Abstract
Crossing-over ensures accurate chromosome segregation during meiosis, and every pair of chromosomes obtains at least one crossover, even though the majority of recombination sites yield non-crossovers. A putative regulator of crossing-over is RNF212, which is associated with variation in crossover rates in humans. We show that mouse RNF212 is essential for crossing-over, functioning to couple chromosome synapsis to the formation of crossover-specific recombination complexes. Selective localization of RNF212 to a subset of recombination sites is shown to be a key early step in the crossover designation process. RNF212 acts at these sites to stabilize meiosis-specific recombination factors, including the MutSγ complex (MSH4-MSH5). We infer that selective stabilization of key recombination proteins is a fundamental feature of meiotic crossover control. Haploinsufficiency indicates that RNF212 is a limiting factor for crossover control and raises the possibility that human alleles may alter the amount or stability of RNF212 and be risk factors for aneuploid conditions.
The pairing, synapsis and segregation of homologous parental chromosomes (homologs) are unique features of the meiotic program. Homologous recombination has essential roles in these processes1. First, homology recognition and DNA strand exchange promote the pairing of homologs and their intimate connection by zipper-like structures called synaptonemal complexes2. Subsequently, a subset of recombination sites form crossovers, resulting in stable interhomolog connections that facilitate homolog bi-orientation on the spindle to promote accurate disjunction at meiosis I (refs. 3,4). Failure to crossover or the suboptimal location of crossovers (proximal to centromeres or telomeres) places homologs at risk for missegregation5. In humans, aneuploidy resulting from meiotic errors is a leading cause of spontaneous abortion and developmental disease6.
Meiotic recombination is initiated by the programmed induction of DNA double-strand breaks (DSBs)7. In mammals, only a minority (~10–25%) of DSBs produce crossovers, with the majority being repaired with a non-crossover outcome. However, regulatory processes ensure that every pair of homologs obtains at least one crossover: that is, in each cell, a nonrandom subset of DSBs is selected to become crossovers, and this fate is then implemented with high efficiency8,9. Although the mechanism of this crossover designation process remains unknown, crossover and non-crossover pathways are highly differentiated with respect to both molecular intermediates and genetic requirements. Most notably, crossing-over involves the formation of double–Holliday junction intermediates and is facilitated by at least a dozen procrossover factors, including the meiosis-specific ZMM proteins10–15.
In humans, the rate of crossing-over varies significantly between individuals and has a strong heritable component16–20. Notably, higher maternal crossover rates have been associated with greater fecundity17,20,21. To date, only three loci have been reproducibly correlated with heritable variation in the mean crossover rate. The first of these, inversion 17q21.31, encompasses a 900-kb segment, and the rarer H2 haplotype is associated with increased crossing-over and fecundity in European females17. The second, PRDM9, encodes a histone methyltransferase that selectively binds DNA sequence motifs via a C-terminal zinc-finger array and is required for DSB formation at these sites22–26. Although the primary effect of PRDM9 variants is the altered localization of recombination hotspots, small but significant effects on recombination rate have been inferred25,27. The third locus, RNF212, encodes a protein with homology to Zip3 and ZHP-3, meiotic procrossover factors, which were identified in Saccharomyces cerevisiae and Caenorhabditis elegans, respectively16,18,19,28,29. All three proteins contain RING-finger domains, the signature of a class of E3 ligase enzymes that catalyze protein modification by ubiquitin-like molecules30. Budding yeast Zip3 has been implicated in the SUMO pathway of post-translational modification31 (although a role in ubiquitin modification has also been suggested; see ref. 32).
In this study, we provide evidence that mouse RNF212 has a central role in designating crossover sites and coupling chromosome synapsis to the formation of crossover-specific recombination complexes. Our analysis also indicates that differential RNF212-dependent stabilization of key recombination proteins is a basic feature of crossover/non-crossover differentiation. Together, these observations provide key insights into the meiotic crossover control process and indicate a central role for RNF212.
RESULTS
Identification and expression of mouse RNF212
We cloned and sequenced full-length Rnf212 cDNA from testis mRNA (Online Methods). Out of 25 clones, 17 encoded a 307-amino-acid protein, designated mouse RNF212 isoform a. The C-terminal 37 amino acids of this protein differ from those predicted by reference genomes but are supported by EST, cDNA and BAC sequence data (Supplementary Fig. 1). Eight out of 25 cDNA clones encoded putative splice variants and are further described in Supplementary Figure 1. Mouse RNF212 isoform a shows extensive alignment with human RNF212 isoform a and more limited identity with C. elegans ZHP-3 and S. cerevisiae Zip3 (Supplementary Fig. 1). All Zip3 and RNF212 homologs have a common tripartite arrangement of domains, with an N-terminal RING motif, a region of 50–100 amino acids predicted to be a coiled-coil domain and a divergent serine-rich C-terminal region. RT-PCR, protein blot analysis and chromosomal localization indicated that mouse RNF212 is expressed exclusively in meiocytes of the gonads (Fig. 1, Supplementary Fig. 2 and data not shown).
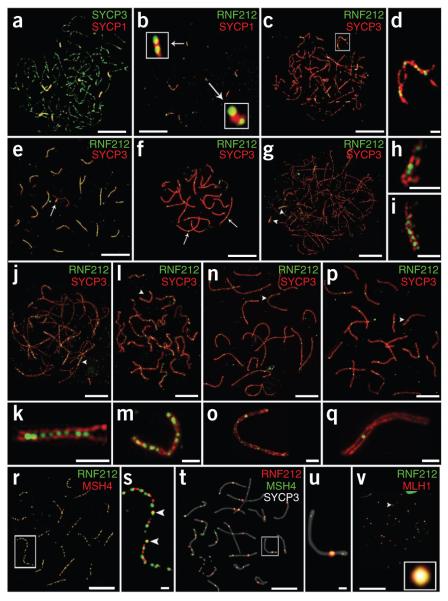
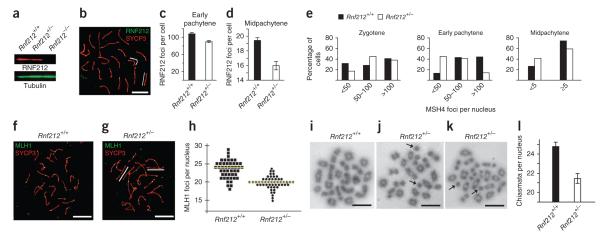
Figure 1.
Dynamic localization of RNF212 to synaptonemal complexes and crossover sites in mouse spermatocytes. (a,b) Nucleus at very early zygonema immunolabeled for SYCP1 and SYCP3 (a) and showing colocalization of SYCP1 and RNF212 (b). Insets show magnified views of two short stretches of synaptonemal complex with overlapping RNF212 foci. (c–f) Representative prophase nuclei immunolabeled for RNF212 and SYCP3. (c,d) Early zygotene nucleus (c) with magnified view of a chromosome pair (d), highlighting the exclusion of RNF212 from unsynapsed regions. (e) Early pachytene nucleus. The X-Y chromosome pair is highlighted by an arrow. (f) Midpachytene nucleus. Two RNF212 foci are highlighted by arrows. (g–q) SIM images of selected prophase nuclei immunolabeled for RNF212 and SYCP3. (g–i) Very early zygonema nucleus (g) with magnified views of synapsed regions indicated by arrowheads: left arrowhead (h), right arrowhead (i). (j,k) Midzygonema nucleus (j), with magnified view of the highlighted region (k). (l,m) Early pachynema nucleus (l), with magnified view of the indicated chromosome (m). (n,o) Early pachynema to midpachynema nucleus (n), with magnified view of the indicated chromosome (o). (p,q) Midpachynema nucleus (p), with magnified view of the chromosome indicated by the arrowhead (q). (r,s) Early pachytene nucleus costained for RNF212 and MSH4 (r), with magnified views of the two chromosome pairs highlighted by the white box (s). Arrowheads highlight RNF212 and MSH4 foci that are fully colocalized. (t,u) Midpachytene nucleus immunolabeled for RNF212, MSH4 and SYCP3 (t), with magnified view of the chromosome pair highlighted by the white box (u). Note the single site of RNF212-MSH4 colocalization. (v) Midpachytene nucleus immunolabeled for RNF212 and MLH1. The inset shows a single RNF212-MLH1 focus. Scale bars, 10 μm in a–c,e,f,r,t,v; 5 μm in g,j,l,n,p and 1 μm in d,h,i,k,m,o,q,s,u.
Dynamic localization of RNF212 to synaptonemal complexes
We analyzed the chromosomal localization of RNF212 during meiosis by immunostaining surface-spread spermatocyte and oocyte nuclei (Fig. 1 and Supplementary Fig. 3). Nuclei were categorized by stage using standard cytological criteria and the meiotic chromosome axis marker SYCP3 (ref. 33). During leptonema, short stretches of SYCP3 staining mark developing homolog axes. SYCP3 axes elaborate into contiguous structures throughout zygonema, and homologs progressively synapse (Fig. 1a,c). Pachynema is defined by complete synapsis of the autosomes (Fig. 1e). Progressive desynapsis of homologs then occurs during diplonema.
RNF212 was first detected at the transition from leptonema to zygonema, localizing specifically to initial sites of homolog synapsis (Fig. 1a,b). In these nuclei, 83% of initial stretches of synaptonemal complex (identified by immunostaining for the synaptonemal complex transverse filament protein SYCP1; Fig. 1b) overlapped with one or more immunostaining focus of RNF212 (118/142 foci; 7 nuclei). The number of synaptonemal complex–associated RNF212 foci increased as synapsis ensued, but RNF212 was notably excluded from unsynapsed homolog axes (Fig. 1c,d). In 5 midzygotene nuclei, only 14 of 96 unsynapsed regions overlapped with RNF212 foci. In early pachynema, as cells completed synapsis, RNF212 was detected as a punctate pattern of irregular foci along the synaptonemal complexes (107 ± 3.5 (s.e.m.) foci per nucleus; 26 nuclei). RNF212 also localized to the synapsed pseudoautosomal regions of the X-Y chromosomes (Fig. 1e).
After early pachynema, an apparently precipitous loss of RNF212 foci was seen such that, by midpachynema, only one or two foci per synaptonemal complex remained (average 19.63 ± 0.4 (s.e.m.) foci per nucleus; n = 42 nuclei; Fig. 1f). These remaining RNF212 foci disappeared in late pachynema and were not detected in early diplotene–stage cells in which homologs begin to desynapse (data not shown).
The RNF212 immunostaining pattern detected in fetal oocytes was very similar to that described for spermatocytes (Supplementary Fig. 3). The only distinction was that the late-stage RNF212 foci were still detected in nuclei in both the late-pachytene and early diplotene stages.
We confirmed and refined RNF212 localization patterns in spermatocytes by super-resolution structured illumination microscopy (SIM; Fig. 1g–q)34. By resolving the two SYCP3 staining axes of synapsed homologs, SIM imaging showed that RNF212 localizes specifically to the central region of developing synaptonemal complexes. In addition, an intermediate RNF212 staining pattern was visualized by SIM. Specifically, transition nuclei containing a small number of large, bright foci in addition to a large number of small, dim foci were observed (Fig. 1n,o). This observation indicates that synaptonemal complex–associated RNF212 complexes undergo selective turnover as prophase progresses.
A minority of RNF212 foci localize to DSB sites
The staining pattern of RNF212 in zygonema and early pachynema resembles that seen for a number of ‘transition nodule’ recombination factors, including the meiosis-specific MutSγ complex MSH4-MSH5 (ref. 35). Current evidence indicates that MutSγ binds and stabilizes DNA strand-exchange intermediates to promote homolog synapsis and crossing-over13,36–38. Therefore, we addressed whether immunostaining foci of RNF212 and MSH4 colocalize (Fig. 1r,s). Unexpectedly, at early pachynema, only 35% of MSH4 foci colocalized with RNF212 staining (34.9 ± 1.8% (s.e.m.); 9 nuclei), corresponding to ~41 MSH4-RNF212 foci per nucleus (41.4 ± 6.0). Conversely, only 33% of RNF212 foci colocalized with MSH4 staining (33.3 ± 4.4%). These estimates included faint and partially colocalizing foci and, as such, likely over-estimate the true colocalization frequency of MSH4 and RNF212 foci along synaptonemal complexes. Thus, RNF212 only marks a minor subset of MutSγ-containing recombination sites at early pachynema.
RNF212 foci mark crossover sites during midpachynema
At midpachynema, when most RNF212 foci have disappeared, the number of MSH4 foci also decreased from > 100 (113.3 ± 4.3 (s.e.m.); 16 nuclei) at early pachynema to ~60 (59.2 ± 4.5 (s.e.m.); 12 nuclei) at midpachynema. At this stage, in contrast to early pachynema, nearly all remaining RNF212 foci localized to recombination sites, as indicated by a high degree of colocalization with MSH4 foci (91.5 ± 1.4% (s.e.m.); 12 nuclei; Fig. 1t,u). However, this still represented a minority of all ongoing recombination events, as shown by the fact that only 41% of MSH4 foci colocalized with RNF212 (40.7 ± 2.9%; 12 nuclei), corresponding to ~24 costaining foci per nucleus.
The RNF212 staining pattern in midpachynema is highly reminiscent of that seen for the crossover-specific markers MLH1 and MLH3, components of the MutLγ complex, which is required for crossing-over39–43. Indeed, RNF212 foci in midpachynema nuclei showed a high degree of colocalization with MLH1 staining (82.0 ± 1.9%; 10 nuclei, Fig. 1v; similar results were obtained for MLH3, data not shown). Thus, RNF212 foci at midpachynema specifically mark crossover sites.
RNF212 localization in the absence of recombination
We examined the genetic requirements for RNF212 localization using several knockout lines. SPO11 belongs to the type II topoisomerase family of transesterases and catalyzes meiotic DSB formation7. Although homolog pairing is severely defective in Spo11−/− spermatocytes, a fraction of cells assemble incomplete synaptonemal complexes, which generally involve non-homologous chromosomes and multiple partners (Fig. 2a)44,45. In these nuclei, RNF212 localized as a punctate staining pattern specifically in regions of synapsis marked by SYCP1 (Fig. 2b). Thus, RNF212 can localize to synaptonemal complexes independent of recombination and even when synaptonemal complexes are formed between non-homologous chromosomes. These observations suggest that RNF212 has a general binding affinity for the synaptonemal complex central element.
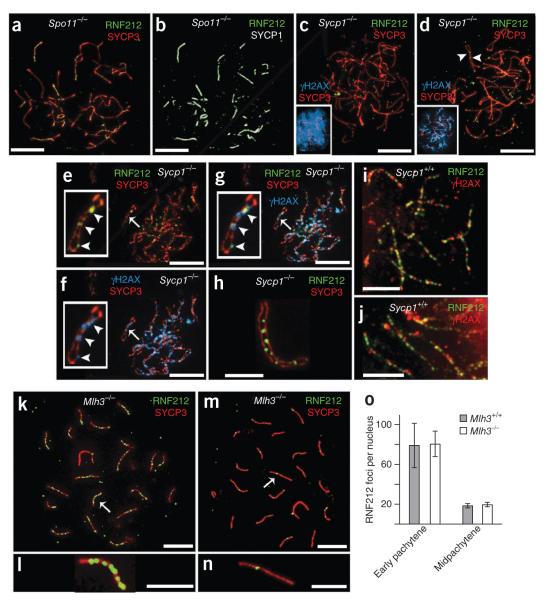
Figure 2.
Genetic requirements for RNF212 localization. (a,b) Spermatocyte nucleus from a Spo11−/− mouse with immunolabeling for SYCP3 and SYCP1 (a) and RNF212 and SYCP1 (b). (c–g) Spermatocyte nuclei from a Sycp1−/− mutant mouse immunolabeled for RNF212, SYCP3 and γH2AX. (c) Zygotene-like nucleus with pannuclear γH2AX staining. (d) Pachytene-like nucleus with diminishing γH2AX staining. Insets in c and d show SYCP3 and γH2AX channels. Arrowheads in d highlight RNF212 foci localized to coaligned homolog axes. (e–g) Late-pachytene/early diplotene nucleus stained for RNF212 and SYCP3 (e) and γH2AX and SYCP3, showing residual γH2AX foci (f). (g) Merged staining of this nucleus for RNF212, SYCP3 and γH2AX. Insets in e–g show a single homolog pair, with RNF212-γH2AX foci highlighted by arrowheads. (h) Selected homolog pair from a Sycp1−/− spermatocyte showing RNF212 foci associated with one of the two SYCP3-staining axes. (i) Representative homolog pairs from a wild-type early pachytene spermatocyte immunolabeled for RNF212 and γH2AX. (j) Independent nucleus immunolabeled as in i. (k–n) Spermatocyte nuclei from an Mlh3−/− knockout mouse, immunolabeled for RNF212 and SYCP3. (k,l) Early pachynema nucleus (k), with magnified view of the chromosome indicated by an arrow (l). (m,n) Midpachynema nucleus (m), with magnified view of the indicated chromosome (n). (o) Quantification of RNF212 foci (± s.d.) in early pachytene and midpachytene spermatocytes from wild-type and Mlh3−/− mice. For wild-type cells, 16 early pachytene and 16 midpachytene nuclei were analyzed. For Mlh3−/− cells, 39 early pachytene and 10 midpachytene nuclei were analyzed. Scale bars, 10 μm in a–g,k,m and 5 μm in h–j,i,n.
Defective localization of RNF212 in the absence of synapsis
The close spatial correlation between RNF212 staining and synaptonemal complexes observed in wild-type and Spo11−/− meiocytes suggests that RNF212 localization requires synapsis. To address this inference, we also analyzed RNF212 localization in spermatocytes from Sycp1 knockout mice, which lack the major component of the synaptonemal complex central region46. In Sycp1−/− meiocytes, recombination initiates normally, and homolog axes coalign, becoming closely associated at multiple positions, which are sites of recombination (Fig. 2c–e). However, synapsis is precluded.
In Sycp1−/− nuclei with zygotene-like and early pachytene–like morphologies, RNF212 generally did not localize to paired chromosomes (Fig. 2c–f). However, weak RNF212 foci were discerned along homologs that were extensively aligned (highlighted by arrowheads in Fig. 2d). In more advanced nuclei, in which the staining pattern of DSB-induced H2AX phosphorylation (γH2AX) had diminished from a pan-nuclear cloud (Fig. 2c) to limited chromatin flares and foci (Fig. 2f), RNF212 staining was more prominent (Fig. 2e). Moreover, in this class of nuclei, RNF212 foci frequently overlapped with γH2AX signals (highlighted by arrowheads in Fig. 2e–g; 77.8 ± 2.1% of RNF212 foci showed some overlap with γH2AX signals; 5 nuclei). Also, RNF212 foci were often associated with only one of the two homolog axes (Fig. 2e,h), in contrast to the central synaptonemal complex localization observed in wild-type cells. Therefore, in the absence of the synaptonemal complex central region, a general propensity of RNF212 to localize to γH2AX-associated recombination sites is observed. This contrasts with the finding in wild-type pachytene cells in which the level of RNF212 -γH2AX colocalization was only 29.5% (± 2.4%; 5 nuclei; Fig. 2i,j). We infer that, despite a general capacity to associate with recombination sites (at least in Sycp1−/− cells), the RNF212 localization pattern seen in wild-type early pachytene cells (with a minority of foci located at γH2AX- and/or MSH4-associated recombination sites) is strongly dependent on the synaptonemal complex central element.
Crossover factor MLH3 is not required for RNF212 localization
The relationship between crossing-over and RNF212 localization was addressed by analyzing Mlh3 knockout mice (Fig. 2k–n)40. The MutLγ complex, MLH1-MLH3, localizes specifically to future crossover sites at midpachynema and is required for the formation of ≥90% of all crossovers39–42. The RNF212 staining patterns detected in Mlh3−/− spermatocytes were very similar to those seen in wild-type cells (Fig. 2k–n): numerous RNF212 foci were initially detected along zygotene and early pachytene synaptonemal complexes, and their numbers were subsequently reduced to 1–2 foci per synaptonemal complex around midpachynema (quantified in Fig. 2o). Thus, RNF212 is able to attain a crossover-specific localization pattern independent of MutLγ and crossing-over: that is, midpachytene RNF212 foci specifically mark crossover precursors not crossover products.
Rnf212 knockout mice are sterile
Our localization studies suggest potential roles for RNF212 in synaptonemal complex function and/or crossing-over. To test these possibilities, we generated a knockout line lacking the Rnf212 promoter and first two exons that encode the RING domain (Fig. 3a–c and Supplementary Fig. 1). Rnf212+/− heterozygous mice appeared normal and produced litters with mendelian ratios of the expected genotypes. Both male and female Rnf212−/− homozygous animals appeared healthy but were sterile. Mature Rnf212−/− males did not make sperm and had testes that were ~70% smaller than those of wild-type animals, a characteristic of mutants with meiotic defects. Histological analysis of Rnf212−/− mutant testes showed an absence of post–anaphase I cells, indicating loss of spermatocytes at this stage (stage XII seminiferous tubules) (Fig. 3d,e). Although females were sterile, ovary size was similar to that of wild-type animals, and high numbers of oocytes were present in mature animals (Fig. 3f,g and Supplementary Fig. 4). These phenotypes contrast with those of synapsis- and recombination-defective mutants, such as Dmc1−/−, Msh5−/− and Sycp1−/− mice, in which spermatocytes undergo apoptosis around the time of pachytene (stage IV seminiferous tubules) and a majority of oocytes are lost shortly after birth46–48. However, testis and ovary morphologies seen in synapsis-proficient but crossover-deficient mutants Mlh1−/−, Mlh3−/− and Hei10−/− closely resemble those of Rnf212−/− mice40,42,49,50.
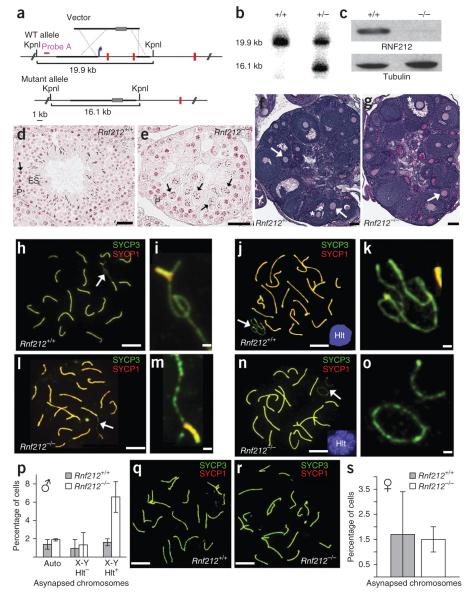
Figure 3.
Gonad morphology and homolog synapsis in Rnf212−/− knockout mice. (a) Rnf212 targeting scheme. Thick lines represent homology arms used for targeting. Gray rectangles represent the neomycin-resistance cassette. The blue arrow indicates the position of the promoter, and red vertical lines represent Rnf212 exons (not to scale). WT, wild type. (b) Southern analysis of KpnI-digested genomic DNA from wild-type and heterozygous embryonic stem cells hybridized with the probe shown in a. (c) Protein blot analysis of RNF212 in protein extracts from whole testes. (d,e) Representative seminiferous tubules stained with hematoxylin and eosin from testis sections from wild-type (d) and Rnf212−/− (e) mice. P, pachytene-stage cells; ES, elongated spermatids. Arrows indicate metaphase I cells. In Rnf212−/− tubules, cells progress to metaphase I (stage XII), but normal chromosome congression is not observed, and postmeiotic spermatogenic cells (spermatids and spermatozoa) are absent. (f,g) Representative ovary sections stained with periodic acid Schiff and hematoxylin from wild-type (f) and Rnf212−/− (g) mice. Asterisks indicate antral follicles with defined antral space. Arrows highlight secondary follicles surrounded by more than one layer of cuboidal granulosa cells. (h–o) Spread spermatocyte nuclei immunostained for SYCP3 and SYCP1. (h,i) Early pachynema nucleus from wild-type mouse (h), with magnified view of the sex chromosomes indicated by the arrow (i). (j,k) Midpachynema nucleus from wild-type cells (j). The inset shows H1t staining. (k) Magnified view of the indicated sex chromosomes in j. (l,m) Early pachynema nucleus from Rnf212−/− cells (l), with magnified view of the sex chromosomes indicated by an arrow (m). (n,o) Midpachynema nucleus from Rnf212−/− cells (n), with magnified view of the sex chromosomes (o). (p) Quantification of synapsis defects in pachytene spermatocytes. Numbers of nuclei analyzed in wild-type and Rnf212−/− cells, respectively: 293 and 521 for autosomal asynapsis (auto) and X-Y asynapsis in H1t-negative pachytene cells; 686 and 622 for X-Y asynapsis in H1t-positive pachytene cells. (q,r) Pachytene-stage fetal oocytes from wild-type (q) and Rnf212−/− mutant (r) animals immunostained for SYCP3 and SYCP1. (s) Quantification of synapsis defects in pachytene oocytes (191 and 272 wild-type and Rnf212−/− nuclei, respectively). Error bars in p and s, s.e.m. Scale bars, 100 μm in d–g, 10 μm in h,j,l,n,q,r and 1 μm in i,k,m,o.
Complete synapsis is achieved in Rnf212 knockout mice
The late-stage loss of spermatocytes and high oocyte numbers seen in Rnf212−/− mice suggest that the synaptonemal complex forms efficiently. To test this inference, we immunostained surface-spread spermatocytes and fetal oocytes for homolog axis (SYCP3) and synaptonemal complex central element (SYCP1) markers (Fig. 3h–s). Apparently normal pachytene nuclei, with fully synapsed autosomes, were observed in both spermatocyte and oocyte nuclei from Rnf212−/− mice (Fig. 3l,n,r). Moreover, the frequencies of synaptic defects, such as unsynapsed autosomes, were not increased in pachytene-stage meiocytes from male or female Rnf212−/− animals, indicating that synapsis occurs efficiently (Fig. 3p,s). This finding contrasts sharply with observations made for budding yeast zip3 mutants, which show severe defects in synaptonemal complex formation28, but is analogous to the apparently normal synapsis seen for C. elegans zhp-3 mutants29.
Temporal analysis of synapsis in spermatocytes from juvenile males, undergoing the first synchronous wave of meiosis, suggests that full synapsis is achieved with a slight delay in Rnf212−/− mutants (Supplementary Fig. 5a; delayed synapsis was not seen in females, Supplementary Fig. 5b). Moreover, in zygotene-stage nuclei from Rnf212−/− spermatocytes, we often observed SYCP1 staining associated with unsynapsed homolog axes (Supplementary Fig. 5c–f). These configurations may result from destabilized synaptonemal complexes that leads to partial desynapsis during zygonema or may represent sites where SYCP1 has associated with one axis, but synapsis has not successfully ensued.
X-Y synapsis is destabilized in Rnf212−/− spermatocytes
In spermatocytes, the X and Y chromosomes undergo recombination-dependent pairing and synapsis between short regions of homology termed the pseudoautosomal regions (PARs)51. Crossing-over subsequently occurs between the PARs to form X-Y chiasmata. At early pachynema, relatively extensive X-Y synapsis is observed that can encompass most of the length of the Y chromosome axis (for example, Fig. 3i,m). However, as pachynema progresses, X-Y synapsis contracts until only end-to-end association is seen (for example, Fig. 3k).
A significant number of adjacent but unsynapsed X-Y chromosomes were observed in Rnf212−/− spermatocytes (Fig. 3o). To quantify and determine the timing of this defect, we divided pachytene-stage cells into early and mid/late substages by immunostaining for the spermatocyte-specific histone H1 variant H1t52. In early pachynema (H1t negative), the efficiencies of X-Y synapsis in Rnf212−/− and wild-type spermatocytes were indistinguishable (Fig. 3p). However, in spermatocytes that had progressed beyond midpachynema (H1t positive), 6.7 ± 1.7% of cells (n = 622) had paired but unsynapsed X and Y chromosomes compared to 1.6 ± 0.3% of wild-type cells (n = 686; P > 0.001; Fig. 3p). We suggest that terminal X-Y synapsis is normally reinforced by nascent crossing-over between the PARs and that the recombination defect of Rnf212−/− mutants can lead to premature desynapsis.
Crossover complexes are absent in Rnf212−/− mutants
Despite contrasting synapsis phenotypes, yeast zip3 and C. elegans zhp-3 mutants share a common defect in crossing-over13,28,29. Given the mild effect of the Rnf212 knockout on synapsis, defective crossing-over is a likely cause of infertility. To analyze crossing-over, we counted chiasmata in chromosome spreads from spermatocytes in diakinesis/metaphase I stages (Fig. 4a–c). In wild-type cells, chiasmata averaged 24.8 ± 0.5 (12 nuclei) per nucleus, consistent with previous estimates53. In marked contrast, 60% of Rnf212−/− cells (30/50 spreads) contained exclusively univalent chromosomes, indicating a complete absence of chiasmata (Fig. 4b). The remaining 40% of metaphase nuclei (20/50) contained 1–3 bivalent chromosomes (Fig. 4c), for an average of only 0.84 ± 0.17 chiasmata per nucleus. Thus, crossing-over is diminished by ≥90% in Rnf212−/− mutants. Notably, chromosomal breaks and fragments were never observed in diakinesis/metaphase I cells from Rnf212−/− mutants, implying that DSBs are efficiently repaired, presumably as non-crossovers.
Figure 4.
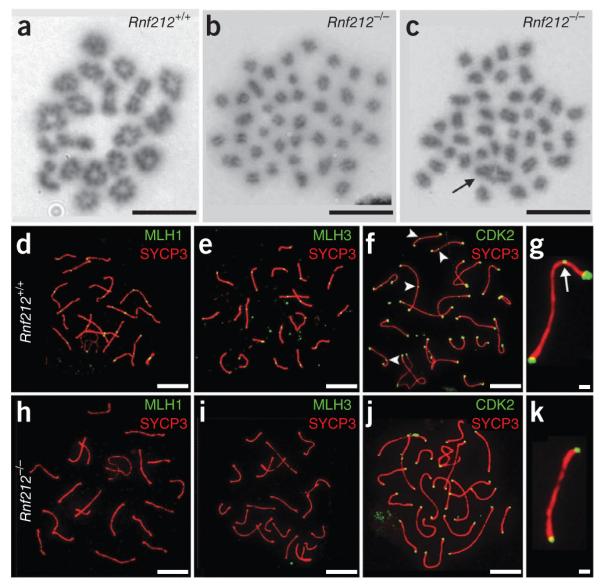
RNF212 is required for chiasma formation and assembly of crossover-specific recombination complexes. (a–c) Chromosome spreads of cells in diakinesis/metaphase I stained with Giemsa from wild-type (a) and Rnf212−/− (b,c) mice. (b) Nucleus with no chiasmata. (c) Nucleus with a single chiasmate bivalent (highlighted by an arrow). (d–k) Pachytene spermatocytes. (d–g) Wild-type cells immunostained for MLH1 and SYCP3 (d), MLH1 and SYCP3 (e) or CDK2 and SYCP3 (f). (g) Magnified view of a chromosome from f. (h–k) Rnf212−/− cells immunostained for MLH1 and SYCP3 (h), MLH3 and SYCP3 (i) or CDK2 and SYCP3 (j). (k) Magnified view of a chromosome from j. The arrowheads and arrow (f,g) highlight interstitial CDK2 foci. Scale bars, 10 μm in a–f,h–j and 1 μm in g,k.
To begin to understand why crossing-over is diminished in the absence of RNF212, we analyzed the chromosomal localization of crossover-specific, late-recombination-nodule components MLH1 and MLH3 (MutLγ) and cyclin-dependent kinase CDK2 (refs. 54–58). None of the three proteins were assembled into crossover-specific immunostaining foci in Rnf212−/− spermatocytes (Fig. 4d–k; see Supplementary Fig. 6 for analysis of MLH1 in fetal oocytes). CDK2, which normally localizes to telomeres as well as crossover sites, was only observed at telomeres in pachytene-stage Rnf212−/− spermatocytes. Given that MutLγ is essential for ~90% of crossovers in mice40,57, the crossover defect of Rnf212−/− mutants is explained by an inability to localize this complex to crossover precursor sites.
RNF212 stabilizes two ZMM factors, MutSγ and TEX11
The MutSγ complex, represented by MSH4 foci, initially localizes to a majority of recombination sites as homologs synapse (Fig. 5a,b). Subsequently, MSH4 foci begin to decrease in number, until ~50–60 remain at midpachynema (Fig. 5c,d). Less than half of these late-stage MSH4 foci colocalize with RNF212 and assemble the crossover-specific MutLγ complex (Fig. 1r–v)58.
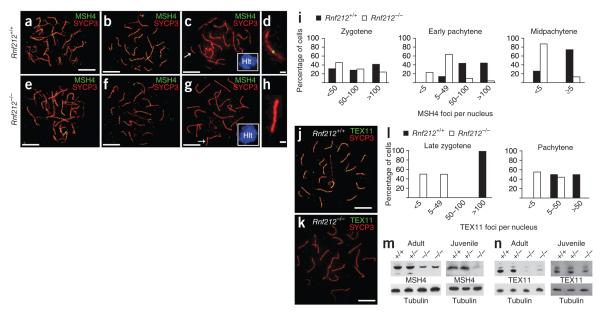
Figure 5.
RNF212 stabilizes two ZMM factors, Mutsγ and TEX11. (a–h) Spermatocyte nuclei immunostained for MSH4 and SYCP3 at successive prophase stages. (a–d) Wild-type cells stained at zygonema (a), early pachynema (b) and midpachynema (c). (d) Magnified chromosome from c. (e–h) Rnf212−/− cells stained at zygonema (e), early pachynema (f) and midpachynema (g). (h) Magnified chromosome from g. Insets in c and g show H1t staining. (i) Quantification of MSH4 foci at successive prophase stages. Statistical comparisons: zygotene, P = 0.1 (G test; n = 83 Rnf212+/+ and 51 Rnf212−/− nuclei); early pachytene, P = 1 × 10−32 (G test; n = 131 Rnf212+/+ and 103 Rnf212−/− nuclei); midpachytene, P = 1 × 10–19 χ-square test; n = 121 Rnf212+/+ and 103 Rnf212−/− nuclei). (j,k) Early pachytene spermatocyte nuclei immunolabeled for TEX11 and SYCP3 from wild-type (j) and Rnf212−/− (k) animals. (l) Quantification of TEX11 foci. In late zygonema, TEX11 foci average 122 ± 11 (s.d.; n = 5) in wild-type nuclei and 21 ± 22 (s.d.; n = 10) in Rnf212−/− mutants (P = 0.0027, Mann-Whitney test). In pachynema, TEX11 foci average 56 ± 27 (s.d., n = 10) in wild-type nuclei and 10 ± 11 (s.d., n = 9) in Rnf212−/− mutants (P = 0.0004, Mann-Whitney test). (m,n) Protein blot analysis of MSH4 (m) and TEX11 (n) in protein extracts from whole testes. In adult testes, the levels of MSH4 and TEX11 are approximately 40% and 17% of wild-type levels, respectively; in juvenile testes (18 d postpartum), protein levels are ~20% and ~63% of wild-type levels. Scale bars, 10 μm in a–c,e–g,j,k and 1 μm in d,h.
In Rnf212−/− spermatocytes, the initial formation of MSH4 foci in zygonema appeared normal, but, in subsequent stages, the numbers of these foci were significantly reduced compared to wild-type cells (Fig. 5e–i; similar observations were made for fetal oocytes, Supplementary Fig. 6). Most notably, the majority of midpachytene Rnf212−/− spermatocytes (positive for H1t staining) completely lacked MSH4 foci (Fig. 5i). These data indicate that RNF212 stabilizes a subset of MutSγ complexes.
We also examined the effects of the Rnf212 knockout on the localization of a second ZMM factor, TEX11. TEX11 is the mammalian homolog of budding yeast Zip4, a large TPR-repeat protein required for normal synapsis and crossing-over32,59–61. In mice, TEX11 localizes as numerous foci along synaptonemal complexes during zygonema and pachynema and colocalizes with transition-nodule components RPA and MSH4 (Fig. 5j)59. In Rnf212−/− spermatocytes, the number of TEX11 foci was markedly lower than in wild-type cells (Fig. 5k,l).
The above analysis indicates that RNF212 stabilizes ZMM proteins at the cytological level. We also examined the effects of the Rnf212 knockout on the stability of MSH4 and TEX11 at the protein level by protein blot analysis of testis extracts. Mirroring the cytological data, MSH4 and TEX11 protein levels were lower in Rnf212−/− mutants (Fig. 5m,n). This effect was not a trivial consequence of the reduced cellularity of adult Rnf212−/− testes because reduced levels of MSH4 and TEX11 were also seen in testes from juvenile animals (Fig. 5m,n).
Rnf212 is haploinsufficient
The contribution of characterized RNF212 alleles to the variance of human recombination rates is estimated to be on the order of two crossovers per meiosis16,18,19. The modest effects of RNF212 alleles in humans prompted us to carefully examine recombination in Rnf212+/− heterozygous mice (Fig. 6). Protein blot analysis of extracts from Rnf212+/− testes showed the expected 50% reduction in the amount of RNF212 protein (Fig. 6a). However, haploinsufficiency was suggested by quantification of RNF212 immunostaining foci, which were reduced by ~16% in early pachynema cells (Fig. 6c; P = 0.0003; n = 26 wild-type and 21 Rnf212−/− nuclei) and by ~13% in midpachynema cells (Fig. 6d; P = 0.001; n = 34 Rnf212+/+ and 38 Rnf212−/− nuclei). Correspondingly, the numbers of MSH4 foci were also significantly lower in Rnf212+/− spermatocytes (Fig. 6e).
Figure 6.
Rnf212 is haploinsufficient. (a) Quantitative protein blot analysis of RNF212 in testis cell extracts from wild-type, Rnf212+/− and Rnf212−/− mice. (b) Midpachytene Rnf212+/− spermatocyte nucleus immunostained for RNF212 and SYCP3. White lines highlight synaptonemal complexes that lack RNF212 foci. (c,d) Numbers of RNF212 foci per nucleus ± s.e.m. in wild-type and Rnf212+/− spermatocytes at early pachynema (c) and midpachynema (d). (e) Numbers of MSH4 foci per nucleus in wild-type and Rnf212+/− spermatocytes at successive prophase substages. Statistical comparisons: zygotene, P = 0.04 (G test; n = 83 Rnf212+/+ and 68 Rnf212+/− nuclei); early pachytene, P = 9 × 10−13 (G test; n = 131 Rnf212+/+ and 270 Rnf212−/− nuclei); midpachytene, P = 0.01 χ-square test; n = 121 Rnf212+/+ and 126 Rnf212−/− nuclei). (f,g) Midpachytene spermatocyte nuclei from wild-type (f) and Rnf212+/− (g) mice immunolabeled for MLH1 and SYCP3. White lines in g highlight synaptonemal complexes that lack MLH1 foci. (h) Numbers of MLH1 foci per nucleus in midpachytene-stage spermatocytes. Each symbol represents a single nucleus. Yellow lines indicate the average numbers of foci. (i–k) Chromosome spreads of spermatocytes in diakinesis/metaphase I from Rnf212+/+ (i) and Rnf212+/− (j,k) mice. Independent examples from Rnf212−/− spermatocytes are shown in (j) and (k). Arrows in j and k highlight achiasmate univalent chromosomes. (l) Numbers of chiasmata per nucleus (± s.e.m.) in wild-type and Rnf212+/− diakinesis/metaphase I spermatocytes. Scale bars, 10 μm.
Although Rnf212+/− males are fertile and have wild-type sperm counts, they showed significantly fewer MLH1 foci relative to wild-type cells (20.1 ± 0.3 in n = 51 Rnf212+/− nuclei versus 23.4 ± 0.3 in n = 47 wild-type nuclei; P < 0.0001; Fig. 6f–h). Similarly lower numbers of MLH1 foci were observed in fetal oocytes (Supplementary Fig. 7). Correspondingly, analysis of chromosome spreads from diakinesis/metaphase I spermatocytes showed significantly fewer chiasmata in Rnf212+/− heterozygotes (21.5 ± 0.4 in n = 13 Rnf212+/− nuclei versus 24.8 ± 0.5 in n = 12 wild-type nuclei; P < 0.0001; Fig. 6i–l). Moreover, 8.5% (5/59) of nuclei contained a pair of achiasmate univalent chromosomes, whereas no achiasmate univalent chromosomes were observed in 50 wild-type spermatocytes (Fig. 6i–k).
DISCUSSION
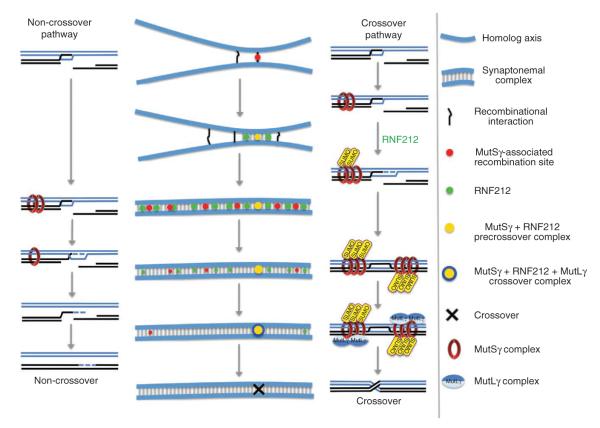
This study identifies RNF212 as an essential crossover factor during mammalian meiosis and provides insights into the molecular processes that underlie the differentiation of crossover and non-crossover recombination. How and when specific recombination sites are designated as having a crossover fate has remained unclear. At the cytological level, crossover-specific MutLγ-CDK2 foci do not appear until mid- to late pachynema and are clearly a secondary manifestation of crossover designation. Our analysis provides cytological evidence that the differentiation of crossover and non-crossover events occurs at least as early as zygonema, as RNF212 becomes localized to a subset of MutSγ-associated recombination complexes. This inference is consonant with DNA studies in budding yeast, which imply that crossover and non-crossover pathways diverge at the onset of zygonema, shortly after the formation of nascent strand-exchange intermediates called D-loops10–15 (Fig. 7). Whereas crossing-over involves the formation of metastable joint molecules (single-end invasions and double Holliday junctions), non-crossovers are inferred to arise from the disassembly of D-loops and annealing of DSB ends in a process termed synthesis-dependent strand annealing10,11,62 (Fig. 7). We infer that this differential stabilization or dissociation of DNA joint molecules is a consequence of selective RNF212-dependent stabilization of recombination factors, such as MutSγ, at precrossover sites.
Figure 7.
Summary and model of RNF212 function. Schematics showing the cytological development of recombination complexes and parallel molecular pathways of crossover and non-crossover recombination. Black and blue lines represent homologous DNA duplexes. Although four chromatids are present at this stage, for simplicity, only the two chromatids involved in recombination are shown. Both crossover and non-crossover pathways initiate from a common D-loop precursor. As synapsis ensues, binding of the MutSγ complex initially stabilizes most or all D-loops. In the absence of RNF212-mediated stabilization, MutSγ dissociates, and D-loops are unwound, resulting in non-crossover formation. At crossover sites, RNF212-dependent SUMOylation enhances the association of MutSγ, D-loops are stabilized, and formation of crossover-specific double Holliday junctions ensues. These crossover precursors become competent to assemble the crossover-specific resolution factor, MutLγ, and crossing-over occurs. Recombination sites that nucleate synaptonemal complex formation have a high probability of being the first sites where MutSγ and RNF212 colocalize. At such sites, a positive feedback loop locally enhances the binding of both MutSγ and RNF212. General binding of RNF212 to sites along the synaptonemal complex central element disfavors stabilization of MutSγ at other recombination sites.
MutSγ is an attractive target for crossover/non-crossover differentiation because in vivo and in vitro studies indicate that it directly binds and stabilizes nascent joint molecules and thereby facilitates the formation of crossover-specific double Holliday juctions13,36,63,64. Current evidence suggests that Zip3 and RNF212 are E3 ligases for SUMO31 (Y.Y. and N.H., unpublished data). We suggest that the association of MutSγ with nascent crossover intermediates may be stabilized via RNF212-mediated SUMOylation. SUMO modification could stabilize MutSγ in a number of ways, for example, by promoting protein-protein interactions, altering ATP binding and hydrolysis (which modulate the binding and dissociation of MutS complexes)65 or antagonizing ubiquitin-dependent protein turnover. Consistent with the latter possibility, MSH4 and TEX11 protein levels are lower in Rnf212−/− testes (Fig. 5m,n).
In budding yeast, the absence of ZMM proteins, including MutSγ, Zip4 and the RNF212 homolog Zip3, causes defects in both crossing-over and synaptonemal complex formation, raising the possibility that the crossover defects of zmm mutants are a secondary consequence of defective synapsis14. In mice, even though Msh4, Msh5 and Tex11 (Zip4) knockouts have synapsis defects37,38,59, full synapsis occurs in the Rnf212 knockout (Fig. 3). This result is unexpected because mice, like budding yeast, require recombination for normal synaptonemal complex formation44,45 (by contrast, synapsis in C. elegans is promoted by specialized pairing centers and does not require DSBs)66. Thus, unlike yeast Zip3, mammalian RNF212 is not essential for synapsis, and loss of RNF212 separates the early roles of MutSγ and TEX11 in promoting synapsis from their later functions in facilitating crossing-over.
Although RNF212 is not essential for synapsis, mild synaptonemal complex defects are detected in Rnf212−/− mice (Fig. 3). These defects are explained by the idea that synapsis is reinforced via RNF212-mediated stabilization of MutSγ-associated recombination complexes. However, it remains possible that RNF212 influences synaptonemal complex dynamics more directly, as inferred for Zip3 and ZHP-3 (refs. 31,67).
Zip3 localizes to recombination sites independent of the synaptonemal complex central region and is inferred to facilitate the initiation of synaptonemal complex formation from these sites28,68. This scenario sharply contrasts with the situation in mice and C. elegans, where timely localization and normal patterning of RNF212 foci are strongly dependent on the synaptonemal complex central element (Fig. 2c–i)29,67,69. Thus, mouse RNF212 couples synapsis to crossover and non-crossover differentiation. Notably, in synapsis-defective Sycp1−/− mutants, RNF212 patterning is both temporally and spatially defective, being greatly delayed and then showing a high degree of colocalization with γH2AX, which is not seen in wild-type cells. To reconcile these data, we propose that the synaptonemal complex central region has a high affinity for RNF212 binding that tends to outcompete binding to MutSγ-associated recombination sites.
In mice, mutation affecting a second RING-family E3 ligase, HEI10, causes a very similar phenotype to that of Rnf212−/− mice, with normal synapsis, failure to assemble crossover-specific recombination complexes and diminished crossing-over50. Intriguingly, recent analysis of HEI10 proteins from plants70,71 shows spatial-temporal patterns of chromosomal localization that are reminiscent of those observed for mouse RNF212. In fact, it has been proposed that rice HEI10 is the functional ortholog of yeast Zip3 and C. elegans ZHP-3 (and, by extension, mammalian RNF212)71. However, human HEI10 was reported to have ubiquitin E3 ligase activity (although indirect evidence has associated it with the SUMO pathway)72,73, whereas Zip3 and RNF212 seem to be SUMO E3 ligases31 (Y.Y. and N.H., unpublished data). Clearly, precisely defining the activities and relationships between RNF212 and HEI10 proteins is an important goal for the future.
The patterns of chromosomal localization for C. elegans ZHP-3 also resemble those of mouse RNF212 but with some important differences. ZHP-3 initially localizes along the lengths of synaptonemal complexes, but staining appears to be continuous, in contrast to the punctate staining of mouse RNF212 (refs. 29,67,69). Consequently, selective localization of ZHP-3 to a subset of early recombination sites cannot be discerned. As prophase progresses, ZHP-3 is selectively retained at crossover sites, but these crossover-specific foci are not apparent until the onset of diplotene, long after crossover designation is manifested by the formation of crossover-specific COSA-1 foci69. In fact, loss of general ZHP-3 staining from synaptonemal complexes seems to be coupled to the global remodeling of chromosomes that occurs as a consequence of crossing-over in C. elegans67,74,75. Thus, it remains unclear whether ZHP-3 has an early function in crossover differentiation analogous to that inferred here for mouse RNF212.
The question remains of how RNF212 localizes to specific recombination sites and not to others. One possibility is that crossover sites are predesignated via an unknown process that licenses the local accumulation of RNF212. A model has been proposed in which crossover designation and patterning occur via the imposition and redistribution of mechanical stress within the chromosomes76. Under this class of model, selective colocalization of RNF212 and MutSγ would occur as a downstream consequence of crossover designation. Alternatively, we suggest that the binding and interaction properties of RNF212 with recombination sites and synaptonemal complexes could form part of a self-organizing system that ultimately leads to the stable accumulation of RNF212 and MutSγ at only one or a few sites per synaptonemal complex (Fig. 7).
It has been proposed that a positive feedback loop, involving phosphorylation of MutSγ by a dedicated cyclin-Cdk complex, functions to reinforce crossover designation in C. elegans69. The data presented here raise the possibility that the process of designating, not just reinforcing, crossover sites may occur via a positive feedback loop involving RNF212 and MutSγ (Fig. 7). We suggest that RNF212-dependent SUMOylation of MutSγ locally enhances its binding to recombination sites, and, in turn, SUMOylated MutSγ locally enhances the association of RNF212. Sequestration of RNF212 by binding to sites along the synaptonemal complex central region could help limit this process to one or two recombination sites per synaptonemal complex (Fig. 7). This idea can also reconcile the observed correlation between synaptonemal complex initiation sites and crossing-over2,77–79 because the underlying recombination complexes at synaptonemal complex initiation sites will be the first to encounter a high local concentration of RNF212 and, as such, will have a high probability of being stabilized beyond early pachynema (Fig. 7).
Haploinsufficiency indicates that RNF212 is a limiting factor for crossover designation and/or reinforcement. We suggest that, when RNF212 levels are lower than some critical threshold, general binding to the synaptonemal complex central region leaves insufficient RNF212 available to accumulate at recombination sites and crossing-over stochastically fails. Haploinsufficiency for mouse Rnf212 also has implications for comprehending human RNF212 variants, which we suggest may alter the effective concentration of RNF212 protein. Our observations also suggest the idea that human RNF212 alleles may interact with variants that alter the levels or stability of recombination factors, such as MutSγ and TEX11. Indeed, an MSH5 allele (encoding p.Cys85Thr) and altered MSH4 expression have been tentatively associated with human male infertility80,81.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Mice
All mice were congenic with the C57BL/6J background. Mice were maintained and used for experimentation according to the guidelines of the Institutional Animal Care and Use Committee (University of California, Davis). The Mlh3, Spo11 and Sycp1 knockout lines were previously described40,44,46. The Rnf212 knockout line was constructed in collaboration with the University of California, Davis, Mouse Biology program. The Rnf212 knockout cassette comprised a 7.5-kb XhoI-NotI genomic fragment located upstream of the Rnf212 promoter and a 2.8-kb SalI-ClaI fragment located downstream of Rnf212 exon 2, flanking a neomycin cassette in the vector pKSloxPNT82. Targeted R1 embryonic stem cells83 were confirmed by Southern analysis of KpnI-digested genomic DNA (Fig. 3a). Chimeras on the 129X1/SvJ background were used to derive a congenic C57BL/6J Rnf212+/− line via marker-assisted breeding84. Primer sequences for genotyping are shown in Supplementary Table 1.
Expression analysis
RNA was isolated from mouse testes using TRIzol (Invitrogen), and cDNA was synthesized using Superscript III (Invitrogen). Rnf212 cDNAs were cloned into the TOPO-TA vector (Invitrogen), and individual clones were sequenced. For RT-PCR analysis (Supplementary Fig. 2), RNA was isolated from various mouse tissues, and, following cDNA synthesis, Rnf212 transcript variant 1 was amplified by PCR together with control cDNA, Gapdh. Primer sequences are shown in Supplementary Table 1.
Antibody production
Antibodies to RNF212 were produced by Antibodies Incorporated. To generate rabbit polyclonal antibody to RNF212, animals were immunized with full-length mouse RNF212 expressed as a calmodulin-binding peptide fusion (CBP-RNF212) in pCAL-n (Agilent Technologies) and purified from inclusion bodies. Antibodies were then affinity purified against immobilized mouse RNF212-6His fusion protein. To generate guinea pig antibody to RNF212, animals were immunized with full-length mouse RNF212 expressed as a 6His fusion (RNF212-6His) in pET28b (Novagen) and purified from inclusion bodies. Antibodies were then affinity purified against immobilized CBP-RNF212. The specificity of RNF212 antibodies was confirmed by protein blot analysis and immunostaining.
Protein blot analysis
Tissues from adult or juvenile mice were sonicated in RIPA buffer, protein concentration was assessed by the Bradford assay and 100–200 μg of protein was separated by SDS-PAGE (10% gels for analysis of RNF212 and 7.5% gels for analysis of MSH4 and TEX11). After protein transfer to PVDF or nitrocellulose membranes (Waterman), blots were incubated overnight with rabbit antibody to RNF212 (1:1,000 dilution), rabbit antibody to MSH4 (1:1,000 dilution; Abcam, ab58666), rabbit antibody to TEX11 (1:500 dilution)59 or mouse antibody to tubulin (1:2,000 dilution; BioLegend, 625902). Secondary antibodies (1:5,000 dilution for detection of MSH4 and 1:10,000 dilutions for all other experiments) were goat antibody to rabbit or anti-mouse IgGs conjugated to horseradish peroxidase (HRP) (SouthernBiotech, 4050-05 and 1031-05, respectively) or to infrared dyes (LI-COR, 926-32221 and 926-32210, respectively). HRP was detected using the ECL reagent (Pierce). Infrared secondary antibodies were imaged and quantified using a LI-COR Odyssey imaging system.
Cytology
Testes and ovaries were dissected from freshly killed animals and processed for surface spreading as described53,85. Immunofluorescence staining was performed as described86, using the following primary antibodies with incubation overnight at room temperature: mouse antibody to SYCP3 (Santa Cruz Biotechnology, sc-74568;1:200 dilution), rabbit antibody to SYCP3 (Santa Cruz Biotechnology, sc-33195; 1:300 dilution), mouse monoclonal antibody to rat SYCP1 (1:400 dilution)87, guinea pig antibody to TEX11 (1:200 dilution)59, rabbit antibody to MSH4 (Abcam, ab58666; 1:100 dilution), mouse antibody to MLH1 (BD Pharmingen, 550838; 1:50 dilution), rabbit antibody to MLH3 (1:500 dilution)55, guinea pig antibody to H1t (a gift from M.A. Handel, The Jackson Laboratory; 1:1,000 dilution)52, mouse monoclonal antibody to γH2AX (Millipore, 05-636; 1:500 dilution) and mouse monoclonal antibody to CDK2 (Santa Cruz Biotechnology, sc-6248; 1:200 dilution). Slides were incubated overnight at room temperature and were subsequently incubated with the following goat secondary antibodies for 1 h at 37 °C: antibody to rabbit 488 (A11070 Molecular Probes; 1:1,000 dilution), antibody to rabbit 568 (Molecular Probes, A11036; 1:2,000 dilution), antibody to mouse 555 (Molecular Probes, A21425; 1:1,000 dilution), antibody to mouse 594 (Molecular Probes, A11020; 1:1,000 dilution) and antibody to guinea pig fluorescein isothiocyanate (FITC; The Jackson Laboratory, 106-096-006; 1:200 dilution). Coverslips were mounted with ProLong Gold antifade reagent (Molecular Probes). For chiasma counts, air-dried preparations of diakinesis/metaphase I–stage cells were prepared as described88 and stained with Giemsa.
Imaging
Immunolabeled chromosome spreads and Giemsa-stained diakinesis/metaphase I nuclei were imaged using a Zeiss AxioPlan II microscope with 63× Plan Apochromat 1.4 objective and EXFO X-Cite metal halide light source. Images were captured by a Hamamatsu ORCA-ER CCD camera and processed using Volocity (PerkinElmer) and Photoshop (Adobe) software packages. SIM analysis was performed using a Nikon N-SIM super-resolution microscope system and NIS-Elements 2 image processing software.
Histology
Testes and ovaries were fixed in Bouins solution for 12 h at 4 °C and embedded in paraffin, and 4-μm (testes) or 8-μm (ovaries) sections were stained with hematoxylin and eosin (testes) or periodic acid Schiff and hematoxylin (ovaries). Follicle counts were performed as described48.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Heyting (Wageningen University) for antibodies to SYCP1 and SYCP3, S. Keeney and M. Jasin (Memorial Sloan-Kettering Cancer Center) for Spo11 knockout mice, M. Paddy for help with SIM imaging, J. Trimmer for help with imaging tissue sections and G. Coop for discussions. Knockout mice were generated in collaboration with the University of California, Davis, Mouse Biology Program (MBP). B.d.M. is supported by grants from the CNRS, Association pour la Recherche sur le Cancer and Agence Nationale de la Recherche (ANR-09-BLAN-0269-01). This work was supported by US National Institutes of Health (NIH) grant R01GM084955 to N.H. N.H. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS A.R., N.J., H.Q., Y.Y., K.B., P.E.C. and N.H. conceived and designed the experiments. A.R., N.J., H.Q., Y.Y., J.K.C., K.B., J.K.H., B.d.M., F.B. and N.H. performed the experiments. A.R., H.Q., Y.Y., J.K.C., K.B., B.d.M. and N.H. analyzed the data. J.W. and C.H. contributed reagents, materials and/or analysis tools. A.R., H.Q., Y.Y., K.B. and N.H. wrote the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Springer-Verlag; Heidelberg, Germany: 2006. pp. 381–442. [Google Scholar]
- 2.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 3.Sakuno T, Tanaka K, Hauf S, Watanabe Y. Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev. Cell. 2011;21:534–545. doi: 10.1016/j.devcel.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Hirose Y, et al. Chiasmata promote monopolar attachment of sister chromatids and their co-segregation toward the proper pole during meiosis I. PLoS Genet. 2011;7:e1001329. doi: 10.1371/journal.pgen.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 2007;16(Spec. No. 2):R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- 6.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 7.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones GH. The control of chiasma distribution. Symp. Soc. Exp. Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 9.Cole F, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 2012;14:424–430. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 11.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double–Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 12.Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 13.Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/ zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 14.Lynn A, Soucek R, Borner GV. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 15.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong A, et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- 17.Stefansson H, et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009;5:e1000648. doi: 10.1371/journal.pgen.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fledel-Alon A, et al. Variation in human recombination rates and its genetic determinants. PLoS ONE. 2011;6:e20321. doi: 10.1371/journal.pone.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fledel-Alon A, et al. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 2009;5:e1000658. doi: 10.1371/journal.pgen.1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong A, et al. Recombination rate and reproductive success in humans. Nat. Genet. 2004;36:1203–1206. doi: 10.1038/ng1445. [DOI] [PubMed] [Google Scholar]
- 22.Ségurel L, Leffler EM, Przeworski M. The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans. PLoS Biol. 2011;9:e1001211. doi: 10.1371/journal.pbio.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grey C, et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinch AG, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- 29.Jantsch V, et al. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol. Cell Biol. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 31.Cheng CH, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry J, Kleckner N, Borner GV. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc. Natl. Acad. Sci. USA. 2005;102:17594–17599. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan L, et al. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- 34.Carlton PM. Three-dimensional structured illumination microscopy and its application to chromosome structure. Chromosome Res. 2008;16:351–365. doi: 10.1007/s10577-008-1231-9. [DOI] [PubMed] [Google Scholar]
- 35.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- 36.Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 37.Edelmann W, et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 1999;21:123–127. doi: 10.1038/5075. [DOI] [PubMed] [Google Scholar]
- 38.Kneitz B, et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 39.Kolas NK, Cohen PE. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet. Genome Res. 2004;107:216–231. doi: 10.1159/000080600. [DOI] [PubMed] [Google Scholar]
- 40.Lipkin SM, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 41.Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker SM, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 43.Edelmann W, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 44.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 45.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 46.de Vries FA, et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barchi M, et al. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell Biol. 2005;25:7203–7215. doi: 10.1128/MCB.25.16.7203-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Giacomo M, et al. Distinct DNA-damage–dependent and –independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc. Natl. Acad. Sci. USA. 2005;102:737–742. doi: 10.1073/pnas.0406212102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kan R, et al. Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biol. Reprod. 2008;78:462–471. doi: 10.1095/biolreprod.107.065771. [DOI] [PubMed] [Google Scholar]
- 50.Ward JO, et al. Mutation in mouse Hei10, an E3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 2007;3:e139. doi: 10.1371/journal.pgen.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otto SP, et al. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 2011;27:358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Cobb J, Cargile B, Handel MA. Acquisition of competence to condense metaphase I chromosomes during spermatogenesis. Dev. Biol. 1999;205:49–64. doi: 10.1006/dbio.1998.9101. [DOI] [PubMed] [Google Scholar]
- 53.Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3–independent crossovers in mammalian meiosis. PLoS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcon E, Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid–treated mouse spermatocytes. Genetics. 2003;165:2283–2287. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolas NK, et al. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 2005;171:447–458. doi: 10.1083/jcb.200506170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashley T, Walpita D, de Rooij DG. Localization of two mammalian cyclin dependent kinases during mammalian meiosis. J. Cell Sci. 2001;114:685–693. doi: 10.1242/jcs.114.4.685. [DOI] [PubMed] [Google Scholar]
- 57.Woods LM, et al. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol. 1999;145:1395–1406. doi: 10.1083/jcb.145.7.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santucci-Darmanin S, et al. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 2000;14:1539–1547. doi: 10.1096/fj.14.11.1539. [DOI] [PubMed] [Google Scholar]
- 59.Yang F, et al. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 2008;22:682–691. doi: 10.1101/gad.1613608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adelman CA, Petrini JH. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 2008;4:e1000042. doi: 10.1371/journal.pgen.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsubouchi T, Zhao H, Roeder GS. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev. Cell. 2006;10:809–819. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 63.Oh SD, et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 66.Bhalla N, Dernburg AF. Prelude to a division. Annu. Rev. Cell Dev. Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macqueen AJ, Roeder GS. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr. Biol. 2009;19:1519–1526. doi: 10.1016/j.cub.2009.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yokoo R, et al. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chelysheva L, et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 2012;8:e1002799. doi: 10.1371/journal.pgen.1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K, et al. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 2012;8:e1002809. doi: 10.1371/journal.pgen.1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh MK, et al. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene. 2007;26:4825–4832. doi: 10.1038/sj.onc.1210282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strong ER, Schimenti JC. Evidence implicating CCNB1IP1, a RING domain–containing protein required for meiotic crossing over in mice, as an E3 SUMO ligase. Genes (Basel) 2010;1:440–451. doi: 10.3390/genes1030440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nabeshima K, Villeneuve AM, Colaiacovo MP. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 2005;168:683–689. doi: 10.1083/jcb.200410144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez-Perez E, et al. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008;22:2886–2901. doi: 10.1101/gad.1694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kleckner N, et al. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- 78.Henderson KA, Keeney S. Synaptonemal complex formation: where does it start? Bioessays. 2005;27:995–998. doi: 10.1002/bies.20310. [DOI] [PubMed] [Google Scholar]
- 79.Anderson LK, Stack SM. Recombination nodules in plants. Cytogenet. Genome Res. 2005;109:198–204. doi: 10.1159/000082400. [DOI] [PubMed] [Google Scholar]
- 80.Xu K, Lu T, Zhou H, Bai L, Xiang Y. The role of MSH5 C85T and MLH3 C2531T polymorphisms in the risk of male infertility with azoospermia or severe oligozoospermia. Clin. Chim. Acta. 2010;411:49–52. doi: 10.1016/j.cca.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 81.Terribas E, et al. Changes in the expression profile of the meiosis-involved mismatch repair genes in impaired human spermatogenesis. J. Androl. 2010;31:346–357. doi: 10.2164/jandrol.109.008805. [DOI] [PubMed] [Google Scholar]
- 82.Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- 83.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture–derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Markel P, et al. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat. Genet. 1997;17:280–284. doi: 10.1038/ng1197-280. [DOI] [PubMed] [Google Scholar]
- 85.Susiarjo M, Rubio C, Hunt P. Analyzing mammalian female meiosis. Methods Mol. Biol. 2009;558:339–354. doi: 10.1007/978-1-60761-103-5_20. [DOI] [PubMed] [Google Scholar]
- 86.Qiao H, Lohmiller L, Anderson L. Cohesin proteins load sequentially during prophase I in tomato primary microsporocytes. Chromosome Res. 2011;19:193–207. doi: 10.1007/s10577-010-9184-1. [DOI] [PubMed] [Google Scholar]
- 87.Meuwissen RL, et al. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J. Cell Biol. 2010;188:779–789. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.