Abstract
Background
Messenger RNA (mRNA) expression levels in blood cells are important in disease diagnosis, prognosis and biomarker discovery research. Accurate measurements of intracellular mRNA levels in blood cells depend upon several pre-analytical factors, including delays in RNA extraction from blood after phlebotomy. Dramatic changes in mRNA expression levels caused by delays in blood sample processing may render such samples unsuitable for gene expression analysis.
Objectives
This study was conducted to evaluate a blood collection tube, cell-free RNA-BCT® (RNA-BCT), for its ability to stabilize mRNA expression level in blood cells post-phlebotomy using indicator mRNAs in reverse transcription quantitative real-time PCR (RT-qPCR) assays.
Methods
Blood samples from presumed healthy donors were drawn into both RNA-BCT and K3EDTA tubes and maintained at room temperature (18–22 °C). The samples were processed to obtain white blood cells (WBCs) at days 0, 1, 2 and 3. Total cellular RNA was extracted from WBCs and mRNA concentrations were quantified by RT-qPCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), c-fos, and p53 transcripts.
Results
While blood cells isolated from K3EDTA tubes showed significant changes in cellular mRNA concentrations for GAPDH, c-fos, and p53, these mRNAs concentrations were stable in blood drawn into RNA-BCT.
Conclusion
The reagent in the RNA-BCT device stabilizes cellular mRNA concentrations for GAPDH, c-fos and p53 for at least three days at room temperature.
Key Points
| A specialized blood collection device, cell-free RNA BCT (RNA-BCT) is investigated for its ability to stabilize RNA in intact blood cells at room temperature post-phlebotomy. |
| The RT-qPCR-based analysis is carried out to study the expression levels of GAPDH, c-fos and p53 mRNA of neutrophils isolated from blood samples collected in K3EDTA and RNA-BCT devices. |
| Results indicate that expression levels of these mRNAs are stable in neutrophils obtained from RNA-BCT blood for up to 3 days at room temperature as compared to the neutrophils obtained from K3EDTA blood. |
Introduction
Messenger RNA (mRNA) profile of a cell reveals real time activity of that cell and provides useful information in disease diagnosis, prognosis or in biomarker discovery [1–4]. Profiling of cellular mRNA expression patterns has been done by use of microarrays, reverse transcription quantitative real-time PCR (RT-qPCR) and molecular beacons [1–6]. Many of these studies have relied on comparison between affected and unaffected tissue samples. However, tissue biopsies often require highly invasive procedures that may not be readily available. As an alternative, researchers have explored using human peripheral blood cells as a source material for gene expression profiling, which are readily available via a relatively noninvasive procedure [2, 7].
Blood samples in collection devices present some inherent challenges to the genetic profiling of stored blood cells. Post-phlebotomy, ex vivo blood cells are vulnerable to metabolic stress due to the lack of glucose and oxygen during sample storage [8]. As such, certain mRNAs may be up-regulated unintentionally while others may be degraded. The primary aim of accurate gene profiling should be to prevent or mitigate changes to ex vivo mRNA expression patterns that may occur during blood sample handling prior to the extraction of mRNA [9, 10]. Methods for isolation of blood cells often include density-gradient centrifugation for which equipment is frequently beyond the scope of a typical clinical setting. This limitation may require that samples be sent out to an external facility, which further delays the process and alters mRNA profiles [7].
The above challenges emphasize the importance of implementing a blood collection device that is capable of stabilizing mRNAs and their expression at the time of a blood draw. This may be accomplished by stabilizing cells immediately, during storage in the tube and minimizing the time elapsed from blood cell harvest to white blood cell (WBC) isolation and RNA extraction. Reliable mRNA expression profiles in blood cells depend on effectively overcoming post-phlebotomy changes by inhibiting cellular metabolism and RNase activity.
Blood collection technologies, including PAXgene (PreAnalytiX) and Tempus (Applied Biosystems) have been designed to inhibit RNase and metabolic activities by lysing blood cells at the point of blood collection and thereby stabilizing the mRNA expression profile [10–12]. However, there are some associated drawbacks when using these blood collection devices. During total cell lysis, reticulocytes release substantial amounts of α- and β-globin mRNA. The presence of excessive amounts of globin mRNA decreases RT-qPCR detection sensitivity of lower abundance mRNA transcripts and increases signal variation on microarrays [13, 14]. To circumvent these problems, additional methods are performed to reduce globin mRNA from whole blood RNA samples obtained using PAXgene or Tempus blood collection devices [15]. These additional globin depletion steps are reportedly inefficient and require additional costs and resources [16]. These RNA stabilizing devices are also not useful when RNA expression profile of a subset is to be investigated because all WBCs are lysed at the point of phlebotomy. Furthermore, another significant disadvantage of these RNA stabilizing devices is their inability to utilize technologies such as molecular beacon technology, where intact cells are required to visualize the gene-specific fluorescence staining by histology or by flow cytometry.
An ideal blood collection device for genetic profiling would maintain blood cells intact while stabilizing mRNA expression after the blood draw. Such a device would allow investigators to isolate WBCs by widely used methodologies without compromising the original gene expression profile and to utilize stabilized intact cells in gene expression profiling using technologies which require intact cells such as molecular beacon technology. Cell-free RNA BCT® (RNA-BCT) is a blood collection device known to stabilize cell-free RNA in blood during storage and shipping at room temperature for 3 days [17–19]. This study is undertaken to investigate the ability of the RNA-BCT device to stabilize mRNA in intact blood cells during room temperature storage for 3 days using mRNAs for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), c-fos and p53 as indicators. These genes are chosen based on their post-phlebotomy responses; GAPDH as a housekeeping gene, c-fos as a fast acting upregulated oncogene and p53 as a downregulated tumor suppressor gene.
Materials and Methods
Recruitment of Blood Donors
Anonymous volunteer blood donors were recruited from Streck, Inc., Omaha, NE, USA. The study was approved by the institutional review board of the Methodist Hospital, Omaha, NE, USA and informed consent was obtained prior to the blood draw. Donors were both male and female and presumed to be healthy. All donor samples were drawn by venipuncture.
Blood Collection
Blood samples were collected from six donors. From each donor, blood was drawn into four 10 mL K3EDTA tubes (BD Vacutainer®, Becton Dickinson, Franklin Lakes, NJ, USA) and four 10 mL Cell-Free RNA BCT® (Streck Inc., Omaha, NE, USA). Blood samples were mixed immediately after the draw by inverting 10 times and maintained at room temperature (18–20 °C).
Isolation of Neutrophils from Whole Blood Using Magnetic Beads
For neutrophil isolation from a whole blood samples, red blood cells (RBCs) were first removed by lysis. Plasma was removed after centrifugation at 350 × g for 10 minutes and the remaining buffy coat/RBC layer was treated with a red cell lysis buffer (150 mM ammonium chloride, 1 mM EDTA, pH 7.2) in a 1:2 ratio (blood:RBC lysis buffer), for 20 minutes at room temperature. Following centrifugation at 350 × g for 10 minutes, the cell pellets were suspended in isotonic phosphate buffered saline (PBS) containing 0.1 % BSA. Cell counts were determined using a Sysmex KX-21 Hematology Analyzer (Sysmex, Kobe, Japan). The WBC suspension was treated with CD15 MicroBeads to isolate neutrophils following the manufacturer’s protocol (Miltenyi Biotec, Inc., Auburn CA, USA). Briefly, 5 µL MicroBeads suspension was added for every 107 cells and incubated for 45 minutes at 4 °C. A QuadroMACS Separation Unit (Miltenyi Biotec) was used that was capable of performing four simultaneous magnetic isolations. The unit was fitted with MACS LS Separation Columns and the cell suspensions were applied to the columns, wherein the MicroBead-bound cells were retained. The columns were washed with PBS and then the magnetic field was removed before elution with two 1 mL volumes of PBS. After enumerating the cells, aliquots of 2 × 106 neutrophils were prepared and frozen at −80 °C. Total cellular RNA was extracted from these neutrophils to quantify c-fos, GAPDH and p53 mRNAs.
Total Cellular RNA Isolation
RNA was extracted from neutrophils (2 × 106) using a commercially available kit, QIAGEN RNeasy® FFPE Kit, (Qiagen, Santa Clarita, CA, USA). The manufacturer’s recommended protocol was modified for extracting the total RNA. Steps 1–5, and 7 of the protocol were skipped and RNA extraction started with step 6 by treatment of a cell suspension containing 2 × 106 cells with 240 µL of buffer PKD. An additional step was introduced by incubating the samples at 95 °C for 10 minutes. The samples were then cooled by placing them in an ice-bath for 3 minutes. Step 8 was modified by increasing the amount of Proteinase K from 10 to 20 µL. In step 9, the first incubation time at 56 °C was extended from 15 minutes to 1 hour. The second incubation time at 80 °C was kept unchanged at 15 minutes. The samples were eluted in 60 µL sterile nuclease-free water and stored at −80 °C until analysis by RT-qPCR.
RNA Yield and Purity
RNA yield and purity was detected using NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, Delaware USA) following manufacture’s’ recommended protocol.
RT-qPCR
Primers and hydrolysis probes for the RT-qPCR quantification of mRNAs for GAPDH, c-fos, and p53 were purchased from Integrated DNA Technologies (Coralville, IA, USA). Primers and probe for GAPDH mRNA were prepared as previously described by Cohrs et al. [20]. Primers and probe for the quantification of c-fos mRNA were prepared as described by Gan et al. [21]. The primers and probes used for the p53 gene are as follows: forward primer 5′-TCAACAAGATGTTTTGCCAACTG-3′; reverse primer 5′-ATGTGCTGTGACTGCTTGTAGATG-3′; probe 5′/56FAM/ATCAACCCACAGCTGCACAGGGCAGGTCTT-BHQ. Plasmid DNA constructs were prepared by cloning a DNA fragment into Zero Blunt TOPO (Invitrogen, Carlsbad, CA), with each construct containing a single copy of human c-fos, GAPDH, or p53 mRNA. The resulting amplicons are 67, 75, and 118 bp long, respectively. These plasmid constructs were used to constitute the standard curves. The highest standard had 300,000 copies in 5 µL which was serially diluted (10-fold) 5 times to get 30,000, 3,000, 300, 30 and 3 copies per 5 µL. GAPDH and c-fos mRNA were quantified by 1-step RT-qPCR method using TaqMan® RNA-to CT™ 1-step Kit (Applied Biosystems, Foster City, CA, USA). For GAPDH and c-fos assays 5 µL of RNA (~242–358 ng of total RNA) was used. The RT-qPCR protocol included a reverse transcription step at 48 °C for 15 minutes, PCR enzyme activation at 95 °C for 10 minutes, followed by 45 cycles of denaturation at 95 °C for 15 s and anneal / extension at 60 °C for 1 minute. p53 mRNA was quantified by 2-step RT-qPCR method, for which iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad Laboratories, Pleasanton, CA, USA) was used for RT step and TaqMan® Universal Master Mix II, with UNG (Applied Biosystems, Foster City, CA, USA) was used for the PCR step. For p53 cDNA synthesis 2 µL of RNA (~97–143 ng of total RNA) was used. From total cDNA volume of 20, 5 µL of cDNA was used for qPCR. The RT and PCR were performed in CFX96TM Touch (Bio-Rad, Pleasanton, CA, USA) and the data were processed using Bio-Rad CFX Manager software.
Statistical Analysis
Statistical analysis was carried out using Microsoft Excel for Office 2007. Analysis was performed using paired, two-tailed Student’s t test and p < 0.05 was considered statistically significant.
Results
We evaluated the effect of RNA-BCT versus K3EDTA on the stabilization of neutrophil mRNA levels in whole blood samples, stored at room temperature over 3 days. RNA yield and purity was determined by a spectrophotometric method. The mRNA levels were determined by RT-qPCR using gene-specific assays for GAPDH, c-fos and p53. Each data was averaged from duplicate set of PCR samples for six donors.
RNA Yield and Purity
Table 1 shows the effect of storage on RNA yield and purity in neutrophils obtained from blood collected into K3EDTA tube and RNA BCT. There was no statistically significant change in RNA yield in neutrophils obtained from both tube types over time. According to Table 1, there was no statistically significant change also in RNA purity over time as indicated by 260/280 and 260/230 ratios.
Table 1.
RNA yield and purity in neutrophils obtained from blood collected into K3EDTA tubes and RNA BCT
| Sample type | Total RNA yield (µg) | 260/280 nm ratio | 260/230 nm ratio |
|---|---|---|---|
| Mean ± SD (p) | Mean ± SD (p) | Mean ± SD (p) | |
| K3EDTA day 0 | 3.7 ± 0.7 | 1.9 ± 0.09 | 1.8 ± 0.51 |
| K3EDTA day 1 | 3.5 ± 1 (0.77) | 1.9 ± 0.06 (1) | 1.6 ± 0.27 (0.25) |
| K3EDTA day 2 | 2.9 ± 1.4 (0.32) | 1.8 ± 0.06 (0.74) | 1.6 ± 0.15 (0.63) |
| K3EDTA day 3 | 3.6 ± 0.9 (0.81) | 1.8 ± 0.11 (0.9) | 1.7 ± 0.77 (0.84) |
| RNA-BCT day 0 | 2.9 ± 0.5 (0.06) | 1.8 ± 0.1 (0.64) | 1.32 ± 0.4 (0.26) |
| RNA-BCT day 1 | 3.4 ± 0.7 (0.58) | 1.8 ± 0.05 (0.80) | 1.5 ± 0.3 (0.35) |
| RNA-BCT day 2 | 4.3 ± 2 (0.58) | 1.8 ± 0.09 (0.43) | 1.5 ± 0.3 (0.50) |
| RNA-BCT day 3 | 3.7 ± 1 (0.91) | 1.7 ± 0.3 (0.43) | 1.7 ± 0.1 (0.69) |
Total RNA was extracted from 2 × 106 neutrophils and eluted in 60 µL of RNase and DNase free water as described in the Sect. 2. Total RNA concentration, 260/280 and 260/230 nm ratios was determined using NanoDrop® ND-1000 spectrophotometer. All time points in both tube types were compared against K3EDTA day 0 value. n = 6
Effect of RNA-BCT Reagent on GAPDH mRNA
Figure 1 shows the effect of K3EDTA tube and RNA-BCT device on GAPDH mRNA level when blood samples drawn into the respective blood collection tubes were stored at room temperature over 3 days. The initial mean GAPDH mRNA concentration in K3EDTA samples (Fig. 1a) was 2.39 × 104 copies/1 × 106 neutrophils. This initial value increased over time by 5.8-, 1.1- and 1.2-fold at days 1, 2 and 3, respectively. The increase observed at day 1 was statistically significant (p = 0.01). A similar initial mean GAPDH mRNA concentration, 2.88 × 104 copies/1 × 106 neutrophils, in RNA-BCT sample (Fig. 1b) was determined. However, the GAPDH level in neutrophils obtained from blood samples drawn into RNA-BCT showed no change at day 1 and a 1.2-fold statistically non-significant decrease at days 2 and 3 (p values 0.7 and 0.8, respectively).
Fig. 1.
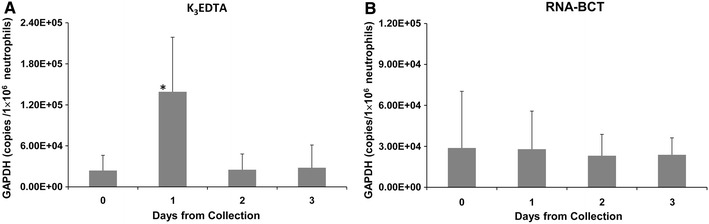
Effect of storage of blood samples on GAPDH mRNA concentration in neutrophils obtained from K3EDTA tubes (a) or RNA-BCTs (b). Blood from normal healthy donors were drawn into K3EDTA tubes and RNA-BCTs and stored at room temperature. Neutrophils were isolated on days 0, 1, 2 and 3, total RNA extracted, and mRNA for GAPDH was quantified by RT-qPCR. The GAPDH mRNA concentration in neutrophils obtained from K3EDTA blood increased over time compared to initial day 0 value. There was 2.2-, 1.1- and 1.2-fold increases at days 1, 2 and 3, respectively. The GAPDH mRNA concentration in neutrophils obtained from RNA-BCTs remained relatively stable over time except for a slight decrease at days 2 and 3. Error bars indicate SD. n = 6. *p = 0.01
Effect of RNA-BCT Reagent on c-Fos mRNA
Figure 2 shows the effect of two blood collection tubes on c-fos mRNA level in neutrophils obtained from whole blood samples. The initial mean c-fos mRNA concentration in neutrophils obtained from K3EDTA blood (Fig. 2a) was 3.60 × 105 copies/1 × 106 neutrophils. This initial value increased over time showing 5.9- and 1.4-fold increases at days 1 and 2, respectively. However, the increases observed at days 1 and 2 were not statistically significant (p values 0.08 and 0.5, respectively). At day 3 there was a slight decrease in c-fos mRNA level in neutrophils isolated from K3EDTA blood. The initial mean c-fos mRNA concentration in neutrophils obtained from RNA-BCT blood (Fig. 2b) was 6.09 × 104 copies/1 × 106 neutrophils, which was significantly low compared to initial value for K3EDTA neutrophils. However c-fos mRNA concentrations in neutrophils obtained from RNA-BCT blood were relatively stable over time showing 1.7-, 1.5- and 1.8-fold statistically non-significant increase at days 1, 2 and 3, respectively with corresponding p values 0.2, 0.1 and 0.2 compared to the initial value.
Fig. 2.
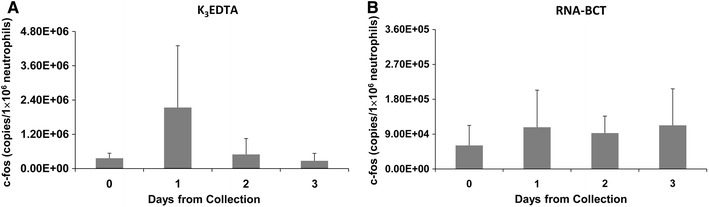
Effect of storage of blood samples on c-fos mRNA concentration in neutrophils obtained from K3EDTA tubes (a) or RNA-BCTs (b). Blood from normal healthy donors were drawn into K3EDTA tubes and RNA-BCTs and stored at room temperature. Neutrophils were isolated on days 0, 1, 2 and 3, total RNA extracted, and mRNA for c-fos was quantified by RT-qPCR. The fos mRNA concentrations in neutrophils obtained from K3EDTA blood showed 3.0 and 1.4-fold increases on days 1 and 2, respectively as compared to initial day 0 value. On day 3 there was a 1.2-fold decrease compared to the day 0 value. The c-fos mRNA concentration in neutrophils obtained from RNA-BCT blood was relatively stable. Error bars indicate SD. n = 6
Effect of RNA-BCT Reagent on p53 mRNA
Figure 3 illustrates the effects of two blood collection tubes on p53 mRNA level. The p53 mRNA level in neutrophils obtained from K3EDTA tubes showed a statistically significant decrease over three days. The initial mean p53 mRNA concentration in neutrophils obtained from K3EDTA blood (Fig. 3a) was 3.20 × 105 copies/1 × 106 neutrophils. On days 1, 2 and 3 the p53 expression levels are decreased by 2.4-, 3.2- and 2.5-fold, respectively. The decreases observed on days 1 and 2 were statistically significant showing p values of 0.005 and 0.04, respectively. The p53 mRNA level in RNA-BCT samples (Fig. 3b) were stable compared to K3EDTA blood samples. The initial mean p53 mRNA concentration in neutrophils obtained from RNA-BCT blood was 3.18 × 105 copies/1 × 106 neutrophils. The p53 mRNA levels in neutrophils obtained from RNA-BCT were relatively stable at all time points even though there was a slight decrease at day 2 compared to initial day 0 value (1.2-fold) which was not statistically significant (p = 0.8).
Fig. 3.
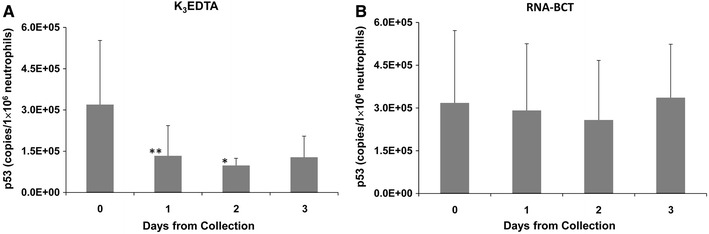
Effect of storage of blood samples on p53 mRNA concentration in neutrophils obtained from K3EDTA tubes (a) or RNA-BCTs (b). Blood from normal healthy donors were drawn into K3EDTA tubes and RNA-BCTs and stored at room temperature. Neutrophils were isolated on days 0, 1, 2 and 3, total RNA extracted, and mRNA for p53 was quantified by RT-qPCR. The p53 mRNA concentration in neutrophils obtained from K3EDTA blood decreased over time compared to initial day 0 value. There were 1.1- and 1.3-fold decrease at days 2 and 3, respectively. The p53 mRNA concentration in neutrophils obtained from RNA-BCTs remained relatively stable over time except for a slight decrease at day 1. Error bars indicate SD. n = 6. * p = 0.04, ** p = 0.005
Discussion
Cellular RNA in human peripheral blood cells can be a valuable source for noninvasive diagnosis and prognosis of various disease conditions and for new biomarker discovery. In vivo changes in expression levels of specific mRNAs can provide important information associated with different disease states. Among the challenges for the accurate detection of mRNA expression levels in human peripheral blood cells, are perturbations caused by delays between phlebotomy and RNA extraction from blood cells [8]. Therefore, there is a need for a blood collection device which can stabilize mRNA levels in human peripheral blood cells at room temperature when RNA extraction from blood is to be delayed. Previous studies demonstrated that the reagent cocktail in a RNA-BCT device can stabilize circulating cell-free RNA in whole blood plasma by preventing cellular RNA release into plasma during sample storage and shipping [17]. This study is undertaken to investigate the ability of a RNA-BCT device to stabilize cellular mRNA level in intact peripheral blood cells using mRNAs for GAPDH, c-fos and p53 in neutrophils as markers.
In our experiments, blood drawn into standard K3EDTA collection tubes showed an increase in GAPDH mRNA level while GAPDH mRNA level in blood drawn into RNA-BCT remained relatively stable for three days at room temperature (Fig. 1a). Salway, Godfrey and their colleagues have also found that GAPDH mRNA concentrations increase with time in white blood cells obtained from non-stabilized whole blood samples [20, 21]. As depicted in Fig. 2a, c-fos mRNA levels in neutrophils obtained from blood drawn into K3EDTA blood increased on day 1 but then decreased on days 2 and 3. This is similar to the c-fos expression pattern observed by Rainen et al. [12]. However, c-fos mRNA level of neutrophils obtained from RNA-BCT blood remained relatively stable for 3 days at room temperature (Fig. 2b). Initial c-fos mRNA levels detected in RNA-BCT sample is lower than the initial c-fos mRNA level detected in K3EDTA. Earlier studies showed that the up-regulation of c-fos in peripheral blood cells, obtained from non-stabilized blood, is very fast and occurs within hours [12, 22, 23]. Our result is consistent with the literature report as c-fos may have been up-regulated between phlebotomy and neutrophil lysis, contributing to the high initial c-fos mRNA copy numbers in the K3EDTA stored sample. Conversely the initial c-fos mRNA copy numbers of RNA-BCT stored samples remained low and possibly represent the actual value. The mRNA level for p53 in neutrophils obtained from K3EDTA tube decreases slowly with time. The expression levels are decreased 1.1-fold on day 2 and 1.4-fold on day 3. This is similar to the results reported by Rainen et al. [12]. On the contrary, p53 mRNA level remains relatively stable in neutrophils obtained from the blood drawn in to the RNA-BCT device. Our results indicate that chemical cocktail present in RNA-BCT can stabilize RNA in peripheral blood cells in a blood sample for 3 days at room temperature.
Unintended alteration in cellular RNA expression that may take place between phlebotomy and RNA extraction can be reduced if blood cells are separated immediately after blood draw and then the RNA extracted. However, immediate processing can be challenging when the blood sample is collected in a remote location and necessary separation equipment is not available. The new RNA-BCT device can be used to circumvent these issues. Since the RNA-BCT device stabilizes cellular RNA in peripheral blood cells, it is possible to store patient blood samples in these vacuum blood collection tubes at ambient temperatures for up to 3 days without any major change to RNA expression profile in blood cells. This may enhance the clinical utility of cellular RNA in blood samples for diagnostic and prognostic assay development and new biomarker discovery efforts.
Conclusion
Our results indicate that the expression patterns of GAPDH, c-fos and p53 genes are stable in the blood samples collected in the RNA-BCT device. Such stability in RNA expression is necessary to rely on RNA-based diagnostic assay results or to accurately develop new RNA-based biomarkers. Although our study is limited by the number of donors and genes, this opens up an encouraging possibility to study the effects on the expression levels of more genes in the blood samples drawn in RNA-BCT.
Acknowledgments
Authors are graciously thankful to Zachari R. Friesen for graphic preparation; Stephanie M. Wigginton and Joel M. Lechner for assisting JRA in neutrophil isolation.
Conflict of interest
TLW declares no conflict of interest. KD, SEN, JRA, GDK and MRF are employees of Streck, Inc.
References
- 1.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. PNAS. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, et al. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- 4.Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homolog of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/S0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 5.Tsuji A, Koshimoto H, Sato Y, Hirano M, Sei-lida Y, Kondo S, Ishibashi K. Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys J. 2000;78:3260–3274. doi: 10.1016/S0006-3495(00)76862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Ernst L, Wagner M, Waggoner A. Sensitive detection of RNAs in single cells by flow cytometry. Nucleic Acids Res. 1992;20:83–88. doi: 10.1093/nar/20.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baechler EC, Batliwala FM, Karypis G, Gaffney PM, Moser K, Ortmann WA, et al. Expression levels for many genes in human peripheral blood cells are highly sensitive to ex vivo incubation. Genes Immun. 2004;5:347–353. doi: 10.1038/sj.gene.6364098. [DOI] [PubMed] [Google Scholar]
- 8.Härtel C, Bein G, Müller-Steinhardt M, Klüter H. Ex vivo induction of cytokine mRNA expression in human blood samples. J Immun Methods. 2001;249:63–71. doi: 10.1016/S0022-1759(00)00334-3. [DOI] [PubMed] [Google Scholar]
- 9.Choi SJ, Shin IJ, Je K-H, Min EK, Kim EJ, Kim H-S, Choe S, Kim D-E, Lee DK. Hypoxia antagonizes glucose deprivation on Interleukin 6 expression in an Akt dependent, bu HIF-1/2α independent manner. PLoS One. 2013;8:e58662. doi: 10.1371/journal.pone.0058662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debey S, Schoenbeck U, Hellmich M, Gathof BS, Pillai R, Zander T, Schultze JL. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and role of different cell types. Pharmacogenomics J. 2004;4:193–207. doi: 10.1038/sj.tpj.6500240. [DOI] [PubMed] [Google Scholar]
- 11.Duale N, Brunborg G, Rønningen KS, Briese T, Aarem J, Aas KA, et al. Human blood RNA stabilization in samples collected and transported for a large biobank. BMC Res Notes. 2012;5:510–519. doi: 10.1186/1756-0500-5-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, et al. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–1890. [PubMed] [Google Scholar]
- 13.Wright C, Bergstrom D, Dai H, Marton M, Morris M, Tokiwa G, et al. Characterization of globin RNA interference in gene expression profiling of whole-blood samples. Clin Chem. 2008;54:396–405. doi: 10.1373/clinchem.2007.093419. [DOI] [PubMed] [Google Scholar]
- 14.Feezor RJ, Baker HV, Mindrinos M, Hayden D, Tannahill CL, Brownstein BH, et al. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19:247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 15.Mastrokolias A, den Dunnen JT, van Ommen GB, Hoen PAC, van Roon-Mom WMC. Increased sensitivity of next generation sequencing-based expression profiling after globin reduction in human blood RNA. BMC Genomics. 2012;13:28–37. doi: 10.1186/1471-2164-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Walter E, Stenger D, Thach D. Effects of globin mRNA reduction methods on gene expression profiles from whole blood. J Mol Diagn. 2006;8:551–558. doi: 10.2353/jmoldx.2006.060021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernando MR, Norton SE, Luna KK, Lechner JM, Qin J. Stabilization of cell-free RNA in blood samples using a new collection device. Clin Biochem. 2012;45:1497–1502. doi: 10.1016/j.clinbiochem.2012.07.090. [DOI] [PubMed] [Google Scholar]
- 18.Qin J, Williams TL, Fernando MR. A novel blood collection device stabilizes cell-free RNA in blood during sample shipping and storage. BMC Res Notes. 2013;6:380. doi: 10.1186/1756-0500-6-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin J, Bassett C, Fernando MR. Preservation of circulating cell-free fetal RNA in maternal blood using a blood collection device containing a stabilizing reagent. J Mol Genet Med. 2014;14:23–8.
- 20.Godfrey TE, Kim SH, Chavira M, Ruff DW, Warrant RS, Gray JW, Jensen RH. Qunatitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5′ nuclease quantitative reverse trasnscription-polymerase chain reaction. J Mol Diagn. 2000;2:84–91. doi: 10.1016/S1525-1578(10)60621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salway F, Day PJR, Ollier WER, Peakman TC. Levels of 5′ RNA tags in plasma and buffy coat from EDTA blood increase with time. Int J Epidemiol. 2008;37:i11–i15. doi: 10.1093/ije/dym279. [DOI] [PubMed] [Google Scholar]
- 22.Gan L, Doroudi R, Hägg U, Johansson AN, Selin-Sjögren L, Jern S. Differential immediate-early gene response to shear stress and intraluminal pressure in intact human conduit vessels. FEBS Lett. 2000;477:89–94. doi: 10.1016/S0014-5793(00)01788-9. [DOI] [PubMed] [Google Scholar]
- 23.Hu E, Mueller E, Oliviero S, Papaionnou VE, Johnson R, Spiegelman BM. Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factors or oncogenes. EMBO J. 1994;13:3094–3103. doi: 10.1002/j.1460-2075.1994.tb06608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


