Abstract
Multi-domain enzymes often rely on large conformational motions to function. However, the conformational setpoints, rates of domain motions, and relationships between these parameters and catalytic activity is not well understood. To address this, we determined and compared the conformational setpoints and the rates of conformational switching between closed unreactive and open reactive states in four mammalian di-flavin NADPH oxidoreductases that catalyze important biological electron transfer reactions: cytochrome P450 reductase (CPR), methionine synthase reductase (MSR), and endothelial and neuronal NO synthase (eNOS & nNOS). We used stopped-flow spectroscopy, single turnover methods, and a kinetic model that relates electron flux through each enzyme to its conformational setpoint and its rates of conformational switching. Results show that the four flavoproteins, when fully-reduced, have a broad range of conformational setpoints (from 12 to 72% open state) and also vary 100-fold regarding their rates of conformational switching between unreactive closed and reactive open states (CPR > nNOS > MSR > eNOS). Furthermore, simulations of the kinetic model could explain how each flavoprotein can support its given rate of electron flux (cytochrome c reductase activity) based on its unique conformational setpoint and switching rates. Our study is the first to quantify these conformational parameters among the di-flavin enzymes, and suggests how the parameters might be manipulated to speed or slow biological electron flux.
Keywords: flavoprotein, electron transfer, conformational equilibrium, kinetic model, nitric oxide, simulation, redox enzymes
Introduction
The diflavin reductases transfer electrons from NADPH to a variety of one-electron acceptors [1–4]. They are comprised of attached NADPH, FAD, and FMN binding domains [1;5] that are analogous to existing proteins: the NADPH and FAD binding domains (NADPH/FAD or FNR) resemble NADP+-ferredoxin reductases while the FMN domains are analogous to flavodoxins [1;6–8]. Either flavin cofactor can exist in three different redox states: oxidized, semiquinone (sq), and hydroquinone (hq). The electron flux through these enzymes follows a linear pathway: NADPH → FAD → FMN → final acceptors [9]. Each flavin has a unique function: the FAD accepts a hydride from NADPH and then transfers reducing equivalents to FMN one at a time [10;11]. Because the mammalian diflavin reductases stabilize their FMNsq, only the FMNhq acts as an electron donor to external acceptors and then cycles between the FMNsq and FMNhq forms. The diflavin reductase family includes mammalian members NADPH-cytochrome P450 reductase (CPR) [6;12], the three nitric oxide synthases (NOS) [13–15], methionine synthase reductase (MSR) [16–18], and novel reductase 1 [19], and includes bacterial members sulfite reductase (SiR) [20] and flavocytochrome P450 BM3 [21;22].
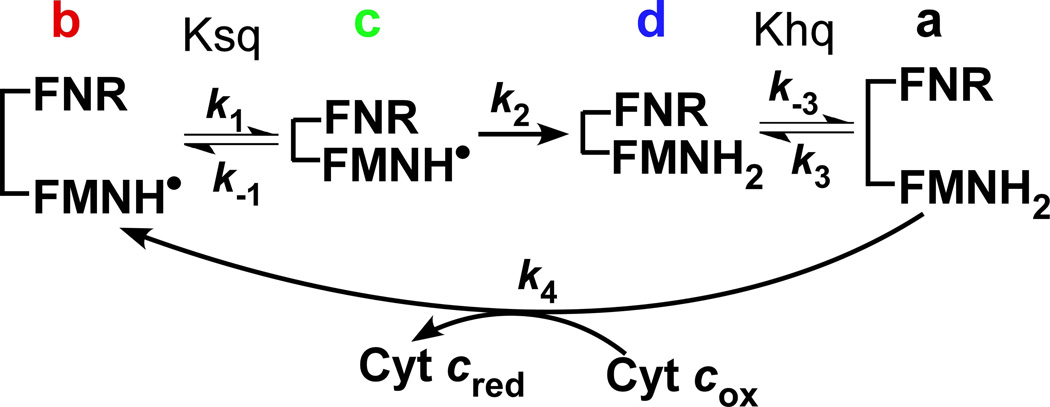
Electron transfer (ET) through diflavin enzymes involves specific domain-domain interactions. An interface that forms between the NADPH/FAD (FNR) and FMN domains allows the interflavin ET that forms FMNhq. Crystal structures of rat and human CPRs [23;24] and rat nNOSred [25] show the isoalloxazine rings of their bound FAD and FMN cofactors are closely positioned for rapid ET. However, in this conformation the FMN cofactor is buried from solvent and is incapable of performing ET with protein redox partners or artificial acceptors like cytochrome c. Transition of the conformationally-closed structures to a more open forms is thought to be required, as was confirmed recently for CPR [26]. Electron flux through all diflavin enzymes is thought to involve conformational shuttling between closed and open states [27–31]. Crystal structures and biophysical studies have increased our understanding of the conformationally-closed and -open states of diflavin reductases and how they may participate in catalyzing electron flux to their natural acceptor proteins or to cytochrome c [15;23;25–27;30;32–39]. However, it is not clear how the conformational equilibria and rates of conformational switching relate to electron flux, or how these parameters compare among the diflavin reductases. To address this, we proposed a simple four-state kinetic model (Fig. 1) [15;34;35] that relies on cytochrome c reductase activity to assess electron flux through the diflavin enzymes. Under typical experimental conditions with excess NADPH and cytochrome c, the rate of flavin reduction or cytochrome c reduction (k4) is fast and not rate-limiting [30;36;37;40;41] and so the electron flux through the diflavin enzyme is only dependent on the rates of FMN domain conformational switching (k1, k−1, k3, k−3) and the rate of interflavin electron transfer between the FAD and FMN cofactors (k2). The model incorporates the one-electron redox cycling of the FMNhq and FMNsq, which occurs naturally during turnover, and therefore includes two temporally-separate equilibrium terms, Ksq and Khq, that differ according to the reduction state of the FMN (FMNsq versus FMNhq). We define Ksq = k−1 / k1 and Khq = k−3 / k3 (Fig. 1) such that higher K values indicate a greater abundance of open conformations. The equilibrium described by Khq involves a conformational opening step that allows the FMNhq to reduce an acceptor (cytochrome c in this case) and consequently become oxidized to FMNsq. The equilibrium described by Ksq involves a conformational closing step that allows the FMNsq to receive another electron from the NADPH/FAD (FNR) domain. For simplicity, the model assumes (i) the interflavin electron transfer step (k2) is irreversible. (ii) the reduction of FAD by NADPH and the reduction of cytochrome c by FMNhq (k4 step) are rapid and irreversible steps relative to all the other kinetic steps, (iii) the conformational equilibrium setpoint is unaffected by the redox state of the FMN (i.e., Ksq = Khq). We previously utilized this approach to determine estimates for the Khq setpoint and rates of conformational switching and interflavin ET in the flavoprotein domains of endothelial and neuronal NOS (eNOSred, nNOSred) [34;36] and in site-specific variants of nNOSred [35].
Figure 1. Kinetic model for electron flux through a dual-flavin enzyme.
The model uses four kinetic rates: association (k1 or k3) and dissociation (k−1 or k−3) of the FMN and FNR domains; the FMNH• reduction rate (k2), and the cytochrome c reduction rate (k4). The fully-reduced enzyme in the open conformation (species a) reduces cytochrome c and generates species b, which then undergoes successive conformational closing, interflavin electron transfer, and conformational opening steps to complete the cycle.
Here, we expanded our studies to investigate CPR and MSR and we re-analyzed nNOSred and eNOSred in parallel, in order to understand the range of conformational behaviors displayed across the mammalian dual-flavin reductase family, and how their conformational parameters may determine their different electron flux profiles. Simulations of the kinetic model show how each enzyme’s unique blend of conformational equilibrium and kinetic parameters help determine its steady-state species distribution and catalytic activity. Our study provides the first measures of conformational setpoints and switching rates, which in turn have protein structural, mechanistic, and functional implications.
Experimental Procedures
Steady-state Cytochrome c Reduction Assays
The cytochrome c reductase activity was determined at 25 °C and 10 °C by monitoring the increase in absorption at 550 nm and using an extinction coefficient ε550 = 21 mM−1 cm−1 as described previously [17;27;35;36].
Reaction of Fully Reduced Proteins with Excess Cytochrome c
The rate of reduction of excess cytochrome c by fully reduced proteins was measured in the stopped-flow instrument under anaerobic conditions at 10 °C as described previously [35;36]. The nNOSred or eNOSred (10–12 µM) proteins in 40 mM EPPS buffer (pH 7.6) with 10% glycerol, and 150 mM NaCl containing EDTA (2 mM) was fully reduced by titrating it with anaerobic sodium dithionite solution. We used 0.1 M Potassium phosphate buffer (pH 7.4) with 10% glycerol for CPR and MSR proteins, otherwise keeping the rest of the procedure the same as that used for the two NOSred proteins. An anaerobic solution of each fully-reduced protein containing NADPH (200 µM) was mixed with an anaerobic solution of cytochrome c (100 µM) while monitoring the changes in absorption at 550 nm. Initially, the solution of cytochrome c was mixed with anaerobic buffer alone to obtain the initial 550 nm absorbance reading at time = 0. All mixing reactions were repeated consecutively 6 to 8 times, and then the individual kinetic traces were averaged. The entire analysis was then repeated using a separately-purified batch of each enzyme. In the reactions of reduced enzyme with cytochrome c, a linear regression analysis of late points in the traces and/or numeric derivatives of the traces were run to determine the deflection points separating the early burst of fast-reacting and late slow-reacting phases (vide infra “Results”).
Simulation of the Kinetic Traces of Fully Reduced Flavoproteins with Excess Cytochrome c
We used the computer program Gepasi v.3.30 [42] to simulate the experimental electron flux to cytochrome c using the kinetic model as outlined in Fig. 1. Details of this type of simulations have been reported earlier [34;35]. Here, we set the reaction rate with cytochrome c (k4 in the kinetic model) at 225 s−1 in all cases, based on a reported second order rate constant (4.5 × 106 M−1s−1) for the reaction of cytochrome c with the reduced conformationally-open nNOSred, or with its reduced isolated FMN domain, at 10 °C [36;37;40]. Values for each of the four conformational rates (k1, k−1, k3 and k−3) and the interflavin electron transfer rate (k2) were input into the software. Each simulated reaction began with 100% of the enzyme in the fully reduced state (represented by species d and a in Fig. 1). The initial concentrations of species d and a were determined based on the experimentally obtained Khq values. Inputs that satisfy the observed conformational equilibrium constant are first used, and then refined in an iterative process using the time of first turnover and overall best fit of the kinetic trace as criteria to identify the best fit rates for FMN conformational motion and interflavin electron transfer.
Structure-based analysis of the domain interfaces
Existing PDB structures were used when available. We used 1TLL [25] for rat nNOS and 1AMO [23] for rat cytochrome P450 reductase. In the case of bovine eNOS and human methionine synthase reductase, structures were modeled using the Swiss Model Server [43]. The available structure of rat nNOS (1TLL) was used as template for the bovine eNOS model, being by far the template with the highest sequence identity. In the case of human methionine synthase reductase, a structure for the FNR domain is available [44] but the structure of the FMN domain has not been determined. As both nNOS and cytochrome P450 reductase have significant sequence identity (≈33%) to methionine synthase reductase, we built two different models for methionine synthase reductase using either nNOS or cytochrome P450 reductase structures as templates.
The interface area and the atom interactions for each structure were calculated using the 2P2I server [45]. The output of the 2P2I server was also used to compile the tables of atom and residue contacts (Supplemental tables S1).
The definition of the FNR-like and FMN domains for each protein was as follows: rat nNOS, FMN domain amino acids 750 to 942, FNR domain amino acids 968 to 1413; CPR, FMN amino acids domain 64 to 235, FNR domain amino acids 243 to 678; eNOS, FMN domain amino acids 519 to 721, FNR domain amino acids 728 to 1180 (segment 722–727 removed for calculations); MSR/1TLL model, FMN domain amino acids 6 to 162, FNR domain amino acids 210 to 697 (segment 163–209 removed for calculations); MSR/1AMO model, FMN domain amino acids 2 to 162, FNR domain amino acids 210 to 698 (segment 163–209 removed for calculations). In the available structures of CPR and nNOS the hinge fragments between the FNR and FMN domains are too flexible and not visible in the crystal structure (236–242 in CPR; 952–958 in nNOS). We considered that the structure of the homologous fragment in the eNOS model (722–727) was not reliable and was removed before analysis. In the case of MSR, the polypeptide including the FNR domain and the hinge domain (amino acids 166–698) has been analyzed, but only the fragment between amino acids 217 and 698 yield a crystal structure [44]. We therefore excluded the fragment from our study. Comparison of the MSR models with the available MSR structure [44] indicates that the estimated location of the segments between amino acids 210 and 270 is not consistent with the reported structure. Nevertheless, we did not remove these fragments as they are far from the FNR/FMN domain interface and do not modify our calculations. The PDB files used for the analysis are available in the supplementary material.
RESULTS
Steady-state electron flux varies widely among family members
We compared the NADPH-dependent cytochrome c reductase activities of the four flavoproteins at 10 °C and 25 °C (Table 1). Steady-state cytochrome c reductase activity indicates the maximal electron flux that can be achieved through the four flavoproteins because cytochrome c reacts quickly and irreversibly to accept an electron from their reduced FMN domains [1;15;17;27;35;36]. The reductase activities we obtained matched with earlier reports [17;27;35;36], and at either temperature gave a rank order of CPR > nNOSred > eNOSred = MSR that spanned almost two orders of magnitude. This difference became the premise for our current study.
Table 1. Steady-state cytochrome c reductase activities of Dual-flavin reductases.
The cytochrome c reductase activities were determined at 25 and 10 °C as described under “Experimental Procedures.” The values are the mean ±S.D. from three measurements done under identical conditions, using two different protein preparations for each enzyme. Rates are expressed as per minute. Activities of nNOSred and eNOSred were measured in CaM-free conditions.
| Proteins | 10 °C | 25 °C |
|---|---|---|
| eNOSred | 27 ± 2.3 | 68 ± 6 |
| nNOSred | 474 ± 29 | 752 ± 42 |
| CPR | 1680 ± 160 | 3629 ± 290 |
| MSR | 37 ± 3 | 56 ± 4.2 |
Conformational Keq setpoints vary among the fully-reduced flavoprotein
We estimated the conformational Khq setpoints ([open-reactive]/[closed-unreactive]) of each fully-reduced flavoprotein by monitoring its reaction with an excess of cytochrome c in a stopped-flow spectrophotometer, as done previously [35;36]. Under our particular experimental conditions (Final cytochrome c concentration was 50 µM after mixing, 2nd order rate constant of 4.5 × 106 M−1s−1 at 10 °C) the portion of reduced enzyme molecules that are in a cytochrome c-reactive conformation (which we define as an open conformation) will transfer an electron from FMNhq to cytochrome c at approximately 225 s−1, and thus this reaction will occur nearly within the instrument mixing dead time (approximately 4 ms). In contrast, the portion of enzyme molecules that exist in a cytochrome c-unreactive conformation (which we define as closed conformation) will only reduce cytochrome c gradually and in relation to the rate of conformational opening. Thus, the extent to which reduction of the first molar equivalent of cytochrome c exhibits a fast reduction (approximately within the mixing dead-time) versus a slower reduction indicates what proportion of the fully-reduced enzyme molecules were in an open versus closed conformational state at the time of mixing, and gives an estimate for the conformational Khq of the fully-reduced enzyme [34–36].
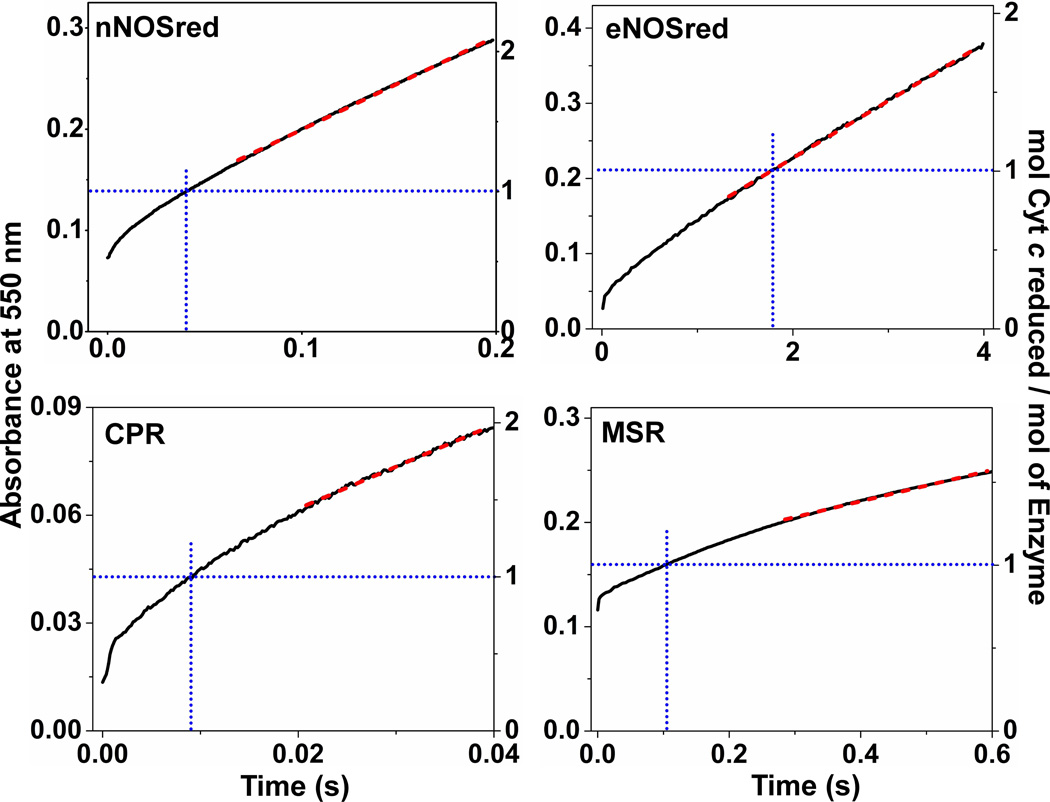
We rapid-mixed the reduced, NADPH-bound forms of each flavoprotein with an excess (100 µM) of cytochrome c in the stopped-flow spectrometer under anaerobic conditions at 10 °C, and monitored the increase in absorption at 550 nm during the first few electron transfers to cytochrome c (i.e. the first few turnovers). Traces that were averaged from 6–8 consecutive mixing reactions are shown in Fig. 2. Initial absorbance readings for time = 0 were determined by rapid-mixing the enzyme-free reaction buffer with the cytochrome c reaction solution. The experimental data are plotted in terms of the increase in absorption from the t = 0 point (a value designated as 0 on the left Y axis in Fig. 2, all panels). The right Y axis of each panel indicates the moles of cytochrome c reduced per mol of total flavoprotein that was present in each reaction. The blue dotted box in each panel indicates the absorbance change that occurred and the time that elapsed during consumption of the first molar equivalent of cytochrome c.
Figure 2. Reaction of fully reduced flavoproteins with excess cytochrome c.
Solutions of pre-reduced proteins (~10–12 µM) containing 200 µM NADPH were rapidly mixed with excess cytochrome c (100 µM) in a stopped-flow instrument under anaerobic conditions at 10 °C. Kinetic traces were recorded at 550 nm during the first few electron transfers to cytochrome c. This was done for nNOSred, eNOSred, CPR, and MSR. The absorbance change representing the first turnover is shown by blue dotted lines according to the right-hand scale in each figure. To calculate slope, tangent lines drawn onto the near-linear portions of the stopped-flow traces (indicated as red dashed lines) in each panel. Data are representative of at least two experiments.
In all four enzyme reactions, we observed an initial burst phase that was followed by a slower and nearly linear rate of cytochrome c reduction, as we had observed previously for nNOSred and eNOSred under similar reaction conditions [34–36]. This behavior suggests the transition to steady-state cycling already occurred within the first few turnovers in all four flavoprotein reactions. This concept was supported by our finding that the enzyme activity calculated from a tangent line drawn onto each kinetic trace within the time taken to reduce the first two equivalents of cytochrome c (tangents are indicated as red dashed lines in Fig. 2) essentially matched the steady-state activity that was determined for each flavoprotein in a standard steady-state assay run in a cuvette for 3 min at 10 °C (table 2 and table 3).
Table 2. Summary of results from reactions mixing Dual-flavin reductases with excess cytochrome c.
An excess of cytochrome c was mixed with fully reduced proteins in a stopped-flow instrument at 10 °C as described under “Experimental Procedures”.
| Protein | Time (1st turnover) |
Fraction Open |
Fraction Closed |
Khq | Steady-state Electron flux (min−1) |
|---|---|---|---|---|---|
| eNOSred | 1.8 s | 0.11 | 0.89 | 0.125 | 29.5 |
| nNOSred | 40 ms | 0.51 | 0.49 | 1.04 | 486 |
| CPR | 9 ms | 0.33 | 0.67 | 0.5 | 1620 |
| MSR | 117 ms | 0.72 | 0.28 | 2.57 | 42 |
Table 3. Kinetic Parameters derived from fitting the experimental traces according to the four-state kinetic model.
See text for details. Data are representative of two or three trials with each protein. Values in parentheses are experimental values.
| Protein | Measured KA |
Best fit k1 = k3 (s−1) |
Best fit k1 = k3 (s−1) |
Best fit k2 (s−1) |
Fitted Steady state electron flux (s−1) (Experimental value) |
Fitted Time required for 1st turnover (Experimental value) |
|---|---|---|---|---|---|---|
| eNOSred | 0.125 | 5 | 0.63 | 5 | 0.49 (0.45) | 1.76 (1.8) s |
| nNOSred | 1.0 | 53 | 53 | 22 | 8.1 (7.9) | 38 (40) ms |
| CPR | 0.5 | 460 | 230 | 52 | 27 (28) | 9 (9) ms |
| MSR | 2.57 | 8 | 20.5 | 2.8 | 0.7 (0.62) | 114 (117) ms |
The data in Fig. 2 suggest that the reduced diflavin enzymes have different conformational equilibrium setpoints (Khq). For example, in Fig. 2 (upper left panel), the t = 0 value of the nNOSred absorbance trace, relative to the Y-axis intercept of the blue dashed line, shows that about 50% of the first equivalent of cytochrome c became reduced within the mixing dead time and the remaining 50% of the first equivalent became reduced during the subsequent observable period (within the dotted blue box). This implies the NADPH-bound, fully-reduced nNOSred molecules equally populated the open reactive and closed unreactive conformations at the time of mixing (species a and d in Fig. 1, respectively), giving an estimated Khq = 1 for the CaM-free, NADPH-bound, fully-reduced nNOSred, which matches with our previous Khq estimate for nNOSred [35;36]. The situation drastically differed for eNOSred (upper right panel, Fig. 2). In this case, only ~11% of the absorbance change occurred in the instrument dead time during reduction of the first equivalent of cytochrome c, leaving ~89% of the absorbance change to be observable. This indicates the CaM-free, NADPH-bound, fully-reduced eNOSred predominantly populates the closed unreactive conformation, according to estimated Khq of ~0.12, again similar to our earlier study [34;36].
For the NADPH-bound, fully-reduced CPR (Fig. 2, lower left panel), the trace showed that 33% of the first equivalent of cytochrome c was reduced within the instrument mixing dead time, indicating that CPR also favors the closed unreactive conformation according to Khq = 0.5. In contrast, the NADPH-bound, fully-reduced MSR (lower right panel, Fig. 2) completed most of its first turnover within the mixing dead time of the instrument, leaving only ~28% of the 1st turnover observable, and giving an estimated Khq = 2.6. Thus, the four flavoproteins displayed a range of conformational Khq setpoints in their NADPH-bound, fully-reduced states, according to the rank order: MSR > nNOSred > CPR > eNOSred (Table 2). MSR predominantly populated a reactive open conformation, nNOS was poised equally between the reactive open and unreactive closed conformations, and CPR and eNOS predominantly populated unreactive closed conformations.
Rates of conformational switching and interflavin ET differ among the four flavoproteins
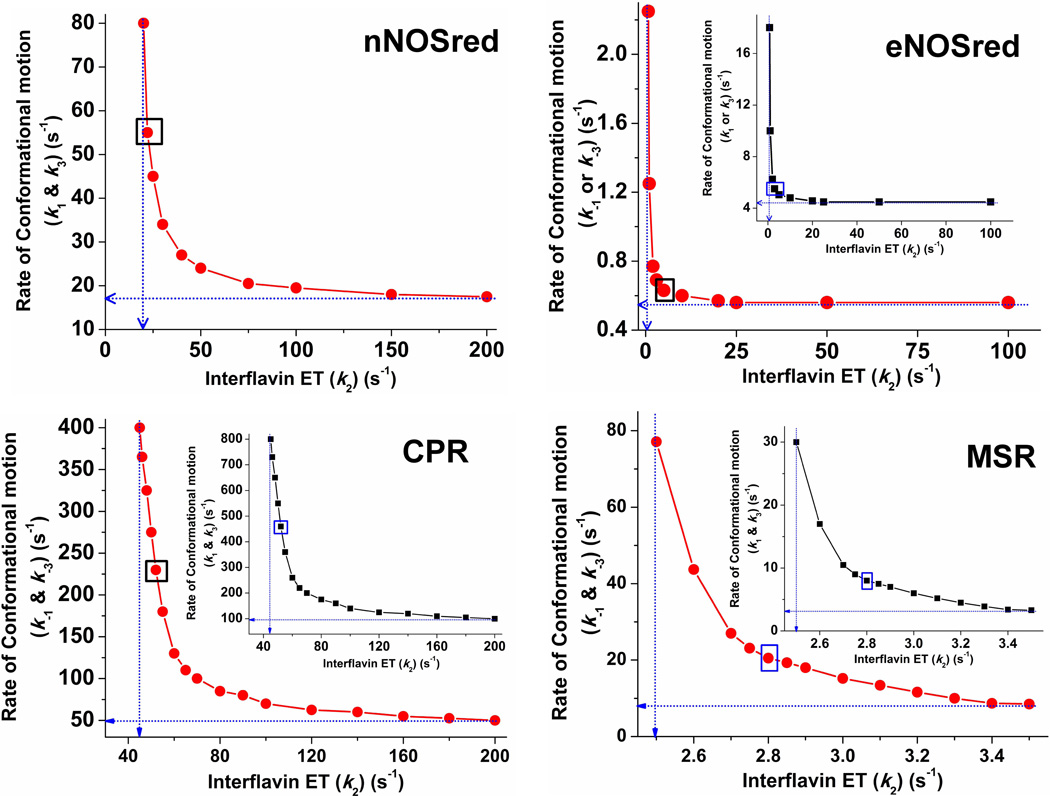
To derive rate estimates for the conformational transitions between the open reactive and closed unreactive states, and to derive an estimated rate of interflavin ET in each flavoprotein, we used a computer simulation to fit the observed kinetic traces of cytochrome c reduction in Fig. 2 according to the four-state kinetic model in Fig. 1 that links the rates of conformational motions and interflavin ET to the electron flux to cytochrome c [34]. We previously used the same approach to derive rate estimates for nNOSred, nNOSred mutants, and eNOSred [34;35].
In the current study, we incorporated the estimated Khq values derived above into the simulations and then used an iterative method to find rate pairs for interflavin ET (k2) and FMN domain conformational transitions (k1, k−1, k3, k−3) that support the experimentally-observed rate of electron flux to cytochrome c for each flavoprotein [34;35]. This analysis generated combinations of rate pairs that can each support the observed steady-state electron flux rate for each enzyme, with the general relationship being that faster rates of interflavin ET can pair with slower rates of FMN conformational switching, and vice-versa. The traces in the graphs of Fig. 3 indicate the allowable rate pairs for each enzyme that we derived from this fitting process. This analysis is instructive because it indicates the lower rate boundaries for the interflavin ET and the conformational opening and closing steps in each enzyme [34;35]. For example, the horizontal blue tangent line drawn in the nNOSred panel of Fig. 3 (upper left panel) indicates the lowest boundary rate of conformational switching (17 s−1) that would be able to maintain the observed electron flux to cytochrome c (~8 s−1 for nNOSred at 10 °C), when paired with a comparatively fast rate of interflavin ET. Similarly, a vertical blue tangent line indicates the lowest boundary rate for interflavin ET (k2 ~20 s−1) that would still be able to maintain the experimentally observed electron flux, when paired with a very fast rate of conformational switching. Because the conformational Khq is 1 in nNOSred, k1 = k−1 = k3 = k−3, and so only one lower boundary rate value is needed to describe all four of the conformational opening and closing steps in the kinetic model. However, for the three other flavoproteins, their conformational Khq values are either greater or less than 1, and so stipulate that k1 ≠ k−1 and k3 ≠ k−3. Thus, two distinct lower boundary values are derived in this case that describe the conformational opening and closing rates (indicated by the blue lines in the main panels and in the insets).
Figure 3. Conformational motion and interflavin electron transfer rate settings that support observed electron flux through dual-flavin enzymes.
Data were obtained from simulations of the kinetic model in Fig. 1. For a given k2 value, rates of conformational motion were screened for a value that yielded the observed electron flux. For all panels, blue dotted lines indicate the lower boundary values for the rates of conformational motion (Y-intercept) and interflavin electron transfer (X-intercept). For eNOSred, CPR, and MSR, the main panel shows the resulting values in terms of the conformational opening rates (k−1 = k−3). In the inset, the same data points are plotted, this time indicating the rates of conformational closing (k1 = k3) on the y axis. The boxed points in each panel indicate the best-fit rate pairs. For nNOSred, rates of conformational motion were screened for a given k2 value that yielded an electron flux of ~8 s−1 when Keq =1 and k1 = k−1 = k3 = k−3. In eNOSred conformational motion rates were set so that Khq = Ksq = 0.125 and the electron flux equaled 0.49 s−1. For CPR to achieve an electron flux of 28 s−1 and a Keq of 0.5, a number of k2 values were simulated in combination with different conformational motion rates. For MSR, rates of conformational motion were screened for a corresponding k2 value that yielded an electron flux of 0.7 s−1, while Khq = Ksq = 2.6, k1 = k3, and k−1 = k−3.
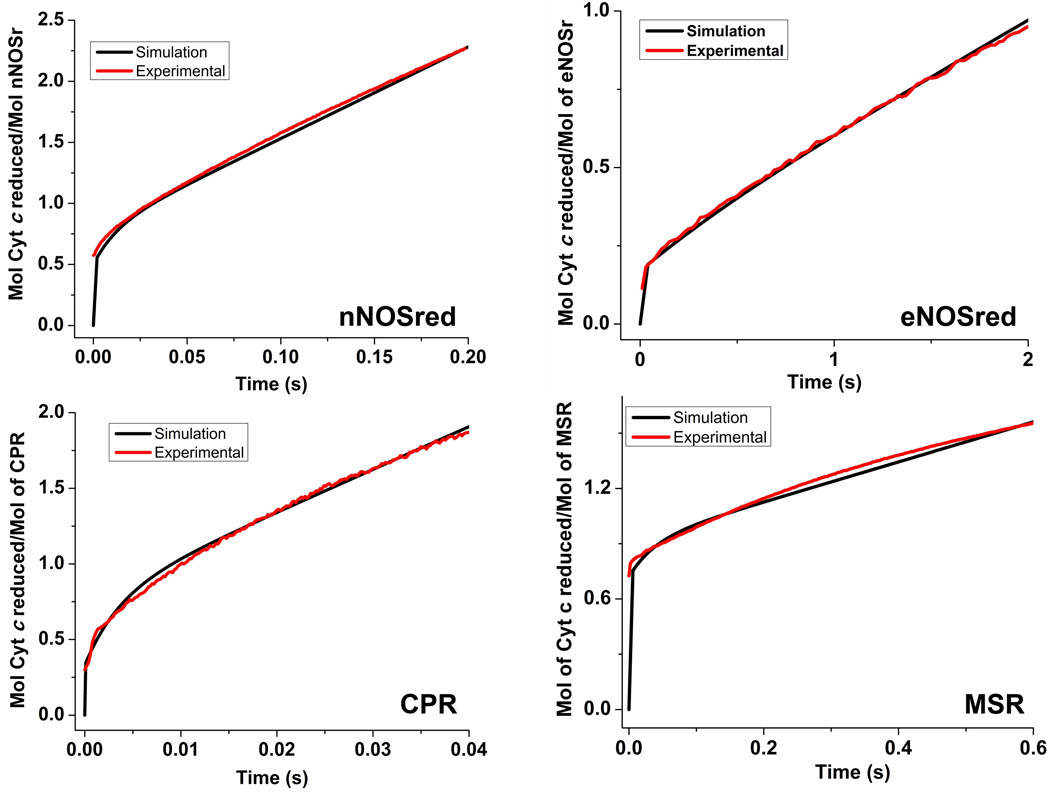
To obtain the best-fit rate pair values for each flavoprotein, we incorporated as an additional fitting parameter the elapsed time that was needed by each enzyme to reduce the first equivalent of cytochrome c [34;35] (obtained from the data in Fig. 2 as discussed above). These simulations provided a best-fit rate pair for each enzyme, whose values are marked by a blue or black box on each trace in Fig. 3, and they are listed in Table 3. The simulated kinetic traces of cytochrome c reduction for each enzyme, using these best-fit rate values, are overlaid with the actual experimental traces in Fig. 4, and show that a reasonable fit was obtained in each case. Together, the analysis suggests that the rates of conformational switching differ by over two orders of magnitude among the four diflavin reductases, according to rank order: CPR > nNOSred > MSR > eNOSred, while their corresponding rates of interflavin ET range over one order of magnitude according to the same rank order.
Figure 4. Simulated reactions of fully-reduced dual-flavin enzymes with excess cytochrome c.
The traces were obtained by simulating the kinetic model in Fig. 1, using different rates of conformational motion and interflavin electron transfer (k2) to match experimental traces for each of the four dual-flavins under study. Total protein concentration is 1.0, and the concentration of enzyme species d + a was set equal to 1.0 at time = 0 in the simulations. For nNOSred, conformational motion rates were set so that Khq = Ksq = 1, and k1 = k−1 = k3 = k−3. With these parameters, the closest match to the experimental traces in Fig. 2 was found when k1 = k−1 = k3 = k−3= 53, and k2 = 22. For eNOSred, conformational motion rates were set so that Khq = Ksq = 0.125, k1 = k3, and k−1 = k−3 = 0.125×k1. The best fit of experimental traces was found when the kinetic settings (s−1) were k1 = k3 = 5.0, k−1 = k−3 = 0.63, k2 = 5.0. In case of CPR the conformational motion rates were set so that Khq = Ksq = 0.5, k1 = k3, and k−1 = k−3 = 0.5×k1. The best fit was found when the kinetic settings (s−1) were k1 = k3 = 460, k−1= k−3 = 230, k2 = 52. For MSR, conformational motion rates were Khq = Ksq = 2.57 and k1 = k3;k−1 = k−3 = 2.57×k1. The best fit was found when the kinetic settings (s−1) were k1 = k3 = 8, k−1= k−3 = 20.5, k2= 2.8. Simulated traces (black lines) are overlayed with experimental traces (red lines).
The approach to and the final steady-state distributions of enzyme species differ among the flavoproteins
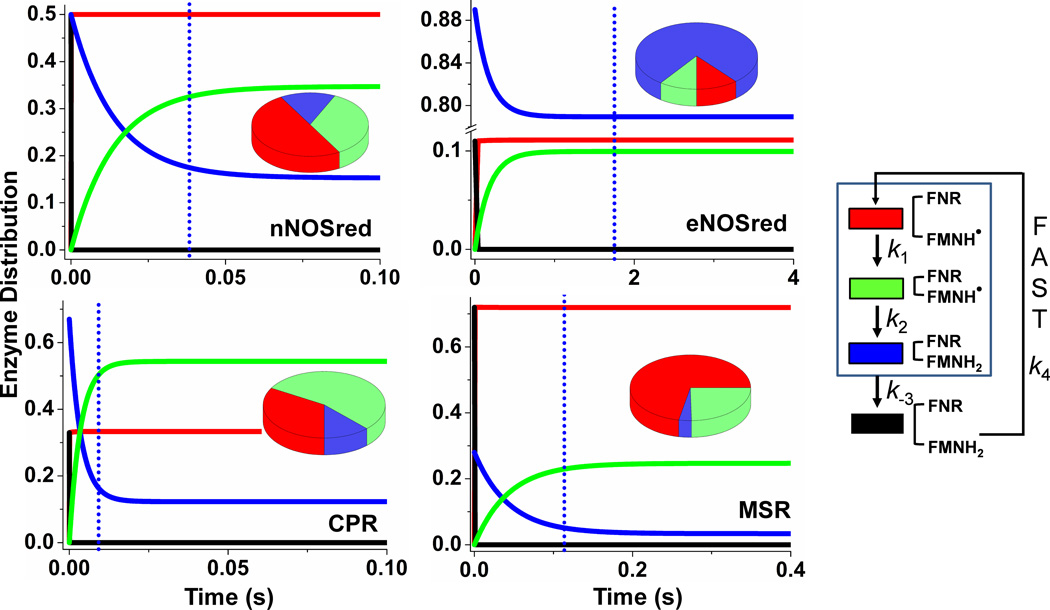
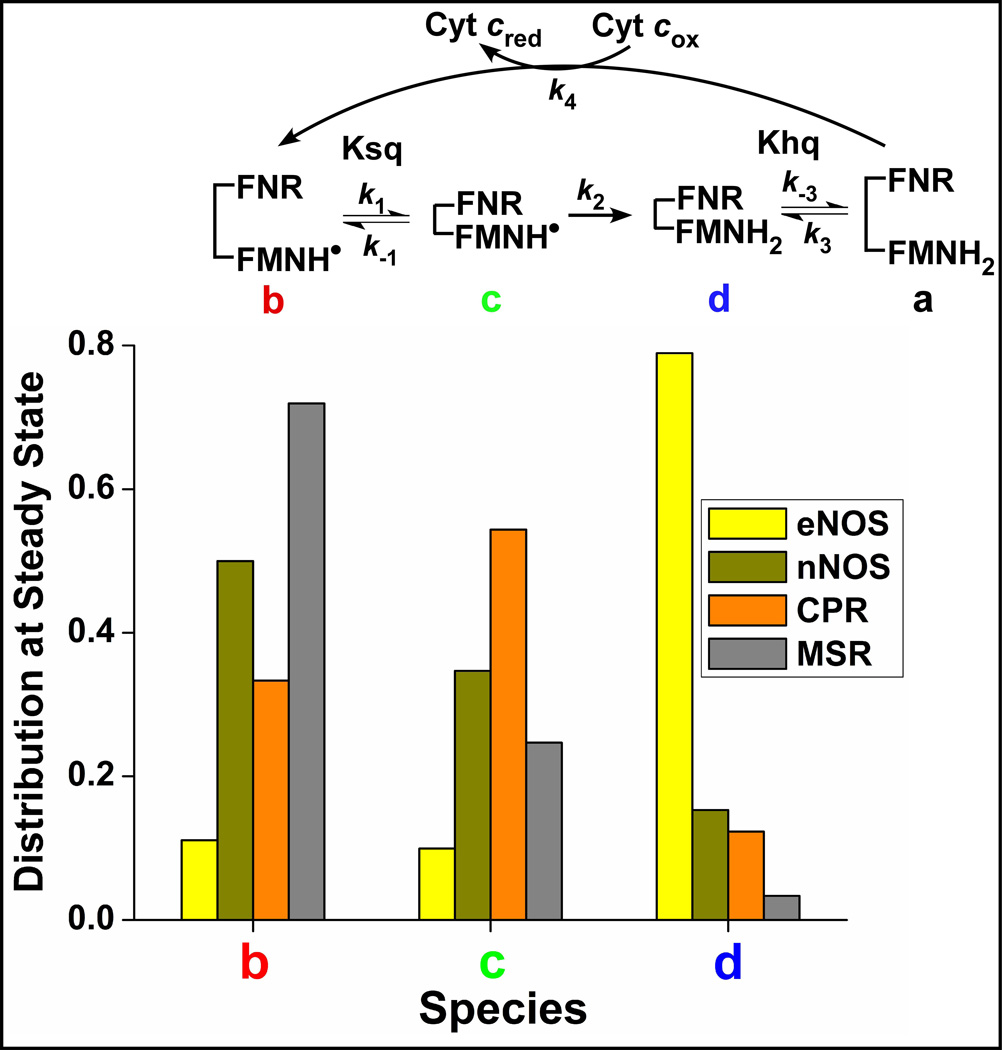
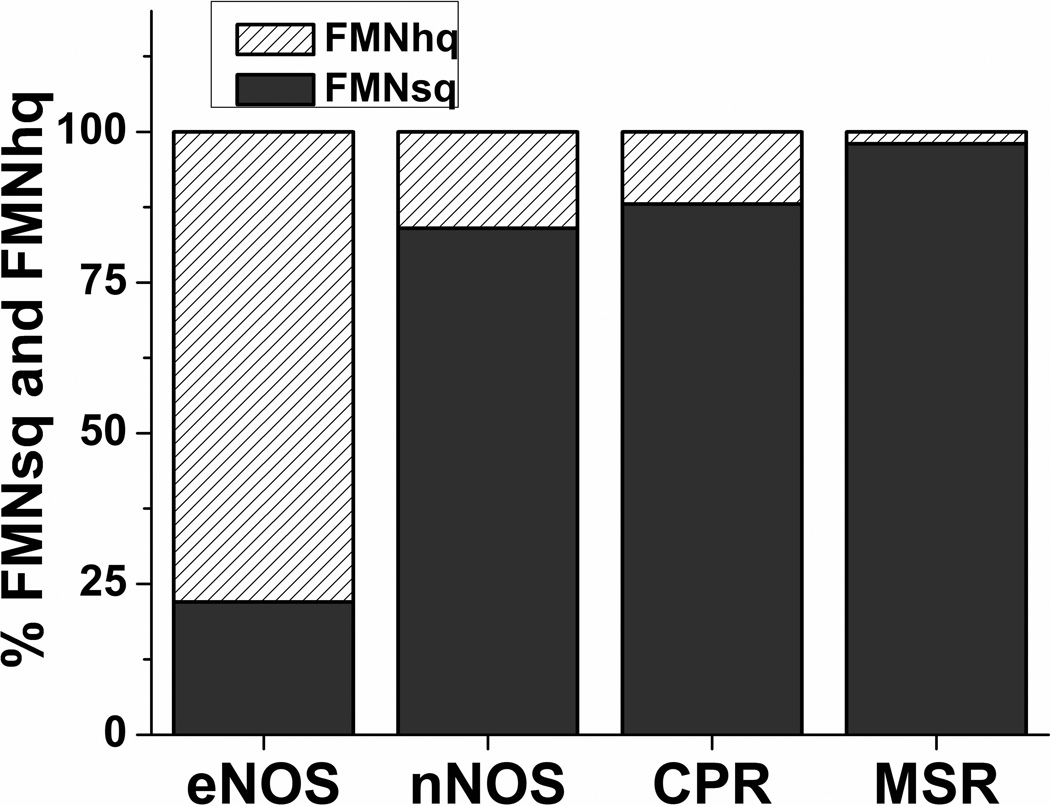
The simulations can also predict how the four enzyme conformational/redox species noted in the kinetic model (species a-d in Fig. 1) populate as each fully-reduced flavoprotein (species d + a) reacts with excess cytochrome c as depicted in the Fig. 2 kinetic traces. Fig. 5, show how the species distribution changes with time in all four enzymes. The kinetics of redistribution differ broadly among the flavoproteins as a direct consequence of their different rates of conformational transitions and interflavin ET. Also, only the eNOSred achieves a steady-state species distribution before the first equivalent of cytochrome c is reduced (time point which is indicated by a dotted vertical line in each panel), whereas the three other flavoproteins only approach their steady-state distributions within the time needed to reduce the first equivalent. All four enzymes also have unique steady-state species distributions, as indicated by the pie graphs in Fig. 5, and Fig. 6. None of the four flavoproteins build up as the conformationally-open FMNhq species (species a) during catalysis because this species reacts so quickly with the cytochrome c, but there are significant differences in the buildup of the species b, c, and d. For example, the fast conformational transitions in CPR cause its closed FMNsq species (species c) to build up, because the subsequent interflavin ET step is relatively slow. In contrast, eNOSred predominantly builds up as its closed FMNhq species d, because it opens relatively slowly to react with cytochrome c. The MSR and nNOSred both buildup mainly as their open FMNsq species b, primarily because their conformational Ksq values provide a relatively poor driving force for the open form to transition to the closed form as is required for the FMNsq to be reduced. These properties also determine what percentage of FMNsq versus FMNhq should be present in each flavoprotein during its steady-state reaction with cytochrome c (Fig. 7). The eNOSred is predicted to contain 80% FMNhq due to its buildup primarily as the closed reduced species d during catalysis. In marked contrast, the CPR, MSR, and nNOSred are predicted to contain between 80 to 95% FMNsq during their steady-state reactions. The difference between eNOSred and MSR is particularly striking, because they have nearly equivalent slow cytochrome c reductase activities (Table 1). Thus, their electron flux rates are similarly limited but for opposite reasons: eNOS has slow opening when it is in the reduced (FMNhq) closed form, while MSR has slow closing when it is in the oxidized (FMNsq) open form.
Figure 5. Patterns of enzyme distribution versus reaction time in dual-flavin enzymes.
Rate pairs of conformational motion and interflavin electron transfer (obtained from the simulated best-fit values (Fig. 4)) were used in each case to simulate and achieve experimentally obtained cytochrome c reduction rates. Lines indicate the relative concentrations of each enzyme species a-d (see Fig. 1), with the total enzyme concentration being 1.0 and the concentration of enzyme species d + a set equal to 1.0 at time = 0 in the simulations. The blue dotted line in each panel marks the time required for the enzyme to reduce one equivalent of cytochrome c. Kinetic settings were: For nNOSred: k1 = k−1= k3 = k−3 = 53, k2 = 22; For eNOSred: k1 = k−3 = 5.0, k−1 = k3 = 0.63, k2 = 5; For CPR: k1 = k3 = 460, k−1 = k−3 = 230, k2 = 52; For MSR k1 = k3 = 8, k−1 = k−3 = 20.5, k2 = 2.8. Pie graphs in each panel are showing different species distributed at steady-state. Color scheme for different species are as follows: species a: black, species b: red, species c: green, and species d: blue.
Figure 6. The steady-state distributions of different species among the four flavoproteins.
Buildup of different species at steady-state. Buildup of the conformationally open FMNhq species (species a) is essentially nil in all enzymes because it reacts so quickly with the excess cytochrome c.
Figure 7.
Percentage proportion of FMNsq versus FMNhq present in each enzyme during its steady-state reaction with cytochrome c.
Discussion
Translating the cytochrome c reduction data to enzyme conformational parameters
It is important to note that our stopped-flow experiments measured reactivity of the reduced FMN domains (FMNhq) toward cytochrome c, and did not directly measure specific changes in the protein conformation. Thus, linking our data to flavoprotein structural changes depends on two assumptions: First, that the FMNhq is unreactive toward cytochrome c when the enzyme is in a conformationally-closed form. A recent study showed that a covalently-cross-linked closed form of CPR (whose crystal structure was also reported) only supported 5% electron flux to cytochrome c relative to the un-linked CPR [26]. We have obtained the same result for a similarly cross-linked nNOSred (Dai, Y and Stuehr DJ, unpublished observation). Thus, the assumption holds for CPR & nNOSred, and we assume it holds for the other two flavoproteins, given their structural and mechanistic similarities. Secondly, we assume that the reduced FMN domain is very reactive toward cytochrome c once it becomes unencumbered enough for its bound FMNhq to come within a distance that allows fast ET to cytochrome c. The cytochrome c reactivity of the reduced, isolated FMN domains of CPR and nNOS [37;46] were sufficiently reactive to support this assumption, and we assume this is also the case for MSR and eNOS.
At equilibrium, the four reduced flavoproteins likely populate a myriad of closed and open forms that distribute across a conformational and energetic landscape as described for example by Scrutton & colleagues [47]. Crystal structures of nNOSred and CPR illustrate their tightly closed forms [23;25], and more recent crystallographic, cryogenic, SAXS, SANS, and solution NMR or EPR studies of CPR and its variants, or of nNOS, have identified conformationally-open forms and shown that multiple states are indeed populated at equilibrium [27;32;48–50]. The FMN domain in each of three open conformer crystal structures of CPR is unencumbered enough to engage in fast ET with cytochrome c. But it is unclear which of these open conformations may actually form during catalysis, or how well they each might cycle to participate in steady-state electron flux to cytochrome c. Obviously, minimal conformational changes that would still allow ET from FMNhq to cytochrome c would be likely to enable the fast conformational cycling and fast electron flux that is seen in CPR. In contrast, the more extended open conformers may participate poorly, given that these conformers were observed in the structure of CPR variants that display very low steady-state cytochrome c reductase activity [27]. Populating extended conformations, or alternatively, undergoing conformational relaxations back to closed conformations, have been suggested to be gradual processes [51;52] and to possibly be artifacts of dilution [30;53] or of the long equilibration times that enzyme molecules experience when being readied for laboratory studies. We do not know if such relaxations redistributed the conformationally-extended or closed forms in the fully-reduced flavoproteins prior to our stopped-flow mixing experiments. However, the traces in Fig. 2 may suggest that this did not happen to any significant extent. Consider that the cytochrome c reductase activities that were calculated from tangents drawn to the stopped-flow traces in Fig. 2 matched fairly well with the steady-state activities that we determined for MSR, CPR, eNOSred, and nNOSred in standard activity assays (table 3) that were run over several minutes and did not involve a pre-reduction of the enzymes before their activity was measured. Indeed, the kinetic traces in Fig. 2 imply that each fully-reduced flavoprotein quickly achieved the conformational and redox cycling that normally occurs during its steady-state reaction with cytochrome c. If instead, a significant relaxation to poorly-cycling open conformers, or a relaxation to closed conformers, had occurred during the time required to prepare the enzymes for the stopped-flow experiments, then we would expect a mismatch between the tangent-based activity derived from the traces in Fig. 2 and the steady state activity measured by the standard method.
Possible relationships between the enzyme redox state and conformational behaviors
Studies with CPR have suggested that the flavin reduction may impact the conformational setpoint by shifting CPR to favor more open forms [48;49;54]. NADPH binding site occupancy can also impact conformational behaviors of CPR and nNOS [13;15;30;35;40;54–56]. Moreover, the interflavin ET step that generates FMNhq (k2 in Fig. 1) has been suggested to gate a subsequent conformational opening step that frees the reduced FMNhq domain in CPR (k−3 in Fig. 1) [47;55;57]. These concepts are fundamentally important but are not directly addressed by our results because we only investigated the conformational behaviors of the fully-reduced flavoproteins. However, some of our observations may shed some light. First, we found that none of the four flavoproteins existed in a completely open conformation when they were fully-reduced (i.e. when they contained FADhq, and FMNhq, and bound NADPH). Indeed, our evidence suggests that two of the four (eNOSred and CPR) exist predominantly in their closed conformations even when fully reduced. This implies that reduction to create FMNhq has at most a partial and variable capacity to trigger conformational opening among the four flavoproteins. This can be viewed within the context of recent SAXS and SANS studies on CPR, which reported that a 2-electron reduction of the fully-oxidized enzyme caused some opening and stabilized about 50% of the enzyme in an open conformation [49]. Note that CPR in the two-electron reduced state would predominantly contain FADsq and FMNsq, and so the conformational change that was reported here was predominantly associated with the transition of FMNox to FMNsq in CPR. The study did not address what effect, if any, that further reduction of FMNsq to FMNhq might cause on the CPR conformation. Given that our study estimated that fully-reduced CPR is 1/3 open and 2/3 closed, our results imply there may be no further opening upon reduction of its FMNsq to FMNhq. Secondly, when one compares the best-fit rate estimate that we derived for the interflavin ET step (k2) and the rate for the conformational opening step of each reduced flavoprotein (k−3), it shows that no correlation exists between these two rates (Table 3). For example, the estimated rate of interflavin ET in eNOSred is eight times faster than its estimated rate of conformational opening. For MSR and CPR the situation is reversed, and their estimated rates of conformational opening are seven and four times faster than their estimated rates of interflavin ET, respectively. In contrast, the estimated conformational closing rates of the FMNsq-containing species (k1 in Fig. 1), which is the step that precedes the interflavin ET, are more similar to the estimated rates of interflavin ET in eNOSred and MSR (Table 3). Further investigation is clearly warranted, but in any case, our study suggests that one need not invoke a conformational sensitivity toward the FMNsq and FMNhq redox states, or a gating of the FMN domain motion to the interflavin ET step, in order to model electron flux through these flavoproteins.
Structural Considerations and Possible Basis for Different Electron Flux among the Diflavin Reductases
Perhaps the most remarkable findings suggested by our study is that the four diflavin reductases differ so widely in their conformational equilibrium setpoints and also exhibit a two order of magnitude range in their rates of FMN domain conformational switching. We found that MSR and eNOSred have conformational equilibrium setpoints that lie on either side of the optimum setting (Khq = 1 being approximately optimal in this model, [15;34]), and their Ksq and Khq set points therefore help to limit their electron flux due to an imbalance between their conformational closing and opening rates. However, on balance it is their slow rates of conformational switching that present the biggest barrier to their electron flux. Consider that the Khq setpoint of CPR is also suboptimal, but its much faster rates of conformational switching supercede this limitation. Interestingly, our results predict that slightly shifting the Khq of CPR toward its open reactive form would increase its activity above wild-type enzyme, and we suspect that this effect might explain a recently-characterized CPR point mutant with a higher than normal activity [49].
The best-fit rates we derived for CPR conformational switching (230 & 460 s−1) are in the middle to high range of similar switching rates reported in other multi-domain redox enzymes [28;55;56;58], while the rates we estimated for nNOSred, MSR, and eNOSred are in the lower to extreme lower end of the range. This suggests that “suppressive” features exist in these three flavoproteins that slow down their rates of conformational cycling relative to CPR or other multi-domain redox enzymes, and for MSR & eNOSred may also give them suboptimal conformational Khq settings, thus preventing them from achieving a faster electron flux as is seen for CPR. What might these features be? We know that calmodulin binding to nNOSred boosts its cytochrome c reductase activity to the level of CPR [15;36;40;59–63] and thus calmodulin likely relieves the suppressive features that are present in the nNOSred [15;40;61;64].
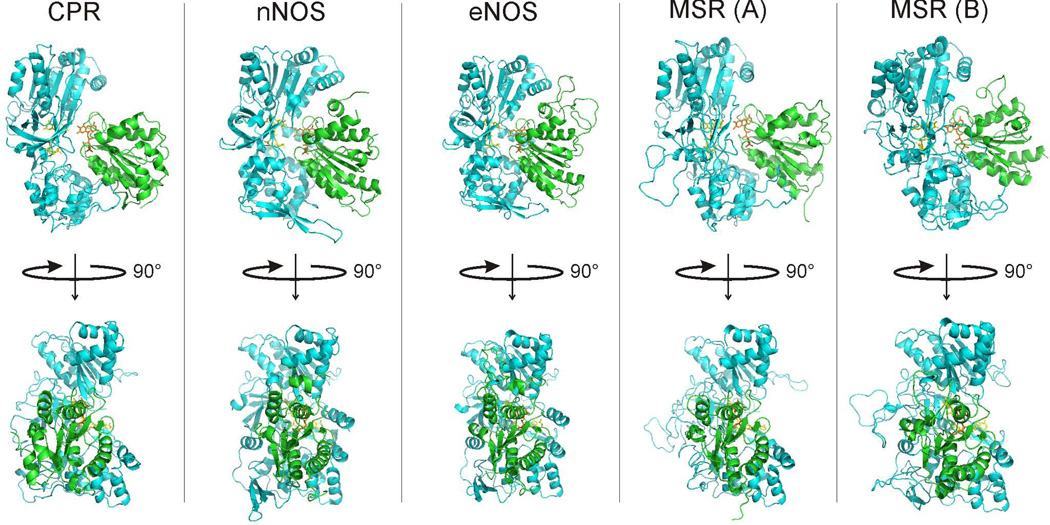
Regarding the protein structural features held in common among diflavin reductases, we may consider the FNR-FMN domain linker that is directly involved in conformational switching and differs considerably with regard to its length and amino acid sequence [18;19;25;27;65], and also consider the domain-domain interface. We have examined the available structural information on dual-flavin reductases. The structure of the diflavin module has been elucidated for nNOS and CPR. We used the PDB files 1TLL [25] for nNOS and 1AMO [23] for CPR. As the structures of the complete diflavin module are not available for eNOS and MSR we have built structural models using available modeling servers (See materials and methods).
The analysis of the FNR/FMN domain interfaces in the structures and models is shown in Table 4. In summary, it appears that the interface area between FNR and FMN domains is very similar for nNOS, eNOS and CPR (~2500 Å2) whereas the interface is smaller in the case of MSR (~2000 Å2). The arrangement of the FNR and FMN domains appears to be very similar in all cases (Fig. 8) and the changes in surface area can be ascribed to the presence of “extra” elements in NOS and CPR as compared to MSR. NOS proteins include an additional autoinhibitory helix element in their FMN domain that is known to modulate the interaction with CaM (amino acids 836–849 in nNOS, 609–621 in eNOS) [25;66] and also contain a C-terminal extension in their FNR domain that modulates the FMN-FNR domain interaction and CaM response [39]. Both elements contribute to their FMN-FNR interface. In the case of CPR, the FMN domain possesses an extra helical segment (amino acids 67–78) that interacts with the FNR domain, including the formation of a salt bridge between amino acids Arg78 of the FMN domain and Asp352 of the FNR domain. The MSR sequence does not incorporate any of these elements, and thus their resulting interaction area is around 500 Å2 smaller than for NOS or CPR (Table 4).
Table 4. Properties of the FNR/FMN domain interaction surfaces of selected di-flavin reductases.
| Protein | Neuronal nitric oxide synthase |
Endothelial nitric oxide synthase |
Cytochrome P450 reductase |
Methionine Synthase reductase |
Methionine Synthase reductase |
|---|---|---|---|---|---|
| PDB file | 1TLL | Model (template: 1TLL) |
1AMO | Model (template: 1AMO) |
Model (template: 1TLL) |
| Total interface area (Å2) |
2510.9 | 2502.2 | 2501.5 | 1970.4 | 1980.8 |
| % Charged residues |
40.0 | 47.4 | 57.1 | 52.9 | 31.2 |
| Non bonded contacts |
83 | 101 | 84 | 134 | 60 |
| Hydrogen bonds |
2 | 3 | 5 | 2 | 0 |
| Salt bridges | 1 | 1 | 4 | 2 | 1 |
Figure 8. Crystal Structures and structure based models of flavoproteins.
We used 1AMO for rat cytochrome P450 reductase and 1TLL for rat nNOS. The available structure of rat nNOS (1TLL) was used as template for the bovine eNOS model. We built two different models for methionine synthase reductase using either nNOS (1TLL) or cytochrome P450 reductase (1AMO) structures as templates. Upper panels are side view and Lower panels are top view of structure. FNR domain is shown in blue, FMN domain is shown in green; FAD (yellow) and FMN (orange) are shown as sticks.
MSR has a long hinge region (~80 amino acids) between the FMN and FNR domains, different from the shorter hinges (14 to 25 amino acids) in NOS or CPR. Due to its location in the primary structure, is reasonable to think that this hinge region could mediate additional FMN/FNR domain interactions in MSR. However, some secondary evidence suggests that this may not be the case. First, crystallization of the hinge and FNR portion of MSR (residues 166–698) was only able to determine the structure of the portion of the hinge between amino acids 217 and 247 [44]. This section stretches away from the putative location of the FMN module and is unlikely to contribute to FNR-FMN binding; the region between residues 166 and 217 was too flexible for the structure to be determined. Secondly, the difficulty to obtain a crystal structure of the whole MSR protein also points towards a transient, not stable FNR-FMN complex in MSR. Thirdly, our experimental data indicates that an open conformation is prevalent in MSR (Table 2). It is possible that the presence of the FMN module helps to arrange the hinge region in a more stable structure that provides additional stability to the FMN-FNR module, but the evidence discussed above seems to argue against this hypothesis. In the absence of more structural data, the contribution of this sequence to interdomain binding remains an open question.
As pointed out before, NOS and CPR include a shorter (~14 amino acids in CPR, ~25 amino acids in NOS), flexible hinge region between the FMN and FNR domains. The composition of the hinge modulates the equilibrium between open and closed conformations, as shown by studies on CPR [27;67] and NOS [65;68]. Due to the flexibility of these regions, they do not appear in the crystal structures available, and we excluded the sequences from our analysis in the modeled structures. As discussed in the case of MSR, the hinge can contribute to the energy of the closed conformation. However, we expect that contribution to be minor in terms of complex stabilization, as these fragments do not form a fixed, “clamp” structure between the FNR and FMN domains, as proven by the crystallography experiments. In turn, they appear to operate on the conformational equilibrium by limiting the degrees of freedom of the open conformations [15;27;65;67;68]. In this way, the effect of the hinges will be more related to the kinetics of the conformational changes between open and closed conformations, whereas the equilibrium setpoint between both conformations will be more related to the strength of the interaction between the FNR and FMN domains in the closed conformation. Thus we consider that the omission of the hinge fragments from the interface analysis will not modify the conclusions of the study.
The comparison of the binding interactions for the structures and models indicates that the number of hydrogen bonds plus salt bridges is higher for eNOS (4) and CPR (9) than for nNOS (3) (Table 4). Qualitatively, and considering that the stability of the complex will be mainly determined by the interface area, the stability of the “closed” conformations could be ordered as eNOS≈CPR>nNOS>MSR. We observe that this ranking correlates with the observed amount of “closed” conformation for each fully-reduced enzyme (Table 2). Although our analysis cannot accurately predict the 40% ability in the closed conformation fractions for eNOS, nNOS and CPR, the lower stability of the MSR closed conformation is in agreement with our calculated parameters. Clearly, how these and other structural features influence interflavin ET, conformational Khq setpoints, and switching rates remains a topic of ongoing study.
In sum, our study provides the first comparative estimates of the conformational equilibria setpoints, interflavin ET rates, and conformational switching rates in four mammalian dual flavin reductases, and shows how differences in these parameters can explain their widely different capacities to support electron flux from NADPH to cytochrome c.
Supplementary Material
Acknowledgements
We thank Deb Durrah for technical assistance and Dr. David Coe for performing initial experiments with CPR. We thank Dr. Ruma Banerjee for the MSR bacterial expression construct. This work was supported by National Institutes of Health Grants GM51491 and HL076491 to D.J.S.
Abbreviations
- NOS
nitric oxide synthase
- eNOS
endothelial nitric oxide synthase
- nNOS
neuronal nitric oxide synthase
- NOSred
reductase domain of NOS
- CaM
Calmodulin
- CPR
cytochrome P450 reductase
- MSR
Methionine synthase reductase
- FAD
flavin-adenine dinucleotide
- FADhq
two-electron reduced (hydroquinone) FAD
- FMN
flavin mononucleotide
- FMNsq
one-electron reduced (semiquinone) FMN
- FMNhq
two-electron reduced (hydroquinone) FMN
- FNR
Ferredoxin-NADP+-Reductase-like domain
- NADPH
nicotinamide adenine dinucleotide phosphate, reduced form
- EPPS
4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
References
- 1.Aigrain L, Fatemi F, Frances O, Lescop E, Truan G. Dynamic control of electron transfers in diflavin reductases. Int. J. Mol. Sci. 2012;13:15012–15041. doi: 10.3390/ijms131115012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De CL, Mattevi A. New frontiers in structural flavoenzymology. Curr. Opin. Struct. Biol. 2006;16:722–728. doi: 10.1016/j.sbi.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Iyanagi T. Structure and function of NADPH-cytochrome P450 reductase and nitric oxide synthase reductase domain. Biochem. Biophys. Res. Commun. 2005;338:520–528. doi: 10.1016/j.bbrc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 4.Joosten V, van Berkel WJ. Flavoenzymes. Curr. Opin. Chem. Biol. 2007;11:195–202. doi: 10.1016/j.cbpa.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Moreno C. New roles of flavoproteins in molecular cell biology. FEBS J. 2009;276:4289. doi: 10.1111/j.1742-4658.2009.07140.x. [DOI] [PubMed] [Google Scholar]
- 6.Iyanagi T, Xia C, Kim JJ. NADPH-cytochrome P450 oxidoreductase: prototypic member of the diflavin reductase family. Arch. Biochem. Biophys. 2012;528:72–89. doi: 10.1016/j.abb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porter TD, Kasper CB. NADPH-cytochrome P-450 oxidoreductase: flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochemistry. 1986;25:1682–1687. doi: 10.1021/bi00355a036. [DOI] [PubMed] [Google Scholar]
- 8.Porter TD. An unusual yet strongly conserved flavoprotein reductase in bacteria and mammals. Trends Biochem. Sci. 1991;16:154–158. doi: 10.1016/0968-0004(91)90059-5. [DOI] [PubMed] [Google Scholar]
- 9.Murataliev MB, Feyereisen R, Walker FA. Electron transfer by diflavin reductases. Biochim. Biophys. Acta. 2004;1698:1–26. doi: 10.1016/j.bbapap.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Iyanagi T, Mason HS. Some properties of hepatic reduced nicotinamide adenine dinucleotide phosphate-cytochrome c reductase. Biochemistry. 1973;12:2297–2308. doi: 10.1021/bi00736a018. [DOI] [PubMed] [Google Scholar]
- 11.Vermilion JL, Coon MJ. Purified liver microsomal NADPH-cytochrome P-450 reductase. Spectral characterization of oxidation-reduction states. J. Biol. Chem. 1978;253:2694–2704. [PubMed] [Google Scholar]
- 12.Shen AL, O’Leary KA, Kasper CB. Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J. Biol. Chem. 2002;277:6536–6541. doi: 10.1074/jbc.M111408200. [DOI] [PubMed] [Google Scholar]
- 13.Daff S. NO synthase: structures and mechanisms. Nitric. Oxide. 2010;23:1–11. doi: 10.1016/j.niox.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Feng C. Mechanism of Nitric Oxide Synthase Regulation: Electron Transfer and Interdomain Interactions. Coord. Chem. Rev. 2012;256:393–411. doi: 10.1016/j.ccr.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuehr DJ, Tejero J, Haque MM. Structural and mechanistic aspects of flavoproteins: electron transfer through the nitric oxide synthase flavoprotein domain. FEBS J. 2009;276:3959–3974. doi: 10.1111/j.1742-4658.2009.07120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leclerc D, Wilson A, Dumas R, Gafuik C, Song D, Watkins D, Heng HH, Rommens JM, Scherer SW, Rosenblatt DS, Gravel RA. Cloning and mapping of a cDNA for methionine synthase reductase, a flavoprotein defective in patients with homocystinuria. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3059–3064. doi: 10.1073/pnas.95.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olteanu H, Banerjee R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J. Biol. Chem. 2001;276:35558–35563. doi: 10.1074/jbc.M103707200. [DOI] [PubMed] [Google Scholar]
- 18.Wolthers KR, Scrutton NS. Protein interactions in the human methionine synthase-methionine synthase reductase complex and implications for the mechanism of enzyme reactivation. Biochemistry. 2007;46:6696–6709. doi: 10.1021/bi700339v. [DOI] [PubMed] [Google Scholar]
- 19.Paine MJ, Garner AP, Powell D, Sibbald J, Sales M, Pratt N, Smith T, Tew DG, Wolf CR. Cloning and characterization of a novel human dual flavin reductase. J. Biol. Chem. 2000;275:1471–1478. doi: 10.1074/jbc.275.2.1471. [DOI] [PubMed] [Google Scholar]
- 20.Ostrowski J, Barber MJ, Rueger DC, Miller BE, Siegel LM, Kredich NM. Characterization of the flavoprotein moieties of NADPH-sulfite reductase from Salmonella typhimurium and Escherichia coli. Physicochemical and catalytic properties, amino acid sequence deduced from DNA sequence of cysJ, and comparison with NADPH-cytochrome P-450 reductase. J. Biol. Chem. 1989;264:15796–15808. [PubMed] [Google Scholar]
- 21.Munro AW, Leys DG, McLean KJ, Marshall KR, Ost TW, Daff S, Miles CS, Chapman SK, Lysek DA, Moser CC, Page CC, Dutton PL. P450 BM3: the very model of a modern flavocytochrome. Trends Biochem. Sci. 2002;27:250–257. doi: 10.1016/s0968-0004(02)02086-8. [DOI] [PubMed] [Google Scholar]
- 22.Munro AW, Girvan HM, McLean KJ. Cytochrome P450--redox partner fusion enzymes. Biochim. Biophys. Acta. 2007;1770:345–359. doi: 10.1016/j.bbagen.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia C, Panda SP, Marohnic CC, Martasek P, Masters BS, Kim JJ. Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13486–13491. doi: 10.1073/pnas.1106632108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J. Biol. Chem. 2004;279:37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 26.Xia C, Hamdane D, Shen AL, Choi V, Kasper CB, Pearl NM, Zhang H, Im SC, Waskell L, Kim JJ. Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding. J. Biol. Chem. 2011;286:16246–16260. doi: 10.1074/jbc.M111.230532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamdane D, Xia C, Im SC, Zhang H, Kim JJ, Waskell L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laursen T, Jensen K, Moller BL. Conformational changes of the NADPH-dependent cytochrome P450 reductase in the course of electron transfer to cytochromes P450. Biochim. Biophys. Acta. 2011;1814:132–138. doi: 10.1016/j.bbapap.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Meints CE, Gustafsson FS, Scrutton NS, Wolthers KR. Tryptophan 697 modulates hydride and interflavin electron transfer in human methionine synthase reductase. Biochemistry. 2011;50:11131–11142. doi: 10.1021/bi2012228. [DOI] [PubMed] [Google Scholar]
- 30.Pudney CR, Heyes DJ, Khara B, Hay S, Rigby SE, Scrutton NS. Kinetic and spectroscopic probes of motions and catalysis in the cytochrome P450 reductase family of enzymes. FEBS J. 2012;279:1534–1544. doi: 10.1111/j.1742-4658.2011.08442.x. [DOI] [PubMed] [Google Scholar]
- 31.Wolthers KR, Scrutton NS. Electron transfer in human methionine synthase reductase studied by stopped-flow spectrophotometry. Biochemistry. 2004;43:490–500. doi: 10.1021/bi0356303. [DOI] [PubMed] [Google Scholar]
- 32.Aigrain L, Pompon D, Morera S, Truan G. Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase. EMBO Rep. 2009;10:742–747. doi: 10.1038/embor.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh DK, Ray K, Rogers AJ, Nahm NJ, Salerno JC. FMN fluorescence in inducible NOS constructs reveals a series of conformational states involved in the reductase catalytic cycle. FEBS J. 2012;279:1306–1317. doi: 10.1111/j.1742-4658.2012.08525.x. [DOI] [PubMed] [Google Scholar]
- 34.Haque MM, Kenney C, Tejero J, Stuehr DJ. A kinetic model linking protein conformational motions, interflavin electron transfer and electron flux through a dual-flavin enzyme-simulating the reductase activity of the endothelial and neuronal nitric oxide synthase flavoprotein domains. FEBS J. 2011;278:4055–4069. doi: 10.1111/j.1742-4658.2011.08310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haque MM, Bayachou M, Fadlalla MA, Durra D, Stuehr DJ. Charge Pairing Interactions Control the Conformational Setpoint and Motions of the FMN Domain in Neuronal Nitric Oxide Synthase. Biochem. J. 2013;450:607–617. doi: 10.1042/BJ20121488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ilagan RP, Tiso M, Konas DW, Hemann C, Durra D, Hille R, Stuehr DJ. Differences in a conformational equilibrium distinguish catalysis by the endothelial and neuronal nitric-oxide synthase flavoproteins. J. Biol. Chem. 2008;283:19603–19615. doi: 10.1074/jbc.M802914200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilagan RP, Tejero J, Aulak KS, Ray SS, Hemann C, Wang ZQ, Gangoda M, Zweier JL, Stuehr DJ. Regulation of FMN subdomain interactions and function in neuronal nitric oxide synthase. Biochemistry. 2009;48:3864–3876. doi: 10.1021/bi8021087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konas DW, Zhu K, Sharma M, Aulak KS, Brudvig GW, Stuehr DJ. The FAD-shielding residue Phe1395 regulates neuronal nitric-oxide synthase catalysis by controlling NADP+ affinity and a conformational equilibrium within the flavoprotein domain. J. Biol. Chem. 2004;279:35412–35425. doi: 10.1074/jbc.M400872200. [DOI] [PubMed] [Google Scholar]
- 39.Tiso M, Tejero J, Panda K, Aulak KS, Stuehr DJ. Versatile regulation of neuronal nitric oxide synthase by specific regions of its C-terminal tail. Biochemistry. 2007;46:14418–14428. doi: 10.1021/bi701646k. [DOI] [PubMed] [Google Scholar]
- 40.Craig DH, Chapman SK, Daff S. Calmodulin activates electron transfer through neuronal nitric-oxide synthase reductase domain by releasing an NADPH-dependent conformational lock. J. Biol. Chem. 2002;277:33987–33994. doi: 10.1074/jbc.M203118200. [DOI] [PubMed] [Google Scholar]
- 41.Gutierrez A, Paine M, Wolf CR, Scrutton NS, Roberts GC. Relaxation kinetics of cytochrome P450 reductase: internal electron transfer is limited by conformational change and regulated by coenzyme binding. Biochemistry. 2002;41:4626–4637. doi: 10.1021/bi0159433. [DOI] [PubMed] [Google Scholar]
- 42.Mendes P. GEPASI: a software package for modelling the dynamics, steady states and control of biochemical and other systems. Comput. Appl. Biosci. 1993;9:563–571. doi: 10.1093/bioinformatics/9.5.563. [DOI] [PubMed] [Google Scholar]
- 43.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolthers KR, Lou X, Toogood HS, Leys D, Scrutton NS. Mechanism of coenzyme binding to human methionine synthase reductase revealed through the crystal structure of the FNR-like module and isothermal titration calorimetry. Biochemistry. 2007;46:11833–11844. doi: 10.1021/bi701209p. [DOI] [PubMed] [Google Scholar]
- 45.Basse MJ, Betzi S, Bourgeas R, Bouzidi S, Chetrit B, Hamon V, Morelli X, Roche P. 2P2Idb: a structural database dedicated to orthosteric modulation of protein-protein interactions. Nucleic Acids Res. 2013;41:D824–D827. doi: 10.1093/nar/gks1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grunau A, Paine MJ, Ladbury JE, Gutierrez A. Global effects of the energetics of coenzyme binding: NADPH controls the protein interaction properties of human cytochrome P450 reductase. Biochemistry. 2006;45:1421–1434. doi: 10.1021/bi052115r. [DOI] [PubMed] [Google Scholar]
- 47.Hay S, Brenner S, Khara B, Quinn AM, Rigby SE, Scrutton NS. Nature of the energy landscape for gated electron transfer in a dynamic redox protein. J. Am. Chem. Soc. 2010;132:9738–9745. doi: 10.1021/ja1016206. [DOI] [PubMed] [Google Scholar]
- 48.Ellis J, Gutierrez A, Barsukov IL, Huang WC, Grossmann JG, Roberts GC. Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering. J. Biol. Chem. 2009;284:36628–36637. doi: 10.1074/jbc.M109.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang WC, Ellis J, Moody PC, Raven EL, Roberts GC. Redox-linked domain movements in the catalytic cycle of cytochrome p450 reductase. Structure. 2013;21:1581–1589. doi: 10.1016/j.str.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Persechini A, Tran QK, Black DJ, Gogol EP. Calmodulin-induced structural changes in endothelial nitric oxide synthase. FEBS Lett. 2013;587:297–301. doi: 10.1016/j.febslet.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leys D, Basran J, Talfournier F, Sutcliffe MJ, Scrutton NS. Extensive conformational sampling in a ternary electron transfer complex. Nat. Struct. Biol. 2003;10:219–225. doi: 10.1038/nsb894. [DOI] [PubMed] [Google Scholar]
- 52.Toogood HS, Leys D, Scrutton NS. Dynamics driving function: new insights from electron transferring flavoproteins and partner complexes. FEBS J. 2007;274:5481–5504. doi: 10.1111/j.1742-4658.2007.06107.x. [DOI] [PubMed] [Google Scholar]
- 53.Brenner S, Hay S, Munro AW, Scrutton NS. Inter-flavin electron transfer in cytochrome P450 reductase - effects of solvent and pH identify hidden complexity in mechanism. FEBS J. 2008;275:4540–4557. doi: 10.1111/j.1742-4658.2008.06597.x. [DOI] [PubMed] [Google Scholar]
- 54.Pudney CR, Khara B, Johannissen LO, Scrutton NS. Coupled motions direct electrons along human microsomal P450 Chains. PLoS. Biol. 2011;9:e1001222. doi: 10.1371/journal.pbio.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez A, Paine M, Wolf CR, Scrutton NS, Roberts GC. Relaxation kinetics of cytochrome P450 reductase: internal electron transfer is limited by conformational change and regulated by coenzyme binding. Biochemistry. 2002;41:4626–4637. doi: 10.1021/bi0159433. [DOI] [PubMed] [Google Scholar]
- 56.Gutierrez A, Munro AW, Grunau A, Wolf CR, Scrutton NS, Roberts GC. Interflavin electron transfer in human cytochrome P450 reductase is enhanced by coenzyme binding. Relaxation kinetic studies with coenzyme analogues. Eur. J. Biochem. 2003;270:2612–2621. doi: 10.1046/j.1432-1033.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 57.Leferink NG, Pudney CR, Brenner S, Heyes DJ, Eady RR, Samar HS, Hay S, Rigby SE, Scrutton NS. Gating mechanisms for biological electron transfer: integrating structure with biophysics reveals the nature of redox control in cytochrome P450 reductase and copper-dependent nitrite reductase. FEBS Lett. 2012;586:578–584. doi: 10.1016/j.febslet.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Gutierrez A, Grunau A, Paine M, Munro AW, Wolf CR, Roberts GC, Scrutton NS. Electron transfer in human cytochrome P450 reductase. Biochem. Soc. Trans. 2003;31:497–501. doi: 10.1042/bst0310497. [DOI] [PubMed] [Google Scholar]
- 59.Abu-Soud HM, Yoho LL, Stuehr DJ. Calmodulin controls neuronal nitric-oxide synthase by a dual mechanism. Activation of intra- and interdomain electron transfer. J. Biol. Chem. 1994;269:32047–32050. [PubMed] [Google Scholar]
- 60.Chen PF, Wu KK. Structural elements contribute to the calcium/calmodulin dependence on enzyme activation in human endothelial nitric-oxide synthase. J. Biol. Chem. 2003;278:52392–52400. doi: 10.1074/jbc.M305469200. [DOI] [PubMed] [Google Scholar]
- 61.Daff S. Calmodulin-dependent regulation of mammalian nitric oxide synthase. Biochem. Soc. Trans. 2003;31:502–505. doi: 10.1042/bst0310502. [DOI] [PubMed] [Google Scholar]
- 62.Matsuda H, Iyanagi T. Calmodulin activates intramolecular electron transfer between the two flavins of neuronal nitric oxide synthase flavin domain. Biochim. Biophys. Acta. 1999;1473:345–355. doi: 10.1016/s0304-4165(99)00193-2. [DOI] [PubMed] [Google Scholar]
- 63.Wu PR, Kuo CC, Yet SF, Liou JY, Wu KK, Chen PF. Lobe-specific calcium binding in calmodulin regulates endothelial nitric oxide synthase activation. PLoS. One. 2012;7:e39851. doi: 10.1371/journal.pone.0039851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roman LJ, Masters BS. Electron transfer by neuronal nitric oxide synthase is regulated by concerted interaction of calmodulin and two intrinsic regulatory elements. J. Biol. Chem. 2006;281:23111–23118. doi: 10.1074/jbc.M603671200. [DOI] [PubMed] [Google Scholar]
- 65.Haque MM, Panda K, Tejero J, Aulak KS, Fadlalla MA, Mustovich AT, Stuehr DJ. A connecting hinge represses the activity of endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9254–9259. doi: 10.1073/pnas.0700332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salerno JC, Harris DE, Irizarry K, Patel B, Morales AJ, Smith SM, Martasek P, Roman LJ, Masters BS, Jones CL, Weissman BA, Lane P, Liu Q, Gross SS. An autoinhibitory control element defines calcium-regulated isoforms of nitric oxide synthase. J. Biol. Chem. 1997;272:29769–29777. doi: 10.1074/jbc.272.47.29769. [DOI] [PubMed] [Google Scholar]
- 67.Grunau A, Geraki K, Grossmann JG, Gutierrez A. Conformational dynamics and the energetics of protein--ligand interactions: role of interdomain loop in human cytochrome P450 reductase. Biochemistry. 2007;46:8244–8255. doi: 10.1021/bi700596s. [DOI] [PubMed] [Google Scholar]
- 68.Haque MM, Fadlalla MA, Aulak KS, Ghosh A, Durra D, Stuehr DJ. Control of electron transfer and catalysis in neuronal nitric-oxide synthase (nNOS) by a hinge connecting its FMN and FAD-NADPH domains. J. Biol. Chem. 2012;287:30105–30116. doi: 10.1074/jbc.M112.339697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.