Abstract
The mammalian odorant receptors (ORs) form a chemical-detecting interface between the atmosphere and the nervous system. This large gene family is composed of hundreds of membrane proteins predicted to form as many unique small molecule binding niches within their G-protein coupled receptor (GPCR) framework, but very little is known about the molecular recognition strategies they use to bind and discriminate between small molecule odorants. Using rationally designed synthetic analogs of a typical aliphatic aldehyde, we report evidence that among the ORs showing specificity for the aldehyde functional group, a significant percentage detect the aldehyde through its ability to react with water to form a 1,1-geminal (gem)-diol. Evidence is presented indicating that the rat OR-I7, an often-studied and modeled OR known to require the aldehyde function of octanal for activation, is likely one of the gem-diol activated receptors. A homology model based on an activated GPCR X-ray structure provides a structural hypothesis for activation of OR-I7 by the gem-diol of octanal.
The mammalian nose is a chemistry–biology interface. Odorant molecules are detected there by specialized cells known as olfactory sensory neurons (OSNs).1,2 Each OSN expresses on its surface a single member of the odorant receptor (OR) family, so that the pharmacologic odorant response of the OSN is determined by the OR it expresses.3,4 The ORs make up the largest family of G-protein coupled receptors (GPCRs) in the mammalian genome. Rodent genomes, for example, are predicted to encode ≈1100 functional ORs,5−7 while in humans about half of the ≈800 GPCRs are odorant receptors.8 Each membrane-bound OR has a different primary sequence, and each is expected to form a unique small-molecule binding niche within the GPCR structural framework. Fewer than 10% of the mouse and human ORs have been matched with an odorant agonist,9 and to date, no olfactory GPCR crystal structures have been solved. The small molecule recognition and discrimination strategies used in mammalian olfaction are therefore largely unexplored. Understanding the molecular details of odorant binding and functional group discrimination by the ORs (i) will improve our understanding of membrane protein–small molecule recognition, (ii) may reveal new strategies for targeting nonolfactory GPCRs of therapeutic interest, and (iii) could lead to high-affinity ligands able to promote the crystallization of odorant-bound GPCRs for pioneering structural studies. Until OR X-ray crystal structures become feasible, less direct approaches such as structure–activity relationships, mutagenesis studies, and computational modeling continue to be needed.10
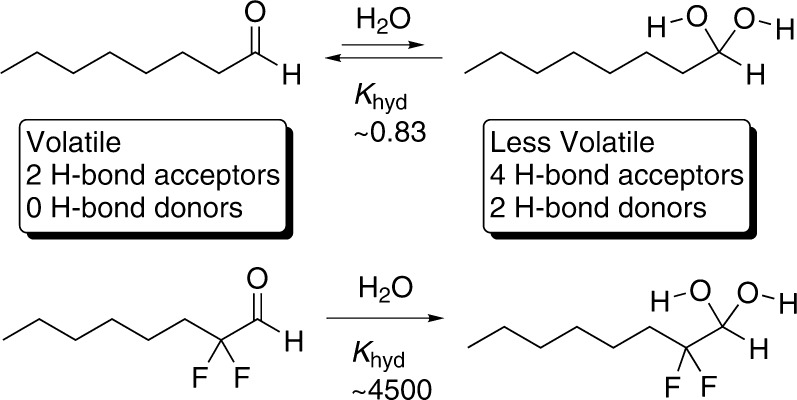
The aldehyde functional group is common among natural product odorants and synthetic fragrances.11 Although to reach the ORs an odorant must first dissolve in the water-based mucus covering the OSN tissue, the possibility that the hydrated form of the aldehyde, that is, the 1,1-geminal-diol or gem-diol (Scheme 1), is the activating ligand for some aldehyde-specific receptors has, to our knowledge, not been investigated. The lack of experimental OR structural information has prompted many computational OR studies, several of which have been carried out on aldehyde-binding ORs. In particular, of the 20 studies we found where at least one OR-odorant complex was computationally modeled, 14 (64%) included the modeling of an aldehyde in its carbonyl form.10,12−24 Clearly, to obtain the most accurate results, it is important to know the physiologically active form of the odorant.
Scheme 1. Aldehyde Hydration Equilibria and H-Bonding Capability.
Hydration of an aldehyde to its corresponding gem-diol dramatically changes the steric and electronic environment around the aldehyde carbon (C-1). First, the geometry rearranges from planar (sp2) to tetrahedral (sp3), reorienting the polar covalent bonds at C-1 (Scheme 1). Second, the hydrogen (H)-bonding capabilities near C-1, which likely play a role in binding aldehyde-specific ORs, are tripled, creating two new H-bond donors and two new acceptor lone pairs, while reorienting the initial two H-bond acceptor pairs. Third, while the net molecular dipoles likely do not differ greatly between the two forms, the individual C–O σ bond dipoles of the gem-diol are weaker and reoriented. Fourth, the gem-diol of an aldehyde can be more extensively solvated than the aldehyde form, making it more amphipathic, a difference that may affect activation by changing the kinetics of entering and leaving the binding niche, or by allowing water molecules to mediate recognition. Overall, hydration changes the aldehyde functional group to such an extent that, among those ORs that are specific for, that is, narrowly tuned to, the aldehyde functional group, it is unlikely that a single activated receptor conformation would recognize and be stabilized by both forms. This idea raises the possibility that for some aldehyde-specific ORs, the aldehyde group may be discriminated from other H-bond accepting functional groups by virtue of its ability to undergo chemical transformation to the gem-diol prior to encountering, or once within, the OR.
In this study, we have aimed to understand the true chemical nature of an activating aldehyde odorant, first among a large collection of rat ORs activated by a common fragrant aldehyde, octanal, and then for a well characterized OR whose activation is known to be rigorously aldehyde-specific. We present pharmacologic evidence supporting the conclusion that among the ORs activated by octanal, approximately 11% are activated by the less volatile but more H-bond-rich octane-1,1-diol. Surprisingly, within the subset of octanal-activated ORs that show specificity for the aldehyde functional group compared to its corresponding alcohol, nearly half appear to be activated by the gem-diol, raising the possibility that carbonyl hydration is a common determinant of aldehyde discrimination.
Results and Discussion
A Strategy to Detect Gem-Diol Dependent Receptor Activation
Our hypothesis is that some ORs appearing to recognize the aldehyde functional group are in fact activated by the gem-diol. To test this hypothesis, our approach is to manipulate the hydration equilibrium constant for a typical aldehyde through derivatization and then to compare the activity of the derivatized and natural compounds on live rat OSNs. The equilibrium hydration constant for n-aldehydes (Khyd) is ≈0.83 (25 °C; 0.62 at 35 °C) (Scheme 1).25,26 Highly electronegative groups such as fluorine on carbon 2 (C-2) upset this equilibrium and lead to near-complete hydration, with for example an estimated Khyd of 4500 (20 °C) for 2,2-difluorononanal.27 We selected octanal to represent a typical aliphatic aldehyde odorant and 2,2-difluorooctanal to represent its fully hydrated form (Scheme 1). We chose octanal because it is a structurally simple, frequently studied aldehyde odorant that activates a large number (≈70 at 30 μM) of different rodent OR family members,28−31 and because it is the primary natural product odorant for the well characterized rat OR-I7 receptor, which is known to require the aldehyde functional group for binding and activation.31,32
Fluorine is strongly electronegative and, with a van der Waals radius of 1.47 Å, only slightly larger than hydrogen (1.2 Å).33 These characteristics should maximize the electronic effect on hydration while minimizing confounding steric effects. To avoid a chirality center at C-2 and the well-known instability of α-monofluoro aldehydes,34 we limited our study to 2,2-difluoro substitution. Beyond altering the hydration behavior, difluoro-substitution can cause other changes and some of these may affect OR binding and activation. For instance, the fluorines introduce two bond dipoles at C-2, and these may dominate the receptor interaction for some ORs apart from the hydration effect. However, since we consider only the subset of cells (and therefore ORs) activated by octanal, ORs responding chiefly to the C–F dipoles will be disregarded because octanal does not contain C–F bonds and most octanal ORs should not be activated directly by them. Moreover, as described in detail below, compound 4 provides an additional control to filter out ORs whose activation depends primarily on fluorine substitution at C-2.
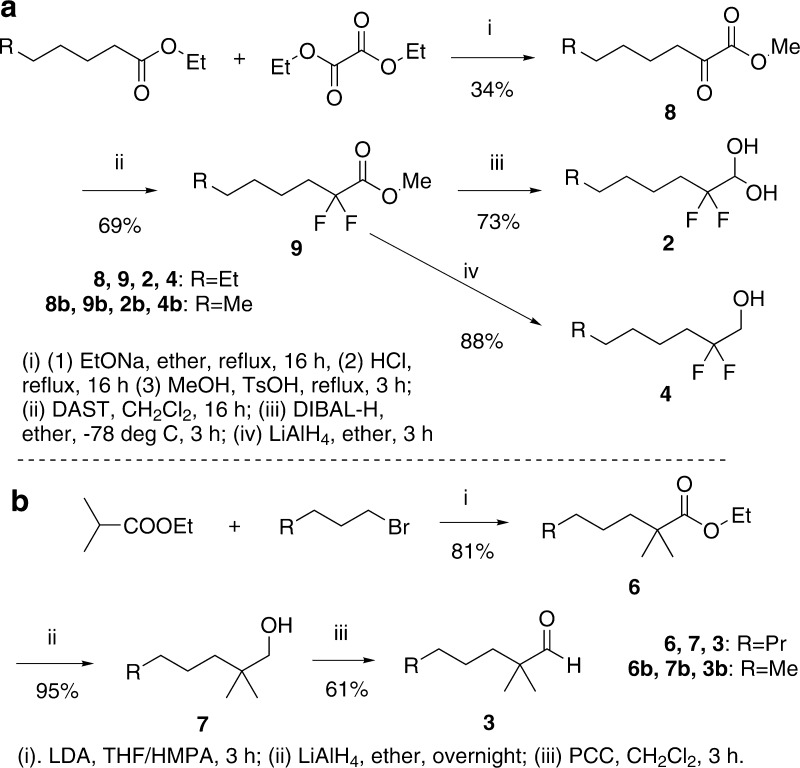
We chose the four additional compounds shown in Figure 1 to interrogate a large sampling of rat octanal ORs for evidence of octane-1,1-diol recognition. We reasoned that cells expressing octanal receptors requiring the gem-diol will respond to octanal 1, which at equilibrium forms ≈40% of the gem-diol and, for those ORs where the fluorines do not interfere, to the 2,2-difluoro analog 2, which forms >99.9% of the gem-diol. However, the corresponding alcohols, 2,2-difluorooctanol 4 and 1-octanol 5, will not activate octanal ORs that require the second hydroxyl of the gem-diol. We thus look for cells whose activation hinges upon the presence of the geminal hydroxyls. Using compound 4 as a control reduces the chances of false positives due to the C–F bond dipoles introduced by using fluorine. For example, consider a cell expressing the rare OR activated by octanal in its carbonyl form, but that also happens to respond to the dipoles of fluorine substitution. The response of such a cell could be dominated by the dipoles to the extent that it is also activated by 2, which forms a negligible amount of the carbonyl, thereby giving a false positive. However, activation of an octanal receptor by 4 would alert us to the possibility that the C–F dipoles are contributing directly to the activation of that OR, and information from that cell would not be taken as evidence for gem-diol recognition. Compound 3, 2,2-dimethyloctanal, serves as a control compound with an inverse inductive effect which should suppress gem-diol formation compared to octanal. Though methyl groups are the smallest electron-releasing groups we can use, they are significantly larger than H and F, and might for steric reasons fail to activate some of the ORs that require the aldehyde carbonyl (i.e., false negatives for carbonyl form). We also considered including octanoic acid in the list of control compounds, but a previous study in rat OSNs reported that 90% of octanal-responding cells that failed to respond to octanol also failed to respond to octanoic acid.35 To minimize the number of test compounds, and therefore maximize the number of cells remaining functional until the end of the assay, it was not included. Overall, in a particular cell, comparably strong activation by compounds 1 and 2, with no activation from compounds 3, 4, and 5 will constitute a pharmacologic signature for gem-diol-specific ORs, and allow us to assess the prevalence of this OR strategy for recognizing the aldehyde functional group. As described above, our approach seeks to minimize false positives resulting from the fluorine substituents, that is, carbonyl-specific cells that appear to be activated by the gem-diol, but false negatives are unavoidable and prevent us from making a complete tally of the carbonyl-specific versus gem-diol-specific octanal ORs. False negatives include gem-diol specific ORs unable to accommodate the two fluorines on compound 2 because they are too large, or incompatibility with the dipoles, and carbonyl-specific ORs unable to accommodate the two methyls of compound 3. The synthesis of compounds 2–4 is outlined in Scheme 2. Experimental details can be found in the Supporting Information.
Figure 1.
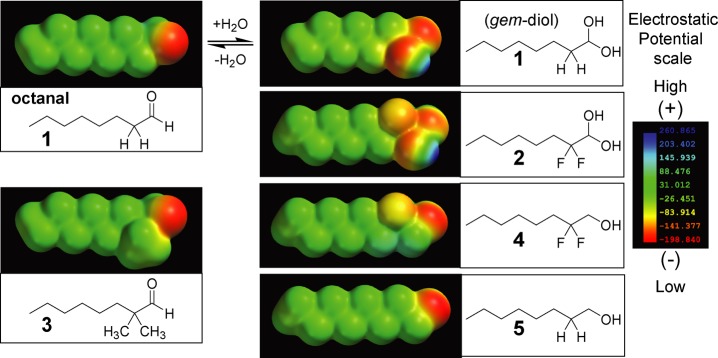
Octanal and structural analogs used to screen rat olfactory sensory neurons for activation by the gem-diol of octanal. Electrostatic potential maps were calculated using Spartan 10 V1.1.0.
Scheme 2. Synthetic Routes to Compounds Used in Odorant Receptor Testing and NMR Hydration Study.
Aldehyde Hydration Equilibria and α-Substitution
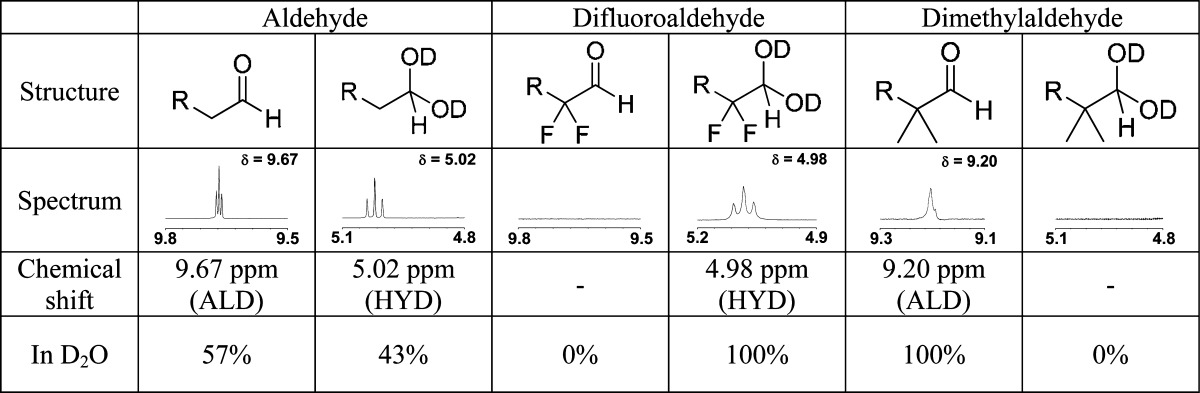
Prior to biological testing, we studied aldehydes similar to 1–3 by 1H NMR to verify the hydration change between n-alkanals and the corresponding 2,2-disubstituted analogs (Table 1; see Supporting Information for full spectra). Due to the low solubility of octanal in water, we compared the shorter congeners hexanal, 2,2-dimethylhexanal, and 2,2-difluoroheptanal. The aldehyde Khyd has been found elsewhere to be unaffected by the number of carbons in an n-alkyl chain.25 The Khyd (23 °C) changed from ≈0.75 for hexanal to ≈5000 for difluoroheptanal. In contrast, 2,2-dimethylhexanal formed no detectable gem-diol.
Table 1. Hydration Equilibrium of Aldehydes Measured by 1H-NMR in D2O at 23 °C.
Octanal Analog Screening in Live Olfactory Sensory Neurons
We used calcium imaging recordings4,28 to profile 1053 functional OSNs following dissociation of the cells from the rat olfactory epithelium and mucus. Since OSNs express a single OR family member,3,4 single-cell activity can be taken to represent a single OR’s response to each of compounds 1–5. In this technique, the OSNs are first loaded with the calcium sensitive fluorescent dye Fura-2 and then exposed to 30 μM ligand solutions in a flow-through perfusion chamber fitted onto a fluorescence microscope. The short lifetime of the dissociated OSNs limits the number of tests that can be done on dissociated OSNs, so we relied on a single concentration that was previously found to be conducive for detecting low and high affinity ORs and for detecting functional group selectivity in OSNs.35 Compounds functioning as agonists activate signal transduction within the cells, leading rapidly to depolarization-driven calcium influx and a reduction of fluorescence at the monitored wavelength. Thus, optical monitoring of the dispersed cells permits the screening of many OSNs while retaining single-cell, and therefore single OR family member, resolution.
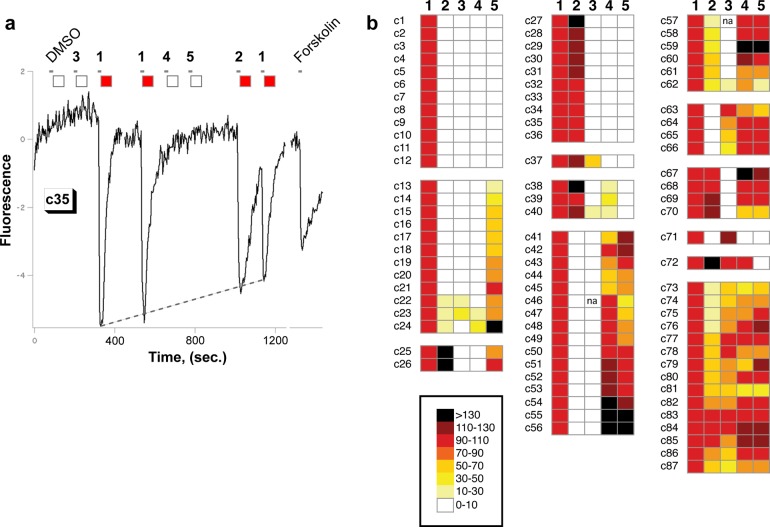
The fluorescence trace of a representative octanal-activated cell is shown in Figure 2a, and a summary of the responses of all octanal-activated cells to the screening compounds is shown in Figure 2b. Responses for each compound are reported relative to the octanal response generated by that cell, which is set to 100% (red in color scale), and the cells are grouped according to similarity of response. Out of 1053 cells, 87 cells (8%, Figure 2b, c1–c87) were activated by octanal and then observed for their response to compounds 2–5. These cells exhibited 59 unique response patterns when the scaled measurements were taken into account, suggesting the presence of a large variety of OR binding niches differentially affected by this group of close analogs. Substitution at C-2 was generally unfavorable for octanal OR activation. Only 28% of octanal-activated cells were activated by 3, and 52% were activated by 2. This trend argues that the loss of activation of these ORs is more steric than electronic, as the smaller fluorine substituent was better tolerated. This experimentally verified bias against C-2 substitution increased our expectation that there would be some false negatives, that is, aldehyde-specific ORs that our approach would not be able to identify as either carbonyl- or gem-diol-specific.
Figure 2.
Calcium imaging results for olfactory sensory neuron responses to compounds 1–5. (a) A representative calcium imaging trace, here depicting the cell c35 response. Broken line shows the octanal trend-line over the course of the experiment (see Methods). Small squares summarize the fluorescence response normalized to that of octanal, according to color scheme shown in panel b. The tick mark below each compound number marks the start of the 4 s injection of odorant solution into buffer stream flowing over cells. (b) Summary of responses for all octanal-activated cells to compounds 1–5 at 30 μM. (na, no data). Fluorescence changes are normalized to each cell’s response to compound 1, which is set to 100%.
Octanal and octanol are natural products that differ only by the oxidation state at C-1. Of the 87 cells activated by octanal, 59 cells (68%; Figure 2B, c13–c26, c41–c70, c73–c87) were also activated by octanol. The ORs expressed in these cells failed to distinguish between octanal and octanol and are therefore not aldehyde group-specific octanal ORs. In contrast, 24 cells (28%, c1–12, c27–37, c71) were activated by octanal but not by alcohols 4 or 5. These cells express ORs appearing to require the aldehyde group for activation. The remaining ≈4% of cells (c38–40, c72) were activated by difluoro alcohol 4 but not by octanol 5. Of these cells, c38–39 appear to have some affinity for the fluorine substituents or their dipoles, and thus, we do not assign them to the gem-diol specific category even though they are strongly activated by gem-diol 2.
The 24 cells appearing to require the aldehyde for activation by octanal fell into four subgroups: those stringently specific for octanal and responding to no other analog (50%, c1–12); those producing the pharmacologic pattern consistent with a requirement for the gem-diol (42%, c27–36; 11% of all octanal-activated cells); one cell producing the pharmacologic pattern consistent with a requirement for the carbonyl form (4%, c71); and one indeterminate cell appearing to require the gem-diol, but also activated by 2,2-dimethyloctanal (4%, c37). Assuming the aldehyde is recognized as either the carbonyl or gem-diol, cells c1–12 could be false negatives for either carbonyl- or gem-diol-specific ORs, but we cannot assign them to either category. The data from cells c27–36 support the surprising conclusion that, among aldehyde-specific cells, about 42% (10/24) appeared to require the gem-diol. Thus, recognition of the gem-diol may be a common means to discriminate the aldehyde functional group from other H-bond accepting functional groups such as the corresponding alcohol. We note that the actual percentages found here apply only to our sampling of 1053 cells which approaches nominal 1× coverage of the ≈1100 rat ORs. At this low level of coverage, some ORs were likely not present, and some may occur more than once. The time- and labor-intensive nature of live neuron screening makes a higher sampling coverage impractical using current methods, and the limited lifetime of the dissociated OSNs precludes the testing of a larger group of compounds on a given OSN.
Dose–Response Curves in the Aldehyde-Specific Receptor OR-I7
Though it is not possible to identify which OR family member is expressed in each of the cells profiled in Figure 2b, the data suggest that gem-diol recognition is common among ORs specific for the aldehyde functional group. Pharmacologically, the rodent OR-I7 is one of the most thoroughly characterized ORs and has been found to have a strict requirement for the aldehyde group in the context of aliphatic chains with 6 to 11 carbons.30−32,36,37 To ask whether OR-I7 detects the gem-diol form of the aldehyde, we probed the mouse and rat OR-I7 with compounds 1–5. Both orthologs are activated by octanal, though with some difference in the preferred chain length.36,38 On the one hand, if OR-I7 is activated by octanal’s carbonyl form, we would expect compound 2 (>99% gem-diol) to be completely inactive. On the other hand, if OR-I7 activation depends on the gem-diol, we would expect 2 to be two- to 3-fold more potent than octanal, due to the greater percentage of the gem-diol form, unless the fluorines have an unfavorable steric or dipole effect. In one type of experiment, we expressed recombinant mouse OR-I7 in Hana3A cells,39−41 an OR heterologous expression system based on HEK293T cells, and probed the cellular response using an assay that responds directly to the cAMP second messenger (Figure 3a, GloSensor Assay). The summed Hana3A/mouse OR-I7 dose response curves are shown over the 3 to 7.5 min time period in Figure 3A. Raw data for the entire 30 min experiment is included in the Supporting Information. Mouse OR-I7 was activated by octanal with an EC50 of about 1.5 μM. Difluorooctanal 2 activated OR-I7, but about 7-fold more weakly (EC50 ≈10 μM). The alcohols and, notably, the other 2,2-disubstituted octanal, 3, did not significantly activate mouse OR-I7. Compounds 1–3 were also tested against the recombinant rat ortholog expressed in rat OSNs with similar results (Figure 3b). Alcohol 5 is known not to activate rat OR-I7.32 In addition, gem-diol 2 was tested against the rat OR-I7 in Hana3A cells using the luciferase reporter gene as an alternative readout system and was also found to have an EC50 of ≈10 μM (not shown). These data support the possibility that the gem-diol is required for activation of this aldehyde-specific receptor, since the corresponding primary alcohols were inactive. The 7-fold lower potency of gem-diol 2 in comparison to octanal is subject to interpretation. In view of our finding in the rat OSN survey that substitution at C-2 is generally unfavorable for octanal ORs, our interpretation is that the fluorines create opposing steric and electronic effects: through their inductive effect, they permit only the gem-diol form, which is favorable, but they are sterically unfavorable, and so, compound 2 requires a higher concentration for binding and activation. In compound 3, both steric and electronic effects are unfavorable. Thus, the OR-I7 receptor appears to be activated by the octanal gem-diol and, given the structural differences between the aldehyde and gem-diol forms described in the Introduction, likely achieves its aldehyde specificity through sensing the gem-diol form.
Figure 3.
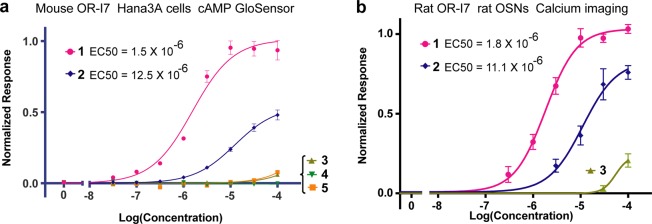
Dose–response curves for compounds 1–5 and rodent OR-I7. (a) Hana3A cells expressing mouse OR-I7 were exposed to odorants while cAMP production was monitored over a 30 min period. The summed response between 3 and 7.5 min is shown versus odorant concentration. (b) Rat olfactory sensory neurons infected with adenovirus expressing rat OR-I7 were assayed using calcium imaging during exposure to odorants 1–3.
Homology Model of Rodent OR-I7 Docked with Octane-1,1-Gem-Diol
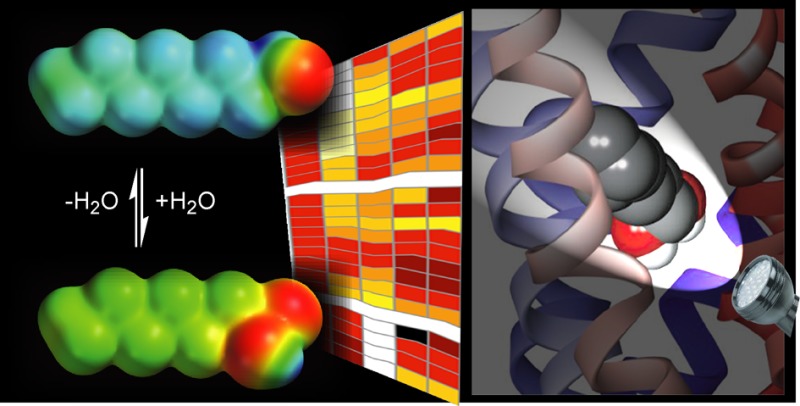
To further evaluate the possibility that rat and mouse OR-I7 might be activated by the gem-diol, we modeled both orthologs with this form of the aldehyde functional group.42 The only high resolution structural information available for odorant receptors has come from homology models, and many have been based on GPCRs crystallized in their inactive form. While these models may prove to be accurate for binding the unactivated ORs, they are less likely to provide direct insight into how odorant ligands stabilize the activated form of the OR to initiate signal transduction. Our two new models are based on the recently solved crystal structure of the activated, ligand-, and G-protein-bound β2-adrenergic receptor (β2AR) (Pdb 3SN6).43 The two ortholog models proved to be closely similar, and representative views of the rat OR-I7 model are presented in Figure 4. We docked into the two models the gem-diol of a conformationally restricted analog of octanal previously found to be as potent as octanal against the rat OR-I731 and evaluated its accommodation in the binding site for the best scored poses (see Methods for details). The more flexible octane-1,1-diol (or octanal in its carbonyl form, see below) was then superposed on and replaced the optimal pose of this ligand. In its most favorable position (Figure 4a, rat OR-I7 model), the gem-diol ligand was found tipping slightly down toward the intracellular side and aiming the gem-diol at TM2 and TM7, while in some previous models the ligand is found slightly higher within the membrane, tipping toward the extracellular side, and aiming at TM4, where it makes a possible contact with Lys164.18,23,24 In our model, TM4 is further from the ligand. A side-view of octane-1,1-diol (Figure 4b) shows the alkyl chain resides in a hydrophobic pocket formed by TMs 3, 5, and 6 with the geminal hydroxyls well oriented to interact through hydrogen bonds with Tyr74 (BW 2.53) and Tyr257 (BW 6.48) (Figure 4c). Interestingly, Tyr257 may be stabilized by a hydrogen bond to Glu116 in such a way as to position the Tyr257 hydroxyl oxygen to act as hydrogen bond acceptor for the gem-diol. The carbonyl form of octanal would be unable to interact with Tyr257 in this way, or with both tyrosines simultaneously, which provides a preliminary explanation for a more favorable interaction between OR-I7 and the gem-diol of octanal compared to the carbonyl form. Nevertheless, both gem-diol and aldehyde were well accommodated in the binding pocket of the receptors, as estimated by interaction energy calculations (rat I7, DS 3.5 Accelrys; mouse I7 DS 4.0, Accelrys). The values of interaction energy with the rat OR-I7 (−18.12 kcal/mol for the gem-diol, −12.05 kcal/mol for the carbonyl form), and the mouse OR-I7 (−7.4 kcal/mol for the gem-diol, −5.5 kcal/mol for the carbonyl form) predict that the gem-diol is superior to the aldehyde by ≈2–6 kcal/mol.
Figure 4.
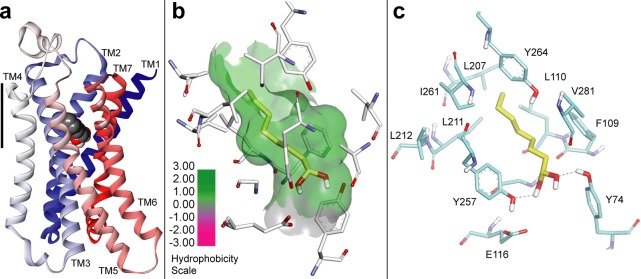
Homology model of rat OR-I7 based on the activated ß2-adrenergic receptor (pdb 3SN6) and bound to octane-1,1-diol. (a) Overall structure showing OR-I7 with the octane-1,1-diol agonist aiming the gem-diol toward trans-membrane helices (TM) 2 and 7. TMs are colored from blue (N-terminus) to red (C-terminus). Ligand membrane depth is shown in relation to TM4 (scale bar, 12.7 Å). (b) The octanal carbon chain is in a hydrophobic pocket formed by TMs 3, 5, and 6. (c) Possible H-bond recognition of the gem-diol by Y74 and Y257. Carbons of octane-1,1-diol are shown in yellow.
Since the carbonyl form of an aldehyde is more volatile than the gem-diol, it is reasonable to expect that most of an aldehyde sample reaching the nose through the air will initially be in the carbonyl form. Aldehydes undergo rapid acid-25 and base-44 catalyzed hydration, but at the slightly acidic pH of the nasal epithelium,45 the uncatalyzed rate of hydration is expected to be slow (k ≈ 3.5 × 10–3 s–1, t1/2 = 3.3 min).25 Although some gem-diol will have formed within the time it takes to perceive an aldehyde, without catalysis the equilibrium concentration will not be achieved within that time. In our live OSN assay, where the mucus is lost during OSN isolation, we avoided any possible kinetic influence by equilibrating compounds 1–5 in aqueous buffer prior to testing. However, in live animals, an aldehyde hydratase activity might be necessary to meet a gem-diol threshold concentration for some aldehyde ORs. Interestingly, carbonic anhydrase, an enzyme known to catalyze the hydration of aliphatic aldehydes46 is found in the nasal mucus47 and, we speculate, might play a role in accelerating gem-diol formation. Evidence supporting the enzymatic conversion of odorants in the mucus has previously been found.48 Phosphate and other solutes have also been found to modestly accelerate aldehyde hydration.49 Since GPCRs can harbor significant numbers of ordered water molecules50 and are predicted to contain even more,51 some aldehyde ORs might mediate aldehyde hydration themselves upon ligand binding. Using simple acid–base catalysis, a mucus catalyst, or the OR itself, might provide the modest rate enhancement necessary to maximize gem-diol formation on the time scale of olfaction.
In conclusion, our data suggest that a significant percentage of aldehyde-specific ORs recognize this functional group through its ability to engage in an equilibrium-based chemical transformation to a different functional group, the gem-diol. We propose that this is one way that aldehyde-specific ORs discriminate aldehydes from similar H-bond accepting functional groups, allowing the OR to contribute unequivocal aldehyde-specific information to the olfactory code.4
Methods
Electrostatic Potential Maps
Models were constructed and EPM calculations made using Spartan 10 V1.1.0 (Wavefunction, Inc.).
Chemical Synthesis and Characterization
Aldehyde Hydration Equilibrium 1H NMR Measurements
Hexanal or difluoroheptanal (3 mg) was dissolved in 1 mL D2O. 64 transients were accumulated. For dimethylhexanal, ≈0.5 mg was used because of its lower solubility in water, and 800 transients were accumulated. Data acquisition was begun at least 15 min after dissolving the compound in D2O.
Olfactory Sensory Neuron Preparation and Calcium Imaging Recordings
All animal procedures were approved by the Columbia University Institutional Animal Care and Use Committee (IACUC) and performed at Columbia University in compliance with relevant national guidelines and regulations.
Procedures for isolating rat OSNs31 and performing calcium imaging recordings35 were done as previously described. Briefly, dissociated cells were washed in rat Ringer’s solution and loaded in the dark with Fura-2AM supplemented with pluronic acid in rat Ringer’s solution for 45 min at room temperature (RT). Fluorescent recordings were made at 380 nm excitation and 510 nm emission. In order to minimize photobleaching, images were only taken every 4 s. The coverslip was placed into a perfusion chamber (200 μL) that pumped fresh rat Ringer’s solution over the cells at 2 mL min–1. Odorant application consisted of injecting 400 μL of solution into the constant perfusion stream over the course of 4 s.
Odorants were stored under argon gas at or under 4 °C and used within 7 days of purification. Freshly made DMSO-odorant stocks were diluted to 30 μM in rat Ringer’s solution31 (pH 7.4) and loaded into stimulus syringes. The diluted odorants were prepared at least 1 h prior to the start of imaging. Plain DMSO in Ringer’s solution at a matched volume was applied as a control; the rare cells that responded to vehicle alone were excluded from further study. Stimuli were given at least 2 min apart to permit complete odorant clearance.
Data in Figure 2 are shown as the fractional change in fluorescent light intensity, (F–F0)/F0, where F is the fluorescent light intensity at each point and F0 is the value for the emitted fluorescent light at the start of each CCD camera movie before the first stimulus application. Responses were measured between the baseline and peak ΔF/F change. To permit within-cell normalization of responses and to correct for any baseline drift due to incomplete recovery or focus shift, octanal applications were provided at the start or soon after the start of compounds testing, and near or at the end. We previously established that when a cell is challenged with three sequential identical stimuli, the magnitude of the response to the second application meets or exceeds 90% (0.90) that predicted from a trend line drawn between the peak magnitudes of the first and third flanking applications. Using this trend-line approach, we calculated the relative response of odorants compared to the response to octanal in each cell by taking the ratio of the measured response to the trend-line predicted response. When a compound is more efficacious than octanal, these ratios exceed 1.0.
At the end of each recording session, cells were challenged with 10 μM forskolin to activate adenylyl cyclase, a component of the signal transduction cascade downstream of the OR. We take the response to forskolin as an indicator that the cell is functionally intact. Only cells that could respond to forskolin were included in Figure 2 data.
Calcium imaging dose response curves for compounds 1–5 against the recombinant rat OR-I7 were done similarly, as previously described,31,32 in rat OSNs expressing OR-I7 and GFP from an adenovirus vector.37 For these experiments, 10 μM octanal, a saturating concentration for rat OR-I7, was used as the flanking stimulus to allow for normalization.
Mouse OR-I7 Hana3A GloSensor Assay
The GloSensor cAMP Assay System (Promega) was used according to manufacturer’s instructions with slight modifications. Briefly, a plasmid encoding Rho-tagged mouse OR-I7 (80 ng/well) was transfected into the Hana3A cell line in 96-well plate format along with plasmids encoding the human receptor trafficking protein, RTP1S40 (10 ng/well), type 3 muscarinic acetylcholine receptor (M3-R)39 (10 ng/well), and pGloSensorTM-22F (10 ng/well). Then, 18 to 24 h following transfection, cells were loaded with 2% GloSensor reagent for 2 h and treated with odorant compounds in a total volume of 74 μL. Luminescence was measured using a Polarstar Optima plate reader (BMG) with a time interval of 90 s per well. Raw data for the first 30 min is shown in Supporting Information. Data was analyzed and EC50s estimated using Prism 5.0 and Microsoft Excel. Responses over t = 3–7.5 min were summed, base-lined, normalized, and plotted vs odorant concentration in Figure 3A.
Rat OR-I7 Hana3A Luciferase Assay (Compound 2 Only)
The Dual-Glo Luciferase Assay System (Promega) was used for the luciferase assay as previously described.41 Briefly, a plasmid encoding Rho-tagged rat OR-I7 (5 ng/well) was transfected into the Hana3A cell line in 96-well plate format along with plasmids encoding the human receptor trafficking protein, RTP1S40 (5 ng/well), pSV40-Renilla (5 ng/well; Promega), CRE-luciferase (10 ng/well; Stratagene), and type 3 muscarinic acetylcholine receptor (M3-R)39 (2.5 ng/well). Then, 18 to 24 h following transfection, cells were treated with compound 2 for 4 h at 37 °C, as described.39 Luminescence was measured using a Polarstar Optima plate reader (BMG). Luciferase measurements were normalized to Renilla luciferase measurements to control for transfection efficiency and cell viability. Fold change values were calculated by the formula (F1–F0)/F0, where F1 is the normalized luminescence response to the odorant and F0 is the normalized luminescence when no odorant was applied. Data were analyzed and the EC50 for 2 (≈10 μM) was estimated using Prism 5.0 and Microsoft Excel. Estimating the EC50s for the other four odorants under the conditions of this assay was not possible because they underwent significant evaporation. For this reason, we used the GloSensor and calcium imaging assays described above to monitor OR-I7 activation in real time.
Homology Model Construction and Ligand Docking
The rat OR-I7 (Uniprot entry: P23270) was aligned with the human β2-AR sequence (3SN6.pdb) using TM Coffee (http://tcoffee.crg.cat/apps/tcoffee/do:tmcoffee) and MAFFT (http://mafft.cbrc.jp/alignment/server/). The manually refined alignment is shown in the Supporting Information. A disulfide bond was maintained between Cys102 and Cys184 as a restriction during model generation. A model of rat OR-I7 was created using the MODELER protocol in Discovery Studio 3.5 (DS3.5, Accelrys). The model was refined using minimization and side-chain optimization using SCWRL (http://dunbrack.fccc.edu/scwrl4). Trp154 (4.50) in OR-I7 was manually changed to a rotamer most similar to the one in β2-AR. This rotamer also has the most favorable energy. Before docking, the extracellular and intracellular loops were removed and a binding site was created using ‘define and edit binding site’ protocol (Discovery Studio 3.5, Accelrys). Ligands were prepared using “prepare ligands” protocol and conformations were generated using “generate conformations” protocol. To minimize ligand flexibility during docking, the gem-diol form of the conformationally restricted octanal analog, trans-2-(4-ethylcyclohexyl)ethanal was used in place of octane-1,1-diol. This aldehyde was previously found to have about the same rat OR-I7 potency as octanal.31 Docking of this ligand was performed using CDocker protocol (all protocols available in Discovery Studio 3.5, Accelrys). Octane-1,1-diol was superposed onto the optimal pose and used to replace the conformationally restricted ligand, and the model was energy minimized. An identical protocol was used to prepare a model of the mouse OR-I7 ortholog (Uniprot entry: Q9QWU6) using Discovery Studio 4.0 (DS4.0, Accelrys).
Acknowledgments
This work was supported by the U.S. Army Research Laboratory and U.S. Army Research Office under grant number W911NF-13-1-0148 (to K.R.). Additional support was provided by the National Institutes of Health under grant numbers 5SC1GM083754 (to K.R.), DC010857 (to H.M.) and DC012095 (to H.M.). Additional infrastructural support at the City College of New York was provided by the National Center for Research Resources (2G12RR03060-26A1) and the National Institute on Minority Health and Health Disparities (8G12MD007603-27). Mass spectrometry instrumentation was supported in part by National Science Foundation grant 0840498.
Supporting Information Available
Complete synthetic procedures, compound characterization including hydration 1H NMR spectra, organoleptic observations, sequence alignments between rodent OR-I7, and human β2AR, and cAMP time course dose–response data for mouse OR-I7 and compounds 1–5 in Hana3A cells. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
⊥ Y.L., Z.P., and J.H. contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- DeMaria S.; Ngai J. (2010) The cell biology of smell. J. Cell Biol. 191, 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein S. (2001) How the olfactory system makes sense of scents. Nature 413, 211–218. [DOI] [PubMed] [Google Scholar]
- Chess A.; Simon I.; Cedar H.; Axel R. (1994) Allelic inactivation regulates olfactory receptor gene expression. Cell 78, 823–834. [DOI] [PubMed] [Google Scholar]
- Malnic B.; Hirono J.; Sato T.; Buck L. B. (1999) Combinatorial receptor codes for odors. Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- Niimura Y.; Nei M. (2005) Evolutionary changes of the number of olfactory receptor genes in the human and mouse lineages. Gene 346, 23–28. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Firestein S. (2002) The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5, 124–133. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Zhang X.; Firestein S. (2007) Comparative genomics of odorant and pheromone receptor genes in rodents. Genomics 89, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A. J.; Deupi X.; Lebon G.; Tate C. G.; Schertler G. F.; Babu M. M. (2013) Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194. [DOI] [PubMed] [Google Scholar]
- Saito H.; Chi Q.; Zhuang H.; Matsunami H.; Mainland J. D. (2009) Odor coding by a mammalian receptor repertoire. Sci. Signal. 2, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S.; Hirokawa T.; Oka Y.; Suwa M.; Touhara K. (2005) Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: Mapping the odorant-binding site. J. Neurosci. 25, 1806–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibka J.; Glinski M. (2006) Synthesis and olfactory properties of 2-alkanals, analogues of 2-methylundecanal. Flavour Fragrance J. 21, 480–483. [Google Scholar]
- Anselmi C.; Buonocore A.; Centini M.; Facino R. M.; Hatt H. (2011) The human olfactory receptor 17–40: Requisites for fitting into the binding pocket. Comput. Biol. Chem. 35, 159–168. [DOI] [PubMed] [Google Scholar]
- Baud O.; Etter S.; Spreafico M.; Bordoli L.; Schwede T.; Vogel H.; Pick H. (2011) The mouse eugenol odorant receptor: Structural and functional plasticity of a broadly tuned odorant binding pocket. Biochemistry 50, 843–853. [DOI] [PubMed] [Google Scholar]
- Charlier L.; Topin J.; Ronin C.; Kim S. K.; Goddard W. A. 3rd; Efremov R.; Golebiowski J. (2012) How broadly tuned olfactory receptors equally recognize their agonists. Human OR1G1 as a test case. Cell. Mol. Life Sci. 69, 4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doszczak L.; Kraft P.; Weber H. P.; Bertermann R.; Triller A.; Hatt H.; Tacke R. (2007) Prediction of perception: Probing the hOR17–4 olfactory receptor model with silicon analogues of bourgeonal and lilial. Angew. Chem., Int. Ed. Engl. 46, 3367–3371. [DOI] [PubMed] [Google Scholar]
- Hall S. E.; Floriano W. B.; Vaidehi N.; Goddard W. A. 3rd (2004) Predicted 3-D structures for mouse I7 and rat I7 olfactory receptors and comparison of predicted odor recognition profiles with experiment. Chem. Senses 29, 595–616. [DOI] [PubMed] [Google Scholar]
- Khafizov K.; Anselmi C.; Menini A.; Carloni P. (2007) Ligand specificity of odorant receptors. J. Mol. Model. 13, 401–409. [DOI] [PubMed] [Google Scholar]
- Kurland M. D.; Newcomer M. B.; Peterlin Z.; Ryan K.; Firestein S.; Batista V. S. (2010) Discrimination of saturated aldehydes by the rat I7 olfactory receptor. Biochemistry 49, 6302–6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai P. C.; Singer M. S.; Crasto C. J. (2005) Structural activation pathways from dynamic olfactory receptor-odorant interactions. Chem. Senses 30, 781–792. [DOI] [PubMed] [Google Scholar]
- Launay G.; Teletchea S.; Wade F.; Pajot-Augy E.; Gibrat J. F.; Sanz G. (2012) Automatic modeling of mammalian olfactory receptors and docking of odorants. Protein Eng., Des. Sel. 25, 377–386. [DOI] [PubMed] [Google Scholar]
- Sanz G.; Thomas-Danguin T.; Hamdani el H.; Le Poupon C.; Briand L.; Pernollet J. C.; Guichard E.; Tromelin A. (2008) Relationships between molecular structure and perceived odor quality of ligands for a human olfactory receptor. Chem. Senses 33, 639–653. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg K.; Shirokova E.; Weber H. P.; Schilling B.; Meyerhof W.; Krautwurst D. (2007) Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J. Struct. Biol. 159, 400–412. [DOI] [PubMed] [Google Scholar]
- Singer M. S. (2000) Analysis of the molecular basis for octanal interactions in the expressed rat 17 olfactory receptor. Chem. Senses 25, 155–165. [DOI] [PubMed] [Google Scholar]
- Vaidehi N.; Floriano W. B.; Trabanino R.; Hall S. E.; Freddolino P.; Choi E. J.; Zamanakos G.; Goddard W. A. 3rd (2002) Prediction of structure and function of G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 99, 12622–12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H.-J.; Dutkiewicsz E.; Knoche W. (1982) The reversible hydration of carbonyl compounds in aqueous solution. Part II: The kinetics of the keto/gem-diol transition. Ber. Bunsen-Ges. Phys. Chem. Chem. Phys. 86, 139–144. [Google Scholar]
- Buschmann H.-J.; Fuldner H.-H.; Knochem W. (1980) The reversible hydration of carbonyl compounds in aqueous solution. Part I: The keto/gem-diol equilibrium. Ber. Bunsen-Ges. Phys. Chem. Chem. Phys. 84, 41–44. [Google Scholar]
- Quero C.; Rosell G.; Jimenez O.; Rodriguez S.; Bosch M. P.; Guerrero A. (2003) New fluorinated derivatives as esterase inhibitors. Synthesis, hydration, and crossed specificity studies. Bioorg. Med. Chem. 11, 1047–1055. [DOI] [PubMed] [Google Scholar]
- Araneda R. C.; Peterlin Z.; Zhang X.; Chesler A.; Firestein S. (2004) A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J. Physiol. 555, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluza J. F.; Breer H. (2000) Responsiveness of olfactory neurons to distinct aliphatic aldehydes. J. Exp. Biol. 203, 927–933. [DOI] [PubMed] [Google Scholar]
- Krautwurst D.; Yau K. W.; Reed R. R. (1998) Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 95, 917–926. [DOI] [PubMed] [Google Scholar]
- Peterlin Z.; Li Y.; Sun G.; Shah R.; Firestein S.; Ryan K. (2008) The importance of odorant conformation to the binding and activation of a representative olfactory receptor. Chem. Biol. 15, 1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araneda R. C.; Kini A. D.; Firestein S. (2000) The molecular receptive range of an odorant receptor. Nat. Neurosci. 3, 1248–1255. [DOI] [PubMed] [Google Scholar]
- O’Hagan D. (2008) Understanding organofluorine chemistry. An introduction to the C–F bond. Chem. Soc. Rev. 37, 308–319. [DOI] [PubMed] [Google Scholar]
- Steiner D. D.; Mase N.; Barbas C. F. 3rd. (2005) Direct asymmetric alpha-fluorination of aldehydes. Angew. Chem., Int. Ed. Engl. 44, 3706–3710. [DOI] [PubMed] [Google Scholar]
- Araneda R. C.; Peterlin Z.; Zhang X.; Chesler A.; Firestein S. (2004) A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J. Physiol. 555, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T.; Feinstein P.; Zheng C.; Mombaerts P. (2002) Odorant receptor expression defines functional units in the mouse olfactory system. J. Neurosci. 22, 3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.; Ivic L.; Otaki J. M.; Hashimoto M.; Mikoshiba K.; Firestein S. (1998) Functional expression of a mammalian odorant receptor. Science 279, 237–242. [DOI] [PubMed] [Google Scholar]
- Connelly T.; Savigner A.; Ma M. (2013) Spontaneous and sensory-evoked activity in mouse olfactory sensory neurons with defined odorant receptors. J. Neurophysiol. 110, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. R.; Matsunami H. (2011) Activation state of the M3 muscarinic acetylcholine receptor modulates mammalian odorant receptor signaling. Sci. Signaling 4, ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang H.; Matsunami H. (2007) Synergism of accessory factors in functional expression of mammalian odorant receptors. J. Biol. Chem. 282, 15284–15293. [DOI] [PubMed] [Google Scholar]
- Zhuang H.; Matsunami H. (2008) Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat. Protoc. 3, 1402–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levit A.; Barak D.; Behrens M.; Meyerhof W.; Niv M. Y. (2012) Homology model-assisted elucidation of binding sites in GPCRs. Methods Mol. Biol. 914, 179–205. [DOI] [PubMed] [Google Scholar]
- Rasmussen S. G.; DeVree B. T.; Zou Y.; Kruse A. C.; Chung K. Y.; Kobilka T. S.; Thian F. S.; Chae P. S.; Pardon E.; Calinski D.; Mathiesen J. M.; Shah S. T.; Lyons J. A.; Caffrey M.; Gellman S. H.; Steyaert J.; Skiniotis G.; Weis W. I.; Sunahara R. K.; Kobilka B. K. (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruen L. C.; McTigue P. T. (1963) Kinetics of hydration of aliphatic aldehydes. J. Chem. Soc. 5224–5229. [Google Scholar]
- Washington N.; Steele R. J.; Jackson S. J.; Bush D.; Mason J.; Gill D. A.; Pitt K.; Rawlins D. A. (2000) Determination of baseline human nasal pH and the effect of intranasally administered buffers. Int. J. Pharm. 198, 139–146. [DOI] [PubMed] [Google Scholar]
- Pocker Y.; Dickerson D. G. (1968) The catalytic versatility of erythrocyte carbonic anhydrase. V. Kinetic studies of enzyme-catalyzed hydrations of aliphatic aldehydes. Biochemistry 7, 1995–2004. [DOI] [PubMed] [Google Scholar]
- Debat H.; Eloit C.; Blon F.; Sarazin B.; Henry C.; Huet J. C.; Trotier D.; Pernollet J. C. (2007) Identification of human olfactory cleft mucus proteins using proteomic analysis. J. Proteome Res. 6, 1985–1996. [DOI] [PubMed] [Google Scholar]
- Nagashima A.; Touhara K. (2010) Enzymatic conversion of odorants in nasal mucus affects olfactory glomerular activation patterns and odor perception. J. Neurosci. 30, 16391–16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocker Y.; Meany J. E. (1967) Acid-base-catalyzed hydration of acetaldehyde. Buffer and metal ion catalysis. J. Phys. Chem. 71, 3113–&. [Google Scholar]
- Okada T.; Fujiyoshi Y.; Silow M.; Navarro J.; Landau E. M.; Shichida Y. (2002) Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 99, 5982–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfield A.; Pitman M. C.; Feller S. E.; Soubias O.; Gawrisch K. (2008) Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J. Mol. Biol. 381, 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.