Abstract
Irisin might play an important role in reducing the risk of obesity, insulin resistance, or several related diseases, and high irisin levels may contribute to successful aging. Thus, the irisin precursor (FNDC5) gene is a candidate to influence exceptional longevity (EL), i.e., being a centenarian. It has been recently shown that two single-nucleotide polymorphisms (SNPs) in the FNDC5 gene, rs16835198 and rs726344, are associated with in vivo insulin sensitivity in adults. We determined luciferase gene reporter activity in the two above-mentioned SNPs and studied genotype distributions among centenarians (n = 175, 144 women) and healthy controls (n = 347, 142 women) from Spain. We also studied an Italian [79 healthy centenarians (40 women) and 316 healthy controls (156 women)] and a Japanese cohort [742 centenarians (623 women) and 499 healthy controls (356 women)]. The rs726344 SNP had functional significance, as shown by differences in luciferase activity between the constructs of this SNP (all P ≤ 0.05), with the variant A-allele having higher luciferase activity compared with the G-allele (P = 0.04). For the rs16835198 SNP, the variant T-allele tended to show higher luciferase activity compared with the G-allele (P = 0.07). However, we found no differences between genotype/allele frequencies of the two SNPs in centenarians versus controls in any cohort, and no significant association (using logistic regression adjusted by sex) between the two SNPs and EL. Further research is needed with different cohorts as well as with additional variants in the FNDC5 gene or in other genes involved in irisin signaling.
Keywords: Myokines, Cardiometabolic, Disorders, Allele, Longevity, Centenarians
Introduction
Myokines are substances that are produced by skeletal muscles, especially induced by exercise, and modulate different metabolic processes locally or in other target tissues (Febbraio and Pedersen 2005; Duzova 2012). The peptidic myokine “irisin,” secreted in response to activation of the peroxisome proliferator-activated receptor gamma coactivator-1 α (PGC-1α), has been recently identified (Bostrom et al. 2012). The precursor of irisin is a type I transmembrane protein of skeletal muscle, the fibronectin type III domain-containing protein 5 (FNDC5) (Bostrom et al. 2012). Irisin acts in the white adipose tissue, promoting the acquisition of a brown adipocyte phenotype prone to energy expenditure (Polyzos et al. 2013). Accordingly, irisin might play an important role in reducing the risk of obesity (Spiegelman 2013), insulin resistance (Bostrom et al. 2012) [included in aged populations (Sanchis-Gomar et al. 2014b)], diabetes (Spiegelman 2013), or several related diseases (Sanchis-Gomar et al. 2012; Hojlund and Bostrom 2013; Sanchis-Gomar 2013; Sanchis-Gomar and Perez-Quilis 2014). It can also help to preserve vascular function and skeletal muscle mass (Spiegelman 2013; Bostrom and Fernandez-Real 2014). On the other hand, circulating irisin levels can predict telomere length in healthy adults (Rana et al. 2014). Great expectations have been created based on the above-mentioned findings (Elbelt et al. 2013; Villarroya 2013) with a new provocative concept, “irisinemia,” been proposed to monitor metabolic disorders such as diabetes, obesity, or the metabolic syndrome (Sanchis-Gomar and Perez-Quilis 2013). We recently hypothesized that high irisinemia may contribute to successful aging (Emanuele et al. 2014). Serum irisin levels were in fact higher in healthy centenarians compared to young healthy controls and, especially, to young adults with precocious myocardial infarction (Emanuele et al. 2014).
For the above-mentioned reasons, the irisin precursor (FNDC5) gene is a candidate to influence the risk of cardiometabolic diseases as well as the aging process. Staiger et al. recently showed that two single-nucleotide polymorphisms (SNPs) in the FNDC5 gene, rs16835198 and rs726344, were associated with in vivo insulin sensitivity (Staiger et al. 2013). In this regard, human exceptional longevity (EL), that is, being a centenarian, is associated with insulin sensitivity/signaling (Barbieri et al. 2008; Brunet 2012), e.g., with genetic variation in the insulin-human forkhead box O3A (FOXO3A) pathway influencing EL (Willcox et al. 2008; Flachsbart et al. 2009). Nonetheless, to the best of our knowledge, no previous research has studied the potential functional consequences of FNDC5 SNPs and their influence on EL. Centenarians are the paradigm of healthy aging, as they have postponed if not avoided major age-related diseases, including cardiometabolic diseases, as well as the onset of disability (Garatachea et al. 2014).
The search for the gene variants that might influence the likelihood of reaching EL is of medical interest, particularly to help identifying potential targets of highly specific “antiaging” drugs. To gain insight into their functionality, here we determined luciferase gene reporter activity in the two above-mentioned FNDC5 SNPs, rs16835198 and rs726344. We then compared allele/genotype frequencies of rs16835198 and rs726344 among centenarians (cases) and disease-free controls of the same ethnic and geographic origin (Spanish cohort) as well as in two other geographically and ethnically independent replication cohorts (from Italy and Japan).
Materials and methods
Functional analysis: luciferase reporter gene
The fragment, including the allele, was directly inserted into the pGL3-promoter at the restriction recognition sites MluI in the 5′ and XhoI in the 3′ (see below—in bold) of the sequences obtained from the genomes of the following:
One individual homozygous for the rs726344 C-allele and one homozygous for the rs726344 T-allele (see below—underlined)
One individual homozygous for the rs16835198 G-allele and one homozygous for the rs16835198 T-allele (see below—underlined)
rs726344-CC
ACGCGTTGTGAGGGCGGGGACAGTGTCTGGTTCACCTGGGAGCCCCGCCTGGCCTGGTAGCCGCTCGCAGGGCCTGTGGATGAGGCCGGGCAGCAATGCCACTGCTCTCCTCATATCACCCATTTCACCCTCACAGACACCCGTGAGGTGGAAGTTAATGTCCACAGCTTATAGGGAGCCAAACTGAGCCTCAGAAAGGCTGACTTGCTCGAG.
rs726344-TT
ACGCGTTGTGAGGGCGGGGACAGTGTCTGGTTCACCTGGGAGCCCCGCCTGGCCTGGTAGCCGCTCGCAGGGCCTGTGGATGAGGCCGGGCAGCAATGCCACTGCTTTCCTCATATCACCCATTTCACCCTCACAGACACCCGTGAGGTGGAAGTTAATGTCCACAGCTTATAGGGAGCCAAACTGAGCCTCAGAAAGGCTGACTTGCTCGAG.
rs16835198-GG
ACGCGTGGAGGATGCACCTTGGCTCCTATTGTAGGAAGTAGGTAGAGTCTGGGAATCTTCTAAATTCTGTTTTTGGCAGATAAGGACACCACGACCTAACCTCAGAGAATTAACTTGCCTGAGGTGTTAGGTGATTACAAGGCAGGGTCTCCTGTCTCTCAGACCAGCATTTCTCCCACAGGCCTGTGGTAGTCTCCAAGTGAGGTCCTCGAG.
rs16835198-TT
ACGCGTGGAGGATGCACCTTGGCTCCTATTGTAGGAAGTAGGTAGAGTCTGGGAATCTTCTAAATTCTGTTTTTGGCAGATAAGGACACCACGACCTAACCTCAGATAATTAACTTGCCTGAGGTGTTAGGTGATTACAAGGCAGGGTCTCCTGTCTCTCAGACCAGCATTTCTCCCACAGGCCTGTGGTAGTCTCCAAGTGAGGTCCTCGAG
Mice skeletal muscle C2C12 cell lines were used to represent muscle-specific expression. Cell cultures, transfections, and dual-luciferase reporter assays were performed as previously described (He et al. 2011). We used the pRL-SV40 vector as an internal control for variations in transfection efficiency. The pGL3-promoter vector without an insert was used as a negative control. The transfected cells were harvested after 48 h and assayed for firefly luciferase activity and renilla luciferase activity using the dual-luciferase reporter assay system (Promega Biotech, Beijing, China), as suggested by the manufacturer using a luminometer (TecanGenios Pro, Männedorf, Switzerland). From each measurement, relative luciferase activity was calculated by dividing the firefly luciferase activity reading by the renilla luciferase activity reading. Experiments were performed in triplicate. Relative luciferase activity values are expressed as the means ± SD of the three different experiments.
Participants
Written consent was obtained from each participant. The study protocol was approved by the corresponding institutional ethics committees [European University of Madrid (Spanish cohort), University of Pavia (Italian cohort), and National Institute of Health and Nutrition, Medical Research Institute and Keio University (Japanese cohort)] and was in accordance with the Declaration of Helsinki for Human Research of 1974 (last modified in 2008).
Spanish cohort
Two groups of Spanish subjects (most born and living in the central area of Spain, Meseta Castellana) were investigated: (i) 175 cases (centenarians, age range 100–111 years, 144 women, 31 men) and (ii) 347 healthy controls (20–50 years, 142 women, 205 men). All the Spanish participants were of the same Caucasian (Spanish) descent for ≥3 generations and all spoke Spanish as mother language. Centenarians were living mainly in nursing residencies of the Spanish central area, where they were recruited during years 2009–2012 after we had access to the ages’ lists of the people living in the residencies. The participants’ ages were ascertained by the dates of birth as stated on identity cards. This group included the oldest European individual (111 years) alive in June 2012 (http://www.grg.org/Adams/E.HTM), and ∼7 % of the cohort was aged ≥105 years. The most prevalent diseases were osteoarthritis (66 %), hypertension (57 %), dementia (51 %), and cardiovascular disease (29 %). Only two centenarians were free of any diagnosed disease.
The DNA of 347 younger controls was collected from saliva samples during 2008–20012 in the European University (Madrid, Spain). This was a convenient sample composed of students and staff from this institution; all of them were free of any major cardiometabolic disease (including cardiovascular disease (CVD) or Alzheimer) and had no known family history of high longevity (90+ years), as reported in a questionnaire.
Italian cohort
Two groups of Italian subjects born and living in Northern Italy were studied: (i) 79 cases (healthy centenarians, 100–104 years, 40 women, 39 men) and (ii) 316 healthy controls (27–81 years, 156 women, 160 men). The participants’ ages were defined by the dates of birth as stated on identity cards. All patients and controls were Caucasian whites ascertained to be of Italian descent. The criterion of “Italian descent” was met when all the parents and grandparents of an individual originated from Italy. The Italian centenarians were ascertained mainly via general practitioners in the community. These centenarians represent a convenience sample that has been previously described (Emanuele et al. 2010). The history of past and current diseases was accurately collected, checking the centenarians’ medical documentation and the current drug therapy. Accordingly, all the Italian centenarians were free of major age-related diseases, i.e., severe cognitive impairment, clinically evident cancer, CVD, renal insufficiency, or severe physical impairment (only, part of this group had decreased visual or auditory acuity). Thus, all of the Italian centenarians were in apparently good health relative to their very advanced age.
Controls were in apparent good physical health, with exclusion criteria being the following: presence of major CVD or cerebrovascular disease, cancer, dementia, chronic autoimmune or inflammatory disorders, renal or hepatic failure, and major psychiatric disorders.
Japanese cohort
Two groups of Japanese subjects [of the same Asian (Japanese) descent] were studied: (i) 742 cases (centenarians, 100–116 years, 623 women, 119 men) and (ii) 499 healthy controls (23–59 years, 356 women, 143 men). The group of cases was gathered from to prospective cohorts: the Tokyo Centenarians Study (TCS) and the Semi-Supercentenarians Study in Japan (SSC-J). A detailed description of population-based recruitments for the TCS has been previously reported (Gondo et al. 2006). The TCS cohort included 304 centenarians (65 men, 239 women) aged 100–108 years. The SSC-J is a nationwide longitudinal survey consisting mainly of individuals aged 105 years or older, which started in 2002 (with n = 135 SSC). After 2002, the recruitment strategy has relied on responses to local governments and nursing homes in the whole country, and direct inquires to our research team. Consequently, a total of 450 centenarians (58 men, 392 women) were enrolled in the SSC-J by the end of November 2011. The phenotype and disease characteristics of the Japanese centenarians are described elsewhere (Takayama et al. 2007), with a prevalence of hypertension, CVD, and dementia of 63.6, 28.8, and 59.4 %, respectively. Inclusion criteria for the control group, which was recruited during years 2008–2012 by advertising, were being man or woman aged <60 years, and free of diagnosed stroke, cardiac disease, and chronic renal failure (as reported in a questionnaire).
Genotype assessment
Spanish cohort
Genomic DNA was extracted from buccal cells according to standard phenol/chloroform procedures followed by alcohol precipitation. Allelic discrimination analysis was performed in the genetics laboratory of the European University of Madrid with predesigned Life Technologies TaqMan® SNP Genotyping Assays on demand for rs16835198 (ID C_34204885_10) and rs726344 (ID C_927694_10). PCR amplification was performed using a StepOne™ Plus Real-Time PCR System (Life Technologies, Foster City, CA) with a denaturation stage at 95 °C for 10 min, 50 cycles of denaturation at 92 °C for 15 s, annealing/extension at 60 °C for 1 min, and a final extension stage of 30 s at 60 °C. Positive and negative controls were used in each run, and a random 20 % of the samples were repeated (where all the results matched with the initial genotyping). Two investigators independently reviewed all the results.
Italian cohort
Genomic DNA was purified from peripheral blood samples using the QiaAmp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Genotyping was performed at the Cellular Pathophysiology and Clinical immunology Laboratory (University of Pavia using the TaqMan® rs726344 and rs16835198 SNP genotyping assays (Applied Biosystems, Foster City, CA, USA)). For quality control, genotyping analyses were done blind with respect to the age of the study participants, and a random 20 % of the samples were repeated. All genotyping repeated for quality control did not differ from the initial genotyping. Two investigators independently reviewed all results.
Japanese cohort
Total DNA was isolated from venous blood by use of QIAamp DNA Blood Maxi Kit (QIAGEN, Hilden, Germany). The rs16865198 was genotyped using TaqMan® SNP Genotyping Assays (Assay ID C__32747086_10) and StepOnePlusTM Real-Time PCR System (Applied Biosystems, Foster City, CA) in the genomics laboratory of the Tokyo Metropolitan Institute of Gerontology (Tokyo, Japan). A total of 5-μl genotyping mixture contained 2.5-μl GTXpressTM master mix, 0.125-μl assay mix (40×), and 1.375-μl distilled water to mix with 1-μl genomic DNA (10 ng/μl) in each reaction. One or two negative controls were included on each plate. TaqMan® assays for genotype calls were analyzed using StepOneTM Software v2.1 (Applied Biosystems).
The rs726344 polymorphism was not genotyped in the totality of the Japanese cohort because this polymorphism has not been detected in the Japanese population according to HapMap data and our preliminary analysis in 96 samples showed no presence of the variant A-allele.
Determination of serum irisin levels in the Italian cohort
Serum irisin levels were measured in Italian cases and controls in duplicate using a commercially available enzyme-linked immunosorbent assay (ELISA) (Aviscera Biosciences, Santa Clara, CA, USA) in accordance with the manufacturer’s instructions. The sensitivity of the assay was 0.2 ng/ml, the linear range of the standard was 5 to 500 ng/ml, and the intra- and inter-assay coefficients of variation were 4.2 and 6.8 %, respectively. The results of the cases’ group have been recently published by our group (Emanuele et al. 2014).
Statistical methods
Allele frequencies of rs726344 and rs16835198 were calculated from genotypes using the gene-counting method. We tested Hardy–Weinberg equilibrium (HWE) and compared the genotype/allele frequencies of cases versus controls within each cohort (Spanish, Italian, and Japanese) using the χ2 test with α set at 0.05. We used logistic regression analysis to analyze the association between genotypes/alleles and EL under the five genetic models (codominant, dominant, recessive, overdominant, and log-additive) within each of the three cohorts after adjustment for sex. In the Italian cohort, we compared mean serum irisin levels among the three genotypes of each SNP using a one-way ANOVA test.
All statistical analyses were performed using the PASW (v. 18.0 for WINDOWS, Chicago), except for statistical power, which we calculated with the StatMate software, version 2.0 (GraphPad, San Diego, CA, USA) and analysis of haplotypes, which was performed using the free Web-based application SNPStats (Sole et al. 2006). Haplotype frequencies were estimated using the implementations of the EM algorithm coded into the haplo.stats package.
Results
Functional analysis
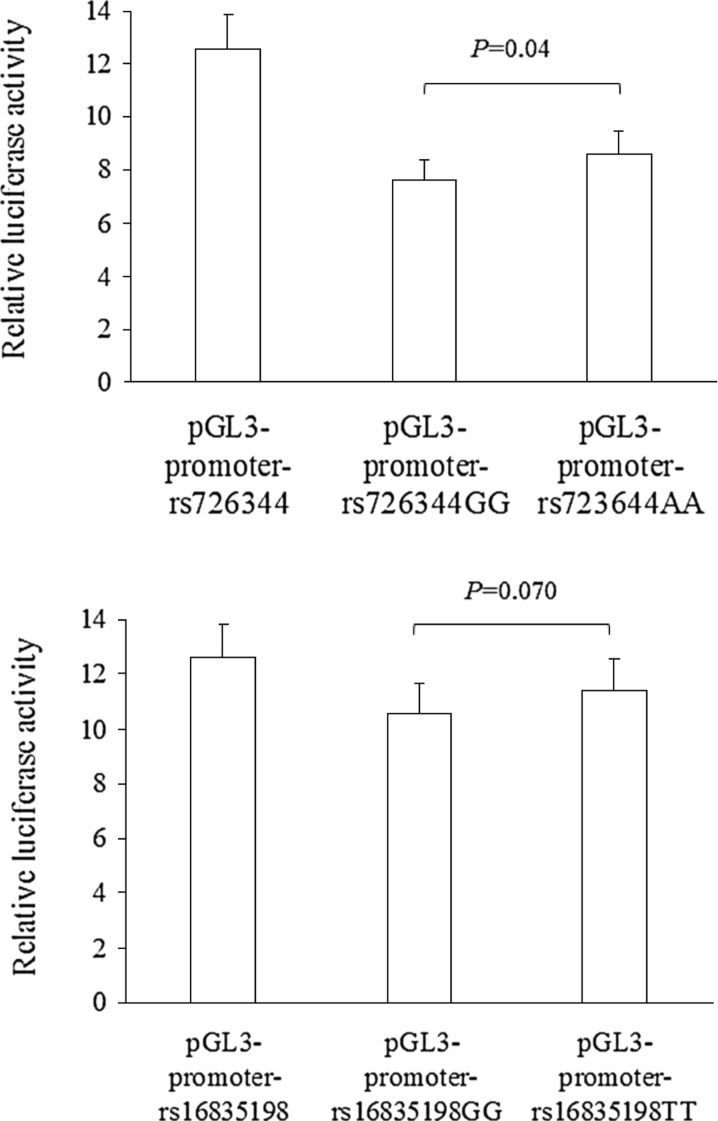
The results of luciferase report analyses are presented in Fig. 1. The rs726344 SNP had functional significance in vitro, as shown by differences in luciferase activity between the constructs of this SNP (all P ≤ 0.05), with the variant A-allele having higher luciferase activity compared with the G-allele (P = 0.04). Statistical significance was almost reached for the rs16835198 SNP (the variant T-allele tended to have higher luciferase activity compared with the G-allele, P = 0.07).
Fig. 1.
Comparison of relative luciferase activity (i.e., firefly luciferase activity divided by renilla luciferase activity) between plasmids for rs726344 (upper figure) and rs16835198 (lower figure). Values are mean ± SD of three different experiments, each performed in triplicate. Upper figure significant differences were found within the rs726344 variant for all comparisons between plasmids (all P < 0.01). Lower figure statistical significance was reached for all the comparisons between plasmids (all P < 0.01), except but for GG vs TT (P = 0.070)
Spanish cohort
Failure rate of genotyping was 21.71 % in cases for both studied genotypes and 0.57 and 0.86 % in controls for rs726344 and rs16835198, respectively. The distribution of rs726344 and rs16835198 genotypes was consistent with the HWE in both groups (P > 0.05). The results of genotype/allele frequency distributions as well as of binary logistic regression are shown in Table 1 and summarized below. The allele frequency distribution of the 2 SNPs did not differ between cases and controls (rs726344 χ2 = 0.671, P = 0.41; rs16835198 χ2 = 1.671, P = 0.20). Likewise, genotype frequencies did not differ between centenarians and controls (rs726344 χ2 = 2.821, P = 0.244; rs16835198 χ2 = 1.540, P = 0.463). Using logistic regression analysis, no significant associations were found between the two SNPs and EL after adjusting for sex. No significant associations were found either when analyzing both sexes separately (data not shown). When the two SNPs were considered together, four haplotypes were estimated, i.e., Grs726344-Grs16835198 (55.1 %), Grs726344-Trs16835198 (34.1 %), Ars726344-Grs16835198 (10.8 %), and Ars726344-Trs16835198 (<1.0 %). We found no association between haplotypes and EL (P > 0.1).
Table 1.
Genotype/allele frequencies of rs726344 and rs16835198 and binary logistic regression adjusted by sex in the Spanish cohort
| rs726344 | rs16835198 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | |||||||||||||
| N | % | N | % | OR | 95%CI | P | N | % | N | % | OR | 95%CI | P | |||
| Codom | GG | 274 | 79.4 | 111 | 81.0 | 1.00 | – | GG | 163 | 47.4 | 58 | 42.3 | 1.00 | 0.48 | ||
| GA | 64 | 19.0 | 26 | 19.0 | 1.19 | 0.69–2.06 | GT | 136 | 39.5 | 56 | 40.9 | 0.81 | 0.51–1.29 | |||
| AA | 7 | 2.0 | 0 | 0.0 | – | – | TT | 45 | 13.1 | 23 | 16.8 | 0.71 | 0.38–1.33 | |||
| Dom | GG | 274 | 79.2 | 111 | 81.0 | 1.00 | 0.23 | GG | 163 | 47.4 | 58 | 42.3 | 1.00 | 0.25 | ||
| GA-AA | 71 | 20.6 | 26 | 19.0 | 1.38 | 0.81–2.36 | GT + TT | 181 | 52.6 | 79 | 57.7 | 0.78 | 0.51–1.20 | |||
| Reces | GG-GA | 338 | 98.0 | 137 | 100.0 | 1.00 | – | GG + GT | 299 | 86.9 | 114 | 83.2 | 1.00 | 0.42 | ||
| AA | 7 | 2.0 | 0 | 0 | – | – | TT | 45 | 13.1 | 23 | 16.8 | 0.78 | 0.43–1.41 | |||
| Overdom | GG-AA | 281 | 81.5 | 111 | 81.0 | 1.00 | 0.63 | GG-TT | 208 | 60.5 | 81 | 59.1 | 1.00 | 0.56 | ||
| GA | 64 | 18.6 | 26 | 19.0 | 1.14 | 0.66–1.96 | GT | 136 | 39.5 | 56 | 40.9 | 0.88 | 0.57–1.36 | |||
| Log-additive | – | – | – | – | – | 1.52 | 0.94–2.48 | 0.08 | – | – | – | – | 0.83 | 0.62–1.12 | 0.23 | |
| Allele | G | 612 | 88.7 | 248 | 90.5 | 1.00 | G | 462 | 67.2 | 172 | 62.8 | 1.00 | ||||
| A | 78 | 11.3 | 26 | 9.5 | 0.65 | 0.39–1.06 | 0.84 | T | 226 | 32.8 | 102 | 37.2 | 0.81 | 0.59–1.12 | 0.20 | |
Codom codominant, Dom dominant, OR odds ratio, Overdom overdominant, Reces recessive;95 % CI 95 % confidence interval
Based on the observed prevalence of the G-allele of the rs726344 and rs16835198 SNPs in the control group, the Spanish cohort’s sample size had an 80 % power to detect a relative likelihood of 1.07 and 1.20, respectively, for being a centenarian between G-allele carriers and noncarriers with a significance level (α) of 0.05 (two-tailed).
Italian cohort
Rate failure of genotyping was 0 %. The distribution of rs726344 and rs16835198 genotypes was consistent with the HWE in both groups (P > 0.05). The results of genotype/allele frequency distributions as well as data of binary logistic regression are shown in Table 2 and summarized below. Allele (rs726344 χ2 = 0.096, P = 0.756; rs16835198 χ2 = 0.810, P = 0.368) and genotype frequencies did not differ between groups (rs726344 χ2 = 0.122, P = 0.941; rs16835198 χ2 = 1.128, P = 0.569), and no significant association was found between the two SNPS and EL using logistic regression adjusted by sex. No significant associations were found either when analyzing both sexes separately (data not shown). When the two SNPs were considered together, four haplotypes were estimated, i.e., Grs726344-Grs16835198 (44.3 %), Grs726344-Trs16835198 (36.7 %), Ars726344-Grs16835198 (18.2 %), and Ars726344-Trs16835198 (<1.0 %). No association between haplotypes and EL was observed (P > 0.1).
Table 2.
Genotype/allele frequenciesof rs726344 and rs16835198 and binary logistic regression adjusted by sex in the Italian cohort
| rs726344 | Irs16835198 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | |||||||||||||
| N | % | N | % | OR | 95%CI | P | N | % | N | % | OR | 95%CI | P | |||
| Codom | GG | 234 | 74.1 | 57 | 72.1 | 1.00 | 0.941 | GG | 104 | 32.9 | 31 | 39.2 | 1.00 | 0.569 | ||
| GA | 78 | 24.7 | 21 | 26.6 | 0.88 | 0.50–1.55 | GT | 151 | 47.8 | 34 | 43.0 | 1.31 | 0.77–2.24 | |||
| AA | 4 | 1.2 | 1 | 1.3 | 0.98 | 0.12–8.47 | TT | 61 | 19.3 | 14 | 17.8 | 1.27 | 0.66–2.64 | |||
| Dom | GG | 234 | 74.1 | 57 | 72.2 | 1.00 | 0.732 | GG | 104 | 32.9 | 31 | 39.2 | 1.00 | 0.288 | ||
| GA-AA | 82 | 25.9 | 22 | 27.8 | 0.86 | 0.54–1.60 | GT + TT | 212 | 67.1 | 48 | 60.8 | 1.33 | 0.81–2.20 | |||
| Reces | GG-GA | 312 | 98.7 | 78 | 98.7 | 1.00 | 1 | GG + GT | 255 | 80.7 | 65 | 82.3 | 1.00 | 0.748 | ||
| AA | 4 | 1.3 | 1 | 1.3 | 1.01 | 0.13–9.11 | TT | 61 | 19.3 | 14 | 17.7 | 1.12 | 0.61–2.11 | |||
| Overdom | GG-AA | 238 | 74.3 | 58 | 73.4 | 1.00 | 0.727 | GG + TT | 165 | 52.2 | 45 | 57.0 | 1.00 | 0.449 | ||
| GA | 78 | 24.7 | 21 | 26.6 | 0.91 | 0.49–1.57 | GT | 151 | 47.8 | 34 | 43.0 | 1.22 | 0.71–2.01 | |||
| Log-additive | – | – | – | – | – | 0.84 | 0.61–1.78 | 0.814 | – | – | – | – | 1.16 | 0.87–1.98 | 0.378 | |
| Allele | G | 546 | 86.4 | 135 | 85.4 | 1.00 | G | 359 | 56.8 | 96 | 60.8 | 1.00 | ||||
| A | 86 | 13.6 | 23 | 14.6 | 0.89 | 0.50–1.59 | 0.756 | T | 273 | 43.3 | 62 | 39.2 | 1.15 | 0.83–1.71 | 0.368 | |
Codom codominant, Dom dominant, OR odd ratio, Overdom overdominant, Reces recessive, 95 % CI 95 % confidence interval
Based on the observed prevalence of the G-allele of the rs726344 and rs16835198 SNPs in the control group, the Italian cohort’s sample size had an 80 % power to detect a relative likelihood of 1.20 and 1.31, respectively, for being a centenarian between G-allele carriers and noncarriers (two-tailed α = 0.05).
We failed to identify a significant association between serum irisin levels and rs726344 and rs16835198 SNPs. Indeed, in the entire Italian cohort, the distribution of serum irisin levels according to the rs726344 genotypes was as follows (mean ± SD): GG, 22.4 ± 6.4 ng/ml; GA, 20.9 ± 7.1 ng/ml; and AA, 24.7 ± 8.2 ng/ml (P = 0.67 for the genotype effect). The concentrations of serum irisin for rs16835198 genotypes were as follows: GG, 20.1 ± 5.7 ng/ml; GT, 23.3 ± 7.4 ng/ml; and TT, 22.9 ± 9.1 ng/ml (P = 0.81). Similarly, no statistical differences were found between genotypes when analyzing cases and controls separately (data not shown).
Japanese cohort
Rate failure of genotyping was 1.21 % in cases and 0.02 % in controls. The distribution of rs16835198 genotypes was consistent with the HWE in both groups (P > 0.05). The results of rs16835198 genotype/allele frequency distributions as well as data of binary logistic regression are shown in Table 3 and summarized below. Allele (χ2 = 1.001, P < 0.317) and genotype frequencies did not different between groups (χ2 = 5.337, P < 0.069), and no significant association was found between rs16835198 and EL using logistic regression adjusted by sex. No significant associations were found when analyzing both sexes separately (data not shown). Haplotype analysis of the two SNPs could not be performed owing to the absence of the variant A-allele of rs726344 in this cohort.
Table 3.
Genotype/allele frequencies of rs16835198 and binary logistic regression adjusted by sex in the Japanese cohort
| Controls | Cases | |||||||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | OR | 95%CI | P | ||
| Codom | TT | 148 | 29.7 | 200 | 27.3 | 1.00 | 0.58 | |
| GT | 239 | 47.9 | 356 | 48.6 | 1.10 | 0.84–1.44 | ||
| GG | 111 | 22.4 | 177 | 24.1 | 1.18 | 0.86–1.62 | 0.35 | |
| Dom | TT | 148 | 29.7 | 200 | 27.3 | 1.00 | ||
| GT-GG | 350 | 70.3 | 533 | 72.7 | 1.13 | 0.88–1.45 | 0.45 | |
| Reces | TT-GT | 387 | 77.6 | 556 | 75.8 | 1.00 | ||
| GG | 111 | 22.3 | 177 | 24.1 | 1.11 | 0.85–1.45 | ||
| Overdom | TT-GG | 259 | 52.1 | 377 | 51.4 | 1.00 | 0.84 | |
| GT | 239 | 47.9 | 356 | 48.6 | 1.02 | 0.93–1.27 | ||
| Log-additive | --- | --- | --- | --- | --- | 1.09 | 0.93–1.27 | 0.30 |
| Allele | T | 535 | 54 | 756 | 52 | 1.00 | 0.29 | |
| G | 461 | 46 | 710 | 48 | 1.09 | 0.93–1.28 | ||
Codom codominant, Dom dominant, OR odd ratio, Overdom overdominant, Reces recessive, 95 % CI 95 % confidence interval
Based on the observed prevalence of the G-allele of the rs16835198 SNP in the control group, the Japanese cohort’s sample size had an 80 % power to detect a relative likelihood of 1.19 for being a centenarian between G-allele carriers and noncarriers (two-tailed α = 0.05).
Discussion
The main findings of our study are two-fold. First, the rs726344 SNP had functional significance in vitro, with the variant A-allele upregulating luciferase activity compared to the common G-allele. On the other hand, we found no major association between FNDC5 rs16835198 or rs726344 and EL. Regarding the results of the functionality analyses (luciferase constructs), to the best of our knowledge, this is the first attempt to gain insights (using in vitro analyses) into the potential functional consequences of these two FNDC5 variants.
The rs16835198 and rs726344 SNPs are not likely to influence the amino acid sequence of the protein product owing to the fact that the rs16835198 and rs726344 SNPs are located in the 3′-flanking region and in intron 5, respectively, of the FNDC5 gene. Alterations in DNA sequence in the 3′-UTR region of a gene (e.g., rs16835198) have the potential to alter the level, location, or timing of gene expression whereas intronic genomic variants (e.g., rs726344) can influence gene expression and thus phenotype, by altering mRNA stability, alternative mRNA splicing, or the binding of transcription factors (Tabor et al. 2002; Knight 2005; Mercado et al. 2005; Sasabe et al. 2007). However, Staiger et al. (2013) found no association between the A-allele of the rs726344 SNP and FNDC5 mRNA expression in human myotubes (P = 0.19). Although the minor A-allele of rs726344 is characterized by higher luciferase activity compared with the wild-type G-allele and has been recently associated with decreased insulin sensitivity in vivo (Staiger et al. 2013), our results indicated that this SNP was neither associated with EL nor with serum irisin levels. We can therefore hypothesize that the previously observed association between serum irisin levels and EL (Emanuele et al. 2014) is not mediated by this specific SNP. This result is only partly surprising because it is known that serum irisin concentrations are regulated by a number of factors (including food intake and exercise) (Lopez-Legarrea et al. 2014) that are not necessarily limited to genetic variants in the irisin gene. On the other hand, although the association between the A-allele of rs726344 and increased luciferase activity shown here in vitro is counterintuitive given the insulin-desensitizing effect of this allele in middle-aged adults at increased risk for type II diabetes recently shown by Staiger and coworkers (Staiger et al. 2013), here we did not measure insulin resistance. Thus, we cannot exclude that the previously demonstrated insulin-desensitizing effect of the A-allele of rs726344 could be either modified or abolished by other factors, including age, drug use, or comorbidities.
There is a strong rationale in postulating that the gene, FNDC5, encoding the precursor of irisin might be associated, at least partly, with healthy aging (Bostrom et al. 2012), based on the beneficial multisystemic effects of this myokine, i.e., browning of white adipocytes (Polyzos et al. 2013), reduced risk of obesity (Spiegelman 2013) and related diseases (Sanchis-Gomar et al. 2012; Hojlund and Bostrom 2013; Sanchis-Gomar 2013), potential preservation of vascular function and skeletal muscle mass ((Huh et al. 2012; Spiegelman 2013; Bostrom and Fernandez-Real 2014), higher aerobic fitness in cardiac patients (Lecker et al. 2012), improved neurogenesis in animal models (Hashemi et al. 2013), and also based on the association between irisin levels and higher telomere length in healthy adults (Rana et al. 2014). On the other hand, inhibition of another myokine, myostatin, can have irisin-like effects, e.g., “browning” of the white adipose tissue (though AMPK-PGC-1α-irisin pathway) and amelioration of muscle weakness (see Fiuza-Luces et al. 2013 for a review). Interestingly, the K153R polymorphism in the gene (MSTN) encoding myostatin could be associated with EL, as shown by previous research from our group (Garatachea et al. 2013). There is some controversy among the numerous studies that have assessed circulating blood levels of irisin in people with obesity, diabetes, and CVD (Huh et al. 2012; Choi et al. 2013; Liu et al. 2013; Moreno-Navarrete et al. 2013; Park et al. 2013; Stengel et al. 2013; Vamvini et al. 2013; Sanchis-Gomar et al. 2014a; Sanchis-Gomar et al. 2014b). Some authors have expressed their concerns about the notion that irisin is a real myokine (i.e., being released by contracting muscles) or that this molecule has actual beneficial effects on health (Timmons et al. 2012; Erickson 2013; Pekkala et al. 2013; Raschke et al. 2013). And yet, the bulk of evidence seems to indicate that irisin plays an overall protective role against cardiometabolic disorders (Sanchis-Gomar et al. 2012; Eckardt et al. 2014) and can be upregulated by muscle exercise (Bostrom et al. 2013). However, despite the overall metabolic-protective effect of irisin, no evidence is available on the potential benefits of irisin to prevent sarcopenia, with recent data showing no differences in irisin levels between subjects with sarcopenia and healthy controls, and also indicating no association between this molecule and skeletal muscle mass index (Choi et al. 2014). Arguably, the strongest support for a potential involvement of irisin on EL comes from a recent study from our group showing that irisin levels were ∼70 and ∼133 % higher in the present Italian cohort of healthy centenarians compared with younger controls who were healthy or had cardiovascular disease, respectively (Emanuele et al. 2014).
Studies on genetic polymorphisms in the irisin (FNDC5) gene are scarce and nonexistent in aged population. In this regard, EL is associated with insulin sensitivity and insulin signaling (Willcox et al. 2006; Barbieri et al. 2008; Guevara-Aguirre et al. 2011; Brunet 2012). Particularly, genetic variation in the insulin-FOXO3A gene pathway influences EL (Willcox et al. 2008; Flachsbart et al. 2009). Long-lived men exhibit several biological markers indicative of greater insulin sensitivity, which is associated with the FOXO3A GG genotype (Willcox et al. 2008). Also, a favorable glucose metabolism is associated with familial longevity (Rozing et al. 2010). Here, we failed to detect any significant association between the two FNDC5 SNPs we studied and EL, suggesting that other factors (e.g., maybe physical activity levels throughout lifespan, nutritional habits or complex gene-environment associations that are yet to be determined) might have a stronger influence on irisinemia, despite the potential functional consequence of at least one of the two SNPs we studied (i.e., rs726344). In fact, we found no FNDC5 genotype effect on the irisin levels of the Italian cohort, which is in line with recent data on the rs16835198 SNP and serum irisin in Japanese men aged 21–79 years) (Tanisawa et al. 2014). On the other hand, the role of epigenetic factors in irisinemia remains to be determined. Indeed, epigenetic modifications might also influence longevity by affecting gene expression without changing the DNA sequence (Wolffe and Matzke 1999), i.e., through DNA methylation, histone modifications, and altered expression of RNAs or small, noncoding RNAs that regulate gene expression profiles associated with longevity (Mango 2011).
Besides the rationale for postulating the FNDC5 gene as a candidate to influence EL and the clear definition we used for the criterion of EL (with all cases being centenarians), a further strength from our study derives from the luciferase construct study we performed as well as from the fact that the analyses were conducted in three independent cohorts, with one of them, the Italian cohort, being composed of apparently healthy centenarians and another one, the Japanese cohort, including a very large sample of centenarians. On the other hand, there are some limitations in our study, such as the use of convenience samples, which increases the risk of bias due to population stratification or the relatively high rate of genotyping failure in the Spanish centenarians. The latter suggests that DNA extraction from saliva in frail individuals (which we were forced to adopt due to ethical reasons) might result in small amounts of DNA available for analyses. Interpretation of gene association studies with centenarians and younger controls as the one we conducted here may be also biased by differences in date of birth (i.e., early 1900s vs 1930 and onward, respectively).
In summary, we found no major association between FNDC5 rs16835198/rs726344 SNPs and EL despite the fact that the latter variation might have functional consequences and despite the rationale for postulating that irisin is a myokine with the potential to promote longevity owing to its overall cardiovascular and metabolic protective effect. Further research is needed with different cohorts as well as with different variants in the FNDC5 gene or in other genes involved in irisin signaling.
Acknowledgments
Acknowledgments
This study was funded by the Fondo de Investigaciones Sanitarias (FIS, ref. # PI12/00914).
Conflicts of interests
The authors declare no conflict of interest.
Footnotes
Fabian Sanchis-Gomar and Nuria Garatachea contributed equally to this paper
References
- Barbieri M, Gambardella A, Paolisso G, Varricchio M. Metabolic aspects of the extreme longevity. Exp Gerontol. 2008;43:74–78. doi: 10.1016/j.exger.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, Kajimura S, Zingaretti MC, Vind BF, Tu H, Cinti S, Hojlund K, Gygi SP, Spiegelman BM. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom PA, Fernandez-Real JM. Metabolism: Irisin, the metabolic syndrome and follistatin in humans. Nat Rev Endocrinol. 2014;10:11–12. doi: 10.1038/nrendo.2013.230. [DOI] [PubMed] [Google Scholar]
- Bostrom PA, Graham EL, Georgiadi A, Ma X. Impact of exercise on muscle and nonmuscle organs. IUBMB Life. 2013;65:845–850. doi: 10.1002/iub.1209. [DOI] [PubMed] [Google Scholar]
- Brunet A. Aging and the control of the insulin-FOXO signaling pathway. Med Sci (Paris) 2012;28:316–320. doi: 10.1051/medsci/2012283021. [DOI] [PubMed] [Google Scholar]
- Choi HY, Kim S, Park JW, Lee NS, Hwang SY, Huh JY, Hong HC, Yoo HJ, Baik SH, Youn BS, Mantzoros CS, Choi KM (2014). Implication of Circulating Irisin Levels with Brown Adipose Tissue and Sarcopenia in Humans. J Clin Endocrinol Metab, jc20141195. [DOI] [PubMed]
- Choi YK, Kim MK, Bae KH, Seo HA, Jeong JY, Lee WK, Kim JG, Lee IK, Park KG. Serum irisin levels in new-onset type 2 diabetes. Diabetes Res Clin Pract. 2013;100:96–101. doi: 10.1016/j.diabres.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Duzova H. Skeletal Muscle. Myokines and Health Med Sci. 2012;1:211–231. [Google Scholar]
- Eckardt K, Gorgens SW, Raschke S, Eckel J (2014). Myokines in insulin resistance and type 2 diabetes. Diabetologia. [DOI] [PubMed]
- Elbelt U, Hofmann T, Stengel A. Irisin: what promise does it hold? Curr Opin Clin Nutr Metab Care. 2013;16:541–547. doi: 10.1097/MCO.0b013e328363bc65. [DOI] [PubMed] [Google Scholar]
- Emanuele E, Fontana JM, Minoretti P, Geroldi D. Preliminary evidence of a genetic association between chromosome 9p21.3 and human longevity. Rejuvenation Res. 2010;13:23–26. doi: 10.1089/rej.2009.0970. [DOI] [PubMed] [Google Scholar]
- Emanuele E, Minoretti P, Pareja-Galeano H, Sanchis-Gomar F, Garatachea N, Lucia A. Serum irisin levels, precocious myocardial infarction, and healthy exceptional longevity. Am: J Med; 2014. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Irisin and FNDC5 in retrospect: An exercise hormone or a transmembrane receptor? Adipocyte. 2013;2:289–293. doi: 10.4161/adip.26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febbraio MA, Pedersen BK. Contraction-induced myokine production and release: is skeletal muscle an endocrine organ? Exerc Sport Sci Rev. 2005;33:114–119. doi: 10.1097/00003677-200507000-00003. [DOI] [PubMed] [Google Scholar]
- Fiuza-Luces C, Garatachea N, Berger NA, Lucia A. Exercise is the real polypill. Physiology (Bethesda) 2013;28:330–358. doi: 10.1152/physiol.00019.2013. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garatachea N, Fuku N, He ZH, Tian Y, Arai Y, Abe Y, Murakami H, Miyachi M, Yvert T, Venturini L, Santiago C, Santos-Lozano A, Rodriguez-Romo G, Ricevuti G, Pareja-Galiano H, Sanchis-Gomar F, Emanuele E, Hirose N, Lucia A (2014). PTK2 rs7460 and rs7843014 polymorphisms and exceptional longevity: a functional replication study. Rejuvenation Res [DOI] [PMC free article] [PubMed]
- Garatachea N, Pinos T, Camara Y, Rodriguez-Romo G, Emanuele E, Ricevuti G, Venturini L, Santos-Lozano A, Santiago-Dorrego C, Fiuza-Luces C, Yvert T, Andreu AL, Lucia A. Association of the K153R polymorphism in the myostatin gene and extreme longevity. Age (Dordr) 2013;35:2445–2454. doi: 10.1007/s11357-013-9513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondo Y, Hirose N, Arai Y, Inagaki H, Masui Y, Yamamura K, Shimizu K, Takayama M, Ebihara Y, Nakazawa S, Kitagawa K. Functional status of centenarians in Tokyo Japan: developing better phenotypes of exceptional longevity. J Gerontol A Biol Sci Med Sci. 2006;61:305–310. doi: 10.1093/gerona/61.3.305. [DOI] [PubMed] [Google Scholar]
- Guevara-Aguirre J, Balasubramanian P, Guevara-Aguirre M, Wei M, Madia F, Cheng CW, Hwang D, Martin-Montalvo A, Saavedra J, Ingles S, de Cabo R, Cohen P, Longo VD (2011). Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med. 3, 70ra13. [DOI] [PMC free article] [PubMed]
- Hashemi MS, Ghaedi K, Salamian A, Karbalaie K, Emadi-Baygi M, Tanhaei S, Nasr-Esfahani MH, Baharvand H. Fndc5 knockdown significantly decreased neural differentiation rate of mouse embryonic stem cells. Neuroscience. 2013;231:296–304. doi: 10.1016/j.neuroscience.2012.11.041. [DOI] [PubMed] [Google Scholar]
- He ZH, Hu Y, Li YC, Yvert T, Santiago C, Gomez-Gallego F, Ruiz JR, Lucia A. Are calcineurin genes associated with athletic status? A function, replication study. Med Sci Sports Exerc. 2011;43:1433–1440. doi: 10.1249/MSS.0b013e31820e7f38. [DOI] [PubMed] [Google Scholar]
- Hojlund K, Bostrom P. Irisin in obesity and type 2 diabetes. J Diabetes Complicate. 2013;27:303–304. doi: 10.1016/j.jdiacomp.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Huh JY, Panagiotou G, Mougios V, Brinkoetter M, Vamvini MT, Schneider BE, Mantzoros CS. FNDC5 and irisin in humans: I Predictors of circulating concentrations in serum and plasma and II mRNA expression and circulating concentrations in response to weight loss and exercise. Metabolism. 2012;61:1725–1738. doi: 10.1016/j.metabol.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med (Berl) 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Zavin A, Cao P, Arena R, Allsup K, Daniels KM, Joseph J, Schulze PC, Forman DE. Expression of the irisin precursor FNDC5 in skeletal muscle correlates with aerobic exercise performance in patients with heart failure. Circ Heart Fail. 2012;5:812–818. doi: 10.1161/CIRCHEARTFAILURE.112.969543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Wong MD, Toy WC, Tan CS, Liu S, Ng XW, Tavintharan S, Sum CF, Lim SC. Lower circulating irisin is associated with type 2 diabetes mellitus. J Diabetes Complica. 2013;27:365–369. doi: 10.1016/j.jdiacomp.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Lopez-Legarrea P, de la Iglesia R, Crujeiras AB, Pardo M, Casanueva FF, Zulet MA, Martinez JA. Higher baseline irisin concentrations are associated with greater reductions in glycemia and insulinemia after weight loss in obese subjects. Nutr Diabetes. 2014;4:e110. doi: 10.1038/nutd.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE. Ageing: generations of longevity. Nature. 2011;479:302–303. doi: 10.1038/479302a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Navarrete JM, Ortega F, Serrano M, Guerra E, Pardo G, Tinahones F, Ricart W, Manuel-Fernandez-Real J. Irisin Is Expressed and Produced by Human Muscle and Adipose Tissue in Association With Obesity and Insulin Resistance. J Clin Endocrinol Metab. 2013;98:E769–778. doi: 10.1210/jc.2012-2749. [DOI] [PubMed] [Google Scholar]
- Park KH, Zaichenko L, Brinkoetter M, Thakkar B, Sahin-Efe A, Joung KE, Tsoukas M, Geladari EV, Young Huh J, Dincer F, Davis CR, Crowell JA, Mantzoros CS (2013). Circulating Irisin in Relation to Insulin Resistance and the Metabolic Syndrome. J Clin Endocrinol Metab. [DOI] [PMC free article] [PubMed]
- Pekkala S, Wiklund P, Hulmi JJ, Ahtiainen JP, Horttanainen M, Pollanen E, Makela KA, Kainulainen H, Hakkinen K, Nyman K, Alen M, Herzig KH, Cheng S. Are Skeletal Muscle FNDC5 Gene Expression and Irisin Release Regulated by Exercise and Related to Health? J Physiol. 2013;591:5393–5400. doi: 10.1113/jphysiol.2013.263707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyzos SA, Kountouras J, Shields K, Mantzoros CS (2013). Irisin: A renaissance in metabolism? Metabolism. [DOI] [PubMed]
- Rana KS, Arif M, Hill EJ, Aldred S, Nagel DA, Nevill A, Randeva HS, Bailey CJ, Bellary S, Brown JE. Plasma irisin levels predict telomere length in healthy adults. Age (Dordr) 2014;36:995–1001. doi: 10.1007/s11357-014-9620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke S, Elsen M, Gassenhuber H, Sommerfeld M, Schwahn U, Brockmann B, Jung R, Wisloff U, Tjonna AE, Raastad T, Hallen J, Norheim F, Drevon CA, Romacho T, Eckardt K, Eckel J. Evidence against a Beneficial Effect of Irisin in Humans. PLoS One. 2013;8:e73680. doi: 10.1371/journal.pone.0073680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozing MP, Westendorp RG, de Craen AJ, Frolich M, de Goeij MC, Heijmans BT, Beekman M, Wijsman CA, Mooijaart SP, Blauw GJ, Slagboom PE, van Heemst D. Favorable glucose tolerance and lower prevalence of metabolic syndrome in offspring without diabetes mellitus of nonagenarian siblings: the Leiden longevity study. J Am Geriatr Soc. 2010;58:564–569. doi: 10.1111/j.1532-5415.2010.02725.x. [DOI] [PubMed] [Google Scholar]
- Sanchis-Gomar F. The skeletal muscle-metabolism axis in prostate-cancer therapy. N Engl J Med. 2013;367:2257–2258. doi: 10.1056/NEJMc1211955. [DOI] [PubMed] [Google Scholar]
- Sanchis-Gomar F, Alis R, Pareja-Galeano H, Romagnoli M, Perez-Quilis C. Inconsistency in circulating Irisin levels: What is really happening? Horm Metab Res. 2014;46:1–6. doi: 10.1055/s-0033-1363283. [DOI] [PubMed] [Google Scholar]
- Sanchis-Gomar F, Alis R, Pareja-Galeano H, Sola E, Victor VM, Rocha M, Hernandez-Mijares A, Romagnoli M (2014b). Circulating irisin levels are not correlated with BMI, age, and other biological parameters in obese and diabetic patients. Endocrine. [DOI] [PubMed]
- Sanchis-Gomar F, Lippi G, Mayero S, Perez-Quilis C, Garcia-Gimenez JL. Irisin: a new potential hormonal target for the treatment of obesity and type 2 diabetes. J Diabetes. 2012;4:196. doi: 10.1111/j.1753-0407.2012.00194.x. [DOI] [PubMed] [Google Scholar]
- Sanchis-Gomar F, Perez-Quilis C. Irisinemia: A Novel Concept to Coin in Clinical Medicine? Ann Nutr Metab. 2013;63:60–61. doi: 10.1159/000354090. [DOI] [PubMed] [Google Scholar]
- Sanchis-Gomar F, Perez-Quilis C. The p38-PGC-1alpha-irisin-betatrophin axis: Exploring new pathways in insulin resistance. Adipocyte. 2014;3:67–68. doi: 10.4161/adip.27370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe T, Furukawa A, Matsusita S, Higuchi S, Ishiura S. Association analysis of the dopamine receptor D2 (DRD2) SNP rs1076560 in alcoholic patients. Neurosci Lett. 2007;412:139–142. doi: 10.1016/j.neulet.2006.10.064. [DOI] [PubMed] [Google Scholar]
- Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM. Banting lecture 2012: regulation of adipogenesis: toward new therapeutics for metabolic disease. Diabetes. 2013;62:1774–1782. doi: 10.2337/db12-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger H, Bohm A, Scheler M, Berti L, Machann J, Schick F, Machicao F, Fritsche A, Stefan N, Weigert C, Krook A, Haring HU, de Angelis MH. Common genetic variation in the human FNDC5 locus, encoding the novel muscle-derived ‘browning’ factor irisin, determines insulin sensitivity. PLoS One. 2013;8:e61903. doi: 10.1371/journal.pone.0061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A, Hofmann T, Goebel-Stengel M, Elbelt U, Kobelt P, Klapp BF. Circulating levels of irisin in patients with anorexia nervosa and different stages of obesity - Correlation with body mass index. Peptides. 2013;39:125–130. doi: 10.1016/j.peptides.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- Takayama M, Hirose N, Arai Y, Gondo Y, Shimizu K, Ebihara Y, Yamamura K, Nakazawa S, Inagaki H, Masui Y, Kitagawa K. Morbidity of Tokyo-area centenarians and its relationship to functional status. J Gerontol A Biol Sci Med Sci. 2007;62:774–782. doi: 10.1093/gerona/62.7.774. [DOI] [PubMed] [Google Scholar]
- Tanisawa K, Taniguchi H, Sun X, Ito T, Cao ZB, Sakamoto S, Higuchi M. Common single nucleotide polymorphisms in the FNDC5 gene are associated with glucose metabolism but do not affect serum irisin levels in Japanese men with low fitness levels. Metabolism. 2014;63:574–583. doi: 10.1016/j.metabol.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Timmons JA, Baar K, Davidsen PK, Atherton PJ (2012). Is irisin a human exercise gene? Nature. 488, E9-10; discussion E10-11. [DOI] [PubMed]
- Vamvini MT, Aronis KN, Panagiotou G, Huh JY, Chamberland JP, Brinkoetter MT, Petrou M, Christophi CA, Kales SN, Christiani DC, Mantzoros CS. Irisin mRNA and circulating levels in relation to other myokines in healthy and morbidly obese humans. Eur J Endocrinol. 2013;169:829–834. doi: 10.1530/EJE-13-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya F. Irisin, turning up the heat. Cell Metab. 2013;15:277–278. doi: 10.1016/j.cmet.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox DC, Willcox BJ, Hsueh WC, Suzuki M. Genetic determinants of exceptional human longevity: insights from the Okinawa Centenarian Study. Age (Dordr) 2006;28:313–332. doi: 10.1007/s11357-006-9020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]