Abstract
Centromeres are specialized domains of heterochromatin that provide the foundation for the kinetochore. Centromeric heterochromatin is characterized by specific histone modifications, a centromere-specific histone H3 variant (CENP-A), and the enrichment of cohesin, condensin, and topo-isomerase II. Centromere DNA varies orders of magnitude in size from 125 bp (budding yeast) to several megabases (human). In metaphase, sister kinetochores on the surface of replicated chromosomes face away from each other, where they establish microtubule attachment and bi-orientation. Despite the disparity in centromere size, the distance between separated sister kinetochores is remarkably conserved (approximately 1 μm) throughout phylogeny. The centromere functions as a molecular spring that resists microtubule-based extensional forces in mitosis. This review explores the physical properties of DNA in order to understand how the molecular spring is built and how it contributes to the fidelity of chromosome segregation.
Keywords: centromere, heterochromatin, chromosome segregation, DNA mechanics, molecular springs
INTRODUCTION
The centromere is the primary constriction observed in condensed chromosomes during mitosis and provides the site of assembly for the kinetochore. The primary constriction on condensed chromosomes was first documented by Walther Flemming (44). Almost 100 years later, a description of the specialized disc-shaped kinetochore emerged: a proteinaceous structure found at the periphery of the centromere derived from electron micrographs of fixed specimens (15, 80). The identity of centromere and kinetochore proteins hinged serendipitously on the discovery of centromere-specific autoantibodies in sera of patients with an autoimmune disease scleroderma (14, 46, 103, 104). More than 100 kinetochore proteins have since been identified (10, 159, 174). They are organized minimally into five to six key complexes within the kinetochore and are largely conserved from yeast to mammals. Centromere DNA was first cloned and sequenced in the early 1980s (24, 43). In contrast to the high degree of protein conservation among diverse kinetochores, centromeres range from 125 bp of unique DNA in the budding yeast Saccharomyces cerevisiae to several megabases of repetitive DNA in Homo sapiens (Figure 1) (reviewed in 147). There are numerous features that distinguish centromeric heterochromatin from chromosome arms; however, the chromatin organization and the divergence of centromeres throughout phylogeny are outstanding questions in the field. In this review, we explore conserved features of centromeric chromatin organization and evidence for the proposal that the centromere is the primordial segregation machine, preceding the evolution of kinetochore and spindle microtubules.
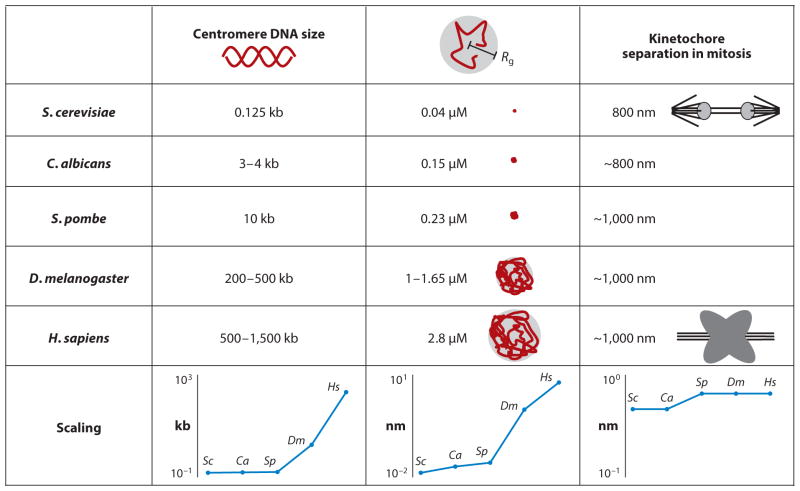
Figure 1.
Physical properties of centromere DNA. Centromere DNA size is defined as the region of DNA required for the segregation function in a variety of organisms. Rg is the radius of the random coil (radius of gyration) defined by Rg2 = n × (2Lp)2/6. Lp is the persistence length of the polymer, and n is the number of segments (total DNA contour length Lc/Lp) (61). Kinetochore separation is the distance between sister kinetochores observed experimentally in a number of organisms [budding yeast, S. cerevisiae (118); fission yeast, S. pombe (32); worm, C. albicans (94); Drosophila melanogaster (160); and human, H. sapiens (132)]. The bottom graphs highlight the scaling (ordinate) of DNA (four orders of magnitude), radius of gyration (three orders of magnitude), and kinetochore separation (constant) throughout phylogeny (abscissa).
Why Heterochromatin versus Euchromatin at Centromeres
Euchromatin is an open chromatin state that enhances accessibility to transcription complexes. Heterochromatin, far from simply being a compaction state, provides a mechanism for recruiting and spreading components across large distances in DNA space (59, 60). To appreciate this aspect of heterochromatin we need to compare DNA length to nuclear volume. In a typical human diploid cell, two meters of DNA reside in an approximately 10-μm-diameter nucleus. The DNA is also very skinny (2 nm), which works in favor of chromatin-packaging strategies. The volume a random coil of DNA adopts in the absence of cellular material can be estimated from its radius, known as the radius of gyration (Rg) (Figure 1). For the mammalian genome, , equivalent to a volume of ~107 μm3 (4/3πRg3). In a typical cell, nuclear volume is approximately 500 μm3 (4/3π 5 μm3). Thus, chromatin is compacted on the order of 2 × 104. The volume of DNA polymer (excluded volume) is approximately 6 μm3 (πr2h = π *1 nm2* 6 m) or only 1% of the nuclear volume. These estimates exclude cellular mechanisms that distinguish euchromatin versus heterochromatin. It is not packaging per se that is challenging to understand, it is the organization of functional domains. The subnuclear organization of genes into various domains, or bodies, is indicative of spatial segregation according to function. Active genes are often clustered to the nuclear periphery, whereas tRNA genes are frequently associated with the nucleolus (73, 84). Mechanistically, chromosome domains entrapped by cohesin bring regulatory domains adjacent to transcription start sites (35, 36, 120), whereas condensin binding to polymerase III transcription factors functions to cluster tRNA genes (65). With respect to the centromere, information about microtubule attachment at one kinetochore is transferred across centromeric heterochromatin to its sister kinetochore as a mechanism for force balance and tension sensing that is critical for chromosome segregation fidelity. In this situation, the pair of sister kinetochores and centromeric heterochromatin make up a single unit with structural/mechanical integrity. Remarkably, centromere DNA spans more than four orders of magnitude (from yeast to human), yet the physical separation between kinetochores is highly conserved (Figure 1). How the transmission of force is managed over the vast range of DNA size is critical for understanding the basis for faithful chromosome segregation. An evolutionarily conserved pathway [spindle assembly checkpoint (SAC)] (105) monitors the status of the kinetochore microtubule attachment site, including the presence or absence of a microtubule and whether tension is generated between sister kinetochores. In the absence of attachment and/or tension, chemical signals are generated that invoke the SAC. These signals relay the status of occupancy and/or tension at the kinetochore to master regulatory kinases that drive cell cycle progression (105). Additionally, there are mechanisms for correcting erroneous attachments, providing the opportunity for sister kinetochores to establish bi-orientation. The error-correction mechanisms and how the cell promotes stable versus unstable microtubule attachment in response to the state of each sister kinetochore is embedded in the structure of the kinetochore and the centromeric heterochromatin. The folding of megabases of centromeric heterochromatin (in the case of humans) into a highly compact and organized structure is better suited for transmitting mechanical force than the more open, disordered euchromatin. Protein machines acting as compactors, loopers, and topology adjusters are integral to mechanisms that convert the DNA random coil into a mechanical tension sensor suitable for transmission of microtubule-based spindle force.
Heterochromatin Organization: Transcription from Repetitive DNA
Heterochromatin is traditionally thought of as a means for setting boundaries between transcribed regions, as well as for providing an environment conducive to centromere function. One of the hallmarks of centromeric heterochromatin is the enrichment of simple sequences and repetitive DNA (68). In mammals, the centromere is marked by hierarchical repeats of α-satellite DNA. α-Satellites are a tandemly repeated array of a 171-bp monomeric unit. In fission yeast, the centromere region is characterized by large repeat structures (imr and otr) surrounding a unique centromere central core (23, 108). The extent of spreading and degree of compaction of centromeric heterochromatin is highly regulated. Transcription from repeat elements is part of this regulatory mechanism and provides a framework for assembly and maintenance of heterochromatin (59, 90). Increasing the stoichiometry of small noncoding RNAs through increased rates of transcription of mouse satellite leads to cell cycle delay, a decrease in heterochromatin compaction, and, likely, errors in chromosome segregation (75). Centromeric heterochromatin also represses transcription at the microtubule attachment site and is crucial for segregation function (50, 66). In budding yeast, modulation of transcription levels through the 125-bp CEN DNA leads to a marked increase in chromosome missegregation (70, 114, 115). In addition to the regulatory role of small RNAs, the act of transcription and topological consequences thereof may contribute to centromere function. However, as discussed below, phylogenetic studies of evolutionary new centromeres (ENC) reveals that satellite DNA accumulation follows centromere formation (112). It is therefore unlikely that number or physical arrangement of DNA repeats contribute to the mechanical coupling between sister chromatids. The accumulation of repeat sequences in the pericentromeres provides mechanisms through RNA function for creating and maintaining heterochromatin boundaries.
Pericentromeres Are Enriched in Condensin and Cohesin
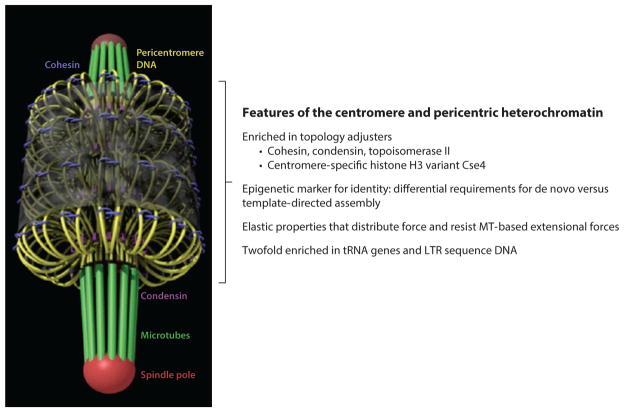
The structural maintenance of chromosome (SMC) proteins cohesin and condensin are enriched at the pericentric region in a number of organisms (27, 39, 86). SMC proteins assemble into complexes that adopt a ring-like conformation (72, 109). The backbone of the ring is formed by the SMC proteins themselves (MukB in bacteria, Smc2 and Smc4 in S. cerevisiae condensin, and Smc1 and Smc3 in S. cerevisiae cohesin). In eukaryotes, the SMC monomer is folded in an antiparallel coiled coil. At one end, the two monomers associate to form a hinge, and at the other end is an ATP-binding head domain. Closure of the ring at the head domain is carried out by proteins known as kleisins, including Scc1 (also known as Mcd1) and Brn1. Each dimer is associated with additional proteins [e.g., Ysc4, Ycg1, Scc3 (also known as Irr1), Rad61, and Pds5] at the head domain to form a functional complex in vivo. Condensin, cohesin, and topoisomerase II are approximately threefold enriched in the 30–50 kb of DNA surrounding the budding yeast centromere (6, 27, 39) relative to the bulk of the genome. An attractive hypothesis is that cohesin, by physically entrapping sister chromatids, is responsible for the resistance between bi-oriented sister kinetochores in mitosis. Consistent with this, most cohesin dissociates from mammalian chromatin via the prophase pathway (93, 165), but a small population, enriched at the centromeric region, remains on chromatin until the onset of anaphase. Studies in budding yeast challenged this hypothesis by observing well-separated sister centromere DNA occurring frequently during metaphase (56, 69, 118, 152). How could proteins that specifically bind DNA together be enriched in a region where the DNA is both separated and transiently together? This apparent contradiction can be resolved if sister kinetochore microtubules stretch the centromeres into a cruciform structure in which each sister centromere lies at the apex of an intramolecular DNA loop while sister chromosome arms are paired via intermolecular cohesion. The cruciform structure serves to orient the centromere (and therefore the kinetochore) on the outer-face of the chromosome, facing toward the spindle pole of the segregation apparatus (142, 175). Cohesin is radially displaced from the microtubule axis, indicating a more complex organization that can be accounted for in simple models (Figure 2).
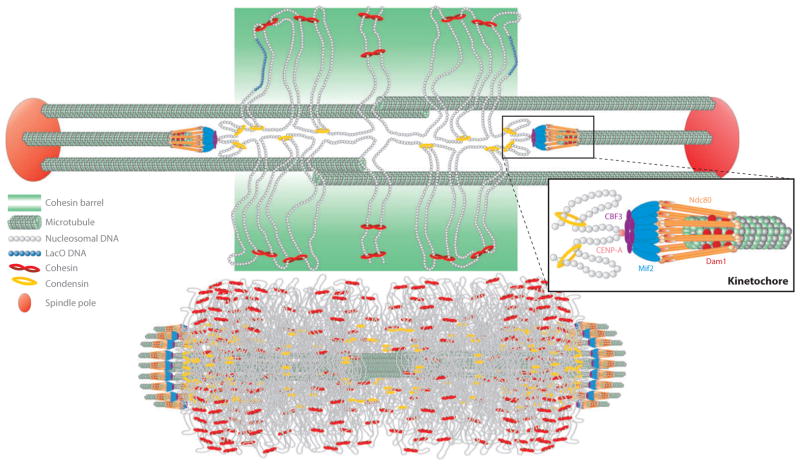
Figure 2.
Organization of pericentric chromatin and cohesin in metaphase in budding yeast. (Top) The yeast segregation apparatus is a composite structure of the kinetochore and interpolar microtubules ( green), the spindle pole body (large red sphere), and pericentric chromatin loops (DNA strands shown as strings of nucleosomes; gray). Centromere DNA (CENP-A nucleosome; pink) is attached to microtubule plus-ends via the kinetochore (orange barbells surrounding the microtubule plus-end). Kinetochore components (right) include Ndc80 (orange barbells), the Dam1 complex (small red spheres interleaved with Ndc80), the Mif2 complex (blue rods), and the DNA-binding complex CBF3 ( purple ovals). Cohesin (red ) and condensin ( yellow) are enriched in the pericentromere and surround the central spindle. Cohesin is radially displaced from the spindle axis, whereas condensin is proximal to the spindle axis (142, 143). One pair of sister chromatids is shown for simplicity. Sister chromatids occupy the left and right half-spindle, respectively. Sister chromatids at the mid-spindle position (perpendicular to the spindle axis) are held via cohesin rings (red ). As sister DNA strands become proximal to the spindle microtubules, they adopt a cruciform-like DNA configuration, with intermolecular sister pairing midway between the spindle poles and intramolecular pairing to the left and right. Condensin rings along the spindle axis contribute to formation of intramolecular loops shown perpendicular to the spindle axis, proximal to the left and right kinetochore. Lac operator DNA (nucleosomes; blue) is radially displaced from the spindle axis when visualized with LacI-GFP (2), similar in dimension to the cohesin barrel. (Bottom) The pericentric chromatin from all 16 chromosomes is shown together with kinetochore microtubules in metaphase. The amount of DNA is drawn to scale and represents the region of DNA from all 16 centromeres that is enriched in the SMC (structural maintenance of chromosome) protein complexes cohesin and condensin. Adapted from figure 4 of Reference 62.
To gain insight into how these SMC architectural proteins might be organized to resist spindle forces in mitosis, we turn to their organization in compacting ribosomal DNA in the actively transcribed nucleolus. A major site of repeat sequences in all organisms is the nucleolus. The budding yeast nucleolus contains 100–200 copies of a 9.1-kb rDNA repeat (~1–2 Mb) together with approximately a threefold greater concentration of cohesin and condensin relative to RNA polymerase II transcribed genes (4, 28, 48). The nucleolus is also a repository of several cell cycle regulatory complexes (7, 129). Condensin plays a fundamental role in rDNA segregation (28) and is coupled to one of these cell cycle regulatory complexes (FEAR) to ensure the timely segregation of rDNA prior to cytokinesis. Condensin recruitment to rDNA depends upon monopolin (including Csm1 and Lrs4) Fob1 and is negatively correlated with levels of RNA polymerase I transcription (78, 79). Condensin also binds tRNA gene transcription complexes and promotes clustering of tRNA genes (65). Furthermore, these tRNA genes are clustered to the nucleolus, dependent on CBF5, a gene implicated in centromere function (84).
The concentration of cohesin is dynamically equilibrated between the nucleolus and pericentric heterochromatin. Reduction of pericentric cohesin (via mcm21 deletion) leads to increased nucleolar levels, whereas reduction of nucleolar cohesin (via sir2 deletion) leads to increased pericentric levels (143). Enrichment to the pericentromere depends upon the kinetochore, but what restricts these proteins to the 30–50-kb region surrounding the centromere is not clear. Because the rDNA repeats are enriched for condensin and cohesin, other repeats might similarly be enriched for these SMC proteins and thereby account for their enrichment in pericentromeric regions. The major repeated DNA sequences in yeast are the subtelomere repeats, LTRs (long terminal repeats of 300–400-bp bracketing retrotransposons, 429 total), and tDNA genes (307 total). LTRs and tDNA genes are enriched 1.8 times in the 50-kb region surrounding the centromere, relative to their concentration in the remainder of the genome. The enrichment of tDNA in the pericentromere and the recruitment of condensin to tRNA transcription factors provide a mechanism for restricting condensin to this functional domain. The enrichment of repeat sequences within the pericentromere may also provide the structural basis for partitioning condensin to the spindle axis and the radial displacement of cohesin (142, 143). Thus, the repeat sequences, although not directly involved in mechanosensing of the kinetochore microtubule attachments, may contribute indirectly to structure via their role in concentrating and partitioning proteins such as cohesin and condensin.
Heterochromatin Function: Making an Elastic Material
Centromeric heterochromatin is distinguished from heterochromatic regions along chromosome arms principally by the degree of compaction and enrichment of cohesin and condensin. Many of the histone modifications are shared with transcriptionally silenced regions (55). The centromere-specific histone H3 variant, CENP-A, resides at the CEN DNA in budding yeast (Figure 2) (145) and is interspersed with methylated histone H3 (mono- and dimethylation of lysine 4 and di- and trimethylation of lysine 36) in centromeres of multicellular organisms (8, 9, 13, 53). Centromere heterochromatin provides three functions in chromosome segregation: to serve as the template for kinetochore formation and microtubule attachment to the mitotic spindle; to link separated sister kinetochores; and to translate the state of kinetochore microtubule attachment between sister kinetochores. The kinetochore constituents and specificity of formation have been addressed in a number of recent reviews (10, 19, 172). Recent progress in genomics and mathematical modeling allows us to address the physical properties of centromere chromatin. For the mitotic spindle, kinesin microtubule-based motor proteins act as force production machines that slide the interpolar microtubules apart (Figure 2), generating an extensional force on the spindle poles (137, 138). The balance of microtubule-based extensional force and a chromatin-spring contractile force is necessary to produce a steady-state spindle length. Tension at the kinetochore satisfies the spindle checkpoint (11). Although the structure and form of the chromatin spring may vary throughout phylogeny, in all organisms the spring is contractile. To build an understanding of the chromatin spring, we start with the mechanical properties of DNA. DNA exhibits properties of an entropic spring, which may contribute to the contractility of centromeric heterochromatin. We can calculate the spring constant for an entropic spring according to polymer theory (12). The short rigid domains of DNA, linked via flexible joints, adopt a state of greatest disorder (entropy). From Hooke’s law, we know that F =κx, where κ is the spring constant (newton/meter) and x is the change in distance (meters). For small forces, F = 3kBTx/n(2Lp)2. The spring constant of this freely jointed chain is equal to 3kBT/n(2Lp)2, where kBT is the Boltzmann constant × T (newton meters), Lp is persistence length (meters), and n is the number of segments. For a DNA length of 10 kb, the spring constant is 0.036 fN/nm, which is small indeed. Note that the larger the DNA [increasing number of segments (n)], the weaker the spring. Thus, the tendency of DNA to adopt a random coil is not sufficient to balance microtubule-based extensional forces in the range of pN (98).
To gain insight into how mechanical force is transmitted between sister centromeres, we must understand the physical organization, starting with the size and shape of the pericentromere and centromere. Upon attachment to kinetochore microtubules, bi-oriented centromeres in everything from budding yeast to flies to mammals exhibit a stereotypic separation ranging from 800 nm to 1,000 nm (Figure 1) (118, 133). Although the yeast centromere is defined by a very small region of DNA (125 bp), the 16 centromeres are clustered around spindle microtubules, where they attach to the plus-ends of 16 kinetochore microtubules (kinetochore microtubule plus-ends shown Figure 2a). On the basis of the enrichment of cohesin and condensin in the 30–50-kb region of DNA flanking the 125-bp centromere, one can consider this domain as functionally equivalent to centromeric heterochromatin in organisms with multiple-kinetochore microtubule attachment sites, such as flies and mammals. Furthermore, the amount of pericentric chromatin encircling the yeast spindle is ~500–800 kb (16 chromosomes × 30–50 kb), comparable to the 1–5 megabase lengths of α-satellite DNA in a mammalian kinetochore. The area between the two clusters of 16 microtubule attachment sites in budding yeast (2πrh ≈ 0.6 μm2) is comparable to estimates of the size of mammalian kinetochores (0.4 μm2) (20). Thus, the distance between sister centromeres as well as the area of centromeric heterochromatin are conserved features in centromere organization.
The next level of understanding of pericentromere function requires the accurate dissection of spatial features, including the DNA and chromatin proteins within the structure. The advantage of the pericentromere in budding yeast is that, unlike in mammals and most other organisms, the site of microtubule attachment is known with base-pair precision. The point centromere provides a critical reference point for localizing components relative to the site of microtubule-based force transduction. Pericentric DNA can be visualized through lac operators and lacI-GFP, which are introduced at specific sites relative to the centromere (146). Using this system, several investigators have found that sister centromeres are precociously separated during mitosis (56, 68, 118, 152). Because separated sister lacO (Escherichia coli lac operator) foci are mobile, their position is best captured as a distribution of statistical probabilities. Surprisingly, lacO foci at centromere proximal positions are found least often on the spindle axis (Figure 2) (2, 142). Pericentric DNA is radially displaced from the position of the kinetochores and kinetochore microtubule plus-ends (Figure 2). Thus, forces at one centromere are not linearly transmitted through the DNA to the sister centromere. The localization of cohesin and condensin helps us understand the distribution of pericentric DNA. Data from chromatin immunoprecipitation (ChIP) studies appear to indicate that cohesin and condensin are mostly uniform across the pericentromere (27, 52, 76, 99, 151, 168). In contrast, live-cell imaging studies indicate that cohesin and condensin adopt stereotypic and spatially segregated structures within the pericentric heterochromatin in mitosis (142, 143). Cohesin is radially distributed into an apparent barrel, displaced from the kinetochore and kinetochore microtubules (500-nm diameter versus 250-nm diameter kinetochore microtubules) (Figure 2). The barrel is uniform in fluorescence intensity, indicative of the even distribution of cohesin. The degree of displacement of the cohesin barrel is comparable to the displacement of the average distribution of pericentric DNA, indicating that both cohesin and pericentric DNA are, on average, displaced from the spindle axis and the main site of microtubule attachment. In contrast, condensin is localized along the spindle axis. Condensin is heterogeneous and appears as single or multiple foci or linear elements, consistent with its biochemical tendency to aggregate (4, 142). The apparent disparity in localization deduced by ChIP versus live-cell fluorescence microscopy is reconciled by considering DNA fluctuations. If the binding sites for pericentric cohesin and condensin are highly variable, a population method such as ChIP averages out the differences and misses the spatial segregation of these complexes observed in live images of single cells. The spatial segregation of chromatin proteins as well as the DNA within the centromeric heterochromatin reveals how little we understand the chromatin spring that resists outward microtubule-based forces.
Ring-like proteins endow synthetic polymers several unique properties (57, 116). Slip-links in place of rigid cross-linkers in a synthetic fiber allow polymers to wiggle through the cross-link, resulting in a “sharing of the load” phenomenon (Figure 3). In the centromeric heterochromatin, condensin and/or cohesin protein rings may function as staples or slip-links (molecular pulleys) to distribute tension from one microtubule attachment site to the entire network (Figure 3). Cohesin and condensin (Figures 2 and 3) have the physical attributes to function as slip rings and regulate centromere elasticity. Cohesin acting distally from the spindle axis promotes looping and/or distributes tension throughout the network as a way of averaging fluctuations from the 32 individual kinetochore microtubules. Condensin localized along the spindle axis could generate a spring force through DNA compaction. Microtubule-extensional forces have a tendency to pull DNA, extending its length, whereas condensin decreases DNA length through compaction. In support of this view, loss of either pericentric cohesin or condensin results in increased spindle length in yeast (142) as well as increased kinetochore-kinetochore distance in vertebrate cells (128, 135).
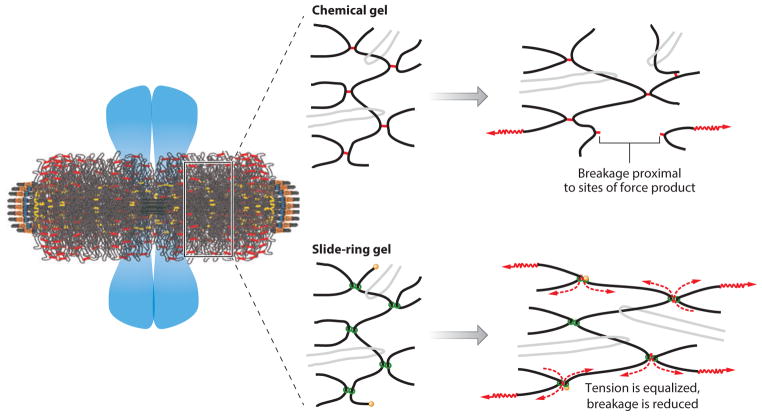
Figure 3.
Strategies for cross-linking centromere DNA in the primary constriction of metaphase chromosomes. (Left) The organization of centromeric heterochromatin relative to chromosome arms in mitosis. The pericentric chromatin from 16 chromosomes surrounding the spindle axis (see key in Figure 2). Sister chromatid arms relative to centromeric heterochromatin are indicated by the blue masses. Pericentric chromatin functions as a contractile element in the spindle, counteracting extensional forces from microtubule-based motor proteins to achieve force balance in metaphase. (Middle) Various ways to generate chromatin cross-links. In a chemical gel (top), inflexible links (red ) connect DNA strands (black). In a slide-ring gel (bottom), molecular pulleys ( green), through which DNA can slide, connect DNA strands. The light gray strands indicate entanglements in which DNA strands are catenated. (Right, top) When force is exerted on DNA strands in a chemical gel, strain is concentrated on inflexible linkages, resulting in rupture. (Right, bottom) When force is exerted on DNA strains that can slip through molecular linkages, tension is distributed throughout the network, equalizing strain among all links, reducing rupture (57, 116).
Unique Features of Polymer Springs
The importance of fidelity in chromosome segregation cannot be overstated. It is not enough to build a robust microtubule attachment site. The ability to detect and correct errors in chromosome alignment is paramount in the development of any organism. In multiple microtubule-attached kinetochores, the error detection/correction system must compensate for stochastic growth and shortening of kinetochore microtubules. Unless the system tolerates a large range of tension at the kinetochore microtubule interface, there are no plausible mechanisms for how natural fluctuations in tension are buffered. We turn to the chromatin spring to gain insight into how tension is managed within the centromere. Long chain polymers such as chromosomes are challenging to model because of the wide range of length and timescales in the system. One of the problems for modeling is the vast number and breadth of polymer configurations, even though the large number of configurations contributes significantly to the entropy and therefore the elastic restoring force. Even with robust algorithms, the complexity in the models must necessarily be reduced relative to what is present in vivo. Representation of the restoring force with springs and modeling the polymer as a bead-spring chain have proven valuable to understanding chromosome behavior (156) (Figure 4). These polymer models do remarkably well in capturing the large-scale folding and organizational principles derived from population studies.
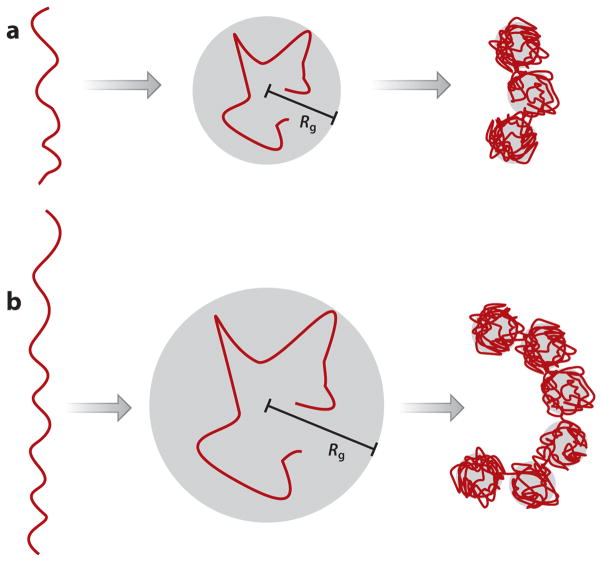
Figure 4.
Polymer model of chromatin provides insight into force transduction through the centromere heterochromatin. (a,b) Linear DNA (left) spontaneously folds into a random coil (middle). The size of the random coil is defined by the radius of gyration (Rg). In a confined and crowded environment such as a cell, the chains adopt smaller random coils within which monomer subunits are free to fluctuate, thus maximizing entropy (right, smaller gray spheres containing red polymer). These are known as thermal blobs and are defined as regions of the polymer chain in which the monomers are unaffected by their environment. If force is exerted at the ends of the chain, the blobs decrease laterally in size, but within each blob, the chains continue to fluctuate with defined statistical properties (164). Tension blobs create significant consequences for the centromere. When force is exerted on the kinetochore, tension can be transmitted over large distances (800 nm) without reducing the entropy of each and every monomer within the chain. The differing lengths of centromere DNA across phylogeny (small to large, as depicted in panels a and b) predict that the number of tension blobs varies considerably, but the mechanism of force transduction is highly conserved. The centroids of blobs can be connected to each other through springs, leading to the bead-spring representation of the chromatin polymer.
Chromosome motion varies in predicted ways along the length of the chromosome. To see how the chromosome behaves, hold a slinky with each hand at an end and shake. The tendency for the middle of the spring to move more and the ends of the spring to move less captures experimental observations (162). The centromere behaves as one of the ends in this toy model. The restricted motion of an end reflects the biological feature that the centromere remains attached to the spindle pole body throughout the cell cycle in budding yeast (162). This is called a tether in the polymer world and is an important physical principle that, together with the bead-spring model, is required to generate the observed variation in chromosome mobility (Figure 4).
During interphase, the Rabl orientation describes a configuration in which centromeres of each chromosome are associated with the spindle pole and telomeres distal to the pole (37). In mitosis, chromosomes are attached to the mitotic spindle and align along the metaphase plate, whereas in meiotic prophase, telomeres are clustered in a bouquet configuration with the centromeres extended distally (21, 22). Microtubule attachment of the centromere to the spindle poles or telomeres to the nuclear envelope provides a physical linkage that significantly restricts the dynamic range of chromatin motion (162). In budding yeast, detached centromeres exhibit the same degree of freedom as a locus hundreds of kilobase pairs away on the chromosome arm (162) that explores threefold more space than the attached centromere configuration (radius of confinement attached 274 nm versus detached 745 nm). The restricted centromere movement in wild-type cells is observed in all stages of the cell cycle. Likewise, telomere tethering to the nuclear envelope must be invoked to predict their observed motion. In a bead-spring model, sister kinetochores function as tethers restricting chromatin motion on the surface of the chromosome (see Figure 2), whereas intervening centromeric heterochromatin fluctuates like the aforementioned slinky. These basic physical principles reveal that in the absence of other features, the chromatin spring constant will be stiffest closest to the kinetochore and softest at the midpoint between sister kinetochores.
A second driving principle to understand how a polymer spring might function involves the excluded volume interaction. Just as we describe the hierarchy of chromosome organization from DNA to nucleosomes to higher-order structures, there are hierarchies in parameter interactions for a polymer. The primary interactions are the covalent bonds. Secondary interactions include the polymer and the solvent or between monomers that are not nearest neighbors. The volume taken up by one monomer is excluded from that available to any other monomer. For a dynamic polymer, the area the polymer explores is larger than the chain itself, giving rise to an exclusion zone unavailable for other molecules to penetrate. This is known as excluded volume, the magnitude of which depends upon the chemical environment. Excluded volumes can overlap when molecules are immersed in poor solvents or crowded conditions, such as are found inside the cell. Considering the enormous length of the chromosome relative to the nucleus, the excluded volume is a significant fraction of the nuclear space. When two monomers come into proximity (i.e., crowding), their excluded volumes overlap. This results in increased available volume for small molecules (here called depletants). The gain in entropy of the system from the increased available volume to the depletants is known as the depletion (entropic) force. The depletion force favors molecular crowding (163) and, depending on the strength of the attraction force between individual segments of a polymer, can lead to collapse of the polymer into a compact structure known as a globule (61). In this way, the depletion force promotes chromosome compaction from first principles. Students of mitosis may recall that counter to intuition, thermodynamics favors the formation of the microtubule polymer (132). It is the loss of ordered water around tubulin dimers that drives this reaction. The critical issue is that the thermodynamics of any system that involves a polymer in solution must be considered in terms of entropy of the whole system, and this consideration can lead to counterintuitive phenomena, such as microtubule polymerization and chromosome condensation.
Finally, cellular geometry confines the DNA to an area much smaller than the thermodynamically favored Rg (Figure 4). For the E. coli chromosome, Rg is ~5 μm. Constraints imposed by cellular geometry mean that DNA is no longer hydrodynamically a spherical ball, but rather is more akin to a deformed or anisotropic configuration. Constraints lead to nonintuitive changes in behavior, such as dynamics and/or segregation of multiple polymer chains in a small space. Entropy drives fluctuations of all chains to adopt a random coil. When the chains overlap, the number of conformational states is reduced (resulting in reduced entropy). This entropic penalty can drive segregation of the two chains in a confined space (82, 83). Such a mechanism has been proposed to operate in prokaryotes (82, 83) and might contribute to bulk chromosome segregation in eukaryotes that undergo closed mitosis (such as fungi and some protists).
Spring Dynamics
Chromosomes are dynamic structures that undergo large spatial fluctuations in interphase and mitosis. In mitosis, centromeres exhibit oscillatory motion toward and away from the poles. The behavior of the chromatin fiber underlying this motion is not resolved in a light microscope. Even with super-resolution microscopy modes, one cannot resolve individual chromatin strands (11–30 nm in diameter). To visualize the chromatin fiber, an array of E. coli lacO can be integrated into a chromosome together with lac repressor–GFP fusion proteins (146). In this way, the underlying chromatin dynamics can be directly visualized in the light microscope. Furthermore, in budding yeast, the operator arrays can be introduced at defined positions along a chromosome. To understand the motion of these DNA arrays, we return to the behavior of the polymer chain. Unlike diffusion of a protein or solute that bounces through the nucleus or cytoplasm with the force of thermal energy (kBT), a polymer wiggles and slithers like a snake across sand. The parameters for describing the motion of a polymer are its persistence length (Lp) and contour length (Lc). The persistence length is the length scale over which the polymer, such as naked DNA, is stiff, with the mathematical definition of Lp being based on the correlation of the ends in space. The contour length is the total chain length. The persistence length of a microtubule is 6 mm as compared with only 50 nm for DNA. The consequence of this ~5 orders of magnitude difference is that microtubules are very stiff and DNA is very floppy. The timescale of motion becomes a variable parameter for a long floppy chain such as DNA. On very short timescales (milliseconds), the motion is dictated by Brownian driven fluctuations of Lp segments. On these timescales, the motion reflects internal conformational changes of the monomers. Even a simple bead-spring model predicts distinct statistical fluctuation of each bead along the chain (162) (Figure 4). This leads to a position-dependent effective spring constant as seen by a particular bead relative to a tether point (i.e., centromere). The stiffer the chromatin spring, the less variance in fluctuations, whereas softer chromatin springs exhibit increased variance.
On longer timescales (seconds to minutes) the chain exhibits translational motion in space. Various models, such as the bead spring (Figure 4), have been invoked to understand the motion. Rouse models include spring and thermal forces, whereas Zimm models add hydrodynamic interactions to the Rouse model (29, 34, 130, 131, 178). The relaxation time of a polymer segment is defined as τR =ζ/kBT(NR2), where ζ is the drag coefficient, kBT is the Boltzmann constant × T, R is the end-end distance of the chain, and N is the number of beads. Motion analysis on timescales less than τR sample viscoelastic modes, whereas timescales greater than τR sample diffusive motion. For purposes of intuition, the diffusion constant of an average protein is 5–15 μm2/s. For a chromosome (bacterial or yeast) the apparent diffusion constant is ~0.0003–0.005 μm2/s (169–171), 3–4 orders of magnitude slower than a protein or small metabolite. The diffusion time for translation of the chromosome across the E. coli cell or a yeast nucleus is ~50 s. This is important when we consider that this is substantially less than the timescale of cell division or mitosis [20 min doubling time in bacteria, ~10–20 min mitosis in yeast (176)]. In other words, polymer diffusion dynamics are sufficient to provide the kinetic motion behind chromosome segregation. From these laws of motion, we infer that the critical function of kinetochore/spindle systems is to direct sisters into daughter cells, ensuring that each cell gets one copy of each chromosome. Thus, consideration of basic physical principles reveals that segregation machines are mainly in the business of accuracy and sorting, and that they just need to act as directional rectifiers to direct kinetic motion toward the spindle pole. As noted by Nicklas (110) many years ago, mitosis is not about speed; rather, it is about fidelity.
Chromatin Proteins that Drive Elasticity
The finding that cohesin is enriched at centromeres (52, 99, 151, 168) raised a paradox, namely how to reconcile sister chromatid cohesion with sister centromere separation in mitosis. The discovery that the pericentromeric region adopts an intramolecular loop structure in vivo provided a solution to this paradox (175). Instead of holding sister chromatids together, cohesin may contribute to the mechanisms that promote centromeric DNA looping and structural features that stiffen the spring and/or promote confinement (Figures 2 and 3). Introducing protein linkages into the system could have a profound effect on the stiffness of the chromatin spring. To understand the contribution of proteins to chromatin stiffness we turn to the relationship between force and chain extension. As discussed above, a fluctuating polymer chain adopts a random coil in the absence of an external force (Figure 4). When forces are exerted on the polymer, as experienced at the kinetochore microtubule attachment site, the polymer (centromere) is stretched. This reduces the number of states available for fluctuation. The reduction in entropy works against external elongation. A model that captures the relationship between force and extension for flexible polymers (random coils, such as unstructured protein or DNA) is known as the worm-like chain (58). The solution can be approximated according to the formula F = (kBT/Lp){[(1/4)(1 − x/Lc)−2]−1/4 + x/Lc}, where F is force, kBT is the Boltzmann constant × T, Lp and Lc are as defined above, and x is the extension. It takes very small forces (on the order of several piconewtons) to reduce the number of available states and extend the chain 60–80% of its contour length. Stretch length (x) approaches the chain length of the polymer (Lc), and extensional forces rise exponentially because of the stretching of covalent bonds. Note that the persistence length for unstructured proteins is less than a nanometer versus 50 nm for DNA (see above). The consequence is that small domains of unstructured proteins in the spring require roughly 50 times the extensional force relative to the same length of DNA. The makeup of centromere heterochromatin is approximately equal mass DNA and histone protein, with protein linkages such as cohesin and condensin distributed in the network. Putting cohesin and/or condensin into the system adds considerable stiffness to the network, analogous to putting pop-rivets along the DNA helix.
Alternatively, the cohesin and condensin rings may contribute in other ways to the elastic properties of the centromere and/or chromosome. When tension is applied to an individual polymer in a cross-linked network, force is exerted on the proximal cross-link that leads to local regions of high stress. Slip rings (or molecular pulleys) provide a mechanism to distribute tension from one location to the entire network (Figure 3) (57, 116). In this scenario, the stretched polymer slips through the ring, equalizing the tension cooperatively. Such a topological distribution of stress was originally proposed in the Doi, Edwards, deGennes reptation tube model (29, 34) and was demonstrated in a recent study of the chemistry of polyrotaxanes (57, 116). The ring-like structure of cohesin and condensin (72, 109) is indicative of the physical attributes expected of slip rings to provide the chemistry for distributing microtubule-based tension throughout centromere heterochromatin. SMC protein rings are ~40 nm [cohesin (64)] or 25 nm [condensin (1)] in diameter, sufficient to encircle one or two 11-nm nucleosome fibers. Cohesin links sister chromatids by encircling two chromatin strands (bracelet model) or dimerization of two rings, each of which encircle a single chromatin strand (handcuff model) or both (109, 117). Condensin also links chromatin strands by encircling one strand and binding to another (25, 26). Both cohesin and condensin are mobile in vivo. In budding yeast, cohesin is loaded at the centromere and slides to locations that cover the 50 kb of pericentric heterochromatin (76), whereas RNA polymerase can slide cohesin to the 3′ end of transcribed genes (112). Likewise, condensin sliding and stopping have been shown to be integral to mechanisms of chromosome condensation during anaphase segregation (26). Slip rings provide a mechanism for distributing tension through centromere heterochromatin that is feeling the ensemble of stochastically growing and shortening kinetochore microtubules.
Another mechanism to regulate motion and the distribution of tension throughout the network is through entanglements between the chains. Entanglements arise when a DNA from one chromosome or topological domain encircles another (see entanglements in Figure 3). This situation may arise in pericentric heterochromatin, where there is a high packing density chromatin. Increasing the number of entanglements throughout a network increases its elasticity (177). This confers a rubber-like behavior to the chromosome. A rubber band stretches because the polymer chains locally slip past one another. As a rubber band is stretched, the number of configurational states decreases, reducing the entropy. The entropic restoring force, which maximizes the number of configurations the chain can explore, is responsible for recoil when the stretch force is released. Cross-links in the polymer prevent chain slippage, providing a mechanism to tune the strength or stiffness of the network. A counter-intuitive property of rubber bands is their contraction when heated. At higher temperature, fluctuations increase and the shorter, random configurations are favored. In the centromere, more energy input in the form of protein machines (e.g., chromatin remodeling complexes and chaperones) increases chain motion, biasing the system toward chain stiffening. Centromere stretch and relaxation are observed in most eukaryotic kinetochores, highlighting this constant interplay between entropic chain fluctuation with its cross-links and entanglements versus the microtubule-based extensional forces that work to stretch the chain and reduce entropy.
Driving Force for Compaction of Pericentric Chromatin
A typical mammalian chromosome is compacted roughly 10,000-fold in mitosis. If we consider the ambient cellular pressure and the energy to pack the chromosome together with molecular crowding, we find that the chromosome is still fairly soft, i.e., loosely packed, even at these seemingly high compaction ratios. Although proteins such as condensin and topoisomerase II play a critical role in compaction, it is important to begin with first principles to determine how much energy is required for compaction. As discussed above, entropic forces favor chromosome compaction because of the increased available volume and therefore gain free energy from the depletants (small molecules) in the cell. Each molecule has an effect of ~1 kBT (3). Given the ~one million proteins in E. coli as an example, the upper limit for the change in free energy available for chromosome compaction is on the order of 106 kBT. It has been estimated that ~105 kBT (~100 pN) free energy is stored mechanically in the in vivo bacterial nucleoid (119). The external pressure required to hold the chromosome at this size is therefore 1,000 times smaller than the turgor pressure inside a cell [~1 atmospheric pressure (ATM) for bacteria (31) and 10 ATM for yeast (100) (1 ATM = 1 × 105 pN/μm2)]. The finding that compaction pressure inside the cell is 1/1,000th of the surrounding pressure indicates that the bacterial chromosome is fundamentally very soft. Similarly, measurements of the Young’s modulus of the chromosome are on the order of a few hundred pascals (pN/μm2) (149). This is very different from compaction of DNA in a virus that is subject to 50 ATM (equivalent to 50 × 105 pN/μm2) (102). Thus, although the degree of compaction (10,000-fold) is large at face value, it is not so large relative to the thermodynamics of the living system. In addition to the role structural proteins play in chromatin compaction, they may also provide internal structure in the form of tethers or entanglements that link chromatin domains (121, 122) and promote distant interactions in heritable ways.
The principles derived from a polymer physics view of centromere heterochromatin impinge on issues central to mechanisms of chromosome segregation. Tension at the kinetochore is derived from a balance of microtubule-based extensional forces and chromatin-based inward force. The shear bulk of centromeric heterochromatin (Figure 1) precludes a simple understanding of the structure of the spring. Furthermore, the stochastic microtubule growth and shortening at the multiple microtubule attachment sites in many organisms, including ourselves, are difficult to integrate into a tension-sensing mechanism that monitors individual kinetochore-microtubule attachment sites. Tackling the polymeric aspect of chromatin provides testable insights into these basic questions. Starting from thermodynamics, considering the entire system, including polymer and solute, we find that chromosome compaction increases the entropy for the solute. Thus, the force to compact the chromosome may be very small relative to what is intuitively a large problem (10,000 × compaction). Likewise, chromosome diffusion is very slow, but not particularly slow on the timescale of chromosome movement, consistent with suggestions made several decades ago (110, 153) that segregation mechanisms are concerned with accuracy rather than strength or speed. Finally, protein complexes that form rings, such as condensin and cohesin, may impart elastic properties to centromere heterochromatin and therefore, together with the entropic nature of a DNA spring, constitute the centromere spring.
Building a Centromere or “Only If You Build It, Do You Understand It”
The concentration of repeated sequences (satellites and/or tDNA), of cohesins, and of condensin distinguish the centromere from chromosome arms and euchromatin but do not reveal the initiating event in centromere formation. As voiced by Richard Feynman in the 1950s, “Only if you build it, do you understand it” (41). We turn to different strategies, including synthetic centromeres, de novo centromere formation, and evolutionary centromere lineages, that allow us to peer into critical nucleation events in centromere formation.
Synthetic centromeres
The elucidation of kinetochore structure in yeast, fly, and human guide strategies for synthesis (10, 125, 150). The kinetochore is the protein-DNA machine built at the centromere. There are more than 100 proteins organized into 6–7 complexes, each of which contain between 4–12 protein subunits (Figure 2). The major force coupler is Ndc80, a tetrameric complex that lies on the microtubule lattice, proximal to its plus-end (158). The Dam1 complex comprises a 12-protein multimer that oligomerizes into a ring encircling the microtubule lattice in vitro (111). The Dam1 complex is essential in budding yeast and is one of the few components that is not evolutionarily conserved. The functional equivalent in mammalian kinetochores may be the Ska1 complex that binds depolymerizing microtubules and enhances the ability of Ndc80 to track microtubule ends (139, 173). The linkage to the chromatin is established via the CCAN complex (45, 150) and the centromere-specific histone H3 variant CENP-A (45, 145). In budding yeast, there is a high-affinity sequence-specific DNA binding complex that recognizes the centromere DNA CBF3 (54, 89). Several recent studies have built microtubule attachment sites for chromosome segregation. A site for a high-affinity DNA-binding protein is one requirement for DNA segregation that has been reconstructed through the use of tandem copies of the lacO. The DNA binding protein lac repressor (product of the LacI gene) is fused to a protein component of the kinetochore essential for microtubule interaction. In one case, fusion of lac repressor to Ask1, a member of the Dam1 complex was used; in another case, lac repressor was fused to Dam1, a member of the same complex (85, 88). In these synthetic kinetochores, the fusion protein recruits sufficient kinetochore proteins to promote chromosome segregation. These systems completely bypass the requirement for the authentic centromere and essential DNA binding components of the natural centromere.
Mammalian centromeres, unlike budding yeast, do not have a specific DNA sequence at the site of microtubule attachment. The first synthesis of an artificial human centromere [human artificial chromosome (HAC)] introduced fragments of cloned centromeres to recruit kinetochore components (67, 95). A clever marriage between the human alphoid repeats and site-specific bacterial binding sites allowed Earnshaw and colleagues (107) to build a conditional human centromere. These investigators introduced a tetracycline operator (TetO) every second alphoid monomer. This system provides the capability to direct a range of proteins into the functional kinetochore as tetracycline repressor-fusion proteins. These synthetic kinetochores can be conditionally inactivated, which has significant implications for gene therapy studies, and can be used to study the chromatin requirements for centromere formation and heritability.
More recently, several groups have used these strategies to address the chromatin requirements for human kinetochores. Gascoigne et al. (49) directed two of the DNA binding components of the mammalian kinetochore to an ectopic site and were able to assemble a microtubule attachment site without CENP-A. An exhaustive study of ectopic targeting by the Fukagawa group revealed two modes of assembly (74). One (using CENP-T or CENP-C N terminus) recruits kinetochore proteins and assembles a functional kinetochore lacking CENP-A. A second strategy that recruits the full complement of kinetochore proteins, including CENP-A, utilized full-length HJURP, CENP-C, CENP-I, or the CENP-C C terminus. Thus, the recruitment of CENP-A is separable from the kinetochore function.
The Fukagawa study (74) is a critical advance in understanding centromeres, as it provides the first functional distinction of centromere versus kinetochore. A microtubule attachment site can be built by tethering DNA to a microtubule component within the kinetochore or via recruitment of a DNA-binding protein that recruits microtubule-binding components. The conceptual advance is that we can formally separate CENP-A chromatin, and potentially alphoid repeats, from kinetochore protein recruitment. The ability to assemble a functional kinetochore without underlying centromeric sequences or chromatin suggests, in turn, that the centromere is not just a landing pad or attachment site for the kinetochore and that the centromere has evolved with unique physical properties distinct from microtubule attachment that contribute to optimal kinetochore function.
The role of the centromere-unique histone H3 variant Cse4 (yeast) or CENP-A (mammals) remains enigmatic (30, 40) because these alternative mechanisms for tethering DNA to a microtubule-binding component of the kinetochore suffice for chromosome segregation. Current understanding of the organization of Cse4 itself within chromatin is equivocal (16). In organisms from yeast to mammal, more Cse4 resides in the centromere than lies at the plus-end of the kinetochore microtubule (62, 92). In budding yeast, several molecules of Cse4 reside in the vicinity of the kinetochore but are not bound in a sequence-specific manner (62). Binding of these noncentromere molecules depends upon Pat1, unlike Cse4 binding at CEN DNA (62, 101), indicating that heterogeneity in nucleosome structure is to be expected. Unlike in histone H3 and canonical nucleosomes, attempts to ascribe a single structure to Cse4-containing nucleosomes has been difficult, with both hemisome and nucleosomal states being identified. A parsimonious view is that the Cse4 nucleosome is dynamic (16). Because molecules closest to the plus-end of the microtubule experience more force than canonical nucleosomes, there may be value for the nucleosome to be adept at switching between hemisome and nucleosomal states. Like the unfolding of transcriptionally active nucleosomes (124), the centromere nucleosome may be adapted to rapid changes in pulling and pushed microtubule-based forces.
Neocentromeres
New centromeres have been found to arise in natural populations in a variety of organisms, providing means to ask how nature builds her own centromeres. These neocentromeres, or ENCs, have been reported in a variety of Equus species (horse, donkey, and zebra) and in Drosophila, Candida albicans (yeast), maize, and humans (17, 47, 140). Several key mechanistic steps in de novo centromere function can be deduced. Using in situ hybridization to detect satellite DNA and immunofluorescence to detect kinetochore proteins, Piras et al. (122) have shown that new centromeres (one million years old; in Equus) arise in regions free of satellite DNA. Through phylogenetic reconstructions, they show that the acquisition of satellite sequences follows kinetochore formation. These satellite-deficient centromeres can function over millions of years, indicating that segregation fidelity must approximate that of endogenous centromeres. The accumulation of repeated sequences over evolutionary time is indicative of their selective advantage. Lessons from the polymer world indicate that satellite sequences, through promotion of entanglements or recruitment of cohesin and condensin, confer elasticity to the pericentromere (Figure 5). Through greater elasticity between sister kinetochores, a higher degree of accuracy in discriminating stochastic microtubule dynamics from bona fide bi-orientation can be achieved.
Figure 5.
Emergent properties of the centromere. (Left) The yeast segregation apparatus, shown in the vertical position, serves as a model for multiple kinetochore-microtubule attachment-site centromeres found in other organisms. The kinetochore microtubules ( green) can be seen at the top and bottom, emanating from the spindle pole body (red ). The pericentromere DNA is organized as a network of loops ( yellow) radially arranged around the spindle axis. Cohesin (blue) and condensin ( purple) are enriched in the pericentromere and surround the central spindle. Abbreviations: LTR, long terminal repeat; MT, microtubule. Adapted from figure 6 in Reference 63.
Neocentromere formation (also known as centromere repositioning) has been studied in systems in which the endogenous centromere can be deleted and replaced with a genetically selectable marker. In C. albicans (yeast) and Gallus gallus (chicken) DT40 cells, new centromeres were found in several positions, most frequently close to the original centromere (141, 154). It is thought that low levels of CENP-A in regions flanking the centromere may seed these new centromeres. The initiating event in these repositioned centromeres remains unknown. In maize, fluid structures dictated by retrotransposons and local repositioning of CenH3 have been reported as well (47, 155).
Centromere Switching: Epigenetic States
The fluidity of centromeres can be readily appreciated from the perspective of a mechanism required to inactivate a centromere in rare chromosome fusion events. As first described by McClintock (96) in maize in the late 1930s, dicentric chromosomes are genetically unstable and undergo a breakage-fusion-bridge reaction. More recently, using directed methods to create dicentric chromosomes (expression of a dominant negative mutant of telomere protein TRF-2), Stimpson et al. (144) demonstrated centromere inactivation in de novo dicentric chromosomes. Studies in human and budding yeast reveal the critical role of CENP-A histone modification in regulating the functional state of the centromere (5, 134). The switchable nature of a centromere highlights the fact that centromere function arises from more than the sum of the parts. There is an emergent structure that is fluid and more likely to be subjected to constant molding and remolding, dependent on cellular and chromosome physiology.
Evidence for centromere remodeling can be found in both fission and budding yeast. In fission yeast, Sato et al. (136) have demonstrated that epigenetic inactivation is initiated by disassembly of kinetochore components. Mutations that compromise kinetochore protein promote centromere inactivation. This is reminiscent of a finding in budding yeast that mutations in centromere DNA binding proteins lead to the stabilization of dicentric chromosomes (33). However, studies have used a centromere that can be conditionally inactivated via transcription from an adjacent promoter to demonstrate that de novo centromere function depends upon both the Swi2/Snf2 chromatin remodeling factor Fun30 (38) and the kinetochore component Chl4 (106) for its activation. In this way, de novo versus template-directed propagation of a centromere state was distinguished. Thus, even in budding yeast, the centromere can exist in two states, and state switching is likely to involve chromatin remodelers. It should be noted that both Fun30 and a second Swi2/Snf2 homolog, Rdh54/Tid1, have been localized to kinetochores (91, 123).
In species other than budding yeasts, the centromere is a dynamic genomic element, and this lack of sequence-specific requirements makes it difficult to identify the chromatin features that predispose centromere formation. The presence of CENP-A (13, 121, 145), reduced histone H2A.Z (113), and a concentration of tRNA genes (77), retrotransposons, or other mobile genetic elements (126) have all been suggested as contributing to centromere-permissive regions. The absence of satellite sequences in neocentromeres is indicative of their nonessential centromere function. The conclusion from a phylogenetically broad spectrum of plants and animals indicates that there are neocentromere-favorable and -unfavorable sites (154, 161). Unlike the kinetochore, with its readily identifiable essential proteins, it is difficult (outside of budding yeast) to identify genetic sequences or modifications that definitively mark or exclude centromere formation.
The capability for neocentromere formation must be balanced against the formation of dicentric chromosomes and potential chromosome instability (71, 97). Nonetheless, the cell retains the capacity to build centromeres de novo in rare instances of centromere loss. It is not surprising that de novo centromere formation is nonrandom, but how sites are chosen is less clear. In addition, centromere complexity evolves with time. The late acquisition of satellite sequences into neocentromeres is indicative of advantageous functions that evolve on timescales slower than those for the microtubule attachment mechanism. Enumerating these functions and considering alternative segregation mechanisms might provide insights into the centromere that transcend kinetochore function.
IMPLICATIONS OF NONCANONICAL SEGREGATION MECHANISMS FOR KINETOCHORE-INDEPENDENT CENTROMERE FUNCTION
Velcro Segregation Mechanisms
The two-micron plasmid in budding yeast is a high-copy extrachromosomal element. The plasmid is partitioned with high fidelity in part because of a highly evolved stabilization element (STB) and the DNA binding proteins Rep1 and Rep2. Recent studies have shown that the two-micron plasmid physically associates with the chromosome and recruits cohesin (51), leading to a proposal for a hitchhiking mechanism of segregation (18). The plasmid recruits substoichiometric quantities of the centromere-specific histone (CENP-A), indicating that the potential site of hitchhiking might be the centromere. Centromere chromatin may provide a similar mechanism of association between the individual centromeres in budding yeast, thereby contributing to the fidelity of segregation.
Structural Maintenance of Chromosome-Based Bacterial Segregation
The timing of condensin-mediated compaction of DNA in bacteria is closely linked to the chromosome segregation cycle and cell division (148, 166). In eukaryotes, condensin and cohesin loading is coupled to DNA replication (27, 167), and chromosomes are condensed well before anaphase chromosome segregation. Linking chromosome compaction with segregation in bacteria may reflect a strategy to harness SMC-mediated compaction forces for segregation. Rudner and colleagues (148, 166) argue that the key feature of SMC-mediated compaction is the orderly folding of DNA, thereby facilitating protein-DNA transactions and that this has a central role in driving segregation. Chromosome individualization (visibly separated sister chromatids) in multicellular organisms likewise reflects the ability of condensation mechanisms to segregate strands. The enrichment of cohesin and condensin in centromeric heterochromatin (27, 86, 99) is indicative of additional functions beyond compaction and cohesion. The analysis of polymer networks makes testable predications for how these proteins contribute to centromere elasticity.
Entropic Recoil in Confinement
Jun, Mulder, and colleagues (82, 83) have shown that the penalty due to polymer repulsion is sufficient to drive chains apart in a confined space, and may drive chromosome segregation in bacterial cells. As soon as the genome becomes fragmented, such as in eukaryotic chromosomes, thermodynamics is completely insufficient, as this mechanism cannot provide a single order or spatial direction. Thus, the polymer repulsion model cannot be the sole mechanism for eukaryotic chromosome segregation but serves as a guide to our intuition. Kleckner and colleagues (81) posit that origins of replication in bacteria are pushed toward the cell poles as a result of intranucleoid confinement. The replicated origins undergo condensation and resolution from each other but remain juxtaposed at specific origin-proximal sites (called snaps). The accumulation of DNA in the confined cellular space generates internal pushing forces. When these forces exceed the strength of the snaps, cohesion is lost, resulting in the abrupt and rapid extrusion of the condensed origin regions toward opposite poles. Because replication probably initiates at the nucleoid periphery, the newly replicated DNA is naturally compartmentalized from the unreplicated chromosome. This helps to prevent entanglements and provides an unimpeded path for segregation of the origins and may explain why yeast centromeres are replicated early. Polymer repulsion may also underlie a mechanism to extrude centromeres and thus prevent them from being buried in the mass of centromeric heterochromatin.
EMERGENT FUNCTION FROM ENRICHMENT AND MOLECULAR ORGANIZATION OF CENTROMERE COMPONENTS
If we build our understanding of the centromere from the biochemical and physical properties of the components, we emerge with a deeper understanding that transcends the kinetochore. The enrichment of topology adjusters, including cohesin, condensin, and topoisomerase II, endows centromeres with elasticity. Entanglement reflects a topological constraint resulting from the fact that DNA strands are unable to cross one another (157) in the absence of topoisomerase II. As a result of entanglements, the motion of one chain influences the other, resulting in an increased elastic modulus of the network (87, 157). In the case of λ DNA at DNA concentrations greater than 0.5 mg/mL, the elastic modulus exceeds the viscous modulus of the network. The network thus exhibits elastic behavior. In the centromere, we can estimate the concentration of DNA on the basis of the region of DNA enriched in SMC proteins (50 kb/chromosome) and the volume of the cylinder between bi-oriented centromeres in mitosis (~0.15 μm3). The DNA concentration is approximately 9 mg/mL (about three times greater than bulk DNA in the nucleus). Thus, it is very likely strands are topologically constrained within the cohesin barrel.
As discussed above, DNA strand sliding within the cohesin ring confers a slip-link property to the network. Tension generated at one strand is distributed among remaining strands because each strand “feels” the other through the equalization of tension via nanopulleys. Gels with slip-links (polyrotaxane gels) have the remarkable property of maintaining their three-dimensional shape over orders of magnitude of size scales (116). The consequence of such a network is that the stochastic growth and shortening of kinetochore microtubules at individual attachment sites can be averaged across the chromatin spring.
Condensin is responsible for chromosome condensation in metaphase as well as recoil of chromosome arms throughout anaphase (127). Condensin is highly enriched in regions of very high transcription, such as the nucleolus, and also at centromeres (4, 27, 28, 86). It has been proposed that condensin in the nucleolus serves to protect the genome from massive rearrangement from recombination between rDNA repeats and potential shuffling thereof. In the centromere, condensin contributes to heterochromatin compaction. However, considering the central role for condensin in prokaryotic chromosome segregation, the enrichment of condensin in eukaryotic chromosomes may be more profound. Compaction of strands naturally (i.e., thermodynamically) promotes strand segregation. Thus, the centromere provides the site of initiation for chromosome strand separation. From this perspective, the spindle apparatus functions as a guidance mechanism to ensure that sisters separate. The driving force for segregation may lie within the centromere proper.
The physical basis for the chromatin spring will ultimately require an understanding of DNA fluctuations, compaction, histone modification and mobility, DNA cross-linking complexes (inter-as well as intramolecular interactions), and topology adjusters. Through DNA strand cross-linking and entanglements, a robust elastic network emerges that maintains mechanical integrity across hundreds to thousands of kilobase pairs of centromere DNA. Polymer physics provide an important framework for integrating protein function with DNA dynamics to comprehend the accuracy of chromosome segregation.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–24. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–39. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asakura S, Oosawa F. On interaction between two bodies immersed in a solution of macromolecules. J Chem Phys. 1954;22:1255. [Google Scholar]
- 4.Bachellier-Bassi S, Gadal O, Bourout G, Nehrbass U. Cell cycle–dependent kinetochore localization of condensin complex in Saccharomyces cerevisiae. J Struct Biol. 2008;162:248–59. doi: 10.1016/j.jsb.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci USA. 2013;110:11827–32. doi: 10.1073/pnas.1300325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldwin M, Warsi T, Bachant J. Analyzing Top2 distribution on yeast chromosomes by chromatin immunoprecipitation. Methods Mol Biol. 2009;582:119–30. doi: 10.1007/978-1-60761-340-4_10. [DOI] [PubMed] [Google Scholar]
- 7.Bardin AJ, Amon A. Men and sin: What’s the difference? Nat Rev Mol Cell Biol. 2001;2:815–26. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- 8.Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, et al. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci. 2012;125:411–21. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–40. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 2013;194:817–46. doi: 10.1534/genetics.112.145276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloom K, Yeh E. Tension management in the kinetochore. Curr Biol. 2010;20:R1040–48. doi: 10.1016/j.cub.2010.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom KS. Beyond the code: the mechanical properties of DNA as they relate to mitosis. Chromosoma. 2008;117:103–10. doi: 10.1007/s00412-007-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–30. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner S, Pepper D, Berns MW, Tan E, Brinkley BR. Kinetochore structure, duplication, and distribution in mammalian cells: analysis by human autoantibodies from scleroderma patients. J Cell Biol. 1981;91:95–102. doi: 10.1083/jcb.91.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- 16.Bui M, Walkiewicz MP, Dimitriadis EK, Dalal Y. The CENP-A nucleosome: a battle between Dr Jekyll and Mr Hyde. Nucleus. 2013;4:37–42. doi: 10.4161/nucl.23588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, et al. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics. 2006;87:777–82. doi: 10.1016/j.ygeno.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Chan KM, Liu YT, Ma CH, Jayaram M, Sau S. The 2 micron plasmid of Saccharomyces cerevisiae: a miniaturized selfish genome with optimized functional competence. Plasmid. 2013;70:2–17. doi: 10.1016/j.plasmid.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Cheerambathur DK, Desai A. Linked in: formation and regulation of microtubule attachments during chromosome segregation. Curr Opin Cell Biol. 2014;26:113–22. doi: 10.1016/j.ceb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherry LM, Faulkner AJ, Grossberg LA, Balczon R. Kinetochore size variation in mammalian chromosomes: an image analysis study with evolutionary implications. J Cell Sci. 1989;92(Pt. 2):281–89. doi: 10.1242/jcs.92.2.281. [DOI] [PubMed] [Google Scholar]
- 21.Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, et al. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–73. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 22.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 23.Clarke L, Baum MP. Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol Cell Biol. 1990;10:1863–72. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–9. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 25.Cuylen S, Metz J, Haering CH. Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol. 2011;18:894–901. doi: 10.1038/nsmb.2087. [DOI] [PubMed] [Google Scholar]
- 26.Cuylen S, Metz J, Hruby A, Haering CH. Entrapment of chromosomes by condensin rings prevents their breakage during cytokinesis. Dev Cell. 2013;27:469–78. doi: 10.1016/j.devcel.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 27.D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–27. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–69. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- 29.de Gennes PG. Scaling Concepts in Polymer Physics. Ithaca, NY: Cornell Univ. Press; 1979. [Google Scholar]
- 30.De Rop V, Padeganeh A, Maddox PS. CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma. 2012;121:527–38. doi: 10.1007/s00412-012-0386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng Y, Sun M, Shaevitz JW. Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys Rev Lett. 2011;107:158101. doi: 10.1103/PhysRevLett.107.158101. [DOI] [PubMed] [Google Scholar]
- 32.Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–51. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–74. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doi M, Edwards SF. The Theory of Polymer Dynamics. Oxford, UK: Oxford Univ. Press; 1986. [Google Scholar]
- 35.Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013;25:327–33. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–67. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durand-Dubief M, Will WR, Petrini E, Theodorou D, Harris RR, et al. SWI/SNF-like chromatin remodeling factor Fun30 supports point centromere function in S. cerevisiae. PLOS Genet. 2012;8:e1002974. doi: 10.1371/journal.pgen.1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–91. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falk SJ, Black BE. Centromeric chromatin and the pathway that drives its propagation. Biochim Biophys Acta. 2013;1819:313–21. [PubMed] [Google Scholar]
- 41.Feynman RP. There’s plenty of room at the bottom. Caltech Eng Sci. 1960;23:22–36. [Google Scholar]
- 42.Fisher JK, Bourniquel A, Witz G, Weiner B, Prentiss M, Kleckner N. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013;153:882–95. doi: 10.1016/j.cell.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–44. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 44.Flemming W. Zellsubstanz, Kern und Zelltheilung. Leipzig, Ger: Vogel; 1882. p. 424. [Google Scholar]
- 45.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–69. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 46.Fritzler MJ, Kinsella TD. The CREST syndrome: a distinct serologic entity with anticentromere antibodies. Am J Med. 1980;69:520–26. doi: 10.1016/0002-9343(80)90462-3. [DOI] [PubMed] [Google Scholar]
- 47.Fu S, Lv Z, Gao Z, Wu H, Pang J, et al. De novo centromere formation on a chromosome fragment in maize. Proc Natl Acad Sci USA. 2013;110:6033–36. doi: 10.1073/pnas.1303944110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gartenberg M. Heterochromatin and the cohesion of sister chromatids. Chromosome Res. 2009;17:229–38. doi: 10.1007/s10577-008-9012-z. [DOI] [PubMed] [Google Scholar]
- 49.Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–22. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gent JI, Dawe RK. RNA as a structural and regulatory component of the centromere. Annu Rev Genet. 2012;46:443–53. doi: 10.1146/annurev-genet-110711-155419. [DOI] [PubMed] [Google Scholar]
- 51.Ghosh SK, Huang CC, Hajra S, Jayaram M. Yeast cohesin complex embraces 2 micron plasmid sisters in a tri-linked catenane complex. Nucleic Acids Res. 2010;38:570–84. doi: 10.1093/nar/gkp993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLOS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glynn M, Kaczmarczyk A, Prendergast L, Quinn N, Sullivan KF. Centromeres: assembling and propagating epigenetic function. Subcell Biochem. 2010;50:223–49. doi: 10.1007/978-90-481-3471-7_12. [DOI] [PubMed] [Google Scholar]
- 54.Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–12. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet. 2009;18:3178–93. doi: 10.1093/hmg/ddp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–33. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 57.Granick S, Rubinstein M. Polymers: a multitude of macromolecules. Nat Mater. 2004;3:586–87. doi: 10.1038/nmat1199. [DOI] [PubMed] [Google Scholar]
- 58.Greulich KO, Wachtel E, Ausio J, Seger D, Eisenberg H. Transition of chromatin from the “10 nm” lower order structure, to the “30 nm” higher order structure as followed by small angle X-ray scattering. J Mol Biol. 1987;193:709–21. doi: 10.1016/0022-2836(87)90353-6. [DOI] [PubMed] [Google Scholar]
- 59.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–41. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 61.Grosberg AY, Khokhlov AR. Giant Molecules Here, There, and Everywhere. San Diego, CA: Acad. Press; 1997. p. 244. [Google Scholar]
- 62.Haase J, Mishra PK, Stephens A, Haggerty R, Quammen C, et al. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr Biol. 2013;23:1939–44. doi: 10.1016/j.cub.2013.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haase J, Stephens A, Verdaasdonk J, Yeh E, Bloom K. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr Biol. 2012;22:471–81. doi: 10.1016/j.cub.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–88. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 65.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–14. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall LE, Mitchell SE, O’Neill RJ. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res. 2012;20:535–46. doi: 10.1007/s10577-012-9297-9. [DOI] [PubMed] [Google Scholar]
- 67.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–55. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 68.Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF. Sequences associated with centromere competency in the human genome. Mol Cell Biol. 2013;33:763–72. doi: 10.1128/MCB.01198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–75. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 70.Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hill A, Bloom K. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1368–70. doi: 10.1128/mcb.9.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–22. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 73.Hopper AK, Pai DA, Engelke DR. Cellular dynamics of tRNAs and their genes. FEBS Lett. 2010;584:310–17. doi: 10.1016/j.febslet.2009.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsieh CL, Lin CL, Liu H, Chang YJ, Shih CJ, et al. WDHD1 modulates the post-transcriptional step of the centromeric silencing pathway. Nucleic Acids Res. 2011;39:4048–62. doi: 10.1093/nar/gkq1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, et al. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011;21:12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell. 2010;21:254–65. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johzuka K, Horiuchi T. RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae. Genes Cells. 2007;12:759–71. doi: 10.1111/j.1365-2443.2007.01085.x. [DOI] [PubMed] [Google Scholar]
- 79.Johzuka K, Horiuchi T. The cis element and factors required for condensin recruitment to chromosomes. Mol Cell. 2009;34:26–35. doi: 10.1016/j.molcel.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 80.Jokelainen PT. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J Ultrastruct Res. 1967;19:19–44. doi: 10.1016/s0022-5320(67)80058-3. [DOI] [PubMed] [Google Scholar]
- 81.Joshi MC, Bourniquel A, Fisher J, Ho BT, Magnan D, et al. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc Natl Acad Sci USA. 2011;108:2765–70. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–93. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol. 2010;8:600–7. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc Natl Acad Sci USA. 2000;97:13108–13. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kiermaier E, Woehrer S, Peng Y, Mechtler K, Westermann S. A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat Cell Biol. 2009;11:1109–15. doi: 10.1038/ncb1924. [DOI] [PubMed] [Google Scholar]
- 86.Kim JH, Zhang T, Wong NC, Davidson N, Maksimovic J, et al. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun. 2013;4:2537. doi: 10.1038/ncomms3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kundukad B, van der Maarel JR. Control of the flow properties of DNA by topoisomerase II and its targeting inhibitor. Biophys J. 2010;99:1906–15. doi: 10.1016/j.bpj.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lacefield S, Lau DT, Murray AW. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat Cell Biol. 2009;11:1116–20. doi: 10.1038/ncb1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–25. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 90.Lejeune E, Bayne EH, Allshire RC. On the connection between RNAi and heterochromatin at centromeres. Cold Spring Harb Symp Quant Biol. 2010;75:275–83. doi: 10.1101/sqb.2010.75.024. [DOI] [PubMed] [Google Scholar]
- 91.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 92.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Losada A, Hirano M, Hirano T. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 2002;16:3004–16. doi: 10.1101/gad.249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maddox PS, Portier N, Desai A, Oegema K. Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay. Proc Natl Acad Sci USA. 2006;103:15097–102. doi: 10.1073/pnas.0606993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Masumoto H, Ikeno M, Nakano M, Okazaki T, Grimes B, et al. Assay of centromere function using a human artificial chromosome. Chromosoma. 1998;107:406–16. doi: 10.1007/s004120050324. [DOI] [PubMed] [Google Scholar]
- 96.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–76. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McClintock B. Induction of instability at selected loci in maize. Genetics. 1953;38:579–99. doi: 10.1093/genetics/38.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McIntosh JR, Molodtsov MI, Ataullakhanov FI. Biophysics of mitosis. Q Rev Biophys. 2012;45:147–207. doi: 10.1017/S0033583512000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–50. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- 100.Minc N, Boudaoud A, Chang F. Mechanical forces of fission yeast growth. Curr Biol. 2009;19:1096–101. doi: 10.1016/j.cub.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mishra PK, Ottmann AR, Basrai M. Structural integrity of centromeric chromatin and faithful chromosome segregation requires Pat1. Genetics. 2013;195(2):369–79. doi: 10.1534/genetics.113.155291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Molineux IJ, Panja D. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol. 2013;11:194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- 103.Moroi Y, Hartman AL, Nakane PK, Tan EM. Distribution of kinetochore (centromere) antigen in mammalian cell nuclei. J Cell Biol. 1981;90:254–59. doi: 10.1083/jcb.90.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moroi Y, Peebles C, Fritzler MJ, Steigerwald J, Tan EM. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc Natl Acad Sci USA. 1980;77:1627–31. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Musacchio A, Ciliberto A. The spindle-assembly checkpoint and the beauty of self-destruction. Nat Struct Mol Biol. 2012;19:1059–61. doi: 10.1038/nsmb.2429. [DOI] [PubMed] [Google Scholar]
- 106.Mythreye K, Bloom KS. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol. 2003;160:833–43. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–22. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986;5:1011–21. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 110.Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–49. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- 111.Nogales E, Ramey VH. Structure-function insights into the yeast Dam1 kinetochore complex. J Cell Sci. 2009;122:3831–36. doi: 10.1242/jcs.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ocampo-Hafalla MT, Uhlmann F. Cohesin loading and sliding. J Cell Sci. 2011;124:685–91. doi: 10.1242/jcs.073866. [DOI] [PubMed] [Google Scholar]
- 113.Ogiyama Y, Ohno Y, Kubota Y, Ishii K. Epigenetically induced paucity of histone H2A.Z stabilizes fission-yeast ectopic centromeres. Nat Struct Mol Biol. 2013;20:1397–406. doi: 10.1038/nsmb.2697. [DOI] [PubMed] [Google Scholar]
- 114.Ohkuni K, Kitagawa K. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr Biol. 2011;21:1695–703. doi: 10.1016/j.cub.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ohkuni K, Kitagawa K. Role of transcription at centromeres in budding yeast. Transcription. 2012;3:193–97. doi: 10.4161/trns.20884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Okumura Y, Ito K. The polyrotaxane gel: a topological gel by figure-of-eight cross-links. Adv Mater. 2001;13:485–87. [Google Scholar]
- 117.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–29. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 118.Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–66. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pelletier J, Halvorsen K, Ha BY, Paparcone R, Sandler SJ, et al. Physical manipulation of the Escherichia coli chromosome reveals its soft nature. Proc Natl Acad Sci USA. 2012;109:E2649–56. doi: 10.1073/pnas.1208689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Phillips-Cremins JE, Corces VG. Chromatin insulators: linking genome organization to cellular function. Mol Cell. 2013;50:461–74. doi: 10.1016/j.molcel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pidoux AL, Allshire RC. The role of heterochromatin in centromere function. Philos Trans R Soc Lond B. 2005;360:569–79. doi: 10.1098/rstb.2004.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Piras FM, Nergadze SG, Magnani E, Bertoni L, Attolini C, et al. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLOS Genet. 2010;6:e1000845. doi: 10.1371/journal.pgen.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J Mol Biol. 2007;369:940–53. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Prior CP, Cantor CR, Johnson EM, Littau VC, Allfrey VG. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983;34:1033–42. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- 125.Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–65. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- 126.Qi LL, Wu JJ, Friebe B, Qian C, Gu YQ, et al. Sequence organization and evolutionary dynamics of Brachypodium-specific centromere retrotransposons. Chromosome Res. 2013;21:507–21. doi: 10.1007/s10577-013-9378-4. [DOI] [PubMed] [Google Scholar]
- 127.Renshaw MJ, Ward JJ, Kanemaki M, Natsume K, Nedelec FJ, Tanaka TU. Condensins promote chromosome recoiling during early anaphase to complete sister chromatid separation. Dev Cell. 2010;19:232–44. doi: 10.1016/j.devcel.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ribeiro SA, Gatlin JC, Dong Y, Joglekar A, Cameron L, et al. Condensin regulates the stiffness of vertebrate centromeres. Mol Biol Cell. 2009;20:2371–80. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rock JM, Amon A. The FEAR network. Curr Biol. 2009;19:R1063–68. doi: 10.1016/j.cub.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rouse PE. A theory of the linear viscoelastic properties of dilute solutions of coiling polymer. J Chem Phys. 1953;21:1272–80. [Google Scholar]
- 131.Rubinstein M, Colby RH. Polymer Physics. Oxford, UK: Oxford Univ. Press; 2003. p. 441. [Google Scholar]
- 132.Salmon ED. Pressure-induced depolymerization of spindle microtubules. II Thermodynamics of in vivo spindle assembly. J Cell Biol. 1975;66:114–27. doi: 10.1083/jcb.66.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Salmon ED, Goode D, Maugel TK, Bonar DB. Pressure-induced depolymerization of spindle microtubules. III Differential stability in HeLa cells. J Cell Biol. 1976;69:443–54. doi: 10.1083/jcb.69.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Samel A, Cuomo A, Bonaldi T, Ehrenhofer-Murray AE. Methylation of CenH3 arginine 37 regulates kinetochore integrity and chromosome segregation. Proc Natl Acad Sci USA. 2012;109:9029–34. doi: 10.1073/pnas.1120968109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Samoshkin A, Arnaoutov A, Jansen LE, Ouspenski I, Dye L, et al. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLOS ONE. 2009;4:e6831. doi: 10.1371/journal.pone.0006831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sato H, Masuda F, Takayama Y, Takahashi K, Saitoh S. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr Biol. 2012;22:658–67. doi: 10.1016/j.cub.2012.02.062. [DOI] [PubMed] [Google Scholar]
- 137.Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–58. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 138.Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–24. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–80. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Scott KC, Sullivan BA. Neocentromeres: a place for everything and everything in its place. Trends Genet. 2014;30:66–74. doi: 10.1016/j.tig.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shang WH, Hori T, Martins NM, Toyoda A, Misu S, et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell. 2013;24:635–48. doi: 10.1016/j.devcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–80. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stephens AD, Quammen CW, Chang B, Haase J, Taylor RM, 2nd, Bloom K. The spatial segregation of pericentric cohesin and condensin in the mitotic spindle. Mol Biol Cell. 2013;24:3909–19. doi: 10.1091/mbc.E13-06-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stimpson KM, Song IY, Jauch A, Holtgreve-Grez H, Hayden KE, et al. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLOS Genet. 2010;6:1–19. doi: 10.1371/journal.pgen.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–86. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 146.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 147.Sullivan BA. The Centromere. New York: Springer; 2009. p. 509. [Google Scholar]
- 148.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun M, Kawamura R, Marko JF. Micromechanics of human mitotic chromosomes. Phys Biol. 2011;8:015003. doi: 10.1088/1478-3975/8/1/015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Takeuchi K, Fukagawa T. Molecular architecture of vertebrate kinetochores. Exp Cell Res. 2012;318:1367–74. doi: 10.1016/j.yexcr.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 151.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–58. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 152.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–99. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 153.Taylor EW. Brownian and saltatory movements of cytoplasmic granules and the movement of anaphase chromosomes. Proc. Int. Congr. Rheol. Symp. Biorheol. 4th; Providence RI. New York: Interscience; 1965. pp. 175–91. [Google Scholar]
- 154.Thakur J, Sanyal K. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 2013;23:638–52. doi: 10.1101/gr.141614.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Topp CN, Okagaki RJ, Melo JR, Kynast RG, Phillips RL, Dawe RK. Identification of a maize neocentromere in an oat-maize addition line. Cytogenet Genome Res. 2009;124:228–38. doi: 10.1159/000218128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Underhill PT, Doyle PS. On the coarse-graining of polymers into bead-spring chains. J Non-Newton Fluid Mech. 2004;122:3–31. [Google Scholar]
- 157.Van der Maarel JR. Introduction to Biopolymer Physics. Hackensack, NJ: World Sci. Publ; 2008. p. 246. [Google Scholar]
- 158.Varma D, Salmon ED. The KMN protein network: chief conductors of the kinetochore orchestra. J Cell Sci. 2012;125:5927–36. doi: 10.1242/jcs.093724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Varma D, Wan X, Cheerambathur D, Gassmann R, Suzuki A, et al. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J Cell Biol. 2013;202:735–46. doi: 10.1083/jcb.201304197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Venkei Z, Przewloka MR, Ladak Y, Albadri S, Sossick A, et al. Spatiotemporal dynamics of Spc105 regulates the assembly of the Drosophila kinetochore. Open Biol. 2012;2:110032. doi: 10.1098/rsob.110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, et al. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Verdaasdonk JS, Vasquez PA, Barry RM, Barry T, Goodwin S, et al. Centromere tethering confines chromosome domains. Mol Cell. 2013;52:819–31. doi: 10.1016/j.molcel.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Verma R, Crocker JC, Lubensky TC, Yodh AG. Entropic colloidal interactions in concentrated DNA solutions. Phys Rev Lett. 1998;81:4004–7. [Google Scholar]
- 164.Waigh TA. Applied Biophysics: A Molecular Approach for Physical Scientists. West Sussex, UK: Wiley; 2007. p. 421. [Google Scholar]
- 165.Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 166.Wang X, Tang OW, Riley EP, Rudner DZ. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol. 2014;24:287–92. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–74. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 168.Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLOS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Weber SC, Spakowitz AJ, Theriot JA. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys Rev Lett. 2010;104:238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc Natl Acad Sci USA. 2012;109:7338–43. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Weber SC, Theriot JA, Spakowitz AJ. Subdiffusive motion of a polymer composed of subdiffusive monomers. Phys Rev E. 2010;82:011913. doi: 10.1103/PhysRevE.82.011913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Welburn JP, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15:645–55. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 173.Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–85. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Westhorpe FG, Straight AF. Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol. 2013;25:334–40. doi: 10.1016/j.ceb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhu X, Kundukad B, van der Maarel JR. Viscoelasticity of entangled-phage DNA solutions. J Chem Phys. 2008;129:185103. doi: 10.1063/1.3009249. [DOI] [PubMed] [Google Scholar]
- 178.Zimm BH. Dynamics of polymer molecules in dilute solution: viscoelasticity, flow birefringence and dielectric loss. J Chem Phys. 1956;24:269–78. [Google Scholar]