Abstract
Objective
Systemic lupus erythematosus (SLE) is a complex and multifactorial autoimmune disease with striking clinical, immunologic and genetic heterogeneity, despite nearly ubiquitous antinuclear antibody (ANA) production. Multiple gene polymorphisms have been associated with the disease, but individually account for only a very small percentage of overall SLE risk. In earlier studies, constitutive expression of the DNA-binding protein, A+T rich interacting domain 3a (ARID3a) in transgenic mouse B lymphocyte lineage cells led to spontaneous ANA production and preferential development of B cells associated with production of polyreactive antibodies. Therefore, we asked if ARID3a was over-expressed in B lymphocytes of SLE patients and if ARID3a expression was associated with disease severity.
Methods
A cross section of SLE patients and age and gender-matched controls were analyzed longitudinally for lupus disease activity, numbers of ARID3a+ peripheral blood mononuclear B cells from multiple B cell subsets, immunoglobulin and cytokine levels.
Results
Fifty of 115 patients (43%) had dramatically increased numbers of ARID3a+ B cells compared to healthy controls. ARID3a is not expressed in naïve B cells of healthy controls, but was abundant in these precursors of antibody-secreting cells in SLE patients. Total numbers of ARID3a+ B cells correlated with increased disease activity as defined by SLE Disease Activity Index scores in individuals assessed at three time points.
Conclusion
These findings identify B cell anomalies in SLE that allow stratification of patient samples based on ARID3a expression and implicate ARID3a as a potential marker of CD19+ B lymphocytes correlated with disease activity.
Systemic lupus erythematosus (SLE) is an autoimmune disease resulting from breaches in immune tolerance and characterized by antinuclear antibody (ANA) production (reviewed in (1)). Although this disease may affect as many as 1 in 2500 individuals, the underlying causes are unknown (2). Environmental factors, hereditary effects and epigenetic variation have all been implicated in SLE pathogenesis (3–6). Therefore, it has been challenging to find a unifying explanation for the complex molecular abnormalities that arise in these patients. The clinically diverse nature of SLE further complicates the identification of new biomarkers that might lead to better treatments (7).
Multiple murine models for lupus exist. In keeping with the complex regulatory mechanisms that control immune responses, these models may involve disruptions in genes expressed in T or B lymphocytes, or may result from combined defects in genes expressed in a variety of immune regulatory cells (reviewed in (8,9)). While each of these models results in ANA production, they all have limitations and differ in the extent to which they mimic the human SLE organ involvement that typically evolves over time within individual patients. We showed that transgenic mice that over-expressed the DNA-binding protein Bright/ARID3a (B cell regulator of immunoglobulin heavy chain transcription/A+T rich interaction domain family protein 3a) in all B lineage cells produced serum ANAs by four weeks of age (10,11). Over-expression also resulted in increased numbers of marginal zone (MZ) B cells which are typically enriched for self-reactive B lymphocytes (11). These data suggest that inappropriate regulation of Bright/ARID3a expression in B lineage cells is sufficient to cause ANA production in these mice. Because constitutive expression of Bright/ARID3a in B cells of transgenic mice resulted in ANA production, a predisposing occurrence for SLE (12), we asked if SLE patients exhibit increased ARID3a expression in their peripheral blood B lymphocytes.
PATIENTS AND METHODS
Participants
Healthy age and gender-matched controls and patients who met a minimum of four American College of Rheumatology Classification Criteria for SLE (13) and for seropositive rheumatoid arthritis (RA) were recruited after informed consent from the Oklahoma Medical Research Foundation Clinical Pharmacology clinic at as part of the Oklahoma Lupus Cohort (IRB compliance #09-07 and #06-19), in accordance with the Declaration of Helsinki. Peripheral blood mononuclear cells from a total of 115 SLE patients (ranging in age from 21 to 72, 94% female), 6 RA patients and 33 healthy controls were analyzed for ARID3a expression. In an effort to monitor changes, forty-four SLE patients, 6 RA patients and 18 controls were randomly recruited into a longitudinal study and provided blood samples for visit 1. The majority of data were obtained from the longitudinal study. Two SLE patient samples were excluded in data analyses due to lymphopenia. Thirty-seven of the 44 SLE patients provided longitudinal samples at 2–3 visits (mean 2.6) over a 36 month period. SLE patients included 42 women and 2 men ranging from 21 to 66 years of age. Age at diagnosis and first blood draw, ethnic background and immunosuppressive medications taken at the first blood draw are given online in Table S-1 for SLE patients in the longitudinal study. Four patients were not taking immunosuppressive medication at their first blood draw. Further details of RA patient characteristics can be found online in Table S-2.
Flow Cytometry
Mononuclear cells were isolated from heparinized peripheral blood (~15 ml) with Ficoll-Paque Plus (GE Healthcare) and stained with the following fluorochrome-labeled antibodies: CD19 PE-Cy5, CD24 APC, IL-10 PE, CD10 Pacific Blue (BioLegend), IgD PerCP-Cy5.5, CD27 PE-Cy7, CD3 Pacific Blue (BD Biosciences), CD38 Alexa Fluor 700 (BD Pharmingen), and IgM APC (Southern Biotech). Cells were fixed with 2% paraformaldehyde, permeabilized with 0.1% Tween-20 and stained with goat anti-human ARID3a antibody (14) followed by rabbit anti-goat IgG FITC (Invitrogen). Gating for individual B cell subsets is shown as described (15,16), and is available online (link). Isotype controls (Caltag, BD Pharmingen, and eBioscience) were used for gating. Data (500,000 events per sample) were collected using an LSRII (BD Biogenics) and FACSDiva (BD Biosciences) software version 4.1 and analyzed using CellQuest Pro (BD Biosciences) and FlowJo (Tree Star) software versions 6.0 and 9.5.2, respectively.
Western Blots and ELISAs
Western blots for ARID3a were performed as previously described (17) using 20,000 peripheral blood mononuclear cells per lane. β-actin (Santa Cruz Biotechnology) was detected as a loading control. Immunoglobulin levels were determined from serially diluted plasma samples using alkaline phosphatase-conjugated anti-human IgG and IgM (Southern Biotech) and a Biotech EL800 plate reader at 405/490nm with GEN5 Biotech software. Autoantibody testing was determined using the Reichlin method as described (18). Levels of plasma TGF-β, IL-17, IL-10, TNFα and MIP1α were detected using a Single-Analyte ELISArray Kit (QIAGEN) according to the manufacturer’s directions.
Statistics
Data were statistically evaluated using Students’ t test or Mann Whitney U tests with (GraphPad) Prism software version 6.0, as indicated. P values of less than 0.05 were considered significant. Association of SLEDAI scores from each visit for each SLE patient with numbers of total ARID3a+ B cells at that time, and with numbers of ARID3a+ B cells of each B cell subset, were performed by best fit linear regression models using the linear models function in R version 3.0.0 (within a Linux operating system). A stepwise modeling approach was used wherein all initial, fully saturated models included adjustment for sex, age, race and overall B cell count. Variables that did not significantly contribute to the model fit (i.e. whose p-value was > 0.05) were removed. The final and most parsimonious models for each of the subsets adjusted only for overall B cell counts since the other covariates (sex, age and race) were found to be noncontributory based on the above criteria.
RESULTS
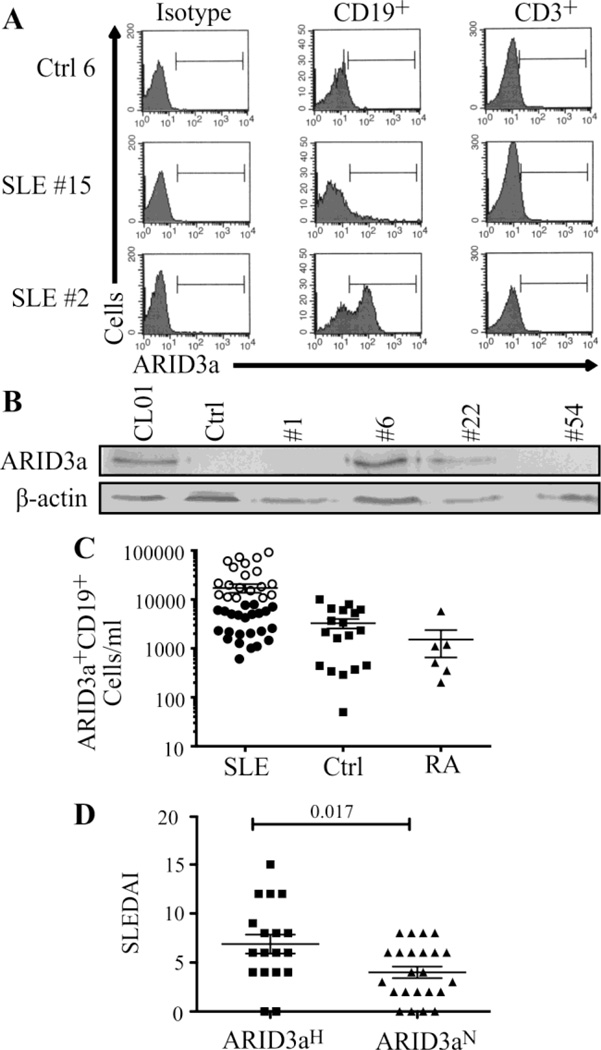
A flow cytometry assay for intracellular ARID3a was developed, validated and used to assess expression in SLE patient B cells. Mononuclear peripheral blood cells from SLE patients and healthy controls were gated for B (CD19+) or T (CD3+) lymphocyte surface markers (Figure 1A). As expected, healthy control samples contained very low numbers of B cells expressing intracellular ARID3a (< 15%). ARID3a expression in T cells was not above background in either control or SLE patients. However, numbers of ARID3a+ B cells were either greatly increased (50 of 115 patients) in a cross section of lupus patient samples, or were similar to healthy controls (e.g. Figure 1A, SLE #2 vs SLE#15). Western blotting confirmed that intracellular staining represented ARID3a protein expression and not an unidentified cross-reactivity; patient samples with abundant numbers of ARID3a+ B cells also showed abundant ARID3a protein (Figure 1B). At their first visit, 45% of SLE patients in our longitudinal study (19 of 42) showed increased numbers of ARID3a+ B cells more than two standard deviations above the mean of the healthy controls (Figure 1C, open circles). We defined those samples as ARID3aH (high), while SLE patient samples with ARID3a+ cells within healthy control ranges were designated ARID3aN (normal). Numbers of ARID3a+ CD19+ B cells in ARID3aH patients ranged from 3- to > 40-fold higher than average numbers of ARID3a+ cells in controls or ARID3aN patients. RA patient samples resembled healthy controls and did not show increased numbers of ARID3a+ B cells. These data indicate that ARID3a+ B cell numbers are dramatically expanded in a large subset of SLE patients.
Figure 1. ARID3a is over-expressed in B lineage cells in a subset of SLE patients.
(A) Immunofluorescent staining of ARID3a+ cells in B (CD19+) and T (CD3+) lymphocytes from a representative control (Ctrl 6) and 2 lupus patients (SLE #15, #2) are shown relative to isotype controls. (B) Mononuclear cells from a representative control and 4 SLE patients were assessed by Western blotting. CL01 is a positive control. Data represent > 6 experiments. (C) Numbers of ARID3a+ B cells/ml are shown for 42 SLE, 6 rheumatoid arthritis (RA) and 18 healthy controls. Open circles indicate patients with numbers of ARID3a+ cells > 2 standard deviations above mean numbers for controls (designated as ARID3aH). (D) The mean SLEDAI score of patients with ARID3aH samples (squares, n=17) was higher than the mean from patients with numbers of ARID3a+ cells within normal range (ARID3aN, circles, n=23). Each symbol represents one SLE patient sample. Statistics were determined by Student’s t test. Means ± SEM are shown.
Disease activity in patients was determined using the SLE Disease Activity Index (SLEDAI) score (19) and spanned a wide range of lupus activity (scores of 0–15). Average SLEDAI scores of patients having ARID3aH samples at the first visit (Figure 1D) were significantly higher than the SLEDAI scores of patients with ARID3aN samples (p = 0.017). No correlations between ARID3a expression and age, race or duration of disease were observed. Likewise, no correlations were observed between numbers of ARID3a+ B cells and medications in use (data available online). Two of four samples designated as ARID3aH were from patients taking no immune-modulatory medications at the first visit. These data suggest a possible correlation with disease activity and numbers of ARID3a+ B cells.
Our previous analyses of human B lineage subpopulations indicated that ARID3a mRNA transcripts were predominantly limited to pre-B, antibody-secreting and memory B cell subpopulations, but were not detectable in immature or naïve mature B cells in healthy individuals (14). Because circulating B cell subpopulations in SLE patients may be skewed compared to healthy populations (20), we examined the possibility that increased numbers of ARID3a+ cells might reflect disproportionate numbers of memory B or plasma cells in the ARID3aH versus the ARID3aN samples. B cell numbers were quantified within eight different B cell subpopulations from ARID3aN and ARID3aH samples by flow cytometry (data available online). Although average numbers of total B cells were slightly increased in the ARID3aH compared to the ARID3aN patient samples, they did not differ significantly from those of healthy controls. In addition, average numbers of most B cell subtypes were similar between control and ARID3aH samples. Therefore, increased numbers of ARID3a+ B cells in the ARID3aH patient samples are not the result of global expansion of all B cells, nor could they be attributed to expansion of B cell subpopulations that normally express ARID3a.
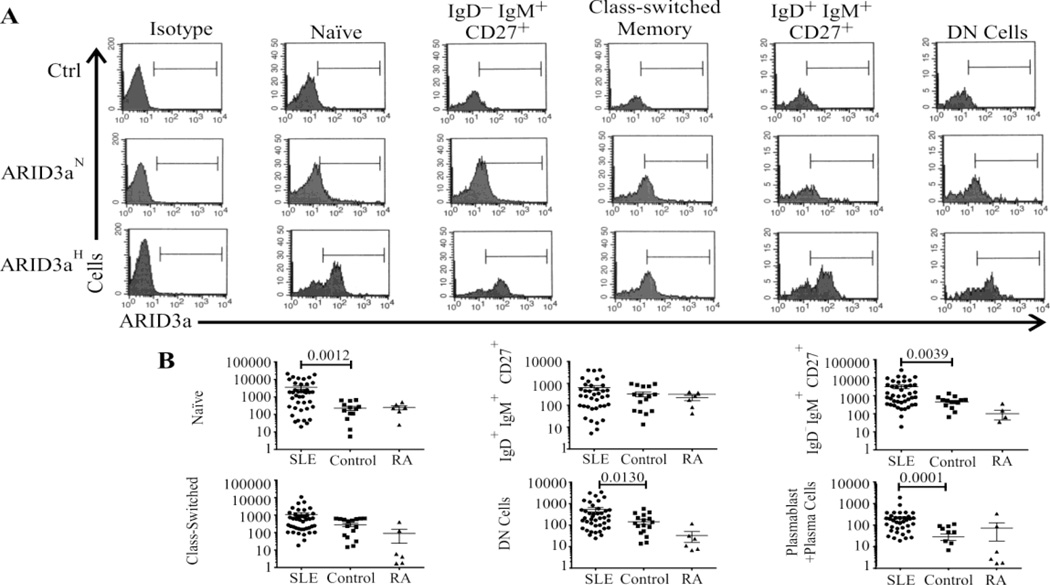
We next compared ARID3a expression levels within each B cell subset from SLE and RA patients to healthy controls (Figure 2) to determine which subset(s) accounted for the increased expression observed in Figure 1C. In healthy controls, ARID3a expression was observed in very small numbers of naïve and transitional (IgM+IgD+CD27−) B cells, but was more evident in the CD27+ memory B cell populations and CD19low/+ plasmablasts/plasma cells (Figure 2A, top panels). ARID3a expression in B cell subsets from ARID3aN samples (Figure 2A, middle panels) closely resembled those of healthy controls. However, ARID3aH samples (Figure 2A, bottom panels) typically showed increased numbers of ARID3a-expressing cells in all subpopulations compared to both healthy control and ARID3aN samples. Unexpectedly, two distinct peaks were evident in many B cell subsets from ARID3aH samples, indicating that only some cells expressed ARID3a while others did not. Enumeration of ARID3a+ B cells in each subset (Figure 2B) indicated that some SLE samples contained more than 100 times the numbers of ARID3a+ naïve B cells found in average healthy control or RA patient samples. Numbers of ARID3a+ IgD−IgM+CD27+, double negative (DN) B cells and CD19low/+ plasmablasts/ plasma cells were also significantly increased (p<0.05) in SLE patients compared to healthy control and RA samples. We previously showed that resting naïve B cells from healthy controls do not transcribe detectable levels of ARID3a (14). However, immature transitional B cells from which the naïve B cells develop, expressed ARID3a in mice (21), and these cells were included within the naïve subpopulation gate. Therefore, this B cell subset was further segregated using CD10 to distinguish transitional cells from naïve B cells. ARID3a+ B cells occurred in both naïve and transitional subsets (data available online), indicating that increased numbers of ARID3a+ transitional B cells within the IgM+IgD+CD27− subset did not account for increases in ARID3a+ naïve B cells in ARID3aH patient samples. These data demonstrate that ARID3a expression in SLE patients can occur in any B cell subset, and that numbers of ARID3a expressing naïve B cells are highly increased in some patients.
Figure 2. ARID3a expressing cells occur in multiple B cell subpopulations in SLE patients.
(A) Histograms show ARID3a expression in 5 B cell subsets for ARID3aN, ARID3aH and control samples compared to isotype controls (left panels). Bars were set relative to the isotype controls. (B) Total numbers of ARID3a+ B cells/ml in each B cell subset from 42 SLE, 6 RA and 17 healthy controls were enumerated. Means ± SEM are shown. Mann-Whitney calculated p values are given. Zero values were included in data analyses.
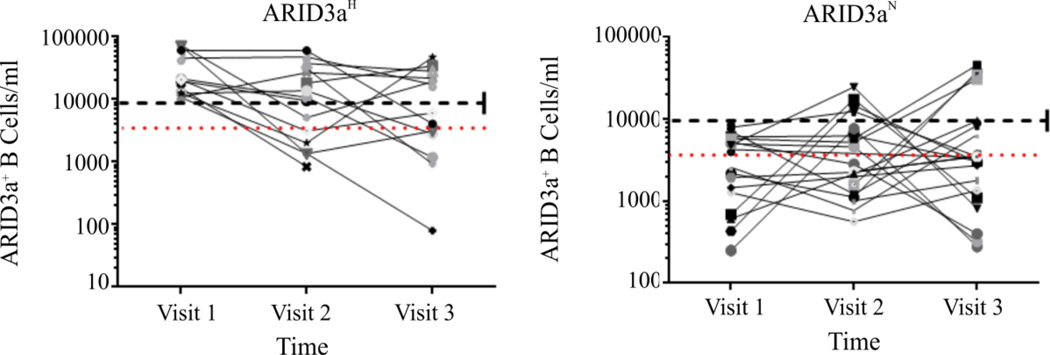
To determine if increased ARID3a expression was a stable phenotype in individual patients over time, patients were divided into ARID3aH and ARID3aN subsets based on analyses of CD19+ ARID3a+ B cell numbers during their first visit and ARID3a+ B cell numbers were plotted for subsequent visits (Figure 3). Ten of 15 ARID3aH patients showed normal numbers of ARID3a+ cells on at least one visit, and several of those were designated ARID3aN on both of their subsequent visits. Eight of 22 patients with normal numbers of ARID3a+ B cells at their first visit showed increased numbers of ARID3a+ cells at a single subsequent visit. Percentages of ARID3a+ cells within individual B cell subsets also varied over time in all individuals (not shown). No correlations between alterations in immunosuppressive drugs and ARID3a+ B cell numbers were observed. Healthy controls were also assessed at multiple time points, and with two exceptions where individuals had recent viral infections, numbers of ARID3a+ B cells varied less than one standard deviation from the mean. Thus, numbers of ARID3a+ B cells in SLE patients vary over time, and individual patients may cycle between different ARID3a phenotypes including periods more consistent with healthy controls.
Figure 3. Numbers of ARID3a+ B cells in individual patients vary with time.
Numbers of ARID3a+ CD19+ B cells/ ml of blood from SLE patients subdivided based on total ARID3a+ B cells at their first visit (ARID3aH, n=15 or ARID3aN, n=22) were plotted for each visit. Each symbol represents one patient sample. Red dotted lines show mean numbers of ARID3a+ B cells from healthy controls; black dashed lines show the value used to designate samples as ARID3aH or ARID3aN.
Higher SLEDAI scores were observed in patients whose samples were categorized as ARID3aH on their first visit (Figure 1D). Because we defined ARID3a phenotypes by absolute numbers of ARID3a+ B cells/ml and patients with fewer B cells might also show a correlation with changes in disease activity over time, we employed a statistical modeling approach. Using data from every patient at all visits, we asked if lupus disease activity correlated with total numbers of ARID3a+ B cells and/or numbers of ARID3a+ B cells in any B cell subset. Associations between ARID3a+ cell numbers for each B cell subset and SLEDAI scores were explored at each visit, adjusting for gender, age, race and total B cell counts. Increased disease activity (SLEDAI scores) is associated with increased total numbers of circulating CD19+ ARID3a+ cells (Table 1, p=0.0039; and by Spearman’s correlation, p=0.006, available online,) and with numbers of ARID3a+ IgD+IgM+CD27+ B cells (p=0.0225) and CD19low/+ plasmablast/plasma cells (p=0.0474), but not with the high numbers of naïve ARID3a+ B cells (Figure 2B and Table 1). Although these data imply that increased numbers of plasmablasts and IgD+IgM+CD27+ mature ARID3a+ B cells may also be associated with increased disease activity in SLE, numbers of cells contained within these subsets are small and preclude firm conclusions. Moreover, alterations in ARID3a expression were associated with changes in disease activity in all patients regardless of segregation into the ARID3aH or ARID3aN SLE phenotype.
Table 1.
Relationship between ARID3a+ cells with SLEDAI for each B cell subset.
| B Cell Subset | Coef. Est. | Std. Error | Pparsimony# | |
|---|---|---|---|---|
| ARID3a CD19 | 0.06624 | 0.02351 | 0.0039 | ** |
| ARID3a MZ Memory | 0.85721 | 0.35807 | 0.0225 | * |
| ARID3a Plasmablasts | 5.32660 | 2.64755 | 0.0474 | * |
| ARID3a Class-switched Memory | 0.72306 | 0.36045 | 0.0514 | |
| ARID3a Naïve | 0.15942 | 0.09129 | 0.0862 | |
| ARID3a DN Memory | 1.34904 | 0.79118 | 0.0990 | |
| ARID3a Memory IgM | 0.16245 | 0.11892 | 0.1180 | |
| ARID3a Naive IgD | 0.27813 | 0.33544 | 0.3886 | |
Note:
P<0.05,
P<0.01.
P-value for the most parsimonious model. All models were adjusted for the total B cell counts for each B cell subset.
To further explore the basis for the association of ARID3a expression with disease activity, we examined plasma Ig levels. Although ARID3aH samples had increased plasma IgM and IgG levels compared to healthy control and ARID3aN samples, and there was little difference in reactivity of the plasma with a large number of common autoantigens (data available online). These data suggest that increased numbers of total ARID3a+ peripheral blood B cells do not correlate with autoantibody reactivity patterns.
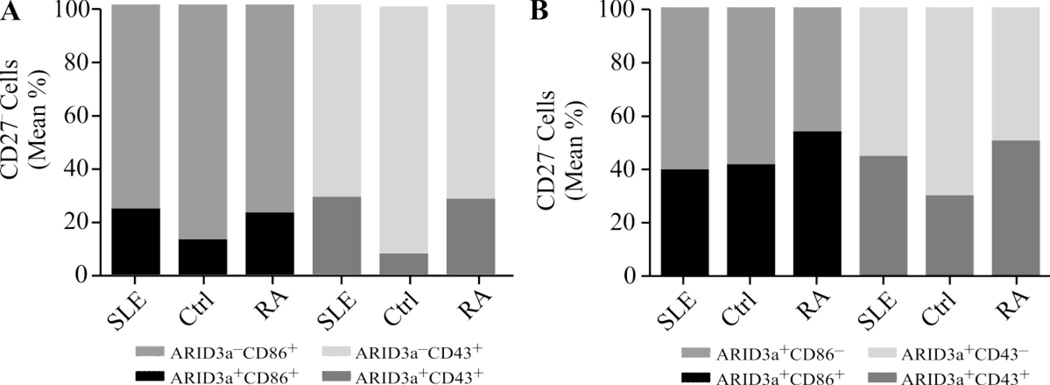
To determine whether ARID3a expression correlated with B cell activation status, we explored expression of the activation marker CD86 and the pre-plasmablast marker CD43 (22–24). Naive B cells are enriched in the CD27− subset which had larger numbers of ARID3a+ cells in many SLE patients, as shown in Figure 2B. Therefore, we examined ARID3a expression in both CD27− CD86+ and CD27−CD43+ B cell subpopulations of SLE, healthy control and RA patient samples (Figure 4A). As expected, a portion of activated CD27− B cells expressed ARID3a (< 30%), with larger percentages occurring in both SLE and RA patient samples compared to healthy controls. However, ARID3a+ cells did not segregate with these markers. The majority of CD27−ARID3a+ SLE B cells (> 56%) did not express CD86 or CD43 (Figure 4B). Therefore, we conclude that ARID3a expression does not correlate with activation status as defined by expression of CD86, or with CD43 expression.
Figure 4. ARID3a expression did not correlate with CD86 and CD43 activation markers in CD27− B cells.
Peripheral blood mononuclear cells were stained for CD27, CD43, CD86 and ARID3a. (A) Mean percentages of CD27−, ARID3a+ and ARID3a− B cells in CD86 and CD43 gates and (B) mean percentages of CD27−, CD86+ and CD86− and CD43+ and CD43− B cells in the ARID3a gate from 42 SLE, 15 controls and 4 RA patient samples are shown.
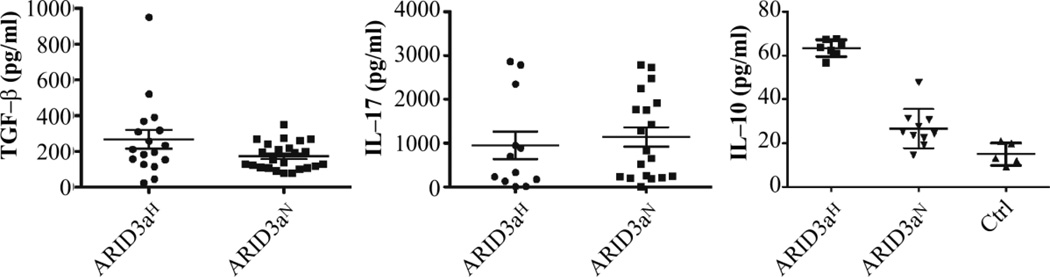
We next evaluated inflammatory cytokine levels previously associated with increased disease severity in SLE. TGF-β, IL-17 (Figure 5) TNF-α and MIP1α levels (not shown) were similar in ARID3aH and ARID3aN samples. Mean IL-10 levels were higher in patients than in healthy controls, with even greater increases in ARID3aH samples (Figure 5). Numbers of IL-10 secreting B10 B cells that act as negative regulators of the immune response (25) were also examined (not shown). No differences in B10 cell numbers between ARID3aH and ARID3aN SLE patients were observed, suggesting that non-circulating B cells or other cell types are responsible for increased IL-10 production in SLE patients.
Figure 5. Circulating IL-10 levels were increased in SLE patients with increased numbers of ARID3a+ B cells.
Plasma levels of TGF-β, IL-17, and IL-10 from ARID3aH (7–15) and ARID3aN (9–24) samples were measured and calculated relative to standard curves. Each symbol represents a different sample. Means and SEMs are indicated.
DISCUSSION
A cross-sectional analysis of 115 samples revealed increased numbers of ARID3a+ peripheral blood B lymphocytes in 43% of lupus patients. ARID3a expression was not limited to B cell differentiation stages that express ARID3a in healthy individuals, but occurred in all patient B cell subsets examined. Total numbers of ARID3a+ CD19+ B cells in 37 patients assessed at multiple time points varied over time, and showed a strong association with SLEDAI scores at each visit by multiple methods of analyses. These data suggest that total numbers of ARID3a+ CD19+ B cells might be tested as a biomarker used to stratify patients in studies of pathogenic mechanisms and/or future trials of relevant targeted treatments.
Subdivision of these SLE patients based on ARID3a expression in B cells did not reveal associations with demographics or age. Two patients receiving no immunosuppressive medications over-expressed ARID3a suggesting that increased numbers of ARID3a+ B cells were not the result of treatment. Rather, ARID3a expression was strongly associated with disease activity. Because total numbers of ARID3a+ CD19+ B cells/ml were used to define ARID3a over-expression, and many patients received immunomodulatory medications during these studies, it is possible that the degree of association between ARID3a expression and disease activity may be an underestimation.
Although ARID3a/Bright enhances transcription of some Ig heavy chains (17), recent data suggest it can also repress transcription of other genes (26,27). Many ARID family proteins are members of large epigenetic regulatory complexes (28), although it is not clear if ARID3a serves similar types of functions in B lymphocytes. Perhaps the most surprising finding was that ARID3aH patient samples displayed bimodal ARID3a expression in most B cell subsets. Because our data show strong associations between total numbers of ARID3a+ CD19+ B cells and disease activity, it will be important to determine if ARID3a expression alters gene expression profiles in those cells that express ARID3a versus those that do not.
Increased plasma IL-10 levels, such as those we observed in ARID3aH patient samples (Figure 5), have been associated with inflammatory responses. We observed increased numbers of ARID3a+ B cells in two controls who were recovering from recent viral infections in our longitudinal studies. This suggested that ARID3a-expressing cells may be recruited as part of normal, and/or inflammatory, immune response. Although some ARID3a+ cells clearly expressed the activation marker CD86, ARID3a expression did not segregate with expression of this marker (Figure 4), an activation marker previously shown to be increased in SLE B lymphocytes (22). Nor did we observe increased numbers of ARID3a+ cells in any of six RA patient samples with similar levels of expression of the activation marker CD86. Therefore, ARID3a expression in SLE B cells did not always coincide with activation.
Expression of ARID3a in SLE naive B lymphocytes that only rarely show detectable ARID3a expression in healthy controls was unexpected. One explanation for the presence of both ARID3a+ and ARID3a− naïve B cells in SLE samples is that the ARID3a+ fraction represents abnormal expansion of B lineage cells that constitutively express it. B1 cells in the mouse may constitute a distinct B cell lineage (29). Mouse B1 B cells constitutively express Bright/ARID3a (21), and these cells were missing in Bright knockout mice (30). Preliminary data recently suggested CD43 was a marker for mature human B1 cells (31); however, others suggested these cells represent an intermediary pre-plasmablast phenotype (24). Regardless, ARID3a expression did not segregate with CD43 expression, so it is not currently possible to distinguish if ARID3a is a marker for a previously undescribed B cell subset.
Alternatively, ARID3a expression in SLE B cells may be a consequence, or a contributing factor of SLE. Although >40% of SLE patient samples showed large numbers of ARID3a+ CD19+ B cells, and increased numbers of these cells were associated with disease activity, RA blood samples did not exhibit expansion of ARID3a+ cells. However, we cannot rule out the possibility that examination of larger numbers of RA samples might show expansion of ARID3a+ cells. Lupus is a polygenic disease, which is thought to arise due to imbalances at multiple regulatory levels of tolerance or immune activity. Immune pathology in lupus is known to be heterogeneous and there are many overlaps with other autoimmune diseases, therefore it is possible that aberrant ARID3a expression is not limited to SLE, or that it might occur transiently in certain healthy immune inflammatory responses. Likewise, ARID3a/Bright over-expression in mouse B lymphocytes caused ANA production, but was insufficient to result in organ-threatening autoimmune pathology or early mortality (10,11). Therefore, dysregulated ARID3a expression could be a predisposing or contributing factor to disease requiring a permissive autoimmune environmental insult or additional genetic predisposition to allow for clinical end-organ impact. Experiments to better define the functions of ARID3a in SLE B cells may shed light on these questions.
Although tremendous progress has been made in identifying genetic associations with SLE, biomarkers that have predictive potential for this diverse disease are essential (7,32,33). Although ARID3a expression in mature B cells was expected, many patients also had dramatically increased numbers of ARID3a+ naïve B cells that are likely precursors to ARID3a+ mature cells. We cannot conclude from the study that numbers of ARID3a+ naïve B cells might be predictive of future disease flares, but it will be important to further explore that idea. The current results showing strong associations with increased numbers of ARID3a+ CD19+ B cells and SLEDAI scores suggest ARID3a may either be useful as a novel biomarker for disease activity, or perhaps eventually be targeted for preventative treatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michelle Ratliff, Katherine Thanou, Hongxia Liu, Michelle Joachims, Thomas F. Tedder and Linda Thompson for technical assistance and helpful discussions, Eliza Chakravarty for reviewing the manuscript, and Beverly Hurt, Beth Mikkola and Shelli Wasson for manuscript preparation.
FUNDING: This work was supported by a Lupus Foundation of America, AI090343 and AI044215 (CFW), and AR053483, GM104938, AI101934, GM103510 and AI082741 (JAJ) from the National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Webb had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analyses.
Study conception and design. Webb. Merrill. James.
Acquisition of data. Ward. Rose.
Analysis and interpretation of data. Webb. Ward. Montgomery. Adrianto.
REFERENCES
- 1.Davidson A, Diamond B. B cells twist and shout. Immunol Cell Biol. 2009;87(7):512–513. doi: 10.1038/icb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum. 2008;58(1):15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 3.Hughes T, Sawalha AH. The role of epigenetic variation in the pathogenesis of systemic lupus erythematosus. Arthritis Res Ther. 2011;13(5):245. doi: 10.1186/ar3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6(12):683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper GS, Gilbert KM, Greidinger EL, James JA, Pfau JC, Reinlib L, et al. Recent advances and opportunities in research on lupus: environmental influences and mechanisms of disease. Environ Health Perspect. 2008;116(6):695–702. doi: 10.1289/ehp.11092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James JA, Harley JB, Scofield RH. Role of viruses in systemic lupus erythematosus and Sjögren syndrome. Curr Opin Rheumatol. 2001;13(5):370–376. doi: 10.1097/00002281-200109000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Liu CC, Kao AH, Manzi S, Ahearn JM. Biomarkers in systemic lupus erythematosus: challenges and prospects for the future. Ther Adv Musculoskelet Dis. 2013;5(4):210–233. doi: 10.1177/1759720X13485503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pathak S, Mohan C. Cellular and molecular pathogenesis of systemic lupus erythematosus: lessons from animal models. Arthritis Res Ther. 2011;13(5):241. doi: 10.1186/ar3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson A, Aranow C. Lupus nephritis: lessons from murine models. Nat Rev Rheumatol. 2010;6(1):13–20. doi: 10.1038/nrrheum.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankar M, Nixon JC, Maier S, Workman J, Farris AD, Webb CF. Anti-nuclear antibody production and autoimmunity in transgenic mice that over-express the transcription factor Bright. J Immunol. 2007;178:2996–3006. doi: 10.4049/jimmunol.178.5.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oldham AL, Miner CA, Wang HC, Webb CF. The transcription factor Bright plays a role in marginal zone B lymphocyte development and autoantibody production. Mol Immunol. 2011;49(1–2):367–379. doi: 10.1016/j.molimm.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349(16):1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 13.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 14.Nixon JC, Rajaiya JB, Ayers N, Evetts S, Webb CF. The transcription factor, Bright, is not expressed in all human B lymphocyte subpopulations. Cell Immunol. 2004;228:42–53. doi: 10.1016/j.cellimm.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Wei C, Anolik J, Cappione A, Zheng B, Pugh-Bernard A, Brooks J, et al. A New Population of Cells Lacking Expression of CD27 Represents a Notable Component of the B Cell Memory Compartment in Systemic Lupus Erythematosus. J Immunol. 2007;178(10):6624–6633. doi: 10.4049/jimmunol.178.10.6624. [DOI] [PubMed] [Google Scholar]
- 16.Kaminski DA, Wei C, Qian Y, Rosenberg AF, Sanz I. Advances in human B cell phenotypic profiling. Front Immunol. 2012;3:302. doi: 10.3389/fimmu.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajaiya J, Nixon JC, Ayers N, Desgranges ZP, Roy AL, Webb CF. Induction of immunoglobulin heavy chain transcription through the transcription factor Bright requires TFII-I. Mol Cell Biol. 2006;26(12):4758–4768. doi: 10.1128/MCB.02009-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruner BF, Guthridge JM, Lu R, Vidal G, Kelly JA, Robertson JM, et al. Comparison of autoantibody specificities between traditional and bead-based assays in a large, diverse collection of patients with systemic lupus erythematosus and family members. Arthritis Rheum. 2012;64(11):3677–3686. doi: 10.1002/art.34651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Touma Z, Urowitz MB, Ibanez D, Gladman DD. SLEDAI-2K 10 days versus SLEDAI-2K 30 days in a longitudinal evaluation. Lupus. 2011;20(1):67–70. doi: 10.1177/0961203310385163. [DOI] [PubMed] [Google Scholar]
- 20.Dörner T, Jacobi AM, Lee J, Lipsky PE. Abnormalities of B cell subsets in patients with systemic lupus erythematosus. J Immunol Methods. 2011;363(187):197. doi: 10.1016/j.jim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Nixon JC, Ferrell S, Miner C, Oldham AL, Hochgeschwender U, Webb CF. Transgenic mice expressing dominant-negative Bright exhibit defects in B1 B cells. J Immunol. 2008;181(10):6913–6922. doi: 10.4049/jimmunol.181.10.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolff S, Wilde B, Patschan S, Durig J, Specker C, Philipp T, et al. Peripheral circulating activated b-cell populations are associated with nephritis and disease activity in patients with systemic lupus erythematosus. Scand J Immunol. 2007;66(5):584–590. doi: 10.1111/j.1365-3083.2007.02008.x. [DOI] [PubMed] [Google Scholar]
- 23.Nagafuchi H, Shimoyama Y, Kashiwakura J, Takeno M, Sakane T, Suzuki N. Preferential expression of B7.2 (CD86), but not B7.1 (CD80), on B cells induced by CD40/CD40L interaction is essential for anti-DNA autoantibody production in patients with systemic lupus erythematosus. Clin Exp Rheumatol. 2003;21(1):71–77. [PubMed] [Google Scholar]
- 24.Covens K, Verbinnen B, Geukens N, Meyts I, Schuit F, Van LL, et al. Characterization of proposed human B-1 cells reveals pre-plasmablast phenotype. Blood. 2013;121(26):5176–5183. doi: 10.1182/blood-2012-12-471953. [DOI] [PubMed] [Google Scholar]
- 25.Iwata Y, Matsushita T, Horikawa M, DiLillo DJ, Yanaba K, Venturi GM, et al. Characterization of a rare IL-10-competent B-cell subset in humans that parallels mouse regulatory B10 cells. Blood. 2011;117(2):530–541. doi: 10.1182/blood-2010-07-294249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An G, Miner CA, Nixon JC, Kincade PW, Bryant J, Tucker PW, et al. Loss of Bright/ARID3a Function Promotes Developmental Plasticity. Stem Cells. 2010;28(9):1560–1567. doi: 10.1002/stem.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popowski M, Templeton TD, Lee BK, Rhee C, Li H, Miner C, et al. Bright/ARID3a acts as a barrier to somatic reprogramming through direct regulation of Oct4, Sox2 and Nanog. Stem Cell Reports. 2014;2:26–35. doi: 10.1016/j.stemcr.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kortschak RD, Tucker PW, Saint R. ARID proteins come in from the desert. Trends Biochem Sci. 2000;25(6):294–299. doi: 10.1016/s0968-0004(00)01597-8. [DOI] [PubMed] [Google Scholar]
- 29.Ghosn EE, Sadate-Ngatchou P, Yang Y, Herzenberg LA, Herzenberg LA. Distinct progenitors for B-1 and B-2 cells are present in adult mouse spleen. Proc Natl Acad Sci U S A. 2011;108(7):2879–2884. doi: 10.1073/pnas.1019764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb CF, Bryant J, Popowski M, Allred L, Kim D, Harriss J, et al. The ARID Family Transcription Factor Bright Is Required for both Hematopoietic Stem Cell and B Lineage Development. Mol Cell Biol. 2011;31(5):1041–1053. doi: 10.1128/MCB.01448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffin DO, Holodick NE, Rothstein TL. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70−. J Exp Med. 2011;208(1):67–80. doi: 10.1084/jem.20101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinman L, Merrill JT, McInnes IB, Peakman M. Optimization of current and future therapy for autoimmune diseases. Nat Med. 2012;18(1):59–65. doi: 10.1038/nm.2625. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Fang X, Li QZ. Biomarker profiling for lupus nephritis. Genomics Proteomics Bioinformatics. 2013;11(3):158–165. doi: 10.1016/j.gpb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.