Abstract
Despite decades of research on the bacterial ribosome, the ribosomal exit tunnel is still poorly understood. Although it has been suggested that the exit tunnel is simply a convenient route of egress for the nascent chain, specific protein sequences serve to slow the rate of translation, suggesting some degree of interaction between the nascent peptide chain and the exit tunnel. To understand how the ribosome interacts with nascent peptide sequences, we synthesized and characterized a novel class of probe molecules. These peptide–macrolide (or “peptolide”) conjugates were designed to present unique peptide sequences to the exit tunnel. Biochemical and X-ray structural analyses of the interactions between these probes and the ribosome reveal interesting insights about the exit tunnel. Using translation inhibition and RNA structure probing assays, we find the exit tunnel has a relaxed preference for the directionality (N → C or C → N orientation) of the nascent peptides. Moreover, the X-ray crystal structure of one peptolide derived from a positively charged, reverse Nuclear Localization Sequence peptide, bound to the 70S bacterial ribosome, reveals that the macrolide ring of the peptolide binds in the same position as other macrolides. However, the peptide tail folds over the macrolide ring, oriented toward the peptidyl transferase center and interacting in a novel manner with 23S rRNA residue C2442 and His69 of ribosomal protein L4. These data suggest that these peptolides are viable probes for interrogating nascent peptide–exit tunnel interaction.
The ribosome, through well-choreographed processes, translates genetically encoded messages on mRNAs to polypeptides. While structural and biochemical studies of prokaryotic 70S ribosome have enhanced our understanding of the role many of these components play during translation,1−6 little is understood about the ribosomal peptide exit tunnel. During elongation of the nascent peptide, the growing peptide chain extends from the peptidyl transferase center (PTC) to the backside of the ribosome through the peptide exit tunnel, an 80 Å long, 20 Å wide exit tunnel that extends across the large subunit of the ribosome from the base of the PTC and opens at the back of the subunit.4,7,8 The role of the peptide exit tunnel is primarily to act as a route of egress for the nascent peptide;4,9,10 however, in some cases, specific interactions between the nascent peptide and the exit tunnel walls have been shown to alter translational regulation.9,11−13 Currently, it is not well understood how the ribosome could distinguish and respond to specific peptide sequences while facilitating an unhindered passage of the vast majority of peptides through the peptide exit tunnel.
Efforts aimed at mapping the paths of the nascent peptide through the ribosome have focused mainly on trapping sequence-specific peptides known to interact directly with the exit tunnel.11,14 Specifically, fluorescence resonance energy transfer (FRET),15 molecular-dynamics simulation,11 and cross-linking experiments16 have furnished biochemical insights into the interactions between a 17 amino acid motif near the C terminus of SecM and the components of the Escherichia coli (E. coli) ribosome exit tunnel, which results in translation arrest. Single particle cryo-EM reconstructions have revealed the presence of a relay mechanism involving direct interactions between the nascent peptide and ribosomal exit tunnel resulting in the ribosome stalling during translation of both the ErmBL leader peptide in the presence of erythromycin17 and the tnaC leader gene.14 Moreover, analysis of primer extension inhibition has led to the postulation of similar peptide-dependent ribosome stalling relay mechanisms at the regulatory cistron of the antibiotic resistance gene ermA.18 While these experiments provide evidence that distinct peptides interact with the exit tunnel, they illustrate the importance of designing molecular probes, which can precisely position any peptide sequence within a defined region of the peptide exit tunnel.
Recently, analogs of 16-membered macrolides tylosin, desmycosin, and 5-O-mycominosyltylonolide incorporating esters of amino acids, di- and tripeptides have been shown to engage the exit tunnel through distinct interactions with ribosomal components, presumably using their amino acid and peptide moieties.19,20 Specifically, molecular dynamics simulations suggest that the N-acylglycyl moiety of the aminoacylated 5-O-mycominosyl-tylonolide derivative could be positioned to form hydrogen bonds with 23S rRNA A752 and Lys 90 of ribosomal protein L22. These hydrogen bonds could potentially disrupt the essential A752:U2609 base pair, which has previously been shown to play a role in both peptide stalling and macrolide antibiotic interactions.20,22,25 However, the peptide moieties of these conjugates are short and are expected to interrogate only a short segment of the exit tunnel, particularly the tunnel entrance.
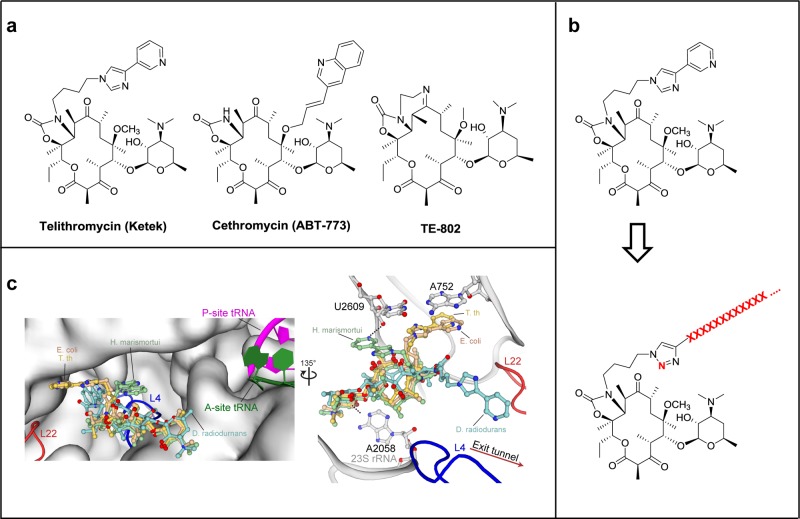
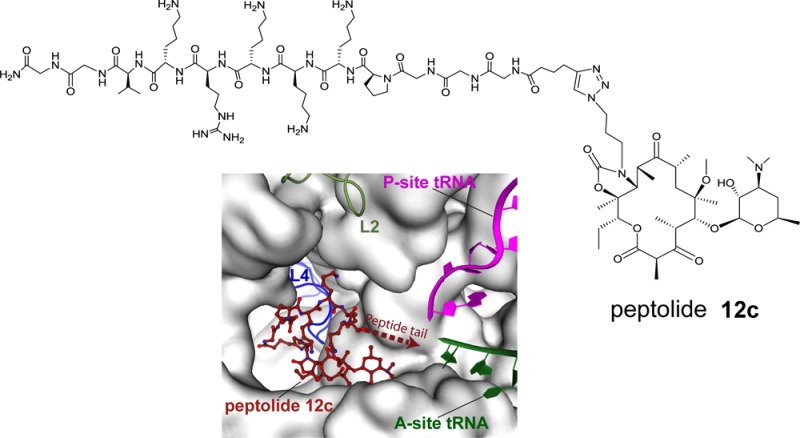
Toward the development of robust molecular probes for exploring the passage of nascent peptide through the exit tunnel, we designed oligopeptides covalently linked to a ketolide analog, which we term “peptolides” (Figure 1). Ketolides are a class of small molecule antibiotics, collectively known as macrolides, which inhibit prokaryotic translation by sterically blocking the exit tunnel near the PTC.21 As an inspiration for the design of these peptolide probes, we looked at telithromycin (TEL), a unique ketolide with a flexible alkyl-aryl arm (Figure 1a). X-ray crystallographic studies have shown that the alkyl-aryl arm of TEL can adopt three distinct conformations: extending toward the A752:U2609 base pair (E. coli and T. thermophilus), down the peptide exit tunnel in the same direction as a nascent chain (D. radiodurans), or folded back over the macrolide ring in the direction of the PTC (H. marismortui) (Figure 1b).22−25
Figure 1.
Design of peptide–ketolide (peptolide) compounds. (a) Structures of representative ketolides: telithromycin (TEL), cethromycin, and TE-802. (b) Binding of TEL to the 50S subunit of the ribosome. (Left) An overlay of TEL shows alternate positioning of the alkyl-aryl arm when bound to T. thermophilus (yellow, PDB ID 3OI3), E. coli (beige, PDB ID 3OAT), H. marismortui (green, PDB ID 1YIJ), and D. radiodurans (cyan, PDB ID 1P9X). TEL binds to the 50S exit tunnel in between the PTC, represented by the P-site and A-site tRNA, and the constriction site in the exit tunnel formed by 50S ribosomal protein L4 (blue) and L22 (red). (Right) In each of the macrolide structures, the desosamine sugar hydrogen bonds with A2058 stabilizing the macrolactone ring. The alkyl-aryl arm stacks against the A752:U2609 base pair in T. thermophilus (yellow) and E. coli (beige). In the absence of the A752:U2609, the alkyl-aryl arm hydrogen bonds to U2609 in H. marismortui (green) or extends down the exit tunnel in D. radiodurans (cyan). There is a 135° orientation between the left and right panel. (c) General structure of peptolide derived from TEL. Changes to the TEL template to yield the target peptolide probes are highlighted in red with ···XXXX··· indicating polypeptide containing any combination of amino acids of interest.
In each of the structures, the alkyl-aryl arm is engaged in different stabilizing interactions. In E. coli and T. thermophilus, the arm is stabilized by stacking against the rRNA A752:U2609 base pair. In H. marismortui, which lacks the A752:U2609 base pair, the arm is stabilized by a hydrogen bond with the O2′ of U2609 while in D. radiodurans, the arm extends down the exit tunnel and is potentially engaged in van der Waals interactions with the 23S rRNA.6 As long as these crucial interactions remain intact, it is feasible that the arm could be modified beyond the aryl group and allow new ligands to be added while preserving the ketolide binding mode. The path of travel through the ribosome by the modified TEL flexible arm could be influenced by the identity of the substituents on its aryl moiety. For the proposed peptolides, it is conceivable that the identity (and/or sequence) of the peptide attached to the modified alkyl-aryl arm could dictate the placement of the peptides within the ribosome through preferential adoption of one of the three distinct alkyl-aryl arm positions (Figure 1b).22−26
We report herein the design, synthesis, and characterization of a set of peptide–ketolide (peptolide) compounds. We demonstrate that these peptolides inhibit translation in cell-free prokaryotic and eukaryotic systems. RNA footprinting experiments suggest that the peptolide macrocycle moiety binds to the parent macrolide binding site on the E. coli ribosome with affinity that is dependent on the overall charge of the peptide tail. Lastly, we solved the X-ray crystal structure of one peptolide bound to the 70S. Our combined data show that the peptolide adopts a conformation with its peptide tail oriented back toward the PTC and the subunit interface close to 23S rRNA residue U1963 located between the A and P sites. Taken together, our results indicate these distinct peptolides could be useful probes for interrogating nascent peptide-exit tunnel interaction between the PTC to the L4/L22 constriction site. This approach could provide a general means for a precise placement of peptides into both the exit tunnel and path from the PTC to the tunnel entrance.
Results and Discussion
Peptolide Design and Synthesis
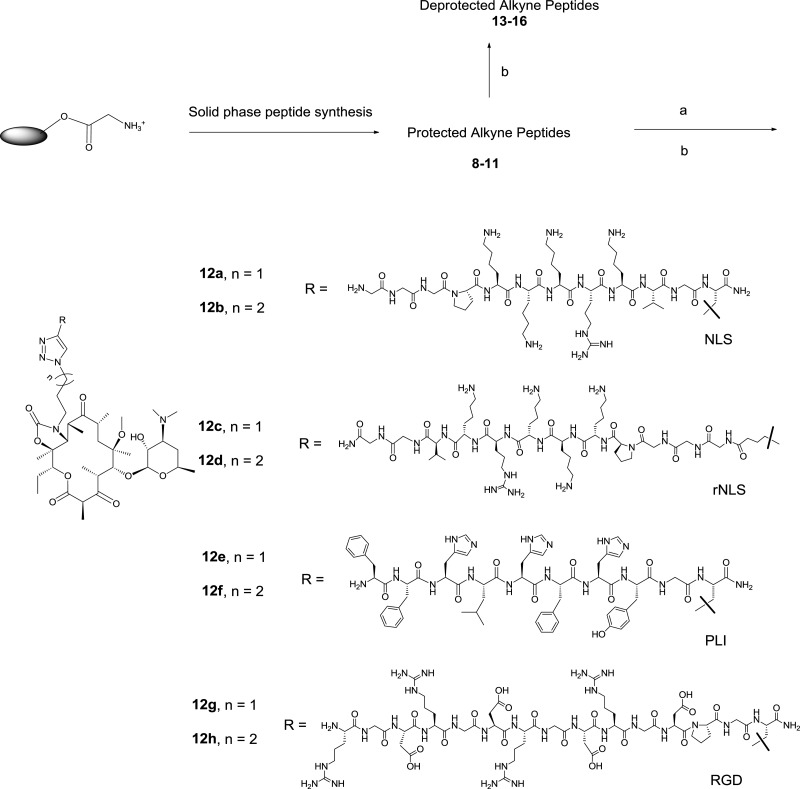
We designed a series of TEL-derived peptolides that have the phenyl and the imidazolyl groups substituted by peptides and 1,2,3-triazole ring, respectively (Figure 1c). The latter modification, achieved through the Cu(I) promoted cycloaddition reaction of crucial azido-ketolides 7a,b (Scheme 1) and appropriately protected terminal alkyne modified peptides, is essential to promote compound synthetic tractability.27,28 These peptolides differ in the (1) direction of attachment (amino or carboxy terminus), (2) number of the amino acids in the peptide moiety, and (3) composition of amino acid of the peptide moiety (Scheme 2). These variations are incorporated into the design in order to investigate the influence of both the nature of individual amino acids and the orientation of the peptide chain within the exit tunnel with respect to the affinity of the peptolides for the ribosome.
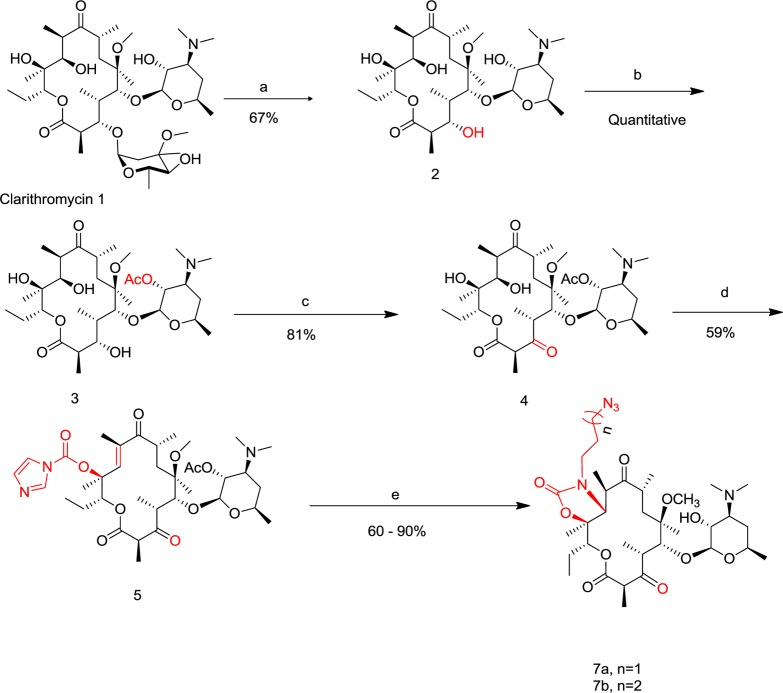
Scheme 1. Synthesis of Azido-Ketolide Intermediates 7a,b.
Reagents and conditions: (a) EtOH, 1M HCl, room temperature, 10–24 h, yield = 67%; (b) acetone, Ac2O, 40°C, 24–48 h, yield = quantitative; (c) dichloromethane (CH2Cl2), N-chlorosuccinimide (NCS), Me2S, Et3N, −20 °C, 3 h, yield = 81%; (d) THF/DMF, CDI, NaHMDS, room temperature, 24 h, yield = 59%; (e) (i) CH3CN/H2O, 6a or 6b, 50°C, 24 h; (ii) MeOH, room temperature, 24 h, yield = 60–90%.
Scheme 2. Synthesis of Peptolides 12a–h.
Reagents and conditions: (a) 7a or 7b, CuI, THF, diisopropylethylamine (DIPEA), room temperature; (b) TFA/TIPS/phenol, room temperature. The structures of deprotected alkyne peptide 13–16 are in the Supporting Information (SI), Figure S1.
The requisite azido-ketolides intermediates 7a,b were obtained from clarithromycin 1 adapting literature produres.27,29 Briefly, selective hydrolysis of the cladinose sugar of clarithromycin 1 in dilute HCl afforded descladinose compound 2 whose 2′-OH group was selectively acylated by treating it with acetic anhydride (Ac2O) in acetone at 40 °C for 24–48 h to give compound 3 in quantitative yield. Corey–Kim oxidation of the 3-OH group of 3 gave 3-keto compound 4, which was subsequently converted to 12-carbamoylimidazolideketolide 5 by treatment with excess carbonyldiimidazole (CDI) and sodium hexamethyldisilazide (NaHMDS) in a mixture of tetrahydrofuran/dimethylformamide (THF/DMF). Compound 5 was converted to the desired intermediates 7a,b through a two-step-one-pot process involving a displacement of the imidazole group followed by an intramolecular Michael addition by azido amine 6a,b to give acetylated 11,12-cyclic carbamate analogs.30 Subsequent methanolysis of the carbamate analogs afforded the desired azido-ketolide 7a,b in 60–90% yield (Scheme 1). The transformation of azido-ketolide intermediates 7a,b into the desired peptolides 12a–h involved Cu (I) catalyzed cycloaddition reaction31 between 7a,b and appropriately protected peptides 8–11 bearing terminal alkynes followed by the removal of the protecting groups by treatment of the fully protected peptolides with trifluoroacetic acid/triisopropylsilane/phenol at room temperature (Scheme 2).32
Translation Inhibition and Footprinting Studies
To characterize the effects of the peptolide probes on ribosomal function, we analyzed the ability of the peptolides to inhibit prokaryotic and eukaryotic translation. The prokaryotic assay we used is based on the whole cell extract from E. coli, while the eukaryotic assay is comprised of rabbit reticulocyte whole cell lysate (RRL). Both assays utilize a luciferase based reporter whereby protein expression is monitored by the amount of luciferase protein product.33,34 Ketolide TE-802 and clarithromycin 1 were used as positive controls. The fully deprotected, unattached peptides 13–16 were also tested to parse out the contribution of the peptolide peptide moieties on translation inhibition activity (Table 1).
Table 1. IC50 Cell Free Assays Translation Inhibition Activity of Peptolides against Prokaryotic and Eukaryotic Ribosomal Preparationsa.
| compds. | E. coli IC50 (μM) | rabbit retic. IC50 (μM) |
|---|---|---|
| 12a | 1.32 ± 0.08 | >250 |
| 12b | 0.67 ± 0.28 | >250 |
| 13 | >250b | >250 |
| 12c | 1.86 ± 0.35 | >250 |
| 12d | 1.10 ± 0.22 | >250 |
| 14 | >250b | >250 |
| 12e | 4.32 ± 0.25 | 140 |
| 12f | 2.73 ± 0.82 | 38 |
| 15 | >1000c | 205 |
| 12g | 56.00 ± 9.60 | >250 |
| 12h | 7.41 ± 1.20 | >250 |
| 16 | >250b | >250 |
| TE-802 | 0.52 ± 0.05 | >250 |
| clarithromycin 1 | 0.32 ± 0.12 | >250 |
Luciferase activity was used as a reporter of translation inhibition in both systems. IC50 values were obtained from an average of three independent experiments.
No inhibition at maximum tested concentration (250 μM).
Maximum tested concentration increased 4-fold to obtain information about the extent of selectivity over RRL.
We observed that each of the peptolide probes retained potent prokaryotic translation inhibition, although the most potent peptolide, 12b, is almost equipotent as TE-802 and 2.0-fold less active than 1, the positive ketolide and macrolide controls, respectively. A striking difference in the translation inhibition activity of these peptolides is evident when comparing the amino acid composition of their peptides. The overall positive charge of the amino acid residues of the deprotected peptides decreases as such: peptides 13 (+6), 14 (+5), 15 (+3), and 16 (zwitterionic) (Supporting Information, Figure S1). Translation inhibition is strongly dependent on the net positive charge on the peptide moieties of each peptolide with 12a–d, peptolides derived from the most positively charged peptides 13 and 14, being most active, while 12g–h, peptolides derived from zwitterion peptide 16 are the least active. This observation suggests that the positively charged peptides such as 13 and 14 have an enhanced affinity for the phosphate backbone of the rRNA, which lines the exit tunnel. Since the peptide tails alone did not cause significant translation inhibition, we believe the ketolide macrocyclic ring of the peptolide acts as an anchor whose affinity for the ribosome is in turn modulated by the extent of accommodation of the peptide moieties within the ribosome.
The effect of peptide direction could be evaluated through the analysis of the translation inhibition activity. Peptides 13 and 14 have net charges of +6 and +5, respectively, and are derived from nearly identical sequences. When conjugated to the ketolide macrocycle, the only major distinction between peptolides derived from 13 and 14 is the direction of peptide attachment to the ketolide ring, with 13 attached through the carboxy terminus and 14 through the amino terminus (Scheme 2). Interestingly, the match pair peptolides 12a/12c and 12b/12d have nearly indistinguishable translation inhibition activity (Table 1). This suggests that the direction in which the peptides travels, N to C or C to N, through the tunnel does not seem to have much of an effect on the translation inhibition of the probes or their ability to interact with the exit tunnel.
We could, however, not rule out from this assay the possibility of the peptide moieties of these peptolides adopting different conformations influenced by the flexibility of the alkyl-triazolyl moiety, akin to what has been structurally observed with TEL (Figure 1b). The consequence of a moderate restriction of the flexibility of the alkyl-triazolyl moiety becomes apparent when comparing the translation inhibition potency of peptolides derived from propyl-linked (n = 1) and butyl-linked (n = 2) azido-ketolides 7a and 7b, respectively. For 13 and 14, peptides with the most positive charge, the effect of reduction of ketolide linker length is negligible (Table 1, compare match pairs 12a/12b and/or 12c/12d). However, linker length reduction further exacerbates the compromised affinity of peptolide derived from a zwitterionic peptide, with approximately 10-fold reduction in translation inhibition activity (Table 1, compare match pairs 12g/12h).
The peptolides were evaluated in a eukaryotic translation system to assess their selectivity for prokaryotic ribosomes over eukaryotic ribosomes. As anticipated, the control TE-802 and Clarithromycin are inactive at 250 μM (Table 1). The majority of the peptolides and the analogous deprotected peptides were unable to inhibit eukaryotic translation. However, the unattached deprotected peptide 15 showed slight translation inhibition with an IC50 of 205 μM, resulting in only a ∼ 4-fold selectivity over the prokaryotic ribosome. When conjugated with the butyl-linked (n = 2) azido-ketolide 7b, the resulting peptolide 12f showed significant inhibition of eukaryotic translation with an IC50 of 38 μM (Table 1). This suggests there are subtle architectural differences between the eukaryotic and prokaryote exit tunnels which allow for more favorable interactions with peptide 15 and its analogous peptolide 12f within the context of the eukaryotic ribosome.
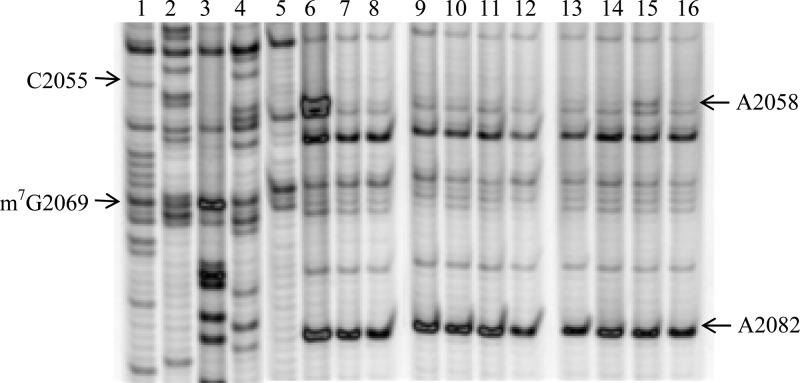
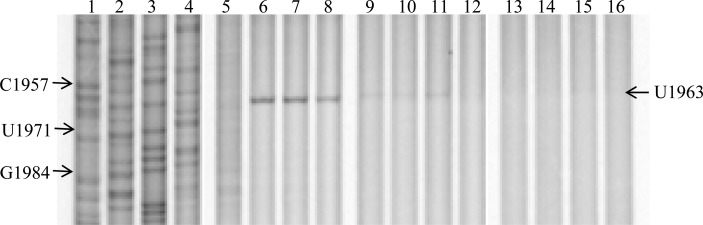
To further characterize the mode of association of the peptolide probes with the ribosome, we performed RNA structure probing analysis of the E. coli 23S rRNA using chemical footprinting of intact 70S. We employed dimethylsulfate (DMS) and N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide (CMCT) as the nucleic acid modifying agents.35,36 DMS footprinting gel in Figure 2 shows a clear peptolide footprint at the residue A2058 of the E. coli 23S rRNA. In lanes 5 and 6 are unmodified and DMS-modified rRNAs, respectively, in the absence of probes. Any differences between these lanes highlight residues that are available for DMS modification. Similarly, any differences between lane 6 and lanes 7–16, lanes containing rRNAs that are DMS-modified after drug or probe binding, will highlight any sites that have changes in DMS availability. The reduction in intensity at nucleotide A2058 in the presence of each peptolide is clear evidence of a drug binding footprint. This is an expected site of drug interaction as A2058 is known to interact with the macrolide ring upon binding to the well-defined macrolide binding pocket of the ribosome.22−25 Thus, this footprinting result verifies that the binding target of the macrocycle moieties of the peptolide probes is indeed the same as the parent macrolide drug.
Figure 2.
DMS footprinting of 23S rRNA. Dideoxy sequencing lanes G, C, A, and T (lanes 1–4) followed by unmodified rRNA (lane 5), DMS modified 23S rRNA (lane 6), DMS modified 23S rRNA in the presence of clarithromycin and 7b intermediate (lanes 7–8, respectively), 23S rRNA DMS modified in the presence of 12a–h (lanes 9–16, respectively). A representative complete gel is shown in SI Figure S2.
For nearly all peptolides, the A2058 band is barely visible except for 12g, the peptolide derived from the propyl-linked (n = 1) azido-ketolide 7a and the zwitterionic peptide 16, which has a relatively more pronounced A2058 band (lane 15). The reduction in peptolide-induced protection from DMS modification at nucleotide A2058 by peptolide 12g, in comparison to the other peptolides, is indicative of a decreased binding affinity of peptolide 12g in the macrolide binding pocket. This decreased binding affinity correlates well with our E. coli translation inhibition study which showed peptolide 12g has the weakest translation inhibition activity (IC50 value of 60 μM, Table 1). DMS probing of the other key residues within the large subunit did not reveal strong peptolide-induced protection (data not shown). Importantly, A752 is protected by TEL presumably due to the placement of the alkyl-aryl arm into this section of the tunnel.37 However, A752 is not protected by any of the peptolides (SI Figure S3) indicating the modified alkyl-aryl arm of the peptolides is not oriented toward the A752:U2609 base pair.
CMCT footprinting reveals information about the binding pocket of the peptolide peptide moieties within the ribosome (Figure 3). In lanes 6–8, a band corresponding to U1963 is clearly visible. This residue is thus available to CMCT modification in the absence and presence of both clarithromycin and the azido-ketolide intermediate 7b. In marked contrast, U1963 is strongly protected from modification by CMCT in the presence of all peptolide probes, evidenced by the near absence of this band in lanes 9–16 (Figure 3, SI Figure S4).
Figure 3.
CMCT footprinting of 23S rRNA. Dideoxy sequencing lanes G, C, A, and T (lanes 1–4) followed by unmodified rRNA (lane 5), CMCT modified 23S rRNA (lane 6), CMCT modified 23S rRNA in the presence of clarithromycin and 7b intermediate (lanes 7 and 8, respectively), 23S rRNA CMCT modified in the presence of 12a–h (lanes 9–16, respectively).
U1963 is part of Helix 71 (H71) located within domain IV of the 23S which forms part of the front rim of the peptidyl transferase active site cleft.4 The CMCT protection of U1963 suggests these peptolides all adopt a similar binding orientation such that their peptide moieties mimic the path a nascent peptide would traverse extending from the PTC to the proximity of the entrance to the exit tunnel. The lack of protection by 7b shows the azido-alkyl moiety is not sufficiently long to block access to U1963. Ribosomal rRNA U1963 is a crucial residue within the overlapping binding sites of RRF, EF-G, and the P-site tRNA38 suggesting a plausible mechanism of translation inhibition by inhibiting the binding of these critical translation components. Both the N- and C-linked peptolides 12a–d cause indistinguishable protection of U1963, indicating the tolerance of the exit tunnel for either the biological N → C or the abiological C → N orientation of nascent peptide. This raised an interesting prospect that the choice of N → C as the direction of travel of the nascent peptides through the exit tunnel may simply be due to the restriction imposed by the peptidyl transfer chemistry.
Structural Studies of Peptolide 12c Bound to the 70S Ribosome
Previous X-ray crystallographic studies of similar ketolides, TEL and CEM101, show these macrolides bind to the macrolide binding pocket in the upper portion of the ribosomal exit tunnel, adjacent to the L22 and L4 constriction site.22−25 In order to determine if these peptolides bind in the same macrolide-binding site with a similar orientation for the modified alkyl-aryl tail, we cocrystallized the peptolide 12c, derived from the propyl-linked (n = 1) azido-ketolide 7a and the positively charged NLS peptide attached at the N-terminal (reverse NLS peptide 14), with T. thermophilus 70S ribosomes programmed with mRNA, P-site tRNAfMet, and A-site tRNAPhe. The peptolide (1 μM final concentration) was incubated with programmed ribosome complexes just prior to crystallization. X-ray diffraction data was collected, processed, and the structure was solved to 3.6 Å using molecular replacement with a 70S ribosome structure where the tRNA and mRNA ligands removed (PDB ID codes 2WDG, 2WDH, 2WDI, and 2WDJ).39 Unbiased Fo–Fc difference electron density maps show clear and connected density of the ketolide macrocyclic ring and eight of the 12 residues in the peptide tail except for Lys5 (SI Figure S5).
The 70S-peptolide 12c structure shows the ketolide macrocyclic ring adopts a similar orientation in the macrolide binding site on the 50S subunit as observed in previous structures.22−25 Specifically, the desosamine sugar at position 5 of the ketolide macrocyclic ring hydrogen bonds with the base of 23S rRNA A2058 and the surface of this ring forms hydrophobic packing interactions with the bases of U2611, A2058, and A2059. These results indicate the addition of the peptide tail does not alter the ketolide macrocyclic ring position as observed in other crystal structures.
Similar to other ketolides, the peptolides described in this study also contain a flexible alkyl-aryl arm. Previous crystal structures have shown three distinct conformations of the alkyl-aryl arm of TEL. When bound to E. coli or T. thermophilus 70S, the alkyl-aryl arm of TEL packs against the U2609:A752 base pair.22,25 In H. marismortui, the alkyl-aryl arm folds back over the top of the macrolactone ring24 while in D. radiodurans, the alkyl-aryl arm extends further down the peptide exit toward the L4/L22 constriction site.23 This altered conformation of the alkyl-aryl arm observed in H. marismortui and D. radiodurans has been hypothesized to be a consequence of the absence of a U2609:A752 base pair.25
Our 70S-peptolide 12c structure indicates the alkyl-aryl arm of the peptolide extends further down the peptide exit tunnel in a similar conformation as observed with D. radiodurans (Figure 1b, Figure 4, and SI Figure S5). However, despite the positioning of the alkyl-aryl arm toward the L4/L22 constriction site, the peptide portion of the peptolide turns and folds back over the top of the ketolide macrocyclic ring oriented toward the PTC with its position stabilized through the formation of two new hydrogen bonds between peptolide residues Lys6 and Lys7 with 23S rRNA C2442 and the side chain of His69 of ribosomal protein L4, respectively (Figure 4). Since all peptolides contain a positively charged residue at an analogous position, it is possible they all interact with C2442 in a similar manner. The electron density of the peptolide tail is disordered beyond the eighth amino acid of the attached peptide chain and therefore the last four residues were not built. While the well-ordered portion of the peptide tail does not extend past the exit tunnel L4/L22 constriction site, the entire exit site tunnel is blocked by the peptide tail folding back on top of the ketolide macrocyclic ring (Figure 4). Although the peptide tail is too short to directly interact with U1963 that is protected in the CMCT footprinting experiments (Figure 3), the peptide tail may cause a rearrangement of the adjacent 23S rRNA residues in the PTC thereby indirectly resulting in U1962 protection.
Figure 4.
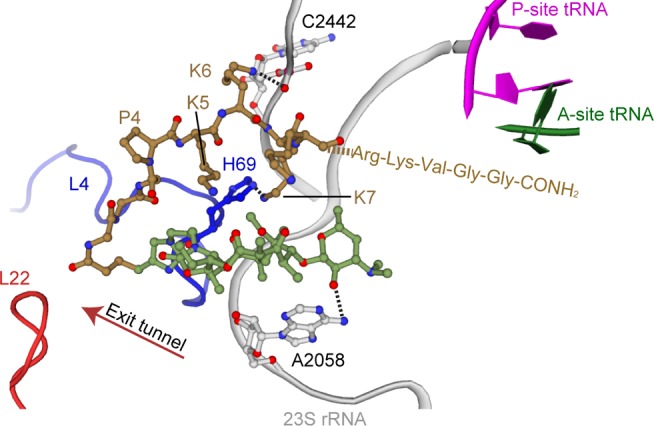
Binding site of peptolide 12c in the 50S subunit of the ribosome. Similar to TEL, peptolide 12c binds in the 50S exit tunnel near the ribosomal protein L4 (blue) constriction site. The peptide tail folds back over the macrolactone ring, extending toward the P-site and A-site tRNA in the PTC. The macrolactone ring (green) is stabilized through a hydrogen bond between A2058 and the desosamine sugar. The peptide tail (gold) is stabilized through hydrogen bonds with 23S rRNA C2442 and His69 of ribosomal protein L4 (blue). The last four amino acids of the peptide tail are not seen in the structure, and instead, their three-letter codes represent their proposed position towards the PTC.
Conclusion
Ribosome structures have given us a molecular understanding of the roles that specific ribosomal components play during translation.1−6,40,41 However, the nascent peptide exit tunnel is a ribosome component that has yet to be fully characterized. The rRNA-lined tunnel serves as the route of travel for nascent peptides from the PTC toward the exterior surface of the ribosome.7,8 Initially, the exit tunnel was proposed to have little impact on specific nascent chain sequences and exist solely to facilitate an unhindered passage of peptides to the exterior of the ribosome. A growing body of research has shown specific amino acid sequences interact with the components of the exit tunnel, concomitantly influencing many aspects of translation such as the translation rate, ribosome stalling and even changes in mRNA frame maintanence.9,13,42−44 Despite these new insights, the molecular mechanisms used by the ribosome to identify and interact with specific nascent peptide sequences are not well understood.
Toward addressing these deficiencies, we designed and validated a series of peptide–ketolide (peptolide) conjugates to probe the interactions between the nascent peptide chain and the ribosome exit tunnel. Translation inhibition and RNA structure probing assays show that all the peptolide probes interact with the macrolide binding pocket in the same manner as the parent macrolide while the peptide tails of the peptolides are oriented toward the PTC. Furthermore, we find the exit tunnel has a relaxed preference for the directionality (N → C or C → N orientation) of the nascent peptides. This observation suggests that the directionality of proteins has not evolved after peptide bond formation for optimization to transit out of the ribosome. In contrast, the overall charge of the peptolide peptide moieties has the strongest influence on translation inhibition.
The location of the peptolide 12c in the exit tunnel in the context of a programmed 70S was determined by X-ray crystallography to 3.6 Å. Our structural data places the ketolide macrocyclic ring in the same place as previous structures.22−25 While the position of the alkyl-aryl arm extends further down the exit tunnel similar to position observed in the D. radiodurans structure, the peptide tail, however, folds back over the ketolide macrocyclic ring, extending in the opposite direction back toward the PTC.
In summary, these data suggest that the peptolides disclosed therein are viable probes for interrogating nascent peptide-exit tunnel interaction from the PTC to the exit tunnel entrance. Ongoing effort is focused on designing a new generation of peptolides, which would be able to insert their peptide tails into the exit tunnel from the PTC entrance to obtain a complete set of probes for the full length of the tunnel.
Methods
In Vitro Cell Extract Inhibition Assays
Cell free translation assays—E. coli (E. coli S30 Extract System for Circular DNA, Promega) and rabbit reticulocyte whole cell extract (Rabbit Reticulocyte Lysate System, Nuclease Treated, Promega)—were performed as recommended by manufacturer.33,34 Briefly, varying concentrations of the compounds of interest were allowed to incubate in a solution of rabbit reticulocyte or E. coli extract and all amino acids for 20 min at room temperature. Following brief centrifugation, 0.95 μL luciferase control template (Promega) was added to each tube. After gentle mixing and spin down, tubes were incubated at 37 °C for 60 min (30 °C for 90 min for eukaryotic samples). Translation was terminated by inactivating on ice for 5 min. Upon returning to ambient temperature, 5 μL per tube (2.5 μL for eukaryotic samples) was delivered to a LUMITRAC 200 96-well plate. Luminescence was immediately read following the addition of the luciferin solution using a Molecular Devices SPECTRAmax GEMINI. IC50 values were fit by logit regression. All compounds were analyzed in triplicate and standardized against an internal vehicle control.
Chemical Footprinting of E. coli 23S rRNA
Chemical footprinting of the E. coli 23S rRNA was performed with dimethyl sulfate (DMS) and 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT) according to the following protocol:45
Peptolide Binding
All peptolides and the azido-macrolide precursor were incubated at a final concentration of 150 μM with 100 pmol E. coli 70S ribosomes (New England Biolabs), whereas clarithromycin was incubated at a concentration of 50 μM. Binding was performed in binding buffer (10 mM HEPES, 10 mM MgCl2, 60 mM NH4Cl) at 37 °C for 30 min.46
DMS Chemical Modification
DMS chemical modification was performed on intact 70S E. coli ribosomes in the absence and presence of bound drug. Approximately 50 μg of intact ribosome, or ribosome-peptolide complex, in 25 μM DMS buffer (80 mM K-HEPES, 10 mM MgCl2, 100 mM NH4Cl) was aliquoted, and to this was added 1 μL DMS Stock (880 mM DMS in abs. EtOH), and the mixture was then incubated for 10 min at 37 °C. The reaction was terminated by the addition of 12.5 μL of DMS stop buffer (1 M Tris-HCl, 0.1 M EDTA, 1 M β-mercaptoethanol, pH 7.5). Ethanol precipitation was followed by RNA extraction. The RNA pellet was resuspended in 400 μL extraction buffer (0.3 M NaOAc (pH 6.5), 0.5% SDS, 5 mM EDTA (pH 8.0)) at room temperature. This was subsequently extracted three times via addition of 400 μL of acid phenol chloroform (pH 4.5, Invitrogen). The final RNA fraction was ethanol precipitated and the pellet was resuspended in H2O (final concentration: 0.4 μM). Aliquots were stored at −80 °C.
CMCT Modification
CMCT chemical modification was performed on intact 70S E. coli ribosomes in the absence and presence of bound drug. Approximately 50 μg of intact ribosome, or ribosome-peptolide complex, in 25 μM CMCT buffer (50 mM potassium borate pH 8.0, 10 mM MgCl2, 100 mM NH4Cl) was aliquoted, and to this was added 12.5 μL CMCT Stock (100 mM CMCT in CMCT buffer), and the mixture was then incubated for 10 min at 37 °C. The reaction was terminated by the addition of 2.5 volumes of −20 °C 95% EtOH. Subsequent ethanol precipitation was followed by RNA extraction as described above.
Reverse Transcription
DNA primers were purchased from Keck Biotechnology (New Haven, CT). An array of primers was initially purchased with the intent of covering as much of the 23S rRNA strand as possible. After footprinting analysis of all primers, the primer yielding the most useful data, named primer 2180, has the following sequence: 5′-GGGTGGTATTTCAAGGTCGG-3′. Primer 2180 is named as such because the 5′ G of the primer anneals to position 2180 (E. coli numbering) of the 23S rRNA. Primer concentration was determined by UV–vis spectroscopy in 1 cm path length quartz cuvettes using molar extinction coefficients determined by OligoAnalyzer (IDT).
A working primer stock (2.0 μM) in H2O was made fresh for each reaction. A hybridization buffer (225 mM K-Hepes (pH 7.0), 450 mM KCl) was used to bring primer concentration to 1.0 μM. This primer mix (2 μL) was added to tubes containing RNA (0.8 μmol). Annealing was proceeded by placing tubes in a water bath at 90 °C for 1 min, and the tubes were allowed to cool to room temperature. While cooling, a master mix was prepared using 20 μL 5× First Strand Buffer (supplied with RT enzyme, Invitrogen), 150 μCi dATPα35S (PerkinElmer), 6.6 μL dNTP mix (110 μL 1 mM d(T, G, C)TP, 6 μL 1 mM dATP, 664 μL H2O), and 1 μL each of RNasin Plus (Promega) and 100 mM dithiothreitol (DTT).
Upon completion of annealing, samples were briefly centrifuged at room temperature. Each tube received 2 μL master mix, and the sequencing tubes received the appropriate ddNTP (1 μL, 9 μM). Added last to each tube was 200U SuperScript II reverse transcripase (Invitrogen). Tubes were gently mixed and centrifuged before being placed in a heating block at 55 °C for 30 min. Chase was initiated by addition of 1 μL dNTP (100 mM) to each tube and 1 μL appropriate ddNTP (402 mM) to sequencing tubes. Tubes were placed back in the heating block at 55 °C for 15 min. The reaction was terminated by the addition of precipitation buffer (75 μL, 95% ethanol, 0.3 M NaOAc pH 6.5, 4 °C) and centrifuged for 90 min (13 200 rpm, 4 °C). The supernatant was removed and the pellets were dried and resuspended in 10 μL loading buffer (95% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue, pH 11).
Analysis of Chemical Footprinting Experiments
Samples were analyzed by running on a 5% polyacrylamide gel in a Sequi-Gen GT system (Bio Rad). The gel and 1× TBE buffer (100 mM Tris, 90 mM Boric Acid, and 1 mM EDTA) were brought to temperature by prerunning at 55 W to 50 °C. Lanes were thoroughly flushed with buffer and loaded with 1 μL of sample each. Migration occurred from 45 to 90 min at 55W and 50 °C. Gels were transferred to Whatman paper and were fixed in 20% EtOH for 20 min. They were then dried under vacuum for 90 min at 80 °C. Once dry, gels were exposed onto a phosphor screen (GE Healthcare) overnight. Screens were scanned on a Typhoon Trio+ (GE Healthcare) and digital images were analyzed with Multi Gauge (FujiFilm).
The footprinting of E. coli 23S rRNA with primer 2180 and DMS modification in the presence of all peptolides can be found in Figure 2. Footprinting of CMCT modified 23S rRNA with primer 2180 can be seen in Figure 3.
X-ray Crystallographic Studies of the Thermus thermophilus 70S Ribosome Bound to the Peptolide 12c
T. thermophilus ribosomes were purified, crystallized, and cryoprotected and the X-ray crystal structure solved as previously described.3Escherichia coli tRNAfMet and tRNAPhe were purchased from Chemical Block. The mRNA oligonucleotide was chemically synthesized by Integrated DNA Technology with a sequence of 5′-GGCAAGGAGGUAAAAAUGUUCAAAA-3′, where the underlined AUG and UUC represent the P- and A-site codons, respectively. Briefly, the programmed 70S complexes were formed using previously established conditions with the additional step being the incubation at 37 °C for 30 min of peptolide 12c prior to crystallization. X-ray diffraction data were collected from four crystals for the 70S-12c complex at the Northeastern Collaborative Access Team (NE-CAT) beamline at the Advanced Proton Source, Argonne National Laboratory. Each diffraction data set was integrated and scaled using the XDS software package.47 The structures were solved by molecular replacement with the PHENIX software suite48 using an initial search model composed of the Tth 70S ribosome (PDB ID codes 2WDG, 2WDH, 2WDI, and 2WDJ) with all ligands and ions removed. An initial round of coordinate refinement was performed using each of the ribosomal subunits as a single rigid group. Additional rounds of rigid and Translation, Liberation, Screw-movement (TLS) refinements were performed using defined rigid and TLS groups comprised of the head, body, platform, and 3′ minor domains for the 30S subunit and the 5S rRNA, L1 arm, A-site finger, central protuberance, and protein L9 domains for the 50S subunit. Modeling of the 12C peptolide, tRNA, mRNA, and placement of the Mg2+ ions was performed using Coot.49 Iterative rounds of model building and positional and atomic displacement parameter (ADP) refinements were performed in PHENIX to yield a final model with the statistics reported in Table 2(51,52). Figures were generated using PyMol.50
Table 2. Summary of Crystallographic Data and Refinementa.
| data collection | |
|---|---|
| space group | P212121 |
| cell dimensions | |
| a, b, c (Å) | 209.2, 443.5, 618.6 |
| α, β, γ (deg) | 90.00, 90.00, 90.00 |
| resolution (Å) | 50.0–3.6 (3.7–3.6) |
| Rmerge (%) | 27.7 (116.9) |
| Rp.i.mb (%) | 10.2 (47.2) |
| I/σI | 7.6 (1.8) |
| completeness (%) | 98.0 (95.8) |
| redundancy | 7.4 (6.1) |
| data collection | |
|---|---|
| space group | P212121 |
| cell dimensions | |
| a, b, c (Å) | 209.2, 443.5, 618.6 |
| α, β, γ (deg) | 90.00, 90.00, 90.00 |
| resolution (Å) | 50.0–3.6 (3.7–3.6) |
| Rmerge (%) | 27.7 (116.9) |
| Rp.i.mb (%) | 10.2 (47.2) |
Values in parentheses are for the highest-resolution shell.
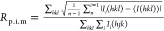
Acknowledgments
We are grateful to A. Mankin for providing us with a list of primer sequences. We thank C. Hsiao for assistance with initial modeling of the peptolides in the PTC of the ribosome at the beginning of this project and S. J. Miles and R. Erdman for technical assistance in the purification of ribosomes.
Glossary
Abbreviations
- CDI
carbonyldiimidazole
- cDNA
complementary DNA
- CMCT
1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate
- DMS
dimethyl sulfate
- FRET
fluorescence resonance energy transfer
- NLS
Nuclear Localization Sequence
- PTC
peptidyl transferase center
- RRL
rabbit reticulocyte lysate
- rNLS
reverse NLS
- rRNA
rRNA
- PLI
plicatamide
- RGD
tetramer-RGD repeat sequence
- RT
reverse transcripase
- TEL
telithromycin
Supporting Information Available
1H spectra information, full gel images, and detailed description of compound synthesis. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4P6F).
Author Present Address
⊥ GlaxoSmithKline, 553 Old Corvallis Road, Hamilton, MT 59840, U.S.A.
Author Contributions
§ A.Z.W., D.B.B., and J.C.C. contributed equally to the manuscript.
This work was financially supported in part by NASA Astrobiology Institute [NNA09DA78A] (A.K.O.) and National Institutes of Health (NIH) Award R01GM093278 (to C.M.D.). J.C.C. and A.Z.W. are thankful recipients of the GAANN predoctoral fellowships from the Georgia Tech Center for Drug Design, Development, and Delivery and from the School of Chemistry and Biochemistry, respectively. CEF support was provided by the Department of Defense through the National Defense Science and Engineering Graduate Fellowship Program (NDSEG) and NIH Training Grant T32 GM8367. C.M.D. is a Pew Scholar in the Biomedical Sciences. This work is based on research conducted at the Advanced Photon Source on the NE-CAT beamlines, which is supported by National Center for Research Resources (NIH) Award RR-15301, and at the SER-CAT beamline. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract DE-AC02-06CH11357.
The authors declare no competing financial interest.
Dedication
This manuscript is dedicated to the memory of Derek B. Benicewicz.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Wimberly B.-T.; Brodersen D.-E.; Clemons W.-M. Jr.; Morgan-Warren R.-J.; Carter A.-P.; Vonrhein C.; Hartsch T.; Ramakrishnan V. (2000) Structure of the 30S ribosomal subunit. Nature 407, 327–329. [DOI] [PubMed] [Google Scholar]
- Schuwirth B.-S.; Borovinskaya M.-A.; Hau C.-W.; Zhang W.; Vila-Sanjurjo A.; Holton J.-M.; Cate J.-H. (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834. [DOI] [PubMed] [Google Scholar]
- Selmer M.; Dunham C.-M.; Murphy F.-V. th; Weixlbaumer A.; Petry S.; Kelley A.-C.; Weir J.-R.; Ramakrishnan V. (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–42. [DOI] [PubMed] [Google Scholar]
- Ban N.; Nissen P.; Hansen J.; Moore P.-B.; Steitz T.-A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Nissen P.; Hansen J.; Ban N.; Moore P.-B.; Steitz T.-A. (2000) The structural basis of ribosome activity in peptide bond formation. Science 289, 920–930. [DOI] [PubMed] [Google Scholar]
- Schluenzen F.; Tocilj A.; Zarivach R.; Harms J.; Gluehmann M.; Janell D.; Bashan A.; Bartels H.; Agmon I.; Franceschi F.; Yonath A. (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102, 615–23. [DOI] [PubMed] [Google Scholar]
- Voss N.-R.; Gerstein M.; Steitz T.-A.; Moore P.-B. (2006) The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 360, 893–906. [DOI] [PubMed] [Google Scholar]
- Jenni S.; Ban N. (2003) The chemistry of protein synthesis and voyage through the ribosomal tunnel. Curr. Opin. Struct. Biol. 13, 212–219. [DOI] [PubMed] [Google Scholar]
- Mankin A. (2006) Nascent Peptide in the ‘birth canal’ of the ribosome. TRENDS Biochem. Sci. 31, 11–13. [DOI] [PubMed] [Google Scholar]
- Cruz-Vera L.-R.; Sachs M.-S.; Sguires C.-L.; Yanofsky C. (2011) Nascent polypeptide sequences that influence ribosome function. Curr. Opin. Micobiol. 14, 160–166. [DOI] [PubMed] [Google Scholar]
- Gumbart J.; Schreiner G.-J.; Wilson D.-N.; Beckmann R.; Schulten K. (2012) Mechanism of SecM-mediated stalling in the ribosome. Biophys. J. 103, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N.; Thum C.; Mankin A.-S. (2008) Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell 2, 190–202. [DOI] [PubMed] [Google Scholar]
- Gupta P.; Sothiselvam S.; Vazquez-Laslop N.; Mankin A.-S. (2013) Deregulation of translation due to post-transcriptional modification of rRNA explains why erm genes are inducible. Nat. Commun. 4, 1984. [DOI] [PubMed] [Google Scholar]
- Seidelt B.; Innis C.-A.; Wilson D.-N.; Gartmann M.; Armache J.-P.; Villa L.; Trabuco L.-G.; Becker T.; Mielke T.; Schulten K.; Steitz T.-A.; Beckmann R. (2009) Structural insight into nascent polypeptide chain–mediated translational stalling. Science 326, 1412–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhead C.-A.; Johnson A.-E.; Bernstein H.-D. (2006) Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell 22, 587–598. [DOI] [PubMed] [Google Scholar]
- Yap M.-N.; Bernstein H.-D. (2009) The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell 34, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenz S.; Ramu H.; Gupta P.; Berninghausen O.; Beckmann R.; Vázquez-Laslop N.; Mankin A.-S.; Wilson D.-N. (2014) Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat. Commun. 5, 3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu H.; Vázquez-Laslop N.; Klepacki D.; Dai Q.; Piccirilli J.; Micura R.; Mankin A.-S. (2011) Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol. Cell 41, 321–330. [DOI] [PubMed] [Google Scholar]
- Starosta A.-L.; Karpenko V.-V.; Shishkina A.-V.; Mikolajka A.; Sumbatyan N.-V.; Schluenzen F.; Korshunova G.-A.; Bogdanov A.-A.; Wilson D.-N. (2010) Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem. Biol. 17, 504–514. [DOI] [PubMed] [Google Scholar]
- Shishkina A.; Makarov G.; Tereshchenkov A.; Korshunova G.; Sumbatyan N.; Golovin A.; Svetlov M.; Bogdanov A. (2013) Conjugates of amino acids and peptides with 5-O-mycaminosyltylonolide and their interaction with the ribosomal exit tunnel. Bioconjugate Chem. 24, 1861–1869 10.1021/bc400236n. [DOI] [PubMed] [Google Scholar]
- Kannan K.; Mankin A.-S. (2011) Macrolide antibiotics in the ribosome exit tunnel: Species-specific binding and action. Ann. N.Y. Acad. Sci. 1241, 33–47. [DOI] [PubMed] [Google Scholar]
- Bulkley D.; Innis C.-A.; Blaha G.; Steitz T.-A. (2010) Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 107, 17158–17163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisio R.; Harms J.; Schluenzen F.; Zarivach R.; Hansen H.-A.; Fucini P.; Yonath A. (2003) Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 185, 4276–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu D.; Blaha G.; Moore P.-B.; Steitz T.-A. (2005) Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 121, 257–270. [DOI] [PubMed] [Google Scholar]
- Dunkle J.-A.; Xiong L.; Mankin A.-S.; Cate J.-H. (2010) Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 107, 17152–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L.-H.; Mauvais P.; Douthwaite S. (1999) The macrolide–ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31, 623–631. [DOI] [PubMed] [Google Scholar]
- Mwakwari S.-C.; Guerrant W.; Patil V.; Khan S.-I.; Tekwani B.-L.; Gurard-Levin Z.-A.; Mrksich M.; Oyelere A.-K. (2010) Nonpeptide macrocyclic histone deacetylase inhibitors derived from tricyclic ketolide skeleton. J. Med. Chem. 53, 6100–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyelere A.-K.; Chen P.-C.; Guerrant W.; Mwakwari S.-C.; Hood R.; Zhang Y.; Fan Y. (2009) Non-peptide macrocyclic histone deacetylase inhibitors. J. Med. Chem. 52, 456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashimura M.; Asaka T.; Misawa Y.; Matsumoto K.; Morimoto S. (2001) Synthesis and antibacterial activity of the tricyclic ketolides TE-802 and its analogs. J. Antibiot. 54(8), 664–678. [DOI] [PubMed] [Google Scholar]
- Agouridas C.; Denis A.; Auger J.-M.; Benedetti Y.; Benefoy A.; Bretin F.; Chantot J.-F.; Dussarat A.; Fromentin C.; D’Ambrières S.-G.; Lachaud S.; Laurin P.; Le Martret O.; Loyau V.; Tessot N. (1998) Synthesis and antibacterial activity of ketolides (6-O-methyl-3-oxoerythromycin derivatives): A new class of antibacterials highly potent against macrolide-resistant and -susceptible respiratory pathogens. J. Med. Chem. 41, 4080–4100. [DOI] [PubMed] [Google Scholar]
- a1 Numerous examples of Cu(I) catalyzed Huigsen cycloaddition reaction have appeared in the literature (a comprehensive list is available at www.scripps.edu/chem/sharpless/click.html (accessed April 28, 2014)). Cited here are three examples:; a Rostovtsev V.-V.; Green L.-G.; Fokin V.-V.; Sharpless K.-B. (2002) A stepwise huigsen cycloaddition process: Copper (I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem., Int. Ed. 41, 2596–2599. [DOI] [PubMed] [Google Scholar]; b Tornoe C.-W.; Christensen C.; Meldal M. (2002) Peptidotriazoles on solid phase: [1,2,3]- Triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 67, 3057–3064. [DOI] [PubMed] [Google Scholar]; c Kolb H.-C.; Finn M.-G.; Sharpless K.-B. (2001) Click Chemistry: Diverse chemical function from a few good reactions. Angw Chem., Int. Ed. 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- Pearson D.-A.; Blanchette M.; Baker M.-L.; Guindon C.-A. (1989) Trialkylsilanes as scavengers for the trifluoroacetic acid deblocking of protecting groups in peptide synthesis. Tetrahedron Lett. 30, 2739–2742. [Google Scholar]
- Pratt S.-D.; David C.-A.; Black-Schaefer C.; Dandliker P.-J.; Xuei X.; Warrior U.; Burns D.-J.; Zhong P.; Cao Z.; Saiki A.-Y.; Lerner C.-G.; Chovan L.-E.; Soni N.-B.; Nilius A.-M.; Wagenaar F.-L.; Merta P.-J.; Traphagen L.-M.; Beutel B.-A. (2004) A strategy for discovery of novel broad-spectrum antibacterials using a high-throughput Streptococcus pneumoniae transcription/translation screen. J. Biomol Screen. 9, 3–11. [DOI] [PubMed] [Google Scholar]
- Thorne C.-A.; Lafleur B.; Lewis M.; Hanson A.-J.; Jernigan K.-K.; Weaver D.-C.; Huppert K.-A.; Chen T.-W.; Wichaidit C.; Cselenyi C.-S.; Tahinci E.; Meyers K.-C.; Waskow E.; Orton D.; Salic A.; Lee L.-A.; Robbins D.-J.; Huppert S.-S.; Lee E. (2011) A biochemical screen for identification of small-molecule regulators of the Wnt pathway using Xenopus egg extracts. J. Biomol. Scr. 16, 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerina P.; Mohr S.; Russell R. (2010) DMS footprinting of structured RNAs and RNA–protein complexes. Nat. Protoc. 2, 2608–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L.-C.; Lewin A.-S. (1995) Protein-induced folding of a group I intron in cytochrome b pre-mRNA. J. Biol. Chem. 270, 21552–62. [DOI] [PubMed] [Google Scholar]
- Xiong L.; Korkhin Y.; Mankin A.-S. (2005) Binding site of the bridged macrolides in the Escherichia coli ribosome. Antimicrob. Agents Chemother. 49, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.-K.; Sharma M.-R.; Kiel M.-C.; Hirokawa G.; Booth T.-M.; Spahn C.-M.-T.; Grassucci R.-A.; Kaji A.; Frank J. (2004) Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc. Natl. Acad. Sci. U.S.A. 101, 8900–8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees R.-M.; Weixlbaumer A.; Loakes D.; Kelley A.-C.; Ramakrishnan V. (2009) Insights into substrate stabilization from snapshots of the peptidyl transferase center for the intact 70S ribosome. NSMB 16, 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A.; Jenner L.; Yusupova G.; Yusupov M. (2010) Crystal structure of the eukaryotic ribosome. Science. 330, 1203–1209. [DOI] [PubMed] [Google Scholar]
- http://apollo.chemistry.gatech.edu/RiboVision/ (accessed Dec. 2, 2013).
- Voss C.; Eyol E.; Frank M.; von der Lieth C.-W.; Berger M.-R. (2006) Identification and characterization of riproximin, a new type II ribosome-inactivating protein with antineoplastic activity from Ximenia americana. FASEB J. 20, 1194–1196. [DOI] [PubMed] [Google Scholar]
- Wilson D.-N.; Beckmann R. (2011) The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 21, 274–282. [DOI] [PubMed] [Google Scholar]
- Ito K.; Chiba S.; Pogliano K. (2010) Divergent stalling sequences sense and control cellular physiology. Biochem. Biophys. Res. Commun. 393, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryman C., and Noller H. F. (1998) Footprinting and modification-interference analysis of binding sites on RNA. In RNA:Protein Interactions, A Practical Approach (Smith C. W. J., Ed.) pp 237–253, Oxford University Press, Oxford, U.K. [Google Scholar]
- Amit M.; Berisio R.; Baram D.; Harms J.; Bashan A.; Yonath A. (2005) A crevice adjoining the ribosome tunnel: Hints for cotranslational folding. FEBS Lett. 579, 3207–13. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010) XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.-V.; Grosse-Kunstleve R.-W.; Echols N.; Headd J.-J.; Mariarty N.-W.; Mustyakimov M.; Terwilliger T.-C.; Urzhumtsev A.; Zwart P.-H.; Adams P.-D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst. D 68, 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W.-G.; Cowtan K. (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC: New York.
- Stinivasan R.; Tan L.-P.; Wu H.; Yang P.-Y.; Kalesh K.; Yao S. (2009) High-throughput synthesis of azide libraries suitable for direct chemistry and in situ screening. Org. Biomol. Chem. 7, 1821–1828. [DOI] [PubMed] [Google Scholar]
- Davidovich C.; Bashan A.; Yonath A. (2008) Structural basis for cross-resistance to ribosomal PTC antibiotics. Proc. Natl. Acad. Sci. U.S.A. 105, 20665–20670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.