SUMMARY
As cells enter mitosis, the two centrosomes separate and grow dramatically, each forming a nascent spindle pole that nucleates a radial array of microtubules. Centrosome growth (and associated microtubule nucleation surge), termed maturation, involves the recruitment of pericentriolar material components via an as yet unknown mechanism. Here we show that Cep192 binds Aurora A and Plk1, targets them to centrosomes in a pericentrin-dependent manner, and promotes sequential activation of both kinases via T-loop phosphorylation. The Cep192-bound Plk1 then phosphorylates Cep192 at several residues to generate the attachment sites for the γ-tubulin ring complex and, possibly, other pericentriolar material components, thus promoting their recruitment and subsequent microtubule nucleation. We further found that the Cep192-dependent Aurora A-Plk1 activity is essential for kinesin-5-mediated centrosome separation, bipolar spindle formation, and equal centrosome/centriole segregation into daughter cells. Thus, our study identifies a Cep192-organized signaling cascade that underlies both centrosome maturation and bipolar spindle assembly.
INTRODUCTION
Centrosomes are non-membrane-bound organelles of animal cells that function as the major microtubule (MT)-organizing centers (MTOCs) and participate in spindle assembly, cell division, polarity, and motility. Centrosomal abnormalities, both numerical and functional, have been linked to cancer and other diseases (Nigg and Raff, 2009). Centrosomes consist of one or two (depending on the cell cycle stage) centrioles surrounded by pericentriolar material (PCM). Centrioles serve as templates for the assembly of new centrosomes and cilia, while PCM nucleates MTs and determines centrosome function (Bettencourt-Dias and Glover, 2007; Mennella et al., 2013; Nigg and Raff, 2009). Prior to mitosis onset, the two centrosomes separate and grow dramatically, forming dense MT arrays involved in spindle assembly and positioning (Bettencourt-Dias and Glover, 2007; Nigg and Raff, 2009). Centrosome growth, termed maturation, is the result of the recruitment, via a hitherto unknown mechanism, of additional PCM components, including MT nucleating and organizing factors. Among them, the most prominent is the multisubunit γ-tubulin ring complex (γ-TuRC), which serves as a MT template and localizes to MTOCs via its interacting protein NEDD1 (Haren et al., 2006; Kollman et al., 2011; Luders et al., 2006).
Centrosome maturation and separation were shown to require the activity of the serine/threonine kinases Aurora A (AurA) and Plk1, which also control mitotic entry (via the adaptor protein Bora) and other aspects of cell division (Archambault and Glover, 2009; Berdnik and Knoblich, 2002; Hannak et al., 2001; Lane and Nigg, 1996; Macurek et al., 2008; Mardin and Schiebel, 2012; Nikonova et al., 2013; Seki et al., 2008). Indeed, the mitotic centrosome-localized AurA and Plk1 are phosphorylated at conserved threonine residues (T288 and T210 in human AurA and Plk1, respectively) within the activation loop (T-loop), indicative of kinase activation (Macurek et al., 2008; Nikonova et al., 2013). However, the mechanism and the role of the AurA and Plk1 activation at centrosomes have been unclear.
Centrosome maturation also requires certain coiled-coil centrosomal proteins, such as Cep192/SPD-2, pericentrin (PCNT)/PLP, and Cep215 (also called Cdk5Rap2)/Cnn, acting in interdependent and, likely, redundant pathways (Kollman et al., 2011; Mennella et al., 2013). Recent studies revealed that these proteins form highly ordered PCM layers, which are conserved from flies to humans. Based on these findings, centrosome maturation can be viewed as an expansion of the PCM inner layer formed around the radially oriented PCNT fibers into an outer matrix that includes Cep192, Cep215, PCNT, and γ-TuRC (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012). How the centrosomal proteins, mitotic kinases, and other regulators work together to form a patterned PCM that nucleates and anchors MTs remains enigmatic (Mahen and Venkitaraman, 2012; Mennella et al., 2013).
Cep192/SPD-2 is a bona fide centrosomal protein, which lies at the top of the hierarchy of protein recruitment during both centrosome maturation and centriole duplication. It determines centrosome size and it is essential for the recruitment to centrosomes of γ-TuRC and other factors and for bipolar spindle assembly (Azimzadeh et al., 2012; Decker et al., 2011; Dix and Raff, 2007; Giansanti et al., 2008; Gomez-Ferreria et al., 2007; Kemp et al., 2004; Pelletier et al., 2004; Zhu et al., 2008). Cep192 was proposed to form a scaffold upon which MT-nucleating and regulatory factors accumulate and become active during mitosis (Gomez-Ferreria et al., 2007; Gomez-Ferreria and Sharp, 2008). This hypothesis, however, has not been experimentally validated and the mechanism by which Cep192 controls PCM protein recruitment and spindle assembly remains elusive.
Using cell-free meiotic Xenopus egg extracts, we previously showed that Cep192 is a centrosome-targeting AurA cofactor and that the accumulation and oligomerization of Cep192/AurA complexes at centrosomes promotes AurA activation and MT assembly (Joukov et al., 2010). Here, we identify a multistep signaling cascade that links Cep192-mediated AurA activation to centrosome maturation and MT nucleation. In this cascade, AurA amplifies the input signal by activating Plk1 and facilitating its binding to Cep192. Plk1, in turn, acts as an effector kinase to generate the attachment sites for γ-TuRC in Cep192. Moreover, we show that the Cep192-organized AurA-Plk1 cascade is conserved in vertebrates and is essential for centrosome cycle and bipolar spindle assembly.
RESULTS
Cep192, in a complex with active AurA, recruits multiple PCM components
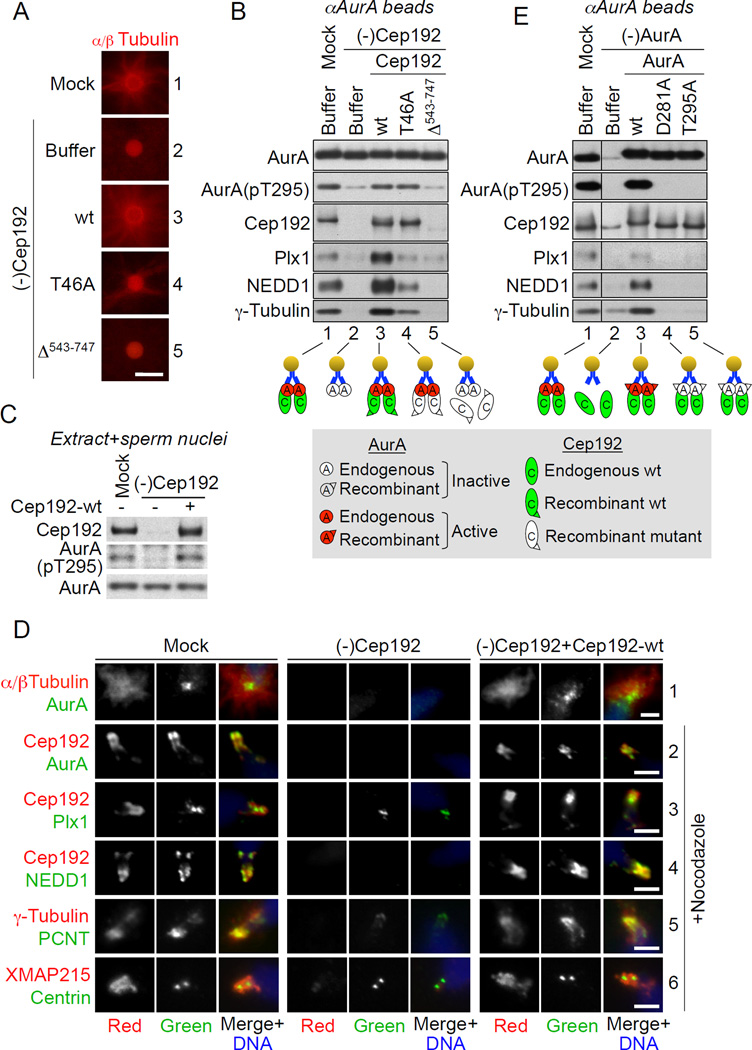
In pursuit of a tractable experimental model to study centrosome maturation, we exploited the observation that anti-AurA antibody (αAurA)-coated beads act as centrosome-like MTOCs, when added to metaphase-arrested (M-phase) Xenopus egg extract (Tsai and Zheng, 2005) (Figure 1A, panel 1). We previously reported that the MT assembly promoted by both centrosomes and αAurA beads depends on the local oligomerization of Cep192/AurA complexes and consequent kinase T-loop phosphorylation at T295 (T288 in human AurA), suggesting that a common mechanism underlies MT assembly in both settings (Joukov, 2011; Joukov et al., 2010). Consistent with this hypothesis, our mass spectrometry analysis revealed that the αAurA beads recruited multiple PCM proteins known to be implicated in centrosomal MT nucleation, including AurA, Cep192, Plx1 (Xenopus ortholog of Plk1), γ-TuRC constituents (e.g. γ-tubulin, GCP2, GCP3, GCP4, GCP6, and Mozart1), and NEDD1 (Archambault and Glover, 2009; Kollman et al., 2011; Mennella et al., 2013). Moreover, the recruitment of most proteins seemed to require the endogenous AurA/Cep192 interaction and/or AurA activity (Figure S1A and S1B). Indeed, Cep192 depletion abrogated or significantly diminished the AurA T295-phosphorylation, the recruitment of γ-TuRC (as assessed by NEDD1 and γ-tubulin) and Plx1, and the MT nucleation promoted by both αAurA beads (Figure 1A, panel 2, and Figure 1B, lane 2) and centrosomes (Figure 1C and 1D). These defects were rescued upon reconstitution of the depleted extract with Cep192-wt but not with Cep192 lacking the AurA-binding domain (AurA-BD) (Cep192-Δ543–747) (Figure 1A-1D and see below). Thus, all three processes - AurA activation, recruitment of PCM components, and MT assembly - are mediated by the Cep192/AurA complexes.
Figure 1. Cep192, in a complex with active AurA, recruits multiple PCM proteins.
(A) Fluorescence microscopy of αAurA beads incubated in mock-treated and Cep192-depleted [(-)Cep192] extracts supplemented with rhodamine tubulin and with extract buffer (XB) or the indicated recombinant full-length Cep192 proteins. Scale bar, 5 µm.
(B) Western blot (W-blot) of aAurA beads retrieved from nocodazole-supplemented extracts treated as in (A). Cep192-T46A and Cep192-Δ543–747 are Cep192 mutants lacking the Plx1-docking threonine 46 and the AurA-BD, respectively.
(C) W-blot of M-phase mock-treated and Cep192-depleted extracts supplemented with XB or recombinant full-length Cep192-wt and with sperm nuclei.
(D) Immunofluorescence (IF) of sperm centrioles in extracts treated as in (C) in the absence/presence of nocodazole. Scale bars, 2.5 µm.
(E) W-blot of aAurA beads incubated in mock-treated and AurA-depleted extracts supplemented with XB or the indicated AurA proteins. AurA depletion was performed using a bead-immobilized AurA-binding Cep192 fragment (Cep192521–757), instead of αAurA beads, to prevent co-depletion of endogenous Cep192.
In (B) and (E), the lower panels schematically illustrate bead-protein interactions. W-blots of extracts used in these experiments are shown in Figures S1C and S1D.
See also Figure S1.
To test whether PCM protein recruitment required Cep192-mediated AurA activation, we performed the aAurA bead assay in M-phase extract in which endogenous AurA was depleted and replaced with its recombinant wild type (AurA-wt), kinase-dead (AurA-D281A), or non-phosphorylatable (AurA-T295A) (Eyers et al., 2003) counterparts. Although the wt and enzymatically-deficient AurA proteins bound equal amounts of Cep192, only AurA-wt was T295-phosphorylated and, when complexed to Cep192, recruited Plk1 and γ-TuRC (Figure 1E, lanes 3–5). Hence, Cep192 enables AurA T295 phosphorylation and, in a complex with active AurA, promotes the recruitment of PCM proteins and, thus, MT assembly.
The N-terminal Cep192 fragment promotes MTOC formation in M-phase extract
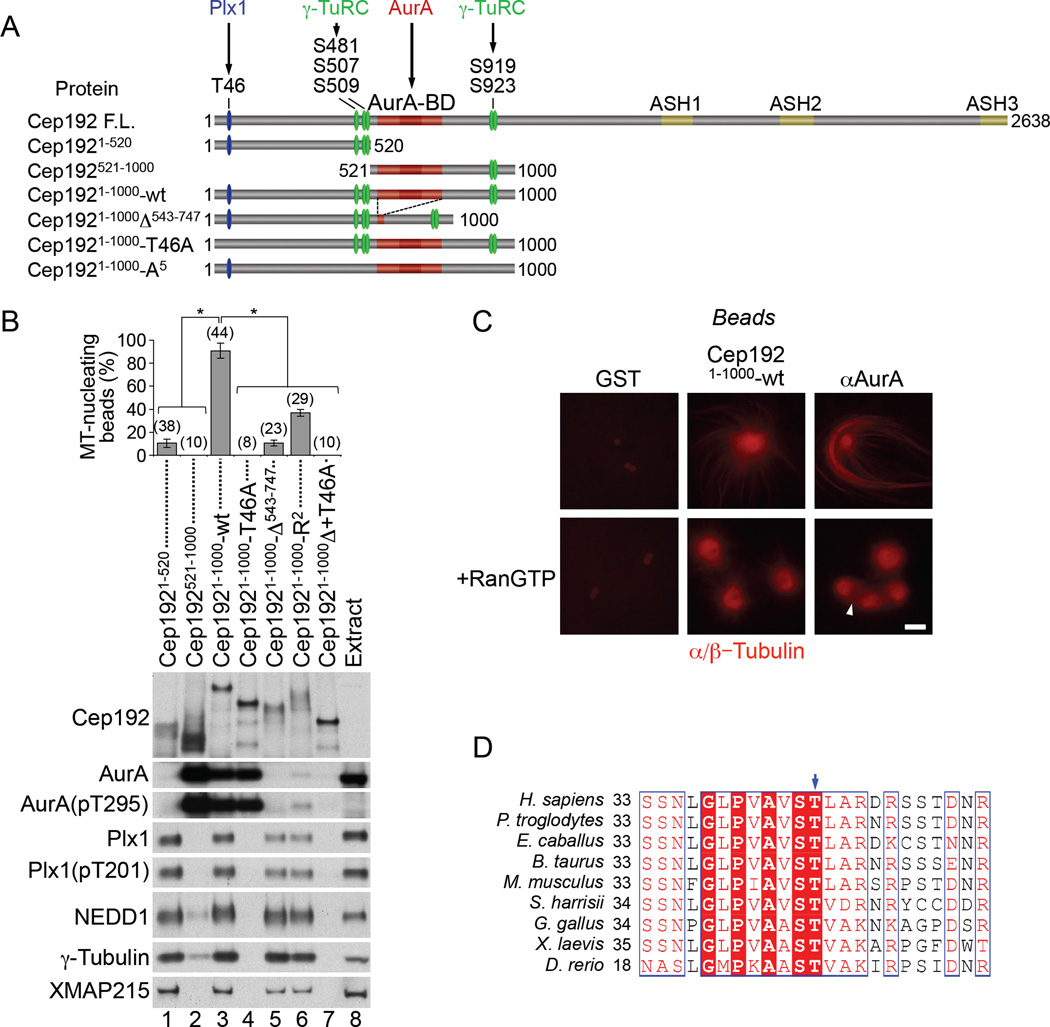
The predominant isoform of Cep192 in human and Xenopus cells is a large (~280 kDa and 290 kDa, respectively) protein (Joukov et al., 2010; Sonnen et al., 2013). To identify the Cep192 region(s) involved in PCM protein recruitment and in MT assembly, we performed pull-down assays in M-phase extract using GST-tagged fragments of the 290-kDa Xenopus Cep192 protein (Joukov et al., 2010) pre-bound to anti-GST antibody-coated beads (Figure S2A and S2B). An N-terminal 1000 amino acid (aa) Cep192 fragment (Cep1921–1000-wt) containing the AurA-BD recruited AurA, Plx1, γ-TuRC, and XMAP215, a processive MT polymerase (Al-Bassam and Chang, 2011) (Figure 2A and 2B, lane 3). Both AurA and Plx1 in Cep1921–1000-wt complexes were T-loop-phosphorylated. However, while Plx1 phosphorylation at T201 (T210 in human Plk1) was already high in M-phase extract and unaffected by the binding of Plx1 to Cep1921–1000-wt beads, AurA phosphorylation at T295 was the result of its oligomerization on the Cep1921–1000-wt bead surface (Figure 2B, lane 8 vs. 3 and Figure S2C, S2D), as already observed for centrosomes and αAurA beads (Joukov et al., 2010). Notably, in M-phase extract, the Cep1921–1000-wt beads also acted as MTOCs, thus behaving like the αAurA beads, except that they did not assemble bipolar spindle-like structures in the presence of an excess of RanGTP (Figure 2C, quantified in Figure S2E). The centrosome-like behavior of the Cep1921–1000-wt beads did not require endogenous Cep192 (Figure S2F and S2G). Thus, Cep192 promotes MT assembly through its N-terminal 1000-aa domain, which binds AurA, Plx1, γ-TuRC, and XMAP215.
Figure 2. The N-terminal domain of Cep192 promotes PCM protein recruitment and MT assembly.
(A) Schematic domain structure of full-length Cep192 and its N-terminal fragments.
(B) W-blot of proteins pulled down from M-phase extract with beads preloaded with GST-tagged Cep192 fragments. The graph shows the mean percentage of bead-induced MT asters +/− SD. The number of beads analyzed is in parentheses. *P<0.001.
(C) Fluorescence microscopy of anti-GST beads (pre-loaded with GST alone or GST-Cep1921–1000-wt) and of αAurA beads incubated in rhodamine tubulin-supplemented M-phase extract without/with Ran(Q69L)GTP. An arrowhead points to a bipolar spindlelike structure. Scale bar, 10 µm.
(D) Multiple amino acid sequence alignment of the Cep192 region surrounding the Plk1-docking threonine (T46 in Xenopus Cep192) (arrow).
See also Figures S2-S6.
Cep192 promotes sequential AurA-Plx1 activation and serves as a substrate for both kinases
Given the above results, we used the Cep1921–1000-wt beads as a defined template to elucidate the mechanism underlying mitotic MTOC formation. We first examined how AurA and Plx1 bind to Cep192. In egg cytoplasm, all Cep192 is bound to a fraction of AurA via a direct and constitutive interaction (Joukov et al., 2010). The Cep192/AurA binding was reduced by substitution to arginine of two conserved hydrophobic Cep192 residues, F629 and I639 (Cep1921–1000-R2) within the AurA-BD (aa 543–747) and abolished by deletion of the entire AurA-BD (Cep1921–1000-Δ543–747) (Figure 2B, lanes 5 and 6 and Figure S3).
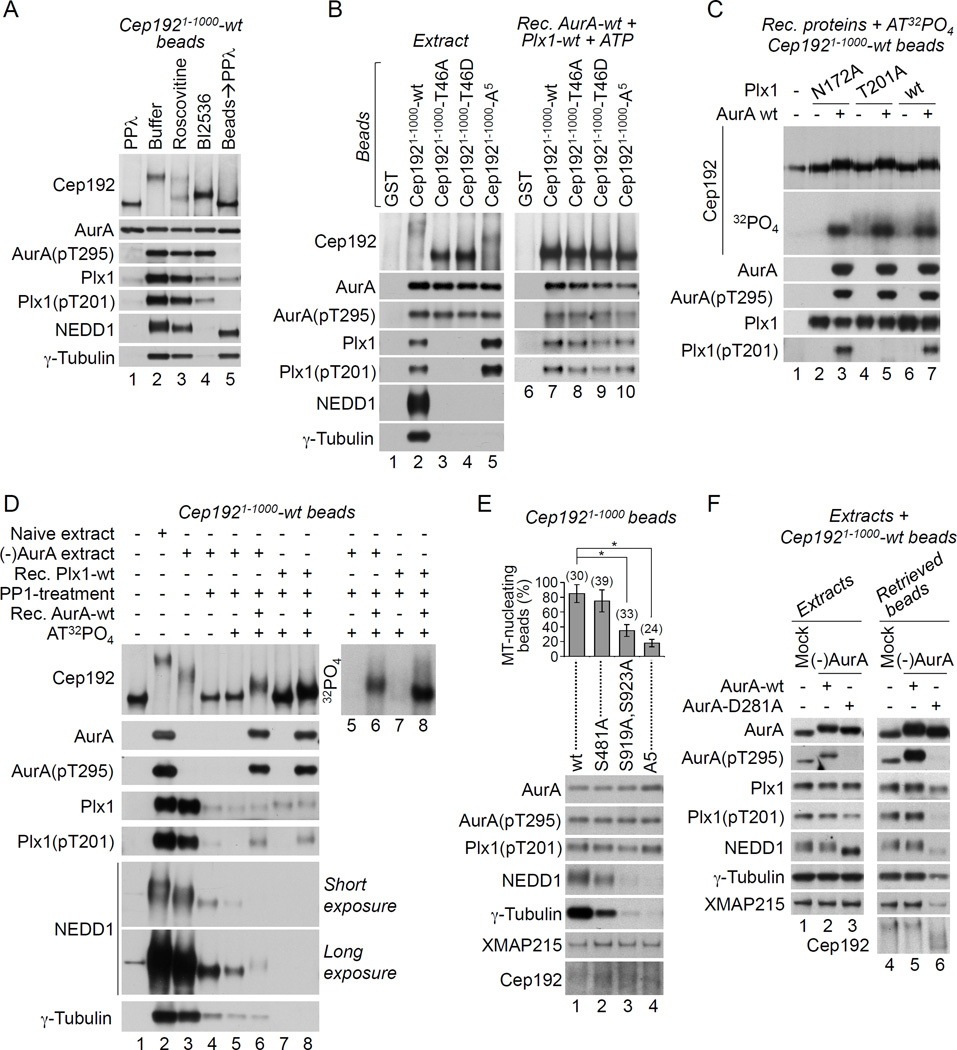
Plk1/Plx1 is known to bind cognate substrates through its phosphopeptide-binding polo-box domain (PBD) that recognizes the core consensus motif S-[pS/pT]-P/X, which is usually phosphorylated by a priming kinase or by Plk1/Plx1 itself (self-priming) (Elia et al., 2003; Park et al., 2010). Using binding assays in egg extract in combination with deletion mapping and site-directed mutagenesis, we identified the highly conserved residue, T46, within a PBD-interacting motif of Cep1921–1000-wt, as Cep192’s Plx1-docking site (Figure 2B, lane 4, 2D, and Figures S4A–S4D). The Plx1/Cep1921–1000-wt interaction was inhibited by λ protein phosphatase (PPλ) treatment (Figure 3A) and it was abolished by the substitution of T46 in Cep1921–1000 with either alanine or aspartate (Figure 3B, lanes 2–4). Notably, the sequence surrounding T46 (Figure 2D) does not conform to the consensus site for Cdk1/2 kinases, which frequently prime Plk1/substrate binding (Park et al., 2010) and were suggested to prime Plk1/SPD-2 interaction in C. elegans (Decker et al., 2011). Moreover, the Plx1/Cep1921–1000-wt interaction in extract was dramatically reduced in the presence of a selective inhibitor of Plk1/Plx1 (BI2536) but not of Cdk1 (roscovitine) (Figure 3A), suggesting that Plx1 itself may phosphorylate Cep192 at T46. Nevertheless, recombinant Plx1 bound Cep1921–1000-wt in vitro independently of either T46 (Figure 3B, lanes 7–9 vs. 2–4) or Plx1 activity (Figure 3C), arguing against this hypothesis. Thus, the selective docking of Plx1 onto T46 of Cep1921–1000-wt requires both Plx1 activity and an additional cytoplasmic factor(s), possibly another kinase that is activated by Plx1 and phosphorylates T46.
Figure 3. Cep192 organizes AurA and Plx1 in a kinase cascade that drives γ-TuRC recruitment.
(A) W-blot of Cep1921–1000-wt beads incubated in M-phase extracts supplemented as indicated (lanes 1–4). Lane 5: the beads in lane 2 were treated with PPλ and washed in XB 0.1% Tween 20.
(B) W-blot of beads pre-loaded with the indicated GST-proteins and incubated in M-phase extracts (left panel) or in XB supplemented with ATP and recombinant AurA-wt and Plx1-wt proteins (right panel).
(C) W-blot and autoradiogram (32PO4) of Cep1921–1000-wt beads incubated with AT32PO4 in the absence/presence of recombinant AurA-wt and of recombinant Plx1-wt or its enzymatycally inactive (N172A) or non-phosphorylatable (T201A) counterparts. Note that diffuse phosphorylated Cep192 forms are generated by Plx1-wt and Plx1-T201A but not by Plx1-N172A (32PO4).
(D) W-blot (left panel) and autoradiogram (right panel) of Cep1921–1000-wt beads incubated in XB (lane 1), in naïve (lane 2) or AurA-depleted (lanes 3–6) M-phase extracts, or in XB supplemented with recombinant Plx1-wt (lanes 7, 8). In lane 4, the retrieved beads were treated with PP1. In lanes 5–8, the beads were retrieved, treated with PP1, washed, and incubated in the presence of AT32PO4 without/with recombinant AurA-wt.
(E) W-blot of beads pre-loaded with Cep1921–1000-wt or with its mutants lacking one (S481), two (S919 and S923), or five (A5) γ-TuRC-binding serines after the incubation in M-phase extract. The graph shows the mean percentage of bead-induced MT asters +/− SD. The number of beads analyzed is in parentheses. *P<0.001.
(F) W-blots of mock-treated and AurA-immunodepleted extracts supplemented with recombinant AurA-wt or AurA-D281A and with Cep1921–1000-wt beads (left panel) and of Cep1921–1000-wt beads retrieved from these extracts (right panel).
See also Figure S5.
Remarkably, upon their binding to the Cep1921–1000-wt beads, the recombinant AurA-wt and Plx1-wt underwent T-loop phosphorylation at T295 and T201, respectively (Figure 3C, lane 7). Both phosphorylation events required the activity of AurA (Figure S5A, lane 3 vs. 4 and S5B) but not of Plx1 (Figure 3C, lane 7 vs. 3). These findings and the fact that the T295 of AurA lies within an optimal consensus sequence for AurA phosphorylation (Cheeseman et al., 2002) indicate that the oligomerization of Cep192 complexes promotes AurA autophosphorylation at T295 followed by the phosphorylation of Plx1 at T201 by AurA.
Next, we asked whether Cep192 is a substrate of the bound, active AurA and/or Plx1. The recombinant AurA and Plx1 phosphorylated Cep192 in vitro (Figure 3C, panel “32PO4”). Moreover, mass spectrometry revealed extensive phosphorylation of endogenous Cep192 in a complex with active AurA and Plx1 (Figure S1A, legend). Likewise, Cep1921–1000-wt was phosphorylated in egg extract, as evidenced by the dramatic increase of its electrophoretic mobility after PPλ treatment (Figure 3A). Furthermore, the electrophoretic mobility of Cep1921–1000-wt was significantly enhanced by the addition of the Plk1 inhibitor BI2536 to the extract (Figure 3A), by point mutations at Cep192’s Plk1- or AurA-binding sites (Figure 3B, lanes 2–4 and 2B, lanes 4 and 6 vs. 3), or by AurA immunodepletion (Figure 3D, lane 3 vs. 2). Thus, following their sequential activation in Cep192 complexes, both AurA and Plx1 phosphorylate Cep192.
To determine whether the T46-specific Plx1/Cep192 docking, which occurs in egg extract, affects protein phosphorylation in Cep192 complexes, we preloaded the Cep1921–1000-wt beads with either the endogenous or the recombinant Plx1 by incubating the beads in AurA-depleted M-phase extract or with recombinant Plx1-wt, respectively (Figure 3D, lanes 5–8). We then dephosphorylated the bead-bound Cep1921–1000-wt/Plx1 complexes with protein phosphatase 1 (PP1) and performed in vitro kinase assays with AT32PO4 in the absence/presence of recombinant AurA-wt. We found that Cep1921–1000-wt promoted AurA and Plx1 activation and itself underwent phosphorylation irrespective of whether it was bound to the endogenous or recombinant Plx1 (i.e. whether Plx1 was docked onto T46 or not) (lanes 6 and 8). Notably, in the presence of endogenous Plx1, Cep1921–1000-wt had a slower electrophoretic mobility (lane 6 vs. 8). Thus, the T46-selective Plx1/Cep192 docking appears to be critical for the modality (e.g. efficiency and/or site-specificity) of Cep192 phosphorylation by Plx1 and not for the AurA-Plx1 activation in Cep192 complexes.
Phosphorylation by Plx1 generates γ-TuRC-binding sites on Cep192
Interestingly, the recruitment of γ-tubulin and NEDD1 by Cep1921–1000-wt was abolished by PPλ or BI2536 (Figure 3A, lanes 1 and 4) or upon mutation of T46 (Figure 3B, lanes 3 and 4). Cep192 lacking T46 also failed to bind XMAP215 (Figure 2B, lane 4). These data suggested that the T46-docked Plx1 phosphorylates Cep192 to generate the binding sites for γ-TuRC and XMAP215. By deletion analysis, we mapped two γ-TuRC-binding domains and an XMAP215-binding region in Cep192 (Figure S4A and S4C-S4E). Using site-directed mutagenesis, we identified five serines (S481, S507, S509, S919, and S923) that specifically and additively contribute to γ-TuRC recruitment (Figures 3E and S4F). Mutation of all five serines to alanines (Cep1921–1000-A5) did not impair AurA and Plx1 binding or activity but abrogated γ-TuRC recruitment by Cep192 complexes (Figure 3B, lane 5 and 3E, lane 4). Consistent with the loss of phosphorylation sites, Cep1921–1000-A5 had a faster electrophoretic migration than Cep1921–1000-wt in egg extract (Figure 3B, lane 5 vs. 2). Collectively, these data indicate that Plx1 docks onto T46 and phosphorylates Cep1921–1000-wt at five serines to generate γ-TuRC attachment sites.
The Cep192-organized AurA-Plx1 cascade drives γ-TuRC recruitment and MT assembly
The above experiments suggest that Cep192, via its N-terminal domain, organizes AurA and Plx1 in a multistep cascade that links AurA activation to γ-TuRC recruitment. Specifically, the oligomerization of Cep192 complexes promotes AurA autophosphorylation at T295. The active AurA then phosphorylates Plx1 at T201. The resulting Plx1 activation enables its docking onto T46 of Cep192. The active, Cep192-bound Plx1, in turn, phosphorylates Cep192 at five serines to generate the attachment sites for γ-TuRC and, possibly, XMAP215, followed by the recruitment of these PCM components. AurA and/or Plx1 may also phosphorylate NEDD1 in Cep192 complexes, as evidenced by the electrophoretic mobility shift of NEDD1 that was sensitive to both phosphatase treatment and the presence of active AurA and/or Plx1 (Figure 3D, lanes 2–6).
Notably, in naïve M-phase extract, the Cep192-mediated γ-TuRC recruitment was critically dependent on the presence in Cep192 of the docking sites for Plx1 and γ-TuRC (Figure 3B, lanes 2–5) but not on the Cep192/AurA binding (Figure 3D, lane 3 and Figure 2B, lanes 1, 5, and 6). Conceivably, in M-phase, AurA is not required for Plx1 activation in Cep192 complexes because the cytoplasmic Plx1 is already extensively phosphorylated at T201. Since Plx1 activity in the cytoplasm fluctuates during the cell cycle, being low in interphase and high in M-phase (see below, Figure 4B), we speculated that AurA might be required for Cep192 function when the activity of Plx1 in the cytoplasm is low (e.g. in G2 phase, when centrosome maturation occurs). To test this hypothesis, we exploited our observation that the level of Plx1(pT201) in M phase extract was reduced by ~50% when AurA-wt was replaced by its kinase-dead counterpart, AurA-D281A (Figure S5C and S5D). In such an extract, in which the Cep192-mediated AurA activation could not occur, all the downstream steps of the Cep192-organized kinase cascade were compromised (Figure 3F, lane 6; see also Figure 1E, lane 4). Specifically, Cep192/AurA-D281A complexes contained reduced levels of unphosphorylated Plx1, as well as of γ-TuRC and XMAP215. Moreover, NEDD1 in these complexes had a faster electrophoretic migration, consistent with impaired phosphorylation (Figure 3F, lane 6). These data imply that the AurA activity in Cep192 complexes drives the Cep192-organized kinase cascade and may, therefore, allow γ-TuRC recruitment when the activity of Plx1 in the cytoplasm is low.
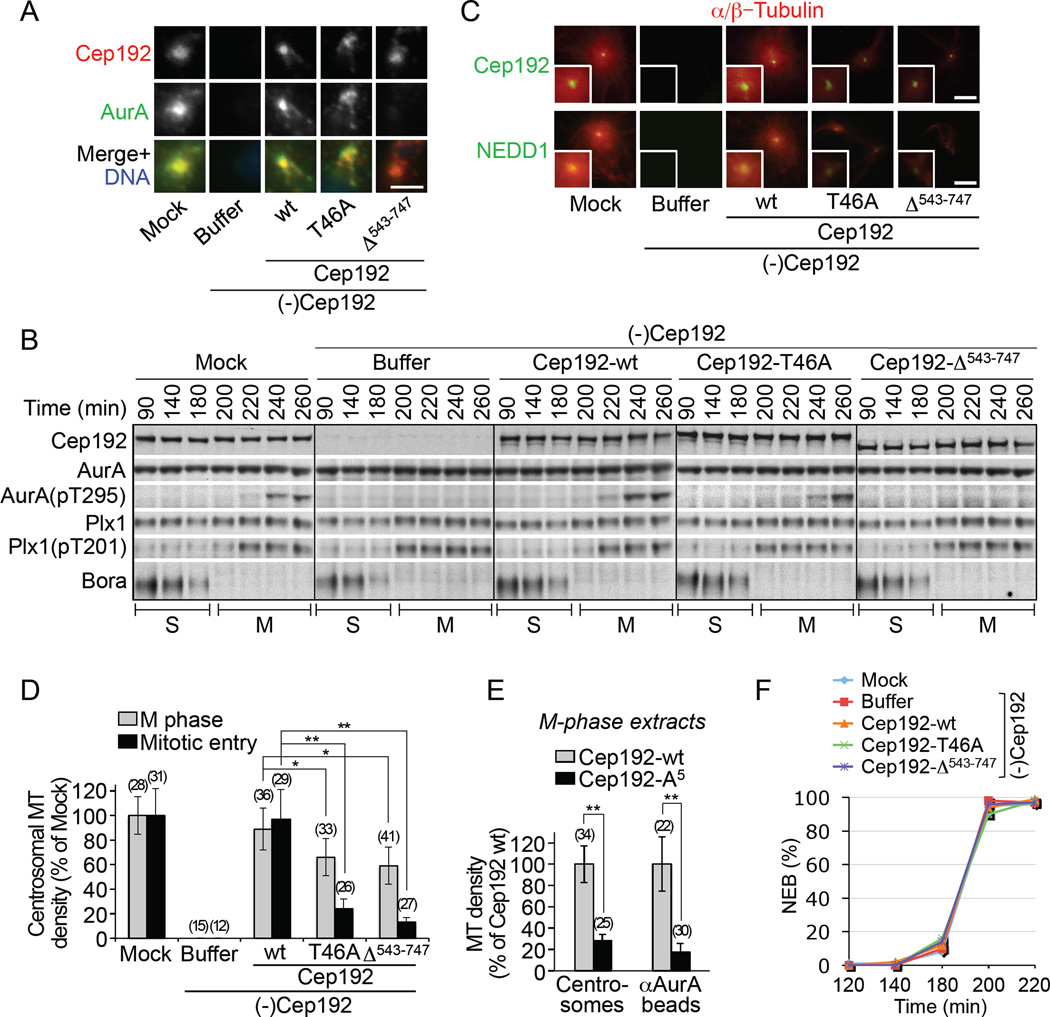
Figure 4. The Cep192-organized AurA-Plx1 cascade is essential for centrosome maturation in cycling egg extracts.
(A) IF of sperm centrioles incubated in M-phase mock-treated and Cep192-depleted extracts supplemented with XB or the indicated full-length Cep192 proteins. Scale bar, 2.5 µm.
(B) W-blots of cycling extracts treated as in (A) and analyzed at the indicated times after Ca2+ addition.
(C) IF of MT asters assembled by sperm centrioles in extracts in (B) entering mitosis (210 min time point). Scale bars, 5 µm.
(D) Average density of centrosomal MTs in egg extracts arrested in M-phase (grey columns) and entering mitosis (black columns) (+/− SD).
(E) Average density of MTs assembled by centrosomes and αAurA beads in Cep192-depleted extract supplemented with full-length Cep192-wt or Cep192-A5 (+/− SD).
In (D) and (E), the number of MT structures analyzed is shown in parentheses. *P<0.05; **P<0.001.
(F) Timing of the nuclear envelope breakdown (NEB) in extracts in (B).
See also Figure S6.
It seemed paradoxical, however, that the Cep192-bound Plx1 lost its T201 phosphorylation when AurA-D281 was present in Cep192 complexes (Figure 3F, lane 6 vs. 3) but not when AurA was absent altogether from these complexes (Figure 3D, lane 3 and Figure 2B, lanes 1 and 5; quantified in Figure S5E). Conceivably, AurA, which was shown to interact with several phosphatases (Nikonova et al, 2013), binds to and counteracts a phosphatase capable of dephosphorylating Plx1(pT201). Such a phosphatase would be absent in Cep192/Plx1 complexes lacking AurA and present, but not counteracted by AurA-D281A, in Cep192/AurA-D281A/Plx1 complexes, thus explaining our results.
We next examined the role of the Cep192-organized AurA-Plx1 cascade in MTOC formation. We found that, unlike Cep1921–1000-wt, its mutant counterparts lacking the binding sites for AurA, Plx1, or γ-TuRC failed to promote MT assembly in M-phase extract (Graphs in Figure 2B and 3E and Figure S6A). The observation that in the absence of AurA binding, Cep192 recruited γ-TuRC and XMAP215 but did not promote MT assembly (Figure 2B, lanes 1 and 5) suggests that the AurA-dependent phosphorylation of NEDD1 (Figures 3D and 3F) and/or of other γ-TuRC subunits renders γ-TuRC capable of MT nucleation in Cep192 complexes. Thus, the Cep192-organized AurA-Plx1 cascade drives γ-TuRC recruitment and MT assembly.
The Cep192-organized AurA-Plx1 cascade is essential for centrosome maturation in cycling egg extracts
To ascertain the role of the Cep192-organized AurA-Plx1 cascade in the centrosomal function of Cep192, we replaced endogenous Cep192 in M-phase extract with recombinant full-length Cep192-wt or with its mutant counterparts lacking the AurA-BD (Cep192-Δ543–747) or the Plx1 -docking T46 (Cep192-T46A) (Figure S6B). We then supplemented the extracts with sperm nuclei without or with Ca2+ [to avoid or induce cell cycle progression, respectively (Murray, 1991)] and analyzed centrosome function during M phase arrest and at mitotic entry. While Cep192-wt, Cep192-T46A, and Cep192Δ543–747 were all recruited to centrosomes, only the former two proteins co-recruited AurA and promoted its phosphorylation at T295 (Figures 4A, 4B, and Figure S6C). These results provide direct proof that the Cep192/AurA binding is essential for both AurA centrosomal localization and its phosphorylation at T295.
Notably, in M-phase extract, Cep192-T46A and Cep192-Δ543–747 retained 74% and 66%, respectively, of the MT-nucleating activity of Cep192-wt (Figure 4D, gray columns and Figure S6D, upper panel), whereas a full-length Cep192 mutant lacking the five γ-TuRC binding serines (Cep192-A5) retained only 28% of it (Figure 4E and Figure S6E). Thus, the centrosomal MT assembly in M-phase seems to rely on Cep192 as a γ-TuRC-anchor rather than as an AurA-Plk1 -activating scaffold. This may be partly explained by the high activity of Plx1 in the mitotic cytoplasm and by the existence of additional, redundant MT assembly mechanisms, including the docking of Plk1 onto Cep192 site(s) other than T46. Such a site(s) may be localized in the Cep192 region C-terminal to aa 1000 because the full-length Cep192-T46A, unlike Cep1921–1000-T46A, was partially functional in the αAurA bead-assay in M-phase extract (Figure 1B, lane 4, compare to Figure 2B, lane 4).
By contrast, both Cep192-T46A and Cep192-Δ543–747 were inactive in promoting centrosomal MT assembly (and γ-TuRC recruitment) upon mitotic entry occurring either naturally, in cycling extracts (Murray, 1991) (Figure 4B, 4C and 4D, black columns), or after the addition of non-degradable cyclin B to S-phase extracts (Figure S6D, lower panel, and S6F–S6H). Thus, the Cep192-organized AurA-Plx1 cascade is required for centrosome maturation in cycling egg extracts.
Cep192 immunodepletion did not significantly affect the abundance in extract of AurA, Plk1, and their other scaffold protein, Bora, the kinetics of Plx1 phosphorylation at T201 and of Bora degradation, and the timing of mitotic entry (Figure 4B and 4F). These results imply that Cep192 controls AurA and Plx1 only locally, at centrosomes, consistent with its centrosome-specific ancestral role (Azimzadeh et al, 2012). By contrast, Bora regulates these kinases in a different spatial and temporal context because it is diffusely distributed in the cytoplasm and is degraded at mitotic entry (Seki et al., 2008) (Figure 4B).
The Cep192-organized AurA-Plx1 cascade operates downstream but independently of PCNT
Cep192 and PCNT are interdependent in their centrosomal localization and function in centrosome maturation (Gomez-Ferreria et al., 2007; Mennella et al., 2013; Zhu et al., 2008). Interestingly, Plk1-mediated phosphorylation of PCNT at mitosis onset was shown to promote centrosome maturation and the recruitment to centrosomes of Cep192, AurA, Plk1, NEDD1 and γ-tubulin (Lee and Rhee, 2011), suggesting that phosphorylated PCNT functions upstream of the Cep192/AurA/Plk1 complex during centrosome maturation. Indeed, we found that PCNT depletion of M phase extract dramatically inhibited the recruitment to centrosomes of Cep192 complexes and γ-tubulin, as well as the centrosome-driven MT assembly (Figure 5A and 5B). By contrast, PCNT depletion did not affect any centrosome-like properties of the αAurA beads, which, as shown above, are mediated by the Cep192/AurA/Plx1 complexes (Figure 5C-5E). Of note, neither PCNT nor centrin were present in these complexes (Figure 5E, lane 2). These data, when taken together with the report by Lee and Rhee (2011), imply that phospho-PCNT recruits Cep192/AurA/Plk1 complexes to centrosomes, whereupon the Cep192-organized signaling cascade drives γ-TuRC recruitment and MT assembly independently of centrioles and PCNT.
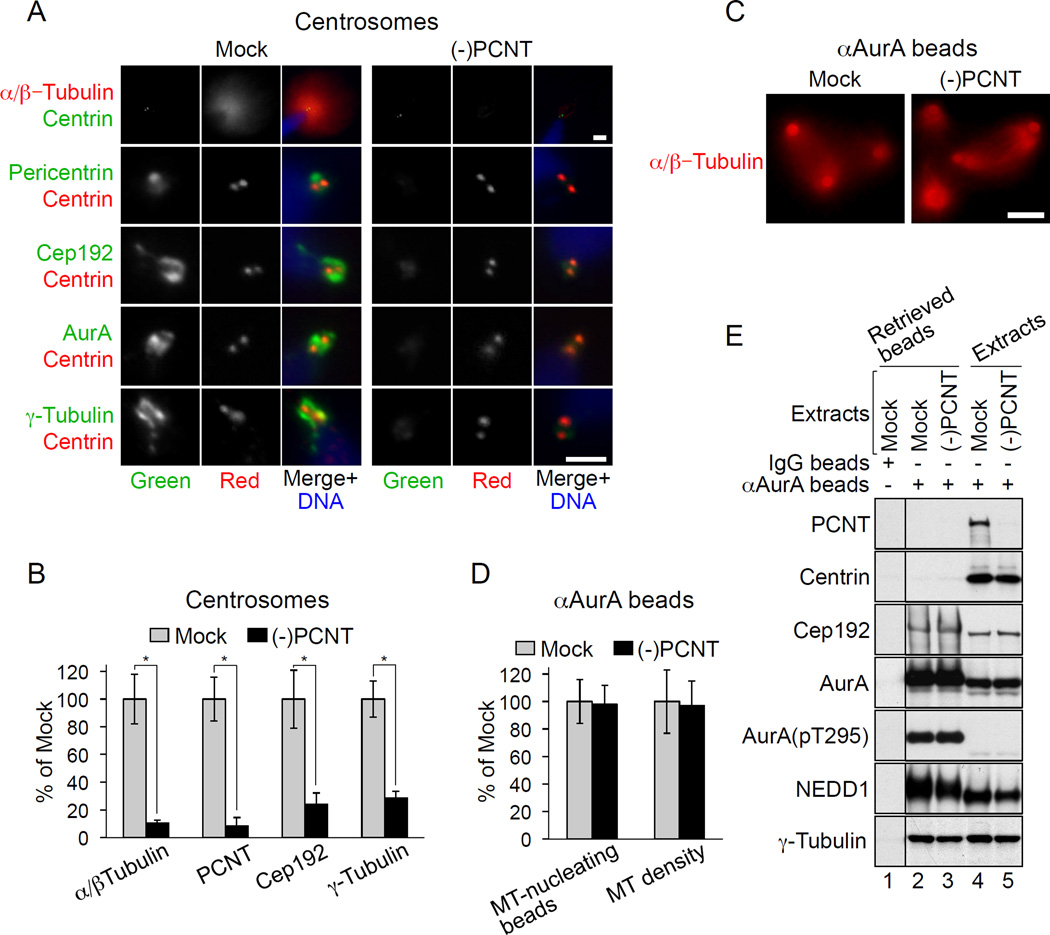
Figure 5. Cep192 promotes MT assembly independently of PCNT.
(A) IF of sperm centrioles in mock-treated and PCNT-depleted M-phase extracts. Scale bars, 2.5 µm.
(B) Quantification of some of the IF signals in (A) (mean +/− SD).
(C) MT structures assembled by αAurA beads in mock-treated and PCNT-depleted M-phase extracts. Scale bar, 10 µm.
(D) Quantification of the proportion of MT-nucleating beads and of MT density in (C) (mean +/− SD).
In (B) and (D), at least 50 MT structures were analyzed for each parameter in each of three independent experiments. *P<0.001.
(E) W-blot of mock-treated and PCNT-depleted extracts and of beads pre-loaded with non-immune IgG or αAurA after their incubation in these extracts.
The Cep192-organized AurA-Plk1 cascade is essential for centrosome maturation in mammalian cells
Next, we asked whether the Cep192-organized signaling cascade is conserved in mammals. In Cep192 siRNA-treated HeLa cells, we expressed siRNA-resistant, HA-tagged human (h) Cep192-wt or its mutant counterparts lacking either the conserved Plk1-docking T44 (hCep192-T44A) (Figure 2D) or the AurA-BD (hCep192-Δ436–634) (Figures S7A and S7B). We then examined whether each recombinant protein rescued the centrosome and spindle assembly defects caused by the Cep192 depletion.
The siRNA-mediated Cep192 silencing abolished AurA phosphorylation at T288 without significantly affecting the abundance of AurA, Plk1, or Plk1(pT210) in the cytoplasm (Figure 6A and Figure S7C). It also abrogated AurA centrosomal localization, which was restored by the expression of hCep192-wt and hCep192-T44A but not of hCep192-Δ436–634 (Figures 6B, panel 1 and S7D). Thus, both in Xenopus and human cells, Cep192 is the major centrosome-targeting and activating cofactor of AurA. Furthermore, Cep192 depletion abolished the centrosomal targeting of Plk1 and this defect was rescued completely by the expression of hCep192-wt and partially by the expression of hCep192-T44A or hCep192-Δ436–634 (Figure 6B, panel 2 and 6C). Hence, the function of Cep192 as a centrosome-specific scaffold for AurA and Plk1 appears to be conserved in mammals.
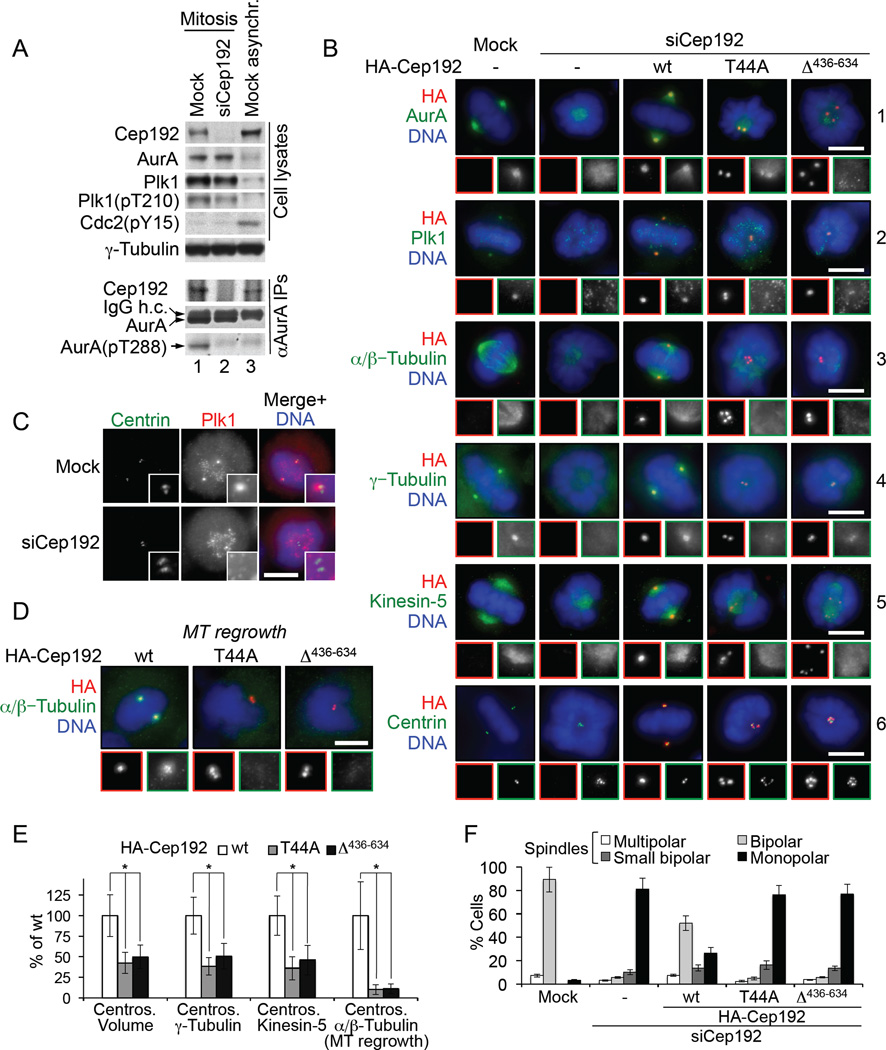
Figure 6. The Cep192-organized AurA-Plk1 cascade is essential for centrosome maturation and bipolar spindle assembly in mammalian cells.
(A) W-blots of lysates and AurA immunoprecipitates from HeLa cells transfected with control siRNA (Mock) or Cep192 siRNA #1. In lanes 1 and 2, cells were synchronized in mitosis by a thymidine-nocodazole block. IgG h.c, heavy chain of IgG.
(B) Representative IF images of cells treated with control siRNA (Mock) or Cep192 siRNA #2. Cells were transfected with an empty vector or with siRNA-resistant cDNAs encoding the indicated HA-tagged hCep192 proteins. Insets show higher-magnification views of the centrosome regions. Scale bars, 10 µm.
(C) IF of Plk1 and centrin in cells transfected with control siRNA (Mock) or Cep192 siRNA #1.
(D) MT regrowth assay in Cep192 siRNA #2-treated cells expressing the indicated HA-tagged hCep1 92 proteins.
(E) Quantification of the centrosome volume in [(B), HA signal] and of the IF intensities of the centrosomal γ-tubulin, kinesin-5 in (B) and α/β-tubulin in (D) (mean +/− SD). At least 50 MT structures were analyzed for each parameter in each of the three independent experiments. *P<0.001.
(F) Quantification of the spindle assembly defects in cells in [(B), panel 3] (mean of three independent experiments +/− SD).
See also Figure S7.
Mitotic centrosomes in Cep192 siRNA-treated cells were depleted of PCM (as assessed by IF for PCNT) and completely devoid of γ-tubulin, consistent with previous reports (Gomez-Ferreria et al., 2007; Zhu et al., 2008) (Figure 6B, panel 4 and Figure S7D). Accordingly, such centrosomes failed to nucleate MTs in a MT regrowth assay (data not shown). The expression of hCep192-wt rescued all the abnormalities seen in Cep192 siRNA-treated cells. By contrast, the expression of hCep192-T44A or hCep192-Δ436–634 did not rescue the MT nucleation defect while partially (by ~40–50%) restoring the centrosome size and γ-tubulin recruitment defects (Figure 6D, 6B, and 6E). These data are consistent with the selective involvement of the Cep192-organized AurA-Plk1 cascade in the formation of the mitotic microtubule-nucleating outer PCM matrix, which is the hallmark of centrosome maturation.
The Cep192-organized AurA-Plk1 cascade is critical for proper centrosome separation and bipolar spindle assembly
Most of the Cep192 siRNA-treated cells arrested in mitosis with monopolar or disorganized spindles (Figure 6B and 6F), as previously reported (Gomez-Ferreria et al., 2007; Zhu et al., 2008). We found that in ~80% of mitotic Cep192 siRNA-treated cells, centrosomes were not separated and clustered at the center of a rosette-like chromatin ring (Figure 6B, panel 6). In those Cep192-depleted cells, which eventually exited mitosis, we observed donut-shaped, multiple, or multilobed nuclei (Figure S7E), a hallmark of a centrosome separation defect (Verstraeten et al., 2011). In ~16% of mitotic Cep192-depleted cells, we observed malformed bipolar spindles and severe chromosome alignment and segregation defects reminiscent of those caused by centriole ablation (Sir et al., 2013) (Figure S7F and S7G). In the latter cells, centrosomes, when present, were partially separated but detached from spindle poles or scattered throughout the spindle. During cell division, such centrosomes were randomly distributed between daughter cells (Figure 7A, S7F, and S7G), resulting in a 2.7 and 4-fold increase in the proportion of mitotic cells containing greater or fewer than four centrioles (including acentriolar cells), respectively (Figure 6B, panel 6, Figure 7B, and Figure S7F–S7H). Centrosome separation is known to depend on at least two processes, the centrosomal linker dissolution and the antiparallel microtubule sliding by the motor protein, kinesin-5 (Eg5), both of which require Plk1 activity (Mardin and Schiebel, 2012). We found that Cep192 depletion did not affect the centrosomal linker dissolution (as assessed by IF for the linker protein rootletin) (Figure S7I) but it significantly decreased the amount of the centrosome-localized kinesin-5 (Figure 6E and Figure 7C). Expression in Cep192 siRNA-treated cells of hCep192-wt (but neither hCep192-T44A nor hCep192-Δ436–634) rescued the defects in spindle assembly and in centrosome anchoring to spindle poles and separation (Figure 6B, 6F, and 7D) and it increased the proportion of mitotic cells with four centrioles (Figure 7B).
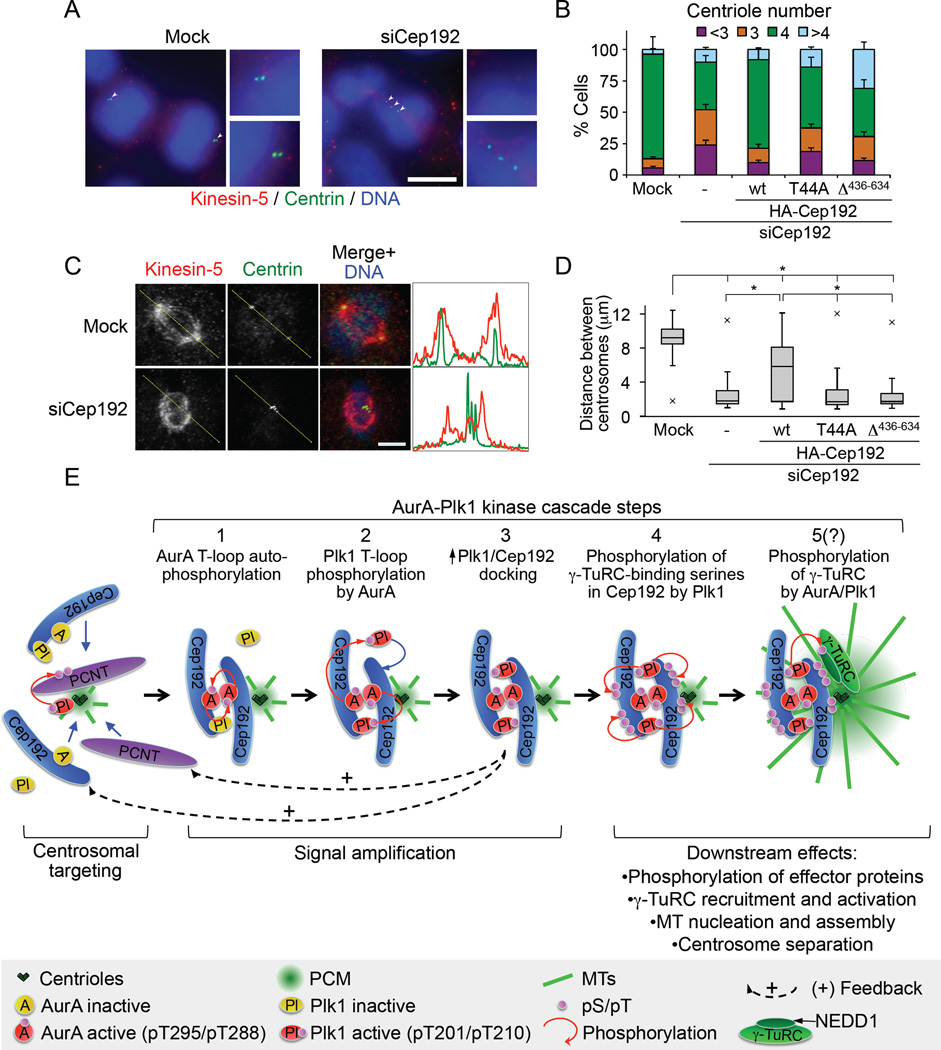
Figure 7. The Cep192-organized AurA-Plk1 cascade is required for proper centrosome separation and segregation into daughter cells.
(A) IF images of control and Cep192 siRNA #1-treated dividing HeLa cells. Centrioles are indicated by arrowheads. Small panels show higher-magnification views of the centrosome regions. Scale bar, 10 µm.
(B) Quantification of the centriole number in mitotic cells treated as in Figure 6B (mean +/− SD of three independent experiments).
(C) Confocal IF staining of kinesin-5 and centrin in control and Cep192 siRNA #1-treated cells. The graphs represent signal intensity scans along the lines drawn. Scale bar, 5µm.
(D) Box-and-whisker plots of the intercentrosomal distances in metaphase cells treated as in Figure 6B. The line represents the median value, while the lower and upper edges of the boxes show the 25th and 75th percentile, respectively. The whiskers extend to the furthest data points within 1.5 times of the interquartile range of the box. The maximum or minimum outliers are shown as crosses. >20 cells were analyzed per each experimental condition in each of three independent experiments. *P<0.0001.
(E) Cep192-centered model of centrosome maturation.
See also Figure S7.
Collectively, these results imply that the Cep192-organized AurA-Plk1 cascade is conserved in vertebrates and underlies the two principal, simultaneously occurring, steps of the centrosome cycle - maturation and separation. This cascade also appears to be essential for proper bipolar spindle assembly and centrosome/centriole segregation into daughter cells.
DISCUSSION
Model of centrosome maturation
Here, we identify a Cep192-organized signaling cascade that underlies centrosome maturation. Based on our findings and previous studies (Decker et al., 2011; Haren et al., 2009; Joukov et al., 2010; Lee and Rhee, 2011), we now propose a model of this process (Figure 7E). In G2 phase, Cep192, in a complex with AurA and Plk1, is recruited to centrosomes by the Plk1-phosphorylated PCNT. The initial level of Plk1 in Cep192 complexes is likely low owing to the low Plk1 activity (which is required for Plk1/Cep192 docking) in the G2 cytoplasm. In mitosis, additional factors may contribute to the centrosomal targeting of Plk1 because Cep192 depletion of M-phase extract did not abolish Plx1 localization to sperm centrioles (Figure 1D). Indeed, at least one other protein, Cenexin 1, was shown to recruit Plk1 to centrosomes (Archambault and Glover, 2009; Park et al., 2010).
By contrast, the recruitment and activation of most, if not all, centrosomal AurA in vertebrates seems to be mediated by Cep192 because both in egg extracts and in mammalian cells, AurA centrosomal localization and T-loop phosphorylation were abolished by the deletion of the AurA-BD of Cep192. A corollary to the above conclusion is that the level of AurA(pT295/pT288), which is commonly used as a readout of the AurA activity in cells (Nikonova et al., 2013), is reflective only of the Cep192-bound (i.e. centrosomal) fraction of active AurA.
The accumulation and consequent oligomerization at centrosomes of Cep192 complexes trigger AurA activation via autophosphorylation at T295/T288 (Figure 7E, Step 1). Active AurA then phosphorylates Plk1 at T201/T210 in its T-loop, thus activating it (Step 2). This event facilitates the docking of Plk1 onto T46/T44 of Cep192 (thus increasing the level of active Plk1 in Cep192 complexes) (Step 3), possibly, with the involvement of a Plk1 -activated kinase that phosphorylates Cep192 at T46/T44. Potential candidates are the NIMA family protein kinases known to be regulated by Plk1 and involved in centrosome functions at the G2/M transition (Mardin and Schiebel, 2012). The active, Cep192-bound Plk1, in turn, phosphorylates Cep192 at several serines to generate the binding sites for γ-TuRC and, possibly, other spindle assembly factors (e.g. XMAP215) (Step 4). The role of Plk1 as an effector kinase in Cep192 complexes is consistent with the evolutionary conserved requirement for polo-like kinases in γ-TuRC recruitment (Archambault and Glover, 2009). Because the five γ-TuRC-binding serines of Xenopus Cep192 are not conserved in its human ortholog, other serines, which are abundantly present in the N-terminal region of hCep192, likely serve as γ-TuRC-binding sites. It remains to be investigated whether the binding of γ-TuRC to the Plk1-phosphorylated Cep192 is direct or dependent on additional factors. Since the active Plx1 did not facilitate the binding between recombinant Cep192 and NEDD1 in vitro (data not shown) and since Cep192 and γ-TuRC do not extensively co-localize in the PCM (Sonnen et al., 2012), it is conceivable that the Plk1 -mediated phosphorylation of Cep192 primes the formation of a “lattice” that captures γ-TuRC.
Furthermore, the active AurA and/or Plk1 in Cep192 complexes may render γ-TuRC capable of MT nucleation by phosphorylating NEDD1 and/or other γ-TuRC subunits (hypothetical Step 5). In keeping with this notion, it was shown that NEDD1 is phosphorylated by AurA and Plk1 (Haren et al., 2009; Pinyol et al., 2013; Zhang et al., 2009) and that its phosphorylation status depends on Cep192 in mammalian cells (Gomez-Ferreria et al., 2012).
Crosstalk between Cep192 and other PCM assembly factors
Based on our findings, the Cep192/AurA/Plk1 complex represents the core component of a signaling cascade that also includes other factors involved in Cep192 centrosomal targeting, AurA activation, Plk1/Cep192 docking, and Cep192/γ-TuRC attachment. Further elucidation of this cascade and its relation to other mechanisms of PCM assembly will be critical for understanding how the mitotic centrosome and spindle form.
The recruitment of Cep192 complexes to centrosomes is likely key to centrosome maturation as it promotes local oligomerization-dependent AurA activation, upon which the triggered AurA-Plk1 cascade then drives MTOC formation independently of the centrosomal mileau. Based on our study and the report by Lee and Rhee (2011), this recruitment appears to be mediated by the Plk1-phosphorylated PCNT, a protein that organizes the inner PCM layer of the interphase centrosomes, upon which the outer mitotic PCM matrix forms (Mennella et al., 2013). Interestingly, subsets of Cep192, Plk1, and AurA also localize to the inner PCM layer in interphase and mitosis, with Cep192 localization being independent of PCNT and Plk1 (Fu and Glover, 2012; Gomez-Ferreria et al., 2007; Lawo et al., 2012; Lee and Rhee, 2011; Mennella et al., 2012; Sonnen et al., 2012; Zhu et al., 2008). One could, therefore, speculate that centrosome maturation is initiated by the activation of Plk1 within the inner layer (with possible involvement of Cep192 and AurA) followed by Plk1 -mediated phosphorylation of PCNT. The consequent phospho-PCNT-mediated recruitment of Cep192 complexes to centrosomes would then unleash the AurA-Plk1 cascade, leading to extensive and persistent Plk1 activation followed by massive recruitment of γ-TuRC and other components that form the outer PCM matrix. The PCM formation may be further facilitated by positive feedback loops that link AurA and Plk1 activities to the centrosomal recruitment of Cep192 and PCNT (Haren et al, 2009; Joukov et al, 2010; Lee and Rhee, 2011; Mennella et al, 2013), as well as by other centrosomal proteins, such as Cep215. Since the centrosomal targeting of both Cep192 and Cep215 is mediated by PCNT (Gomez-Ferreria et al, 2007; Lee and Rhee, 2011; Mennella et al., 2013; Zhu et al, 2008) and since Cep215 was not detected in Cep192/AurA/Plk1 complexes (Figure S1B), Cep192 and Cep215 may function on parallel, rather than linear, pathways. If so, these pathways may contribute differently to PCM assembly depending on the species and the cell types. Consistent with this hypothesis, it has recently been reported that Cnn, the fly ortholog of Cep215, upon phosphorylation by Polo/Plk1 in the inner PCM layer, forms a scaffold that spreads outward (Conduit et al., 2014).
Role of Cep192 in the centrosome cycle and spindle assembly
Remarkably, the loss of the AurA-Plk1 activation module (i.e of the AurA-BD or the N-terminal Plk1-docking threonine) in Cep192 led to the centrosome and spindle assembly defects similar in nature and severity to those seen in cells lacking Cep192, NEDD1, or active AurA or Plk1 (Archambault and Glover, 2009; Gomez-Ferreria et al., 2007; Haren et al., 2006; Luders et al., 2006; Nikonova et al., 2013; Zhu et al., 2008). This fact indicates that the function of Cep192 as a scaffold for the AurA-Plk1 cascade is integral to its role in centrosome biogenesis and spindle formation.
Cep192-depleted cells were shown to display a decrease in centriole number, which was attributed to a centriole duplication defect (Kemp et al., 2004; Pelletier et al., 2004; Zhu et al., 2008). Consistent with these reports, Cep192 was implicated in the recruitment to centrioles of the major centriole duplication factor, the Polo-like kinase 4 (Kim et al., 2013; Sonnen et al., 2013). We found that, in mammalian cells, the loss of Cep192 or of its AurA-Plk1 activation module led to numerical centrosome/centriole abnormalities (i.e. either a decrease or an increase in centriole number), which could clearly be traced to a centrosome segregation defect caused by impaired centrosome separation and anchoring to spindle poles (Figure 7A and Figure S7). These abnormalities may result from the loss of Cep192 function in centrosome maturation because centrosome/spindle pole attachment, centrosome separation, and equal centriole segregation into daughter cells were all shown to require centrosomal MTs (Mardin and Schiebel, 2012; Wang et al., 2011). Thus, Cep192 appears to control the centriole number not only via its involvement in centriole duplication in S phase but also via its mitotic function as a scaffold for the AurA-Plk1 cascade. Importantly, the latter Cep192 function does not depend on the former one because the Cep192-organized kinase cascade operates in M phase-arrested egg extracts that have not been passaged through S phase (Figure 1–Figure 3). In fact, the function of Cep192 in centriole duplication may depend on the Cep192-organized cascade because the initiation step for centriole duplication (which is completed in S-phase) was shown to require NEDD1 and γ-tubulin, as well as Plk1 activity at mitotis onset (Haren et al., 2006; Loncarek et al., 2010; Wang et al., 2011). Consistent with this hypothesis, the ancestral function of Cep192/SPD-2 was to recruit PCM components rather than to assemble centrioles (Azimzadeh et al., 2012).
The multifaceted scaffolding properties of Cep192 revealed in this study validate and confirm a recent theoretical model of PCM formation that invokes a combination of template-based and self-organization principles (Mahen and Venkitaraman, 2012). Moreover, these properties may help explain the severe spindle abnormalities seen in Cep192-depleted cells. Cep192 may control mitotic spindle assembly by promoting both centrosome maturation [which enables centrosomal MT nucleation and proper spindle geometry (Mennella et al., 2013; Sir et al., 2013)] and kinesin-5-mediated centrosome separation [which drives spindle bipolarity (Mardin and Schiebel, 2012)]. The latter Cep192 activity may be a consequence of the former one since both the centrosomal targeting and the function of kinesin-5 (unlike those of Cep192) require MTs (Mardin and Schiebel, 2012). Cep192 may also contribute to spindle bipolarity independently of MTs because the full length Cep192 promoted bipolar spindle formation, while its N-terminal fragment, which assembled MTs, did not (Figure 2C and S2E). In addition, as a centrosome-targeting and activating scaffold for AurA and Plk1, Cep192 may influence spindle assembly by regulating the availability of both kinases to other cofactors/substrates, as well as the spatial gradient of active AurA and Plk1. In keeping with this notion, we observed an inverse correlation between the levels of AurA complexed to Cep192 and to TPX2 in egg extracts (data not shown), consistent with the mutually exclusive binding to AurA of these two cofactors (Joukov et al., 2010).
In conclusion, our study reveals a Cep192-organized signaling cascade that underlies centrosome maturation and that is integral to the centrosome cycle and bipolar spindle assembly in vertebrates. It also offers a framework for further dissection of the mechanisms of mitotic MTOC formation in a cell-free system.
EXPERIMENTAL PROCEDURES
Generation of cDNA constructs
The cDNAs encoding tagged Xenopus and human Cep192 and other proteins were generated by PCR and cloned into pFastBac1 (Life Technologies), pcDNA3.1 (Invitrogen), pGEX6p-1 (GE Healthcare), or pSP64 poly(A) (Promega) vectors. For detailed cloning procedures, see Supplemental Experimental Procedures.
Recombinant proteins and antibodies
The recombinant Xenopus full-length Cep192 proteins were produced using the Bac-to-Bac baculovirus expression system (Life Technologies) and purified by affinity chromatography. The recombinant GST-tagged proteins were produced as previously described (Joukov et al., 2010). Further information on the proteins and antibodies is provided in the Supplemental Experimental Procedures.
Experiments in Xenopus egg extracts
The research with X. laevis was performed under a protocol approved by the Harvard Medical Area Institutional Animal Care and Use Committee. Preparation of M-phase (CSF-arrested) egg extracts and analysis of the MT assembly promoted by centrosomes and αAurA beads were performed as previously described (Joukov et al., 2010; Murray, 1991; Tsai and Zheng, 2005). For further experimental details, see Supplemental Experimental Procedures.
Cell culture experiments
HeLa cells were transfected using oligofectamine (Life Technologies), with control non-targeting siRNA or with either of two different, non-overlapping Cep192-targeting siRNA preparations. Transfection of hCep192 cDNAs was performed using Fugene HD (Promega). A detailed description of the experiments is available in the Supplemental Experimental Procedures.
Fluorescence microscopy and image analysis
Fluorescence microscopy and image analysis were performed using an Axioskop 2 with the AxioVision software (Zeiss) and a Nikon Ti inverted fluorescence microscope with the Perfect Focus System and the Metamorph software. Quantitative analysis of fluorescence signals, centrosome distances, and W-blots was performed using ImageJ 1.46r (http://imagej.nih.gov/ij/) and Excel. For further details, see Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Cep192 is a centrosome-specific activating scaffold for Aurora A and Plk1
Cep192 is an anchoring platform for Plk1 phosphorylation-mediated γ-TuRC recruitment
Cep192 locally organizes Aurora A and Plk1 in a multistep kinase cascade
The cascade is essential for centrosome maturation and bipolar spindle assembly
ACKNOWLEDGEMENTS
We thank D.M. Livingston for his support and comments on the manuscript, M. Budzowska, A. Groen, C.D. Hu, J. Maller, A. Merdes, and T. Mitchison for providing reagents, M. Eck for providing the baculovirus expression facility, S. Gygi and R. Tomaino for mass spectrometry analysis, E. Parisini for help with sequence alignments, and J. Higgins for critical reading of the manuscript. We also thank L. Piedmont, J. Waters, and the Nikon Imaging Center at Harvard Medical School for help with light microscopy. J.C.W. was supported by the NIH Grant GM080676. V.J. conceived the project, performed the experiments, analyzed the data, and wrote the manuscript. J.C.W. contributed with reagents, data analysis, and writing the manuscript. A.D.N. performed the experiments in mammalian cells, provided reagents, and contributed significantly to data analysis and to writing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The full-length human Cep192 cDNA was deposited in GenBank (accession number KJ567064).
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and Supplemental Experimental Procedures.
REFERENCES
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Wong ML, Downhour DM, Sanchez Alvarado A, Marshall WF. Centrosome loss in the evolution of planarians. Science. 2012;335:461–463. doi: 10.1126/science.1214457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Knoblich JA. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 2002;12:640–647. doi: 10.1016/s0960-9822(02)00766-2. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, Chan CS, 3rd, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Conduit PT, Feng Z, Richens JH, Baumbach J, Wainman A, Bakshi SD, Dobbelaere J, Johnson S, Lea SM, Raff JW. The centrosome-specific phosphorylation of cnn by polo/plk1 drives cnn scaffold assembly and centrosome maturation. Dev. Cell. 2014;28:659–669. doi: 10.1016/j.devcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker M, Jaensch S, Pozniakovsky A, Zinke A, O’Connell KF, Zachariae W, Myers E, Hyman AA. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 2011;21:1259–1267. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Dix CI, Raff JW. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 2007;17:1759–1764. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 2003;13:691–697. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 2008;18:303–309. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria M, Bashkurov M, Helbig A, Larsen B, Pawson T, Gingras AC, Pelletier L. Novel NEDD1 phosphorylation sites regulate gamma-tubulin binding and mitotic spindle assembly. J. Cell Sci. 2012;125:3745–3751. doi: 10.1242/jcs.105130. [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, Sharp DJ. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 2007;17:1960–1966. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria MA, Sharp DJ. Cep192 and the generation of the mitotic spindle. Cell Cycle. 2008;7:1507–1510. doi: 10.4161/cc.7.11.5957. [DOI] [PubMed] [Google Scholar]
- Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 2001;155:1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 2006;172:505–515. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Stearns T, Luders J. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 2009;4:e5976. doi: 10.1371/journal.pone.0005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V. Aurora kinases and spindle assembly: Variations on a common theme? Cell Cycle. 2011;10:895–903. doi: 10.4161/cc.10.6.14909. [DOI] [PubMed] [Google Scholar]
- Joukov V, De Nicolo A, Rodriguez A, Walter JC, Livingston DM. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc. Natl. Acad. Sci. USA. 2010;107:21022–21027. doi: 10.1073/pnas.1014664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- Kim TS, Park JE, Shukla A, Choi S, Murugan RN, Lee JH, Ahn M, Rhee K, Bang JK, Kim BY, et al. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 2013;110:E4849–E4857. doi: 10.1073/pnas.1319656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell Biol. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Lee K, Rhee K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 2011;195:1093–1101. doi: 10.1083/jcb.201106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Khodjakov A. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr. Biol. 2010;20:1277–1282. doi: 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Patel UK, Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- Mahen R, Venkitaraman AR. Pattern formation in centrosome assembly. Curr. Opin. Cell Biol. 2012;24:14–23. doi: 10.1016/j.ceb.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Mardin BR, Schiebel E. Breaking the ties that bind: new advances in centrosome biology. J. Cell Biol. 2012;197:11–18. doi: 10.1083/jcb.201108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Agard DA, Huang B, Pelletier L. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 2013;24:188–197. doi: 10.1016/j.tcb.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL, Jr, Golemis EA. Aurora A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 2013;70:661–687. doi: 10.1007/s00018-012-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Soung NK, Johmura Y, Kang YH, Liao C, Lee KH, Park CH, Nicklaus MC, Lee KS. Polo-box domain: a versatile mediator of polo-like kinase function. Cell. Mol. Life Sci. 2010;67:1957–1970. doi: 10.1007/s00018-010-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Pinyol R, Scrofani J, Vernos I. The role of NEDD1 phosphorylation by Aurora A in chromosomal microtubule nucleation and spindle function. Curr. Biol. 2013;23:143–149. doi: 10.1016/j.cub.2012.11.046. [DOI] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir JH, Putz M, Daly O, Morrison CG, Dunning M, Kilmartin JV, Gergely F. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 2013;203:747–756. doi: 10.1083/jcb.201309038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 2013;126:3223–3233. doi: 10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D–structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MY, Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced Spindle Assembly. Curr. Biol. 2005;15:2156–2163. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- Verstraeten VL, Peckham LA, Olive M, Capell BC, Collins FS, Nabel EG, Young SG, Fong LG, Lammerding J. Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc. Natl. Acad. Sci. USA. 2011;108:4997–5002. doi: 10.1073/pnas.1019532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen Q, Feng J, Hou J, Yang F, Liu J, Jiang Q, Zhang C. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J. Cell Sci. 2009;122:2240–2251. doi: 10.1242/jcs.042747. [DOI] [PubMed] [Google Scholar]
- Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, Muller-Reichert T, Kittler R, Hyman AA, Pelletier L. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 2008;18:136–141. doi: 10.1016/j.cub.2007.12.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.