Abstract
Identifying new biomarkers and therapeutic targets for podocytopathies such as focal segmental glomerulosclerosis (FSGS) requires a detailed analysis of transcriptional changes in podocytes over the course of disease. Here we used translating ribosome affinity purification (TRAP) to isolate and profile podocyte-specific mRNA in two different models of FSGS. Expressed eGFP-tagged ribosomal protein L10a in podocytes under the control of the Collagen-1α1 promoter enabled podocyte-specific mRNA isolation in a one-step process over the course of disease. This TRAP protocol robustly enriched known podocyte-specific mRNAs. We crossed col1α1-L10a mice with the actn4−/− and actn4+/K256E models of FSGS and analyzed podocyte transcriptional profiles at 2, 6 and 44 weeks of age. Two upregulated podocyte genes in murine FSGS (CXCL1 and DMPK) were found to be upregulated at the protein level in biopsies from patients with FSGS, validating this approach. There was no dilution of podocyte-specific transcripts during disease. These are the first podocyte-specific RNA expression datasets during aging and in two models of FSGS. This approach identified new podocyte proteins that are upregulated in FSGS and help define novel biomarkers and therapeutic targets for human glomerular disease.
Introduction
Glomerular diseases are an important cause of chronic kidney disease (CKD) and end stage renal disease (ESRD) in both pediatric and adult populations. Primary glomerulonephritis accounts for 14% of childhood CKD in the United States, and is the leading cause of CKD in children older than 12 years of age (1). The main cause of glomerulonephritis in this group is FSGS, which is three times more common in blacks than whites (2). FSGS is the most common cause of ESRD from glomerulonephritis in adults, and its incidence has risen substantially over the last two decades (3).
Although great progress has been made in understanding the genetic and pathophysiologic basis of FSGS, disease biomarkers and targeted therapies are still lacking. Efforts to elucidate the biological properties of podocytes have included microarray analysis of whole kidney, isolated glomeruli and recently, FACS-sorted individual podocytes. Laser capture microdissection (LCM) has also been used for isolation of glomerular mRNA (4, 5). Dynabead perfusion combined with enzymatic digestion and sieving methods has proven effective in isolating glomeruli from mouse kidney at a large scale in vivo and has greatly facilitated the establishment of glomerular transcription profiles (6–8). Very recently, magnetic bead and transgenic models for FACS purification have been successfully applied to separate podocytes from other glomerular cells improving enrichment of podocyte specific RNA (9–11). These techniques however require enzymatic and mechanical disaggregation of kidney tissue for the creation of single cell suspensions that introduce a stress-response gene signature.
To overcome these limitations we applied Translating Ribosome Affinity Purification (TRAP), a novel method to isolate polysomal mRNA from defined cell populations in vivo (12) to extract podocyte specific mRNA from the kidney. This approach has been validated in detecting distinct expression profiles and their changes in rare cell populations (12, 13), and it has two major advantages over other techniques: First, TRAP does not require enzymatic digestion and cell disaggregation, and second, it isolates polysomal RNA which is being translated, and this correlates better with actual protein expression (14). Here, we report the isolation of podocyte specific mRNA in vivo from a Collagen-1α1-eGFP-L10a transgenic mouse by affinity purification. We use this tool to (i) define the translational signature of adult podocytes, (ii) study translational profiles of podocytes over time, and (iii) investigate changes in podocyte gene expression in two genetic mouse models of FSGS.
Results
Generation and characterization of podocyte-specific transgenic TRAP mice
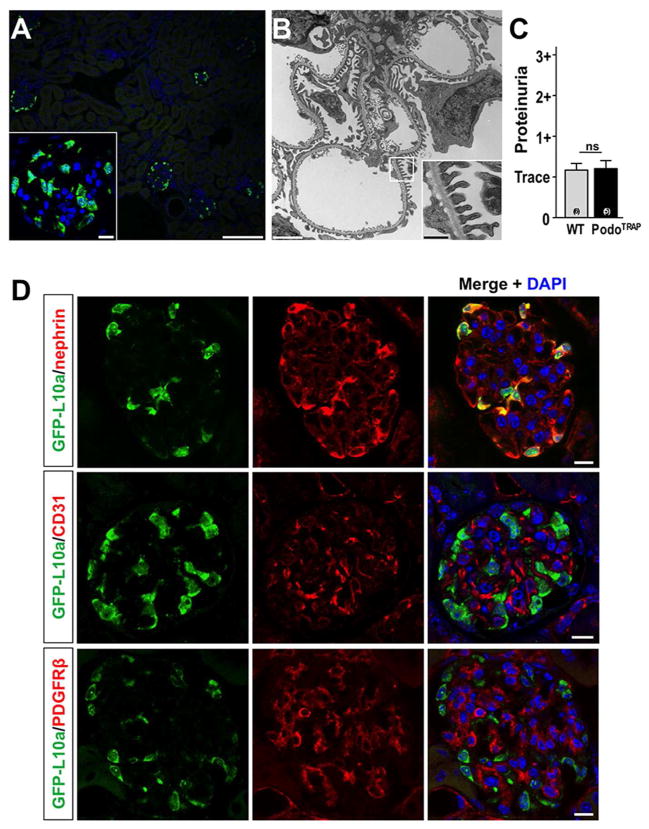
We first confirmed that kidneys from Collagen-1a1-eGFP-L10a (Col1α1-eGFP-L10a; PodoTRAP) mice expressed the eGFP-L10a transgene in podocytes. EGFP-L10a epifluorescence was restricted to glomeruli in kidney cortex (Fig. 1A) and tubulointerstitial cells in the medulla (data not shown). EGFP-L10a expressing glomerular cells were located at the outer aspect of the glomerular tuft and positive cells exhibited marked fluorescence of the perinuclear region and nucleoli, characteristic of a ribosomal distribution (12) (Fig. 1A, insert). Podocyte foot processes in Col1α1-eGFP-L10a podocytes appeared normal by electron microscopy and there was no difference in proteinuria between adult transgenic mice and their littermate controls (Fig. 1, B and C). Cell-type specific immunostaining on Col1α1-eGFP-L10a kidney sections confirmed that the fusion protein was expressed only in podocytes and not in endothelial or mesangial cells (Fig. 1D).
Figure 1. Validation of Col1α1-eGFP-L10a podocyte expression.
(A) eGFP-L10a epifluorescence in kidney cortex is localized to the outer aspect of the glomerular tuft in Col1α1-eGFP-L10a mice. Insert: nucleolar and perinuclear localization of epifluorescence, consistent with ribosomal expression pattern. Scale bars: 100μm and 10μm (insert), respectively. (B) Electron microscopic analysis reveals normal podocyte ultrastructure in Col1α1-eGFP-L10a kidneys. Insert: Foot processes appear normal. Scale bars: 2μm and 0,5μm (insert), respectively. (C) Semiquantitative analysis of proteinuria by dipstick shows no difference between adult Col1α1-eGFP-L10a and wild-type control mice. (D) Immunostaining for podocyte-specific marker nephrin (red) co-labels cells that express the eGFP-tagged L10a (green). By contrast, immunostaining for endothelial cells (anti-CD31) or mesangial cells (anti-PDGFRβ) shows no overlap with cells expressing eGFP-L10a. Scale bars: 10μm.
Extraction of podocyte-specific polysomal mRNA by TRAP
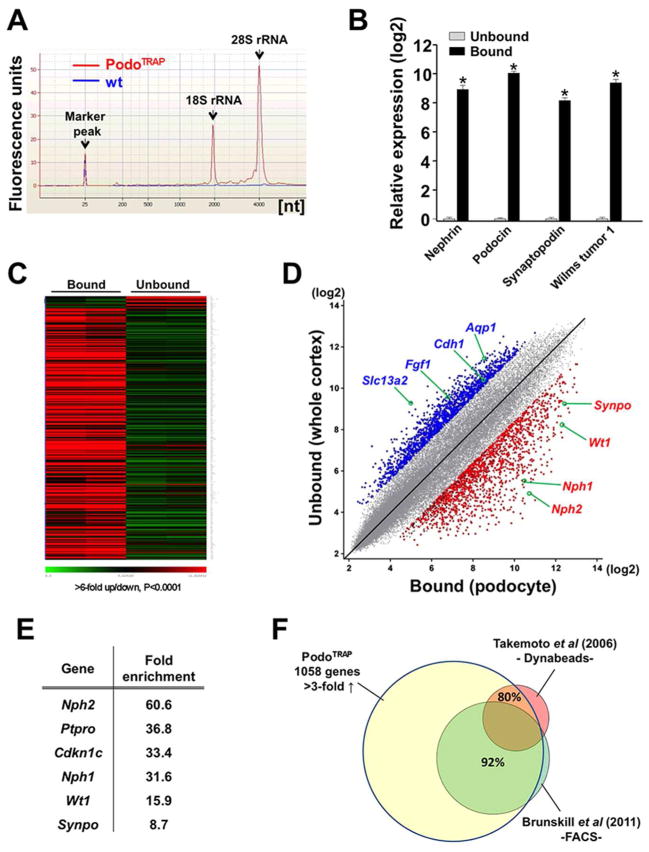
We performed TRAP on kidney cortex from adult Col1α1-eGFP-L10a mice and analyzed RNA quality and quantity by Bioanalyzer Pico-chip. To assess the background we routinely processed tissue from wild-type controls in parallel. Bound fractions of Col1α1-eGFP-L10a mice yielded a total of 50–200ng of high quality RNA with RINs >9 whereas samples of wild-type controls showed only traces of RNA, indicating minimal nonspecific RNA carryover (Fig. 2A). To validate that bound mRNA was podocyte-specific and unbound mRNA reflected whole kidney cortex, we compared expression of several podocyte marker genes in the two fractions by quantitative RT-PCR analysis. Remarkably, we measured at least 200-fold enrichment for established podocyte genes including nephrin, podocin, synaptopodin and WT1 in the bound fraction compared to the unbound fraction (Fig. 2B).
Figure 2. Podocyte TRAP in Col1α1-eGFP-L10a (PodoTRAP) mouse.
(A) Representative Bioanalyzer run with PicoChip (Agilent Technologies) detects high quality RNA (RIN 9.5) purified from a bound fraction of Col1α1-eGFP-L10a kidney cortex (red) but not in a wild-type littermate control (blue). (B) RT-PCR analysis indicates strong enrichment of podocyte-specific transcripts in bound vs. unbound fractions. Logarithmic scale on y axis; *P<0.01. (C) Heatmap of 341 probesets with >6-fold over-/underrepresentation (P<0.0001) in bound (podocyte) versus unbound (whole cortex) fractions of two adult Col1α1-eGFP-L10a animals. Green marks low and red marks high expression levels. Probesets were sorted by level of over-/underrepresentation of RNA messages. (D) Scatterplot of normalized expression values of bound ‘TRAP’ RNA (podocyte) versus unbound RNA (whole cortex) illustrates robust enrichment of podocyte-specific messages (red dots; >3-fold up, P<0.05) and depletion of non-specific messages (blue dots; >3-fold down, P<0.05) in the immunoprecipitated, bound fraction. Selected probesets of known podocyte genes (red) or non-podocyte genes (blue) are highlighted. (E) Selection of widely accepted podocyte marker genes and their fold-enrichment in the immunoprecipitated fraction of PodoTRAP kidney cortex versus whole cortex as determined by microarray analysis. (F) Comparison of TRAP-generated transcriptome of adult podocytes (1058 genes, >3-fold up, P<0.05) with published podocyte-enriched expression profiles using alternative enrichment methods. Approximately 80% of podocyte marker genes proposed by Takemoto et al (7) were represented in the translational profile. 92% of annotated genes in the podocyte transcriptome with 3-fold or higher overrepresentation compared to total cortex as reported by Brunskill et al (10) were also found in the translational profile.
Next, we performed microarray analysis (Affymetrix Mouse Genome 430 2.0) on RNA in both fractions and generated heatmaps as well as scatterplots with the log signal intensity for the bound (B) fraction (‘podocyte-specific’) on the x-axis and the unbound (UB) fraction (‘whole cortex’) on the y-axis (Fig. 2, C and D). The ratio B/UB reflects the level of podocyte RNA enrichment which aids in identifying podocyte-specific messages (Supplemental Figure S1). Thus the probability of a gene being specific to the cell type in question grows as one travels from the upper left to the lower right corner of this plot (15). Consistent with the qPCR analysis, the scatterplot revealed that signal intensities for podocyte-specific genes were multiple times stronger in bound compared to unbound fractions of Col1α1-eGFP-L10a renal cortex, further validating our approach (Fig. 2D). Those highly enriched messages in the bound fraction include Nphs2 (podocin, 60.6-fold), Nes (nestin, 39.5-fold), Angptl2 (angiopoietin-like 2, 36.8-fold), Ptpro (protein tyrosine phosphatase, receptor type, O, 36.8-fold), Nphs1 (nephrin, 31.6-fold), Wt1 (Wilms tumor 1, 15.9-fold) and Synpo (synaptopodin, 8.7-fold) (Figure 2E). Genes typically expressed in other kidney cell types such as endothelial cells (e.g. Pecam1, Vwf, Tie1), mesangial cells (e.g. Thy1), proximal tubule epithelial cells (e.g. Cdh1, Aqp1) or collecting duct cells (e.g. Aqp2) were not enriched, confirming cell specificity of the data set.
Generation and systematic analysis of the podocyte translational profile
To define the baseline adult mouse podocyte translational profile we filtered probesets that met a > 3-fold enrichment and P value < 0.05. After removal of redundancies and unannotated probesets, 1058 individual genes were identified in the podocyte-specific fraction compared to the whole cortex (Supplemental Table 1). We cross-referenced this rather inclusive gene list with two previously published data sets generated by microarray analysis of podocyte-enriched RNA obtained by alternative enrichment methods (7, 10). For instance, Takemoto et al. have used the magnetic bead perfusion method to collect and specifically extract RNA from isolated glomeruli (7). Nearly 80% of genes defined by Takemoto and colleagues as podocyte marker candidates were contained in our translational podocyte profile. Brunskill et al have used the MafB-GFP mouse line to FACS-sort GFP-expressing podocytes, extract RNA and perform microarray analysis (10). When matching the list of genes reported by Brunskill and colleagues to define adult podocytes (>3-fold up) with our translational profile we found an even higher concordance of 92% (Fig. 2F). Genes that were not significantly enriched in our translational profile and thus discordant comprised a number of genes involved in cell-matrix adhesion processes and the integrin-mediated signaling pathway (e.g. Itga1, Itga2, Itgav, Itgb1bp1, Ctgf) as well as in locomotion (e.g. Robo2, Col4a4) and the positive regulation of cell proliferation (e.g.Ccnd2, Cyr61, Mll2). While speculative, it is possible that the stresses of cell isolation and FACS sorting prior to RNA extraction may have induced these damage-response genes.
We performed gene ontology (GO) analysis using DAVID (the database for annotation, visualization and integrated discovery) bioinformatics resources to identify biological processes more relevant to podocyte function (16, 17). Using a gene functional classification tool we first defined 37 gene groups (Supplemental Table 2). Next, we created a functional annotation chart reporting 487 overrepresented (fold enrichment ≥1.5) and statistically significant (P<0.05) biological terms in our profile such as actin cytoskeleton organization (3.7 fold, P=6.9E-10), cytoskeletal protein binding (3.2 fold, P=1.7E-17), actin binding (3.2 fold, P=4.6E-12), wnt signaling pathway (2.9 fold, P=1.2E-4), vasculature development (2.6 fold, P=1.0E-6), and neuron differentiation (2.0 fold, P=2.4E-5) (Supplemental Table 2). To further organize and simplify the annotation results we performed functional annotation clustering and compressed the profile list into smaller, term-centric annotation modules. This pooling also provides more comprehensive gene lists than gene selections from individual terms only. We found a total of 85 clusters with a group enrichment score (GES) of ≥ 1, which can be used as a measure to rank their biological significance. Annotation clusters such as cytoskeletal protein/actin-binding (GES: 11.97), actin cytoskeleton organization (GES: 7.6), and blood vessel development/morphogenesis (GES: 4.52) ranked high in the output (Supplemental Table 2, Supplemental Figure S2).
We sorted genes for those most highly enriched using an 8-fold cutoff for the podocyte-specific fraction compared to whole cortex (Supplemental Table 1). We used MetaCore (GeneGo) to analyze this subset of highly enriched genes and found the top three enriched process networks to be cytoskeleton/actin filaments (P=3.3E-8), development/ neurogenesis/axonal guidance (P=1.9E-7) and cell adhesion/synaptic contact (P=2.5E-6) (Supplemental Table 3), consistent with the highly complex architecture and cytoskeletal organization of podocytes.
Application of podocyte TRAP in two genetic models of FSGS
In order to define the podocyte transcriptome over the course of FSGS, we crossed the Col1a1-eGFP-L10a transgenic mouse to two models of FSGS: the α-actinin 4-knockout (Actn4−/−), which develops a severe renal phenotype with high perinatal mortality (18), and the α-actinin 4 K256E point mutant knockin (Actn4+/K256E), which harbors a human disease-associated point mutation and is characterized by only very minor irregularities in podocyte ultrastructure at an older age (19, 20) (Fig. 3A). We were particularly interested in the latter model, as it would allow us to measure early gene changes, potentially before the appearance of clear morphologic hallmarks of FSGS. In line with previous reports, the number of offspring with a Col1α1-eGFP-L10a;actn4−/− genotype was lower than expected by Mendelian ratios as a consequence of increased perinatal mortality in mice deficient in α-actinin 4 (18).
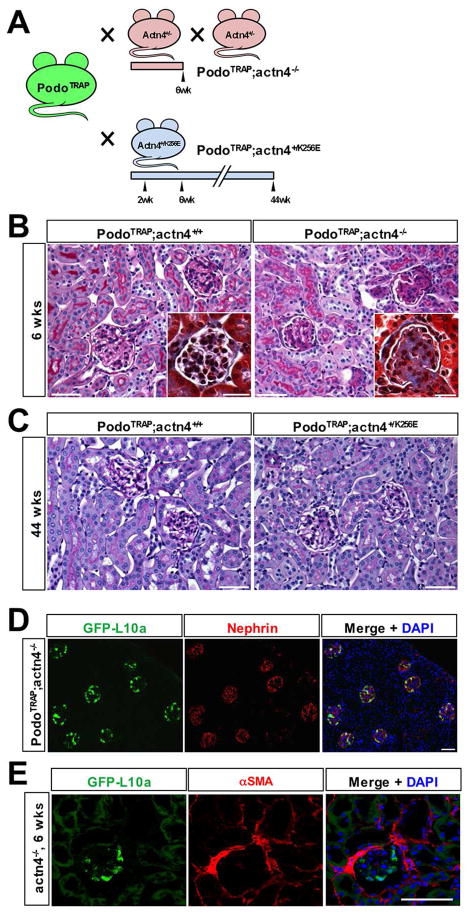
Figure 3. FSGS models used for podocyte-TRAP.
(A) Breeding strategy and time points of kidney harvest: Col1α1-eGFP-L10a (PodoTRAP) mice were crossed to heterozygous Actn4+/− mice to obtain bigenic Col1α1-eGFP-L10a; actn4−/− offspring with a rapidly progressive, severe FSGS phenotype and to mutant Actn4+/K256E mice to produce bigenic Col1α1-eGFP-L10a;actn4+/K256E offspring with a subtle phenotype, respectively. Kidneys of FSGS-prone and Actn4+/+ littermate control Col1α1-eGFP-L10a mice were harvested for TRAP and histological examination at 2, 6 and 44 weeks of age. (B) PAS and Masson-trichrome (inserts) stained kidney sections. As expected, kidneys from Col1α1-eGFP-L10a ;actn4−/− but not from PodoTRAP;actn4+/+ control mice showed overt signs of focal and segmental glomerulosclerotic changes validating the severity of this glomerular disease model. Scale bars: 50μm and 20μm (inserts). (C) PAS stained kidney sections of 44-week-old Col1α1-eGFP-L10a;actn4+/K256E and Col1α1-eGFP-L10a;actn4+/+ control mice. Even at an advanced age mutant Col1α1-eGFP-L10a;actn4+/K256E maintain a normal appearing kidney architecture. Scale bars: 50μm. (D) Anti-nephrin staining (red) of Col1α1-eGFP-L10a;actn4−/− kidney cortex at 6 weeks of age. EGFP-L10a expression (green) is confined to podocytes indicating that kidney disease does not alter transgene expression in cortex. Scale bar: 50μm. (E) αSMA staining (red) and eGFPL10a expression (green) in 6 week Col1α1-eGFP-L10a;actn4−/− kidney cortex shows that eGFPL10a is not induced in fibrotic interstitium but remains localized to podocytes. Scale bar: 50μm.
As expected, Col1α1-eGFP-L10a;actn4−/− mice developed spontaneous proteinuria (data not shown) and histological examination revealed overt signs of focal and segmental glomerulosclerosis at six weeks of age (Fig. 3B). By contrast, bigenic Col1α1-eGFP-L10a;actn4+/K256E mice expressing a mutated, disease-associated variant of α-actinin 4 showed no histological abnormalities at up to 44 weeks of age when compared to Col1α1-eGFP-L10a;actn4+/+ controls (Fig. 3C). Importantly, eGFP-L10a continued to be strongly and specifically expressed in podocytes of Col1α1-eGFP-L10a; actn4−/− and Col1α1-eGFP-L10a;actn4+/K256E mice and no leaky expression was observed in tubules, the cortical tubulointerstitium or other glomerular cells as validated by immunostaining (Fig. 3D, Supplemental Figure S3). TRAP was performed at 6 weeks of age for Col1α1-eGFP-L10a;actn4−/− and at 2, 6 and 44 weeks of age for Col1α1-eGFP-L10a;actn4+/K256E mice, along with their wild-type controls (Col1α1-eGFP-L10a;actn4+/+). RNA yields and quality were comparable between the different genotypes. Only samples with a RIN of ~9 or higher were processed for microarray analysis. Importantly, there was no upregulation of the eGFPL10a transgene in cortical myofibroblasts in 6 week kidneys from eGFP-L10a;actn4−/− mice, confirming that TRAP from cortex is specific to podocytes. By contrast, we have recently shown that the eGFPL10a transgene is expressed in medullary pericytes and myofibroblasts during fibrosis (21).
Podocyte transcriptome changes with age
After data normalization we first performed a principal component analysis (PCA) on all array data from Col1α1-eGFP-L10a;actn4+/K256E (knockin, ‘ki’) and wild-type controls (‘wt’) of different age to test whether variances in the data could be captured independently. The first principal components explained ~72% (P<0.005) of the variance due to differences in age and there was good clustering within groups (Supplemental Figure S4). Notably, expression levels of most top podocyte marker genes were largely comparable between the different age groups and genotypes. We identified 611 probesets that were differentially regulated (2-fold up or down) between 2- and 6-week-old wild-type podocytes (Supplemental Table 4A). Among the downregulated genes were neurotrophin-3 (Ntf3, a.k.a. nerve growth factor 2), which has been reported to promote survival and axonal projection of sensory neurons (22), angiotensin I converting enzyme 2 (Ace2), which has been shown to be expressed in podocytes (23) and to attenuate diabetic nephropathy when overexpressed in podocytes (24), angiotensinogen (Agt), angiopoietin 2 (Angpt2), the serine proteinase inhibitor Serpina 6, and the matrix components fibronectin 1 (Fn1) and collagen, type IV, alpha 1 (Col4a1). Among the upregulated genes were tropomodulin-2 (Tmod2, a.k.a. neuronal tropomodulin), which blocks the elongation and depolymerization of actin filaments at the pointed ends in neurons (25), neuroglobin (Ngb), a novel hypoxia-inducible globin first discovered in brain (26) and recently identified in podocytes with postulated protective roles (27), growth arrest-specific protein 7 (Gas7), which has been shown to regulate neuronal maturation and neurite outgrowth (28), and toll-like receptor 7 (Tlr7) (Fig. 4A).
Figure 4. Podocyte transcriptome changes with age.
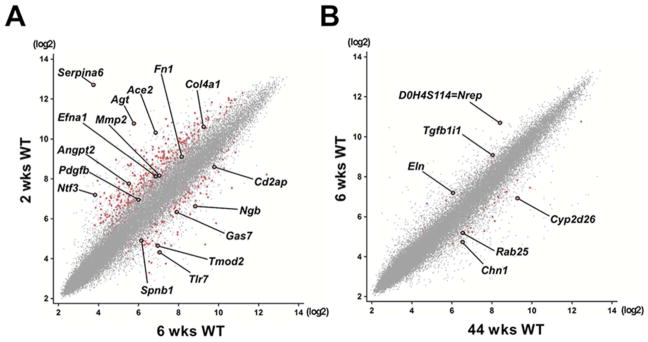
Scatterplots of normalized expression values illustrate distinct differences in translated RNA levels between wild-type podocytes of different age. Axes represent probeset-specific signal intensities (log2) captured in one representative individual of each age group. Each dot represents one probeset, differentially regulated probesets are marked in red. A selection of differentially regulated probesets is highlighted and identified by gene symbol. (A) TRAP profile comparing podocytes at 2 and 6 weeks. Note that a larger number of genes is downregulated than upregulated going from 2 weeks to 6 weeks. (B) Between 6 weeks and 44 weeks, there are very few gene expression changes in podocytes identified by TRAP.
GO analysis of the 2 vs. 6 week samples showed that significantly enriched processes included anatomical structure morphogenesis (9.4E-23), cell differentiation (9.3E-14), vascular development (1.7E-11), locomotion (1.0E-10), and neuron projection morphogenesis (1.8E-10). Significantly enriched localizations included the extracellular matrix (P=4.4E-14), basement membrane (1.8E-10), and microfibril (P=1.4E-5). Together, these terms point to changes in podocyte motility during the transition from postnatal to juvenile age. Interestingly, among the most significantly enriched molecular functions were type 2 angiotensin receptor binding (P=5.6E-14), acetyltransferase activator activity (P=4.9E-13), alpha1-adrenergic receptor activity (8.7E-12), extracellular matrix structural constituent (4.0E-8), and G-protein coupled receptor binding (2.8E-6) (Supplemental Table 4B). These terms highlight podocyte regulation by extracellular ligands and matrix in the first six weeks postnatally. By contrast, the comparison between the translational profiles of 6- and 44-week-old wild-type podocytes yielded only 37 probesets with ≥2-fold up- or downregulation (Fig. 4B, Supplemental Tables 5, A and B). Particularly interesting among this limited geneset was TGFB1l1, also called HIC-5, which was downregulated 2 fold with age. TGFB1i1 is a focal adhesion protein that binds directly to Pleckstrin homology domain-containing, family H (with MyTH4 domain), member 2 (Plekhh2). Plekhh2 was recently identified as a novel podocyte protein, present in foot processes, and downregulated in human FSGS (29).
Translational profile of podocytes from Actn4−/− mice with FSGS differs importantly from wild-type control
Next, we compared the translational profiles of Actn4−/− and wild-type control podocytes at 6 weeks of age and found 503 probe sets that were differentially regulated ≥2-fold (Supplemental Table 6A). Interestingly, gene expression levels of accepted podocyte markers such as Wt1, Nphs1, Nphs2, Synpo and Podxl were not downregulated in podocytes from Col1α1-eGFP-L10a;actn4−/− animals despite histologic evidence of FSGS, in contrast to a recent report from Hodgin and colleagues, that showed lower expression levels for these genes in human kidney samples with histologically proven FSGS (30). However, this study used laser-capture microdissection to isolate RNA from whole glomeruli and since podocyte loss is a hallmark of FSGS (31, 32), the likely explanation for this finding is dilution of podocyte-specific transcripts in whole glomerulus caused by selective podocyte loss. Genes with ≥ 2-fold upregulation in Actn4−/− podocytes included the matrix metalloproteinases (MMP) 10 and 2, latter of which has been recently associated with glomerular injury in patients with chronic humoral rejection of kidney allografts (33), the connective tissue growth factor (Ctgf), which has been demonstrated to be upregulated in podocytes in diabetic nephropathy (34) and to play a detrimental role when overexpressed in podocytes (35), as well as the NF-kappa-B inhibitor alpha (Nfkbia) and the proinflammatory chemokines Cxcl1 and Cxcl10, all of which show induction in podocytes upon exposure to TNFα and are implicated in a NF-κB-mediated response (36). The 25 most highly upregulated genes in Actn4−/− podocytes at 6 weeks compared to wildtype are presented in Table 1.
Table 1.
Top 25 podocyte-specific upregulated genes in Actn4−/− vs. wildtype podocytes at 6 weeks of age1
| Symbol | Name | Fold Change | P-value |
|---|---|---|---|
| Mmp10 | Matrix metalloproteinase-10 | 26.5 | 0.014 |
| Cxcl1 | Chemokine (C-X-C Motif) Ligand 1 | 13.8 | <0.001 |
| Adamts4 | ADAM metallopeptidase with thrombospondin type 1 motif, 4 | 10.6 | 0.009 |
| Timp1 | Tissue Inhibitor of Metalloproteinases 1 | 9.9 | 0.016 |
| Aldh1a7 | Aldehyde dehydrogenase family 1, subfamily A7 | 8.1 | 0.012 |
| Pp11r | Endonuclease, polyU-specific | 7.7 | <0.001 |
| Fam46b | Family with sequence similarity 46, member B | 7.5 | <0.001 |
| Col8a1 | Collagen, type VIII, alpha 1 | 6.9 | 0.012 |
| Clca1 /// Clca2 | Chloride channel calcium activated 1/2 | 6.4 | 0.011 |
| Rcan2 | Regulator of calcineurin 2 | 6.3 | 0.003 |
| Gadd45b | Growth arrest and DNA-damage-inducible 45 beta | 6.3 | 0.003 |
| Ifi202b | Interferon activated gene 202B | 6.2 | 0.046 |
| Glipr2 | GLI pathogenesis-related 2 | 6.1 | <0.001 |
| Crct1 | Cysteine-rich C-terminal 1 | 6.1 | 0.035 |
| Crygn | Crystallin, gamma N | 6.0 | <0.001 |
| Prss23 | Protease, serine 23 | 5.9 | <0.001 |
| Rnd1 | Rho family GTPase 1 | 5.6 | <0.001 |
| Adamts8 | ADAM metallopeptidase with thrombospondin type 1 motif, 8 | 5.5 | <0.001 |
| Col3a1 | Collagen, type III, alpha 1 | 5.3 | 0.026 |
| Pamr1 | Peptidase domain containing associated with muscle regeneration 1 | 5.3 | <0.001 |
| Mfap5 | Microfibrillar associated protein 5 | 5.1 | 0.004 |
| Mmp19 | Matrix metallopeptidase 19 | 5.0 | <0.001 |
| Cd44 | CD44 antigen | 4.9 | 0.012 |
| Birc5 | Baculoviral IAP repeat-containing 5 | 4.9 | 0.015 |
| Il33 | Interleukin 33 | 4.7 | 0.022 |
Fold change is the ratio of Actn4−/− vs. wildtype expression at 6 weeks.
Other notable, strongly induced genes in Actn4−/− podocytes included crystalline, gamma N (Crygn), a member of the crystallin superfamily that shows neurite-promoting properties through putative autocrine and paracrine effects (37, 38), the regulatory protein ‘growth arrest and DNA-damage-inducible 45 beta’ (Gadd45b), which has only recently been demonstrated to promote adult neurogenesis via epigenetic DNA demethylation (39), and toll-like receptor 4 (Tlr4), which has been suggested to directly associate the podocyte with the innate immune system to mediate glomerular injury in membranoproliferative glomerulonephritis (40) and which may offer a mechanistic link to the observed upregulation of Cxcl1 and other NF-κB-regulated genes. Among genes that were ≥2-fold downregulated in Actn4−/− podocytes we identified e.g. cyclase-associated protein 1 (Cap1), a highly conserved monomeric actin binding protein which has been attributed a key role in speeding up the turnover of actin filaments via recycling cofilin and actin and which furthermore seems to provide a direct link from the actin cytoskeleton to the mitochondria by operating as an actin shuttle (41–43), the transcription factor Zfp423, which has been demonstrated to control proliferation and differentiation of neural precursors (44) and to regulate the transition from differentiation to maturation in olfactory neurogenesis (45), and neural cell adhesion molecule 1 (Ncam1) that is known to be involved in neuron-neuron adhesion, neurite fasciculation and outgrowth of neurites (46, 47). Other genes with ≥2-fold decreased expression levels Actn4−/− podocytes included the inositol 1,4,5-triphosphate receptor 3 (Itpr3), which mediates the release of intracellular calcium, and the cytoskeletal protein spectrin beta 1 (Spnb1) (Fig. 5A).
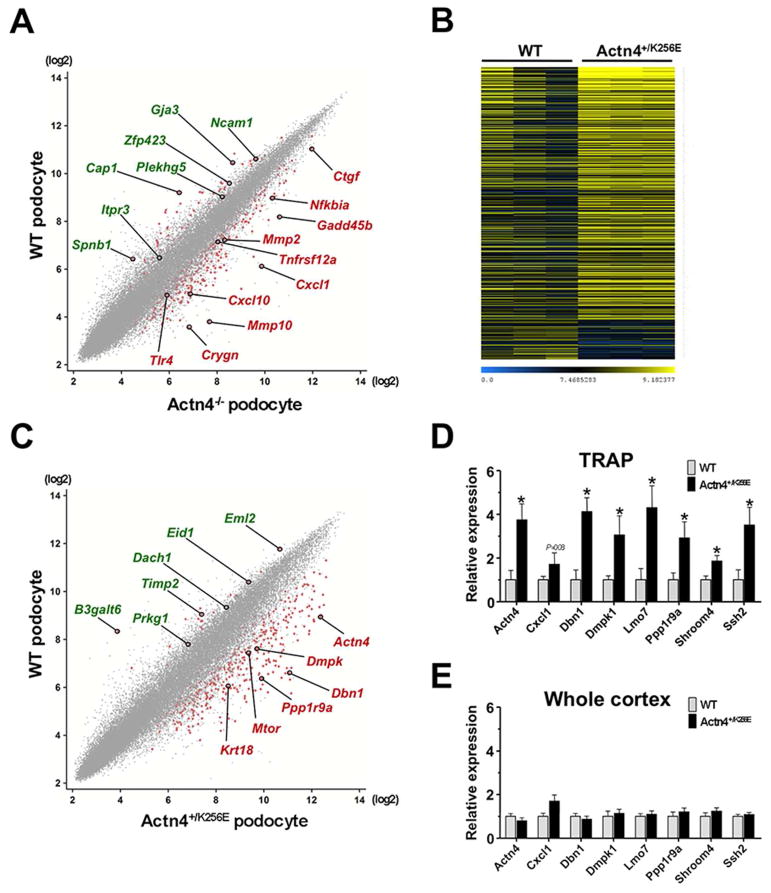
Figure 5. Podocyte TRAP in models of genetic FSGS.
(A) Scatterplot of normalized gene expression values demonstrating differences in podocyte-specific translational profiles between Actn4 knockout and wild-type control mice at 6 weeks. Red dots represent significantly up- or downregulated genes. A selection of differentially regulated probesets including Cxcl1, Tlr4 and Mmp10 is highlighted and identified by gene symbol (red symbols = upregulated; green symbols = downregulated). (B) Heatmap of 725 probesets with >2-fold up-/downregulation (P<0.05) of translated podocyte messages in six week old Col1α1-eGFP-L10a;actn4+/K256E vs. Col1α1-eGFP-L10a;actn4+/+ (WT) control mice (three representative animals per group). Blue marks low and yellow high expression levels. Probesets were sorted according to fold increase of expression. (C) Scatterplot of normalized gene expression values illustrating differences in podocyte-specific translational profiles between mutant Actn4+/K256E and wild-type control mice at 6 weeks. Red dots represent significantly up- or downregulated genes. Selected differentially regulated probesets including Dmpk, Actn4 and Ppp1r9a (Neurabin 1) are pointed out and identified by gene symbol (red symbols = upregulated; green symbols = downregulated). (D and E) Quantitative PCR analysis of reverse transcribed RNA from immunoprecipitated (IP) podocyte-specific fractions and unbound fractions representing the whole cortex. PCR results confirm a significant upregulation of selected genes in IP fractions of Actn4+/K256E compared to wild-type control as previously identified by microarray analysis. Note that these changes are not detected in whole cortex indicating a dilution of podocyte messages by the aggregate of unspecific transcripts from the rest of the cells constituting the kidney cortex. Data are shown as mean ± SEM, n=4–8 for each data point; *P<0.05.
GO-based gene enrichment analysis showed that some of the most significantly overrepresented processes included extracellular matrix organization (P=1.7E-19), response to wounding (P=9.5E-19), response to stress (P=8.0E-16), and locomotion (P=1.5E-12). Among enriched molecular functions were polysaccharide binding (P=4.6E-10), pattern binding (P=4.6E-10), integrin binding (P=1.3E-8), and metalloendopeptidase activity (P=5.5E-6). Significantly enriched localizations included extracellular region part (P=2.8E-22), basement membrane (P=3.0E-10), and microfibril (P=5.0E-6). The top ten enriched pathway maps included Cell adhesion - ECM remodeling (P=1.7E-8), Development - TGF-beta-dependent induction of EMT via SMADs (P=2.0E-6), and Immune response – TLR signaling pathways (P=4.0E-5) and the top ten enriched process networks included Cell adhesion – cell-matrix interactions (P=2.0E-14), Proteolysis – ECM remodeling (P=1.3E-6), Signal transduction – Wnt signaling (1.3E-6), and Proteolysis – connective tissue degradation (P=2.4E-6). Collectively, these GO terms highlight increased podocyte motility and interaction with basement membrane during FSGS.
In addition, we conducted an interactome analysis to identify likely interactions partners of the genes in the data. Overconnected transcription factors included SP1, RelA (p65 NF-κB subunit), NF-κB1 (p50), and c-Rel (NF-κB subunit), overconnected receptors included toll-like receptor 6, Calcitonin receptor, Syndecan-4, and Integrin alpha V. Overconnected ligands included Decorin, Fibrillin 1, Wnt3a, Thrombospondin 1, Fibronectin, and TGFβ1. Overconnected proteases included amongst others several MMPs (1, 2, 7, 8, 11, 12, 13, 14, 15, 17, 24, 25), Stromelysin-2, and Caspase3. (Supplemental Table 6B).
Finally, we performed network analysis to relate gene expression changes during FSGS to generalized podocyte functions (Supplemental Table 7). Two clear gene ontologies emerged from this network analysis, those of response to wounding, and regulation of cellular processes. For example, pathways regulated by ligands or pathways regulating transcription factors both contained multiple NF-κB network terms, as well as other injury response networks such as STAT5, SP1 and c-Fos. Key transcription factors were largely overlapping, including NF-κB, STAT, SP1 and p53 pathways. Cellular process terms were present in both pathways regulated by ligands or pathways regulating transcriptional networks. Examples of genes within these networks include TGF-beta receptor type I and II, alpha-V/β-3, 5 and 6-integrins, Syndecan-4 and Tcf(Lef). Overall, this analysis further highlights the idea that FSGS presents a cellular stress to podocytes which respond with a coordinated cellular injury response.
Translational profile of podocytes from FSGS-prone Actn4+/K256E mice is distinct from wild-type
Next, we compared the translational profiles of Actn4+/K256E with wild-type control podocytes across different time points: at 2 weeks (juvenile), 6 weeks (young adult) and 44 weeks (old adult) of age. Again, gene expression levels of most established, podocyte defining markers were comparable between genotypes. Taking all age groups into account, we found 403 probe sets that were significantly ≥1.5-fold up- or downregulated between genotypes (P<0.005; Supplemental Table 8A). Upregulated genes in Actn4+/K256E mutant podocytes included myotonic dystrophy protein kinase (Dmpk), a Rho-associated serine-threonine protein kinase which has been shown to be prominently expressed in myogenic and neural cell lineages and to play a role in actin-based cell morphodynamics (48, 49), the actin-binding protein filamin B (Flnb) which has been demonstrated to assemble in a protein complex with Ship2 and lamellipodin through nephrin-mediated recruitment in podocytes and contribute to focal adhesion dynamics and lamellipodia formation (50), and the tight junction protein cingulin (Cgn) that has recently been identified as a component of slit diaphragms and shown to be upgregulated in glomeruli in PAN induced nephrosis in rat (51). Of note, alpha-actinin 4 (Actn4) was also among the top 100 upregulated genes in Actn4+/K256E mutant podocytes which is in line with our prior observations suggesting an increase in transcription and translation to compensate for enhanced degradation of the mutant Actn4 protein (19). Additional robustly induced probe sets included genes that encode for neurabin-1 (Ppp1r9a), an F-actin binding protein that recruits protein phosphatase 1a to the cytoskeleton, regulates actin dynamics and neurite outgrowth and instructs synapse assembly (52–54), actin-binding protein drebrin-1 which once was thought to be neuron specific but is also enriched in processes of podocytes (55), slingshot-2 (Ssh2), a phosphatase that dephosphorylates cofilin which is necessary in maintaining podocyte architecture (56), and a number of myosin members such as Myh9, Myh10, Myo1b, Myo1e, Myo6, Myh14, Myo5a, Myo5b, Myo7a, Myo7b, Myo15b and Myo18a, some of which have been described as putative components of the actin-associated cytoskeletal protein network in glomerular podocytes or associated with FSGS (57, 58). Interestingly, downregulated genes in Actn4+/K256E podocytes included tissue inhibitor of metalloproteinase 2 (Timp2) which, when knocked down, was shown to promote cell migration in cultured podocytes (59), and the nuclear factor dachshund 1 (Dach1) that has been recently identified as eGFR-related locus by genome-wide association studies and shown to be associated with incident CKD (60) (Fig. 5, B and C).
To validate the microarray data we performed quantitative PCR on reverse transcribed RNA from bound (podocyte-specific) and unbound (whole cortex) fractions of both genotypes and compared expression values of selected genes that - according to the unbiased microarray approach - were increased in podocytes with the heterozygous K256E mutation in the Actn4 gene. Consistent with the microarray results we found a robust, podocyte-specific upregulation of all tested genes in Actn4+/K256E animals compared to wild-type controls (Fig. 5D). Of note, these differences were not captured when we compared samples from whole cortex illustrating inherent dilution of podocyte-specific messages by other renal cell types (Fig. 5E).
Enrichment analysis on the data set of differentially regulated genes in podocytes of FSGS-prone Actn4+/K256E animals found highly overrepresented biological processes which included the actin filament-based process (P=3.3E-28), actin cytoskeleton organization (P=2.1E-24), cell morphogenesis (1.8E-10), neuron projection development (6.1E-10), and a number of catabolic processes. Among highly enriched molecular functions were actin binding (P=2.7E-28), cytoskeletal protein binding (7.1E-25), motor activity (P=1.8E-16), and calmodulin binding (P=3.2E-11). Cell localizations with very strong enrichment included terms such as actin cytoskeleton (P=1.6E-21), myosin complex (P=6.2E-19), adherens junction (P=4.7E-10), cell projection (P=7.0E-8), and focal adhesion (P=6.2E-6). The top ten process networks included Cytoskeleton – Actin filaments (P=7.5E-19), Cytoskeleton – Regulation of cytoskeleton rearrangement (P=2.7E-11), Cytoskeleton – Intermediate filaments (P=1.1E-6), Cell adhesion – Cell junctions (P=3.7E-6), and Cell adhesion – Integrin-mediated cell-matrix adhesion (P=7.2E-5). The interactome analysis identified a number of strongly overconnected proteins such as transcription factors CREB-H, PPAR-gamma and SP1, receptors mGluR4A and VEGFR-2, ligands angiopoietin 4 and CSF-1, kinases MAP3K3 and RIPK3, the protease MMP23B, tyrosine phosphatase SHP-2, and the enzyme angiogenin. Other overconnected but uncategorized proteins included the adaptor protein GRB2 and the calcium-permeable cation channel TRPC5 (Supplemental Table 8B).
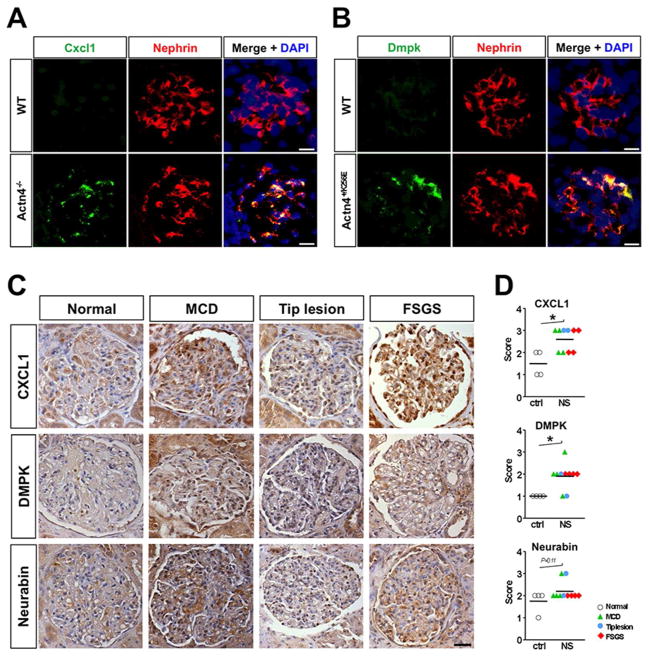
Analysis of protein expression of CXCL1 and DMPK in mouse renal tissue of genetic FSGS and human proteinuric kidney disease
To test whether mRNA upregulation of novel genes identified by TRAP was paralleled by an induction of the respective proteins we performed immunostaining on kidney tissue from Actn4 knockout, Actn4 mutant and wild-type control mice. In line with the microarray data we observed a strong presence of chemokine ligand Cxcl1 protein in podocytes of Actn4 deficient animals with manifest FSGS when compared to wild-type controls (Fig. 6A). We also found evidence for the induction of Dmpk protein in podocytes of FSGS-prone Actn+/K256K and not in wild-type control mice, further validating the TRAP data (Fig. 6B).
Figure 6. Cxcl1 and Dmpk protein upregulation in podocytes of FSGS-prone Actn4−/− and Actn4+/K256E mice and patients with nephrotic syndrome.
(A) Immunostaining showing induction of chemokine ligand Cxcl1 protein in podocytes of Actn4 deficient (Actn4−/−) animals. (B) Immunostaining demonstrating increased protein expression of protein kinase Dmpk in podocytes of mutant Actn4+/K256E mice when compared to wild-type controls. Scale bars: 10μm. (C) Immunohistochemical staining for CXCL1, DMPK, and Neurabin (PPP1R9A) in human kidney biopsies. Representative micrographs of stained glomeruli in normal renal tissue (Ctrl) and in kidney biopsies from patients with nephrotic syndrome (NS) and histologically proven minimal change disease (MCD), tip lesion and primary focal segmental glomerulosclerosis (FSGS). Scale bar: 10μm. (D) Semiquantitative scoring of glomerular staining intensity with focus on the outer aspect of the glomerular tuft. 0- none, 1- low, 2- medium, 3- high, 4- very high; *P<0.01.
Finally, to validate the utility of TRAP to identify new genes upregulated in human FSGS, we next conducted immunohistochemical analyses on kidney biopsies from patients with known proteinuric diagnoses: minimal change disease (MCD), tip lesion, FSGS, and normal control tissue. Sections were scored for staining intensity in a blinded fashion by a renal pathologist. A significant upregulation of podocyte expression of CXCL1 and DMPK as well as a positive trend for neurabin-1 induction were observed in patients with nephrotic-range proteinuria and histologically proven podocytopathy when compared to non-proteinuric controls. (Fig. 6, C and D).
Discussion
Our work provides the first comprehensive description of the podocyte transcriptome during aging and in experimental FSGS. We used a novel transgenic mouse to perform TRAP of podocytes that utilizes a simple, one-step affinity purification procedure for isolation of polysomal RNA. Advantages of this approach include its simplicity, low cost and no need for specialized equipment such as FACS or laser capture microdissection machines. Most importantly, this approach eliminates the risk of transcriptional responses induced by enzymatic dissociation at 37°C required for FACS analysis. We validated this approach both by comparison to other reports of the podocyte transcriptome and by determining the enrichment of known podocyte genes compared to whole kidney. Finally, we demonstrated that two novel podocyte genes identified as upregulated in podocytes during experimental FSGS, CXCL1 and DMPK, are also upregulated in human podocytes during proteinuric glomerular disease. We anticipate that podocyte-TRAP will have broad application among the numerous genetic mouse models of glomerular disease. The Col1α1-eGFP-L10a transgenic mice can be maintained as a homozygous line, simplifying breeding strategies, and one allele is sufficient for podocyte-TRAP due to very strong podocyte expression.
Analysis of the podocyte transcriptome in health confirmed that these cells share most in common with neurons, as previsoulsy described (10). During aging, we noted a relatively high number of gene expression changes between 2 and 6 weeks, likely reflecting important adaptations to the rapid growth in kidney and mouse size, with correspondingly increased single nephron GFR. In contrast, there were very few gene expression changes between 6 and 44 weeks. In the future, it would be interesting to follow the podocyte transcriptome out to later timepoints, since marked glomerular changes are seen in very old (36 months) mice (61), and these may correspond to age-related GFR loss observed in older humans.
Analysis of gene expression changes during FSGS revealed a striking overrepresentation of terms such as podocyte shape change, movement and interaction with matrix. Together, these expression changes paint a picture of dynamic movement during the response of podocytes to FSGS. This is consistent with the morphological observation of progressive foot process effacement observed over the course of disease. Our analysis has identified many new genes including extracellular ligands, proteases and transcription factors that likely play roles in this process. Overall, the analysis confirms the view of the podocyte as a cell that is responding to injury during injury, activating conserved cellular response pathways that result in podocyte morphogenesis and cytoskeletal reorganization.
Several novel applications of podocyte-TRAP can be envisioned. Clearly this approach will be well suited for RNA sequencing which is far more sensitive than microarray, and also detects non-coding RNAs as well as splice variants - recently reported to occur commonly in podocytes (11). Because we show that podocyte-TRAP is highly sensitive, it may also be useful for detecting subclinical toxicities of new drugs during preclinical toxicity testing. Certainly podocyte-TRAP can be easily applied to other podocytopathy models, such as Alport’s syndrome, LPS-induced proteinuria, podocyte depletion and others. Finally, podocyte-TRAP isolates not only polysomal mRNA, but also other molecules that are bound to the polysomal complex including targeting microRNAs (Grgic and Humphreys, unpublished observation) -- a feature that e.g. may be helpful to study posttranscriptional regulation in a cell- and gene-specific manner in vivo.
The dataset described here should serve as a rich resource for researchers studying podocyte function and disease. There is increasing interest in bioinformatics and systems approaches to studying podocytopathies. PodNet is a recently described protein-protein interaction network that maps trancriptome data onto protein interaction networks (62). The podocyte-specific transcriptomes described here should add to such an effort, as well as the other glomerular and kidney expression databases such as Nephromine and the Genitourinary Development Molecular Anatomy Project (GUDMAP) (63, 64).
In conclusion, we have developed a genetic model for defining the translational profiles of podocytes in health and disease in vivo. This approach will improve our understanding of podocyte biology and help define new targets for the treatment of podocytopathies.
Materials and Methods
Mouse maintenance and experiments
Mice were maintained in barrier and all studies performed according to the animal experimental guidelines issued by the Animal Care and Use Committee at Harvard University. Generation of transgenic Col1α1-eGFP-L10a (PodoTRAP) mice will be described in a separate publication. Mice were maintained on a C57BL/6J x DBA/2 mixed background and were crossed to heterozygous actinin 4A+/− (actn4+/−) or heterozygous actinin 4A +/K256E (actn4+/K256E) mice to obtain bigenic PodoTRAP;actn4+/− and PodoTRAP;actn4+/K256E mice, respectively. To generate mice null for actinin 4A, PodoTRAP;actn4+/− mice were crossed again to actn4+/− animals. PodoTRAP;actn4−/− mice were obtained at a substantially lower ratio than the expected Mendelian ratio of 1:8, which is in line with previous reports and likely due to the severe phenotype of actn4-deficient mice with significant perinatal lethality (Kos et al., 2003). Genomic DNA was obtained from tail biopsies and presence of the collagen1α1-eGFP-L10a transgene tested by PCR using the following primers: 5′-GGC ATC GAC TTC AAG GAG GA-3′ (F); and 5′-GGT CGT AGT TCT TCA GGC TGA-3′ (R). For allelic discrimination of actinin 4A we used a standard TaqMan-based assay. Primers and allele specific probes were designed by Applied Biosystems Custom SNP Genotyping Assay and are as follows: 5′-TCC ACT TAC AGA CAT CGT GAA CAC A-3′ (F); 5′-GCA TGG TAG AAG CTG GAC ACA TAT-3′ (R); actn4 wild-type probe: AGG ACA TCG TGA ACA CAG CCC GGC CCG ACG AGA AGG CCA TAA TGA CAT ATG TGT CCA GCT TCT ACC ATG CCT TTT CAG GAG CGC AGA AG; and actn4 del/K256E probe: AGG ACA TCG TGA ACA CAG CCC GGC CCG ACG AAG AGG CCA TAA TGA CAT ATG TGT CCA GCT TCT ACC ATG CCT TTT CAG GAG CGC AGA AG.
Mouse tissue preparation and histology
Mouse kidneys were processed for histological analyses as previously described. Three biologic replicates were used for every experimental condition. Briefly, mice were killed under deep anesthesia and immediately perfused via the left ventricle with ice-cold PBS for 1 min. Kidneys were harvested and hemisectioned, fixed in 4% paraformaldehyde for 1–2 hrs at 4°C, placed in 30% sucrose overnight for cryoprotection, and snap-frozen in optimal cutting temperature compound (OCT, Sakura FineTek). 7μm thick cryosections were mounted on Fisher Superfrost Plus (Fisher) microscope slides, immunostained and mounted in ProLong Gold Antifade containing 4′, 6-diamino-2-phenylindole (Invitrogen). The following primary antibodies were used for immunostaining: guinea pig anti-nephrin (Progen Biotechnik, 1:100), rat anti-CD31 (eBioscience, 1:100), rat anti-PDGFRβ (eBioscience, 1:200), rabbit anti-CXCL1 (Protein Tech, 1:100), and goat anti-DMPK (Santa Cruz, 1:25). Sections were subsequently exposed to corresponding Cy3-conjugated secondary antibodies (Jackson ImmunoResearch). For paraffin-embedding renal tissue was fixed in 10% neutral buffered formalin overnight. Pathohistological examination was performed on paraffin-embedded, 3μm thick kidney sections stained with hematoxylin and eosin (H&E), periodic acid-Schiff (PAS), and Masson’s trichrome. Staining was imaged on a Nikon C1 confocal, Nikon eclipse 90i and Olympus BX41 microscope, respectively.
Electron microscopy
Portions of kidney cortex were fixed in Karnovsky’s fixative and processed for electron microscopic studies by standard procedures. Briefly, semithin sections were prepared and stained with toluidine-blue stain for light microscopic inspection and selection for ultrathin sectioning. Ultrathin sections were stained with uranyl acetate and lead citrate and examined by electron microscopy.
Urine analysis
For gross assessment of overall podocyte function and integrity mouse urine was examined by Multistix 8 SG Urinalysis Strips (Siemens) and proteinuria determined in a semiquantitative fashion.
Immunohistochemical staining of human tissue
All IHC staining was performed on formalin-fixed and paraffin-embedded tissue sections, 4μm in thickness. Sections were deparaffinized, rehydrated and endogenous peroxidase activity blocked by incubation with 3% hydrogen peroxide in 100% alcohol (1:1) for 15 min. Antigen retrieval (for CXCL1) was performed using target retrieval solution (pH 6.0; Dako) and heat induction (120°C at 15psi) for 30 sec. Sections were incubated with following primary antibodies: rabbit anti-CXCL1 (Protein Tech, 1:100), goat anti-DMPK (Santa Cruz, 1:25), and rabbit anti-Neurabin 1 (Abcam, 1:500). Exposure to primary antibodies was followed by incubation with appropriate HRP-labeled polymer secondary antibodies (Dako) and staining visualized with a 3,3′-diaminobenzidine (DAB)/substrate-chromogen system (Dako). Gill’s hematoxylin was used for counterstaining. For anti-DMPK staining sections were treated separately with avidin, biotin and 3% rabbit serum for 10 min followed by primary antibody incubation and subsequent exposure to biotinylated rabbit anti-goat secondary antibody/streptavidin-HRP for 30 min. Control tissues included macroscopically normal renal cortex taken from kidneys resected for localized neoplasia, brain, skin, intestine, heart, tonsil, skeletal muscle, and ovarian carcinoma. Semiquantitative scoring of stained kidney sections was performed in a blinded fashion.
RT-PCR
For quantitative PCR cDNA was produced from RNA using iScript reverse transcriptase (BioRad). Real-time PCR was performed using the iQ-SYBR Green supermix (BioRad) and the iQ5 Multicolor Real-Time PCR Detection System (BioRad) for determination of gene expression levels of Nph1, Nph2, Wt1, Synpo, Actn4, Cxcl1, Dbn1, Dmpk, Lmo7, Ppp1r9a, Shroom4 and Ssh2. Housekeeping gene GAPDH was used as the internal control.
Translational ribosome affinity purification
Extraction and purification of polysomally bound mRNA from whole organ lysate was conducted as described in detail elsewhere with some modifications (Heiman et al, Cell, 2008). In brief, mice were perfused with ice-cold PBS under deep anesthesia, kidneys removed and decapsulated, kidney cortex rapidly microdissected and minced on ice and transferred to 1mL of ice-cold polysome extraction buffer. Dynabeads (MyOne T1 Dynabeads, Invitrogen, Carlsbad, CA) coated with monoclonal anti-GFP antibodies (clones 19F7 and 19C8, 50Yg each per IP; Rockefeller) were added to the post-mitochondrial supernatant of renal cortical extract and together incubated at 4°C with end-over-end rotation for 4 hours. Following incubation, beads were collected on a magnetic rack, repeatedly washed with ice-cold high-salt wash buffer and resuspended in Quiagen RNeasy lysis buffer. RNA was purified using a RNeasy MinElute Cleanup Kit (QIAGEN, Valencia, CA) with in-column DNase digestion (RNase-Free DNase Set, QIAGEN, Valencia, CA).
RNA quality control and microarray
For assessment of RNA quality and quantity, purified mRNA samples were analyzed by chip-based capillary electrophoresis using an Agilent RNA 6000 PicoChip kit and the Agilent 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA). RNA samples of satisfactory quality were processed and amplified (NuGEN Technologies Inc, San Carlos, CA), and subsequently subjected to microarray analysis on a whole Mouse Genome 430 2.0 Array platform (Affymetrix, Santa Clara, CA). Microarrays have been deposited in the GEO database (Geo ID GSM1283090 through GSM1283114) with series ID GSE53156. They are also available from GUDMAP (www.gudmap.org, series ID 49).
Computational analysis of microarray data
Affymetrix array data were processed and analyzed using MATLAB R2011b (7.13.0.564) software and the Bioinformatics Toolbox. For normalization of the data the Robust Multi-array Average (RMA) procedure was applied using the ‘affyrma’ function (Bolstad B: “affy: Built-in Processing Methods” (2005);http://www.bioconductor.org/packages/2.1/bioc/vignettes/affy/inst/doc/builtinMethods.pdf). Principal component analysis (PCA) was performed using the matlab Statistics Toolbox. Distances between different samples were computed using the pdist function which determines the Euclidean distance between pairs of vectors. Vectors were projected over their principal components applying the cmdscale function. For comparison of wild-type vs. Actn4+/K256E podocyte transcriptome expression levels were adjusted using age as a covariate. Trend graphs were generated for selected genes to illustrate expression levels in podocytes in relation to age and genotype. DAVID (the database for annotation, visualization and integrated discovery) was utilized for functional interpretation and visualization of overrepresented genes. GeneGo MetaCore (Thomson Reuters) was used as a bioinformatics resources for differentially regulated genes and network analysis.
Statistics
Data are given as mean ± SEM if not otherwise indicated. Statistical analysis was performed using the unpaired Student’s t test to determine differences between groups. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We wish to thank Reddy Gali (C3 Bioinformatics, Harvard Countway) for help in the initial microarray data management. I.G. was supported by a fellowship from the Deutsche Forschungsgemeinschaft (GR 3301/4-1) and grants from the German Kidney Foundation, the Philipps-University Marburg Innovation Fund and the Von Behring-Röntgen-Foundation. A.F.H. was supported by the Deutsche Forschungsgemeinschaft (SFB593). This work was supported by National Institutes of Health grants DK54931 (M.R.P.), DK88923 and DK087389 (B.D.H.) and an Established Investigator Award from the American Heart Association (B.D.H.).
Footnotes
Disclosures
None.
References
- 1.Harambat J, van Stralen KJ, Kim JJ, Tizard EJ. Epidemiology of chronic kidney disease in children. Pediatr Nephrol. 2011;27:363–373. doi: 10.1007/s00467-011-1939-1. published online EpubMar ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annual Report. The EMMES Corporation; Rockville, MD: 2008. North American Pediatric Renal Transplant Cooperative Study (NAPRTCS) [Google Scholar]
- 3.Kitiyakara C, Eggers P, Kopp JB. Twenty-one-year trend in ESRD due to focal segmental glomerulosclerosis in the United States. Am J Kidney Dis. 2004;44:815–825. published online EpubNov (S0272638604010819 [pii]) [PubMed] [Google Scholar]
- 4.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. published online EpubNov 8 ( [DOI] [PubMed] [Google Scholar]
- 5.Cohen CD, Grone HJ, Grone EF, Nelson PJ, Schlondorff D, Kretzler M. Laser microdissection and gene expression analysis on formaldehyde-fixed archival tissue. Kidney Int. 2002;61:125–132. doi: 10.1046/j.1523-1755.2002.00112.x. published online EpubJan ( [DOI] [PubMed] [Google Scholar]
- 6.Takemoto M, Asker N, Gerhardt H, Lundkvist A, Johansson BR, Saito Y, Betsholtz C. A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol. 2002;161:799–805. doi: 10.1016/S0002-9440(10)64239-3. published online EpubSep ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takemoto M, He L, Norlin J, Patrakka J, Xiao Z, Petrova T, Bondjers C, Asp J, Wallgard E, Sun Y, Samuelsson T, Mostad P, Lundin S, Miura N, Sado Y, Alitalo K, Quaggin SE, Tryggvason K, Betsholtz C. Large-scale identification of genes implicated in kidney glomerulus development and function. Embo J. 2006;25:1160–1174. doi: 10.1038/sj.emboj.7601014. published online EpubMar 8 (7601014 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L, Sun Y, Takemoto M, Norlin J, Tryggvason K, Samuelsson T, Betsholtz C. The glomerular transcriptome and a predicted protein-protein interaction network. J Am Soc Nephrol. 2008;19:260–268. doi: 10.1681/ASN.2007050588. published online EpubFeb ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami A, Oshiro H, Kanzaki S, Yamaguchi A, Yamanaka S, Furuya M, Miura S, Kanno H, Nagashima Y, Aoki I, Nagahama K. A novel method for isolating podocytes using magnetic activated cell sorting. Nephrol Dial Transplant. 2010;25:3884–3890. doi: 10.1093/ndt/gfq323. published online EpubDec ( [DOI] [PubMed] [Google Scholar]
- 10.Brunskill EW, Georgas K, Rumballe B, Little MH, Potter SS. Defining the molecular character of the developing and adult kidney podocyte. PLoS ONE. 2011;6:e24640. doi: 10.1371/journal.pone.0024640. PONE-D-11-10202 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boerries M, Grahammer F, Eiselein S, Buck M, Meyer C, Goedel M, Bechtel W, Zschiedrich S, Pfeifer D, Laloe D, Arrondel C, Goncalves S, Kruger M, Harvey SJ, Busch H, Dengjel J, Huber TB. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int. 2013;83:1052–1064. doi: 10.1038/ki.2012.487. published online EpubJun ( [DOI] [PubMed] [Google Scholar]
- 12.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. published online EpubNov 14 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. published online EpubNov 14 (S0092-8674(08)01366-4 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. published online EpubApr 10 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–4230. doi: 10.1093/nar/gkq130. published online EpubJul (gkq130 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Kos CH, Le TC, Sinha S, Henderson JM, Kim SH, Sugimoto H, Kalluri R, Gerszten RE, Pollak MR. Mice deficient in alpha-actinin-4 have severe glomerular disease. J Clin Invest. 2003;111:1683–1690. doi: 10.1172/JCI17988. published online EpubJun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao J, Le TC, Kos CH, Henderson JM, Allen PG, Denker BM, Pollak MR. Alpha-actinin-4-mediated FSGS: an inherited kidney disease caused by an aggregated and rapidly degraded cytoskeletal protein. PLoS Biol. 2004;2:e167. doi: 10.1371/journal.pbio.0020167. published online EpubJun ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson JM, Al-Waheeb S, Weins A, Dandapani SV, Pollak MR. Mice with altered alpha-actinin-4 expression have distinct morphologic patterns of glomerular disease. Kidney Int. 2008;73:741–750. doi: 10.1038/sj.ki.5002751. published online EpubMar (5002751 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grgic I, Krautzberger AM, Hofmeister AF, Lalli M, Dirocco D, Fleig SV, Liu J, Duffield JS, McMahon AP, Aronow B, Humphreys BD. Translational profiles of medullary myofibroblasts during kidney fibrosis. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2013101143. ePub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usui N, Watanabe K, Ono K, Tomita K, Tamamaki N, Ikenaka K, Takebayashi H. Role of motoneuron-derived neurotrophin 3 in survival and axonal projection of sensory neurons during neural circuit formation. Development. 2012;139:1125–1132. doi: 10.1242/dev.069997. published online EpubMar ( [DOI] [PubMed] [Google Scholar]
- 23.Ye M, Wysocki J, William J, Soler MJ, Cokic I, Batlle D. Glomerular localization and expression of Angiotensin-converting enzyme 2 and Angiotensin-converting enzyme: implications for albuminuria in diabetes. J Am Soc Nephrol. 2006;17:3067–3075. doi: 10.1681/ASN.2006050423. published online EpubNov ( [DOI] [PubMed] [Google Scholar]
- 24.Nadarajah R, Milagres R, Dilauro M, Gutsol A, Xiao F, Zimpelmann J, Kennedy C, Wysocki J, Batlle D, Burns KD. Podocyte-specific overexpression of human angiotensin-converting enzyme 2 attenuates diabetic nephropathy in mice. Kidney Int. 2012;82:292–303. doi: 10.1038/ki.2012.83. published online EpubAug ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fath T, Fischer RS, Dehmelt L, Halpain S, Fowler VM. Tropomodulins are negative regulators of neurite outgrowth. Eur J Cell Biol. 2011;90:291–300. doi: 10.1016/j.ejcb.2010.10.014. published online EpubApr ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. published online EpubSep 28 ( [DOI] [PubMed] [Google Scholar]
- 27.Steenhard BM, Isom K, Stroganova L, St John PL, Zelenchuk A, Freeburg PB, Holzman LB, Abrahamson DR. Deletion of von Hippel-Lindau in glomerular podocytes results in glomerular basement membrane thickening, ectopic subepithelial deposition of collagen {alpha}1{alpha}2{alpha}1(IV), expression of neuroglobin, and proteinuria. Am J Pathol. 2010;177:84–96. doi: 10.2353/ajpath.2010.090767. published online EpubJul ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You JJ, Lin-Chao S. Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J Biol Chem. 2010;285:11652–11666. doi: 10.1074/jbc.M109.051094. published online EpubApr 9 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perisic L, Lal M, Hulkko J, Hultenby K, Onfelt B, Sun Y, Duner F, Patrakka J, Betsholtz C, Uhlen M, Brismar H, Tryggvason K, Wernerson A, Pikkarainen T. Plekhh2, a novel podocyte protein downregulated in human focal segmental glomerulosclerosis, is involved in matrix adhesion and actin dynamics. Kidney Int. 2012;82:1071–1083. doi: 10.1038/ki.2012.252. published online EpubNov. [DOI] [PubMed] [Google Scholar]
- 30.Hodgin JB, Borczuk AC, Nasr SH, Markowitz GS, Nair V, Martini S, Eichinger F, Vining C, Berthier CC, Kretzler M, D’Agati VD. A molecular profile of focal segmental glomerulosclerosis from formalin-fixed, paraffin-embedded tissue. Am J Pathol. 2010;177:1674–1686. doi: 10.2353/ajpath.2010.090746. published online EpubOct (S0002-9440(10)60221-0 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Agati VD, Kaskel FJ, Falk RJ. Focal segmental glomerulosclerosis. N Engl J Med. 2011;365:2398–2411. doi: 10.1056/NEJMra1106556. published online EpubDec 22 ( [DOI] [PubMed] [Google Scholar]
- 32.Kriz W, Gretz N, Lemley KV. Progression of glomerular diseases: is the podocyte the culprit? Kidney Int. 1998;54:687–697. doi: 10.1046/j.1523-1755.1998.00044.x. published online EpubSep ( [DOI] [PubMed] [Google Scholar]
- 33.Wong W, DeVito J, Nguyen H, Sarracino D, Porcheray F, Dargon I, Pelle PD, Collins AB, Tolkoff-Rubin N, Smith RN, Colvin R, Zorn E. Chronic humoral rejection of human kidney allografts is associated with MMP-2 accumulation in podocytes and its release in the urine. Am J Transplant. 2010;10:2463–2471. doi: 10.1111/j.1600-6143.2010.03290.x. published online EpubNov ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roestenberg P, van Nieuwenhoven FA, Joles JA, Trischberger C, Martens PP, Oliver N, Aten J, Hoppener JW, Goldschmeding R. Temporal expression profile and distribution pattern indicate a role of connective tissue growth factor (CTGF/CCN-2) in diabetic nephropathy in mice. Am J Physiol Renal Physiol. 2006;290:F1344–1354. doi: 10.1152/ajprenal.00174.2005. published online EpubJun ( [DOI] [PubMed] [Google Scholar]
- 35.Yokoi H, Mukoyama M, Mori K, Kasahara M, Suganami T, Sawai K, Yoshioka T, Saito Y, Ogawa Y, Kuwabara T, Sugawara A, Nakao K. Overexpression of connective tissue growth factor in podocytes worsens diabetic nephropathy in mice. Kidney Int. 2008;73:446–455. doi: 10.1038/sj.ki.5002722. published online EpubFeb ( [DOI] [PubMed] [Google Scholar]
- 36.Bruggeman LA, Drawz PE, Kahoud N, Lin K, Barisoni L, Nelson PJ. TNFR2 interposes the proliferative and NF-kappaB-mediated inflammatory response by podocytes to TNF-alpha. Laboratory investigation; a journal of technical methods and pathology. 91:413–425. doi: 10.1038/labinvest.2010.199. published online EpubMar ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer D, Hauk TG, Muller A, Thanos S. Crystallins of the beta/gamma-superfamily mimic the effects of lens injury and promote axon regeneration. Mol Cell Neurosci. 2008;37:471–479. doi: 10.1016/j.mcn.2007.11.002. published online EpubMar ( [DOI] [PubMed] [Google Scholar]
- 38.Thanos S, Bohm MR, Schallenberg M, Oellers P. Traumatology of the optic nerve and contribution of crystallins to axonal regeneration. Cell Tissue Res. 2012;349:49–69. doi: 10.1007/s00441-012-1442-4. published online EpubJul ( [DOI] [PubMed] [Google Scholar]
- 39.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. published online EpubFeb 20 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banas MC, Banas B, Hudkins KL, Wietecha TA, Iyoda M, Bock E, Hauser P, Pippin JW, Shankland SJ, Smith KD, Stoelcker B, Liu G, Grone HJ, Kramer BK, Alpers CE. TLR4 links podocytes with the innate immune system to mediate glomerular injury. J Am Soc Nephrol. 2008;19:704–713. doi: 10.1681/ASN.2007040395. published online EpubApr ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubberstey AV, Mottillo EP. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. Faseb J. 2002;16:487–499. doi: 10.1096/fj.01-0659rev. published online EpubApr ( [DOI] [PubMed] [Google Scholar]
- 42.Moriyama K, Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J Cell Sci. 2002;115:1591–1601. doi: 10.1242/jcs.115.8.1591. published online EpubApr 15 ( [DOI] [PubMed] [Google Scholar]
- 43.Wang C, Zhou GL, Vedantam S, Li P, Field J. Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J Cell Sci. 2008;121:2913–2920. doi: 10.1242/jcs.023911. published online EpubSep 1 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alcaraz WA, Gold DA, Raponi E, Gent PM, Concepcion D, Hamilton BA. Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation. Proc Natl Acad Sci U S A. 2006;103:19424–19429. doi: 10.1073/pnas.0609184103. published online EpubDec 19 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng LE, Reed RR. Zfp423/OAZ participates in a developmental switch during olfactory neurogenesis. Neuron. 2007;54:547–557. doi: 10.1016/j.neuron.2007.04.029. published online EpubMay 24 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sytnyk V, Leshchyns’ka I, Delling M, Dityateva G, Dityatev A, Schachner M. Neural cell adhesion molecule promotes accumulation of TGN organelles at sites of neuron-to-neuron contacts. J Cell Biol. 2002;159:649–661. doi: 10.1083/jcb.200205098. published online EpubNov 25 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westphal D, Sytnyk V, Schachner M, Leshchyns’ka I. Clustering of the neural cell adhesion molecule (NCAM) at the neuronal cell surface induces caspase-8- and -3-dependent changes of the spectrin meshwork required for NCAM-mediated neurite outgrowth. J Biol Chem. 285:42046–42057. doi: 10.1074/jbc.M110.177147. published online EpubDec 31 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oude Ophuis RJ, Mulders SA, van Herpen RE, van de Vorstenbosch R, Wieringa B, Wansink DG. DMPK protein isoforms are differentially expressed in myogenic and neural cell lineages. Muscle Nerve. 2009;40:545–555. doi: 10.1002/mus.21352. published online EpubOct ( [DOI] [PubMed] [Google Scholar]
- 49.Mulders SA, van Horssen R, Gerrits L, Bennink MB, Pluk H, de Boer-van Huizen RT, Croes HJ, Wijers M, van de Loo FA, Fransen J, Wieringa B, Wansink DG. Abnormal actomyosin assembly in proliferating and differentiating myoblasts upon expression of a cytosolic DMPK isoform. Biochim Biophys Acta. 2011;1813:867–877. doi: 10.1016/j.bbamcr.2011.01.024. published online EpubMay ( [DOI] [PubMed] [Google Scholar]
- 50.Venkatareddy M, Cook L, Abuarquob K, Verma R, Garg P. Nephrin regulates lamellipodia formation by assembling a protein complex that includes Ship2, filamin and lamellipodin. PLoS One. 2011;6:e28710. doi: 10.1371/journal.pone.0028710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukasawa H, Bornheimer S, Kudlicka K, Farquhar MG. Slit diaphragms contain tight junction proteins. J Am Soc Nephrol. 2009;20:1491–1503. doi: 10.1681/ASN.2008101117. published online EpubJul ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chia PH, Patel MR, Shen K. NAB-1 instructs synapse assembly by linking adhesion molecules and F-actin to active zone proteins. Nat Neurosci. 2012;15:234–242. doi: 10.1038/nn.2991. published online EpubFeb ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakanishi H, Obaishi H, Satoh A, Wada M, Mandai K, Satoh K, Nishioka H, Matsuura Y, Mizoguchi A, Takai Y. Neurabin: a novel neural tissue-specific actin filament-binding protein involved in neurite formation. J Cell Biol. 1997;139:951–961. doi: 10.1083/jcb.139.4.951. published online EpubNov 17 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliver CJ, Terry-Lorenzo RT, Elliott E, Bloomer WA, Li S, Brautigan DL, Colbran RJ, Shenolikar S. Targeting protein phosphatase 1 (PP1) to the actin cytoskeleton: the neurabin I/PP1 complex regulates cell morphology. Mol Cell Biol. 2002;22:4690–4701. doi: 10.1128/MCB.22.13.4690-4701.2002. published online EpubJul ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peitsch WK, Hofmann I, Endlich N, Pratzel S, Kuhn C, Spring H, Grone HJ, Kriz W, Franke WW. Cell biological and biochemical characterization of drebrin complexes in mesangial cells and podocytes of renal glomeruli. J Am Soc Nephrol. 2003;14:1452–1463. doi: 10.1097/01.asn.0000069222.63700.de. published online EpubJun ( [DOI] [PubMed] [Google Scholar]
- 56.Garg P, Verma R, Cook L, Soofi A, Venkatareddy M, George B, Mizuno K, Gurniak C, Witke W, Holzman LB. Actin-depolymerizing factor cofilin-1 is necessary in maintaining mature podocyte architecture. J Biol Chem. 2010;285:22676–22688. doi: 10.1074/jbc.M110.122929. published online EpubJul 16 (M110.122929 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nabet B, Tsai A, Tobias JW, Carstens RP. Identification of a putative network of actin-associated cytoskeletal proteins in glomerular podocytes defined by co-purified mRNAs. PLoS ONE. 2009;4:e6491. doi: 10.1371/journal.pone.0006491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mele C, Iatropoulos P, Donadelli R, Calabria A, Maranta R, Cassis P, Buelli S, Tomasoni S, Piras R, Krendel M, Bettoni S, Morigi M, Delledonne M, Pecoraro C, Abbate I, Capobianchi MR, Hildebrandt F, Otto E, Schaefer F, Macciardi F, Ozaltin F, Emre S, Ibsirlioglu T, Benigni A, Remuzzi G, Noris M. MYO1E Mutations and Childhood Familial Focal Segmental Glomerulosclerosis. N Engl J Med. 2011 doi: 10.1056/NEJMoa1101273. published online EpubJul 14 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma H, Togawa A, Soda K, Zhang J, Lee S, Ma M, Yu Z, Ardito T, Czyzyk J, Diggs L, Joly D, Hatakeyama S, Kawahara E, Holzman L, Guan JL, Ishibe S. Inhibition of podocyte FAK protects against proteinuria and foot process effacement. J Am Soc Nephrol. 2010;21:1145–1156. doi: 10.1681/ASN.2009090991. published online EpubJul ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boger CA, Gorski M, Li M, Hoffmann MM, Huang C, Yang Q, Teumer A, Krane V, O’Seaghdha CM, Kutalik Z, Wichmann HE, Haak T, Boes E, Coassin S, Coresh J, Kollerits B, Haun M, Paulweber B, Kottgen A, Li G, Shlipak MG, Powe N, Hwang SJ, Dehghan A, Rivadeneira F, Uitterlinden A, Hofman A, Beckmann JS, Kramer BK, Witteman J, Bochud M, Siscovick D, Rettig R, Kronenberg F, Wanner C, Thadhani RI, Heid IM, Fox CS, Kao WH. Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD. PLoS Genet. 2011;7:e1002292. doi: 10.1371/journal.pgen.1002292. published online EpubSep ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yabuki A, Tanaka S, Matsumoto M, Suzuki S. Morphometric study of gender differences with regard to age-related changes in the C57BL/6 mouse kidney. Experimental animals / Japanese Association for Laboratory Animal Science. 2006;55:399–404. doi: 10.1538/expanim.55.399. published online EpubJul ( [DOI] [PubMed] [Google Scholar]
- 62.Warsow G, Endlich N, Schordan E, Schordan S, Chilukoti RK, Homuth G, Moeller MJ, Fuellen G, Endlich K. PodNet, a protein-protein interaction network of the podocyte. Kidney Int. 2013;84:104–115. doi: 10.1038/ki.2013.64. published online EpubJul. [DOI] [PubMed] [Google Scholar]
- 63.Martini S, Eichinger F, Nair V, Kretzler M. Defining human diabetic nephropathy on the molecular level: integration of transcriptomic profiles with biological knowledge. Rev Endocr Metab Disord. 2008;9:267–274. doi: 10.1007/s11154-008-9103-3. published online EpubDec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. published online EpubApr ( [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.