Abstract
Despite the considerable successes of highly active antiretroviral therapy (HAART) for the treatment of HIV/AIDS, cumulative drug toxicities and the development of multidrug-resistant virus necessitate the search for new classes of antiretroviral agents with novel modes of action. The HIV-1 capsid (CA) protein has been structurally and functionally characterized as a druggable target. We have recently designed a novel small molecule inhibitor I-XW-053 using the hybrid structure based method to block the interface between CA N-terminal domains (NTD–NTD interface) with micromolar affinity. In an effort to optimize and improve the efficacy of I-XW-053, we have developed the structure activity relationship of I-XW-053 compound series using ligand efficiency methods. Fifty-six analogues of I-XW-053 were designed that could be subclassified into four different core domains based on their ligand efficiency values computed as the ratio of binding efficiency (BEI) and surface efficiency (SEI) indices. Compound 34 belonging to subcore-3 showed an 11-fold improvement over I-XW-053 in blocking HIV-1 replication in primary human peripheral blood mononuclear cells (PBMCs). Surface plasmon resonance experiments confirmed the binding of compound 34 to purified HIV-1 CA protein. Molecular docking studies on compound 34 and I-XW-053 to HIV-1 CA protein suggested that they both bind to NTD–NTD interface region but with different binding modes, which was further validated using site-directed mutagenesis studies.
Introduction
Owing to its pivotal roles in HIV-1 replication (structural and regulatory), the capsid (CA) protein has gained attention as a promising therapeutic target. Moreover, the seminal finding that retroviral species-specific host cell restriction factors target the incoming capsid core highlights the enormous therapeutic potential of targeting the capsid protein.1 The HIV-1 CA, which is translated as the central region of the Gag polyprotein, has primary functions in viral assembly and in packaging the cellular protein prolyl isomerase, cyclophilin A (CypA).2 After the capsid protein has been liberated by proteolytic processing, it rearranges into the conical core structure that surrounds the viral genome at the center of the mature virus.3 The HIV-1 capsid shell is composed of approximately 250 CA hexamers and 12 CA pentamers, comprising about 1500 monomeric CA proteins in all. The multimers interact noncovalently to form the shell’s curved surface. CA itself is composed of two domains: the N-terminal domain (CANTD) and the C-terminal domain (CACTD). Several structures of CA protein constructs have been determined including the NTD hexamer, the single CA protein, and the CANTD linked to MA, as well as several structures of a disulfide-linked CA hexamer.4 These structures reveal that six NTDs form the rigid core of hexameric CA, and six CTDs form the hexamer’s much more flexible outer ring, with dimeric interactions between CTDs of neighboring hexamers holding the capsid together.5 The structural arrangement in the disulfide-constrained hexamer has been echoed in a recent cryo-electron microscopic study of the mature capsid, with the NTD interface between the studies being extremely similar.6 In contrast, the CTD interfaces displayed much more variation between the two studies indicating an inherent flexibility in this region.
The wealth of structural information available on the HIV-1 CA has prompted the search for small molecule inhibitors of this key viral protein. Since the initial discovery of CAP-1 (N-(3-chloro-4-methylphenyl)-N′-[2-[([5-[(dimethylamino)-methyl]-2-furyl]-methyl)-sulfanyl]ethyl]urea),7 several other small molecule inhibitors of the assembly and functions of HIV-1 have been discovered including multiple CAP-1 derivatives,8 several benzodiazepine- and benzimidazole-based inhibitors,9,10 a number of diverse chemical scaffolds that target the hydrophobic cavity of the CTD of HIV-1 capsid,11 and the potent compound PF-3450074 (PF74).12 Interestingly, the binding site of PF74 partially overlaps with that of a capsid host binding protein, cleavage and polyadenylation specific factor 6 (CPSF6), which functions in pre-mRNA processing.13 Moreover, a very recent study by Lamorte et al.14 has identified an inhibitor class that binds to capsid assemblies and artificially increases its stability. This mechanism of inhibition is in contrast to other inhibitors of CA that primarily function by destabilization. Taken together, these studies demonstrate that the CA is an attractive therapeutic target and many of its intrinsic properties can be modulated by small molecules to achieve an antiviral effect.
We have previously used a structure-based drug design strategy to utilize structural and biochemical information about the capsid NTD to design inhibitors of the NTD–NTD interface that would interfere with HIV-1 replication at an early, preintegration stage. This effort yielded compound I-XW-053 (1), which was found to be specific for HIV-1, to have a large therapeutic spectrum, and to bind to HIV-1 CA with micromolar affinity.15
Despite its high micromolar activity against HIV-1, the novel chemotype and binding site on HIV-1 CA, coupled with its broad-spectrum anti-HIV activity, suggests that compound 1 would serve as a good starting point for the development of high-efficacy analogues through chemical optimization. Therefore, in this study we performed a first-stage structure–activity analysis in order to improve the efficacy of this compound while determining groups critical for its anti-HIV activity. This endeavor yielded the identification of compound 34 (5-iodo-2-(3-nitrophenyl)-1H-phenanthro[9,10-d]imidazole), which represents an 11-fold improvement in antiviral potency over the parental I-XW-053 compound.
Results and Discussion
Biological Evaluation of Compound 1 Analogues
We have previously described compound 1 as a novel antiviral that targets a unique site on the HIV-1 CA protein and inhibits by affecting virus uncoating. However, compound 1 inhibits the replication of HIV-1 in the high micromolar range. Therefore, we sought to improve upon this potency while still retaining the novel qualities of this compound. Screening for analogues of compound 1 in commercial vendor chemical libraries resulted in the identification of 56 hit molecules with a unique set of functional groups attached to the core imidazole structure (Table 1). These compounds were purchased for biological testing in a PBMC-based HIV-1 replication assay as previously described.15 We chose to evaluate the compounds against HIV-189BZ19716 as this isolate in our hands is one of the most refractory to inhibition using Gag-targeting small molecules.15,17 Compound toxicity against the peripheral blood mononuclear cells (PBMCs) was assessed in parallel using an MTS assay as previously described and was used to determine the therapeutic index (TI value; CC50/IC50) of the test molecules.15 The results from this analysis are shown in Table 1.
Table 1. Antiviral Activity and SAR of Compound 1 and its Analogues against HIV-189BZ167 Replication in PBMCsa.
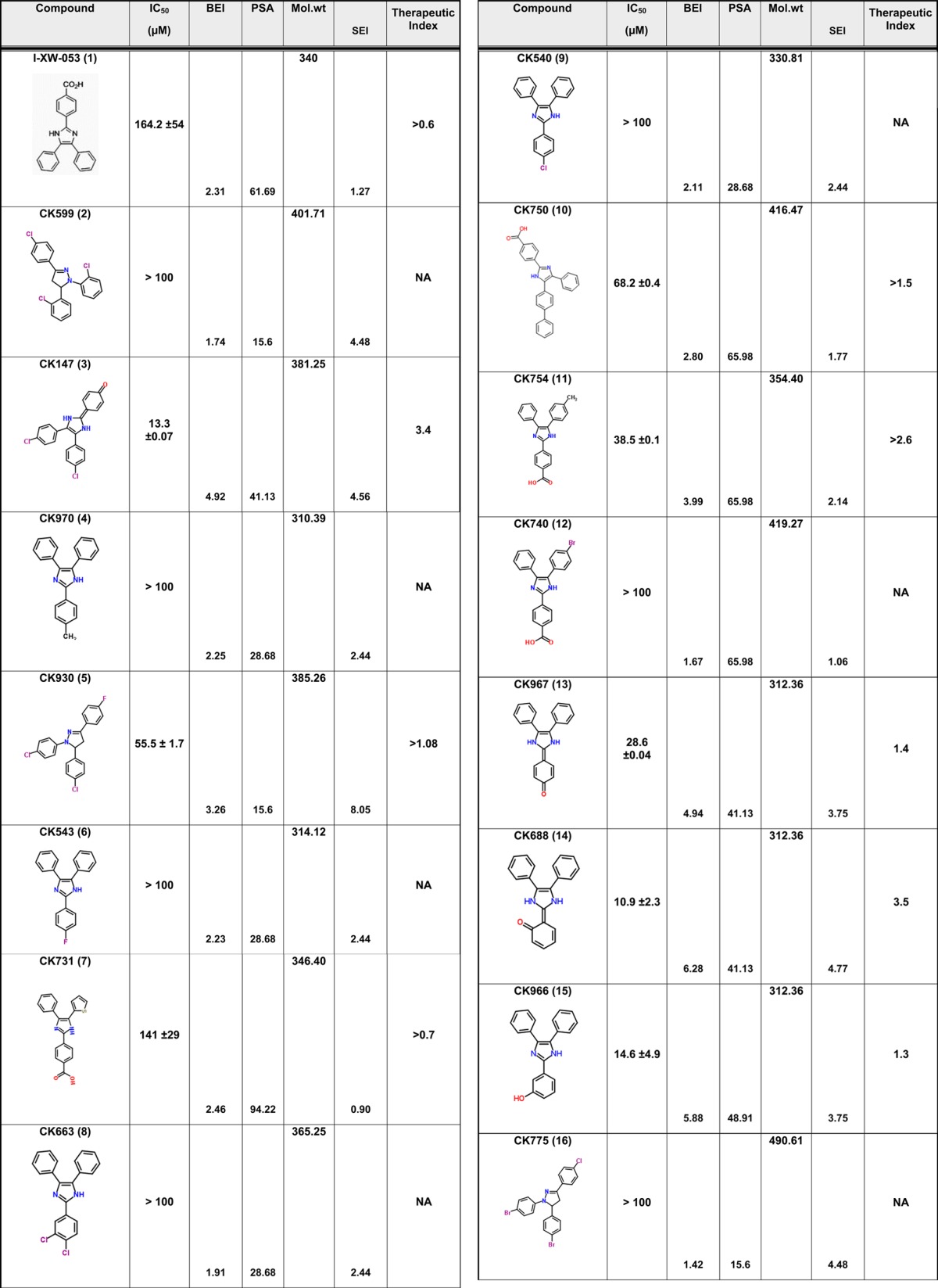
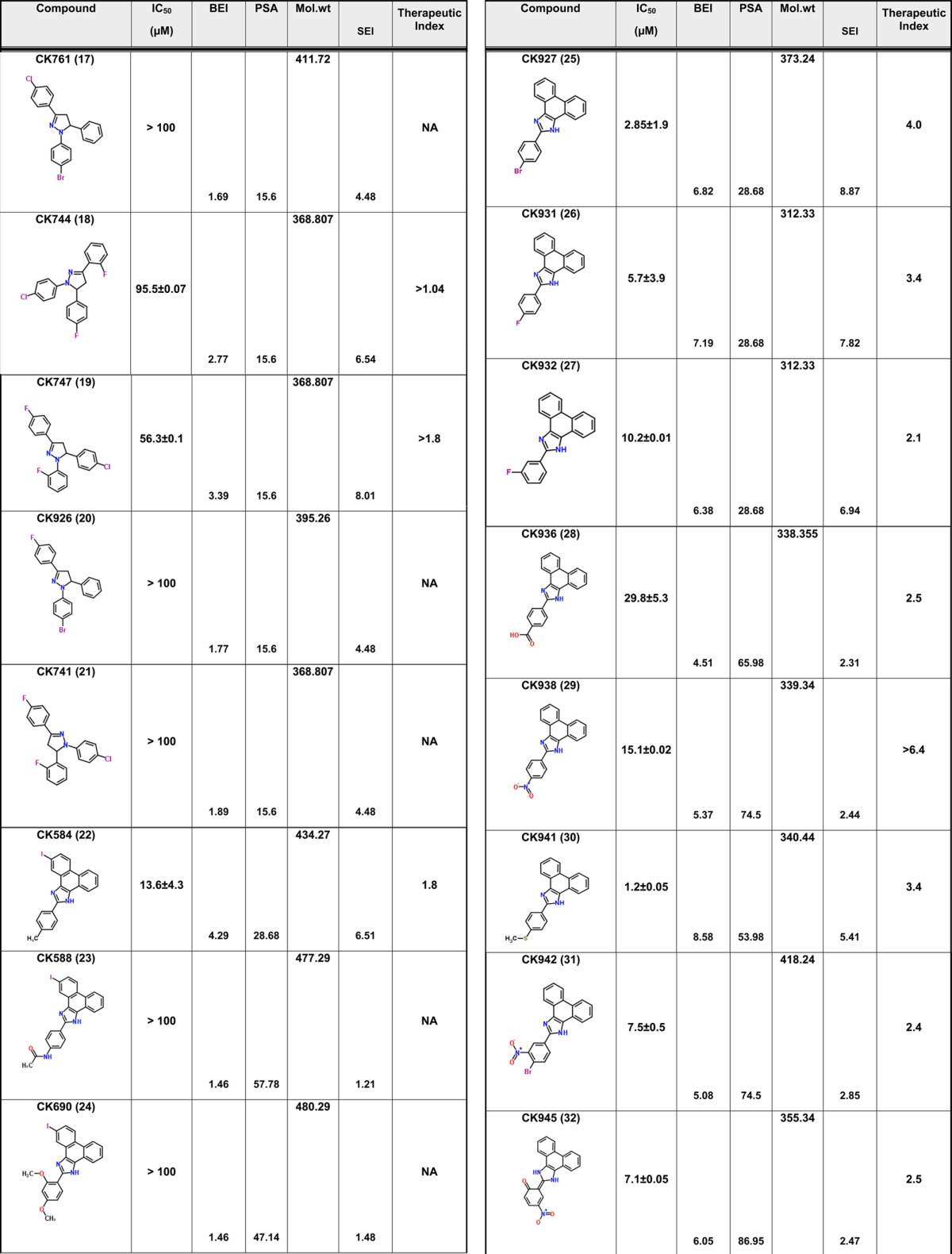
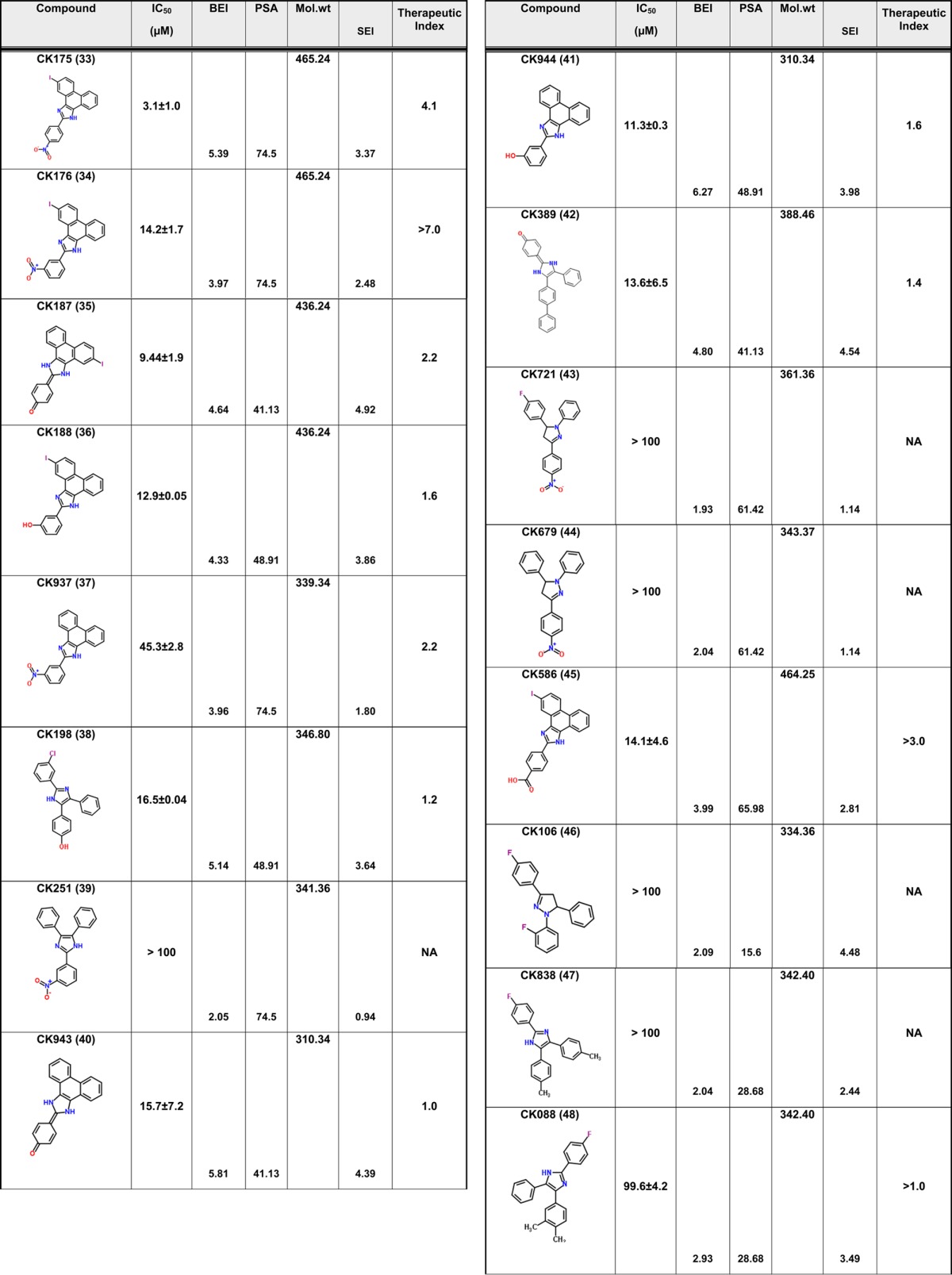
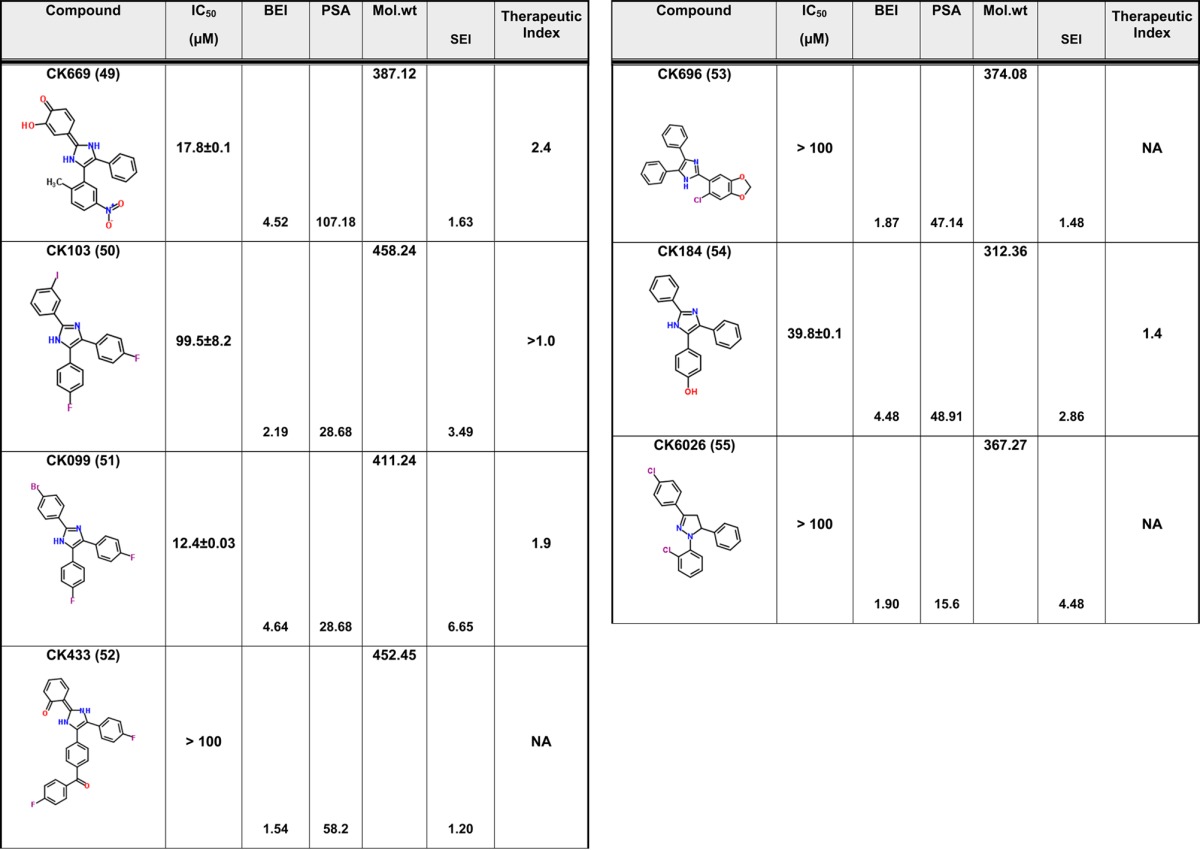
NA = not applicable. Toxicity was tested up to a maximum concentration of 100 μM.
The focused library of compound 1 analogues all share an imidazole core structure. However, they could be further subclassified into four categories: Core-1 triphenyl imidazole core (containing 31 molecules), Core-2 an imidazol-2(3H)-ylidene core with six compounds, Core-3 tetrahydro phenanthro imidazole with 16 compounds, and Core-4 phenanthro[9,10-d]imidazol-2(3H,4H,7H)-ylidene containing three molecules that had a combination of the second and third core groups. In the triphenyl imidazole core containing molecules, chemical modifications at either the ortho, para, or meta positions on any of the three phenyl rings lead to only a modest increase in potency. These compounds also had low TI values indicating that the increase in efficacy could be attributed to a concomitant increase in the toxicity of the compounds. Among the imidazol-2(3H)-ylidene core compounds, a clear SAR was observed with halogen modifications to the basal phenyl groups at the para positions leading to a 2-fold increase in potency and a >3 TI value (compound 3). Similarly, an ester group at the ortho position on one of the basal phenyl group lead to a 3-fold increase in potency and >3 TI value (compound 14). However, addition of large aryl or heteroaryl groups lead to no significant change in efficacy or toxicity (compound 42) and a loss of efficacy (compound 52). Compound 49 containing a nitro substitution on one of the basal phenyl groups had a minor effect on both efficacy and toxicity. Among the tetrahydro phenanthrolines, halogen, nitro, or thiomethyl modifications at the para position of the apical phenyl ring lead to an increase in potency with low toxicity values leading to an improvement in the TI value. In addition, the efficacy of these compounds was further improved with a halogen modification at the para position on the phenanthroline group. The nitro substitution at the para or meta position lead to a marked increase in efficacy and low toxicity as in compounds 29, 31, 33, 37, and 34. Compound 30 with the thiomethyl modification had the best efficacy but with a TI value of 3.4 indicating an increase in toxicity too. However, compound 34 demonstrated an IC50 value of 14.2 ± 1.7 μM and no toxicity up to the highest test concentration of 100 μM (Figure 1). While compound 29 had similar efficacy as compound 34, it exhibited 46.3% toxicity in cells at 100 μM. The last group that had a phenanthro[9,10-d]imidazol-2(3H,4H,7H)-ylidene core structure did not show any gain in potency or TI value. The identification of compound 34 represents an 11-fold improvement in potency over the parental compound 1 (I-XW-053).
Figure 1.
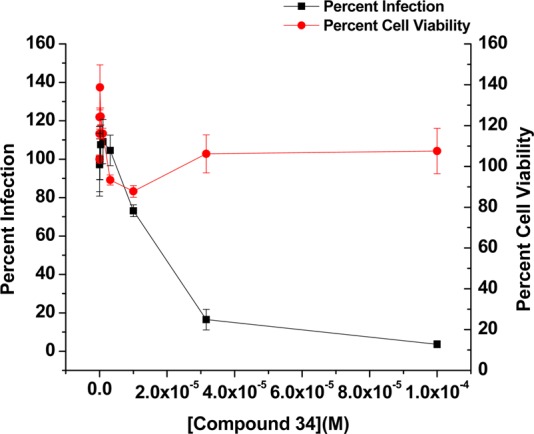
Effect of compound 34 on the replication of HIV-189BZ167 and the viability of primary human PBMCs. Black squares show the effect of compound 34 on the infection of primary human PBMCs by the clade B primary isolate HIV-189BZ167. Virus infection is expressed as the percentage of infection (measured by reverse transcriptase activity) observed in the presence of compound relative to the level of infection observed in the absence of the compound. The data from three replicates are shown. Red circles show the effect of the compound on the viability of the PBMCs and demonstrate that compound 34 is not toxic up to the highest concentration tested (100 μM). The IC50 value for compound 34 against HIV-189BZ167 = 14.2 ± 1.7 μM.
Direct Interaction of Compound 34 with HIV-1NL4-3 CA
Given the favorable properties of compound 34, we next sought to demonstrate its interaction with HIV-1 CA. We therefore employed surface plasmon resonance (SPR) analysis. HIV-1NL4-3 CA protein was purified as previously described15 and attached to the surface of a high density GLH sensor chip (Bio-Rad Laboratories, Hercules, CA) using standard amine coupling. A reference surface was created in a similar fashion using an irrelevant protein. Compound dilutions were injected simultaneously over both these surfaces and the responses recorded. Figure 2 shows the resulting sensorgrams. The compound 34–CA interaction follows a similar kinetic profile as the parental compound 1 with rapid association and rapid dissociation rates. However, unlike the parental compound, 34 appears to interact with CA with a 1:1 stoichiometry. The equilibrium dissociation constant for the compound 34–CA interaction is approximately 5- to 6-fold stronger than the compound 1–CA interaction. Moreover, there is remarkable agreement between the IC50 values and the KD values for compound 34.
Figure 2.
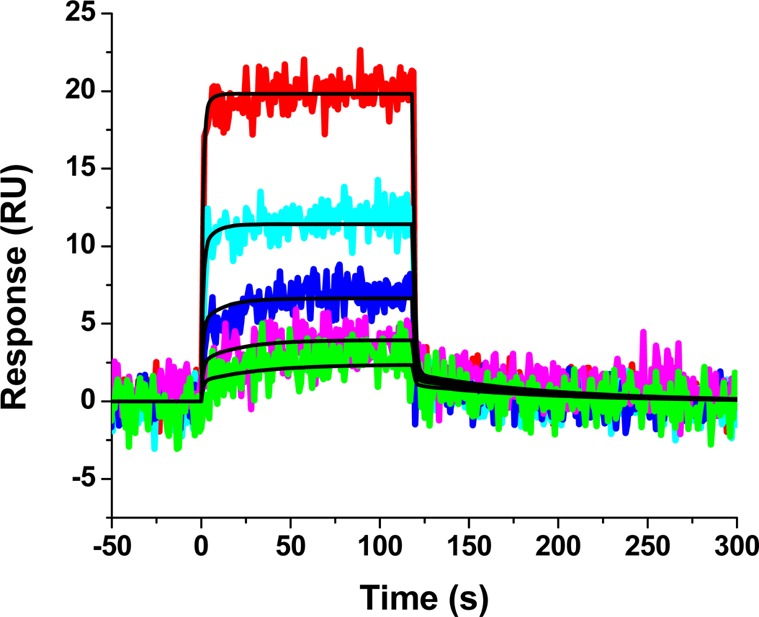
Sensorgrams depicting the interaction of compound 34 with sensorchip-immobilized HIV-1NL4-3 CA. Compound 34 at concentrations in the range 18.75–300 μM are shown. Colored lines indicate experimental data, whereas black lines indicate fitting to a simple 1:1 binding model. The individual rate constants were marginally out of the dynamic range of the instrument and are therefore not reported. The equilibrium dissociation rate constant (KD) for this interaction was determined to be 11.8 ± 4.7 μM.
Structure–Activity Relationships
Having demonstrated potency and maintenance of target specificity for compound 34, we next sought to extract structure–activity relationships (SAR) from our total data. To derive the SAR of these molecules, we calculated two ligand efficiency indices, namely binding efficiency index (BEI) and surface efficiency index (SEI),18 using the experimentally derived IC50 values and other molecular descriptors such as molecular weight (MW), p(IC50) which is computed as −log10(IC50) and polar surface area (PSA) (Table 1). BEI is defined as the ratio of p(IC50) to MW per kilodalton and SEI as the ratio of p(IC50) to PSA per 100 Å2. Since non-hydrogen atoms play a major role in both hydrophobic and hydrophilic interactions, their contributions to ligand efficiency can be measured directly in terms of the MW and the PSA of the compound. Hence the parameters BEI, which correlates binding efficiency with MW, and SEI, which correlates binding efficiency with PSA, have been hypothesized to be important measures of indices for deriving SAR of compounds.
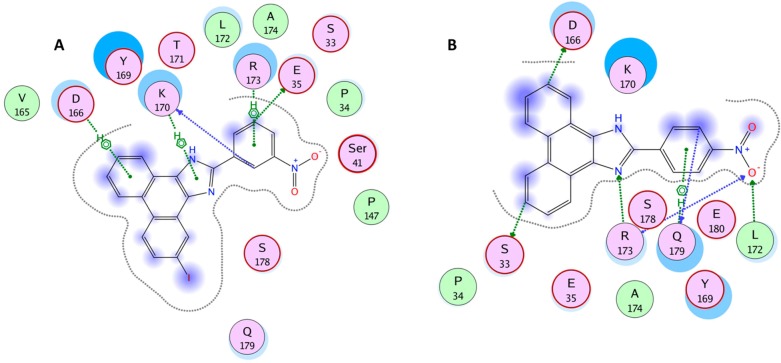
However, in our study on determining the SAR of the compounds, the BEI and the SEI values by themselves were not very confirmatory. In contrast, a ratio of BEI to SEI when plotted against TI values (Figure 3) shows a clear demarcation when a cutoff value of 1 and 3 for the BEI to SEI ratio and TI values respectively are chosen. These cutoff values were chosen based on a hypothetical molecule that has an IC50 value of 20 μM, molecular weight of 500 Da, and a PSA of 50, which resulted in a BEI to SEI ratio of 3.39. A 3-fold difference between effective versus toxic dose is a pharmacologically relevant TI value. Based on these cutoff values, seven molecules were chosen that included two molecules from core-2 (compounds 3 and 14) and five from core-3 (compounds 29, 30, 33, 34, 45). None of the molecules from core-1 or 4 were chosen under this stringent criteria suggesting that BEI, SEI, and TI can be used in this combination to derive lead compounds from a data set that seems to have a bottomless SAR. Additionally, combining core-2 and 3 did not improve the druggability of the molecules. Based on the features described in Figure 3, compounds 29 and 34 were chosen as lead compounds with compound 34 as a representative member of the core. Molecular docking studies suggested that compounds 34 and 29 bind in a similar mode with conserved interactions at the same preferred binding site as the parent compound 1(14) but with a slightly modified binding mode (Figure 4). This binding mode suggests that compounds 29 and 34 have interactions with the binding pocket residues that include strong electrostatic interactions such as arene-cation interactions with Gln179, Lys170, Arg173, Asp166 and other residues such as Tyr169, Glu35, Ser178, and Ser41. In addition, the pocket is lined by hydrophobic residues such as Ala174, Leu172, Val165, Pro34, and Pro147 that are predicted to contribute favorably to the binding of these compounds. These putative binding pocket residues are located in the regions that are important for CA NTD–NTD interaction or NTD–CTD interaction.5,19 Extensive mutagenesis of the NTD domain has been performed previous to this study. A recent study by Manocheewa et al.19a has also confirmed the sensitivity of the NTD–NTD interface to substitutions. Mutations in this area of the capsid fall broadly into two categories: (1) those that destabilized the capsid assembly and (2) those that stabilized the structure of the CA hexamer. Both of these categories have adverse effects on viral replication.20 Several of the above-mentioned residues that may constitute the binding site for compound 34 have been previously mutated and their effect on viral replication ascertained.15,20−25 Many of the residues that participate in the NTD–NTD interface when mutated have adverse effect on HIV-1 and therefore could potentially constitute a novel site of vulnerability for small molecule targeting and disruption of CA protein function.
Figure 3.
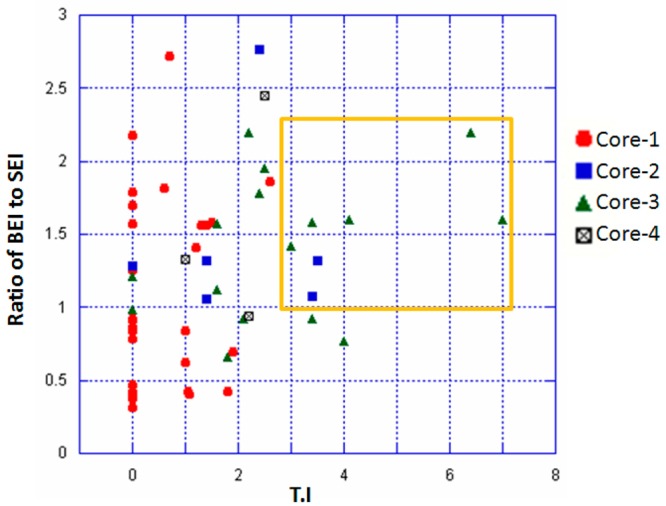
Scatter plot of the numerical values of ratio of BEI to SEI against TI values for all the compounds is shown with color coding for the various core structures. Compounds with best TI values correlating with higher BEI to SEI ratio cluster together and are represented inside an orange rectangle.
Figure 4.
Schematic representation of the binding mode of compounds 34 (A) and 29 (B) are shown. The binding site residues are colored by their nature, with hydrophobic residues in green, polar residues in purple, and charged residues highlighted with bold contours. Blue spheres and contours indicate matching regions between ligand and receptors. Hydrogen bonded interactions are shown by green arrows, ionic interactions in magenta lines, and arene–H interactions in green lines extending across the aryl rings. The figures were generated using the LIGX module of MOE program.
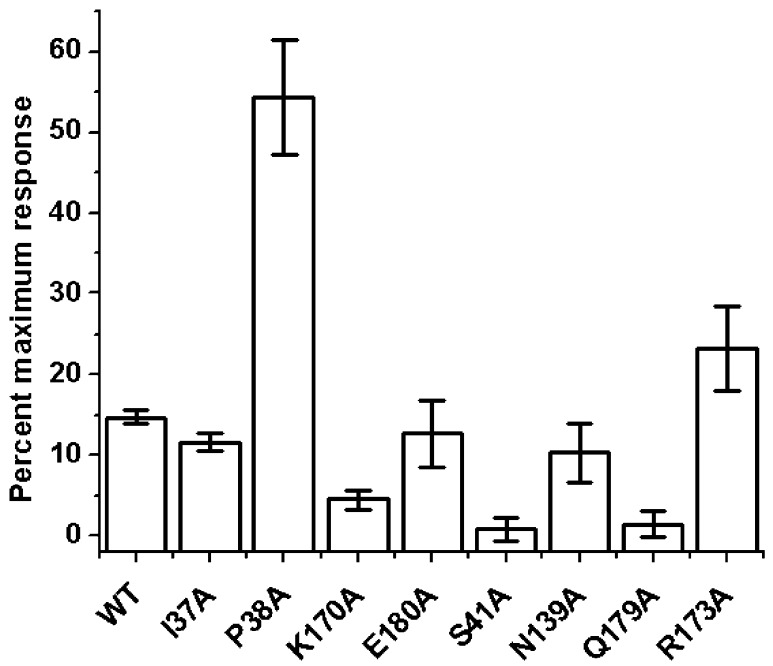
Using a combination of alanine mutagenesis and SPR, we sought to investigate this proposed binding mode. Eight mutant CA proteins containing Ile37Ala, Lys170Ala, Glu180Ala, Asn139Ala, Gln179Ala, Ser41Ala, Pro38Ala, and Arg173Ala, as well as the wild-type HIV-1NL4-3 CA protein were immobilized on a sensor chip and exposed to a single concentration of compound 34. The results of this analysis are shown in Figure 5 and suggest that residues Lys170, Ser41, and Gln179 when mutated to alanine lead to loss of favorable interactions with compound 34. However, residues Arg173 and Pro38 when mutated to alanine appeared to increase the interaction of compound 34 with HIV-1 CA. From the docking model this increase may be due to a strengthening of arene-H interactions with the ligand. Residues Ile37, Glu180, and Asn139 when mutated to alanine did not produce any significant change in binding capacity when compared to wild-type, suggesting these interactions could be compensated by other residues in the binding pocket.
Figure 5.
Effect of mutation of capsid residues in and around the proposed compound 34 binding site on compound binding. The interaction of compound 34 at a concentration of 50 μM with wild-type and mutant versions of the CA protein was assessed using SPR. To allow comparison, responses at equilibrium were normalized to the theoretical Rmax, assuming a 1:1 interaction.
The residues Lys170 and Ser41, which altered compound 34 interactions when mutated, have been studied previously.20−24a Mutation of Lys170Ala was shown by Chang et al. to have no effect on the production of virus-like particles or on the in vitro assembly of the purified protein.20 Mutation of Ser41Ala by Cartier et al. also had no effect on HIV-1 viral particle production and release.25 Similarly, mutation of Gln179 to alanine had no effect on viral replication kinetics.26 Taken together, this indicates that the reduction in binding of compound 34 to these mutations is not due to an overall gross perturbation of the native wild-type structure of the CA protein but most likely reflects disruption of individual contacts.
The mutation data also correlates with the difference in binding mode observed between the parent compound 1 and compounds 29 and 34. Mutation of residues Ile37, Asn139, and Arg173 abrogated the binding of compound 1 but had no effect on compound 34, while Lys170 had a major effect on compound 34 but not compound 1. However, unlike both compound 1 and CK026 (compound 1’s parent),15 whose modeling suggested the possibility of two potential binding sites, both compounds 29 and 34 had a strong preference for one of these binding sites.14 These “binding site preferences” suggested from the modeling studies are reflected in different stoichiometric binding ratios for compound 1 and compound 34 demonstrated using SPR and ITC.
In conclusion, we have designed a novel HIV-1 inhibitor, compound 34, which binds to a previously untargeted site on the HIV-1 CA protein. Compound 34 offers an 11-fold increase in efficacy to its parent compound 1 and was designed by utilizing insightful medicinal and computational chemistry methods. Furthermore, using biomolecular docking and SPR methods we validated that compound 34 may bind to the same site as compound 1 but with a different orientation. Compound 34 is a polycyclic compound and often there are concerns that such planar polycyclic compounds could intercalate with DNA and cause genotoxicity. However, compound 34 contains a nitro substitution that may be utilized to overcome the mutagenic potential and also add other substituents to remove the planarity of the fused rings. Similar studies were performed on lead compounds with acridine core by changing the position of the nitro group substitution that reduced the mutagenic potential of those compounds.27 Future studies will involve focused medicinal chemistry based optimization of compound 34 to improve these metabolic and toxicity liabilities. In addition, cocrystallization of these compounds with CA protein will confirm the differences in binding mode between compounds 1 and 34, to aid in structure based optimization to further improve potency.
Experimental Section
Chemicals
All chemicals were purchased from Asinex Corporation USA and used without further purification. All compounds were at a purity of 95% or greater.
Anti-HIV-1 Efficacy Evaluation in Human Peripheral Blood Mononuclear Cells
Assays involving HIV-1 infection of human peripheral blood mononuclear cells (PBMCs) were performed as described previously.15,28 Briefly, fresh PBMCs, seronegative for HIV-1 and HBV, isolated from blood samples of the screened donors (Biological Specialty Corp., Colmar, PA) by using lymphocyte separation medium (Cellgro; Mediatech, Inc., Manassas, VA), were stimulated by incubation in 4 μg/mL phytohemagglutinin (PHA; Sigma) for 48 to 72 h. Mitogenic stimulation was maintained with the addition of 20 U/mL recombinant human interleukin-2 (rhIL-2; R&D Systems, Minneapolis, MN) to the culture medium. PHA-stimulated PBMCs from at least two donors were pooled, diluted in fresh medium, and added to 96-well plates at 5 × 104 cells/well. Cells were infected with the HIV-1 group M isolate 89BZ167 (subtype B, CXCR4-tropic;16 obtained courtesy of Dr. Nelson Michael through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH [catalog no. 7692]) at a final multiplicity of infection [MOI] of ≅ 0.1 in the presence of nine different concentrations of the test compounds (triplicate wells/concentration) and incubated for 7 days. Viral replication in the presence and absence of the test compounds was determined by analysis of RT activity in the cell free supernatants.29 Concomitantly, the cytotoxicity of the test compounds was measured by the addition of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (CellTiter 96 reagent, MTS; Promega) according to the manufacturer’s instructions. The therapeutic index (TI) of the test molecules was computed as the ratio of the half maximal cytotoxicity concentration (CC50) to the half maximal inhibitory concentration (IC50).
Molecular Modeling
All molecular descriptors were computed using MOE (ver 10.1) and the IC50 values were obtained from experimental methods described above. Docking studies with the 56 test compounds along with compound 1 were performed as previously described and targeting the NTD–NTD binding interface pocket.15 Briefly, three-dimensional structures of all 56 test compounds were modeled using the ligand builder module of MOE. The compounds were optimized using semiempirical MOPAC using AM1 parameters as adapted in MOE. The crystal structure of HIV-1 capsid was obtained from protein databank (PDB code 3H4E) and was prepared for docking by adding hydrogen atoms and energy minimizing the structure as described previously.15 Molecular dockings were performed using GOLD (ver 4.1) using 20 independent runs for each ligand and the protein–ligand complexes were scored using goldscore, chemscore, and a customized consensus scoring scheme.15
Surface Plasmon Resonance Assays
All binding assays were performed on a ProteOn XPR36 SPR interaction array (Bio-Rad, Hercules, CA). ProteOn GLH sensor chips were preconditioned with two 10-s pulses each of 50 mM NaOH, 100 mM HCl, and 0.5% SDS. The system was then equilibrated with 0.005% Tween-20 in phosphate-buffered saline (PBS). Individual ligand flow channels were activated for 5 min at 25 °C with a mixture of 1-ethyl-3-[3-(dimethylamino)propyl carbodiimide hydrochloride) (0.2 M) and sulfo-N-hydroxysuccinimide (0.05 M). Immediately after chip activation, the CA protein, purified as previously described,15 was prepared at 100 μg/mL in sodium acetate pH 4.5 and injected across ligand flow channels for 5 min at a flow rate of 30 μL min–1. Then, after unreacted protein had been washed out, excess active ester groups on the sensor surface were capped by the injection of 50 μL of 1 M ethanolamine (pH 8.0) at a flow rate of 5 μL/min. A reference surface was similarly created by immobilizing a nonspecific antibody ARC4033 (antimouse/rat interferon-γ: BioSource; Invitrogen, Carlsbad, CA) and was used as background to correct nonspecific binding and for instrument and buffer artifacts. The standard errors in the immobilization levels from the six spots within each channel were less than 4%.
Direct Binding of Compounds to HIV-1 CA
A stock solution of compound 34 was prepared by dissolving in 100% DMSO to a final concentration of 10 mM. To prepare the sample for analysis, 20 μL of DMSO was added to 30 μL of the compound stock solution, which was then subsequently added to PBS-T buffer (20 mM Na-phosphate, 150 mM NaCl, and 0.005% polysorbate 20, pH 7.4) to a final volume of 1 mL and mixed thoroughly. Preparation of analyte in this manner ensured that the concentration of DMSO was matched with that of running buffer with 5% DMSO. Lower concentrations of the compound were then prepared by 2-fold serial dilutions into running buffer (PBS-T buffer (20 mM Na-phosphate, 150 mM NaCl, and 0.005% polysorbate 20, pH 7.4. 5% DMSO). These compound dilutions were then injected over the control and CA surfaces at a flow rate of 100 μL/min, for a 2 min association phase, followed by a 2 min dissociation phase using the “one-shot kinetics” functionality of the ProteOn XPR36.30 Specific regeneration of the surfaces between injections was not needed due to the nature of the interaction.
SPR Data Analysis
Data were analyzed using the ProteOn Manager Software version 3.0 (Bio-Rad). The responses of a buffer injection and responses from the reference flow cell were subtracted to account for injection artifacts and nonspecific binding.
Binding Site Analysis via SPR
Wild-type and mutant HIV-1 CA proteins were attached to the surface of a GLH sensor chip by standard amine chemistry as described above. Compound 34 was injected over these surfaces at a concentration of 50 μM at a flow rate of 100 μL/min, for a 2 min association phase, and the response at equilibrium recorded. For comparison, and to take into account minor differences in the ligand density of the mutant surfaces, responses were normalized to the theoretical maximum response (Rmax) for a given surface, assuming a 1:1 interaction. The responses from 12 replicate injections over each CA surface were analyzed.
Acknowledgments
This work was supported in part by NIH/NIAID grant 1R21AI087388-01A1 (Cocklin, PI) and laboratory start-up funds from the Department of Microbiology and Immunology (SK) and Biochemistry & Molecular Biology (SC), Drexel University College of Medicine. We would also like to acknowledge funding from the Pennsylvania department of health (Tobacco settlement funds to SK and SC).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- a Ganser-Pornillos B. K.; Chandrasekaran V.; Pornillos O.; Sodroski J. G.; Sundquist W. I.; Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U. S. A. 2011, 1082534–9. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Pertel T.; Hausmann S.; Morger D.; Zuger S.; Guerra J.; Lascano J.; Reinhard C.; Santoni F. A.; Uchil P. D.; Chatel L.; Bisiaux A.; Albert M. L.; Strambio-De-Castillia C.; Mothes W.; Pizzato M.; Grutter M. G.; Luban J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 2011, 4727343361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Stremlau M.; Owens C. M.; Perron M. J.; Kiessling M.; Autissier P.; Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004, 4276977848–53. [DOI] [PubMed] [Google Scholar]; d Stremlau M.; Perron M.; Lee M.; Li Y.; Song B.; Javanbakht H.; Diaz-Griffero F.; Anderson D. J.; Sundquist W. I.; Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U.S.A. 2006, 103145514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Hill C. P.; Sundquist W. I.; Finch J. T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 2000, 4076802409–13. [DOI] [PubMed] [Google Scholar]
- Freed E. O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 1998, 25111–15. [DOI] [PubMed] [Google Scholar]
- Pornillos O.; Ganser-Pornillos B. K.; Banumathi S.; Hua Y.; Yeager M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. Journal of molecular biology 2010, 4015985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O.; Ganser-Pornillos B. K.; Kelly B. N.; Hua Y.; Whitby F. G.; Stout C. D.; Sundquist W. I.; Hill C. P.; Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell 2009, 13771282–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.; Perilla J. R.; Yufenyuy E. L.; Meng X.; Chen B.; Ning J.; Ahn J.; Gronenborn A. M.; Schulten K.; Aiken C.; Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 4977451643–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.; Loeliger E.; Kinde I.; Kyere S.; Mayo K.; Barklis E.; Sun Y.; Huang M.; Summers M. F. Antiviral inhibition of the HIV-1 capsid protein. Journal of molecular biology 2003, 32751013–20. [DOI] [PubMed] [Google Scholar]
- a Jin Y.; Tan Z.; He M.; Tian B.; Tang S.; Hewlett I.; Yang M. SAR and molecular mechanism study of novel acylhydrazone compounds targeting HIV-1 CA. Bioorg. Med. Chem. 2010, 1862135–40. [DOI] [PubMed] [Google Scholar]; b Tian B.; He M.; Tang S.; Hewlett I.; Tan Z.; Li J.; Jin Y.; Yang M. Synthesis and antiviral activities of novel acylhydrazone derivatives targeting HIV-1 capsid protein. Bioorg. Med. Chem. Lett. 2009, 1982162–7. [DOI] [PubMed] [Google Scholar]
- a Fader L. D.; Bethell R.; Bonneau P.; Bos M.; Bousquet Y.; Cordingley M. G.; Coulombe R.; Deroy P.; Faucher A. M.; Gagnon A.; Goudreau N.; Grand-Maitre C.; Guse I.; Hucke O.; Kawai S. H.; Lacoste J. E.; Landry S.; Lemke C. T.; Malenfant E.; Mason S.; Morin S.; O’Meara J.; Simoneau B.; Titolo S.; Yoakim C. Discovery of a 1,5-dihydrobenzo[b][1,4]diazepine-2,4-dione series of inhibitors of HIV-1 capsid assembly. Bioorg. Med. Chem. Lett. 2011, 211398–404. [DOI] [PubMed] [Google Scholar]; b Titolo S.; Mercier J.-F.; Wardrop E.; von Schwedler U.; Goudreau N.; Lemke C.; Faucher A.-M.; Yoakim C.; Sundquist W.; Mason S.. Discovery of Potent HIV-1 Capsid Assembly Inhibitors. In 17th Conference on Retroviruses and Opportunistic Infections, San Francisco, Feb 16–19, 2010.
- a Fader L. D.; Landry S.; Goulet S.; Morin S.; Kawai S. H.; Bousquet Y.; Dion I.; Hucke O.; Goudreau N.; Lemke C. T.; Rancourt J.; Bonneau P.; Titolo S.; Amad M.; Garneau M.; Duan J.; Mason S.; Simoneau B. Optimization of a 1,5-dihydrobenzo[b][1,4]diazepine-2,4-dione series of HIV capsid assembly inhibitors 2: structure-activity relationships (SAR) of the C3-phenyl moiety. Bioorg. Med. Chem. Lett. 2013, 23113401–5. [DOI] [PubMed] [Google Scholar]; b Fader L. D.; Landry S.; Morin S.; Kawai S. H.; Bousquet Y.; Hucke O.; Goudreau N.; Lemke C. T.; Bonneau P.; Titolo S.; Mason S.; Simoneau B. Optimization of a 1,5-dihydrobenzo[b][1,4]diazepine-2,4-dione series of HIV capsid assembly inhibitors 1: addressing configurational instability through scaffold modification. Bioorg. Med. Chem. Lett. 2013, 23113396–400. [DOI] [PubMed] [Google Scholar]
- Curreli F.; Zhang H.; Zhang X.; Pyatkin I.; Victor Z.; Altieri A.; Debnath A. K. Virtual screening based identification of novel small-molecule inhibitors targeted to the HIV-1 capsid. Bioorg. Med. Chem. 2011, 19177–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair W. S.; Pickford C.; Irving S. L.; Brown D. G.; Anderson M.; Bazin R.; Cao J.; Ciaramella G.; Isaacson J.; Jackson L.; Hunt R.; Kjerrstrom A.; Nieman J. A.; Patick A. K.; Perros M.; Scott A. D.; Whitby K.; Wu H.; Butler S. L. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog 2010, 612e1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. J.; Fletcher A. J.; Schaller T.; Elliott T.; Lee K.; KewalRamani V. N.; Chin J. W.; Towers G. J.; James L. C. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS pathogens 2012, 88e1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamorte L.; Titolo S.; Lemke C. T.; Goudreau N.; Mercier J. F.; Wardrop E.; Shah V. B.; von Schwedler U. K.; Langelier C.; Banik S. S.; Aiken C.; Sundquist W. I.; Mason S. W. Discovery of Novel Small-Molecule HIV-1 Replication Inhibitors That Stabilize Capsid Complexes. Antimicrob. Agents Chemother. 2013, 57104622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortagere S.; Madani N.; Mankowski M. K.; Schon A.; Zentner I.; Swaminathan G.; Princiotto A.; Anthony K.; Oza A.; Sierra L. J.; Passic S. R.; Wang X.; Jones D. M.; Stavale E.; Krebs F. C.; Martin-Garcia J.; Freire E.; Ptak R. G.; Sodroski J.; Cocklin S.; Smith A. B. 3rd. Inhibiting Early-Stage Events in HIV-1 Replication by Small-Molecule Targeting of the HIV-1 Capsid. Journal of virology 2012, 86168472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Brown B. K.; Darden J. M.; Tovanabutra S.; Oblander T.; Frost J.; Sanders-Buell E.; de Souza M. S.; Birx D. L.; McCutchan F. E.; Polonis V. R. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J. Virol 2005, 79106089–101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Jagodzinski L. L.; Wiggins D. L.; McManis J. L.; Emery S.; Overbaugh J.; Robb M.; Bodrug S.; Michael N. L. Use of calibrated viral load standards for group m subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J. Clin Microbiol 2000, 3831247–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Michael N. L.; Herman S. A.; Kwok S.; Dreyer K.; Wang J.; Christopherson C.; Spadoro J. P.; Young K. K. Y.; Polonis V.; McCutchan F. E.; Carr J.; Mascola J. R.; Jagodzinsi L. L.; Robb M. L. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 1999, 3782557–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Vahey M.; Nau M. E.; Barrick S.; Cooley J. D.; Sawyer R.; Sleeker A. A.; Vickerman P.; Bloor S.; Larder B.; Michael N. L.; Wegner S. A. Performance of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping of human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J. Clin. Microbiol. 1999, 3782533–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zentner I.; Sierra L. J.; Fraser A. K.; Maciunas L.; Mankowski M. K.; Vinnik A.; Fedichev P.; Ptak R. G.; Martin-Garcia J.; Cocklin S. Identification of a small-molecule inhibitor of HIV-1 assembly that targets the phosphatidylinositol (4,5)-bisphosphate binding site of the HIV-1 matrix protein. ChemMedChem. 2013, 83426–32. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zentner I.; Sierra L. J.; Maciunas L.; Vinnik A.; Fedichev P.; Mankowski M. K.; Ptak R. G.; Martin-Garcia J.; Cocklin S. Discovery of a small-molecule antiviral targeting the HIV-1 matrix protein. Bioorg. Med. Chem. Lett. 2013, 2341132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad-Zapatero C. Ligand efficiency indices for effective drug discovery. Expert opinion on drug discovery 2007, 24469–88. [DOI] [PubMed] [Google Scholar]
- a Manocheewa S.; Swain J. V.; Lanxon-Cookson E.; Rolland M.; Mullins J. I. Fitness Costs of Mutations at the HIV-1 Capsid Hexamerization Interface. PloS one 2013, 86e66065. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Pornillos O.; Ganser-Pornillos B. K.; Yeager M. Atomic-level modelling of the HIV capsid. Nature 2011, 4697330424–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. F.; Wang S. M.; Huang K. J.; Wang C. T. Mutations in capsid major homology region affect assembly and membrane affinity of HIV-1 Gag. Journal of molecular biology 2007, 3703585–97. [DOI] [PubMed] [Google Scholar]
- Han Y.; Ahn J.; Concel J.; Byeon I. J.; Gronenborn A. M.; Yang J.; Polenova T. Solid-state NMR studies of HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 2010, 13261976–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien A. I.; Liao W. H.; Yang D. M.; Wang C. T. A domain directly C-terminal to the major homology region of human immunodeficiency type 1 capsid protein plays a crucial role in directing both virus assembly and incorporation of Gag-Pol. Virology 2006, 348184–95. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos B. K.; von Schwedler U. K.; Stray K. M.; Aiken C.; Sundquist W. I. Assembly properties of the human immunodeficiency virus type 1 CA protein. Journal of virology 2004, 7852545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a von Schwedler U. K.; Stray K. M.; Garrus J. E.; Sundquist W. I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. Journal of virology 2003, 7795439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hatziioannou T.; Cowan S.; Von Schwedler U. K.; Sundquist W. I.; Bieniasz P. D. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. Journal of virology 2004, 78116005–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier C.; Sivard P.; Tranchat C.; Decimo D.; Desgranges C.; Boyer V. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. J. Biol. Chem. 1999, 2742719434–40. [DOI] [PubMed] [Google Scholar]
- Yufenyuy E. L.; Aiken C. The NTD-CTD intersubunit interface plays a critical role in assembly and stabilization of the HIV-1 capsid. Retrovirology 2013, 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L. R.; Denny W. A. The genetic toxicology of acridines. Mutat. Res. 1991, 2582123–60. [DOI] [PubMed] [Google Scholar]
- a Lanier E. R.; Ptak R. G.; Lampert B. M.; Keilholz L.; Hartman T.; Buckheit R. W. Jr.; Mankowski M. K.; Osterling M. C.; Almond M. R.; Painter G. R. Development of hexadecyloxypropyl tenofovir (CMX157) for treatment of infection caused by wild-type and nucleoside/nucleotide-resistant HIV. Antimicrob. Agents Chemother. 2010, 5472901–9. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ptak R. G.; Gallay P. A.; Jochmans D.; Halestrap A. P.; Ruegg U. T.; Pallansch L. A.; Bobardt M. D.; de Bethune M. P.; Neyts J.; De Clercq E.; Dumont J. M.; Scalfaro P.; Besseghir K.; Wenger R. M.; Rosenwirth B. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob. Agents Chemother. 2008, 5241302–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckheit R. W. Jr.; Swanstrom R. Characterization of an HIV-1 isolate displaying an apparent absence of virion-associated reverse transcriptase activity. AIDS research and human retroviruses 1991, 73295–302. [DOI] [PubMed] [Google Scholar]
- Bravman T.; Bronner V.; Lavie K.; Notcovich A.; Papalia G. A.; Myszka D. G. Exploring ″one-shot″ kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal. Biochem. 2006, 3582281–8. [DOI] [PubMed] [Google Scholar]