Abstract
Neurofibromatosis type 1 (NF1) is a common neurodevelopmental disorder in which affected individuals are prone to learning, attention and behavioral problems. Previous studies in mice and flies have yielded conflicting results regarding the specific effector pathways responsible for NF1 protein (neurofibromin) regulation of neuronal function, with both cyclic AMP (cAMP)- and RAS-dependent mechanisms described. Herein, we leverage a combination of induced pluripotent stem cell-derived NF1 patient neural progenitor cells and Nf1 genetically engineered mice to establish, for the first time, that neurofibromin regulation of cAMP requires RAS activation in human and mouse neurons. However, instead of involving RAS-mediated MEK/AKT signaling, RAS regulation of cAMP homeostasis operates through the activation of atypical protein kinase C zeta, leading to GRK2-driven Gαs inactivation. These findings reveal a novel mechanism by which RAS can regulate cAMP levels in the mammalian brain.
INTRODUCTION
Neurofibromatosis type 1 (NF1; OMIM#162200) is one of the most common single gene disorders in which individuals are prone to neurodevelopmental abnormalities. As such, over 60% of children with NF1 develop specific learning disabilities, difficulties with visual-spatial tasks, attention deficits and motor delays (1–5). Moreover, there is an increased prevalence of sleep disturbances (6), autism spectrum abnormalities (7) and impairments in social interactions (8). Collectively, these clinical observations suggest that the NF1 gene is a critical regulator of brain neuronal function.
The NF1 gene encodes a large 220 kDa cytoplasmic protein (neurofibromin) which contains a 300 amino acid domain that functions as a GTPase-activating protein (GAP) for p21-Ras (RAS) (9,10). In both human and mouse cells with a germline NF1 gene mutation, reduced neurofibromin expression is associated with increased RAS and RAS pathway activation (11,12). In support of a primary role for neurofibromin as a negative RAS regulator, pioneering work by Silva and colleagues has shown that neurofibromin controls mouse central nervous system (CNS) neuron function in vivo in a RAS-dependent manner. In these studies, genetic or pharmacologic inhibition of RAS activity ameliorated the learning and memory deficits observed in Nf1+/− mice (13,14).
In contrast, previous studies from our laboratory and others have demonstrated that CNS neuron axonal length, growth cone diameter and survival are dependent on neurofibromin positive regulation of cyclic AMP (cAMP) levels, which cannot be reversed by inhibition of RAS–MEK or RAS–PI3K downstream signaling (15–17). Moreover, additional investigations in Nf1 mutant Drosophila have revealed that the observed learning deficits depend on neurofibromin regulation of cAMP homeostasis, which may operate in either a RAS-dependent (18,19) or RAS-independent (20,21) manner.
Leveraging a combination of pharmacologic and genetic strategies in both human NF1 patient-derived induced pluripotent stem cell (iPSC)-neural progenitor cells (NPCs) and mouse Nf1+/− neurons, we establish that neurofibromin controls cAMP homeostasis in a RAS-dependent manner. In contrast to other cell types, RAS/cAMP regulation does not involve MEK/AKT signaling, but rather operates through the activation of atypical protein kinase C zeta (PKCζ), leading to GRK2-driven Gαs inactivation.
RESULTS
Neurofibromin controls cAMP generation in CNS neurons via Gαs activation
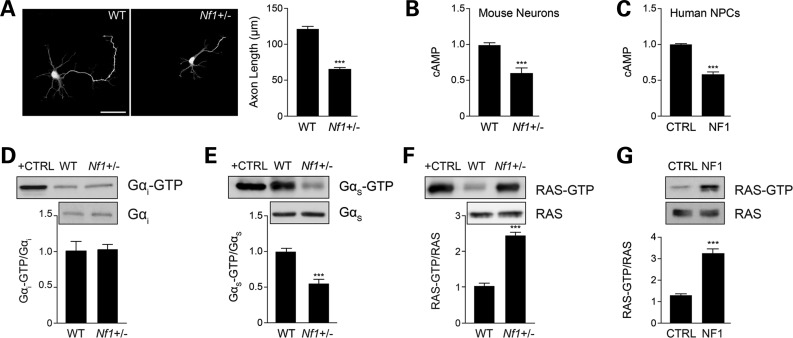
Similarly to previous reports using striatal and retinal ganglion neurons (15,16), mouse primary Nf1+/− hippocampal neurons exhibit shorter axonal lengths relative to their wild-type (WT) counterparts (46% reduction; Fig. 1A) concomitant with reduced intracellular cAMP levels (45% reduction; Fig. 1B). To gain insights into the mechanism underlying neurofibromin cAMP regulation in human NF1 patients, we employed iPSC technology to reprogram and subsequently differentiate NF1 patient as well as sex- and age-matched control skin fibroblasts into NPCs (Anastasaki et al., manuscript in preparation). Similar to the mouse Nf1+/− neurons, NPCs from patients with NF1 (hNF1-NPCs) also have reduced cAMP levels relative to age- and sex-matched control NPCs (47% reduction; Fig. 1C). Next, we examined the activation of the two major heterotrimeric Gα proteins, Gαi (inhibitory) and Gαs (activating), responsible for regulating adenylyl cyclase (AC) activation and cAMP production following G protein-coupled receptor (GPCR) stimulation. While there was no significant change in Gαi activity (Fig. 1D), Nf1+/− hippocampal neurons exhibited decreased Gαs activity (42% reduction; Fig. 1E) compared with WT controls. These data suggest that neurofibromin promotes cAMP production through Gαs activation.
Figure 1.
Neurofibromin regulates cAMP in a RAS/Gαs-dependent manner. (A) Quantification of hippocampal neuron axons lengths by Smi-312 immunostaining. Nf1+/− mouse hippocampal neuron axons are significantly shorter than WT neurons (P < 0.001; n = 200). (B and C) Measurement of cAMP generation in mouse hippocampal neurons and human NF1 patient-derived NPCs (hNF1-NPCs). (B) Nf1+/− neurons have lower cAMP levels relative to their WT counterparts (P = 0.0002; n = 5). (C) hNF1-NPCs have reduced cAMP levels compared with age- and sex-matched controls (P < 0.0001; n = 3). (D and E) Quantification of Gαi and Gαs activation of mouse embryonic hippocampal preparations. (D) Nf1+/− mouse hippocampal preparations show no difference in Gαi activation relative to WT neurons (P = 0.7638; n = 5). (E) Nf1+/− mouse embryonic hippocampal preparations exhibit significantly lower Gαs activity (Gαs-GTP) than their WT counterparts (P = 0.0001; n = 8). (F and G) Measurement of RAS activation in mouse neurons and human NPCs. (F) Nf1+/− mouse hippocampal neurons exhibit higher levels of RAS activation (P = 0.0027; n = 5). (G) hNF1-NPCs (NF1) exhibit higher levels of RAS activation than control (CTRL) NPCs (P = 0.0002; n = 3). Data are presented as means ± SEM (n ≥ 3). ***P < 0.001; Student's t-test.
Neurofibromin-regulated Gαs activation is RAS-dependent
Since neurofibromin functions as an endogenous inhibitor of RAS activity by catalyzing the conversion of RAS from its active GTP-bound to its inactive GDP-bound form (13,22), we next examined RAS-GTP levels in Nf1+/− hippocampal preparations and hNF1-NPCs. In these experiments, Nf1+/− mouse hippocampal neurons had 2.5-fold increased RAS activity relative to WT neurons (Fig. 1F), whereas hNF1-NPCs had 3-fold higher RAS activity relative to age- and sex-matched control NPCs (Fig. 1G).
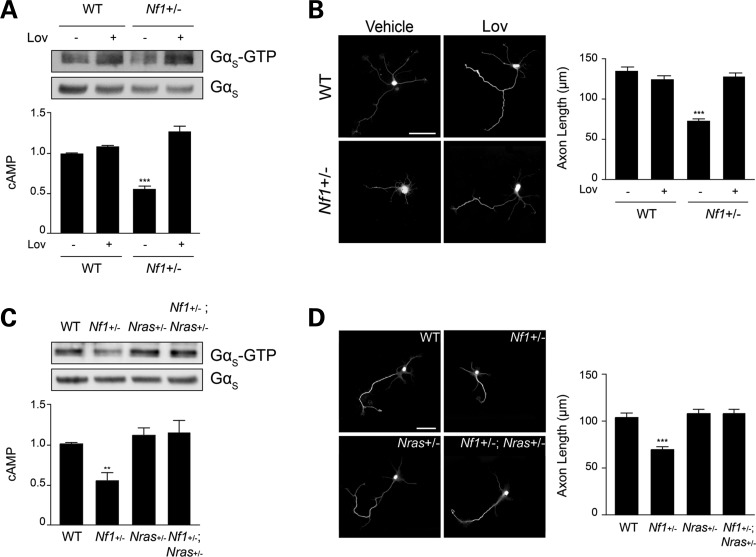
To determine whether RAS is responsible for neurofibromin regulation of cAMP-dependent axonal length, we employed a combination of pharmacologic and genetic approaches. First, we inhibited RAS function with the farnesyltransferase inhibitor lovastatin (Supplementary Material, Fig. S1A), previously employed to reverse the cognitive deficits in Nf1 mutant mice (14). Following continuous lovastatin exposure in vitro, the attenuated Gαs activity, lower cAMP levels and reduced axonal lengths observed in Nf1+/− neurons were corrected (Fig. 2A and B). Secondly, since hippocampal neurons only express Nras and Kras, but not Hras (Supplementary Material, Fig. S1B–D), we focused on genetically engineered mouse strains in which one allele of the Nras (LSL-NrasG12D) or Kras (LSL-KrasG12D) genes were inactivated (23,24). Intercrossing Nf1+/− mice with LSL-NrasG12D or LSL-KrasG12D mice yielded Nf1+/− mice with reduced Nras or Kras expression, respectively. Consistent with the lovastatin results, neurons from Nf1+/− mice with reduced Nras (Nf1+/−;LSL-NrasG12D; Fig. 2C–D) or Kras (Nf1+/−;LSL-KrasG12D; Supplementary Material, Fig. S1E and F) expression exhibited Gαs activity, cAMP levels and axonal lengths similar to those observed in WT mice. Collectively, these data establish RAS as a primary regulator of CNS neuron cAMP generation.
Figure 2.
Pharmacologic and genetic reduction of RAS activity corrects Nf1+/− neuronal defects. (A) Quantification of Gαs activation and cAMP levels in mouse hippocampal neuron preparations. Nf1+/− mouse neuronal Gαs activity (P < 0.0001; n = 6) and cAMP generation (P < 0.0001; n = 5) are restored to WT levels after lovastatin (Lov) treatment. (B) Measurement of axonal lengths by Smi-312 immunostaining. Lov administration restores Nf1+/− mouse hippocampal neuron axonal lengths to WT levels (P < 0.0001; n = 150). (C) Genetic Nras reduction restores Gαs activation (P < 0.0001; n = 6) and cAMP levels (P < 0.005; n = 4) in Nf1+/− mouse hippocampal preparations to WT levels. (D) Smi-312 immunostaining of mouse hippocampal neurons. Nf1+/−; Nras+/− neurons exhibit axonal lengths indistinguishable from WT neurons (P < 0.0001; n = 73). Data are presented as means ± SEM (n ≥ 5). **P < 0.01; ***P < 0.001; One-way ANOVA with Bonferroni post-test correction. Scale bars 50 µm.
Neurofibromin regulates atypical PKC activity in a RAS-dependent manner
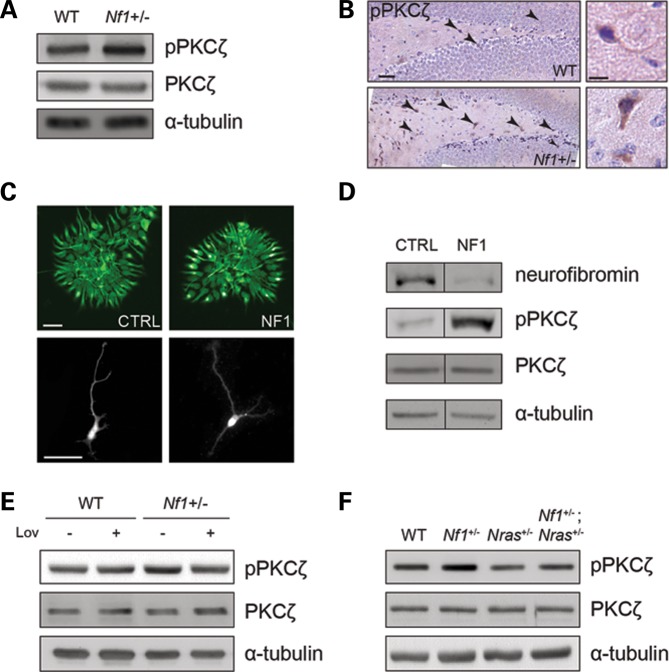
To further define the mechanism underlying RAS regulation of Gαs activation, we initially examined the activity of the major downstream RAS effectors. Employing both embryonic Nf1+/− mouse hippocampi and hNF1-NPCs, no differences in AKT, ERK, p38 MAPK or JNK activation were identified (Supplementary Material, Fig. S2). In contrast, there was a 2-fold increase in atypical PKCζ Thr-403 phosphorylation in Nf1+/− primary hippocampal neuron cultures in vitro (Fig. 3A) as well as in adult Nf1+/− mice in vivo relative to their WT counterparts (Fig. 3B; Supplementary Material, Fig. S3A). Importantly, 2.5-fold increased PKCζ phosphorylation was also observed in hNF1-NPCs relative to age- and sex-matched controls (Fig. 3D), thereby implicating PKCζ as a potential novel effector of neurofibromin/RAS signaling in the brain.
Figure 3.
RAS regulates Gαs/cAMP activity in a PKCζ-dependent manner. (A) Immunoblot analysis of PKCζ activity in mouse hippocampal neuron preparations. PKCζ activity (phosphorylation) is increased in Nf1+/− neurons in vitro (n = 5). (B) Immunohistochemical detection of active PKCζ in adult mouse hippocampi. PKCζ activity is increased in Nf1+/− mouse hippocampi in vivo (n = 3). Insets depict representative immunopositive neurons. Scale bar left: 50 µm; right: 12.5 µm. (C) Smi-312 immunostaining of human NPC colony cultures. Representative images of control (CTRL) and NF1 patient (NF1) NPC colonies (top panels) and their derivative differentiated neurons (bottom panels). Scale bars: 50 µm. (D) Immunoblot analysis of PKCζ activity in human NPCs. PKCζ is activated in human NF1 patient-derived NPCs (NF1) relative to those from age- and sex-matched control (CTRL) individuals (n = 2). (E and F) Immunoblot analysis of PKCζ phosphorylation in Nf1+/− mouse neurons after pharmacologic or genetic reduction of RAS activity. (E) Lov treatment or (F) genetic Nras reduction restores PKCζ activity to WT levels (n = 3).
Since previous studies have shown that RAS and PKCζ physically interact to result in RAS-mediated atypical PKC activation (25), we sought to determine whether RAS activation was required for PKCζ phosphorylation. Consistent with a model in which RAS regulates PKCζ function, both pharmacologic and genetic reduction of RAS activity restored mouse Nf1+/− neuron PKCζ activation to WT levels (Fig. 3E and F). Together these experiments establish RAS as a critical regulator of PKCζ activity.
RAS inhibits Gαs-GTP through PKCζ phosphorylation
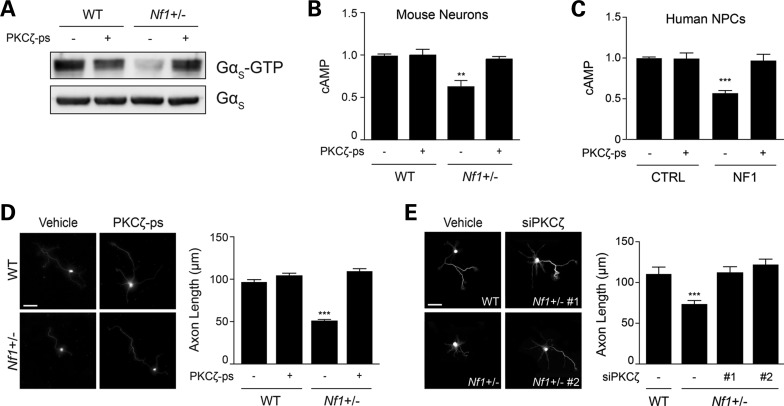
We next sought to determine whether PKCζ activation is responsible for regulating cAMP homeostasis in Nf1+/− CNS neurons and hNF1-NPCs. First, we employed two independent small molecule inhibitors of PKCζ activity (PKCζ pseudosubstrate, PKCζ-ps and PITenin7, PIT7). Following PKCζ-ps or PIT7 treatment, PKCζ activation was decreased in both Nf1+/− hippocampal neurons (2.2- and 2-fold reduction, respectively; Supplementary Material, Fig. S3B and D) and hNF1-NPCs (2.5- and 1.8-fold reduction, respectively; Supplementary Material, Fig. S3C and E). Moreover, after PKCζ-ps (Fig. 4A, B and D) or PIT7 (Supplementary Material, Fig. S3D and F) administration, Gαs activity, cAMP levels and axonal lengths in Nf1+/− neurons were indistinguishable from WT controls. Similarly, in hNF1-NPCs, PKCζ-ps (Fig. 4C) or PIT-7 (Supplementary Material, Fig. S3E) treatment increased cAMP levels by 2-fold.
Figure 4.
Pharmacologic and genetic inhibition of PKCζ restores neuronal defects. (A) Immunoblot analysis of Gαs activity of mouse hippocampal neurons following administration of the PKCζ pseudosubstrate (PKCζ-ps). Treatment of Nf1+/− mouse hippocampal neurons with PKCζ-ps increases Gαs activity (P < 0.0001; n = 3). (B and C) Quantification of cAMP levels in mouse neurons and human-derived NPCs after PKCζ-ps treatment. PKCζ-ps administration restores cAMP in (B) Nf1+/− mouse hippocampal neurons (P < 0.005; n = 4) and (C) human NF1 patient-derived NPCs (P < 0.0001; n = 3) to control levels. (D and E) Smi-312 immunostaining of mouse hippocampal neurons and measurements of axonal length. (D) PKCζ-ps treatment corrects the axonal length defects of Nf1+/− mouse neurons to WT levels (P < 0.0001; n = 200). (E) Genetic inhibition of PKCζ with siRNA in Nf1+/− mouse neurons restores axonal length to WT levels (P < 0.001; n = 150). Data are presented as means ± SEM. **P < 0.01; ***P < 0.001; One-way ANOVA with Bonferroni post-test correction. Scale bars: 50 µm.
Secondly, to establish PKCζ as a key regulator of cAMP-driven axonal length, we reduced PKCζ protein expression in Nf1+/− neurons using two independent siRNA constructs (46 and 54% reduction in protein expression; Supplementary Material, Fig. S3G). In these experiments, both siRNA constructs restored Nf1+/− neuron axonal lengths to WT levels (Fig. 4E). These results demonstrate that PKCζ activation is necessary for Gαs-modulated cAMP homeostasis in mouse and human CNS neuronal cultures.
Thirdly, to mimic the increased PKCζ activation observed in Nf1+/− neurons, WT neurons were treated with phosphatidic acid (PA) dioleoyl to activate PKCζ (2-fold). Following PA administration, WT neurons had lower cAMP levels and shorter axonal lengths (Supplementary Material, Fig. S3H and I). Taken together, these data reveal that neurofibromin-regulated cAMP generation requires PKCζ function in mammalian neurons.
PKCζ regulates cAMP homeostasis through GRK2
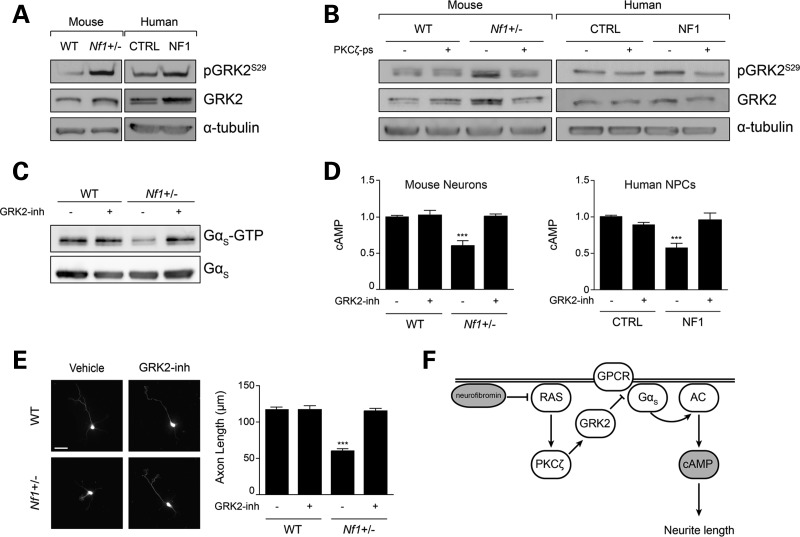
Based on these findings, we explored the possibility that PKCζ blocks Gαs activation by modulating GPCR function. GPCR signal transduction is controlled by GPCR kinases (GRKs), such that agonist-bound GPCRs are phosphorylated by GRKs to cause receptor de-sensitization (26). Since chronic PKC activity can activate GRK2 (27), we hypothesized that PKCζ blocks Gαs activation by modulating GRK2-dependent GPCR signaling. Consistent with this hypothesis, we found 2- and 3-fold increases in GRK2 phosphorylation (at Ser-29 and Ser-685, respectively) in both Nf1+/− mouse CNS neurons and hNF1-NPCs (Fig. 5A, Supplementary Material, Fig. S4C and D). In addition, there was increased total GRK2 expression in Nf1+/− mouse CNS neurons (2-fold) and hNF1-NPCs (2-fold; Fig. 5A). Since GRK2-mediated GPCR phosphorylation uncouples Gαs from AC, we reasoned that PKCζ downstream signaling might activate GRK2 to attenuate Gαs activation. To evaluate this, primary mouse CNS neuronal cultures and hNF1-NPCs were treated with PKCζ-ps. Following PKCζ inhibition, GRK2 phosphorylation was reduced by 2-fold (near WT levels; Fig. 5B), indicating that PKCζ signaling is necessary for GRK2 activation.
Figure 5.
PKCζ inhibits Gαs/cAMP in a GRK2-dependent manner. (A) Immunoblot analysis of GRK2 in mouse neurons and human-derived NPCs. GRK2 activation (phosphorylation) and total protein expression is increased in both Nf1+/− mouse neurons and human NF1 patient-derived NPCs. (B) Immunoblot analysis of GRK2 activity in mouse neurons and human NPCs after PKCζ-ps treatment. PKCζ-ps administration normalizes GRK2 activity in mouse Nf1+/− neurons and human NF1 patient-derived NPCs. (C) Immunoblot analysis of Gαs activity following GRK2 inhibition (GRK2-inh) in mouse hippocampal preparations. GRK2-inh treatment restores Gαs activity to WT levels (P < 0.0001; n = 3). (D) Quantification of cAMP levels in mouse neurons and human NPCs after GRK2-inh treatment. GRK2-inh corrects cAMP in Nf1+/− mouse neurons (P = 0.0011; n = 3) and human NF1 patient-derived NPCs (P < 0.0001; n = 3) to WT levels. (E) Measurement of mouse neuron axonal length by Smi-312 immunoblotting following GRK2-inh treatment. GRK2-inh treatment corrects Nf1+/− mouse neuron axonal lengths to WT levels (P < 0.0001; n = 132). (F) Schematic representation of the proposed mechanism underlying neurofibromin cAMP regulation in mammalian CNS neurons. Data are presented as means ± SEM. ***P < 0.001; One-way ANOVA with Bonferroni post-test correction. Scale bar: 50 µm.
Lastly, to determine whether GRK2 mediates neurofibromin/RAS/PKCζ regulation of Gαs activity, cAMP levels and axonal length, we employed the GRK2 inhibitor, βARK1 inhibitor (GRK2-inh). Following GRK2-inh treatment of either Nf1+/− mouse neurons or hNF1-NPCs, GRK2 phosphorylation was decreased by 2- or 2.3-fold, respectively (Supplementary Material, Fig. S4A and B). Moreover, GRK2 inhibition normalized Nf1+/− mouse neuronal Gαs activity, cAMP levels and axonal lengths (Fig. 5C–E) as well as hNF1-NPCs cAMP levels (Fig. 5D). Together, these findings demonstrate that neurofibromin/RAS regulate mammalian CNS neuron cAMP homeostasis through GRK2-mediated attenuation of GPCR-Gαs activation (Fig. 5F).
DISCUSSION
The majority of what is known about neurofibromin regulation of cAMP regulation derives from studies in Drosophila. In the fly, Nf1 loss leads to a neuromuscular junction overgrowth phenotype (28,29), olfactory learning and memory defects (21,30,31), reduced lifespan (32) and somatic growth deficits (17,19). The use of human NF1 gene mutants lacking a functional RAS regulatory GAP-related domain (GRD) have revealed that some of these Nf1 fly phenotypes could be rescued (31), while others were not (19,31). These experimental observations establish both RAS-dependent and RAS-independent functions for neurofibromin in the Drosophila nervous system. In support of a RAS-independent neurofibromin mechanism of action, subsequent studies demonstrated that many of these abnormal Nf1 mutant phenotypes were rescued by manipulations that elevated cAMP and PKA activity, rather than by acting through RAS/RAF (19,20,28–30,32). However, to complicate matters, in Drosophila Nf1-dependent cAMP regulation does not always require RAS-GRD activity. In this regard, neurofibromin-controlled long-term memory and reduced somatic growth require GRD function (19,21,31), whereas immediate memory, lifespan, resistance to oxidative stress and neuromuscular junction overgrowth are GRD-independent (21,29,31,32). Moreover, these non-GRD mechanisms may involve either amino terminal (29) or carboxyl terminal (31) neurofibromin domains not primarily involved in RAS-GRD function.
To determine how neurofibromin regulates cAMP in the mammalian brain, we leveraged iPSC-NPCs from human NF1 patients in combination with Nf1 GEM neurons. In the current study, we employed a complementary combination of genetic and pharmacologic strategies to establish a novel mechanism for neurofibromin regulation of cAMP generation in CNS neurons. Specifically, we demonstrate that neurofibromin/cAMP homeostasis operates in a RAS-dependent manner, but functions through PKCζ phosphorylation of GRK2 and suppression of Gα activity, rather than through the canonical RAS/RAF/MEK or RAS/AKT effector pathways. Collectively, these experimental observations raise several important points relevant to neurofibromin function in the vertebrate brain.
First, our findings reconcile a series of seemingly contradictory reports demonstrating that RAS inhibition, using either farnesyltransferase inhibitors (14) or genetic knockdown (13), restores Nf1 mutant mouse neuronal dysfunction, but that impaired cAMP generation in Nf1+/− mouse neurons could not be rescued by PI3-Kinase or MEK pharmacologic inhibition (15). The discovery that neurofibromin regulates cAMP in a RAS-dependent manner, but involving a distinct downstream effector pathway (PKCζ-GRK2) separable from RAS/PI3K and RAS/MEK signaling, brings some mechanistic clarity to this issue.
Secondly, the studies in CNS neurons reported herein also demonstrate that neurofibromin regulation of RAS downstream signaling can be cell type-specific. In this regard, RAS activation resulting from reduced or absent neurofibromin expression in neointimal cells (33), leukemic cells (34), some NPCs (35), hematopoietic cells (36) and osteoblasts (37) operates through a MEK-dependent pathway. In astrocytes and Schwann cells, Nf1 loss leads to both MEK/ERK and AKT/mTOR hyperactivation to result in dysregulated cell growth (38–40). However, abnormal neurofibromin function in microglia (41) and osteoblast progenitors (42) involves JNK signaling. As such, the observation that neurofibromin/RAS transmits its regulatory signal through an atypical PKC in CNS neurons and in human patient-derived NPCs provides further experimental evidence for the use of distinct RAS downstream effectors in different cell types. This cell type specificity should be considered when extrapolating results from other tissues relevant to therapeutic drug design for NF1 patient neuronal dysfunction.
Thirdly, neurofibromin regulation of cAMP functions at the level of GPCR-Gα signaling to AC. In this manner, pharmacologic treatments that activate AC ameliorate the neuronal defects observed in Nf1+/− mouse neurons (15). However, rather than involving Gαi activation, as observed in Nf1-deficient astrocytes (43,44), neurofibromin regulation of cAMP in CNS neurons involves GPCR suppression of Gαs activation. Similarly, the engagement of Gαs in neurons is also required for neurotransmitter-induced neurofibromin/cAMP generation and learning in flies (18,31). Neurofibromin control of cAMP in Drosophila operates in a RAS-dependent manner, thus reinforcing both cell type- and species-related differences in the mechanisms underlying neurofibromin/cAMP signaling. Further studies will be required to more precisely define how GPCR-Gα protein coupling to AC is regulated by neurofibromin in astrocytes and neurons in the CNS.
Fourthly, we show that neurofibromin control of cAMP generation involves RAS-mediated PKCζ engagement to modulate GPCR signaling through GRK2. Previous studies have demonstrated that RAS can activate PKCζ (45,46) as well as regulate vascular endothelial growth factor transcription in a PKCζ-dependent manner (47). In addition, PKCζ can physically bind to RAS (25,48), supporting a direct interaction underlying RAS/PKCζ activation. While atypical PKC molecules have not been previously shown to phosphorylate GRK2, our studies demonstrate that inhibition of PKCζ function impairs GRK phosphorylation and expression. Since GRK2 lacks a consensus PKC phosphorylation motif, additional investigations will be required to determine whether PKCζ directly phosphorylates GRK2 or operates indirectly through another, currently unidentified, kinase molecule. Similarly, while GRK2 can regulate cAMP homeostasis in cardiac fibroblasts (49), it is not known how GRK2 regulates GPCR function, which could operate at the level of desensitization or resensitization (50). Nonetheless, the fact that neurofibromin modulates cAMP generation downstream of cell type-specific GPCRs (20,51,52) responsive to distinct ligands and extracellular signals provides a novel way for neurofibromin to regulate a diverse number of GPCRs in distinct CNS cell populations and suggests previously unexplored strategies for correcting NF1-related CNS deficits.
Finally, the availability of human NF1-NPCs as a complementary resource to study CNS neuronal function in this common neurogenetic condition provides unprecedented opportunities to validate observations initially made in rodent systems relevant to future translation to the treatment of children and adults with NF1.
MATERIALS AND METHODS
Mice
Nf1+/− (53), LSL-KRasG12D (24), LSL-NRasG12D (23) mice were generated as previously described. All mice were maintained on an inbred C57BL/6 background and used in accordance with an approved Animal Studies protocol at the Washington University School of Medicine. All mice had ad libitum access to food and water. Littermate controls were used for all experiments.
Human iPSC generation and NPC differentiation
Primary fibroblast cultures from individuals with an established diagnosis of NF1 (NIH Consensus Development Conference 1988) or age- and sex-matched controls were collected (54) reprogrammed into iPSCs as previously described (55). In brief, confluent fibroblast cultures were reprogrammed into iPSCs using Cyto-Tune technology (Invitrogen). Cultures were infected once with integration-free Sendai virus carrying the four Yamanaka stem cell reprogramming factors (OCT4, KLF4, SOX2, C-MYC) and cultured for ∼6 weeks. iPSC colonies were isolated, and their pluripotency was confirmed by assessing their morphology and expression of stem cell markers (OCT4, SSEA-3, TRA-1-60/81). Chromosomal analysis ensured normal karyotype (Anastasaki et al., manuscript in preparation). Two clones from each iPSC line were cultured in Neural Induction Medium (NIM; STEMCell Technologies) as previously described (56), to form embryoid bodies (EBs) for 5 days. EB aggregates were then plated in NIM on adhesive plates pre-coated with poly-ornithine/laminin to allow for rosette formation. Once established, neural rosettes were collected, gently dissociated and re-plated in PLO/laminin-coated plates to differentiate into NPCs. A portion of the NPCs spontaneously differentiated into neurons, which were then analyzed by immunofluorescence. NPC pellets were snap-frozen in liquid nitrogen for western blot analysis.
Primary neuronal cultures
Primary hippocampal neuronal cultures were generated from E12.5–13 mouse embryos as previously described (15). Lovastatin (6.25 µM; Sigma), PKCζ pseudosubstrate (PKCζ-ps), myristoylated (0.5 µm; Enzo Life Sciences), PITenin-7 (PIT7, 5 µm; Millipore), Phosphatidic Acid, dioleoyl (PA dioleoyl, 5 µm; Enzo Life Sciences) or βARK1 inhibitor (GRK2-inh, Methyl 5-[2-(5-nitro-2-furyl)vinyl]-2-furoate), 5 µm; Millipore) were added to the culture media 1h after initial neuronal plating for the entire 3-day culture period. A minimum of three animals per genotype were used and experiments were repeated at least three times with identical results.
Lentivirus generation and lentiviral infection of primary hippocampal neurons
HEK-293T (293T) cells were cultured in DMEM (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% Pen/Strep solution (Gibco). Twenty-four hours before transfection, 293T cells were seeded at 300 000 cells/well in 6-well plates in antibiotic-free media. Three micrograms of total DNA was transfected per well (1.5 µg shPKCζ or pLKO-GFP control; 1.5 µg lentiviral packaging constructs) using Fugene HD reagent (Promega) following manufacturer's instructions. The media was replaced with fresh DMEM supplemented with 10% FBS and 1% antibiotics 12 h post-transfection. Viral preparations were collected at 48 and 72 h post-transfection, aliquoted and frozen at −80°C. Viral titers of 108–109 were used for primary neuronal infections. The sequences of the siRNA constructs employed are shown below:
| Sample | TRC identifier | Vector | Sequence |
|---|---|---|---|
| siPKCζ #1 | TRCN0000022871 | pLKO.1 | 5′-GCACTGGGTGTCCTTATGTTT-3′ |
| siPKCζ #2 | TRCN0000022873 | pLKO.1 | 5′-GAAGTGCTCATCATTCATGTT-3′ |
Primary hippocampal neurons were cultured in 10 cm diameter Poly-d-Lysine/Laminin-coated culture plates as previously described (15) for 1 day. Viral dilutions of 1:1000 v/v were prepared in Neurobasal media with added Polybrene (1:10 000 v/v), and were subsequently administered directly to the neurons for 3 h. Following infection, the media was completely aspirated and replaced with fresh Neurobasal media. The cultures were allowed to grow for a total of 3 days, at which time the cells were collected for further immunocytochemical or protein analysis.
Immunocytochemistry and immunohistochemistry
Immunocytochemistry was performed as previously described (15). Images were acquired on an inverted Olympus FV-500 confocal microscope and analyzed using ImageJ software (http://rsbweb.nih.gov/ij/; Wayne Rasband, National Institute of Mental Health, Bethesda, MD). Immunohistochemistry was performed on mice perfused transcardially with 4% paraformaldehyde (PFA) in 0.1 M sodium phosphate buffer (pH 7.4) and post-fixed in 4% PFA prior to paraffin embedding, as previously described (57). Appropriate primary antibodies (Table 1) and secondary antibodies were used.
Table 1.
Primary antibodies
| Antibody | Source | Host | Dilution | Application |
|---|---|---|---|---|
| Active-Gαi-GTP | NewEast Biosciences | Mouse | 1:1000 | IP |
| Active-GαS-GTP | NewEast Biosciences | Mouse | 1:1000 | IP |
| Akt | Cell Signaling | Rabbit | 1:1000 | WB |
| GRK2 | Cell Signaling | Rabbit | 1:500 | WB |
| Gαi | NewEast Biosciences | Mouse | 1:1000 | WB |
| GαS | NewEast Biosciences | Mouse | 1:1000 | WB |
| Hras (F235) | Santa Cruz Biotechnology | Mouse | 1:500 | WB |
| Kras (F234) | Santa Cruz Biotechnology | Mouse | 1:500 | WB |
| Nras (F155) | Santa Cruz Biotechnology | mouse | 1:500 | WB |
| p38 MAPK (D13E1) | Cell Signaling | Rabbit | 1:1000 | WB |
| p44/42 MAPK | Cell Signaling | Rabbit | 1:1000 | WB |
| Phospho-Akt (Ser473) | Cell Signaling | Rabbit | 1:1000 | WB |
| Phospho-GRK2 (Ser29) | Sigma Aldrich | Rabbit | 1:500 | WB |
| Phospho-GRK2 (Ser670) | Millipore | mouse | 1:250 | WB |
| Phospho-GRK2 (Ser685) | Abcam | Rabbit | 1:250 | WB |
| Phospho-p38 MAPK (Thr180/Thr182) | Cell Signaling | Rabbit | 1:1000 | WB |
| Phospho-p44/42 MAPK (Thr202/Thr204) | Cell Signaling | Rabbit | 1:1000 | WB |
| Phospho-PKCζ (Thr410) | Assay BioTech | Rabbit | 1:100 | IHC |
| Phospho-PKCζ (Thr410/403) | Cell Signaling | Rabbit | 1:500 | WB |
| Phospho-SAPK/JNK (Thr183/Thr185) | Cell Signaling | Rabbit | 1:1000 | WB |
| PKCζ C24E6 | Cell Signaling | Rabbit | 1:500 | WB |
| RAS (Clone RAS10) | Millipore | mouse | 1:1000 | WB |
| SAPK/JNK | Cell Signaling | Rabbit | 1:1000 | WB |
| Smi-312 | Covance | mouse | 1:1000 | IF |
| α-Tubulin | Life Technologies | mouse | 1:10000 | WB |
WB, western blot; IHC, immunohistochemistry; ICC, immunocytochemistry; IP, immunoprecipitation.
Western blotting
Western blotting was performed as previously described (15) using appropriate primary antibodies (Table 1), secondary horseradish peroxidase-conjugated antibodies (Sigma) and ECL (Fisher) chemiluminescence.
cAMP and activity assays
All assays were performed on dissected embryonic hippocampi, snap-frozen in liquid nitrogen. cAMP levels were quantitated from tissue homogenized in 0.1 M HCl, using a cAMP ELISA immunoassay kit (Enzo Life Sciences) following manufacturer's instructions. GαS and Gαi activity were determined using commercially available activation kits (NewEast Biosciences) following the manufacturer's instructions. Active Ras (Ras-GTP) was detected by Raf1-RBD immunoprecipitation using the RAS activation kit (Millipore) following manufacturer's instructions.
Statistical analyses
All statistical analyses were performed using GraphPad Prism 5 software. Unpaired two-tailed Student's T-tests were used for experiments analyzing data between two groups. One-way analysis of variance (ANOVA) with Bonferroni post-test correction analyses were employed for multiple comparisons.
SUPPLEMENTARY MATERIAL
FUNDING
This work was partly funded by a grant from the National Cancer Institute (CA141549-01 to D.H.G.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Matthew Harms for providing us with the age- and sex-matched control patient fibroblasts, generated with support from the P30 Neuroscience Blueprint Interdisciplinary Center Core award to Washington University (P30 NS057105), which was used for iPSC reprogramming.
Conflict of Interest statement. The authors declare that they have no conflict of interest.
REFERENCES
- 1.Dilts C.V., Carey J.C., Kircher J.C., Hoffman R.O., Creel D., Ward K., Clark E., Leonard C.O. Children and adolescents with neurofibromatosis 1: a behavioral phenotype. J. Dev. Behav. Pediatr. 1996;17:229–239. [PubMed] [Google Scholar]
- 2.Hyman S.L., Shores A., North K.N. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology. 2005;65:1037–1044. doi: 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- 3.North K., Joy P., Yuille D., Cocks N., Hutchins P. Cognitive function and academic performance in children with neurofibromatosis type 1. Dev. Med. Child Neurol. 1995;37:427–436. doi: 10.1111/j.1469-8749.1995.tb12026.x. [DOI] [PubMed] [Google Scholar]
- 4.Ozonoff S. Cognitive impairment in neurofibromatosis type 1. Am. J. Med. Genet. 1999;89:45–52. [PubMed] [Google Scholar]
- 5.Soucy E.A., Gao F., Gutmann D.H., Dunn C.M. Developmental delays in children with neurofibromatosis type 1. J. Child Neurol. 2012;27:641–644. doi: 10.1177/0883073811423974. [DOI] [PubMed] [Google Scholar]
- 6.Licis A.K., Vallorani A., Gao F., Chen C., Lenox J., Yamada K.A., Duntley S.P., Gutmann D.H. Prevalence of sleep disturbances in children with neurofibromatosis type 1. J. Child Neurol. 2013;28:1400–1405. doi: 10.1177/0883073813500849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg S., Green J., Leadbitter K., Emsley R., Lehtonen A., Evans D.G., Huson S.M. Neurofibromatosis type 1 and autism spectrum disorder. Pediatrics. 2013;132:e1642–e1648. doi: 10.1542/peds.2013-1868. [DOI] [PubMed] [Google Scholar]
- 8.Lehtonen A., Howie E., Trump D., Huson S.M. Behaviour in children with neurofibromatosis type 1: cognition, executive function, attention, emotion, and social competence. Dev. Med. Child Neurol. 2013;55:111–125. doi: 10.1111/j.1469-8749.2012.04399.x. [DOI] [PubMed] [Google Scholar]
- 9.Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63:851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- 10.Xu G.F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990;63:835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- 11.Basu T.N., Gutmann D.H., Fletcher J.A., Glover T.W., Collins F.S., Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 12.DeClue J.E., Cohen B.D., Lowy D.R. Identification and characterization of the neurofibromatosis type 1 protein product. Proc. Natl Acad. Sci. USA. 1991;88:9914–9918. doi: 10.1073/pnas.88.22.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa R.M., Federov N.B., Kogan J.H., Murphy G.G., Stern J., Ohno M., Kucherlapati R., Jacks T., Silva A.J. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 14.Li W., Cui Y., Kushner S.A., Brown R.A., Jentsch J.D., Frankland P.W., Cannon T.D., Silva A.J. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr. Biol. 2005;15:1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 15.Brown J.A., Gianino S.M., Gutmann D.H. Defective cAMP generation underlies the sensitivity of CNS neurons to neurofibromatosis-1 heterozygosity. J. Neurosci. 2010;30:5579–5589. doi: 10.1523/JNEUROSCI.3994-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown J.A., Diggs-Andrews K.A., Gianino S.M., Gutmann D.H. Neurofibromatosis-1 heterozygosity impairs CNS neuronal morphology in a cAMP/PKA/ROCK-dependent manner. Mol. Cell Neurosci. 2012;49:13–22. doi: 10.1016/j.mcn.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong J., Hannan F., Zhu Y., Bernards A., Zhong Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat. Neurosci. 2002;5:95–96. doi: 10.1038/nn792. [DOI] [PubMed] [Google Scholar]
- 18.Hannan F., Ho I., Tong J.J., Zhu Y., Nurnberg P., Zhong Y. Effect of neurofibromatosis type I mutations on a novel pathway for adenylyl cyclase activation requiring neurofibromin and Ras. Hum. Mol. Genet. 2006;15:1087–1098. doi: 10.1093/hmg/ddl023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker J.A., Tchoudakova A.V., McKenney P.T., Brill S., Wu D., Cowley G.S., Hariharan I.K., Bernards A. Reduced growth of Drosophila neurofibromatosis 1 mutants reflects a non-cell-autonomous requirement for GTPase-activating protein activity in larval neurons. Genes Dev. 2006;20:3311–3323. doi: 10.1101/gad.1466806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H.F., The I., Hannan F., Bernards A., Zhong Y. Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science. 1997;276:795–798. doi: 10.1126/science.276.5313.795. [DOI] [PubMed] [Google Scholar]
- 21.Guo H.F., Tong J., Hannan F., Luo L., Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature. 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- 22.Jacks T., Shih T.S., Schmitt E.M., Bronson R.T., Bernards A., Weinberg R.A. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat. Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 23.Haigis K.M., Kendall K.R., Wang Y., Cheung A., Haigis M.C., Glickman J.N., Niwa-Kawakita M., Sweet-Cordero A., Sebolt-Leopold J., Shannon K.M., et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braun B.S., Tuveson D.A., Kong N., Le D.T., Kogan S.C., Rozmus J., Le Beau M.M., Jacks T.E., Shannon K.M. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc. Natl Acad. Sci. USA. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz-Meco M.T., Lozano J., Municio M.M., Berra E., Frutos S., Sanz L., Moscat J. Evidence for the in vitro and in vivo interaction of Ras with protein kinase C zeta. J. Biol. Chem. 1994;269:31706–31710. [PubMed] [Google Scholar]
- 26.Winstel R., Freund S., Krasel C., Hoppe E., Lohse M.J. Protein kinase cross-talk: membrane targeting of the beta-adrenergic receptor kinase by protein kinase C. Proc. Natl Acad. Sci. USA. 1996;93:2105–2109. doi: 10.1073/pnas.93.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Blasi A., Parruti G., Sallese M. Regulation of G protein-coupled receptor kinase subtypes in activated T lymphocytes. Selective increase of beta-adrenergic receptor kinase 1 and 2. J. Clin. Invest. 1995;95:203–210. doi: 10.1172/JCI117641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker J.A., Gouzi J.Y., Long J.B., Huang S., Maher R.C., Xia H., Khalil K., Ray A., Van Vactor D., Bernards R., et al. Genetic and functional studies implicate synaptic overgrowth and ring gland cAMP/PKA signaling defects in the Drosophila melanogaster neurofibromatosis-1 growth deficiency. PLoS Genet. 2013;9:e1003958. doi: 10.1371/journal.pgen.1003958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai P.I., Wang M., Kao H.H., Cheng Y.J., Walker J.A., Chen R.H., Chien C.T. Neurofibromin mediates FAK signaling in confining synapse growth at Drosophila neuromuscular junctions. J. Neurosci. 2012;32:16971–16981. doi: 10.1523/JNEUROSCI.1756-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchanan M.E., Davis R.L. A distinct set of Drosophila brain neurons required for neurofibromatosis type 1-dependent learning and memory. J. Neurosci. 2010;30:10135–10143. doi: 10.1523/JNEUROSCI.0283-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho I.S., Hannan F., Guo H.F., Hakker I., Zhong Y. Distinct functional domains of neurofibromatosis type 1 regulate immediate versus long-term memory formation. J. Neurosci. 2007;27:6852–6857. doi: 10.1523/JNEUROSCI.0933-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong J.J., Schriner S.E., McCleary D., Day B.J., Wallace D.C. Life extension through neurofibromin mitochondrial regulation and antioxidant therapy for neurofibromatosis-1 in Drosophila melanogaster. Nat. Genet. 2007;39:476–485. doi: 10.1038/ng2004. [DOI] [PubMed] [Google Scholar]
- 33.Stansfield B.K., Bessler W.K., Mali R., Mund J.A., Downing B.D., Kapur R., Ingram D.A., Jr Ras–Mek–Erk signaling regulates Nf1 heterozygous neointima formation. Am. J. Pathol. 2014;184:79–85. doi: 10.1016/j.ajpath.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauchle J.O., Kim D., Le D.T., Akagi K., Crone M., Krisman K., Warner K., Bonifas J.M., Li Q., Coakley K.M., et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461:411–414. doi: 10.1038/nature08279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Kim E., Wang X., Novitch B.G., Yoshikawa K., Chang L.S., Zhu Y. ERK inhibition rescues defects in fate specification of Nf1-deficient neural progenitors and brain abnormalities. Cell. 2012;150:816–830. doi: 10.1016/j.cell.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staser K., Park S.J., Rhodes S.D., Zeng Y., He Y.Z., Shew M.A., Gehlhausen J.R., Cerabona D., Menon K., Chen S., et al. Normal hematopoiesis and neurofibromin-deficient myeloproliferative disease require Erk. J. Clin. Invest. 2013;123:329–334. doi: 10.1172/JCI66167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma R., Wu X., Rhodes S.D., Chen S., He Y., Yuan J., Li J., Yang X., Li X., Jiang L., et al. Hyperactive Ras/MAPK signaling is critical for tibial nonunion fracture in neurofibromin-deficient mice. Hum. Mol. Genet. 2013;22:4818–4828. doi: 10.1093/hmg/ddt333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dasgupta B., Yi Y., Chen D.Y., Weber J.D., Gutmann D.H. Proteomic analysis reveals hyperactivation of the mammalian target of rapamycin pathway in neurofibromatosis 1-associated human and mouse brain tumors. Cancer Res. 2005;65:2755–2760. doi: 10.1158/0008-5472.CAN-04-4058. [DOI] [PubMed] [Google Scholar]
- 39.Johannessen C.M., Reczek E.E., James M.F., Brems H., Legius E., Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl Acad. Sci. USA. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jessen W.J., Miller S.J., Jousma E., Wu J., Rizvi T.A., Brundage M.E., Eaves D., Widemann B., Kim M.O., Dombi E., et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J. Clin. Invest. 2013;123:340–347. doi: 10.1172/JCI60578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daginakatte G.C., Gianino S.M., Zhao N.W., Parsadanian A.S., Gutmann D.H. Increased c-Jun-NH2-kinase signaling in neurofibromatosis-1 heterozygous microglia drives microglia activation and promotes optic glioma proliferation. Cancer Res. 2008;68:10358–10366. doi: 10.1158/0008-5472.CAN-08-2506. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan K., El-Hoss J., Little D.G., Schindeler A. JNK inhibitors increase osteogenesis in Nf1-deficient cells. Bone. 2011;49:1311–1316. doi: 10.1016/j.bone.2011.09.043. [DOI] [PubMed] [Google Scholar]
- 43.Warrington N.M., Gianino S.M., Jackson E., Goldhoff P., Garbow J.R., Piwnica-Worms D., Gutmann D.H., Rubin J.B. Cyclic AMP suppression is sufficient to induce gliomagenesis in a mouse model of neurofibromatosis-1. Cancer Res. 2010;70:5717–5727. doi: 10.1158/0008-5472.CAN-09-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warrington N.M., Woerner B.M., Daginakatte G.C., Dasgupta B., Perry A., Gutmann D.H., Rubin J.B. Spatiotemporal differences in CXCL12 expression and cyclic AMP underlie the unique pattern of optic glioma growth in neurofibromatosis type 1. Cancer Res. 2007;67:8588–8595. doi: 10.1158/0008-5472.CAN-06-2220. [DOI] [PubMed] [Google Scholar]
- 45.Valverde A.M., Teruel T., Lorenzo M., Benito M. Involvement of Raf-1 kinase and protein kinase C zeta in insulin-like growth factor I-induced brown adipocyte mitogenic signaling cascades: inhibition by cyclic adenosine 3′,5′-monophosphate. Endocrinology. 1996;137:3832–3841. doi: 10.1210/endo.137.9.8756554. [DOI] [PubMed] [Google Scholar]
- 46.Uberall F., Hellbert K., Kampfer S., Maly K., Villunger A., Spitaler M., Mwanjewe J., Baier-Bitterlich G., Baier G., Grunicke H.H. Evidence that atypical protein kinase C-lambda and atypical protein kinase C-zeta participate in Ras-mediated reorganization of the F-actin cytoskeleton. J. Cell Biol. 1999;144:413–425. doi: 10.1083/jcb.144.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pal S., Datta K., Khosravi-Far R., Mukhopadhyay D. Role of protein kinase Czeta in Ras-mediated transcriptional activation of vascular permeability factor/vascular endothelial growth factor expression. J. Biol. Chem. 2001;276:2395–2403. doi: 10.1074/jbc.M007818200. [DOI] [PubMed] [Google Scholar]
- 48.Marshall M.S. Ras target proteins in eukaryotic cells. FASEB J. 1995;9:1311–1318. doi: 10.1096/fasebj.9.13.7557021. [DOI] [PubMed] [Google Scholar]
- 49.D'Souza K.M., Malhotra R., Philip J.L., Staron M.L., Theccanat T., Jeevanandam V., Akhter S.A. G protein-coupled receptor kinase-2 is a novel regulator of collagen synthesis in adult human cardiac fibroblasts. J. Biol. Chem. 2011;286:15507–15516. doi: 10.1074/jbc.M111.218263. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Aragay A.M., Ruiz-Gomez A., Penela P., Sarnago S., Elorza A., Jimenez-Sainz M.C., Mayor F., Jr G protein-coupled receptor kinase 2 (GRK2): mechanisms of regulation and physiological functions. FEBS Lett. 1998;430:37–40. doi: 10.1016/s0014-5793(98)00495-5. [DOI] [PubMed] [Google Scholar]
- 51.Mertens I., Vandingenen A., Johnson E.C., Shafer O.T., Li W., Trigg J.S., De Loof A., Schoofs L., Taghert P.H. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 52.Dasgupta B., Dugan L.L., Gutmann D.H. The neurofibromatosis 1 gene product neurofibromin regulates pituitary adenylate cyclase-activating polypeptide-mediated signaling in astrocytes. J. Neurosci. 2003;23:8949–8954. doi: 10.1523/JNEUROSCI.23-26-08949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brannan C.I., Perkins A.S., Vogel K.S., Ratner N., Nordlund M.L., Reid S.W., Buchberg A.M., Jenkins N.A., Parada L.F., Copeland N.G. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 54.Rittie L., Fisher G.J. Isolation and culture of skin fibroblasts. Methods Mol. Med. 2005;117:83–98. doi: 10.1385/1-59259-940-0:083. [DOI] [PubMed] [Google Scholar]
- 55.Yagi T., Ito D., Okada Y., Akamatsu W., Nihei Y., Yoshizaki T., Yamanaka S., Okano H., Suzuki N. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 56.Xia G., Santostefano K., Hamazaki T., Liu J., Subramony S.H., Terada N., Ashizawa T. Generation of human-induced pluripotent stem cells to model spinocerebellar ataxia type 2 in vitro. J. Mol. Neurosci. 2013;51:237–248. doi: 10.1007/s12031-012-9930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hegedus B., Dasgupta B., Shin J.E., Emnett R.J., Hart-Mahon E.K., Elghazi L., Bernal-Mizrachi E., Gutmann D.H. Neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by both cAMP- and Ras-dependent mechanisms. Cell Stem Cell. 2007;1:443–457. doi: 10.1016/j.stem.2007.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.