Abstract
Objective
The Fli-1 transcription factor is implicated in the pathogenesis of systemic lupus erythematosus (SLE) in both human patients and animal models. Dysregulation of interleukin 6 (IL-6) is also associated with SLE. We investigated whether Fli-1 directly regulates the expression of IL-6.
Methods
Sera were collected from wild-type and Fli-1 heterozygous (Fli-1+/−) MRL/lpr mice and the concentration of IL-6 was measured by ELISA. Expression of IL-6 in the kidney was measured by real-time PCR. T cells were isolated from wild-type and Fli-1+/− MRL/lpr mice and stimulated with CD3/CD28 beads, and the concentration of IL-6 in the supernatants was measured by ELISA. MS1 endothelial cells were transfected with Fli-1 and control siRNA, and the production of IL-6 was compared after lipopolysaccharides (LPS) stimulation. A chromatin immunoprecipitation (ChIP) assay was performed to determine whether Fli-1binds to the IL-6 promoter region. Transient transfections with the NIH 3T3 cell line were performed to study if Fli-1 regulates the expression of IL-6.
Results
Fli-1+/− MRL/lpr mice had significantly decreased IL-6 in sera and reduced expression of IL-6 in kidneys compared to wild-type littermates. The T cells isolated from Fli-1+/− MRL/lpr mice produced less IL-6. Inhibiting the expression of Fli-1 in endothelial cells resulted in reduced production of IL-6. The ChIP assay revealed direct binding of Fli-1 to three regions within the IL-6 promoter. Fli-1 activated transcription from the IL-6 promoter in a dose-dependent manner.
Conclusion
Fli-1 directly regulates IL-6 expression as one of possible mechanisms for the protective effect in lupus of decreased Fli-1 expression.
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that often affects multiple organs with inflammation (1–2). Lupus nephritis in SLE patients is a major cause of mortality; nearly 60% of SLE patients develop lupus nephritis in the course of their illness (1–2). During lupus nephritis disease development, many inflammatory cells including T cells, B cells, monocytes and macrophages infiltrate into the glomerular and tubulointerstitial area of the kidneys and generate inflammatory cytokines and chemokines (3–4). The infiltration of inflammatory cells into the kidney has a critical role in lupus nephritis progression (5–7).
It is well documented that inflammatory cytokines and chemokines have a significant role in the development of SLE and lupus nephritis (7–9). In SLE development, Type I and II interferons (IFNs), interleukin 6 (IL-6), interleukin 1 (IL-1), tumor necrosis factor α (TNF-α), interleukin 10 (IL-10), interleukin 17 (IL-17), interleukin 21 (IL-21), and transforming growth factor β (TGF-β) are all key players (10). IL-6 has immunomodulatory effects on a wide range of biological activities (11). Previous studies have demonstrated that IL-6 is associated with T cell activation, γ-globulin production by B cells, osteoclast activation, hematopoiesis (platelet production), acute-phase protein induction in the liver, and mesangial cell proliferation in the kidney (11–16). Elevated serum IL-6 levels were observed in human SLE patients and correlated with disease activity (17–18). Additionally, high IL-6 expression in the kidney is reported in lupus nephritis patients (19). In murine models of lupus, elevation of serum IL-6 concentration are found in MRL/MpJ Faslpr/lpr (MRL/lpr) mice, and IL-6-deficient MRL/lpr mice showed delayed progression of lupus nephritis and prolonged survival (20–21). However it is not well known how IL-6 expression is regulated in a lupus-like proinflammatory environment.
Similar to IL-6, high levels of the transcription factor, Fli-1, in both patients and murine models, has been associated with the pathogenesis of lupus and dysfunction of the immune system (22, 23). Fli-1 belongs to the Ets gene family (24), which has been very well conserved; members have been discovered in Drosophila, sea urchin, Xenopus, chicken, mouse, and human (25–26). The consensus GGA(A/T) core motif, also known as the Ets-binding site, is the target region that Fli-1 and all other Ets family members can bind. Members of this large gene family affect gene transcription through both activation and suppression of target genes (25–26), many of which are critical for the control of cellular proliferation, differentiation, and apoptosis. In particular, endothelial cells, fibroblasts and hematopoietic lineages, including T and B cells possess high expression levels of Fli-1 (25–27). Mice die during the embryonic stage due to hemorrhages in the neural tube after targeted constitutive disruption of the Fli-1 gene likely due to thrombocytopenia and vascular deficiency (28). Development of a lupus-like disease has been observed in Fli-1 transgenic mice that overexpress the Fli-1 protein (23). Previously, we generated Fli-1 heterozygous (Fli-1+/−) mice in murine models of lupus, MRL/lpr and NZM2410, and demonstrated that mice with decreased expression of Fli-1 have profound, prolonged survival with significantly reduced lupus nephritis (29–30). Additionally it has been observed that peripheral blood lymphocytes (PBLs) from SLE patients have significantly increased expression of Fli-1, which has been linked to activity of the disease (22). We recently discovered that expression of the inflammatory chemokine Chemokine (C-C motif) ligand 2 (CCL2, also known as monocyte chemotactic protein-1, MCP-1) in endothelial cells is directly regulated by Fli-1 (31). In this study we investigated whether Fli-1 affects lupus disease development by regulating the expression of IL-6 in a murine model of lupus. We found that Fli-1+/− MRL/lpr mice had significantly decreased IL-6 protein concentrations in serum and reduced IL-6 mRNA expression in kidneys compared to wild-type littermates. Inhibiting the expression of Fli-1 with siRNA resulted in decreased IL-6 production in endothelial cells after lipopolysaccharide (LPS) stimulation. Furthermore, we found that Fli-1 directly binds to the IL-6 promoter region by chromatin immunoprecipitation (ChIP) assay and Fli-1 activated transcription from the IL-6 promoter in a dose-dependent manner. Thus, Fli-1 is an essential regulator for the expression of IL-6 and expression of Fli-1 impacts lupus disease development by regulating IL-6 expression in a murine model of lupus.
Materials and Methods
Mice
MRL/lpr mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Fli-1+/− MRL/lpr mice were generated by backcrossing with Fli-1+/− C57BL/6 (B6) mice for more than 12 generations as previously reported and used together with wild-type littermates in this study (29). All mice were housed under pathogen-free conditions at the animal facility of the Ralph H. Johnson Veterans Affairs Medical Center and all animal experiments were approved by the Institutional Animal Care and Use Committee.
Cells
Murine endothelial MS1 cells were purchased from the American Type Culture Collection (ATCC) and maintained with Dulbecco’s Modified Eagle’s Medium (DMEM) medium (Mediatech Inc., Manassas, VA) with 10% fetal bovine serum (FBS). The mouse embryonic fibroblast NIH3T3 cell line was grown in DMEM with 10% FBS and 1% penicillin/streptomycin. Human primary endothelial cells were purchased from Lonza Inc. (Allendale, NJ).
Genotyping of the mice by PCR
For genotyping, PCR was used to detect fragments of the wild-type Fli-1 and Fli-1+/− allele, as previously described (29). The primers for PCR were as follows: Fli-1 exon IX /forward primer (positions 1156 to 1180), GACCAACGGGGAGTTCAAAATGACG; Fli-1 exon IX/reverse primer (positions 1441 to 1465), GGAGGATGGGTGAGACGGGACAAAG; and Pol II /reverse primer, GGAAGTAGCCGTTATTAGTGGAGAGG. DNA was isolated from tail snips (4-week old mice) using a QIAamp Tissue Kit (Qiagen, Germantown, MD). PCR analyses were performed under the following conditions: 1 cycle at 95°C for 5 min, followed by 36 repeating cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min followed by 72°C for 7 min. A 309-bp fragment indicates the presence of the wild-type allele, and a 406-bp fragment is amplified from the mutant allele.
Measurement of cytokine IL-6
IL-6 concentrations in mouse serum or medium supernatants were determined by ELISA (eBioscience, San Diego, CA). The assays were performed using the manufacturer’s instructions.
Fli-1 specific siRNA transfection into endothelial cells
Fli-1 specific siRNA and negative control siRNA were purchased from Invitrogen and transfected into the MS1 cells using lipofectamine according to the manufacturer’s instructions (Life Technologies/Invitrogen, Grand Island, NY). After transfection, MS1 cells were stimulated by LPS (1μg/ml, Sigma-Aldrich, St. Louis, MO) and the supernatant was collected at the following time points 0, 2, 4, 6 and 24 hours after LPS stimulation. Collected supernatants were analyzed by ELISA to determine IL-6 concentration.
Measurement of IL-6 expression in kidneys by real-time PCR
Total RNA was prepared from part of a fresh kidney for real-time PCR analysis. A total of 1μg of RNA was used to synthesize cDNA using the RT2 First Strand Kit (Qiagen). Real-time PCR was performed according to the manufacturer’s instructions, in triplicate using the RT2 SYBR Green Rox qPCR Mastermix (Qiagen). The IL-6 primers used were: forward, 5′-GTTCTCTGGGAAATCGTGGA-3′, reverse, 5′-TGTACTCCAGGTAGCTATGG-3′. PCR was performed using the CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA), the housekeeping gene GAPDH was used as the control, and relative expression analysis was conducted using the program provided by SABiosciences. The cycling conditions for all genes were: pre-incubation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 seconds, and annealing and extension at 60°C for 1 min, with a single data acquisition at the end of each extension.
T cell isolation and stimulation by CD3/CD28 activator
T cells were negatively isolated from Fli-1+/− and wild-type MRL/lpr mice using the T cell negative Dynabeads isolation kit (Life Technologies, Grand Island, NY) following instructions from the manufacture. Purified T cells were seeded in 24-well plates at a concentration of 1×106 cells in RPMI 1640 medium with 10% fetal bovine serum and stimulated with CD3/CD28 activator microbeads (Life Technologies). The supernatants were collected 1 to 3 days after stimulation and ELISA was used to determine the IL-6 concentrations.
ChIP assay
ChIP assay was performed as we described previously using an anti-Fli-1 rabbit polyclonal antibody and control IgG (32). The primers used in the ChIP assay are not shown, but can be made available upon request to the corresponding author. After immunoprecipitation by Fli-1 specific antibody and normal IgG controls, the DNA was purified and amplified by PCR according to manufacturer’s instructions (Qiagen).
Reporter and Expression Constructs
PCR primers were designed to directionally clone the full length IL-6 promoter region, -1231bp upstream of the transcription start site, into the pGL3 basic vector upstream of the Luciferase gene (Promega, Madison, WI). The primers were designed using the 5′UTR of the murine IL-6 gene obtained from the Ensembl genome browser (Ensembl Gene ID ENSMUSG00000025746) (33). The forward primer 5′ AAG GTA CCA TAT CTG GAG ACA GG 3′ contains a KpnI restriction enzyme site (underlined) and the reverse primer 5′ AAT AAA GCT TAG CGG T TT CTG GAA TTG 3′ a HindIII restriction enzyme site (underlined). The promoter was PCR amplified from genomic DNA isolated from a B6 mouse. The PCR program used to amplify the 1231bp sequence was as follows: 94°C for 3 min.; thirty cycles of 94°C for 30 sec, 52°C for 30 sec, 72°C for 1 min; 72°C for 5 min; held at 4°C until the program was stopped. The mouse Fli-1 expression vector was obtained from Dr. Dennis Watson (Medical University of South Carolina) and confirmed by DNA sequencing. Briefly, the Fli-1 gene containing a 5′ kozak sequence and Flag tag, was cloned into the pcDNA3.0 expression vector (Life Technologies), which is under the control of a CMV promoter. The Fli-1 expression vector has been described previously (34). All of the constructs were confirmed by DNA sequencing (Genewiz, South Plainfield, NJ).
DNA Transfection
NIH3T3 cells were seeded at 4×105 cells per well in 6 well plates, one day prior to transfection. Transfections were performed using the Fugene 6 transfection reagent (Promega) following the manufacturer’s instructions. For all of the transfection experiments 2μg of the reporter constructs pGL3/basic and pGL3/IL6 were used. For the Fli-1 dose response, increasing amounts (0.025μg, 0.05μg, 0.1μg, 0.25μg, 0.5μg, and 1μg) of the Fli-1 expression construct were transfected into the cells. The pcDNA3.0 construct was added to all transfection experiments when necessary to ensure that equimolar amounts of total DNA were transfected into the cells. Luciferase activity was measured using the Dual-luciferase reporter assay system (Promega) and the Luminoskan Ascent Microplate Luminometer (Thermo Fisher Scientific, Waltham, MA). Transfection activity was normalized to the activity of the co-transfected Renilla luciferase construct. Fold activation was calculated based on the activity over the pGL3/basic vector and the values are reported as mean ± SE.
Statistical analysis
The unpaired Student’s t test was used to determine significant differences between two groups. A p < 0.05 was considered to be statistically significant. The Mann-Whitney U-test was also used when appropriate.
Results
Significantly reduced serum IL-6 concentrations in Fli-1+/− MRL/lpr mice compared to wild-type littermates
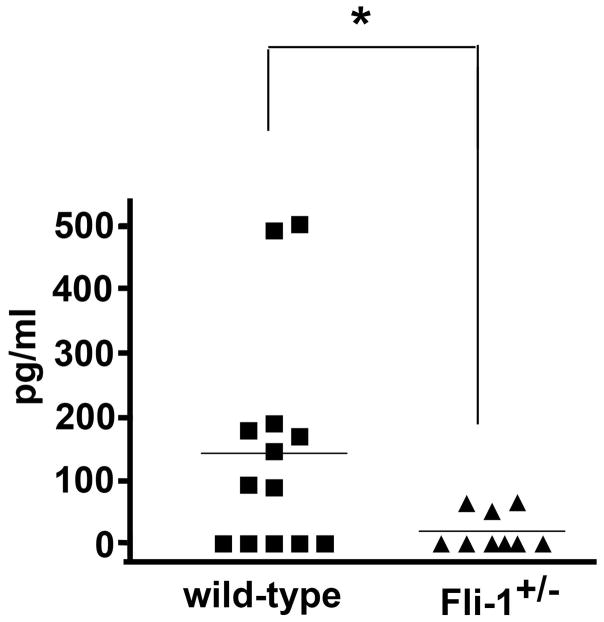
To assess whether the expression level of the transcription factor Fli-1 affects IL-6 production, we measured serum IL-6 concentrations in Fli-1+/− (n=9) and wild-type (n=13) littermates at the age of 22 weeks. As shown in Fig.1, the serum IL-6 concentrations in the Fli-1+/− MRL/lpr mice were significantly lower compared to wild-type littermates.
Fig. 1. Decreased IL-6 concentration in serum from Fli-1+/− MRL/lpr mice compared to wild-type littermates.
The serum was collected from Fli-1+/− MRL/lpr mice (n=9) and wild-type littermates (n=13) at the age of 22 weeks. IL-6 concentrations were detected by ELISA and are presented as pg/ml. * p< 0.05.
Reduced IL-6 mRNA expression in kidneys from Fli-1+/− MRL/lpr mice compared to wild-type littermates
Since lupus nephritis is the major cause of death in human patients and murine models of lupus, including MRL/lpr mice, and we previously reported that Fli-1+/− MRL/lpr mice have significant, prolonged survival with reduced renal scores (29), we next examined the mRNA expression of IL-6 in kidneys from Fli-1+/− and wild-type MRL/lpr mice. Kidneys were removed from mice at the age of 22 weeks and expression of IL-6 was measured by RT-PCR. As shown in Fig. 2, the expression of IL-6 in kidneys from Fli-1+/− MRL/lpr mice was significantly decreased compared to that from wild-type littermates (p<0.05). We examined IL-6 expression in kidneys using immunohistochemical staining. The expression of IL-6 was mainly observed in the glomerulus of MRL/lpr mice. Interstitial IL-6 expression was also observed but to a lesser extent, along with expression in small arteries and capillaries (data not shown).
Fig. 2. Reduced expression of IL-6 in kidneys from Fli-1+/− MRL/lpr mice compared to wild-type littermates.
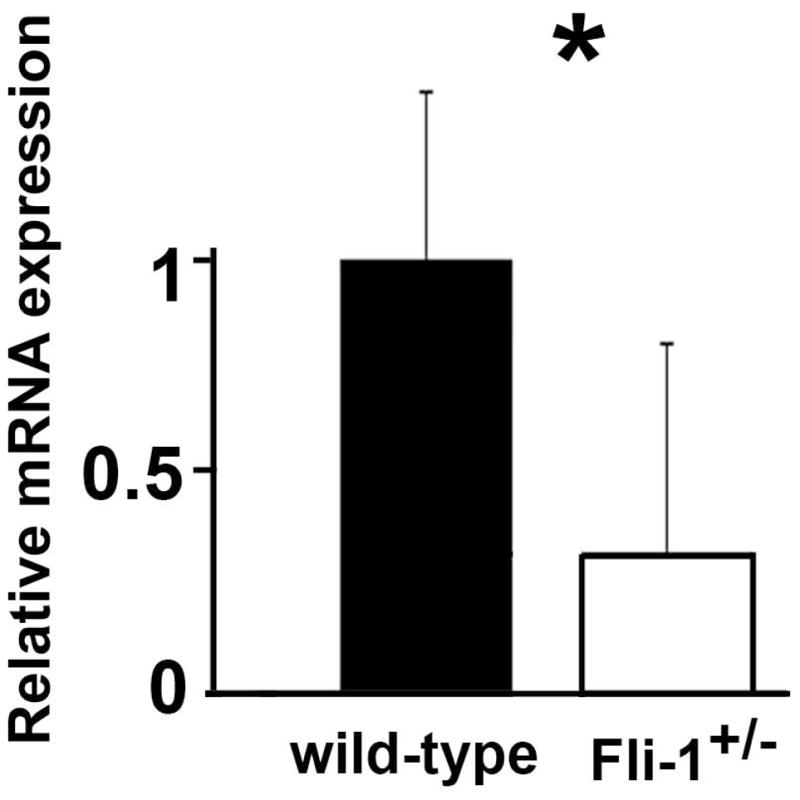
Total RNA was isolated from kidneys of mice at the age of 22 weeks (Fli-1+/−, n = 9, wild-type, n=13). Total RNA was transcribed to cDNA using the SuperScript First-Strand Synthesis System (Life Technologies/Invitrogen). Real-time polymerase chain reaction was performed in triplicate with the appropriate primers. The data presented are the mean ± SD. *p < 0.05.
T cells isolated from Fli-1+/− MRL/lpr mice produced significantly lower IL-6 after stimulation
To further verify that Fli-1 affects IL-6 expression, we isolated T cells including CD4+ and CD8+ cells from Fli-1+/− and wild-type MRL/lpr mice at the age of 16 weeks by negative selection as described in the Materials and Methods section. Double negative T cells (B220+) were excluded by this isolation. As we have reported previously, there were no differences in T cell subtype populations between Fli-1+/− and wild-type MRL/lpr mice (29). The T cells were stimulated with CD3/CD28 beads and the supernatants were collected. IL-6 concentrations were measured by ELISA. As shown in Fig. 3, T cells isolated from Fli-1+/− MRL/lpr mice produced significantly less IL-6, 24, 48 and 72 hours after stimulation compared to the T cells from wild-type littermates. To test if expression of Fli-1 affects IL-6 production in T cells from a normal strain of mice, we isolated T cells from Fli-1+/− B6 mice and their wild-type littermates at the age of 8 weeks. T cells from Fli-1+/− B6 mice produced reduced IL-6 after CD3/CD28 bead stimulation (at 2 days, T cells from wild-type, 39.5±3.6 pg/ml versus T cells from Fli-1+/− mice, 28.0±1.4 pg/ml, p< 0.05; at 3 days, T cells from wild-type, 55.9±3.3 pg/ml versus T cells from Fli-1+/− mice, 28.9±4.5 pg/ml, p< 0.05; at 7 days, T cells from wild-type, 146.0±26.4 pg/ml versus T cells from Fli-1+/− mice, 75.4±9.2 pg/ml, p<0.05; n=5 in each group).
Fig. 3. Decreased IL-6 production in T cells isolated from Fli-1+/− MRL/lpr mice compared to wild-type littermates.
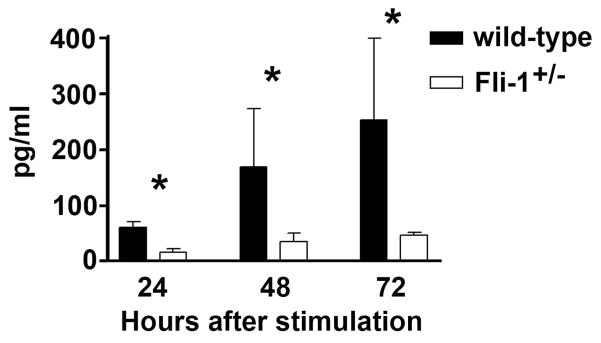
T cells were negatively isolated from spleens of Fli-1+/− (n=6) or wild-type littermates (n=6) at the age of 16 weeks using the T cell negative Dynabead isolation kit. The T cells were cultured in 24-well plates at a concentration of 1×106/ml cells. The supernatants were collected 1 to 3 days after stimulation with CD3/CD28 activator microbeads and ELISA was used to determine the IL-6 concentrations. The data presented are the mean ± SD. * p<0.05.
Inhibition of Fli-1 by siRNA led to decreased secretion of IL-6 in endothelial cells
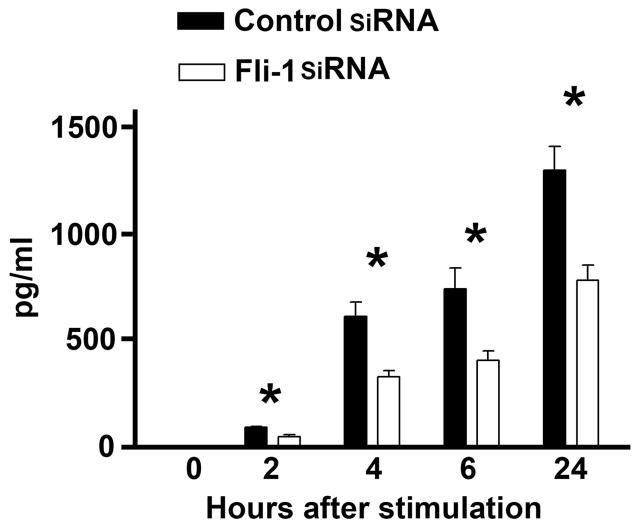
Since IL-6 expression was predominantly located in glomeruli and the expression of Fli-1 was observed only in endothelial cells located in the glomeruli, we investigated whether Fli-1 regulates the expression of IL-6 in endothelial cells (31, 35). Fli-1 specific siRNA was transfected into the MS1 endothelial cell line and expression of the Fli-1 protein was reduced around 90% as reported previously (31). As shown in Fig. 4, IL-6 expression levels were undetectable prior to LPS stimulation. MS1 cells transfected with Fli-1 siRNA produced significantly lower levels of IL-6 compared to MS1 cells transfected with control siRNA 2, 4, 6 and 24 hours after LPS stimulation (Fig. 4). To determine whether Fli-1 also regulates IL-6 expression in human endothelial cells, we transfected Fli-1 specific siRNA and negative control siRNA into human primary endothelial cells. The endothelial cells transfected with Fli-1 siRNA demonstrated significantly decreased expression of Fli-1 and produced less IL-6 compared to the cells transfected with negative siRNA after stimulation with 0.1 μg/ml LPS (before the stimulation, control siRNA treated cells 203.8±15.5 pg/ml versus Fli-1 siRNA treated cells, 153.9±8.0 pg/ml, p<0.05; at 2 hours after LPS stimulation, control siRNA, 285.0±33.53 pg/ml versus Fli-1 siRNA, 168.4±4.6 pg/ml, p<0.05, n=4 in each group, p<0.05).
Fig. 4. Inhibition of Fli-1 expression using siRNA resulted in significantly decreased production of IL-6 in endothelial cells.
MS1 endothelial cells were transfected with Fli-1 specific siRNA or negative control siRNA. Supernatants were collected after stimulation with LPS at different time points as indicated in figure. IL-6 concentrations were measured by ELISA. The data presented are the mean ± SD. * indicates p< 0.05.
Fli-1 binds directly to the promoter region of IL-6
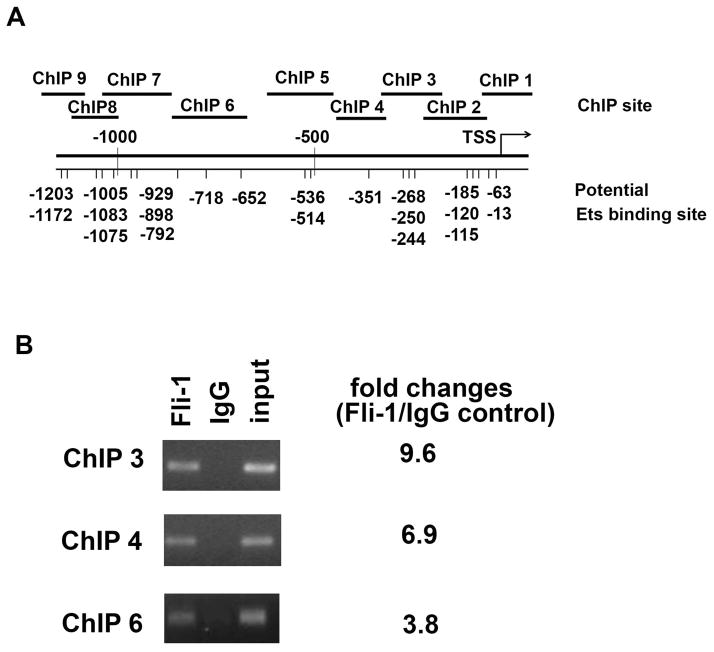
Next, we performed a ChIP assay to determine whether Fli-1 binds directly to the promoter region of IL-6. As shown in Fig. 5A, many putative Fli-1 binding sites were identified in the mouse IL-6 gene promoter. Nine primer pairs were designed to cover the putative Fli-1 binding sites as shown in Fig. 5A. After immunoprecipitating DNA from the MS1 endothelial cell line by either the Fli-1 specific antibody or the control IgG antibody, the crosslinked protein/DNA complexes were amplified by RT-PCR to calculate amount of ChIP DNA isolated. As shown in Fig. 5B, ChIP3, ChIP4 and ChIP 6 sites were significantly enriched with specific Fli-1 antibodies compared to normal IgG controls. All other ChIP sites were not significantly enriched with specific Fli-1 antibodies (fold change was less than 1.3).
Fig. 5. ChIP analysis of Fli-1 binding to the IL-6 promoter.
The putative Fli-1 binding sites in the promoter region of the mouse IL-6 gene and the location of the primers used in the ChIP assay are shown in (A). MS1 endothelial cells were cross-linked with formaldehyde, chromatin was isolated from the cells and immunoprecipitated with specific Fli-1 or control IgG antibodies. The genomic fragments associated with the immunoprecipitated DNA were amplified by RT-PCR using primers specifically designed to cover putative Ets binding sites (The sequences of primers are available upon request to the corresponding author). PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Input represents 1% total cross-linked, reversed chromatin before immunoprecipitation. Fold changes were calculated by RT-PCR, ChIP1, ChIP2, ChIP5, and ChIP 7–9 sites were not significantly enriched with a specific antibody against Fli-1 (less than 1.3 fold change, not shown in the figure) (B). Data are representative of five independent experiments.
Fli-1 drives transcription from the IL-6 promoter
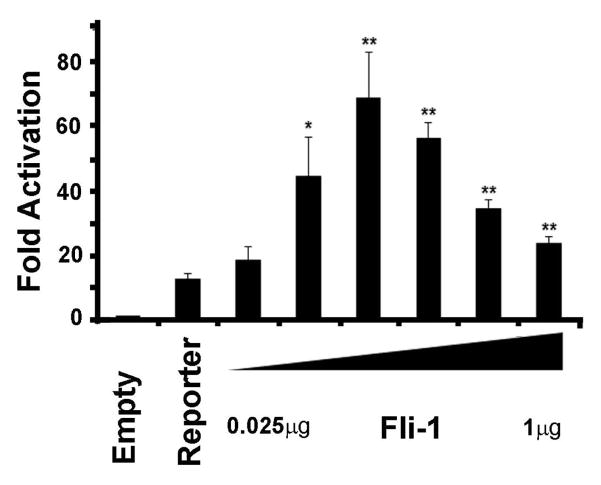
To confirm that Fli-1 directly regulates the expression of IL-6, the murine IL-6 promoter was PCR amplified from genomic DNA and cloned into the pGL3 basic reporter construct. Transient transfection assays were performed in order to demonstrate that Fli-1 regulates the expression of IL-6. As shown in Fig. 6, the transfection results clearly demonstrate that Fli-1 drives transcription from the IL-6 promoter in a dose-dependent manner. As little as 50ng of Fli-1 is needed to significantly activate transcription from the IL-6 promoter.
Fig. 6. Fli-1 drives transcription from the IL-6 promoter.
NIH3T3 cells were transfected with a Fli-1 expression construct and /the pGL3/IL6 reporter construct using the Fugene 6 transfection reagent (Promega). Transcriptional activation of the pGL3/IL6 reporter construct is shown as fold activation over the empty pGL3 basic vector. All transfections were carried out at equimolar concentrations. Increasing amounts of the Fli-1 expression construct (0.025, 0.05, 0.1, 0.25, 0.5, to 1μg) were transfected into NIH3T3 cells. Values shown are the mean + SE for three replicate experiments (n=9).* p<0.05, **p<0.01.
Discussion
We previously demonstrated that reduced expression of Fli-1 had a profound impact on disease development in murine models of lupus, including MRL/lpr and NZM2410 mice (29–30). Fli-1+/− MRL/lpr mice had significantly prolonged survival with decreased renal pathology (29). Enhanced expression of IL-6 is associated with the development of SLE in both human patients and murine models of lupus (11, 17–21). In this study, for the first time, we demonstrated that Fli-1 is a critical regulator of IL-6 in a murine model of lupus and likely affects disease development by regulating this critical cytokine.
Expression of IL-6 is associated with autoimmune disease progression in rheumatoid arthritis and SLE. Elevated serum IL-6 levels were found in human lupus patients and concentrations correlated with disease activity (18, 36–37). Similarly, MRL/lpr mice showed high serum IL-6 levels (20). In this report, we show that Fli-1+/− MRL/lpr mice, with decreased expression of Fli-1 protein, had significantly lower serum IL-6 compared to wild-type littermates (Fig. 1). IL-6 is implicated in development of lupus nephritis (37–38). Higher expression of IL-6 reported in lupus nephritis and expression of IL-6 is correlated with interstitial immune cell infiltration (19). Furthermore, administration of the anti-IL-6 receptor antibody (tocilizumab) improves lupus disease in lupus prone mice (39–41). We also found the expression of IL-6 was significantly deceased in the kidneys from Fli-1+/− MRL/lpr mice compared to wild-type littermates (Fig. 2). We have previously reported that kidneys from Fli-1+/− MRL/lpr mice had significantly less inflammatory infiltrating inflammatory cells (29). By comparison, both Fli-1+/− MRL/lpr mice and IL-6-deficient MRL/lpr have prolonged survival, significantly reduced proteinuria and renal pathological scores with decreased infiltration of inflammatory cells (21, 29). Furthermore, we demonstrated recently that a significantly increased number of green florescence protein (GFP) expressing inflammatory cells infiltrated the kidneys of wild-type MRL/lpr mice compared to Fli-1+/− MRL/lpr mice after injection of inflammatory cells from congenic GFP transgenic MRL/lpr mice (42). Lower IL-6 expression in kidneys from Fli-1+/− MRL/lpr mice likely contributes to the decreased infiltration of inflammatory cells as lupus mice deficient in IL-6 had a significant reduction in renal inflammatory cells.
Many types of cells produce IL-6 including osteoblasts, macrophages, B cells, T cells, synoviocytes, endothelial cells and fibroblasts (11). In a previous report, IL-6 expression was increased in the area of glomerular and tubular inflammation and caused tubular atrophy (43). We found strong expression of IL-6 localized to the glomerulus in MRL/lpr mice, while lower levels of expression was observed in the interstitium, small arteries and capillaries (data not shown). Since previous results have shown that Fli-1 is mainly expressed in glomerular endothelial cells, arterioles and interstitial capillaries, but not in mesangial cells and podocytes (31, 35), we inhibited the expression of Fli-1 in endothelial cells to investigate the effect on production of IL-6 after LPS stimulation. As shown in Fig. 4, inhibiting Fli-1 in endothelial resulted in significantly decreased production of IL-6 after LPS stimulation.
The transcriptional regulation of IL-6 is complex and several transcription factors are involved (44–45). In the human IL-6 promoter, functional cis-regulatory elements include interferon regulatory factor 1 (IRF-1), activator protein 1 (AP-1), nuclear factor kappa B (NFkB), CCAAT-enhancer binding proteins (C/EBP), specificity protein 1 (Sp1), nuclear factor IL-6 (NF-IL6), the multiple response element (MRE) and the cAMP response element (CRE) (44–45). Clear evidence of transcription regulation of IL-6 by several of these transcription factors was demonstrated in mesangial cells (45), where Fli-1 is not expressed. The promoter region of the mouse IL-6 gene is highly conserved (>80% similarity to the human promoter) and transcription factor binding sites, including CRE and AP-1, were also conserved between the two promoters (46). Within the mouse promoter there are two conserved motifs that may be associated with interferon inducibility that have the cognate motif of Ets binding sites GGAA (46). In this study we demonstrated that Fli-1 binds to three regions identified by ChIP assay within the murine promoter (Fig. 5, ChIP 3, -344 to -195; ChIP 4, -419 to -281; ChIP 6, -820- -613). Since the ChIP 3 and ChIP 4 sites are very close, further detailed analysis into the precise sites within the IL-6 promoter to which Fli-1 binds will be performed in future studies. Interestingly, the highest binding is within the ChIP3 region, the region that corresponds to the conserved GGAA motifs described above (Fig. 5, 46). Figure 6 demonstrates that Fli-1 activates transcription from the IL-6 promoter, validating that Fli-1 is involved in the regulation of the IL-6 gene. At higher doses, activation of the IL-6 promoter decreases, although even at the highest dose, Fli-1 still significantly activates transcription above control (Fig.5). This effect may be due to increased competition for the available DNA binding sites or possibly the recruitment of other cofactors. Ets family members are known to regulate transcription through protein-protein interactions (26). Fli-1 has been shown to activate or repress transcription from both the collagen α2 promoter and the HIV-1 core enhancer depending on interactions with different cofactors (47–48). That Fli-1 regulates transcription of the IL-6 promoter is particularly interesting given that Fli-1 expression is up-regulated by IL-6 through the JAK2 signaling pathway in a pre-osteoblastic cell line (49). This may suggest a possible feedback loop between the two factors. Transcriptional regulation of IL-6 is reported to be cell-specific (45). In this study, we found that T cells isolated from Fli-1+/− MRL/lpr mice produced significantly reduced IL-6 compared to T cells isolated from wild-type littermates (Fig. 3). Additionally, inhibiting expression of Fli-1 in endothelial cells led to decreased IL-6 production (Fig. 4). Thus, Fli-1 is likely a positive regulator of IL-6 in T cells and endothelial cells. We also demonstrated that Fli-1 regulates IL-6 production in human endothelial cells, which suggests expression of Fli-1 is likely play a role in human disease development.
CCL2 may upregulate the production of IL-6 in kidney tubular epithelial cells (50). We previously reported that significantly decreased expression of CCL2 was observed in Fli-1+/− MRL/lpr mice and Fli-1+/− NZM2410 mice compared to wild-type littermates (30, 31). Furthermore, we demonstrated that Fli-1 directly binds to the promoter of CCL2 in endothelial cells isolated from kidneys of Fli-1+/− NZM2410 mice. These mice also had significantly lower production of CCL2 compared to wild-type NZM2410 mice (31). Thus, decreased expression of CCL2 in kidneys from Fli-1+/− MRL/lpr may also contribute to the decreased expression of IL-6 compared to wild-type littermates.
In conclusion, we present here the first evidence that the Fli-1 transcription factor regulates the expression of IL-6. We believe the decreased production of IL-6 in Fli-1+/− MRL/lpr mice compared to wild-type MRL/lpr mice contributes, in part, to the prolonged survival and significantly reduced renal disease observed in Fli-1+/− MRL/lpr mice. The results of this study identify the Fli-1 transcription factor as a novel, critical regulator of IL-6 transcription and bring new insight into understanding the complex mechanisms involved in the pathogenesis of lupus disease.
Acknowledgments
Supported in part by grant from National Institutes of Health grants (AR056670 to X.Z.) and by the Medical Research Service, Department of Veterans Affairs (to G.G. and X. Z.)
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content. All authors have approved the final version to be published. Dr. Zhang had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Sato, Lennard Richard, Zhang.
Acquisition of data. Sato, Lennard Richard, Brandon, Buie.
Analysis and interpretation of data. Sato, Lennard Richard, Brandon, Buie, Oates, Gilkeson, Zhang.
References
- 1.D’Cruz DP, Khamashta MA, Hughes GR. Systmeic lupus erythematosus. Lancet. 2007;369:587–96. doi: 10.1016/S0140-6736(07)60279-7. [DOI] [PubMed] [Google Scholar]
- 2.Borchers AT, Leibushor N, Naguwa SM, Cheema GS, Shoenfeld Y, Gershwin ME. Lupus nephritis: A critical review. Autoimmun Rev. 2012;12:174–94. doi: 10.1016/j.autrev.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 3.Boucher A, Droz D, Adafer E, Noël LH. Characterization of mononuclear cell subsets in renal cellular interstitial infiltrates. Kidney Int. 1986;29:1043–9. doi: 10.1038/ki.1986.105. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulos E, Seron D, Hartley RB, Cameron JS. Lupus nephritis: correlation of interstitial cells with glomerular function. Kidney Int. 1990;37:100–9. doi: 10.1038/ki.1990.14. [DOI] [PubMed] [Google Scholar]
- 5.D’Amico G, Ferrario F, Rastaldi MP. Tubulointerstitial damage in glomerular diseases: its role in the progression of renal damage. Am J Kidney Dis. 1995;26:124–32. doi: 10.1016/0272-6386(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 6.Hill GS, Delahousse M, Nochy D, Thervet E, Vrtovsnik F, Rémy P, et al. Outcome of relapse in lupus nephritis: roles of reversal of renal fibrosis and response of inflammation to therapy. Kidney Int. 2002;61:2176–86. doi: 10.1046/j.1523-1755.2002.00357.x. [DOI] [PubMed] [Google Scholar]
- 7.Castiglione A, Bucci A, Fellin G, d’Amico G, Atkins RC. The relationship of infiltrating renal leucocytes to disease activity in lupus and cryoglobulinaemic glomerulonephritis. Nephron. 1988;50:14–23. doi: 10.1159/000185110. [DOI] [PubMed] [Google Scholar]
- 8.Nowling TK, Gilkeson GS. Mechanisms of tissue injury in lupus nephritis. Arthritis Res Ther. 2011;13:250–9. doi: 10.1186/ar3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apostolidis SA, Lieberman LA, Kis-Toth K, Crispín JC, Tsokos GC. The dysregulation of cytokine networks in systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31:769–79. doi: 10.1089/jir.2011.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohl K, Tenbrock K. Inflammatory cytokines in systemic lupus erythematosus. J Biomed Biotechnol. 2011;2011:Article ID 432595. doi: 10.1155/2011/432595. URL: http://www.hindawi.com/journals/bmri/2011/432595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kishimoto T. IL-6: from its discovery to clinical applications. Int Immunol. 2010;22:347–52. doi: 10.1093/intimm/dxq030. [DOI] [PubMed] [Google Scholar]
- 12.Andus T, Geiger T, Hirano T, Northoff H, Ganter U, Bauer J, et al. Recombinant human B cell stimulatory factor 2 (BSF-2/IFN-beta 2) regulates beta-fibrinogen and albumin mRNA levels in Fao-9 cells. FEBS Lett. 1987;221:18–22. doi: 10.1016/0014-5793(87)80344-7. [DOI] [PubMed] [Google Scholar]
- 13.Hirano T, Matsuda T, Hosoi K, Okano A, Matsui H, Kishimoto T. Absence of antiviral activity in recombinant B cell stimulatory factor 2 (BSF-2) Immunol Lett. 1988;17:41–5. doi: 10.1016/0165-2478(88)90099-5. [DOI] [PubMed] [Google Scholar]
- 14.Okada M, Kitahara M, Kishimoto S, Matsuda T, Hirano T, Kishimoto T. IL-6/BSF-2 functions as a killer helper factor in the in vitro induction of cytotoxic T cells. J Immunol. 1988;141:1543–9. [PubMed] [Google Scholar]
- 15.Kopf M, Baumann H, Freer G, Freudenberg M, Lamers M, Kishimoto T, et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–42. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- 16.Yoshizaki K, Nishimoto N, Mihara M, Kishimoto T. Therapy of rheumatoid arthritis by blocking IL-6 signal transduction with a humanized anti-IL-6 receptor antibody. Springer Semin Immunopathol. 1998;20:247–59. doi: 10.1007/BF00832010. [DOI] [PubMed] [Google Scholar]
- 17.Linker-Israeli M, Deans RJ, Wallace DJ, Prehn J, Ozeri-Chen T, Klinenberg JR. Elevated levels of endogenous IL-6 in systemic lupus erythematosus. A putative role in pathogenesis. J Immunol. 1991;147:117–23. [PubMed] [Google Scholar]
- 18.Chun HY, Chung JW, Kim HA, Yun JM, Jeon JY, Ye YM, et al. Cytokine IL-6 and IL-10 as biomarkers in systemic lupus erythematosus. J Clin Immunol. 2007;27:461–6. doi: 10.1007/s10875-007-9104-0. [DOI] [PubMed] [Google Scholar]
- 19.Takemura T, Yoshioka K, Murakami K, Akano N, Okada M, Aya N, et al. Cellular localization of inflammatory cytokines in human glomerulonephritis. Virchows Arch. 1994;424:459–64. doi: 10.1007/BF00191429. [DOI] [PubMed] [Google Scholar]
- 20.Tang B, Matsuda T, Akira S, Nagata N, Ikehara S, Hirano T, Kishimoto T. Age-associated increase in interleukin 6 in MRL/lpr mice. Int Immunol. 1991;3:273–8. doi: 10.1093/intimm/3.3.273. [DOI] [PubMed] [Google Scholar]
- 21.Cash H, Relle M, Menke J, Brochhausen C, Jones SA, Topley N, et al. Interleukin 6 (IL-6) deficiency delays lupus nephritis in MRL-Faslpr mice: the IL-6 pathway as a new therapeutic target in treatment of autoimmune kidney disease in systemic lupus erythematosus. J Rheumatol. 2010;37:60–70. doi: 10.3899/jrheum.090194. [DOI] [PubMed] [Google Scholar]
- 22.Georgiou P, Maroulakou I, Green J, Dantis P, Romanospica V, Kottaridis S, et al. Expression of ets family of genes in systemic lupus erythematosus and Sjogren’s syndrome. Int J Oncol. 1996;9:9–18. [PubMed] [Google Scholar]
- 23.Zhang L, Eddy A, Teng YT, Fritzler M, Kluppel M, Melet F, et al. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol Cell Biol. 1995;5:6961–70. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-David Y, Giddens EB, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–18. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 25.Papas TS, Bhat NK, Spyropoulos DD, Mjaatvedt AE, Vournakis J, Seth A, et al. Functional relationships among ETS gene family members. Leukemia. 1997;11:557–66. [PubMed] [Google Scholar]
- 26.Li R, Pei H, Watson D. Regulation of Ets function by protein-protein interactions. Oncogene. 2000;19:6514–6523. doi: 10.1038/sj.onc.1204035. [DOI] [PubMed] [Google Scholar]
- 27.Anderson MK, Hernandez-Hoyos G, Diamond RA, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–48. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 28.Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, et al. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–52. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang XK, Gallant S, Molano I, Moussa OM, Ruiz P, Spyropoulos DD, et al. Decreased expression of the Ets family transcription factor Fli-1 markedly prolongs survival and significantly reduces renal disease in MRL/lpr mice. J Immunol. 2004;173:6481–9. doi: 10.4049/jimmunol.173.10.6481. [DOI] [PubMed] [Google Scholar]
- 30.Mathenia J, Reyes-Cortes E, Williams S, Molano I, Ruiz P, Watson DK, et al. Impact of Fli-1 transcription factor on autoantibody and lupus nephritis in NZM2410 mice. Clin Exp Immunol. 2010;162:362–71. doi: 10.1111/j.1365-2249.2010.04245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki E, Karam E, Williams S, Watson DK, Gilkeson G, Zhang XK. Fli-1 transcription factor affects glomerulonephritis development by regulating expression of monocyte chemoattractant protein-1 in endothelial cells in the kidney. Clin Immunol. 2012;145:201–8. doi: 10.1016/j.clim.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang XK, Moussa O, LaRue A, Bradshaw S, Molano I, Spyropoulos DD, et al. The transcription factor Fli-1 modulates marginal zone and follicular B cell development in mice. J Immunol. 2008;181:1644–54. doi: 10.4049/jimmunol.181.3.1644. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent s, et al. Ensembl 2013. Nucleic Acids Research. 2013;41:D48–55. doi: 10.1093/nar/gks1236. URL: http://nar.oxfordjournals.org/content/41/D1/D48.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svenson JL, Chike-Harris K, Amria MY, Nowling TK. The mouse and human Fli1 genes are similarly regulated by Ets factors in T cells. Genes Immun. 2010;11:161–72. doi: 10.1038/gene.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J Histochem Cytochem. 2006;54:385–95. doi: 10.1369/jhc.4A6514.2005. [DOI] [PubMed] [Google Scholar]
- 36.Sabry A, Sheashaa H, El-Husseini A, Mahmoud K, Eldahshan KF, George SK, et al. Proinflammatory cytokines (TNF-alpha and IL-6) in Egyptian patients with SLE: its correlation with disease activity. Cytokine. 2006;35:148–53. doi: 10.1016/j.cyto.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 37.Fukatsu A, Matsuo S, Tamai H, Sakamoto N, Matsuda T, Hirano T. Distribution of interleukin-6 in normal and diseased human kidney. Lab Invest. 1991;65:61–6. [PubMed] [Google Scholar]
- 38.Fukatsu A, Matsuo S, Yuzawa Y, Miyai H, Futenma A, Kato K. Expression of interleukin 6 and major histocompatibility complex molecules in tubular epithelial cells of diseased human kidneys. Lab Invest. 1993;69:58–67. [PubMed] [Google Scholar]
- 39.Illei GG, Shirota Y, Yarboro CH, Daruwalla J, Tackey E, Takada K, et al. Tocilizumab in systemic lupus erythematosus: data on safety, preliminary efficacy, and impact on circulating plasma cells from an open-label phase I dosage-escalation study. Arthritis Rheum. 2010;62:542–52. doi: 10.1002/art.27221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiberd BA. Interleukin-6 receptor blockage ameliorates murine lupus nephritis. J Am Soc Nephrol. 1993;4:58–61. doi: 10.1681/ASN.V4158. [DOI] [PubMed] [Google Scholar]
- 41.Liang B, Gardner DB, Griswold DE, Bugelski PJ, Song XY. Anti-interleukin-6 monoclonal antibody inhibits autoimmune responses in a murine model of systemic lupus erythematosus. Immunology. 2006;119:296–305. doi: 10.1111/j.1365-2567.2006.02433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato S, Zhang XK. The Friend leukaemia virus integration 1 (Fli-1) transcription factor affects lupus nephritis development by regulating inflammatory cell infiltration into the kidney. Clin Exp Immunol. 2014;177:102–9. doi: 10.1111/cei.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romano M, Sironi M, Toniatti C, Polentarutti N, Fruscella P, Ghezzi P, et al. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity. 1997;6:315–25. doi: 10.1016/s1074-7613(00)80334-9. [DOI] [PubMed] [Google Scholar]
- 44.Keller ET, Wanagat J, Ershler WB. Molecular and Cellular Biology of interleukin-6 and its receptor. Front Biosci. 1996;1:340–57. doi: 10.2741/a136. [DOI] [PubMed] [Google Scholar]
- 45.Grassl C, Luckow B, Schlondorff D, Dendorfer U. Transcriptional Regulation of the Interleukin-6 Gene in Mesangial Cells. J Am Soc Nephrol. 1999;10:1466–77. doi: 10.1681/ASN.V1071466. [DOI] [PubMed] [Google Scholar]
- 46.Tanabe O, Akira S, Kamiya T, Wong GG, Hirano T, Kishimoto T. Genomic structure of the murine IL-6 gene. High degree conservation of potential regulatory sequences between mouse and human. J Immunol. 1988;141:3875–81. [PubMed] [Google Scholar]
- 47.Czuwara-Ladykowska J, Shirasaki F, Jackers P, Watson D, Trojanowska M. Fli-1 Inhibits Collagen Type I Production in Dermal Fibroblasts via an Sp1-dependent Pathway. J Biol Chem. 2001;276:20839–48. doi: 10.1074/jbc.M010133200. [DOI] [PubMed] [Google Scholar]
- 48.Hodge D, Robinson L, Watson D, Lautenberger J, Zhang X, Venanzoni M, Seth A. Interaction of ETS-1 and ERGB/FLI-1 proteins with DNA is modulated by spacing between multiple binding sites as well as phosphorylation. Oncogene. 1996;12:11–18. [PubMed] [Google Scholar]
- 49.Thaler R, Agsten M, Spitzer S, Paschalis EP, Karlic H, Klaushofer K, et al. Homocysteine suppresses the expression of the collagen cross-linker lysyl oxidase involving IL-6, Fli1, and epigenetic DNA methylation. J Biol Chem. 2011;286:5578–88. doi: 10.1074/jbc.M110.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viedt C, Dechend R, Fei J, Hänsch GM, Kreuzer J, Orth SR. MCP-1 induces inflammatory activation of human tubular epithelial cells: involvement of the transcription factors, nuclear factor-kappaB and activating protein-1. J Am Soc Nephrol. 2002;13:1534–47. doi: 10.1097/01.asn.0000015609.31253.7f. [DOI] [PubMed] [Google Scholar]