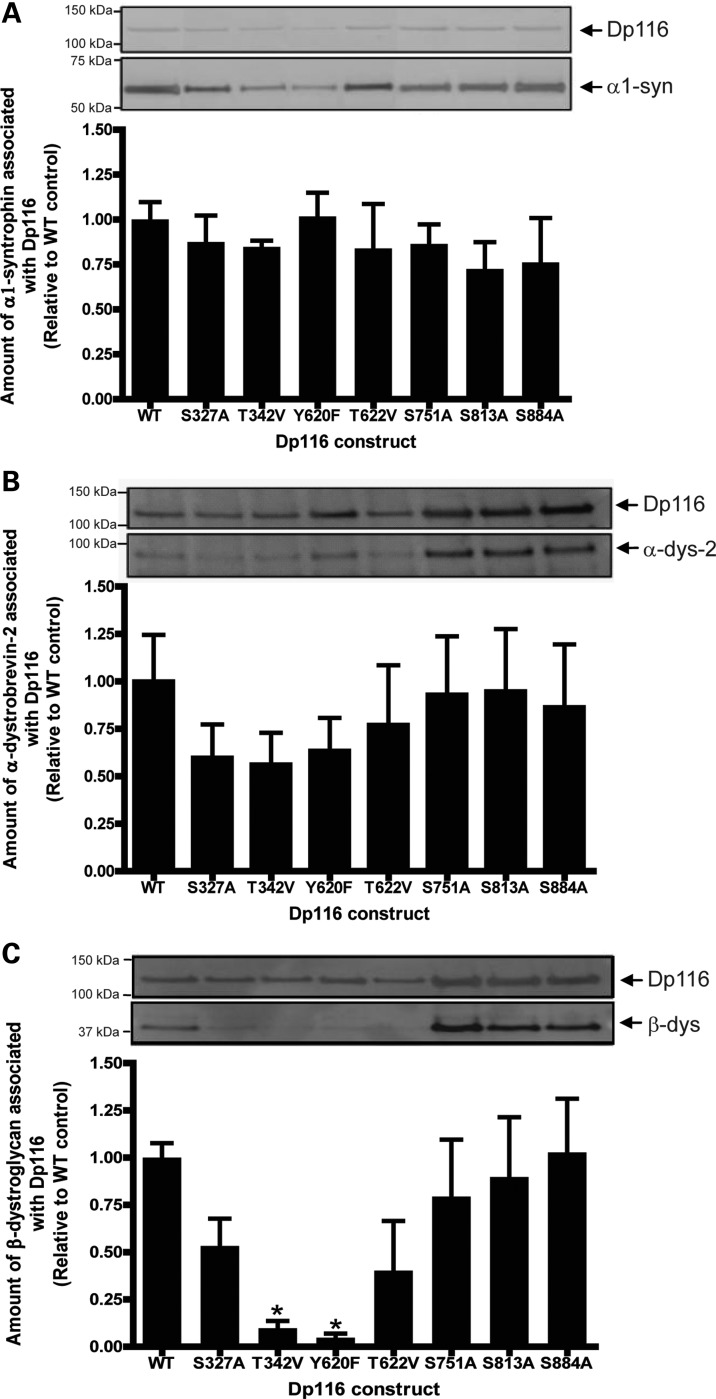
Phosphorylation of amino acids within the dystrophin protein has been hypothesized to regulate interactions with other DGC members (7,26,31). Therefore, the role of phosphorylation at seven sites was examined by introducing mutations to substitute in non-phosphorylatable amino acids (serine to alanine, threonine to valine and tyrosine to phenylalanine) within a Dp116 DNA construct. Four amino acids in the β-dystroglycan-binding domain region, including two amino acids within the WW domain (S327 and T342) and two amino acids downstream of the ZZ domain (Y620 and T622), were investigated as this domain is a known site of protein–protein interaction (6). Three amino acids in the CT domain (S751, S813 and S884) were also chosen as they were also identified in the full-length dystrophin protein. Mass spectrometry analysis of the transfected wild-type (WT) Dp116 protein confirmed that phosphorylation of the transfected protein occurred in human embryonic kidney (HEK)-293 cells, thereby establishing that this system contains the appropriate kinases for phosphorylation of the WT construct (Supplementary Material, Table S3). Co-transfection of each of the Dp116 mutant constructs alongside a construct encoding α1-syntrophin into HEK-293 cells revealed no significant effect in the binding of α1-syntrophin to Dp116, despite S751 lying within the syntrophin-binding domain (Fig. 3A).
Figure 3.
Co-immunoprecipitation studies indicate a requirement for phosphorylation at specific dystrophin residues for protein–protein interactions. HEK-293D cells were transfected with pAAV6:CMV-Dp116WT, pAAV6:CMV-Dp116S327A, pAAV6:CMV-Dp116T342V, pAAV6:CMV-Dp116Y620F, pAAV6:CMV-Dp116T622V, pAAV6:CMV-Dp116S751A, pAAV6:CMV-Dp116S813A or pAAV6:CMV-Dp116S884A in conjunction with pSport:CMV-V5-α1-syntrophin (A), pAAV2:CMV-GFP-α-dystrobrevin-2 (B) or pAAV2:CMV-V5-β-dystroglycan (C). Cells were lysed and the amount of co-transfected protein associating with the Dp116 protein was examined by immunoprecipitation and western immunoblotting. Representative immunoblots are shown. Bands were quantitated from three separate experiments and the amount of protein associated with Dp116 was graphed as a percentage of the control (pAAV6:CMV-Dp116WT). *P < 0.05 compared with WT.
Cotransfection of each of the Dp116 mutant constructs alongside a construct encoding α-dystrobrevin-2 into HEK-293 cells revealed a trend towards reduction in the binding of α-dystrobrevin-2 to Dp116 containing either S327A, T342V or Y620F mutations; however, this did not reach significance (Fig. 3B). However, cotransfection of the Dp116 mutant constructs S327A, T342V, Y620F and T622V alongside a construct encoding β-dystroglycan into HEK-293 cells resulted in a decrease in Dp116 binding to β-dystroglycan, with a significant decrease observed for the T342V and Y620F constructs (Fig. 3C). Phosphorylation at these sites therefore may be important for Dp116 binding to β-dystroglycan.
Phosphorylation at S327 modulates the dystrophin interaction with β-dystroglycan and α-dystrobrevin-2 in vitro
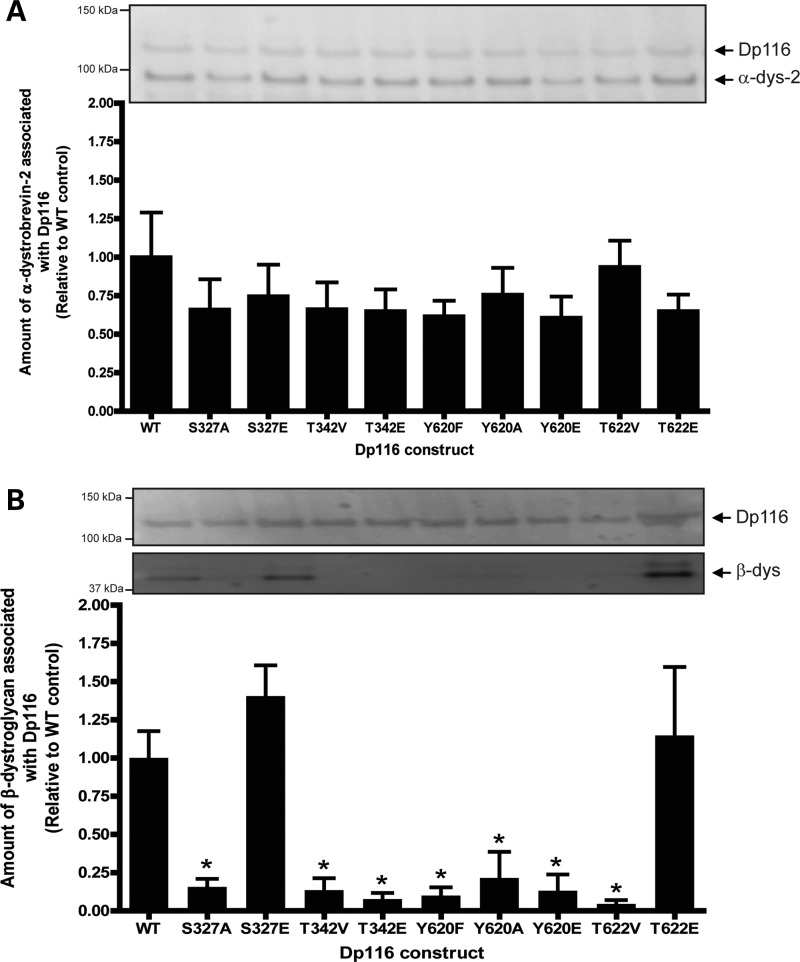
While deficits in protein–protein interactions in the non-phosphorylatable Dp116 mutants is indicative of a requirement for phosphorylation, the possibility exists that the observed changes in binding affinity may be due simply to the presence of a different amino acid at this site. To confirm that the changes in protein–protein interaction were due to the altered phosphorylation status, S327, T342, Y620 and T622 were mutated to the negatively charged amino acid, glutamate, to mimic the negative charge of the phosphate group. We added S327 and T622 for continued analysis even though there was no significant change in its association with β-dystroglycan due to their location in the dystroglycan-binding domain. To control for any change in amino acid structure as well as the lack of charge, Y620 was also mutated to alanine.
Cotransfection of the Dp116 mutants with α-dystrobrevin-2 into HEK-293 cells revealed that the presence of a negative charge at S327, T342, Y620 or T622 did not restore binding of α-dystrobrevin-2 to Dp116 to that of WT control Dp116 levels (Fig. 4A). This suggests that the reduced interaction observed previously was likely an effect of the amino acid change and not the loss of a negative charge at these sites. However, mutation of S327 or T622, but not T342 or Y620, to glutamate completely restored the interaction between β-dystroglycan and Dp116 (Fig. 4B). Therefore, phosphorylation at S327 may be beneficial for binding of β-dystroglycan to Dp116 when coexpressed in HEK-293 cells.
Figure 4.
The introduction of a phosphomimetic mutation at S327 in Dp116 enhances the association of Dp116 with α-dystrobrevin-2 and β-dystroglycan in vitro. HEK-293D cells were transfected with pAAV6:CMV-Dp116WT, pAAV6:CMV-Dp116S327A, pAAV6:CMV-Dp116S327E, pAAV6:CMV-Dp116T342V, pAAV6:CMV-Dp116T342E, pAAV6:CMV-Dp116Y620F, pAAV6:CMV-Dp116Y620A, pAAV6:CMV-Dp116Y620E, pAAV6:CMV-Dp116T622V or pAAV6:CMV-Dp116T622E in conjunction with pAAV2:CMV-GFP-α-dystrobrevin-2 (A) or pAAV2:CMV-V5-β-dystroglycan (B). Cells were lysed and the amount of cotransfected protein associating with the Dp116 protein was examined by immunoprecipitation and western immunoblotting. Bands were quantitated from three separate experiments and the amount of protein associated with Dp116 was graphed as a percentage of the control (pAAV6:CMV-Dp116-WT). *P < 0.01 compared with non-phosphorylated amino acid, **P < 0.05 compared with WT.
Phosphorylation at S327 or T622 restores Dp116 expression and interaction with endogenous α1-syntrophin in murine C2C12 cells
To determine whether the differences in Dp116 construct function were true changes or simply an artifact of overexpression, we also overexpressed Dp116WT, Dp116S327A, Dp116S327E, Dp116T622V and Dp116T622E in C2C12 cells to determine their ability to form the DGC with endogenous binding proteins. Neither Dp116S327A nor Dp116T622V bound α1-syntrophin but this interaction was restored when either site was mutated to glutamate (Fig. 5A). However, in contrast to overexpression in HEK-293 cells, reprobing for the FLAG-tagged Dp116 constructs revealed little to no expression of the non-phosphorylatable mutant Dp116 constructs in the C2C12 system. This suggests that the lack of phosphorylation at S327 or T622 may render the Dp116 protein unstable, resulting in its rapid degradation in skeletal muscle cells in vitro.
Figure 5.
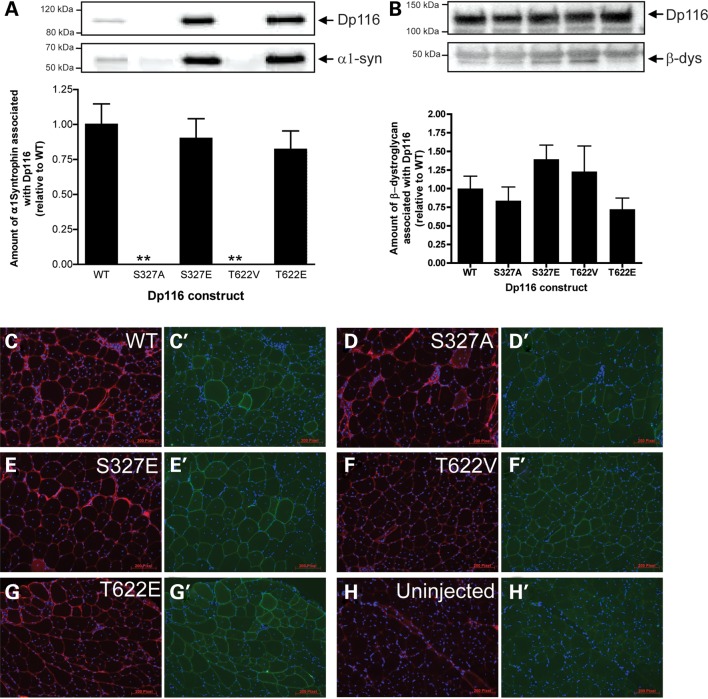
Phosphorylation of S327 modulates the interaction between Dp116 and endogenous DGC proteins. (A) C2C12 cells were transfected with pAAV6:CMV-Dp116WT, pAAV6:CMV-Dp116S327A, pAAV6:CMV-Dp116S327E, pAAV6:CMV-Dp116T622V or pAAV6:CMV-Dp116T622E. Cells were lysed and the amount of endogenous α1-syntrophin associated with the Dp116 protein was examined by immunoprecipitation and western blotting. Bands were quantitated from three separate experiments and the amount of α1-syntrophin protein associated with Dp116 was graphed as a percentage of the control (pAAV6:CMV-Dp116WT). (B) Mdx mice received a single intramuscular injection of vehicle alone (HBSS), or 1 × 1010 vector genomes (vg) of rAAV6:Dp116WT, rAAV6:Dp116S327A, rAAV6:Dp116S327E, rAAV6:Dp116T622V or rAAV6:Dp116T622E into the TA muscle. In 4 weeks post-injection, the TA muscle was excised, homogenized and Dp116 protein was immunoprecipitated. The amount of β-dystroglycan associated with Dp116 was determined by western immunoblotting. Bands were quantitated and the ratio of β-dystroglycan associated with Dp116 was graphed as a percentage of the control (rAAV6:Dp116WT). The sarcolemmal localization of the β-dystroglycan (C–H) and Dp116 (FLAG; C′–H′) proteins was confirmed by immunofluorescence.
Phosphorylation at S327 modulates the Dp116/β-dystroglycan interaction in vivo
Cell culture studies demonstrated that phosphorylation of S327 may regulate binding of the dystrophin Dp116 isoform to β-dystroglycan (Figs 3–5). To determine whether phosphorylation at these sites also regulates the Dp116/β-dystroglycan interaction in vivo, recombinant AAV vectors carrying the different Dp116 constructs (rAAV6:CMV-Dp116WT, rAAV6:CMV-Dp116S327A, rAAV6:CMV-Dp116S327E, rAAV6:CMV-Dp116T622V and rAAV6:CMV-Dp116T622E) were injected into the tibialis anterior (TA) muscles of dystrophic mdx mice. In contrast to in vitro studies, no significant difference was observed for the binding of β-dystroglycan by any mutated construct in vivo (Fig. 5B). However, immunoprecipitation of the Dp116 protein from each injected muscle confirmed that the presence of a negative charge at S327 (S327E) showed a trend towards increased binding to β-dystroglycan which was reduced in the absence of a negative charge (Fig. 5B). Immunofluorescence demonstrated that β-dystroglycan was restored to the sarcolemma in all Dp116 transduced muscle fibers (Fig. 5C–H).
3D Modeling predicts an increased affinity for dystrophin binding to β-dystroglycan when dystrophin S3059 is phosphorylated
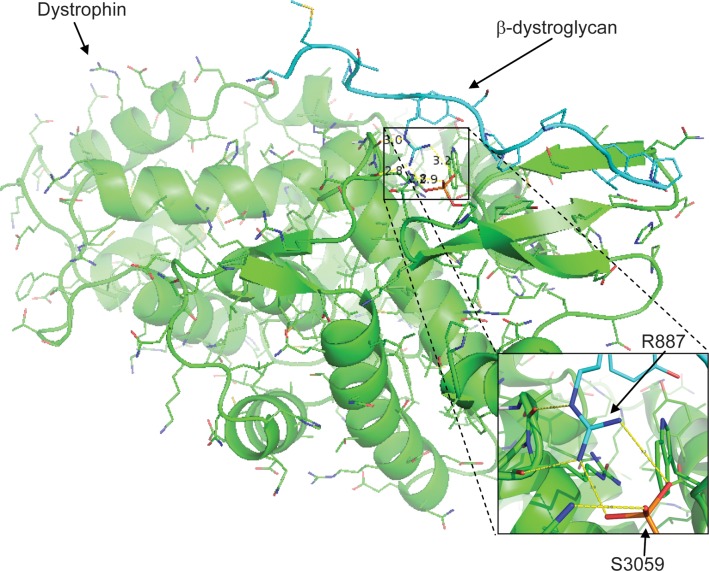
The crystal structure of dystrophin bound to β-dystroglycan [PDB ID 1eg4; (36)] was modeled using the Rosetta3 macromolecular modeling suite (37), where serine 3059 was modeled as a serine or as a phosphorylated serine (Fig. 6). Following minimization of both structures, the Rosetta score was calculated in the peptide-bound and unbound states for each structure, resulting in the binding energy of the peptide to dystrophin in Rosetta energy units (REUs). The unphosphorylated complex gave a binding energy of −17.2 REUs, whereas the phosphorylated complex gave a binding energy of −19.5 REUs. The difference of −2.3 REU corresponds to an ∼1 kcal/mol gain in binding energy due to phosphorylation. The minimized model of the phosphorylated S3059 reveals that the phosphate can make a bidentate salt bridge to R7 of β-dystroglycan, proving a mechanism for how phosphorylation of S3059 increases the affinity of β-dystroglycan for dystrophin.
Figure 6.
3D modeling of the interaction between dystrophin and a β-dystroglycan peptide indicate an enhanced association in the presence of dystrophin S3059 phosphorylation. The interaction between dystrophin (green) and a β-dystroglycan peptide (cyan) as previously demonstrated by Huang et al. (36) was modeled in Rosetta. No constraints were made on the rotation of the phosphate group, it was allowed to rotate freely about all rotatable bonds and is modeled in the position having the lowest energy in Rosetta. This model indicates that the presence of a phosphate at S3059 (orange) on dystrophin creates a negative charge opposite R887 on β-dystroglycan that increases the binding affinity of the interaction between the two proteins. This interaction is shown in greater detail in the inset.
DISCUSSION
Increasing evidence demonstrates that posttranslational modification is essential for proper function of the DGC. The interaction between laminin and the dystroglycan complex is dependent on the glycosylation of α-dystroglycan (38–43). Furthermore, the correct expression and function of β-dystroglycan and dystrobrevin has been demonstrated recently to be dependent on phosphorylation (18,20). While several studies have previously examined the phosphorylation state of the dystrophin protein, all but one (7) have relied on in vitro systems with all the attendant caveats associated with such studies. In an attempt to identify a role for phosphorylation of specific amino acids in the dystrophin protein, we used mass spectrometry to study the phosphorylation of dystrophin isolated from its endogenous environment. Our findings revealed that both the full-length dystrophin protein (Dp427) and the Dp116 isoform are phosphorylated on multiple amino acids in skeletal muscle. Furthermore, phosphorylation at a specific site within the CR region modulated the interaction between dystrophin and β-dystroglycan.
The identification of phosphorylation sites within the endogenous dystrophin protein in healthy skeletal muscle indicates that phosphorylation at these amino acids occurs as part of normal muscle function. While the large size of dystrophin prohibited an in depth analysis of the entire protein, peptides covering >50% of the 427 kDa protein were detected and identified five sites of phosphorylation. Phosphorylation in the dystrophin CT domain has been suggested to regulate binding to α1-syntrophin in vitro (22,26). In these studies, in vitro kinase assays using dystrophin fusion proteins showed that S3616 can be phosphorylated by CaM kinase, reducing association with α1-syntrophin (26). We observed phosphorylation at S3483, S3545 and S3616 in the CT domain of Dp427 in vivo, but mutagenesis of these sites did not reveal any significant changes in the ability of dystrophin to bind either α1-syntrophin or α-dystrobrevin-2 in vitro, despite the proximity of these sites to the known binding regions for α1-syntrophin and α-dystrobrevin-2 (44,45). It is possible that these conflicting results reflect a difference in binding requirements of the Dp116 isoform compared with the full-length 427 kDa dystrophin protein. However, the various syntrophin isoforms interact with the DGC by binding with both dystrophin and α-dystrobrevin (6). We previously showed that deleting the entire syntrophin-binding domain from dystrophin did not prevent the syntrophins from associating with the DGC in vivo, although the relative ratio of the different syntrophin isoforms was altered (46). Thus, the presence or posttranslational modification of the syntrophin-binding domain may be important for modulating the affinity of specific isoforms of syntrophin rather than determining absolute binding.
The Dp116 dystrophin isoform is composed of the CR and CT domains of the dystrophin protein fused to a unique N-terminal domain and is specifically expressed by Schwann cells within the peripheral nervous system (33). Overexpression of Dp116 preserves functional muscle mass and prolongs the lifespan of severely dystrophic mdx/utrn−/− mice without preventing the characteristic degeneration/regeneration cycles of dystrophic skeletal muscle (8). Therefore, the phosphorylation sites observed in the Dp116 protein that were not observed in the Dp427 protein are potentially a consequence of the dystrophic pathology and may reflect changes in the stability and/or function of the dystrophin protein in diseased muscle. In total, we observed phosphorylation of 17 amino acids unique to the Dp116 protein. However, it is important to note that due to the low sequence coverage obtained for the full-length Dp427 protein, many of these sites may not be unique to Dp116. Continued mass spectrometric studies on the Dp427 protein may identify more of these sites.
Phosphorylation has been suggested to regulate the interaction between dystrophin and β-dystroglycan. Early in vitro investigations demonstrated that phosphorylation of tyrosine residues within and adjacent to the WW domain binding motif of β-dystroglycan disrupted its interaction with dystrophin (19). More recently, in vivo studies have confirmed that β-dystroglycan is phosphorylated on Y890 upon disruption of its interaction with dystrophin, resulting in the subsequent degradation of β-dystroglycan (20). Mutagenesis studies of the phosphorylation sites observed in the present study revealed that phosphorylation of dystrophin at S3059, at least in the context of Dp116 (S327), enhanced the interaction with β-dystroglycan. Crystallography studies have demonstrated that S3059, which is located in the WW domain of dystrophin, is in direct contact with β-dystroglycan (36) and therefore it is not surprising that phosphorylation might modulate this interaction. Based on the peptide sequence (ERAISPNKVPY), we predict S3059 to be a substrate for phosphorylation by Erk1, ERK2, CDK2 or CDK5; however, this remains to be determined (34,35). Determination of the kinase responsible for phosphorylation of S3059 in vivo would greatly enhance the ability to alter the activity of the dystrophin protein in skeletal muscle.
Coexpression of Dp116 constructs with either an alanine or glutamate at S327 with β-dystroglycan in HEK-293 cells indicated a lack of Dp116 association with β-dystroglycan in the absence of phosphorylation (S327A) that was restored upon constitutive phosphorylation at this site (S327E). However, viral overexpression of these two proteins in the muscles of mdx mice, while reflecting the in vitro result, demonstrated a more modest effect. This raises the question of whether the changes in β-dystroglycan binding with each mutation results from a physical change in the ability to bind β-dystroglycan, or as a result of increased Dp116 protein expression by the S327E mutant. While it is understood that dystrophin is a long-lived protein and the turnover rate is very low, the half-life of the dystrophin protein in healthy skeletal muscle remains to be precisely determined; however, studies utilizing transgenic mice or exon skipping have suggested the half-life of dystrophin to be ∼2 months (47,48).
The exact mechanism(s) underlying the increased association between Dp116 and β-dystroglycan in the S327E mutant has yet to be determined. However, in support of our data, 3D modeling based on crystallography input (PDB 1eg4) demonstrating the interaction between dystrophin and the β-dystroglycan peptide indicates that phosphorylation of S3059 would create a negative charge precisely opposite the positively charged arginine residue on β-dystroglycan, thereby increasing the affinity of the association between the two proteins. Therefore, we propose a model in which phosphorylation at S327 enhances the association between dystrophin and β-dystroglycan to stabilize the DGC at the sarcolemma. It is unclear whether this increased association has a beneficial or negative effect on the functional capacity of skeletal muscle, but we contend that an increased presence of stable DGC at the sarcolemma would result in increased membrane protection during contraction. Support for this theory comes from the accepted function of the sarcoglycan complex as a stabilizer of the DGC. In the absence of the sarcoglycan complex, associations between dystrophin and α-dystroglycan with β-dystroglycan are weakened, resulting in impaired DGC function at the sarcolemma (49–52). This weakness may also be contributed by interactions with dystrobrevin and the sarcoglycan subcomplex (53). Indeed, in patients with sarcoglycan mutations resulting in limb girdle muscular dystrophy, which also express normal levels of dystrophin, α-dystrobrevin localization to the sarcolemma is dramatically reduced (54). The biological implications of increasing DGC stability through the modulation of a single amino acid within the dystrophin protein are significant. In addition to allowing a better understanding of the mechanistic function of dystrophin within the DGC, this knowledge could inform the development of novel therapeutics for the muscular dystrophies.
In conclusion, the identification of novel phosphorylation sites within the dystrophin protein from skeletal muscles of healthy and dystrophic mice has provided new insights into the regulation of the dystrophin protein in its endogenous state. This is the first study to demonstrate that endogenous dystrophin is phosphorylated on specific amino acids in skeletal muscle. Furthermore, we have provided the first evidence that phosphorylation of an amino acid in the dystrophin WW domain (S3059) enhances the interaction between dystrophin and β-dystroglycan. Not only do these findings provide new information about dystrophin protein regulation, they also suggest approaches to advance therapeutic strategies for muscle wasting and weakness. With specific reference to DMD, current gene replacement therapies involve the delivery of truncated dystrophin proteins to dystrophic skeletal muscle. Studies have shown that the truncated dystrophin proteins are functional at the sarcolemma, but not to the extent of the full-length protein and therefore do not restore full functional capacity (12,13,55–81). Therefore, considerable effort has been invested in searching for microdystrophins with enhanced function (12,14,15,65,82). We have shown that addition of a negative charge to a single amino acid within the β-dystroglycan-binding domain might stabilize dystrophin protein expression and/or increase the β-dystroglycan-binding affinity of truncated dystrophins. Therefore, the mutagenesis of amino acids may increase the therapeutic efficacy of mini- and microdystrophin gene replacement strategies for DMD.
MATERIALS AND METHODS
Animals
C57BL/6 mice were bred in the Department of Comparative Medicine at the University of Washington, Seattle, WA, USA. Mdx4cv:Utrn−/−/Dp116 transgenic mice were generated as previously described (8). C57BL/10ScSnmdx (mdx) mice were obtained from the Animal Resources Centre, WA, Australia. All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Washington and the Animal Ethics Committee of The University of Melbourne and conducted in accordance with the Australian code of practice for the care and use of animals for scientific purposes as stipulated by the National Health and Medical Research Council (Australia).
Cell culture
HEK-293 cells (Invitrogen) were maintained in Dulbecco's modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% l-glutamine on tissue culture plates at 37°C + 5% CO2 and subcultured upon reaching ∼60–70% confluency. The C2C12 myoblast cell line(ATCC) was maintained in DMEM/10% FBS/1% l-glutamine on tissue culture plates at 37°C + 5% CO2 and subcultured upon reaching ∼70% confluency. For experiments utilizing myotubes, C2C12 cells were seeded into 6-well tissue culture plates and grown to >90% confluency. At this point, cells were incubated in DMEM/2% horse serum (HS)/1% l-glutamine for 4 days at 37°C + 5% CO2 to induce differentiation during which time the media was changed every 48 h. Experiments were performed on Day 4 of differentiation at which point healthy myotubes were observed.
Antibodies
The following primary antibodies were used throughout the experiments in 5% skim milk powder/PBS/0.5% Tween-20: horseradish peroxidase (HRP)-conjugated Mouse-anti-M2 FLAG (Sigma-Aldrich; 1:10 000), HRP-conjugated mouse-anti-V5 (Sigma-Aldrich; 1:1000), rabbit-anti GFP (Santa Cruz Biotechnology; 1:1000), mouse-anti-β-dystroglycan (Novocastra Laboratories; 1:500), mouse-anti-α-dystrobrevin-2 (BD Transduction Laboratories; 1:500), rabbit-anti-α1-syntrophin (kind gift from Stan Froehner; 1:500) and goat-anti-α1-syntrophin (Sigma-Aldrich; 1:1000). HRP-conjugated donkey-anti-rabbit immunoglobulin (GE Healthcare Life Sciences) secondary antibody was used at 1:10 000 in 5% skim milk powder/PBS/0.5% Tween-20. HRP-conjugated donkey-anti-mouse immunoglobulin (GE Healthcare Life Sciences) secondary antibody was used at 1:10 000 in 5% skim milk powder/PBS/0.5% Tween-20. HRP-conjugated donkey-anti-goat immunoglobulin (Sigma-Aldrich) secondary antibody was used at 1:10 000 in 5% skim milk powder/PBS/0.5% Tween-20.
Cotransfection assays
For in vitro interaction analyses, pAAV-CMV-FLAG-Dp116 and the various mutant forms were co-transfected into HEK-293 cells alongside pAAV-CMV-V5-β-dystroglycan, pAAV-CMV-α-dystrobrevin-2-GFP or pCMVsport6-V5-α1-syntrophin. 3.7 × 106 HEK-293 cells were seeded into 6-well tissue culture dishes and incubated overnight (O/N) at 37°C + 5% CO2. Upon reaching ∼70% confluency, cells were transfected by calcium phosphate precipitation with 1–2 µg DNA and incubated O/N at 37°C + 5% CO2.
Endogenous pull-down assays
C2C12 (2 × 105) cells were seeded in each well of a 6-well tissue culture plate in 2 ml growth media (DMEM/10% FCS/1% glut) and incubated overnight at 37°C + 5% CO2 to 75% confluency. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol with a few modifications. Briefly, 4 µg DNA and 8 µl Lipofectamine 2000 reagent were each diluted in 0.25 ml OptiMEM (Invitrogen) and incubated at room temperature (RT) for 5 min. Diluted lipofectamine was then gently mixed with the DNA mixture by pipetting and the solution was incubated at RT for 20 min. Growth media was aspirated from C2C12 cells, and the cells rinsed with PBS, and media replaced with 0.5 ml OptiMEM. DNA : lipofectamine complexes were added to each plate, swirled to mix, and incubated at 37°C + 5% CO2 for 4 h. After 4 h, 1 ml growth media was added to each plate and plates were returned to the incubator overnight. The next morning, all media was aspirated from the plate and replaced with 2 ml of fresh growth media. Cells were incubated for 24 h at 37°C + 5% CO2 to recover and induced to differentiate in DMEM/2% HS/1% l-glutamine for 5 days, with media supplemented every 48 h.
Immunoprecipitation and immunoblotting
For in vitro Dp116 transfection analysis, cells were lysed in lysis buffer [1%, v/v, Triton X-100, 50 mm Tris–HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 2 mm Na3VO4, 10 mm NaF, complete protease inhibitor cocktail (Roche Applied Science)] and lysates were cleared by centrifugation at 13 000 rpm for 15 min at 4°C. Cleared lysates were subject to immunoprecipitation with M2 agarose for 12 h at 4°C. M2 beads were washed three times with high salt lysis buffer [1%, v/v, Triton X-100, 50 mm Tris–HCl, pH 7.5, 300 mm NaCl, 1 mm EDTA, 2 mm Na3VO4, 10 mm NaF, complete protease inhibitor cocktail (Roche Applied Science)] and immunocaptured proteins were eluted by incubation for 5 min at 95°C. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) under reducing conditions and electrophorectically transferred onto PVDF membranes. Membranes were blocked for 1 h in 10% (w/v) skim milk and incubated with primary antibody overnight at 4°C. Antibody binding was visualized with peroxidase-conjugated immunoglobulin, and the enhanced chemiluminescence system (Millipore). To reprobe, membranes were stripped of antibodies with 0.1 m glycine, pH 2.9. To quantitate the western blots, the amount of interacting protein was expressed as an amount of the immunoprecipitated Dp116 protein. This was subsequently determined relative to the WT Dp116 construct.
Mass spectrometry
For mass spectrometric analyses, mouse hind-limb muscles (quadriceps, hamstrings, TA and gastrocnemius) were dissected and snap-frozen in liquid N2. Frozen muscles were pulverized in a liquid N2-cooled mortar and pestle and subsequently lysed by dounce homogenization in lysis buffer [1%, v/v, Triton X-100, 50 mm Tris–HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 2 mm Na3VO4, 50 mm NaF, 10 mm β-glycerophosphate and complete protease inhibitor cocktail (Roche Applied Science)]. Lysates were cleared by centrifugation at 13 000 rpm for 15 min at 4°C. For the full-length dystrophin protein, cleared lysates were subject to immunoprecipitation with an antibody directed to the dystrophin protein C-terminus (83) at 4°C overnight and immune complexes were captured the next day with Protein A Sepharose for 1 h at 4°C. For immunoprecipitation of Dp116 from transgenic tissue, cleared lysates were incubated with M2 agarose (Sigma-Aldrich) overnight at 4°C. All beads were washed three times with high salt lysis buffer [1%, v/v, Triton X-100, 50 mm Tris–HCl, pH 7.5, 300 mm NaCl, 1 mm EDTA, 2 mm Na3VO4, 50 mm NaF, 10 mm β-glycerophosphate and complete protease inhibitor cocktail (Roche)], and immunocaptured proteins were eluted by incubation for 5 min at 95°C. Eluted proteins were separated by SDS–PAGE on a 5% Tris–HCl gel and gel bands were visualized by staining with Imperial protein stain (Thermo scientific). Bands corresponding to the dystrophin protein (Dp427 or Dp116) were excised and subject to in-gel digestion with chymotrypsin (25 µg/ml in 0.01% glacial acetic acid) overnight in 50 mm ammonium bicarbonate.
Peptide digests were desalted and separated by nanoflow liquid chromatography using a Waters NanoAcquity UPLC and analyzed by electrospray ionization in the positive ion mode on a Thermo Scientific LTQ Orbitrap hybrid mass spectrometer. Peptides were loaded and washed using a 100 µm i.d. fused silica capillary with a sintered glass frit (Lichrosorb 60 Ǻ (5 µm) Si; Varian) packed with ∼2 cm of 200 Ǻ 5 µm C18AQ particles (Michrom Bioresources) at 4 µl/min with water/acetonitrile (95/05) containing 0.1% (v/v) formic acid. Peptides were then separated and eluted on an analytical column consisting of a 75 µm i.d. fused silica capillary with a gravity-pulled tapered tip packed with ∼30 cm of 100 Ǻ 5 µ C18AQ particles (Michrom). Peptides were eluted using an acetonitrile gradient at a flow rate of ∼200 nl/min and a mobile phase consisting of: A, water (0.1% formic acid) and B, acetonitrile (0.1% formic acid). In brief, the gradient program was: 0–1 min, B (5%); 1–55 min, B (5–35%); 65–75 min, B (35–90%); 80 min, B (5%).
The electrospray voltage was applied via a liquid junction using a gold wire inserted into a micro-tee union (Upchurch Scientific) located in between the precolumn and analytical column. Instrument calibration and ion source conditions were optimized using a tuning solution composed of caffeine (Sigma), MRFA (Bachem) and Ultramark 1621 (Lancaster Synthesis). The heated capillary inlet was maintained at 200°C and injection waveforms for the LTQ linear ion trap and Orbitrap analyzers were kept on for all acquisitions. For the MS ‘survey’ scan, Orbitrap resolution was set to 60 000 (m/z 400) and ion populations were held at 5e5 (2.0 s maximum fill time) through the use of automatic gain control. For MS–MS, precursor ions (minimum signal threshold, 1e3 counts) were isolated and fragmented in the linear ion trap and the corresponding fragments transmitted to the Orbitrap for detection. For this, the ion population in the Orbitrap was set to 2e5 (2.0 s maximum fill time), the LTQ precursor isolation width was set to 4.0 Da, the LTQ collision energy set to 40% (activation Q, 0.25; activation time, 30 ms), and the Orbitrap resolution set to 7500. All data were acquired in the profile mode using an MS ‘survey’ scan over the m/z 350–2000 range followed by MS–MS data-dependent selection of the five most abundant precursor ions contained in the spectrum. For the runs collecting data on doubly and triply charged precursor ions, singularly charged ions and ions with charge-states 4+ or greater were excluded from data-dependent selection using the charge-state exclusion feature. Data redundancy were minimized further by excluding previously selected precursor ions (−0.1 Da/+1.1 Da) for 45 s (140 list maximum) following their selection for MS–MS. All data were acquired using Xcalibur 2.0.2 (Thermo). Raw data files containing tandem mass spectrometry data were converted into peak lists (.dta files) using the instrument vendor's software (extract_msn.exe; Thermo) and searched using SEQUEST (University of Washington). All phosphopeptide calls were further annotated manually for quality and site localization.
Viral vector preparation and in vivo analyses
Flag-tagged Dp116 was generated as described previously (8). Site-directed mutagenesis was performed using the Stratagene Quikchange XL mutagenesis kit as per the manufacturer instructions. For non-phosphorylatable mutations, serine was mutated to alanine, threonine to valine and tyrosine to phenylalanine. For phosphomimetic mutations amino acids were mutated to the negatively charged amino acid, glutamate. All mutations were confirmed by DNA sequencing in the Department of Biochemistry at the University of Washington, Seattle, WA, USA. All vectors were prepared as described previously, using a two-plasmid co-transfection method into HEK-293 cells followed by heparin-affinity purification (84). Vector titer was evaluated via qPCR using the following primers and probe: probe-6FAM-actcatcaatgtatcttatcatg-MGBNFQ; forward primer: ttttcactgcattctagttgtggtt; Reverse primer: catgctctagtcgaggtcgagat. rAAV6 (1 × 1010 vg) or HBSS (control) was injected into the TA muscles of 12-week-old male mdx mice. Four weeks post-injection mice were killed and the TA muscles were dissected and either snap frozen in liquid nitrogen for biochemical analysis or embedded in OCT for histological analysis.
Immunofluorescence
Serial sections (5 μm) were cut transversely through the TA muscle using a refrigerated (−20°C) cryostat (HM525 Cyrostat; Microm International GmbH, Germany). To detect the FLAG-conjugated Dp116 protein, sections were stained with the FITC-conjugated M2 antibody (Sigma-Aldrich). To detect β-dystroglycan, sections were stained with the Novocastra mouse monoclonal β-dystroglycan antibody (Leica Biosystems) with an Alexa Fluor® 555 Goat anti Mouse IgG2a (γ2a) secondary applied. All sections were counterstained with DAPI to visualize nuclei. Digital images were obtained using an upright microscope with camera (Axio Imager D1, Carl Zeiss, Wrek, Göttingen, Germany), controlled by AxioVision AC software (AxioVision AC Rel. 4.7.1, Carl Zeiss).
Statistical analyses
Data were analyzed with the GraphPad Prism software. Statistical significance was determined using a one-way ANOVA with a Dunnett's multiple comparison test to determine significance compared with the WT control. Values are presented as the mean ± SEM.
SUPPLEMENTARY MATERIAL
FUNDING
Supported by National Institutes of Health (NIH) grants AR40864 and AR44533 (to J.S.C.). K.S. was supported by an Early Career Fellowship from the National Health & Medical Research Council of Australia (NH&MRC). P.G. was supported by a Career Development Fellowship (1046782) from the NH&MRC, and previously, a Senior Research Fellowship sponsored by Pfizer Australia. A.L.A. was supported by the Medical Scientist Training Program, and a National Research Service Award (NIH F30NS068005). J.M. was supported by NIH grant GM103533.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Hongwei Qian (Baker IDI Heart and Diabetes Institute) for assistance with manufacture of recombinant AAV vectors. The Baker IDI Heart & Diabetes Institute is supported in part by the Operational Infrastructure Support Program of the Victorian Government.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ervasti J.M., Ohlendieck K., Kahl S.D., Gaver M.G., Campbell K.P. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 2.Ozawa E., Yoshida M., Suzuki A., Mizuni Y., Hagiwara Y., Noguchi S. Dystrophin-associated proteins in muscular dystrophy. Hum. Mol. Genet. 1996;4:1711–1716. doi: 10.1093/hmg/4.suppl_1.1711. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida M., Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. J. Biochem. 1990;108:748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- 4.Ohlendieck K., Campbell K.P. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J. Cell Biol. 1991;115:1685–1694. doi: 10.1083/jcb.115.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramaswamy K.S., Palmer M.L., van der Meulen J.H., Renoux A., Kostrominova T.Y., Michele D.E., Faulkner J.A. Lateral transmission of force is impaired in skeletal muscles of dystrophic mice and very old rats. J. Physiol. 2011;589:1195–1208. doi: 10.1113/jphysiol.2010.201921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abmayr S., Chamberlain J.S. In: Molecular Mechanisms of Muscular Dystrophies. Winder S.J., editor. Gorgetown: Landes Biosciences; 2006. pp. 14–34. [Google Scholar]
- 7.Cox G.A., Sunada Y., Campbell K.P., Chamberlain J.S. Dp71 can restore the dystrophin-associated glycoprotein complex in muscle but fails to prevent dystrophy. Nat. Genet. 1994;8:333–339. doi: 10.1038/ng1294-333. [DOI] [PubMed] [Google Scholar]
- 8.Judge L.M., Arnett A.L., Banks G.B., Chamberlain J.S. Expression of the dystrophin isoform Dp116 preserves functional muscle mass and extends lifespan without preventing dystrophy in severely dystrophic mice. Hum. Mol. Genet. 2011;20:4978–4990. doi: 10.1093/hmg/ddr433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ervasti J.M., Campbell K.P. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.England S.B., Nicholson L.V., Johnson M.A., Forrest S.M., Love D.R., Zubrzycka-Gaarn E.E., Bulman D.E., Harris J.B., Davies K.E. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 11.Yazaki M., Yoshida K., Nakamura A., Koyama J., Nanba T., Ohori N., Ikeda S. Clinical characteristics of aged Becker muscular dystrophy patients with onset after 30 years. Eur. Neurol. 1999;42:145–149. doi: 10.1159/000008089. [DOI] [PubMed] [Google Scholar]
- 12.Harper S.Q., Hauser M.A., DelloRusso C., Duan D., Crawford R.W., Phelps S.F., Harper H.A., Robinson A.S., Engelhardt J.F., Brooks S.V., et al. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 13.Gregorevic P., Allen J.M., Minami E., Blankinship M.J., Haraguchi M., Meuse L., Finn E., Adams M.E., Froehner S.C., Murry C.E., et al. rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat. Med. 2006;12:787–789. doi: 10.1038/nm1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banks G.B., Judge L.M., Allen J.M., Chamberlain J.S. The polyproline site in hinge 2 influences the functional capacity of truncated dystrophins. PLoS Genet. 2010;6:e1000958. doi: 10.1371/journal.pgen.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai Y., Thomas G.D., Yue Y., Yang H.T., Li D., Long C., Judge L., Bostick B., Chamberlain J.S., Terjung R.L., et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J. Clin. Invest. 2009;119:624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koo T., Malerba A., Athanasopoulos T., Trollet C., Boldrin L., Ferry A., Popplewell L., Foster H., Foster K., Dickson G. Delivery of AAV2/9-microdystrophin genes incorporating helix 1 of the coiled-coil motif in the C-terminal domain of dystrophin improves muscle pathology and restores the level of alpha1-syntrophin and alpha-dystrobrevin in skeletal muscles of mdx mice. Hum. Gene Ther. 2011;22:1379–1388. doi: 10.1089/hum.2011.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balasubramanian S., Fung E.T., Huganir R.L. Characterization of the tyrosine phosphorylation and distribution of dystrobrevin isoforms. FEBS Lett. 1998;432:133–140. doi: 10.1016/s0014-5793(98)00804-7. [DOI] [PubMed] [Google Scholar]
- 18.Fratini F., Macchia G., Torreri P., Matteucci A., Salzano A.M., Crescenzi M., Macioce P., Petrucci T.C., Ceccarini M. Phosphorylation on threonine 11 of beta-dystrobrevin alters its interaction with kinesin heavy chain. FEBS J. 2012;279:4131–4144. doi: 10.1111/febs.12006. [DOI] [PubMed] [Google Scholar]
- 19.Ilsley J.L., Sudol M., Winder S.J. The interaction of dystrophin with beta-dystroglycan is regulated by tyrosine phosphorylation. Cell Signal. 2001;13:625–632. doi: 10.1016/s0898-6568(01)00188-7. [DOI] [PubMed] [Google Scholar]
- 20.Miller G., Moore C.J., Terry R., La Riviere T., Mitchell A., Piggott R., Dear T.N., Wells D.J., Winder S.J. Preventing phosphorylation of dystroglycan ameliorates the dystrophic phenotype in mdx mouse. Hum. Mol. Genet. 2012;21:4508–4520. doi: 10.1093/hmg/dds293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sotgia F., Lee H., Bedford M.T., Petrucci T.C., Sudol M., Lisanti M.P. Tyrosine phosphorylation of beta-dystroglycan at its WW domain binding motif, PPxY, recruits SH2 domain containing proteins. Biochemistry. 2001;40:14585–14592. doi: 10.1021/bi011247r. [DOI] [PubMed] [Google Scholar]
- 22.Wagner K.R., Huganir R.L. Tyrosine and serine phosphorylation of dystrophin and the 58-kDa protein in the postsynaptic membrane of Torpedo electric organ. J. Neurochem. 1994;62:1947–1952. doi: 10.1046/j.1471-4159.1994.62051947.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y.W., Thomason D.B., Bullberg D., Jarrett H.W. Binding of laminin alpha1-chain LG4–5 domain to alpha-dystroglycan causes tyrosine phosphorylation of syntrophin to initiate Rac1 signaling. Biochemistry. 2006;45:2042–2052. doi: 10.1021/bi0519957. [DOI] [PubMed] [Google Scholar]
- 24.Calderilla-Barbosa L., Ortega A., Cisneros B. Phosphorylation of dystrophin Dp71d by Ca2+/calmodulin-dependent protein kinase II modulates the Dp71d nuclear localization in PC12 cells. J. Neurochem. 2006;98:713–722. doi: 10.1111/j.1471-4159.2006.03904.x. [DOI] [PubMed] [Google Scholar]
- 25.Madhavan R., Jarrett H.W. Calmodulin-activated phosphorylation of dystrophin. Biochemistry. 1994;33:5797–5804. doi: 10.1021/bi00185a018. [DOI] [PubMed] [Google Scholar]
- 26.Madhavan R., Jarrett H.W. Phosphorylation of dystrophin and alpha-syntrophin by Ca(2+)-calmodulin dependent protein kinase II. Biochim. Biophys. Acta. 1999;1434:260–274. doi: 10.1016/s0167-4838(99)00193-4. [DOI] [PubMed] [Google Scholar]
- 27.Michalak M., Fu S.Y., Milner R.E., Busaan J.L., Hance J.E. Phosphorylation of the carboxyl-terminal region of dystrophin. Biochem. Cell Biol. 1996;74:431–437. doi: 10.1139/o96-047. [DOI] [PubMed] [Google Scholar]
- 28.Milner R.E., Busaan J.L., Holmes C.F., Wang J.H., Michalak M. Phosphorylation of dystrophin. The carboxyl-terminal region of dystrophin is a substrate for in vitro phosphorylation by p34cdc2 protein kinase. J. Biol. Chem. 1993;268:21901–21905. [PubMed] [Google Scholar]
- 29.Shemanko C.S., Sanghera J.S., Milner R.E., Pelech S., Michalak M. Phosphorylation of the carboxyl terminal region of dystrophin by mitogen-activated protein (MAP) kinase. Mol. Cell Biochem. 1995;152:63–70. doi: 10.1007/BF01076464. [DOI] [PubMed] [Google Scholar]
- 30.Walsh M.P., Busaan J.L., Fraser E.D., Fu S.Y., Pato M.D., Michalak M. Characterization of the recombinant C-terminal domain of dystrophin: phosphorylation by calmodulin-dependent protein kinase II and dephosphorylation by type 2B protein phosphatase. Biochemistry. 1995;34:5561–5568. doi: 10.1021/bi00016a030. [DOI] [PubMed] [Google Scholar]
- 31.Senter L., Ceoldo S., Petrusa M.M., Salviati G. Phosphorylation of dystrophin: effects on actin binding. Biochem. Biophys. Res. Commun. 1995;206:57–63. doi: 10.1006/bbrc.1995.1009. [DOI] [PubMed] [Google Scholar]
- 32.Acharyya S., Butchbach M.E., Sahenk Z., Wang H., Saji M., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., et al. Dystrophin glycoprotein complex dysfunction: a regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 2005;8:421–432. doi: 10.1016/j.ccr.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Byers T.J., Lidov H.G., Kunkel L.M. An alternative dystrophin transcript specific to peripheral nerve. Nat. Genet. 1993;4:77–81. doi: 10.1038/ng0593-77. [DOI] [PubMed] [Google Scholar]
- 34.Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S.G., Pandey A. A curated compendium of phosphorylation motifs. Nat. Biotech. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 35.Tagliabracci V.S., Engel J.L., Wen J., Wiley S.E., Worby C.A., Kinch L.N., Xiao J., Grishin N.V., Dixon J.E. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science. 2012;336:1150–1153. doi: 10.1126/science.1217817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X., Poy F., Zhang R., Joachimiak A., Sudol M., Eck M.J. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat. Struct. Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 37.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Combs A.C., Ervasti J.M. Enhanced laminin binding by alpha-dystroglycan after enzymatic deglycosylation. Biochem. J. 2005;390:303–309. doi: 10.1042/BJ20050375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patnaik S.K., Stanley P. Mouse large can modify complex N- and mucin O-glycans on alpha-dystroglycan to induce laminin binding. J. Biol. Chem. 2005;280:20851–20859. doi: 10.1074/jbc.M500069200. [DOI] [PubMed] [Google Scholar]
- 40.Michele D.E., Barresi R., Kanagawa M., Saito F., Cohn R.D., Satz J.S., Dollar J., Nishino I., Kelley R.I., Somer H., et al. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 2002;418:417–422. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]
- 41.Brockington M., Blake D.J., Prandini P., Brown S.C., Torelli S., Benson M.A., Ponting C.P., Estournet B., Romero N.B., Mercuri E., et al. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. Am. J. Hum. Genet. 2001;69:1198–1209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito F., Blank M., Schroder J., Manya H., Shimizu T., Campbell K.P., Endo T., Mizutani M., Kroger S., Matsumura K. Aberrant glycosylation of alpha-dystroglycan causes defective binding of laminin in the muscle of chicken muscular dystrophy. FEBS Lett. 2005;579:2359–2363. doi: 10.1016/j.febslet.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 43.Goddeeris M.M., Wu B., Venzke D., Yoshida-Moriguchi T., Saito F., Matsumura K., Moore S.A., Campbell K.P. LARGE glycans on dystroglycan function as a tunable matrix scaffold to prevent dystrophy. Nature. 2013;503:136–140. doi: 10.1038/nature12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newey S.E., Benson M.A., Ponting C.P., Davies K.E., Blake D.J. Alternative splicing of dystrobrevin regulates the stoichiometry of syntrophin binding to the dystrophin protein complex. Curr. Biol. 2000;10:1295–1298. doi: 10.1016/s0960-9822(00)00760-0. [DOI] [PubMed] [Google Scholar]
- 45.Sadoulet-Puccio H.M., Rajala M., Kunkel L.M. Dystrobrevin and dystrophin: an interaction through coiled-coil motifs. Proc. Natl. Acad. Sci. USA. 1997;94:12413–12418. doi: 10.1073/pnas.94.23.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford G.E., Faulkner J.A., Crosbie R.H., Campbell K.P., Froehner S.C., Chamberlain J.S. Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J. Cell Biol. 2000;150:1399–1410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmad A., Brinson M., Hodges B.L., Chamberlain J.S., Amalfitano A. Mdx mice inducibly expressing dystrophin provide insights into the potential of gene therapy for Duchenne muscular dystrophy. Hum. Mol. Genet. 2000;9:2507–2515. doi: 10.1093/hmg/9.17.2507. [DOI] [PubMed] [Google Scholar]
- 48.Wu B., Lu P., Cloer C., Shaban M., Grewal S., Milazi S., Shah S.N., Moulton H.M., Lu Q.L. Long-term rescue of dystrophin expression and improvement in muscle pathology and function in dystrophic mdx mice by peptide-conjugated morpholino. Am. J. Pathol. 2012;181:392–400. doi: 10.1016/j.ajpath.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Araishi K., Sasaoka T., Imamura M., Noguchi S., Hama H., Wakabayashi E., Yoshida M., Hori T., Ozawa E. Loss of the sarcoglycan complex and sarcospan leads to muscular dystrophy in beta-sarcoglycan-deficient mice. Hum. Mol. Genet. 1999;8:1589–1598. doi: 10.1093/hmg/8.9.1589. [DOI] [PubMed] [Google Scholar]
- 50.Ishikawa-Sakurai M., Yoshida M., Imamura M., Davies K.E., Ozawa E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to beta-dystroglycan. Hum. Mol. Genet. 2004;13:693–702. doi: 10.1093/hmg/ddh087. [DOI] [PubMed] [Google Scholar]
- 51.Iwata Y., Nakamura H., Fujiwara K., Shigekawa M. Altered membrane-dystrophin association in the cardiomyopathic hamster heart muscle. Biochem. Biophys. Res. Commun. 1993;190:589–595. doi: 10.1006/bbrc.1993.1089. [DOI] [PubMed] [Google Scholar]
- 52.Roberds S.L., Ervasti J.M., Anderson R.D., Ohlendieck K., Kahl S.D., Zoloto D., Campbell K.P. Disruption of the dystrophin-glycoprotein complex in the cardiomyopathic hamster. J. Biol. Chem. 1993;268:11496–11499. [PubMed] [Google Scholar]
- 53.Yoshida M., Hama H., Ishikawa-Sakurai M., Imamura M., Mizuno Y., Araishi K., Wakabayashi-Takai E., Noguchi S., Sasaoka T., Ozawa E. Biochemical evidence for association of dystrobrevin with the sarcoglycan-sarcospan complex as a basis for understanding sarcoglycanopathy. Hum. Mol. Genet. 2000;9:1033–1040. doi: 10.1093/hmg/9.7.1033. [DOI] [PubMed] [Google Scholar]
- 54.Metzinger L., Blake D.J., Squier M.V., Anderson L.V., Deconinck A.E., Nawrotzki R., Hilton-Jones D., Davies K.E. Dystrobrevin deficiency at the sarcolemma of patients with muscular dystrophy. Hum. Mol. Genet. 1997;6:1185–1191. doi: 10.1093/hmg/6.7.1185. [DOI] [PubMed] [Google Scholar]
- 55.Abmayr S., Gregorevic P., Allen J.M., Chamberlain J.S. Phenotypic improvement of dystrophic muscles by rAAV/microdystrophin vectors is augmented by Igf1 codelivery. Mol. Ther. 2005;12:441–450. doi: 10.1016/j.ymthe.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Anderson C.L., De Repentigny Y., Cifelli C., Marshall P., Renaud J.M., Worton R.G., Kothary R. The mouse dystrophin muscle promoter/enhancer drives expression of mini-dystrophin in transgenic mdx mice and rescues the dystrophy in these mice. Mol. Ther. 2006;14:724–734. doi: 10.1016/j.ymthe.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 57.Balghi H., Sebille S., Constantin B., Patri S., Thoreau V., Mondin L., Mok E., Kitzis A., Raymond G., Cognard C. Mini-dystrophin expression down-regulates overactivation of G protein-mediated IP3 signaling pathway in dystrophin-deficient muscle cells. J. Gen. Physiol. 2006;127:171–182. doi: 10.1085/jgp.200509456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balghi H., Sebille S., Mondin L., Cantereau A., Constantin B., Raymond G., Cognard C. Mini-dystrophin expression down-regulates IP3-mediated calcium release events in resting dystrophin-deficient muscle cells. J. Gen. Physiol. 2006;128:219–230. doi: 10.1085/jgp.200609559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banks G.B., Combs A.C., Chamberlain J.R., Chamberlain J.S. Molecular and cellular adaptations to chronic myotendinous strain injury in mdx mice expressing a truncated dystrophin. Hum. Mol. Genet. 2008;17:3975–3986. doi: 10.1093/hmg/ddn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bostick B., Shin J.H., Yue Y., Wasala N.B., Lai Y., Duan D. AAV micro-dystrophin gene therapy alleviates stress-induced cardiac death but not myocardial fibrosis in >21-m-old mdx mice, an end-stage model of Duchenne muscular dystrophy cardiomyopathy. J. Mol. Cell Cardiol. 2012;53:217–222. doi: 10.1016/j.yjmcc.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bostick B., Yue Y., Long C., Marschalk N., Fine D.M., Chen J., Duan D. Cardiac expression of a mini-dystrophin that normalizes skeletal muscle force only partially restores heart function in aged mdx mice. Mol. Ther. 2009;17:253–261. doi: 10.1038/mt.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Decrouy A., Renaud J.M., Davis H.L., Lunde J.A., Dickson G., Jasmin B.J. Mini-dystrophin gene transfer in mdx4cv diaphragm muscle fibers increases sarcolemmal stability. Gene Ther. 1997;4:401–408. doi: 10.1038/sj.gt.3300407. [DOI] [PubMed] [Google Scholar]
- 63.Decrouy A., Renaud J.M., Lunde J.A., Dickson G., Jasmin B.J. Mini- and full-length dystrophin gene transfer induces the recovery of nitric oxide synthase at the sarcolemma of mdx4cv skeletal muscle fibers. Gene Ther. 1998;5:59–64. doi: 10.1038/sj.gt.3300553. [DOI] [PubMed] [Google Scholar]
- 64.Dickson G., Roberts M.L., Wells D.J., Fabb S.A. Recombinant micro-genes and dystrophin viral vectors. Neuromusc. Disord. 2002;12(Suppl. 1)):40–44. doi: 10.1016/s0960-8966(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 65.Draviam R.A., Wang B., Li J., Xiao X., Watkins S.C. Mini-dystrophin efficiently incorporates into the dystrophin protein complex in living cells. J. Mus. Res. Cell Motil. 2006;27:53–67. doi: 10.1007/s10974-006-9055-8. [DOI] [PubMed] [Google Scholar]
- 66.Fabb S.A., Wells D.J., Serpente P., Dickson G. Adeno-associated virus vector gene transfer and sarcolemmal expression of a 144 kDa micro-dystrophin effectively restores the dystrophin-associated protein complex and inhibits myofibre degeneration in nude/mdx mice. Hum. Mol. Genet. 2002;11:733–741. doi: 10.1093/hmg/11.7.733. [DOI] [PubMed] [Google Scholar]
- 67.Friedrich O., Both M., Gillis J.M., Chamberlain J.S., Fink R.H. Mini-dystrophin restores L-type calcium currents in skeletal muscle of transgenic mdx mice. J. Physiol. 2004;555:251–265. doi: 10.1113/jphysiol.2003.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gregorevic P., Blankinship M.J., Allen J.M., Chamberlain J.S. Systemic microdystrophin gene delivery improves skeletal muscle structure and function in old dystrophic mdx mice. Mol. Ther. 2008;16:657–664. doi: 10.1038/mt.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimura E., Li S., Gregorevic P., Fall B.M., Chamberlain J.S. Dystrophin delivery to muscles of mdx mice using lentiviral vectors leads to myogenic progenitor targeting and stable gene expression. Mol. Ther. 2010;18:206–213. doi: 10.1038/mt.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kornegay J.N., Li J., Bogan J.R., Bogan D.J., Chen C., Zheng H., Wang B., Qiao C., Howard J.F., Jr, Xiao X. Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs. Mol. Ther. 2010;18:1501–1508. doi: 10.1038/mt.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Odom G.L., Gregorevic P., Allen J.M., Chamberlain J.S. Gene therapy of mdx mice with large truncated dystrophins generated by recombination using rAAV6. Mol. Ther. 2011;19:36–45. doi: 10.1038/mt.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Percival J.M., Gregorevic P., Odom G.L., Banks G.B., Chamberlain J.S., Froehner S.C. rAAV6-microdystrophin rescues aberrant Golgi complex organization in mdx skeletal muscles. Traffic. 2007;8:1424–1439. doi: 10.1111/j.1600-0854.2007.00622.x. [DOI] [PubMed] [Google Scholar]
- 73.Phelps S.F., Hauser M.A., Cole N.M., Rafael J.A., Hinkle R.T., Faulkner J.A., Chamberlain J.S. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum. Mol. Genet. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- 74.Roberts M.L., Wells D.J., Graham I.R., Fabb S.A., Hill V.J., Duisit G., Yuasa K., Takeda S., Cosset F.L., Dickson G. Stable micro-dystrophin gene transfer using an integrating adeno-retroviral hybrid vector ameliorates the dystrophic pathology in mdx mouse muscle. Hum. Mol. Genet. 2002;11:1719–1730. doi: 10.1093/hmg/11.15.1719. [DOI] [PubMed] [Google Scholar]
- 75.Rodino-Klapac L.R., Montgomery C.L., Bremer W.G., Shontz K.M., Malik V., Davis N., Sprinkle S., Campbell K.J., Sahenk Z., Clark K.R., et al. Persistent expression of FLAG-tagged micro dystrophin in nonhuman primates following intramuscular and vascular delivery. Mol. Ther. 2010;18:109–117. doi: 10.1038/mt.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakamoto M., Yuasa K., Yoshimura M., Yokota T., Ikemoto T., Suzuki M., Dickson G., Miyagoe-Suzuki Y., Takeda S. Micro-dystrophin cDNA ameliorates dystrophic phenotypes when introduced into mdx mice as a transgene. Biochem. Biophys. Res. Commun. 2002;293:1265–1272. doi: 10.1016/S0006-291X(02)00362-5. [DOI] [PubMed] [Google Scholar]
- 77.Townsend D., Blankinship M.J., Allen J.M., Gregorevic P., Chamberlain J.S., Metzger J.M. Systemic administration of micro-dystrophin restores cardiac geometry and prevents dobutamine-induced cardiac pump failure. Mol. Ther. 2007;15:1086–1092. doi: 10.1038/sj.mt.6300144. [DOI] [PubMed] [Google Scholar]
- 78.Tsukamoto H., Wells D., Brown S., Serpente P., Strong P., Drew J., Inui K., Okada S., Dickson G. Enhanced expression of recombinant dystrophin following intramuscular injection of Epstein-Barr virus (EBV)-based mini-chromosome vectors in mdx mice. Gene Ther. 1999;6:1331–1335. doi: 10.1038/sj.gt.3300944. [DOI] [PubMed] [Google Scholar]
- 79.Wang Z., Kuhr C.S., Allen J.M., Blankinship M., Gregorevic P., Chamberlain J.S., Tapscott S.J., Storb R. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol. Ther. 2007;15:1160–1166. doi: 10.1038/sj.mt.6300161. [DOI] [PubMed] [Google Scholar]
- 80.Wang Z., Storb R., Halbert C.L., Banks G.B., Butts T.M., Finn E.E., Allen J.M., Miller A.D., Chamberlain J.S., Tapscott S.J. Successful regional delivery and long-term expression of a dystrophin gene in canine muscular dystrophy: a preclinical model for human therapies. Mol. Ther. 2012;20:1501–1507. doi: 10.1038/mt.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y., Duan D. Novel mini-dystrophin gene dual adeno-associated virus vectors restore neuronal nitric oxide synthase expression at the sarcolemma. Hum. Gene Ther. 2012;23:98–103. doi: 10.1089/hum.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Banks G.B., Gregorevic P., Allen J.M., Finn E.E., Chamberlain J.S. Functional capacity of dystrophins carrying deletions in the N-terminal actin-binding domain. Hum. Mol. Genet. 2007;16:2105–2113. doi: 10.1093/hmg/ddm158. [DOI] [PubMed] [Google Scholar]
- 83.Rafael J.A., Cox G.A., Corrado K., Jung D., Campbell K.P., Chamberlain J.S. Forced expression of dystrophin deletion constructs reveals structure-function correlations. J. Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gregorevic P., Blankinship M.J., Allen J.M., Crawford R.W., Meuse L., Miller D.G., Russell D.W., Chamberlain J.S. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat. Med. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.