Abstract
The chemical features that impact small-molecule permeability across bacterial membranes are poorly understood, and the resulting lack of tools to predict permeability presents a major obstacle to the discovery and development of novel antibiotics. Antibacterials are known to have vastly different structural and physicochemical properties compared to nonantiinfective drugs, as illustrated herein by principal component analysis (PCA). To understand how these properties influence bacterial permeability, we have developed a systematic approach to evaluate the penetration of diverse compounds into bacteria with distinct cellular envelopes. Intracellular compound accumulation is quantitated using LC-MS/MS, then PCA and Pearson pairwise correlations are used to identify structural and physicochemical parameters that correlate with accumulation. An initial study using 10 sulfonyladenosines in Escherichia coli, Bacillus subtilis, and Mycobacterium smegmatis has identified nonobvious correlations between chemical structure and permeability that differ among the various bacteria. Effects of cotreatment with efflux pump inhibitors were also investigated. This sets the stage for use of this platform in larger prospective analyses of diverse chemotypes to identify global relationships between chemical structure and bacterial permeability that would enable the development of predictive tools to accelerate antibiotic drug discovery.
Understanding the permeability of small molecules across bacterial cell envelopes represents a major current challenge in antibiotic drug discovery and development. While a variety of empirical guidelines have been developed to predict oral bioavailability1,2 and, by extension, cell permeability, most of the drugs that served as the basis for these rules address targets in human eukaryotic cells. In contrast, bacteria have vastly different membrane architectures compared to those of eukaryotic cells, suggesting that the structural and physicochemical properties that govern compound permeability may also differ greatly. Indeed, antibacterials typically have different physicochemical properties compared to other drug classes, such as higher molecular weight and increased polarity, and often violate rules established for oral bioavailability.3 As a result, the structural bias in current small-molecule screening collections toward compounds that address human targets may contribute to the low success rates of such collections in antibacterial drug discovery.4 Thus, the development of quantitative tools to predict small-molecule permeability specifically in bacteria would enable rational chemical approaches to improve screening collections and facilitate lead optimization in the antibacterial arena.5
The bacterial cell envelope is a major barrier that limits the passage of small molecules into the cytoplasm and contributes to intrinsic antibiotic resistance.6 Bacterial membranes vary in complexity depending on lipid composition and embedded channels. Gram-positive bacteria have a relatively simple membrane that is composed of lipoteichoic acids and generally considered to allow passage of nutrients and small molecules.7 The outer membrane of Gram-negative bacteria is composed of anionic lipid polysaccharides, which limits permeation of hydrophobic drugs.6 However, Gram-negative bacteria are permeable to hydrophilic small molecules via nonspecific porins; to bile salts, quaternary ammonium salts, and other cations via self-promoted uptake; and to specific compounds such as vitamin B12 and ferric siderophore complexes via dedicated transporters.6,8 Mycobacteria have a cellular envelope high in lipid content and composed of mycolic acids. The mycobacterial envelope is somewhat permeable to hydrophobic molecules via passive diffusion and to hydrophilic molecules through porins that are smaller and less abundant than those in Gram-negative bacteria.9 In addition, efflux pumps are ubiquitous throughout bacteria and expel a wide array of structurally distinct substrates, further contributing to decreased drug accumulation and increased antibiotic resistance.10
Permeability of small molecules through bacterial cell envelopes remains enigmatic. Previous studies have been limited to only a few classes of known antibiotics and have typically focused on the influences of hydrophobicity and molecular size. For example, an early investigation demonstrated that more hydrophobic β-lactams showed decreased rates of diffusion in Escherichia coli.11 More hydrophobic quinolones have been shown to accumulate at lower levels in Pseudomonas aeruginosa,12E. coli,12Streptococcus pneumoniae,13 and Mycobacterium tuberculosis(14) but at higher levels in Staphylococcus aureus.12 Lower molecular-weight quinolones have also been reported to accumulate more readily in S. pneumoniae,13Bacteroides fragilis,15 and M. tuberculosis.14
Given this limited information, we envisioned that evaluation of a wider range of structural and physicochemical properties that might influence bacterial penetration would enable the development of robust, predictive rules for antibacterial design. Toward this end, we report herein an integrated platform for quantitative analysis of small-molecule permeability in bacteria. As an initial demonstration of this platform, we synthesized a panel of 10 sulfonyladenosine probes and prospectively quantitated their accumulation in Gram-negative, Gram-positive, and mycobacteria using LC-MS/MS.16 Inspired by the sulfamoyladenosine natural products, ascamycin and nucleocidin,17,18 sulfonyladenosines were originally developed as inhibitors of bacterial aminoacyl-tRNA synthetases.19−21 More recently, we and others have advanced this class as potential antibacterials targeting adenylation enzymes involved in bacterial siderophore biosynthesis,22−24 menaquinone biosynthesis,25,26 phenolic glycolipid biosynthesis,27 pantothenate biosynthesis,28 and lipid metabolism.29,100 Notably, the cellular activity of these compounds varies widely, presumably due to poor bacterial penetration in some cases. We then used Pearson pairwise correlations and principal component analysis (PCA) to identify associations between 20 structural and physicochemical properties and sulfonyladenosine accumulation and efflux sensitivity, revealing nonobvious correlations that varied between three bacteria. The development of this analysis platform sets the stage for larger, systematic, prospective studies of these and other compound classes across a broader array of bacteria to identify robust correlations that will ultimately enable the development of predictive rules for small-molecule permeability in bacteria.
Results and Discussion
Cheminformatic Analysis of Antibacterial and Nonantiinfective Drugs
Antibacterials are known to differ greatly in molecular size and polarity compared to other drug classes.3,4 To evaluate a wider range of structural and physicochemical parameters that might provide the basis for a multivariate model, we used PCA, a mathematical method for reducing the dimensionality of multivariable data sets with minimal loss of information.30 In PCA, the original variables are rotated onto new orthogonal, uncorrelated axes called principal components (PC) that are linear combinations of the original variables and represent decreasing proportions of the total variance in the complete data set. We and others have used PCA to evaluate the structural diversity of natural-product-inspired libraries, natural products, and synthetic drugs,31,32 and to correlate biological activity with chemical structure.33
We analyzed 91 structurally diverse antibacterials and 50 top-selling, brand-name, nonantiinfective drugs (Table S1, Supporting Information) for 20 structural and physicochemical parameters (Table S2, Supporting Information) in PCA (Figure 1A and Figure S1, Supporting Information).32 This multivariate analysis indicated that antibacterials have substantially different and more varied structural and physicochemical properties compared to other drug classes. On average, the antibacterials were larger and more hydrophilic than the nonantiinfectives (Table S3, Supporting Information), consistent with a previous report by O’Shea and Moser,3 and also had more stereochemical content compared to the nonantiinfectives. In the PCA plot, quinolones, oxazolidinones, and sulfa drugs were most closely aligned with the nonantiinfective drugs, while β-lactams were positioned at the interface between the nonantiinfectives and the majority of other antibacterials. In contrast, larger glycopeptide, lipopeptide, aminoglycoside, and macrolide antibiotics occupied extreme regions of the plot. Examination of loading plots (Figure 1B and Figure S2, Supporting Information) revealed that parameters associated with size (molecular weight (MW) and surface area (SA)) shift molecules in the positive direction (right and up) along the PC1 and PC2 axes. Hydrophobicity (LogD, ALogPs) shifts molecules left and up along PC1 and PC2, while solubility (ALogpS, relative polar surface area (relPSA)) has opposite effects. Parameters associated with three-dimensional structure (stereocenter count (nStereo), stereochemical density (nStMW), and sp3 content (Fsp3)) shift molecules to the right along PC1 and in the negative direction along PC3. Thus, the observed differences in chemical property space between antibacterials and nonantiinfectives explain why rules for predicting oral bioavailability for drugs with eukaryotic targets are insufficient for predicting cell permeability in bacteria.
Figure 1.
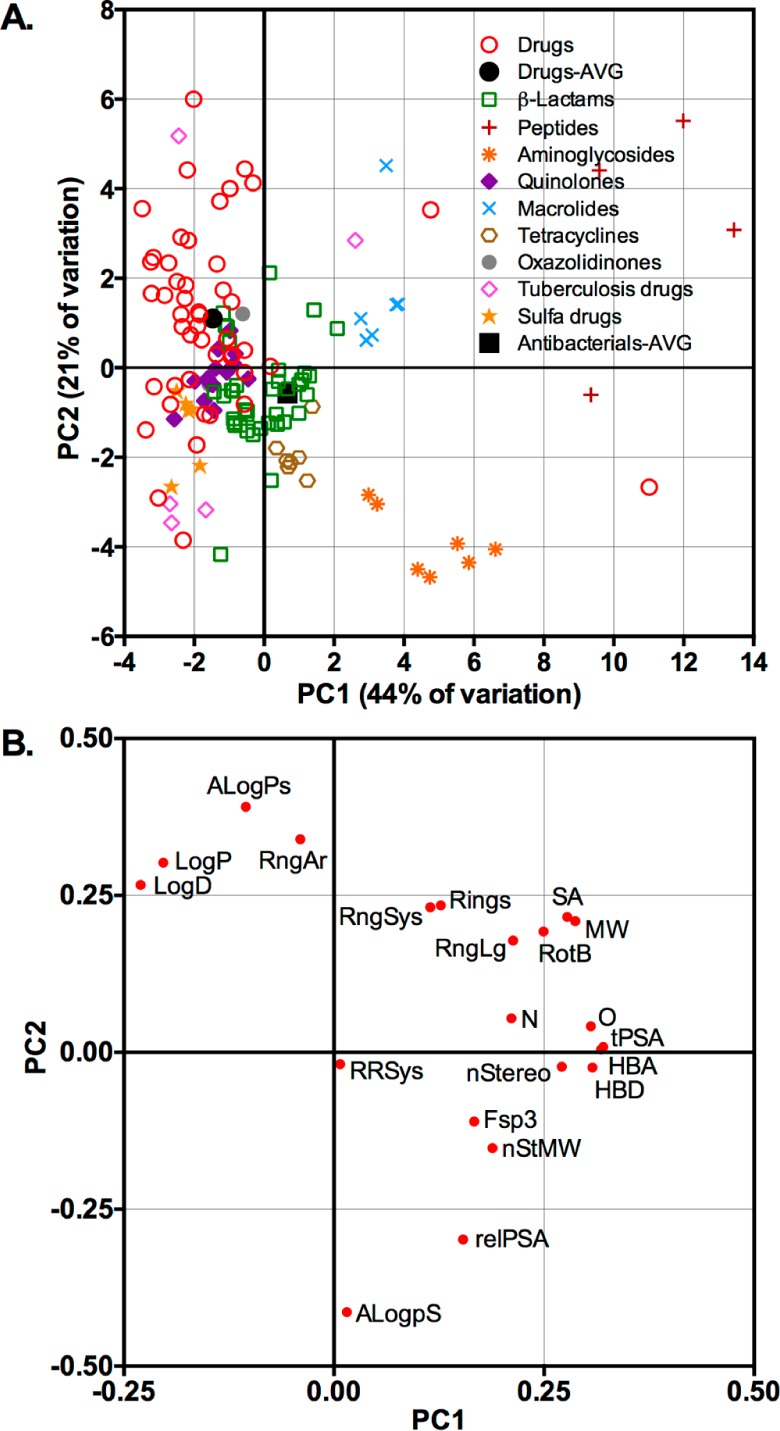
Antibacterials have distinct structural and physicochemical properties compared to those of nonantiinfective drugs. (A) Principal component analysis (PCA) of 91 antibacterials and 50 top-selling nonantiinfective drugs (Drugs) using 20 structural and physicochemical parameters; percent contribution for each principal component indicated on the axes; AVG = hypothetical average for compound class. (B) PCA loading plot showing component loadings of 20 structural/physicochemical parameters used in the PCA; ALogPs = log P; ALogpS = log S; Fsp3 = fraction sp3 carbons; HBA = hydrogen-bond acceptors; HBD = hydrogen-bond donors; MW = molecular weight; N = nitrogens; nStereo = stereocenters; nStMW = stereochemical density (nStereo ÷ MW); O = oxygens; relPSA = relative polar surface area; RngAr = aryl rings; RngLg = largest ring size; RngSys = ring systems; RRSys = rings per ring system, SA = van der Waals surface area; tPSA = topological polar surface area. See Tables S1 and S2 (Supporting Information) for lists of compounds and parameters and Figures S1 and S2 (Supporting Information) for PCA and loading plots with PC3.
LC-MS/MS Quantitation of Compound Accumulation
To begin investigating the physicochemical properties that influence small-molecule permeability specifically in bacteria, we used LC-MS/MS to quantitate compound accumulation in bacterial cells.16 Several other methods have been used previously for this purpose but are restricted to specific drug classes or require the synthesis of labeled variants of the analyte of interest.9,11−15,34−37 In contrast, LC-MS/MS provides a general method to measure compound concentrations relative to an internal standard.16 Importantly, in contrast to stable-isotope dilution MS,38 an isotopically labeled variant of the analyte is not required, and an unlabeled analogue can instead be used as the internal standard.
We initially evaluated this LC-MS/MS quantitation method using salicyl-AMS (1) as an analyte (Figure 2). We have previously demonstrated that this compound inhibits intracellular enzymes in M. tuberculosis and Yersinia pestis that are required for siderophore biosynthesis.22 For these pilot studies, we measured salicyl-AMS accumulation in E. coli because this bacterium has been used frequently in previous analyses of compound permeability.12,34,35 We determined that salicyl-AMS was quantifiable from 0.0025–100 μM (4.6 logs) in PBS (Figure S3, Supporting Information). We then treated E. coli with salicyl-AMS (100 μM, 30 min, tryptic soy broth). The cells were centrifuged, washed, and lysed, then salicyl-AMS concentrations were determined in all fractions by LC-MS/MS. The intracellular concentration was calculated from the lysate concentration based on CFU determination. Under these conditions, E. coli accumulated salicyl-AMS at 25 μM intracellular concentration (Figure 2A). We note that Aldrich and co-workers have reported that salicyl-AMS is inactive against E. coli,51 citing unpublished results, so it is possible that this may be due to effects other than permeability.
Figure 2.
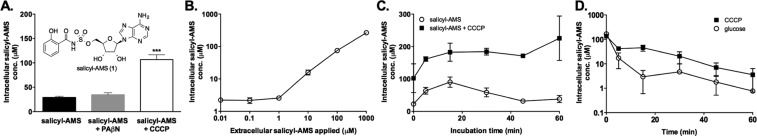
Pilot experiments with salicyl-AMS accumulation in E. coli validate the feasibility of the LC-MS/MS method for quantitating small-molecule uptake in bacteria. (A) Accumulation of salicyl-AMS in E. coli (100 μM extracellular, 30 min, tryptic soy broth) and impacts of efflux pump inhibitors. Statistical significance compared to salicyl-AMS alone assessed using one-way ANOVA and Tukey’s multiple comparison test with 95% confidence intervals: ***p < 0.001. CCCP = carbonyl cyanide m-chlorophenylhydrazone; PAβN = phenylalanine arginine-β-naphthylamide. (B) Concentration of intracellular salicyl-AMS as a function of extracellular salicyl-AMS concentration applied (20 min, PBS). (C) Kinetics of salicyl-AMS accumulation (100 μM extracellular, PBS) in the presence and absence of CCCP (100 μM). (D) Kinetics of salicyl-AMS export from preloaded cells (100 μM extracellular, 15 min, PBS) via passive diffusion (CCCP, 100 μM) and active efflux (glucose, 0.2%). Data reported as the mean ± SD for 4 experiments (panel A) or 3 experiments (panels B–D).
Impact of Efflux Pumps on Salicyl-AMS Accumulation in E. coli
The E. coli genome encodes an estimated 37 efflux transporters.39 The AcrAB-TolC efflux pump is constitutively expressed in E. coli and is regarded as a major contributor to multidrug resistance.39 To assess the role that this pump may play in salicyl-AMS accumulation levels, we pretreated E. coli with phenylalanine arginine-β-naphthylamide (PAβN, 38 μM, tryptic soy broth) to inhibit AcrAB-TolC,40 followed by the addition of salicyl-AMS (100 μM). Intracellular levels of salicyl-AMS did not increase significantly in the presence of PAβN, suggesting that this compound may not be a substrate for AcrAB-TolC (Figure 2A). To assess the roles that other efflux pumps may play in salicyl-AMS accumulation levels, we pretreated E. coli with carbonyl cyanide m-chlorophenylhydrazone (CCCP, 100 μM) to collapse the proton-motive force (pmf) that energizes most efflux pumps.41 Under these conditions, intracellular levels of salicyl-AMS increased 4-fold (107 μM), suggesting that this compound is a substrate for other efflux pumps (Figure 2A). Importantly, this observed differential accumulation supports our assumption that our assay protocol measures intracellular salicyl-AMS rather than the residual membrane-associated compound since the latter would not be impacted by efflux pump activity.14
Concentration Dependence of Salicyl-AMS Accumulation in E. coli
Next, to investigate the concentration dependence of compound accumulation we treated E. coli with varying concentrations of salicyl-AMS (0.01–1000 μM, 20 min, PBS). Treating cells with as little as 0.01 μM extracellular salicyl-AMS resulted in the detection of 2.0 μM intracellular salicyl-AMS (Figure 2B). Interestingly, the intracellular levels of salicyl-AMS remained constant up to 1 μM extracellular concentration, then increased linearly at extracellular concentrations between 10–1000 μM. Notably, at 100 μM extracellular concentration, salicyl-AMS accumulated to much higher levels in PBS (75 μM) than in the nutrient rich media above. It is possible that depriving cells of nutrients may promote salicyl-AMS uptake via upregulation of transporters or other unknown mechanisms. The unexpected observation that intracellular levels exceeded extracellular levels at the low end of the concentration curve suggests that active import or Donnan potential-driven passive diffusion may play a role in compound accumulation at these lower concentrations.16,39,41−43
Kinetics of Salicyl-AMS Accumulation in E. coli
To assess the kinetics of salicyl-AMS uptake in E. coli, we treated cells with salicyl-AMS (100 μM) for varying times (0–60 min), then measured intracellular compound levels. These experiments were carried out in PBS to eliminate consideration of cell doubling. Salicyl-AMS rapidly accumulated within the first 5 min, peaked at 15 min, and declined thereafter (Figure 2C). This general trend is consistent with that reported for ciprofloxacin accumulation in P. aeruginosa.16 Having shown above that CCCP increases salicyl-AMS accumulation in the single time point experiment above (Figure 2A), we assessed the effect of pretreating cells with CCCP (100 μM) on uptake kinetics. Under these conditions, salicyl-AMS accumulated more quickly and to higher concentrations than in the absence of CCCP, reaching a maximum within 5–15 min and remaining at this level after longer incubation times, again consistent with a role for efflux pumps in determining the rate and levels of salicyl-AMS accumulation in E. coli.
Kinetics of Salicyl-AMS Passive Diffusion and Active Efflux out of E. coli
To assess the rate of salicyl-AMS passive diffusion out of E. coli, we preloaded cells with salicyl-AMS (100 μM) in the presence of CCCP (100 μM). These cells were then resuspended in PBS containing CCCP (100 μM) for various times (5–60 min, 37 °C) to allow salicyl-AMS to diffuse out passively. Analysis of intracellular salicyl-AMS levels over time revealed rapid passive diffusion (31% remaining after 5 min) (Figure 2D). Next, to assess the impact of active, pump-mediated efflux on the rate of compound export, we resuspended salicyl-AMS–preloaded cells in PBS containing 0.2% glucose instead of CCCP, to reactivate pmf-driven efflux pumps.41 Under these conditions, the compound was expelled more rapidly (10% remaining after 5 min). The observed time course is consistent with that reported for active efflux of norfloxacin from E. coli (∼60% cell-associated norfloxacin remaining after 1.5 min).44
Systematic Analysis of Sulfonyladenosine Accumulation in Bacteria
Next, we sought to test a broader array of sulfonyladenosines to assess the impacts of structural and physicochemical properties on bacterial accumulation. Toward this end, we synthesized a panel of nine additional sulfonyladenosines to provide a structurally diverse panel (Figure 3 and Table S4, Supporting Information). Unsubstituted sulfamoyladenosine (2) is a known cytotoxic antibiotic45 and served as a basis for comparison of the other analogues, which were all N-acylated. Along with salicyl-AMS (1), benzoyl-AMS (8), anthranilyl-AMS (6), and OSB-AMS (7) were also of interest because they have been investigated as potential antibacterials.25,46 Meanwhile, the zwitterionic l-alanyl-AMS (3) and the anionic l-lactyl-AMS (4) were selected to probe the specific influence of charge on bacterial permeability41 since they have otherwise similar physicochemical properties. 4-Phenylbenzoyl-AMS (9) and decanoyl-AMS (10) have similar hydrophobicity and polarity and were selected to test the specific influence of rotatable bonds on bacterial permeability because such molecular flexibility has been inversely correlated with oral bioavailability in eukaryotic cells.2 Finally, methyl-succinyl-AMS (5) was designed as a compound with a combination of intermediate properties compared to the rest of the panel. In a PCA with the collection of diverse antibacterials and nonantiinfectives using the 20 structural and physicochemical parameters discussed earlier, the 10 sulfonyladenosines clustered just outside the region of the plot occupied by nonantiinfectives, overlapping with β-lactam antibiotics (Figures S4 and S5, Supporting Information).
Figure 3.
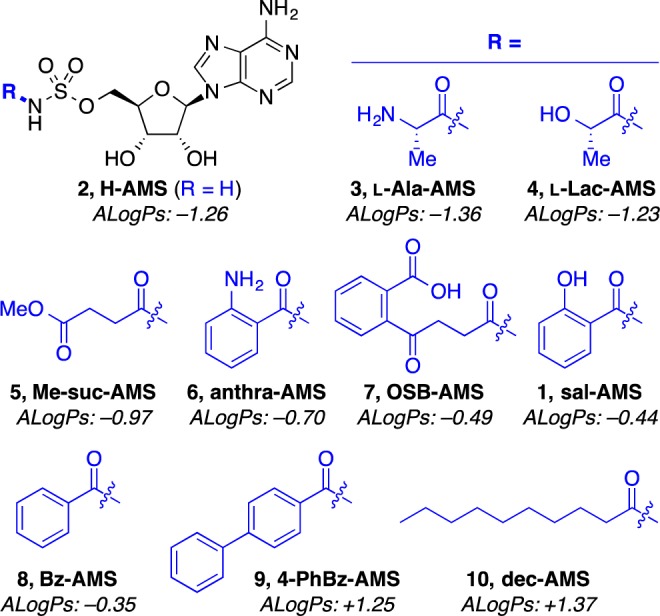
Structures of 10 sulfonyladenosines with different structural and physicochemical properties for bacterial accumulation studies. Compounds are arranged in order of increasing ALogPs, except for H-AMS. See Table S4 (Supporting Information) for a complete list of 20 structural and physicochemical properties for each compound; H-AMS = sulfamoyladenosine (adenosine monosulfamate), l-Ala = l-alanyl, anthra = anthranilyl, Bz = benzoyl, dec = decanoyl, l-Lac = l-lactyl, Me-suc = methyl succinyl, OSB = o-succinylbenzoate, 4-PhBz = 4-phenylbenzoyl, and sal = salicyl.
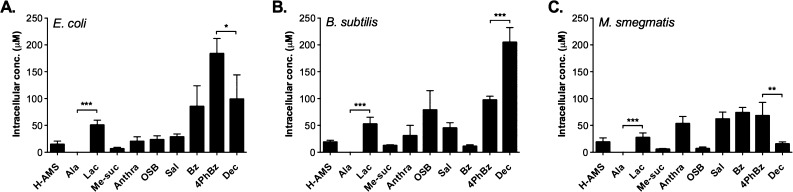
We then assayed compound accumulation of this panel of sulfonyladenosines in three bacteria with distinct cellular envelopes: E. coli (Gram negative), Bacillus subtilis (Gram positive), and Mycobacterium smegmatis (Figure 4A–C). In E. coli, sulfamoyladenosine (2) accumulated to 25 μM intracellular concentration (Figure 4A). Meanwhile, among the other two most polar analogues, l-alanyl-AMS (3) did not accumulate to detectable levels (linear range of detection 0.05–100 μM) (Figure 4A), but l-lactyl-AMS (4) accumulated to 51 μM, even higher than sulfamoyladenosine. Among the most hydrophobic analogues, decanoyl-AMS (10) accumulated to 99 μM, 2-fold lower than 4-phenylbenzoyl-AMS (9) at 184 μM, which is slightly less hydrophobic. These results indicate that polarity alone is insufficient to predict permeability in E. coli accurately.
Figure 4.
Accumulation of 10 sulfonyladenosines in bacteria with distinct membrane architectures. Uptake of sulfonyladenosines (100 μM) in (A) E. coli, (B) B. subtilis, and (C) M. smegmatis. Compounds are arranged left-to-right in order of increasing ALogPs values, except H-AMS. Data are reported as the mean ± SD for 4 experiments. Statistical significance was assessed using two-tailed unpaired t-test with 95% confidence intervals: *p < 0.05, **p < 0.01, and ***p < 0.001. See Figure S6 (Supporting Information) for pairwise comparisons between the three bacteria for each compound.
In B. subtilis, sulfamoyladenosine (2) and l-lactyl-AMS (4) accumulated to levels comparable to those observed in E. coli (Figure 4B and Figure S6, Supporting Information), while l-alanyl-AMS (3) again did not accumulate to detectable levels (<50 nM). In marked contrast to E. coli, decanoyl-AMS (10) accumulated to 205 μM, 2-fold higher than 4-phenylbenzoyl-AMS (9) at 98 μM. Compared to E. coli, the accumulation levels of OSB-AMS (7) were also significantly higher, while the levels of benzoyl-AMS (8) were significantly lower, highlighting the differences in permeability patterns between bacteria with different membrane structures (Figure S6, Supporting Information).
In M. smegmatis, sulfamoyladenosine (2) accumulated to levels comparable to those observed in E. coli (Figure 4C and Figure S6, Supporting Information), while l-lactyl-AMS (4) accumulated to a similar concentration, which was 2-fold lower than that in E. coli. l-Alanyl-AMS (3) again did not accumulate to detectable levels (<50 nM). Among the hydrophobic analogues, decanoyl-AMS (10) accumulated to 4-fold lower levels than 4-phenylbenzoyl-AMS (9), comparable to the much more polar sulfamoyladenosine (2). Interestingly, the aroyl-AMS compounds benzoyl-AMS (8), salicyl-AMS (1), anthranilyl-AMS (6), and 4-phenylbenzoyl-AMS (9) accumulated to comparable cellular concentrations, despite their differing hydrophobicities. These results again highlight the differences in permeability patterns between bacteria and the inability to predict permeability based on polarity alone.
Taken together, these data indicate that hydrophobicity is insufficient to predict compound accumulation in bacteria and that other structural and physicochemical parameters likely influence compound uptake. Indeed, some of the more polar analogues accumulated to levels comparable to some of the more hydrophobic analogues. Further, small-molecule permeability patterns clearly differ across bacteria with distinct membrane architectures, necessitating directed studies of each bacterium of interest. No general trends toward greater or lesser permeability were apparent between the different bacteria, with accumulation levels varying on a compound-by-compound basis (cf. benzoyl-AMS (8) vs decanoyl-AMS (10)).
Multivariate Analyses of Sulfonyladenosine Structural Properties and Accumulation
To identify nonobvious structural and physicochemical properties that correlate with accumulation of the sulfonyladenosines, we conducted PCA of the 10-compound panel using the same 20 chemical parameters in the larger analysis above (Figure 1) plus the average intracellular concentrations determined in the compound accumulation studies for each bacterium (Figure 4). Because PCA is sensitive to outliers, particularly for small data sets,30 we confirmed in a separate PCA that the 10 sulfonyladenosines alone (Figure S7, Supporting Information) had similar relative positions compared to those in the larger PCA above that included nonantiinfectives and other antibiotics (Figure S4–S5, Supporting Information). Moreover, removal of l-alanyl-AMS (3) did not affect the relative positions of the remaining compounds in the PCA. Addition of the compound accumulation parameters to the PCAs likewise did not substantially alter the relative positions of the compounds compared to the PCA carried out using only the 20 chemical parameters. Taken together, these results support the robustness of the 10-compound analyses.
In E. coli, the top three accumulators (4-phenylbenzoyl-AMS (9) > decanoyl-AMS (10) > benzoyl-AMS (8)) clustered in the bottom right quadrant of the PC1 vs PC2 biplot (Figure 5A). The E. coli accumulation parameter also clustered with hydrophobicity parameters (ALogPs, LogP, and LogD) in this quadrant of the biplot, far from parameters associated with polarity (tPSA, HBA, HBD, O, ALogpS, and relPSA).
Figure 5.
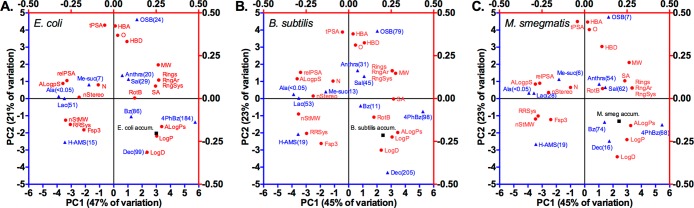
Multivariate analyses reveal correlations between sulfonyladenosine bacterial accumulation with structural and physicochemical parameters. PCA biplots showing relationships between 10 sulfonyladenosines, 20 structural and physicochemical descriptors, and accumulation in (A) E. coli, (B) B. subtilis, and (C) M. smegmatis; average sulfonyladenosine cellular concentration (μM) is noted in parentheses; ▲ = sulfonyladenosines; ● = physicochemical parameters; ■ = accumulation parameter; expanded PCA biplots for each bacterium are in Figures S8–S10 (Supporting Information).
While PCA allows qualitative assessment of correlations between parameters based on their proximity in the component loading plots, some information is inherently lost during the process of dimensionality reduction. Thus, we also analyzed the data set for Pearson pairwise correlations to provide a quantitative assessment of the impact of each of the 20 chemical parameters individually upon permeability. Consistent with the qualitative PCA, accumulation in E. coli was significantly positively correlated with hydrophobicity (LogD, LogP, and ALogPs) and significantly negatively correlated with polarity (ALogpS and relPSA) (Figure 6). Notably, however, the Pearson correlations also revealed additional positive correlations with ring content (Rings, RngAr and RngSys) and size (MW and SA), and additional negative correlations with ring complexity (RRSys), hydrogen bonding capacity (HBA and HBD), heteroatom counts (O and N), and three-dimensional topology (nStereo, nStMW, and Fsp3).
Figure 6.
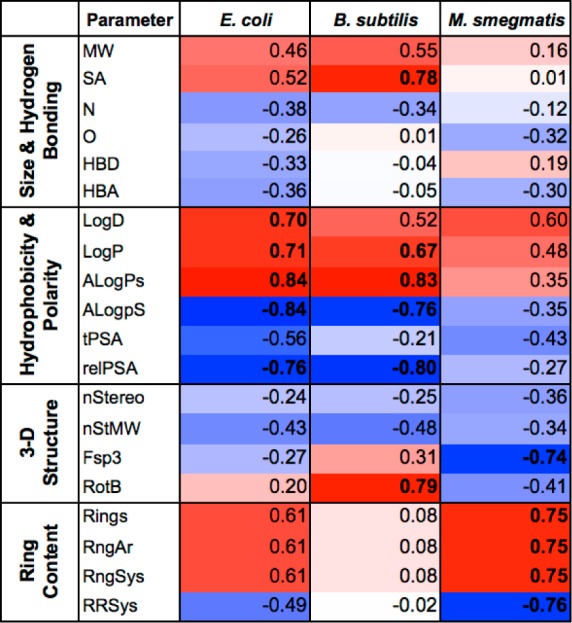
Heat map of Pearson pairwise correlation coefficients of bacterial accumulation and structural/physicochemical properties. Positive correlations are in red and negative correlations are in blue; correlations in bold are statistically significant as assessed using a two-tailed unpaired t-test and 95% confidence intervals (p < 0.05).
In B. subtilis, the top three accumulators (decanoyl-AMS (10) > 4-phenylbenzoyl-AMS (9) > OSB-AMS (7)) fell furthest to the right side of the PC1 vs PC2 biplot but were widely scattered (Figure 5B). The B. subtilis accumulation parameter again fell in the lower right quadrant of the biplot, clustering with hydrophobicity parameters (ALogPs, LogP, and LogD), as well as rotatable bonds (RotB) and surface area (SA). Parameters associated with polarity (ALogpS, relPSA, and tPSA) and hydrogen bonding capacity (HBA, HBD, and O) were again distant. Examination of Pearson correlations confirmed these observations, with accumulation in B. subtilis being significantly positively correlated with not only hydrophobicity (LogP and ALogPs) but also size (SA) and rotatable bonds (RotB), while still being significantly negatively correlated with polarity (ALogpS and relPSA) (Figure 6). Further, in contrast to E. coli accumulation, B. subtilis accumulation was essentially uncorrelated with hydrogen bonding capacity (HBA and HBD) and ring content (Rings, RngAr, and RngSys).
The Pearson analysis also revealed that certain physicochemical parameters are inherently correlated with each other (Figure S11, Supporting Information). For example, there were strong positive correlations between descriptors for size, hydrophobicity, conformational flexibility, and ring content. In contrast, each of these descriptors was negatively correlated with descriptors for polarity. Given the correlations between descriptors themselves, some of the observed correlations between accumulation in E. coli and B. subtilis and physicochemical parameters are not surprising. However, accumulation in B. subtilis did not correlate with ring content (Rings, RngAr, and RngSys), in spite of inherent positive correlations among ring content, hydrophobicity, and size. Based on this result, the correlations with accumulation are unlikely to be a mere consequence of the inherent correlations between the descriptors themselves.
In M. smegmatis, the top four accumulators (benzoyl-AMS (8) > 4-phenylbenzoyl-AMS (9) > salicyl-AMS (1) > anthranilyl-AMS (6)) clustered on the right side of the PC1 vs PC2 biplot (Figure 5C). The M. smegmatis accumulation parameter fell in the lower right quadrant of the biplot, clustering again with hydrophobicity parameters (ALogPs, LogP, and LogD), as well as parameters for ring content (Rings, RngAr, and RngSys) and rotatable bonds (RotB). The Pearson pairwise analysis confirmed significant positive correlations between accumulation and ring content, as well as positive correlations with hydrophobicity (Figure 6). However, accumulation was actually negatively correlated with rotatable bonds due to opposite positioning along the PC3 axis (Figure S10, Supporting Information). This was not apparent from the PC1 vs PC2 biplot, highlighting the inherent loss of information due to dimensionality reduction in PCA and the value of Pearson analysis in assessing all 20 parameters individually. Significant negative correlations with ring system complexity (RRSys) and sp3 content (Fsp3) were also identified that were not obvious from the PCA biplot. In contrast to B. subtilis and E. coli, the Pearson coefficients revealed that accumulation in M. smegmatis was not strongly correlated with size (MW and SA) and was also generally more weakly correlated with hydrophobicity and polarity.
Importantly, these multivariate analyses help to rationalize the observed differences in sulfonyladenosine accumulation across the bacteria. It is not surprising that salicyl-AMS (1) and anthranilyl-AMS (6) accumulated to similar levels in each bacterium since they have similar physicochemical properties. However, benzoyl-AMS (8) also appears to have generally similar physicochemical properties but accumulated to higher levels in E. coli but lower levels in B. subtilis. The increased accumulation of benzoyl-AMS in E. coli relative to salicyl-AMS and anthranilyl-AMS may be due to its lower heteroatom count and decreased hydrogen bonding capacity since these properties negatively correlated with accumulation.
l-Alanyl-AMS (3) and l-lactyl-AMS (4) also have generally similar physicochemical properties, but only the latter penetrated all three bacteria, highlighting the influence of charge on sulfonyladenosine permeability, with the former compound being zwitterionic, while the latter is anionic at the sulfamate nitrogen. It is also possible that l-lactyl-AMS may perturb the outer membrane in a manner similar to lactic acid, a known permeabilizer of the outer membrane in Gram-negative bacteria.47
Moreover, OSB-AMS (7) and methyl-succinyl-AMS (5) are similar in solubility, polar surface area, relative polar surface area, and rotatable bonds, but the higher intracellular levels of OSB-AMS in E. coli and B. subtilis may be explained by its larger size, with which accumulation correlated positively. The higher accumulation of OSB-AMS in E. coli compared to that of methyl-succinyl-AMS may also be influenced by its higher ring content, with which accumulation also correlated positively. In M. smegmatis, these two compounds accumulated to comparable levels, highlighting a counterbalance between multiple physicochemical properties in this bacterium: correlations with ring content and sp3 content would predict higher accumulation of OSB-AMS than methyl-succinyl-AMS, but correlations with oxygen content and hydrogen bond acceptors would predict the converse.
Decanoyl-AMS (10) and 4-phenylbenzoyl-AMS (9) have similar hydrophobicities but are substantially different in terms of sp3 content, rotatable bond count, and ring content. Decanoyl-AMS has more three-dimensional structure (RotB, Fsp3), which correlates positively with accumulation in B. subtilis but negatively with accumulation in M. smegmatis and has mixed correlations in E. coli, consistent with the observed intracellular concentrations. Meanwhile, 4-phenylbenzoyl-AMS has higher ring content (Rings, RngAr, and RngSys), which may be associated with its higher accumulation in E. coli and M. smegmatis.
To assess whether the correlations identified in the PCA and Pearson pairwise analyses would also apply to sulfonyladenosines with structural variations at a different position, we synthesized two additional salicyl-AMS analogues with phenyl and phenylamino substituents at the C2 position of the adenine ring (Figure S12, Supporting Information).48 Both analogues accumulated to significantly higher intracellular levels in E. coli than the parent salicyl-AMS compound, consistent with correlations between accumulation and size, hydrophobicity, and aromatic ring content identified using variants in the acyl region above. This suggests that accumulation correlates with structural and physicochemical properties independent of the exact chemical structure.
Overall, these multivariate analyses demonstrate that nonobvious factors other than hydrophobicity influence sulfonyladenosine accumulation in bacteria and that these correlations differ between bacteria with distinct membrane architectures. It is important to note that correlations between structural/physicochemical properties and accumulation may vary depending on compound class and thus, larger systematic analyses will be required to derive broadly predictive models.
Role of Efflux Pumps on Sulfonyladenosine Accumulation
To probe the role of efflux on accumulation of sulfonyladenosines across the three bacteria, cells were de-energized with CCCP prior to exposure to the compounds. To investigate the roles of specific transporters, E. coli was also treated with PAβN to inhibit the AcrAB-TolC transporter, and B. subtilis and M. smegmatis were treated with reserpine to inhibit the Bmr transporter and ATP-dependent transporters, respectively.36,49
In E. coli, methyl succinyl-AMS (5), anthranilyl-AMS (6), and salicyl-AMS (1) accumulated to significantly higher levels in the presence of CCCP (Figure 7A), but there were no obvious correlations between physicochemical properties and sensitivity to pmf-driven efflux. Intracellular levels of decanoyl-AMS (10) and 4-phenylbenzoyl-AMS (9) increased in the presence of the AcrAB-TolC specific inhibitor, PAβN. Since AcrAB-TolC is pmf-energized, we were surprised that the intracellular levels of these compounds did not increase in the presence of CCCP. Previous studies have demonstrated that, in addition to the specific inhibition of Mex-Opr transporters, PAβN permeabilizes the outer membrane of wild-type- and Mex-Opr-deficient strains of P. aeruginosa.40,50 Thus, enhanced accumulation of the most hydrophobic sulfonyladenosines may involve this alternative mechanism in which PAβN permeabilizes E. coli to passive diffusion.
Figure 7.
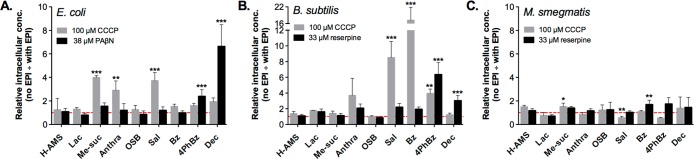
Effect of efflux on the accumulation of 10 sulfonyladenosines in bacteria with distinct membrane architectures. Pretreatment of bacteria with efflux pump inhibitors (EPI) prior to incubation with the sulfonyladenosine (100 μM) in (A) E. coli, (B) B. subtilis, and (C) M. smegmatis. Compounds are arranged left to right in order of increasing ALogPs values. For each compound, data is normalized with respect to cellular concentration in the absence of the efflux pump inhibitor (no EPI). Statistical significance was assessed relative to bacteria not treated with EPI using one-way ANOVA and Tukey’s multiple comparison test and 95% confidence intervals: *p < 0.05, **p < 0.01, and ***p < 0.001.
In B. subtilis, there was a clear correlation between aromatic ring content and sensitivity to pmf-driven efflux (Figure 7B). CCCP significantly enhanced the accumulation of aroyl-AMS compounds (benzoyl-AMS (8) > salicyl-AMS (1) > 4-phenylbenzoyl-AMS (9)) but not of any of the aliphatic acyl-AMS congeners. Interestingly, while the accumulation of decanoyl-AMS (10) was unaltered by CCCP, it increased significantly in the presence of the Bmr-specific inhibitor reserpine. This is surprising because the Bmr transporter is energized by pmf, suggesting an alternative mode of action for reserpine. It is possible that reserpine inhibits an unidentified ATP-dependent efflux transporter in B. subtilis since it is a known inhibitor of ATP-dependent P-glycoprotein and PstB transporters found in mammalian cells and M. smegmatis, respectively.49
In M. smegmatis, most compounds showed only modest increases (less than 2-fold) in accumulation when treated with CCCP, and in a few cases, accumulation surprisingly decreased (Figure 7C). Similarly, most compounds displayed small increases in accumulation when ABC efflux pumps were inhibited with reserpine.
Taken together, these results highlight some of the complications associated with the use of efflux pump inhibitors due to their potential nonspecific and off-target effects. Studies with individual pump mutant strains may be more effective for probing the efflux sensitivity of various compounds in the future.
Multivariate Analyses of Sulfonyladenosine Structural Properties and Efflux Sensitivity
To determine if certain structural and physicochemical properties may sensitize sulfonyladenosines to efflux pump activity, we carried out PCA (Figures S13–S15, Supporting Information) and Pearson analyses (Figure 8) incorporating the normalized accumulation levels in the presence of efflux pump inhibitors.
Figure 8.
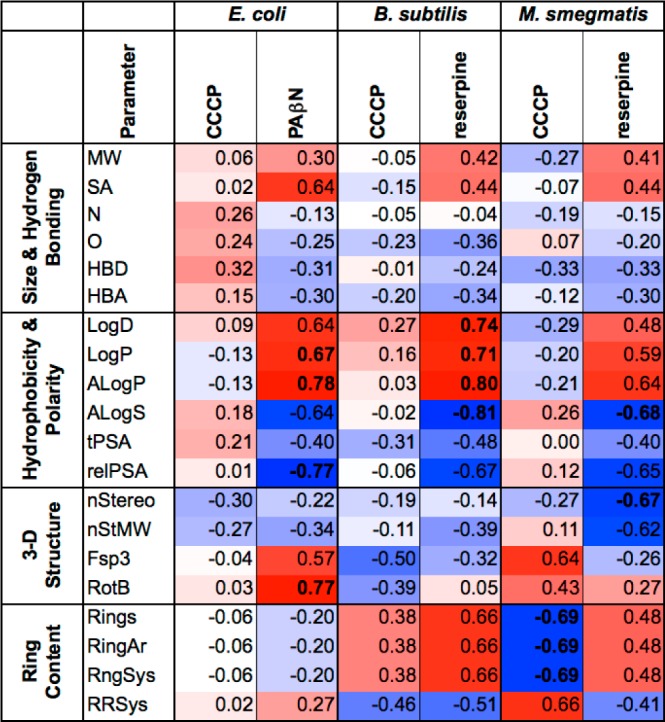
Heat map of Pearson pairwise correlation coefficients of efflux sensitivity and physicochemical properties. Positive correlations are in red and negative correlations are in blue; correlations in bold are statistically significant as assessed using a two-tailed unpaired t-test and 95% confidence intervals (p < 0.05). See Figures S13–S15 (Supporting Information) for corresponding PCA biplots.
In E. coli, there were no significant correlations between physicochemical parameters and sensitivity to pmf-driven efflux transporters (CCCP) (Figure 8 and Figure S13, Supporting Information). Hydrophobicity and rotatable bonds correlated positively with sensitivity to PAβN, whereas polarity correlated negatively. In B. subtilis, PCA and Pearson analysis identified a positive correlation between ring content and sensitivity to pmf-driven efflux transporters (CCCP), whereas hydrophobicity positively correlated with sensitivity to the Bmr transporter (reserpine) (Figure 8 and Figure S14, Supporting Information). In M. smegmatis, ring content correlated negatively with sensitivity to pmf-driven efflux pumps (CCCP) (Figures 8 and S15, Supporting Information). Interestingly, ring content and hydrophobicity correlated positively with sensitivity to ATP-dependent transporters (reserpine).
Conclusions
The permeability of small molecules across bacterial membranes remains poorly understood, and the development of computational tools to predict compound penetration would represent a major advance in antibiotic drug discovery. Antibacterials have vastly different structural and physicochemical properties compared to those of nonantiinfective drugs, presenting an enigma to currently available tools developed for eukaryotic systems. Herein, we have described a quantitative, systematic approach to evaluate compound permeability prospectively in bacteria using a panel of sulfonyladenosines as an initial demonstration. Our cheminformatic analyses revealed nonobvious correlations between the uptake of sulfonyladenosines and physicochemical parameters that vary across bacteria with diverse membrane architectures. Within this sulfonyladenosine panel, hydrophobicity, ring content, and size positively correlated with E. coli accumulation, size, hydrophobicity, and molecular flexibility positively correlated with B. subtilis accumulation, and ring content positively correlated with M. smegmatis accumulation. Additionally, certain physicochemical descriptors correlated with the sensitivity of these sulfonyladenosines to efflux pumps, and these correlations also differed between bacteria.
Correlations between chemical structure and bacterial permeability may vary depending on compound class. Thus, while the results described herein are restricted in scope to the small panel of sulfonyladenosines evaluated, this platform can readily be extended to the prospective analysis of larger collections of compounds in diverse chemical classes and across other bacteria, including efflux and permeability mutant strains. Noting that l-lactyl-AMS and l-alanyl-AMS have similar properties but disparate accumulation levels, it may also be necessary to incorporate additional structural and physicochemical parameters beyond the 20 used herein and to consider potential idiosyncratic or chemotype-specific mechanisms for accumulation of specific functionalities that may lead to outliers.47 Indeed, it is possible that nucleoside transporters may play a role in the intracellular accumulation of the sulfonyladenosines investigated herein. Ultimately, large-scale cheminformatic analysis of these data may enable the development of computational tools such as quantitative structure–accumulation relationship models to predict the permeability and efflux sensitivity of small molecules in pathogenic bacteria that represent a growing threat to human health.
Acknowledgments
This work is dedicated to the memory of our mentor and colleague, Professor David Y. Gin (1967–2011). We thank L. Quadri (Brooklyn College), P. Tonge (Stony Brook University), V. Knight-Connoni, B. Pandya, and D. Ryan (Cubist Pharmaceuticals), and N. Wu, R. Khanin, and T. Taldone (MSKCC) for helpful discussions, J. Cisar (MSKCC) for carrying out preliminary compound accumulation experiments, I. Sharma and C. Ji (MSKCC) for providing OSB-AMS and C2-substituted-salicyl-AMS analogues, C. Stratton (MSKCC) for assistance with cheminformatic analyses and critically reading the manuscript, G. Chiosis (MSKCC) for access to LC-MS/MS instrumentation, G. Sukenick, R. Wang, H. Liu, H. Fang, and S. Rusli (MSKCC) for expert NMR and mass spectral support, and F. Varodayan (The Scripps Research Institute) for critically reading the manuscript. Instant JChem was generously provided by ChemAxon. Financial support from the National Institutes of Health (R01 GM100477 and R21 AI098802 to D.S.T. and T32 CA062948-Gudas to T.D.D.) is gratefully acknowledged.
Supporting Information Available
Complete experimental procedures and analytical data for all new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): D.S.T. is a coinventor of an issued US Patent on sulfonyladenosines as antibacterial agents and has intellectual property interests in these compounds.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 23, 3–25. [DOI] [PubMed] [Google Scholar]
- Veber D. F.; Johnson S. R.; Cheng H. Y.; Smith B. R.; Ward K. W.; Kopple K. D. (2002) Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45, 2615–2623. [DOI] [PubMed] [Google Scholar]
- O’Shea R.; Moser H. E. (2008) Physicochemical properties of antibacterial compounds: implications for drug discovery. J. Med. Chem. 51, 2871–2878. [DOI] [PubMed] [Google Scholar]
- Payne D. J.; Gwynn M. N.; Holmes D. J.; Pompliano D. L. (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discovery 6, 29–40. [DOI] [PubMed] [Google Scholar]
- Lewis K. (2013) Platforms for antibiotic discovery. Nat. Rev. Drug Discovery 12, 371–387. [DOI] [PubMed] [Google Scholar]
- Nikaido H. (1994) Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264, 382–388. [DOI] [PubMed] [Google Scholar]
- Lambert P. A. (2002) Cellular impermeability and uptake of biocides and antibiotics in gram-positive bacteria and mycobacteria. Symp. Ser. Soc. Appl. Microbiol. 46S–54S. [PubMed] [Google Scholar]
- Nikaido H. (1989) Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob. Agents Chemother. 33, 1831–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z.; Zhang L.; Nikaido H. (2004) Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 2415–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Schweizer H. P. (2005) Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv. Drug Delivery Rev. 57, 1486–1513. [DOI] [PubMed] [Google Scholar]
- Zimmermann W.; Rosselet A. (1977) Function of the outer membrane of Escherichia coli as a permeability barrier to beta-lactam antibiotics. Antimicrob. Agents Chemother. 12, 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey C.; Bertasso A.; Pace J.; Georgopapadakou N. H. (1992) Quinolone accumulation in Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus. Antimicrob. Agents Chemother. 36, 1601–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J.; Johnson M. M. (2002) Accumulation of 10 fluoroquinolones by wild-type or efflux mutant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J.; Ricci V. (2001) Accumulation of five fluoroquinolones by Mycobacterium tuberculosis H37Rv. J. Antimicrob. Chemother. 48, 787–791. [DOI] [PubMed] [Google Scholar]
- Ricci V.; Piddock L. (2003) Accumulation of garenoxacin by Bacteroides fragilis compared with that of five fluoroquinolones. J. Antimicrob. Chemother. 52, 605–609. [DOI] [PubMed] [Google Scholar]
- Cai H.; Rose K.; Liang L. H.; Dunham S.; Stover C. (2009) Development of a liquid chromatography/mass spectrometry-based drug accumulation assay in Pseudomonas aeruginosa. Anal. Biochem. 385, 321–325. [DOI] [PubMed] [Google Scholar]
- Isono K.; Uramoto M.; Kusakabe H.; Miyata N.; Koyama T.; Ubukata M.; Sethi S. K.; McCloskey J. A. (1984) Ascamycin and dealanylascamycin, nucleoside antibiotics from Streptomyces sp. J. Antibiot. (Tokyo) 37, 670–672. [DOI] [PubMed] [Google Scholar]
- Takahashi E.; Beppu T. (1982) A new nucleosidic antibiotic AT-265. J. Antibiot. (Tokyo) 35, 939–947. [DOI] [PubMed] [Google Scholar]
- Ueda H.; Shoku Y.; Hayashi N.; Mitsunaga J.; In Y.; Doi M.; Inoue M.; Ishida T. (1991) X-ray crystallographic conformational study of 5′-O-[N-(L-alanyl)-sulfamoyl]adenosine, a substrate analogue for alanyl-tRNA synthetase. Biochim. Biophys. Acta 1080, 126–134. [DOI] [PubMed] [Google Scholar]
- Heacock D.; Forsyth C.; Shiba K.; Musier-Forsyth K. (1996) Synthesis and aminoacyl-tRNA synthetase inhibitory activity of prolyl adenylate analogs. Bioorg. Chem. 24, 273–289. [Google Scholar]
- Tao J.; Schimmel P. (2000) Inhibitors of aminoacyl-tRNA synthetases as novel anti-infectives. Expert Opin. Invest. Drugs 9, 1767–1775. [DOI] [PubMed] [Google Scholar]
- Ferreras J. A.; Ryu J. S.; Di Lello F.; Tan D. S.; Quadri L. E. (2005) Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 1, 29–32. [DOI] [PubMed] [Google Scholar]
- Cisar J. S.; Ferreras J. A.; Soni R. K.; Quadri L. E.; Tan D. S. (2007) Exploiting ligand conformation in selective inhibition of non-ribosomal peptide synthetase amino acid adenylation with designed macrocyclic small molecules. J. Am. Chem. Soc. 129, 7752–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somu R. V.; Boshoff H.; Qiao C.; Bennett E. M.; Barry C. E.; Aldrich C. C. (2006) Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. J. Med. Chem. 49, 31–34. [DOI] [PubMed] [Google Scholar]
- Lu X.; Zhou R.; Sharma I.; Li X.; Kumar G.; Swaminathan S.; Tonge P. J.; Tan D. S. (2012) Stable analogues of OSB-AMP: potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis. ChemBioChem 13, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Suk D. H.; Cai F.; Crich D.; Mesecar A. D. (2008) Bacillus anthracis o-succinylbenzoyl-CoA synthetase: reaction kinetics and a novel inhibitor mimicking its reaction intermediate. Biochemistry 47, 12434–12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras J. A.; Stirrett K. L.; Lu X.; Ryu J. S.; Soll C. E.; Tan D. S.; Quadri L. E. (2008) Mycobacterial phenolic glycolipid virulence factor biosynthesis: mechanism and small-molecule inhibition of polyketide chain initiation. Chem. Biol. 15, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciulli A.; Scott D. E.; Ando M.; Reyes F.; Saldanha S. A.; Tuck K. L.; Chirgadze D. Y.; Blundell T. L.; Abell C. (2008) Inhibition of Mycobacterium tuberculosis pantothenate synthetase by analogues of the reaction intermediate. ChemBioChem 9, 2606–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.; Tiwari D.; Wilson D. J.; Seiler C. L.; Schnappinger D.; Aldrich C. C. (2013) Bisubstrate inhibitors of biotin protein ligase in Mycobacterium tuberculosis resistant to cyclonucleoside formation. ACS Med. Chem. Lett. 4, 1213–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P.; Goyal A.; Natarajan V. T.; Rajakumara E.; Verma P.; Gupta R.; Yousuf M.; Trivedi O. A.; Mohanty D.; Tyagi A.; Sankaranarayanan R.; Gokhale R. S. (2009) Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat. Chem. Biol. 5, 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliffe I. T.; Morgan B. J. (1992) Principal component analysis and exploratory factor analysis. Stat. Methods Med. Res. 1, 69–95. [DOI] [PubMed] [Google Scholar]
- Feher M.; Schmidt J. M. (2003) Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 43, 218–227. [DOI] [PubMed] [Google Scholar]
- Wenderski T. A., Stratton C. F., Bauer R. A., Kopp F., and Tan D. S.. Principal component analysis as a tool for library design: a case study investigating natural products, brand-name drugs, natural product-like libraries, and drug-like libraries. Methods Mol. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty S. J.; Clemons P. A.; Wong J. C.; Schreiber S. L. (2004) Mapping chemical space using molecular descriptors and chemical genetics: deacetylase inhibitors. Comb. Chem. High Throughput Screening 7, 669–676. [DOI] [PubMed] [Google Scholar]
- Kojima S.; Nikaido H. (2013) Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proc. Natl. Acad. Sci. U.S.A. 110, E2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. S.; Georgopapadakou N. H. (1988) Routes of quinolone permeation in Escherichia coli. Antimicrob. Agents Chemother. 32, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyfakh A. A.; Bidnenko V. E.; Chen L. B. (1991) Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc. Natl. Acad. Sci. U.S.A. 88, 4781–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J.; Williams K. J.; Ricci V. (2000) Accumulation of rifampicin by Mycobacterium aurum, Mycobacterium smegmatis and Mycobacterium tuberculosis. J. Antimicrob. Chemother. 45, 159–165. [DOI] [PubMed] [Google Scholar]
- Yu K. H.; Barry C. G.; Austin D.; Busch C. M.; Sangar V.; Rustgi A. K.; Blair I. A. (2009) Stable isotope dilution multidimensional liquid chromatography-tandem mass spectrometry for pancreatic cancer serum biomarker discovery. J. Proteome Res. 8, 1565–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z.; Nikaido H. (2004) Efflux-mediated drug resistance in bacteria. Drugs 64, 159–204. [DOI] [PubMed] [Google Scholar]
- Lomovskaya O.; Warren M. S.; Lee A.; Galazzo J.; Fronko R.; Lee M.; Bliais J.; Cho D.; Chamberland S.; Renau T.; Leger R.; Hecker S.; Watkins W.; Hoshino K.; Ishida H.; Lee V. J. (2001) Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H.; Thanassi D. G. (1993) Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob. Agents Chemother. 37, 1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard J.; Wong S.; Bryan L. E. (1987) Accumulation of enoxacin by Escherichia coli and Bacillus subtilis. Antimicrob. Agents Chemother. 31, 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z.; Livermore D. M.; Nikaido H. (1994) Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 38, 1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer P. G.; Piddock L. J. (1991) A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28, 639–653. [DOI] [PubMed] [Google Scholar]
- Shuman D.; Robbins R. K.; Robins M. J. (1969) The synthesis of adenine 5′-O-sulfamoyl nucleosides related to nucleocidin. J. Am. Chem. Soc. 91, 3391–3392. [DOI] [PubMed] [Google Scholar]
- Qiao C.; Gupte A.; Boshoff H. I.; Wilson D. J.; Bennett E. M.; Somu R. V.; Barry C. E.; Aldrich C. C. (2007) 5′-O-[(N-acyl)sulfamoyl]adenosines as antitubercular agents that inhibit MbtA: an adenylation enzyme required for siderophore biosynthesis of the mycobactins. J. Med. Chem. 50, 6080–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakomi H. L.; Skyttä E.; Saarela M.; Mattila-Sandholm T.; Latva-Kala K.; Helander I. M. (2000) Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 66, 2001–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neres J.; Labello N. P.; Somu R. V.; Boshoff H. I.; Wilson D. J.; Vannada J.; Chen L.; Barry C. E.; Bennett E. M.; Aldrich C. C. (2008) Inhibition of siderophore biosynthesis in Mycobacterium tuberculosis with nucleoside bisubstrate analogues: structure-activity relationships of the nucleobase domain of 5′-O-[N-(salicyl)sulfamoyl]adenosine. J. Med. Chem. 51, 5349–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rossi E.; Aínsa J. A.; Riccardi G. (2006) Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30, 36–52. [DOI] [PubMed] [Google Scholar]
- Lamers R. P.; Cavallari J. F.; Burrows L. L. (2013) The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAβN) permeabilizes the outer membrane of gram-negative bacteria. PLoS One 8, e60666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neres J.; Engelhart C. A.; Drake E. J.; Wilson D. J.; Fu P.; Boshoff H. I.; Barry C. E.; Gulick A. M.; Aldrich C. C. (2013) Non-nucleoside inhibitors of BasE, an adenylating enzyme in the siderophore biosynthetic pathway of the opportunistic pathogen Acinetobacter baumannii. J. Med. Chem. 56, 2385–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.