Nearly all tissues have evolved a complex internal vasculature that is distinct in architecture but in constant communication with the parenchymal compartment wherein the tissue-specific cells reside. The main function of the vasculature is to provide oxygen to the tissue-specific cells, which stay alive by diffusion only when they reside within 150 to 200 μm of a perfusable conduit.[1] A tissue engineered microvasculature has proven challenging to create, especially if the goal is a multi-compartmental tissue with robust conduits that can support fluid flow and mass transport in three dimensions (3D).[2] Here, we describe a novel device comprising distinct vascular and parenchymal compartments and fabricated from biodegradable elastomeric polymers that can provide a basis for building tissue engineered constructs of increasing scale and clinical relevance. Scalability is achieved by pairwise stacking of microvessels and heart cell scaffolds in a parallel configuration. We demonstrate the power of this process by showing that perfused devices support scalable flow rates and numbers of viable heart cells.
A major factor that currently limits the clinical translation of engineered tissues comprised of functional cells is the thickness of viable tissue that can be produced in vitro and can survive implantation in vivo.[2–5] A transport system is required to maintain viable cells within thick tissue, such as heart muscle, during in vitro culture and the initial phase post-implantation because of the slow rate of new blood vessel growth.[6–8] Previously proposed strategies for creating a microvasculature can be broadly subdivided into two groups: biological strategies based on native vascular tissue and its component endothelial cells, and engineering strategies based on synthetically manufactured scaffolds.[2] Biological pre-vascularization strategies[9–11] have produced capillary-sized networks with limited flow capacity. As one recent example, heart cell sheets were sequentially added to a native vascular bed excised from a donor rat, pre-vascularized during in vitro culture, and surgically connected to the circulatory system of a recipient rat to produce viable heart tissue in vivo.[12] However, the final thickness of the heart tissue produced did not increase linearly with the number of heart cell sheets added, wherein perfusion capacity was limited by the small diameter flow networks connecting sequentially added heart cell sheets to the excised vascular bed. To date, biological approaches have not convincingly shown that pre-vascularization can support the viability of tissue-specific cells residing at high density in an adjacent parenchymal compartment.
In contrast, engineering approaches use a materials-based paradigm and offer the potential advantages of a hierarchical fluid flow configuration with increased perfusion capacity. Laser microablation,[13] micromolding,[14–17] microtemplating,[18, 19] layer-by-layer assembly,[20] and 3D printing[21–23] technologies have enabled the microfabrication of tissue engineering scaffolds from a variety of materials. However, previous studies of perfusable microfluidic (μfluidic) devices required non-degradable materials,[21, 22, 24–26] and/or focused only on the microvascular compartment.[16, 22, 25, 27–29] Poly(glycerol-co-sebacate)(PGS) has been utilized in tissue engineering studies for more than a decade.[30–32] PGS[33, 34] and PGS-inspired materials[35] were strategically designed to combine rubber-like elasticity, mechanical robustness and controllable biodegradation by surface erosion to enable tissue remodeling and repair.[13, 36]
In a recent study,[16] a PGS μfluidic device provided flow and transport functions that mimicked some aspects of the vasculature. The design supported flow at rates of 10-to-100 μL per min through a large (4 square cm) parallel array of arteriole-sized (~100 μm high, ~100 μm wide) channels separated by ~30 μm wide ribs. In contrast with previous designs,[21, 22, 25, 28] both 2D (in-plane) fluid flow and the interfacial (out-of-plane) area available for transport to an adjacent parenchymal space were considered. In vitro studies showed transport of a drug, doxorubicin, from the perfusate in the vascular compartment across a solid PGS membrane interface into an adjacent parenchymal compartment by demonstrating a 90% reduction in viability of muscle cells cultured in the parenchymal compartment.[16] In vivo studies showed thinning of the interface and neovascularization of the engineered microvessels by vascular tissue ingrowth from surrounding host tissues at one week, and complete degradation of the polymer scaffold within two weeks.[16]
In another study,[20] PGS heart cell scaffolds with sinusoidal internal pore architectures enabled the development of engineered heart muscle in vitro. More specifically, offset stacking of 2D scaffold layers, each with ~125 μm × 250 μm rectangular through-pores separated by 50 μm wide struts, guided the directional growth of neonatal rat heart cells into oriented muscle-like tissue muscle bundles. Also, one and two layer (2L) heart cell scaffolds were mechanically anisotropic, which has been previously shown to improve cardiac function by mitigating deleterious fibrosis after myocardial infarction.[37–39] However, in vivo studies[16, 40] have shown that rapidly degrading PGS cannot provide heart cell retention or mechanical support over a time frame of approximately four weeks during which healing of a myocardial infarction typically occurs.[37] Together, these data suggest a need for and potential benefit of creating implantable multi-component devices with more customizable properties, such as tunable biodegradability and permeability, through the assembly of distinct layers with different polymer formulations and architectures.
The primary objective of the present study was to develop an implantable scaffold unit for building vascularized cardiac grafts, wherein layers of engineered heart tissue were paired with engineered microvessels that mimicked aspects of vascular function in vitro and provided 3D templates to accelerate neovascularization in vivo. In the present study, all material components were biodegradable elastomeric polymers (PGS and PGS-inspired materials). Polymer scaffolds were designed to deliver exogenous heart cells at high density and guide vascular tissue in-growth from surrounding host tissues, while also meeting the application-specific requirements of suturability, tunable biodegradability, elastomeric mechanical properties, and perfusion-mediated support of myocyte viability prior to implantation.
The primary novelty of the proposed device is its modular design wherein multiple units, each comprising a μfluidic base, vascular-parenchymal interface and heart cell scaffold, are stackable in a parallel configuration to enable scalable flow rate and implant thickness. A second novelty is the incorporation of a rapidly degrading PGS porous interface that is expected to enable perfusion-mediated transport in vitro and then rapidly degrade in vivo, thereby quickly bringing heart cells into contact with nascent host-derived blood vessels. A third novelty is the incorporation of a more slowly degrading polymer, APS, as the heart cell scaffold and μfluidic base components that are respectively expected to provide anisotropic mechanical support and template for neovascularization in vivo. While it was not an objective of the present study, it can be reasonably expected that further device development (i.e., multiplexing to combine a number of smaller inlets and a number of smaller outlets into one single inlet and one single outlet) could result in an engineered microvasculature supplied by conduits of sufficient diameter to allow direct surgical connection with the circulatory system of the recipient.
In an attempt to slow in vivo degradation of the μfluidic base component as compared to our previous study,[16] amide linkages were substituted for a fraction of the ester linkages in PGS. A poly(ester-amide),1:2 poly(1,3-diamino-2-hydroxypropane-co-polyol sebacate) (APS), was selected because it retained approximately 50% of its initial mass over six weeks in vivo.[35] The 2L heart cell scaffold was also modified to delay its degradation as compared to previous designs[13–15, 20] by changing both the polymer formulation and its architecture. The polymer was changed from PGS to 1:8 APS, nominal strut width was increased from 50 to 100 μm, and the sinusoidal internal pore channels were created by using a layer-by-layer stacking pattern with both struts offset (BSoff). In order to increase in vitro permeability and in vivo degradation rate of the vascular-parenchymal interface as compared to our previous study,[16] porosity was introduced while maintaining the same polymer formulation (PGS) and overall interface thickness. The interface pore architecture was inspired by a sphere-templating method used for poly(2-hydroxyethyl methacrylate).[41]
The scalable unit design is diagrammed in Figure 1 and demonstrated in Figure 2. Four layer (4L) devices were assembled by sequential stacking and solvent bonding of the μfluidic base, a membrane with a central cut-out, a spin coated porous interface, and a 2L BSoff heart cell scaffold (Figure 2 A–C) (Supporting Information). First, PGS porous membrane (PM) was prepared by infiltrating a sintered acrylic template with PGS pre-polymer, curing the PGS, leaching out the template, and cryosectioning. The PM was solvent bonded[16] to a thin layer of PGS to create a spin coated PGS PM (sc-PGS PM) bonded pair (Figure 1). This fabrication process yielded ~120 μm thick membranes that preserved the pore morphology of the template (30 μm diameter spherical pores joined by 10 μm diameter necks) with a ~5 μm thick underlying layer in sizes needed for the scalable unit (4 square cm) (Figure 2 D–F). Robustly perfusable devices were created by solvent bonding the sc-PGS PM interface to the μfluidic base, similar to our previous report.[16]
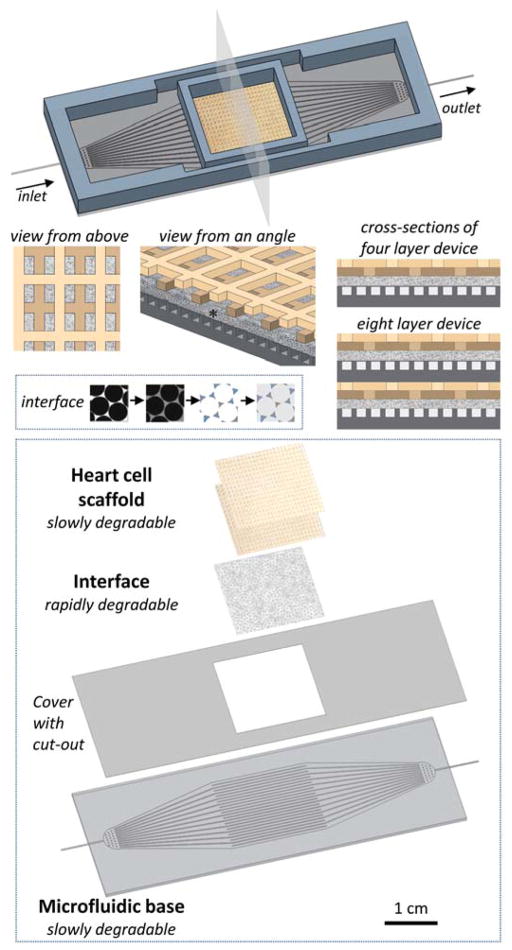
Figure 1.
The scalable unit for building vascularized myocardial grafts meets several criteria including a slowly degrading, porous, structurally and mechanically anisotropic heart cell scaffold (2L BSoff APS, two tan colors), a rapidly degrading, porous vascular-parenchymal interface (sc-PGS PM, stippled grey, *), and a slowly degrading μfluidic base (APS, dark grey). These components are stacked pair-wise in a parallel configuration to produce either 4L or 8L devices. Inset 1 shows fabrication of the interface by sphere pore templating and spin coating. Inset 2 shows layer-by-layer fabrication of a 4L device.
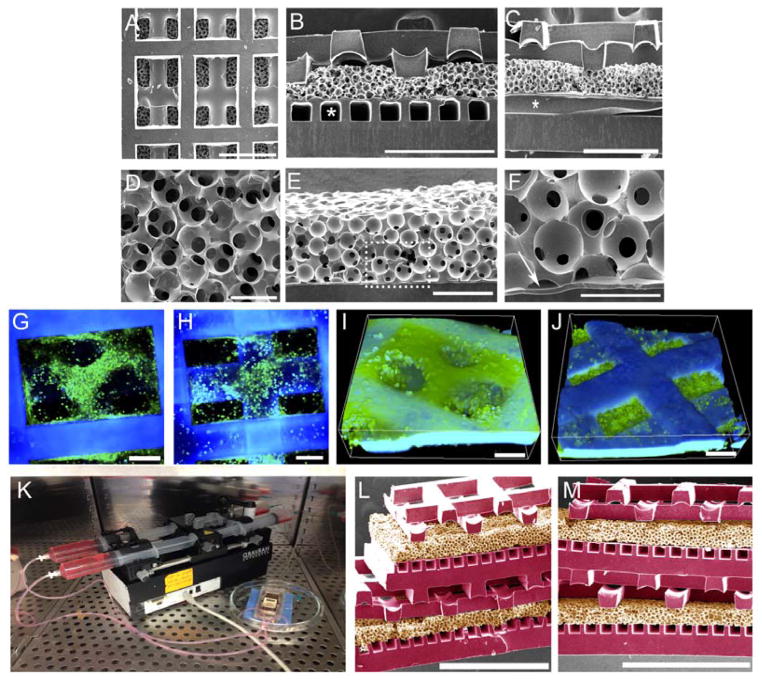
Figure 2.
Demonstration of scalable units made of biomaterial elastomers. (A–C) SEM images of 4L devices viewed (A) from above or (B,C) in cross-sections taken perpendicular and parallel to the microvessels (*), respectively, and showing sinusoidal internal pore architectures of the heart cell scaffolds. Scale bars 500 μm. (D–F) SEM images of interfaces viewed (D) from above or (E,F) in cross-section. Arrow (F) points to spin-coated PGS. Scale bars (D,F) 50 μm; (E) 100 μm. (G–J) Confocal images and 3D renderings of actin-stained heart cells cultured on 4L devices with perfusion for 4 days (G,I) at surface; (H,J) at intersection between the two layers of the heart cell scaffold. Scale bars 100 μm; cells appear green; scaffold appears blue. (K) Perfusion culture of an 8L device; (L,M) SEM images of 8L devices viewed from different angles to which faux coloration was added to distinguish APS μfluidic base and heart cell components (purple) from PGS vascular-parenchymal interfaces (tan). Scale bars 1.0 mm.
Since individual heart cell scaffold layers had large (>100 μm), low aspect ratio (1:1) features, these layers were fabricated from machined poly(methyl methacrylate) (PMMA) molds (Table S1) and then aligned, stacked and solvent bonded to create the BSoffset internal pore architecture (Figure 2 B–C). Use of PMMA molds facilitated the delamination of cured APS by brief immersion in acetone at room temperature (RT). Previously the delamination of PGS, albeit with smaller and higher aspect ratio features, from micropatterned silicon wafers coated with a sacrificial layer of maltose required water immersion for 72 h at 70°C followed by tedious manual labor.[15, 20]
After assembly, the 4L devices were cellularized by pipetting neonatal rat heart cells onto the upper 2L heart cell scaffold, and then perfused by applying continuous unidirectional flow of culture medium through the microvessel network. The heart cells were obtained from heart tissue, itself composed of a mixed population of myocytes, fibroblasts and endothelial and smooth muscle cells.[3] Within four days of in vitro culture, cells were attached and distributed throughout the internal pore architecture of the 2L BSoff heart cell scaffold (Figure 2 G–J). Eight layer (8L) devices were also assembled, cellularized, and perfused (Figure 1 and 2K). Each microvascular layer within these devices was perfusable at 10 μL per min, such that overall flow rate for 8L devices was twice that of 4L devices demonstrating scalable flow capacity. In the current prototype (Figure 2L and 2M), upper and lower interfaces were respectively made of sc-PGS PM and PGS PM in an attempt to allow interstitial flow within the device, similar to a previous report.[27] In the present study, heart cells were injected into the sinusoidal pore channels of the middle compartment using a syringe connected to a short segment of silica tubing that was integrated at this location during device assembly. Additional studies to explore the co-cultivation of heart cells and endothelial cells in 4L and 8L devices are currently ongoing.
Other testing was performed on individual device components and/or multilayer devices to evaluate and compare (i) suturability, (ii) mechanical properties, (iii) in vivo biodegradability, (iv) in vitro myocyte viability, and (v) in vitro heart cell morphology and yield (Figure 3 and 4; Table S2). Microfluidic base components comprised of two different polymer formulations (1:2 APS and PGS, each cured for 24 h) were subjected to qualitative and quantitative mechanical characterizations.[13, 15, 20] The APS base was readily suturable in contrast to the PGS base which tore during suturing (Figure 3 A versus B). Tensile strain-to-failure tests were generally consistent with the qualitative suturability results in that APS exhibited significantly higher toughness and strain-to-failure (1.56 ± 0.46 MJ/m3, 1.42 ± 0.08 mm/mm) than PGS (0.29 ± 0.02 MJ/m3, 0.46 ± 0.05 mm/mm) (Figure 3 C–D) and stiffness tended to be higher for the PGS component (Figure 3G). The sc-PGS PM was less tough than the APS base and less stiff than the PGS base (Figure 3 C,D,G,H). An in vivo study showed the APS base to be present in its original geometry 25 days after subcutaneous implantation in rodents, whereas the PGS base lost its mechanical integrity (Figure 3F versus 3E). Based on these mechanical and biodegradation data, APS was the material selected for the μfluidic base and heart cell scaffolds, whereas PGS was the material selected for the interfaces in subsequent studies.
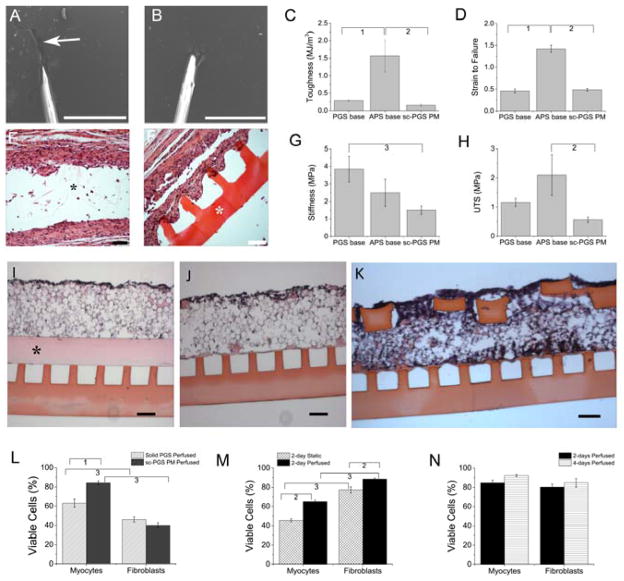
Figure 3.
Tough, permeable microvessels support viable heart cell culture. (A,B) SEMs of needles pulling sutures through microfluidic bases made of (A) PGS or (B) 1:2 APS. Arrow (A) shows PGS tearing. Scale bars: 500 μm. (C,D,G,H) Mechanical properties (C) toughness, (D) strain-to-failure, (G) stiffness, and (H) ultimate tensile strength (UTS) of the PGS base, 1:2 APS base, and sc-PGS PM interface. Data show Average +/− SE. 1Significant difference between PGS base and APS base; 2Significant difference between APS base and sc-PGS PM; 3Significant difference between PGS base and sc-PGS PM. (E,F) Biodegradation, 25 days after implantation and H&E staining of (E) PGS base or (F) 1:2 APS base. Asterisks show non-degraded polymer remnants. Scale bars 100 μm. (I,J,K) H&E stained cross sections of heart cells cultured on microvessel bases covered by (I) solid PGS membrane plus sc-PGS PM, (J) sc-PGS PM, or (K) sc-PGS PM plus 2L BSoff heart cell scaffold. Asterisk in (I) shows solid PGS membrane. Scale bars 100 μm. (L–N) Viable cardiac myocytes and fibroblasts cultured (L) on devices with solid PGS membrane plus sc-PGS PM or sc-PGS PM alone with perfusion for 2 days; (M) on 4L devices without flow (static) or with perfusion for 2 days; (N) on 4L devices with perfusion for 2 or 4 days. Data show Average +/− SE; 1Significant difference due to solid PGS membrane; 2Significant difference due to perfusion; 3Significant difference due to cell type. (Table S2).
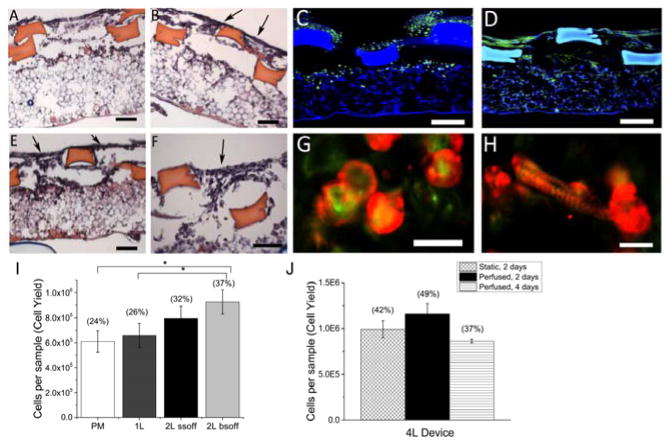
Figure 4.
Heart cell scaffolds support cell attachment and elongation. (A,B,E,F) Distributions of heart cells cultured statically for 2 days on (A) 2L SSoff plus PM or (B,E,F) 2L BSoff plus PM scaffolds in H&E stained cross-sections taken either perpendicular (A,B) or parallel (E,F) to the long axis of the rectangular pores. Arrows (B,E,F) show gaps between struts. Scale bars 100 μm. (C,D,G,H) Morphologies of heart cells cultured statically for (C,G) 2 days or (D,H) 4 days in confocal micrographs after staining for (C,D) actin (green) or co-staining for (G,H) actin (green) and sarcomeric-α-actinin (red). Scale bars (C,D) 100 μm; (G,H) 20 μm. (I) Cell delivery capacities of 0.2 square cm scaffolds with four architectures, PM, 1L plus PM, 2L SSoff plus PM, and 2L BSoff plus PM, after two days of static culture; (J) Cell delivery capacities of 0.2 square cm scaffolds punched from 4 square cm 4L devices and cultured statically or with perfusion for 2 or 4 days. Data show Average +/− SE; parentheses indicate corresponding cell seeding yields (%). *Significantly lower than 2L BSoff plus PM. (Table S2).
Effects of interface architecture on myocyte viability were studied in vitro by comparing two different interfaces: solid PGS membrane plus sc-PGS PM versus sc-PGS PM (Figure 3 I, J, L and Table S2). Heart cells were seeded on top of each interface at an initial concentration of 2 million cells per square cm, cultured with perfusion at 10 μL per min for two days, re-isolated, and analyzed by flow cytometry after staining for viability. Myocyte viability was significantly lower for the solid PGS membrane than the sc-PGS PM (63 +/− 4.3 % versus 85 +/− 2.0 %, p <0.001, Figure 3L). These data suggest sc-PGS PM, which is predominately a porous layer, offers lower resistance than solid PGS membrane to mass and/or hydraulic permeation of oxygen and thereby enable perfusion-mediated support of myocyte viability. Another study showed the viable myocyte fraction was also significantly higher for heart cells seeded at 12 million cells per square cm on 4L devices that were perfused two days, as compared with statically cultured controls (Figure 3M). Moreover, high myocyte viability was maintained between culture days two and four (Figure 3N). Together, these data suggest that convective-diffusive transport through the sc-PGS PM supports myocyte viability during in vitro culture.
In the present study, offset stacking of scaffold layers with wide struts and large rectangular pores enabled the development of tissue-like structures within four days (Figure 3K), similar to offset stacking of scaffold layers with smaller feature sizes.[20] Heart cell delivery capacity, assessed as DNA per square cm of scaffold, increased sequentially as scaffold architecture progressed from PGS PM, to PM bonded to a one layered (1L) APS scaffold, to PM bonded to a 2L scaffolds stacked with either the short strut offset (SSoff) or BSoff alignment pattern (Figure 4I). Heart cells more readily penetrated and distributed throughout BSoff scaffolds, where pores were interconnected along all three axes, than SSoff scaffolds, where pores were interconnected in only two axes (Figure 4 B,D–F versus 4A). Myocyte morphology was mainly rounded on culture day two (Figure 4C and 4G); some elongated cells were present by day four (Figure 4 D–H). A rough estimate of the heart cell delivery capacity of small (~0.2 square cm, ~0.2 mm thick) 2L BSoff specimens was one million cells, and this delivery capacity was scalable in length and width to larger (4 square cm, ~0.2 mm thick) 2L BSoff specimens (Figure 4 I,J). By comparison, Hirt et al. [3] estimated that the addition of two million heart cells in a 0.44 mm thick implant could give rise to an improvement in ventricular function of 20 to 30% in a rat model of myocardial infarction assuming a cell survival rate of 50%. In the present study, cell yields were in the range of 37 to 49% for heart cells cultured on 2L BSoff APS bonded to sc-PGS PM. These relatively high yields can be attributed to the structure of the heart cell scaffold, wherein offset APS layers enabled cell penetration of and distribution within sinusoidal pore channels while the underlying sc-PGS PM permitted cell attachment and blocked cell egress.
In conclusion, implantable scalable units for building vascularized cardiac grafts were fabricated by solvent bonding of elastomeric polymers with different in vivo degradation rates and different pore architectures, and demonstrated by in vitro culture of neonatal rat heart cells. The present work focused on certain design features we considered important to the success of the grafts, including scalable design based on a parallel configuration of heart cell scaffolds with sinusoidal pore architectures and dedicated microvessel conduits, perfusion-mediated support of myocyte viability in vitro, microchannels to enable host vascular tissue in-growth in vivo, elastomeric mechanical properties, and suturability. The heart cell scaffold and μfluidic base components, which were intended to serve as 3D templates for heart muscle and vascular tissue development, were made from slowly degrading polymers expected to maintain their architectures for many weeks to match the typical time frame of myocardial infarct healing.[37] The interface component, which was intended to provide transport during perfusion culture and degrade rapidly after implantation, was made from porous, rapidly degrading PGS. Ongoing work is focused on endothelialization of the microvasculature and induction of host blood vessel in-growth by adding angiogenic growth factors such as VEGF and PDGF to the microchannels of devices prior to their implantation.
Experimental Section
Fabrication and characterization of scaffolds
To prepare PGS pre-polymers, 1.0 mol glycerol (G) and 1.0 mol sebacic acid (SA) were stirred vigorously in a 500 mL round bottom flask using a two stage synthesis under argon: Stage 1 (130°C, 6 h, atmospheric pressure), Stage 2 (125°C, 30 h, vacuum ~50 mTorr).[15, 30]. To prepare APS pre-polymers,[35] consisting of 1,3-diamino-2-hydroxypropane (DAHP), G and SA, the same general two-stage synthesis as PGS was used. For the 1:2 APS formulation (0.33 mol DAHP, 0.66 mol G and 1.0 mol SA), Stage 1 (120°C, 3 h, atmospheric pressure), Stage 2 (130°C, 16 h, vacuum ~50mTorr). For the 1:8 APS formulation (0.125 mol DAHP, 0.875 mol G and 1.0 mol SA), Stage 1 (120°C, 3 h, atmospheric pressure), Stage 2 (130°C, 22 h, vacuum ~50mTorr).
To make the interface, PGS (50% w/v in ethanol) was infiltrated into a sintered sphere pore template (Healionics Co., Seattle, WA) at RT and vacuum level of 20 In Hg for 24 h, then cured at 155°C and 20 In Hg for 24 h. The template was leached out by immersion in dichloromethane and then acetone. The templated PGS was washed with ethanol and water, embedded in Tissue Tek O.C.T. (Sakura, Finetek), frozen and sectioned (CM1900 Leica cryotome) to produce a PM. The sc-PGS PM was made by spin-coating a silicon wafer first with maltose[15] then with PGS (25% w/v in ethanol), curing the PGS (165°C and ~15 mTorr for 6.5 h), adding a PGS PM by solvent bonding,[16] re-curing (165°C and ~15 mTorr for 2 h), and delaminating by immersion in 60°C water. To make heart cell scaffold layers, 1:8 APS was cast (in 50% w/v in ethanol) and cured (155°C and ~15 mTorr for 8 h) in a mold made of PMMA (Interstate Plastics ACRCLCP PMMA), itself made using a 0.004 inch Endmill and route cutter (T-Tech Quick Circuit QC7000, Norcross, GA), and delaminated by immersion in acetone at RT.
To make the μfluidic base, 1:2 APS was cast (in 50% w/v in ethanol) and cured (165°C and ~15 mTorr for 24 h) on a layer of maltose in a micropatterned silicon wafer, and delaminated by immersion in 70°C water.[16] To make a membrane with cut-out to cover the microfluidic base, PGS was cast and cured in a PDMS gasket, [16] then a 1.75 cm × 1.75 cm central area was removed using a razor (Figure 1). The 4L devices were assembled by PGS solvent bonding[16] of a 1:2 APS microfluidic base fitted with coated silica tubing, a PGS membrane with a central cut-out, a sc-PGS PM patch, oriented with its spin-coated surface facing the PGS membrane, a two-layer assembly of 1:8 APS heart cell scaffolds with the BSoff alignment and stacking pattern, and a PDMS gasket for culturing heart cells surrounded by a larger PDMS gasket to hold culture medium and prevent its evaporative loss (Supporting Information). The 8L devices were assembled by combining a 4L device with a sc-PGS PM interface and a 4L device with a PGS PM interface. Mechanical properties of device components were measured as previously described (Supporting Information).[13, 15, 20] Scaffolds were imaged without sputter-coating, using a S3500 SEM (Hitachi High Technologies America).[20]
In vitro and in vivo studies
All experiments involving animals were performed according to an Institute-approved protocol. Cells were obtained from 1-to-2 day old neonatal rat hearts using a GentleMACS Dissociator (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). After fibronectin coating[16], cells were seeded on heart cell scaffolds at initial concentrations of two or twelve million cells per square cm and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen). 4L devices were operated at a flow rate of 10 μL per min using a programmable pump (PHD RS485, Harvard Apparatus), while 8L devices were perfused at 10 μL per min per microvessel network, using two push and two pull syringes mounted on the same pump (Supporting Information). Full thickness specimens were harvested from the center of the device using a 6 mm diameter dermal punch (Acuderm, Ft. Lauderdale, FL). Subcutaneous implantations in immunodeficient rats were carried out as previously described[16] (Supporting Information). Samples to be analyzed by immunohistochemistry or histology were embedded and sectioned as previously described.[16]
To visualize actin, specimens were stained with fluorescein isothiocyanate (FITC)-phalloidin conjugate (Invitrogen) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI).[13] To locate the cardiac-specific marker sarcomeric α-actinin, primary antibody (Sigma) was used in conjunction with FITC-phalloidin followed by a blocking step, Texas-red conjugated secondary antibody staining and DAPI counterstaining.[20] Sp[16]ecimens were examined with a Zeiss Axiovert or a Nikon 1AR spectral scanning confocal microscope.
Heart cell viability was quantified using a flow cytometer (LSR II, Becton Dickinson) as follows. Cells were dissociated from freshly harvested cell-polymer constructs at high (~85%) viability by gentle mixing in 0.5 mg/mL collagenase II (Worthington Biomedical, LS004176) and 1.5 mg/mL dispase (Sigma, D4693) in Hank’s buffered saline solution (HBSS) at 37°C with periodic gentle pipetting (Table S3). Cells were then labeled with allophycocyanin (APC) fixable viability stain (L10120, Invitrogen), fixed in buffered formalin, and subjected to flow cytometry. Heart cell populations corresponding to either myocytes or non-myocytes could be identified on the basis of their light scattering properties (i.e. forward scatter, FSC, versus side scatter, SSC). These populations were verified by selectively staining the myocytes with sarcomeric α-actinin, which confirmed that the two populations could be identified on the basis of FSC versus SSC. Populations were further gated based on side scatter width versus area to remove doublets. Cell viability, which was quantified independently for myocytes and non-myocytes, was determined based on APC fluorescence where cells with higher fluorescence than an unstained control were classified as dead. DNA was quantified after digesting specimens in papainase using the PicoGreen dsDNA assay (Molecular Probes).[13] Cell seeding yields were estimated as the ratio of final specimen DNA content and initial number of cells seeded, assuming 8 pg of DNA per cell[42] and mononuclear neonatal rat heart cells during two-to-four days of culture.
Supplementary Material
Acknowledgments
This work is funded by the National Heart, Lung and Blood Institute (NHLBI), Award 1-R01-HL107503 (LEF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or the NIH. We thank the core facilities at the MIT Koch Institute, A.J. Marshall from Healionics Co. (Seattle, WA) for the STAR® biomaterial pore templates, R. Langer and G. Vunjak-Novakovic for general advice, E. Kim for helpful discussions and engineering drawings, A.J. Spencer for mold fabrication.
Glossary of non-standard acronyms and abbreviations
- 1L, 2L, 4L or 8L
refers to the numbers of layers in the component or device (1, 2, 4 or 8)
- APS
is the acronym for 1:2 poly(1,3-diamino-2-hydroxypropane-co-polyol sebacate)
- PGS
is the acronym for poly(glycerol-co-sebacate)
- BSoff
refers to a both strut offset layer-by-layer stacking pattern for 2L heart cell scaffolds, which are shown in Figure 1, 2, 3 & 4
- SSoff
refers to short strut offset stacking pattern for 2L heart cell scaffolds, shown in Figure 4
- μfluidic base
refers to the microfluidic base part shown in Figure 1, 2 and 3
- sc-PGS PM
refers to a spin-coated PGS porous membrane interface shown in Figure 1, 2 & 3
- PM
refers to a PGS porous membrane without spin coating shown in Figure 4
Contributor Information
Dr. Xiaofeng Ye, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Liang Lu, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Martin E. Kolewe, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Keith Hearon, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Kristin M. Fischer, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Jonathan Coppeta, Biomedical Microsystems Development and Microfabrication Design Groups, Charles Stark Draper Laboratory, Cambridge, MA 02139, USA
Dr. Lisa E. Freed, Email: lfreed@mit.edu, lfreed@draper.com, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Biomedical Microsystems Development and Microfabrication Design Groups, Charles Stark Draper Laboratory, Cambridge, MA 02139, USA.
References
- 1.Cassell OC, Hofer SO, Morrison WA, Knight KR. Br J Plast Surg. 2002;55:603. doi: 10.1054/bjps.2002.3950. [DOI] [PubMed] [Google Scholar]
- 2.Novosel EC, Kleinhans C, Kluger PJ. Adv Drug Deliv Rev. 2011;63:300. doi: 10.1016/j.addr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Hirt MN, Hansen A, Eschenhagen T. Circ Res. 2014;114:354. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]
- 4.Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA, Murry CE. Nature. 2014;510:273. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laflamme MA, Murry CE. Nature. 2011;473:326. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bae H, Puranik AS, Gauvin R, Edalat F, Carrillo-Conde B, Peppas NA, Khademhosseini A. Sci Transl Med. 2012;4:160ps23. doi: 10.1126/scitranslmed.3003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Nature. 2010;464:549. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laschke MW, Vollmar B, Menger MD. Tissue Eng Part B Rev. 2009;15:455. doi: 10.1089/ten.TEB.2009.0252. [DOI] [PubMed] [Google Scholar]
- 10.Lesman A, Koffler J, Atlas R, Blinder YJ, Kam Z, Levenberg S. Biomaterials. 2011;32:7856. doi: 10.1016/j.biomaterials.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Koffler J, Kaufman-Francis K, Shandalov Y, Egozi D, Pavlov DA, Landesberg A, Levenberg S. Proc Natl Acad Sci U S A. 2011;108:14789. doi: 10.1073/pnas.1017825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekine H, Shimizu T, Sakaguchi K, Dobashi I, Wada M, Yamato M, Kobayashi E, Umezu M, Okano T. Nat Commun. 2013;4:1399. doi: 10.1038/ncomms2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelmayr GC, Jr, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Nat Mater. 2008;7:1003. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park H, Larson BL, Guillemette MD, Jain SR, Hua C, Engelmayr GC, Jr, Freed LE. Biomaterials. 2011;32:1856. doi: 10.1016/j.biomaterials.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neal RA, Jean A, Park H, Wu PB, Hsiao J, Engelmayr GC, Jr, Langer R, Freed LE. Tissue Eng Part A. 2013;19:793. doi: 10.1089/ten.tea.2012.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye X, Lu L, Kolewe ME, Park H, Larson BL, Kim ES, Freed LE. Biomaterials. 2013;34:10007. doi: 10.1016/j.biomaterials.2013.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bian W, Jackman CP, Bursac N. Biofabrication. 2014;6:024109. doi: 10.1088/1758-5082/6/2/024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madden LR, Mortisen DJ, Sussman EM, Dupras SK, Fugate JA, Cuy JL, Hauch KD, Laflamme MA, Murry CE, Ratner BD. Proc Natl Acad Sci U S A. 2010;107:15211. doi: 10.1073/pnas.1006442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson KS, Korte FS, Giachelli CM, Ratner BD, Regnier M, Scatena M. Tissue Eng Part A. 2013;19:967. doi: 10.1089/ten.tea.2012.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolewe ME, Park H, Gray C, Ye X, Langer R, Freed LE. Adv Mater. 2013;25:4459. doi: 10.1002/adma.201301016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen CJ, Saksena R, Kolesky DB, Vericella JJ, Kranz SJ, Muldowney GP, Christensen KT, Lewis JA. Adv Mater. 2013;25:96. doi: 10.1002/adma.201203321. [DOI] [PubMed] [Google Scholar]
- 22.Wu W, DeConinck A, Lewis JA. Adv Mater. 2011;23:H178. doi: 10.1002/adma.201004625. [DOI] [PubMed] [Google Scholar]
- 23.Temple JP, Hutton DL, Hung BP, Huri PY, Cook CA, Kondragunta R, Jia X, Grayson WL. J Biomed Mater Res A. 2014 doi: 10.1002/jbm.a.35107. [DOI] [PubMed] [Google Scholar]
- 24.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. Sci Transl Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chau LT, Rolfe BE, Cooper-White JJ. Biomicrofluidics. 2011;5:34115. doi: 10.1063/1.3609264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carraro A, Hsu WM, KKM, Cheung WS, Miller ML, Weinberg EJ, Swart EF, Kaazempur-Mofrad M, Borenstein JT, Vacanti JP, Neville C. Biomed Microdevices. 2008;10:795. doi: 10.1007/s10544-008-9194-3. [DOI] [PubMed] [Google Scholar]
- 27.Wong KH, Truslow JG, Khankhel AH, Chan KL, Tien J. J Biomed Mater Res A. 2013;101:2181. doi: 10.1002/jbm.a.34524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fidkowski C, Kaazempur-Mofrad MR, Borenstein J, Vacanti JP, Langer R, Wang Y. Tissue Eng. 2005;11:302. doi: 10.1089/ten.2005.11.302. [DOI] [PubMed] [Google Scholar]
- 29.King KR, Wang CCJ, Kaazempur-Mofrad M, Vacanti JP, Borenstein JT. Adv Materials. 2004;16:2007. [Google Scholar]
- 30.Wang Y, Ameer GA, Sheppard BJ, Langer R. Nat Biotechnol. 2002;20:602. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- 31.Rai R, Tallawi M, Grigore A, Boccaccini AR. Prog Polyme Sci. 2012;37:1051. [Google Scholar]
- 32.Chen Q, Liang S, Thouas GA. Prog Polym Sci. 2013;38:584. [Google Scholar]
- 33.Wang Y, Kim YM, Langer R. J Biomed Mater Res A. 2003;66:192. doi: 10.1002/jbm.a.10534. [DOI] [PubMed] [Google Scholar]
- 34.Wu W, Allen RA, Wang Y. Nat Med. 2012;18:1148. doi: 10.1038/nm.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bettinger CJ, Bruggeman JP, Borenstein JT, Langer R. J Biomed Mater Res A. 2009;91:1077. doi: 10.1002/jbm.a.32306. [DOI] [PubMed] [Google Scholar]
- 36.Pomerantseva I, Krebs N, Hart A, Neville CM, Huang AY, Sundback CA. J Biomed Mater Res A. 2009;91:1038. doi: 10.1002/jbm.a.32327. [DOI] [PubMed] [Google Scholar]
- 37.Holmes JW, Borg TK, Covell JW. Annu Rev Biomed Eng. 2005;7:223. doi: 10.1146/annurev.bioeng.7.060804.100453. [DOI] [PubMed] [Google Scholar]
- 38.Fomovsky GM, Clark SA, Parker KM, Ailawadi G, Holmes JW. Circ Heart Fail. 2012;5:515. doi: 10.1161/CIRCHEARTFAILURE.111.965731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fomovsky GM, Macadangdang JR, Ailawadi G, Holmes JW. J Cardiovasc Transl Res. 2011;4:82. doi: 10.1007/s12265-010-9241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuckey DJ, Ishii H, Chen QZ, Boccaccini AR, Hansen U, Carr CA, Roether JA, Jawad H, Tyler DJ, Ali NN, Clarke K, Harding SE. Tissue Eng Part A. 2010;16:3395. doi: 10.1089/ten.TEA.2010.0213. [DOI] [PubMed] [Google Scholar]
- 41.Fukano Y, Usui ML, Underwood RA, Isenhath S, Marshall AJ, Hauch KD, Ratner BD, Olerud JE, Fleckman P. J Biomed Mater Res A. 2010;94:1172. doi: 10.1002/jbm.a.32798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alsberg E, Anderson KW, Albeiruti A, Rowley JA, Mooney DJ. Proc Natl Acad Sci U S A. 2002;99:12025. doi: 10.1073/pnas.192291499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.