Abstract
β-N-methylamino-l-alanine (BMAA) is a neurotoxic non-protein amino acid suggested to be involved in neurodegenerative diseases. It was reported to be produced by cyanobacteria, but also found in edible aquatic organisms, thus raising concern of a widespread human exposure. However, the chemical analysis of BMAA and its isomers are controversial, mainly due to the lack of selectivity of the analytical methods. Using factorial design, we have optimized the chromatographic separation of underivatized analogues by a hydrophilic interaction chromatography coupled to tandem mass spectrometry (HILIC-MS/MS) method. A combination of an effective solid phase extraction (SPE) clean-up, appropriate chromatographic resolution and the use of specific mass spectral transitions allowed for the development of a highly selective and sensitive analytical procedure to identify and quantify BMAA and its isomers (in both free and total form) in cyanobacteria and mollusk matrices (LOQ of 0.225 and 0.15 µg/g dry weight, respectively). Ten species of cyanobacteria (six are reported to be BMAA producers) were screened with this method, and neither free nor bound BMAA could be found, while both free and bound DAB were present in almost all samples. Mussels and oysters collected in 2009 in the Thau Lagoon, France, were also screened, and bound BMAA and its two isomers, DAB and AEG, were observed in all samples (from 0.6 to 14.4 µg/g DW), while only several samples contained quantifiable free BMAA.
Keywords: cyanotoxins, BMAA, DAB, AEG, HILIC-MS/MS, cyanobacteria, bivalve mollusks, French Mediterranean
1. Introduction
β-N-methylamino-l-alanine (BMAA) is a non-protein amino acid that was discovered in 1967 from the seeds of Cycas micronesica on the island of Guam [1]. This neurotoxic compound [2] was suggested to be linked to the high incidence of amyotrophic lateral sclerosis/Parkinsonism-dementia complex (ALS/PDC) observed among the native Chamorro people. This hypothesis was later criticized and rejected [3] and subsequently reinforced in the early 2000s. At this time, a group demonstrated successively that BMAA can be produced by a symbiotic cyanobacteria (Nostoc sp.) [4], that BMAA can be biomagnified within the Guam ecosystem, from cyanobacteria to the brains of people who died from ALS/PDC [4,5,6], and that large amounts of BMAA can be released from proteins after total acid hydrolysis of samples [5].
After Cox et al. [7] reported that almost all cyanobacteria can produce BMAA, other groups have found BMAA in different ecosystems all around the world [8,9,10,11,12], and three patterns of biomagnification were also suggested, respectively, in the Baltic Sea [13], in the Florida Bay [14] and in Lake Taihu, China [15]. Taken together with the observation of BMAA in the brains of Canadian patients who suffered from several neurodegenerative diseases [16], these findings suggested a possible widespread human exposure to the BMAA neurotoxin and its global implication in ALS and also in Alzheimer’s disease and Parkinson’s disease.
However, the presence of BMAA in cyanobacteria and other matrices is controversial [17,18]. Indeed, the classically-used fluorescent mode of detection after derivatization of samples is known to lack selectivity [19], and high amounts of BMAA were reported with this method (up to 7000 µg/g dry weight). On the opposite side, when more selective methods, like LC-MS/MS, were employed, concentrations or even the presence of BMAA were not confirmed [12,19,20,21,22]. The derivatization allows for an increase of the molecular weight of molecules, thus reducing the background signal while improving the ionization efficiency with electrospray ionization systems [23]. Nevertheless, the derivatization is not specific; it affects the chromatographic separation and leads to the indirect detection of BMAA [17]. Discrepancies observed among the BMAA concentrations reported likely originate from the lack of selectivity of the methods employed and not because of the use of a derivatization step [18]. Indeed, highly selective methods have been validated for BMAA and analogues [24,25].
The existence of at least three natural isomers, 2,4-diaminobutyric acid (DAB), N-2-aminoethylglycine (AEG) and β-amino-N-methyl-alanine (BAMA), can be partly involved in BMAA controversy and highlighted the requirement of highly selective methods to unambiguously detect and quantify BMAA from its isomers. DAB is a neurotoxic isomer of BMAA [17] that was first found in cyanobacteria in 2008 [22], but it has also been widely reported in prokaryotes and eukaryotes [23]. Among all known isomers of BMAA, AEG and BAMA were selected by two groups for method development, because they could potentially interfere with BMAA analysis [26,27]. Indeed, AEG was found in cyanobacteria, and its production was suggested to be highly conserved [28], while BAMA was observed in mollusks of the Baltic Sea [27]. The toxicity of these two isomers has not been studied so far, especially for AEG.
To date, two papers have reported BMAA in cyanobacteria with selective MS/MS methods using a derivatization of samples [25,29]. Recently, BMAA was also reported to be produced by diatoms and dinoflagellates [30,31,32]. However, the free form of BMAA was not considered. It is unclear why the free form of BMAA is no longer analyzed, since it seems reasonable to assume that biomagnification could more easily originate from the free rather than the bound form of BMAA. Indeed, Dunlop et al. [33] have reported that free BMAA can be misincorporated into human neuroproteins instead of serine in an in vitro cell line, and this incorporation into proteins had been initially postulated as the mechanism of the bioaccumulation of BMAA in the Guam ecosystem [5].
BMAA is a small molecule that can be directly detected by mass spectrometry (MS), but the use of a derivatization step is also chosen by some groups, followed by either fluorescent (FLD) or MS detection. To cope with the complexity of biological matrices and the controversy arising from detecting trace amounts of BMAA, we aimed at the development of a highly selective and sensitive LC-MS/MS method to confidently quantify both free and bound forms of BMAA, DAB and AEG. Thus, following the recommendations of Cohen [34], we included a solid phase extraction (SPE) clean-up step in the sample preparation procedure and the use of an isotopically labeled molecule (i.e., deuterated D5DAB) as the internal standard. There are ongoing debates about the use of a derivatization step [17,20,23]. While the derivatization approach is interesting in combination with mass spectrometry, there is little information in the literature on the derivatization yield for BMAA [34]. A lot of compounds can react with the 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) reagent during the derivatization process [17]. Furthermore, the analysis of BMAA and analogues is already rather complex, and any additional step could contribute to recovery losses and add variability to the method. Additionally, the chromatographic separation of the derivatized analogues is poor compared to that of the underivatized compounds [17]. We therefore chose not to use a derivatization step in our analytical approach. We decided to optimize the analysis of underivatized BMAA and isomers (DAB and AEG) with HILIC chromatography coupled to a sensitive triple quadrupole mass spectrometer (QTRAP 5500 system). This optimized procedure was then applied to screen ten species of cyanobacteria and nineteen samples of mollusks collected in 2009 in Thau Lagoon, French Mediterranean, where BMAA and DAB were recently reported in mussels and oysters with an LC-MS/MS method employing derivatization of the samples [35].
2. Results and Discussion
2.1. Optimization of the Analysis of BMAA and Isomers
The analysis of BMAA requires several steps. After performing cell lysis in an adapted solvent, samples should be cleaned up using an SPE procedure to remove interfering compounds and then injected into an LC-MS/MS system to take advantage of the higher selectivity of this hyphenated technique. BMAA can be extracted either as the free or as the bound form. To release the BMAA bound to proteins, an acid hydrolysis step has to be added before SPE clean-up.
2.1.1. Cell Lysis and Solvent Extraction
To extract cyanotoxins, including BMAA (both in free and protein-bound form), cells have to be disrupted in a solvent facilitating the solubilization of the analytes. Generally, freeze-thawing and maceration [36] or sonication [37] in an appropriate solvent were employed. However, the cell lysis of cyanobacteria can be hard to achieve [38]. Using a mixer mill, excellent disruption of cyanobacteria was observed (checked by microscopic observation). The protocol, proposed by Serive et al. [39], was adapted to extract polar and basic compounds from freeze-dried material.
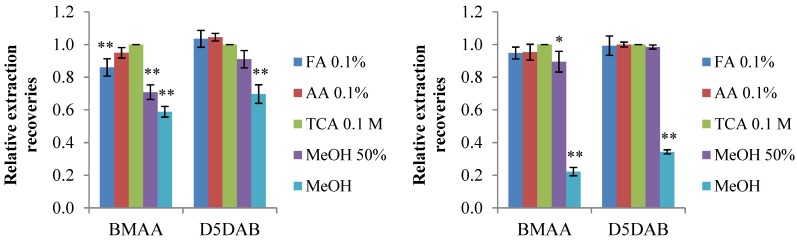
To select an adapted solvent of extraction, both freeze-dried cyanobacteria Leptolyngbya PCC 73110 and mussel (Mytilus galloprovincialis) were spiked in triplicate with BMAA and D5DAB before grinding and extracted with five solvents. To date, different solvents have been used to extract free, bound or total BMAA [34]. We compared the recoveries for free BMAA and its isomers between formic acid (FA) 0.1%, acetic acid (AA) 0.1%, trichloroacetic acid (TCA) 0.1 M, methanol (MeOH) and MeOH/water-FA 0.1% (50:50, v/v). The solvents were chosen accordingly to the literature: e.g., TCA 0.1 M is classically used to extract BMAA [34]; acetic acid is employed to extract paralytic shellfish poisoning (PSP) toxins (basic polar compounds) [40]; while MeOH is known to be able to extract marine toxins of a wide range of lipophilicity from wet algae [41]. The samples were analyzed after SPE clean-up as detailed in the Experimental Section (Section 3.3.2). The results obtained were expressed relatively to the recoveries in the TCA 0.1 M for cyanobacteria and mussel matrices (Figure 1), because it is widely used to extract free BMAA.
Figure 1.
Extraction recoveries of BMAA and D5DAB spiked at 30 ng/mL in (A) cyanobacteria and (B) mussel matrix with five different solvents (n = 3, error bars represent standard deviations). Results are expressed relatively to the recoveries in trichloroacetic acid (TCA) 0.1 M. * p < 0.01 and ** p < 0.001 compared to TCA 0.1 M. FA, formic acid; AA, acetic acid.
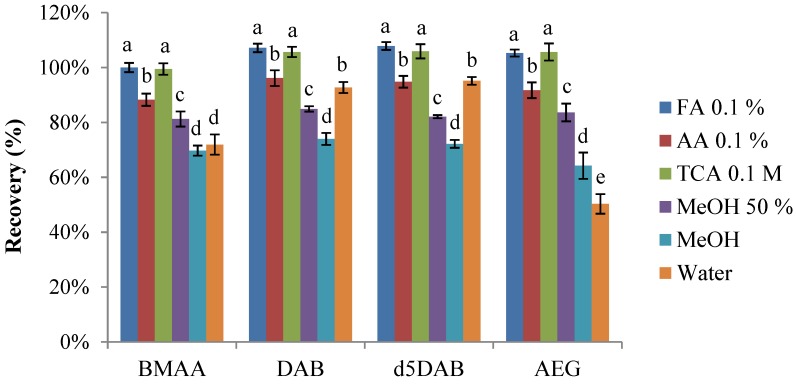
For D5DAB, all solvents, but pure MeOH (p < 0.001), gave similar recoveries. For BMAA, the performances of acidic solvents are alike (except FA 0.1% with cyanobacteria), but lower recoveries were observed in the presence of methanol. The poor recoveries obtained with pure methanol for both matrices were expected, as BMAA and D5DAB are polar compounds. Indeed, the recoveries of pure standards were only about 70% (Figure 2) when using methanol as the extraction solvent, which may indicate low solubility and/or adsorption to glass vials, as suggested by Cohen [34]. Considering the recoveries of standards (Figure 2) and the recoveries obtained with spiked matrices (Figure 1), we chose TCA 0.1 M as the solvent for the extraction of free BMAA and isomers.
Figure 2.
Recoveries of pure standards spiked at 30 ng/mL in different extraction solvents (n = 3, error bars represent standard deviations). Replicates with the same letter are not significantly different, while replicates with different letters are significantly different (p < 0.05).
Subsequently, the protocol was adapted to extract total rather than the bound form of analytes, since the precipitation of proteins is time-consuming and difficult [34]. The classical acid hydrolysis in boiling hydrochloric acid (HCl) 6 M [5] was performed following extraction with TCA 0.1 M.
2.1.2. Comparison of Solid Phase Extraction (SPE) Sorbents
The SPE clean-up step is necessary to remove co-eluting interfering compounds that cause matrix effects during LC-MS/MS analysis while concentrating samples [42,43,44,45]. As basic amino acids, BMAA and isomers (DAB, D5DAB and AEG) can be purified with cation-exchange sorbents, such as Strata SCX (Strong cation exchange) [46], or mixed-mode sorbents, like Oasis® MCX [21] or Isolute® HCX-3 [25]. Here, the widely-used Oasis® MCX [21,24,45] was compared to its Agilent equivalent, Bond Elut® Plexa PCX sorbent (Agilent Technologies, Les Ulis, France), for the recovery of the standards (Figure 3).
Figure 3.
Recoveries of mixed solutions of standards spiked in TCA 0.1 M at a final concentration of 30 ng/mL. Comparison between 60 mg/3cc Plexa PCX and Oasis® MCX cartridges (n = 3, error bars represent standard deviations). * p < 0.05 compared to recoveries with Plexa PCX. # p < 0.005 compared to D5DAB and DAB with Oasis® MCX.
Recoveries with Plexa PCX were better than those obtained with Oasis® MCX cartridges (p < 0.05), except for BMAA. The recoveries obtained here with Oasis® MCX were lower than those obtained for BMAA and DAB by other groups [24,45], possibly related to small differences in protocols. As extraction was performed in TCA 0.1 M, we decided to directly percolate samples in TCA after activation of the cartridges (2 mL of MeOH, then 1 mL of TCA 0.1 M). Indeed, due to their pKa (>6.5) [45], the pH of TCA 0.1 M allows for the ionization of ammonia groups and, hence, retention of positively-charged molecules on the sorbent. To increase recoveries (especially for AEG; data not shown), we used an elution solvent consisting of a mixture of MeOH/NH4OH (93:7 instead of 95:5, v/v). Finally, as trace levels of BMAA and isomers (about 15% of the concentration of the spiking solution) were found in fractions eluting after the 3 mL used by Combes et al. [24], an additional 1 mL was used for the elution step.
It has to be noted that an unknown peak eluted close to the AEG retention time in blank (1 mL TCA 0.1 M) passed through both types of cartridges during LC-MS/MS analysis. However, this unknown compound originating from sorbents is different from the one reported by Li et al. with the Strata-X-C cartridge [45], since only the transition m/z 119 > 102 gave a signal. To accurately quantify AEG, the peak area of this interfering compound was subtracted from the peak area of AEG in samples.
The SPE clean-up procedure with PCX cartridges, as described in the Experimental Section (Section 3.2.2), was applied to all samples after extraction of both free and total BMAA and its isomers.
2.1.3. Optimization of Selectivity: Chromatographic Resolution and Mass Spectral Transitions
Among cyanobacterial toxins, BMAA is a compound that has caused controversy, due to the confusion arising from interferences in the analytical determination [18]. Indeed, at least three isomers of BMAA—DAB, AEG and BAMA—can be found in biological matrices [12,25,30,32]. The large discrepancies of BMAA concentrations observed between studies were suggested to come mainly from the lack of selectivity of the methods that have been used [18]. Therefore, we decided to maximize both chromatographic resolution and the selectivity of mass spectral transitions during the method development.
To have a proper identification of BMAA and isomers, chromatographic resolution has to be reached, since the common mass spectral transition m/z 119 > 102 is used to quantify underivatized samples. For this purpose, a gradient elution with a ZIC®-HILIC column was optimized using a 23 factorial design. The main effect and interaction of three variables, namely acetonitrile (ACN) at the start of gradient (60%–70%), oven temperature (25–35 °C) and the slope of the gradient (0.8%–1.5% ACN/min), were studied on BMAA/DAB and DAB/AEG resolutions. Injections were made twice, which led to 16 experiments, and then, a response surface was generated (Figure 4).
Figure 4.
Chromatographic resolution between BMAA/DAB, optimized using a 23 factorial design. Since temperature did not influence the resolution significantly, it was set to 30 °C. A resolution of 1.5 corresponds approximately to the baseline resolution. ACN, acetonitrile.
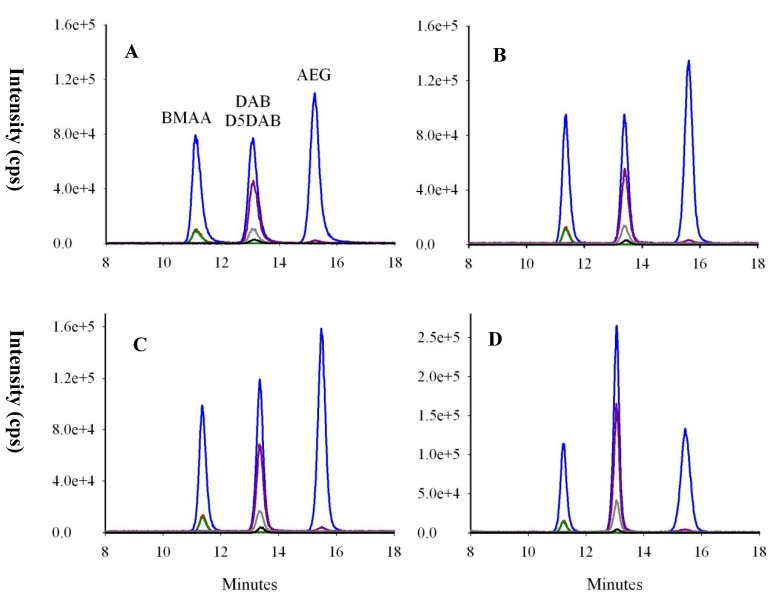
Hydrophilic interaction liquid chromatography (HILIC) columns are designed to efficiently separate polar compounds [47]. The ZIC®-HILIC column has been successfully used to separate BMAA and DAB [11,19,22,24], and the response surface obtained after the factorial design here clearly shows that excellent resolutions can be achieved with this column for underivatized molecules (BMAA, DAB and also AEG). Conditions were set to have a baseline resolution of 1.5 between molecules, as can be seen in Figure 5. Peak maxima were ca. 2 min apart, with a baseline separation of 1 min typically being achieved, thus reducing the risk of misidentification of BMAA, DAB and AEG, while limiting the co-elution of interfering compounds causing matrix effects.
Figure 5.
Chromatograms of BMAA, D5DAB and AEG spiked at 50 ng/mL into (A) standards; (B) cyanobacteria; (C) oyster and (D) mussel matrices after extraction of free analytes. Colored lines represent mass spectral transitions at m/z 119 to m/z 102 (blue), 88 (red), 76 (green), 101 (purple), 74 (grey) and m/z 124 to m/z 47 (dark).
Classically, LC-MS/MS analysis is preferred to quantify BMAA, because of the four criteria for the identification of molecules (retention times, parent ion, product ion(s) and ion ratios) [18]. For maximum selectivity, we decided to consider not only one (as methods involving derivatization do [25]), but two specific (qualitative) transitions for BMAA and DAB and the ratios between qualitative and quantitative ions (respectively, m/z 119 to m/z 88 and 76 for BMAA and m/z 119 to m/z 101 and 74 for DAB; ratios 88/102, 76/102, 101/102 and 74/102). For AEG, no specific mass spectral transition was found (m/z 119 > 41 was specific, but suffered from a high background signal). Generally, the ratios of the qualifier ion(s) to the quantifier ion that are used to confirm the identity of molecules in samples should be within a 10% error range of the ratios of standards [27], which was the case in our study (Table 2). Finally, we have developed a highly selective method to confidently identify BMAA and isomers in biological samples.
Table 2.
Retention times, correlation coefficients (R2) and ion ratios of standards’ curves and spiked cyanobacteria (Leptolyngbya PCC73110), oyster and mussel matrices (mean ± SD) for the extraction of free and total amino acids. For standards, the results came from 33 injections of the five-point calibration curves. For matrices, results originate from the established three-point standards’ curves (3–50 ng/mL) for matrix effects injected twice (for r2, n = 2) and both matrix effects and SPE recoveries for RT and ion ratios (n = 6).
| Samples | RT (min) | R2 | Ion Ratios (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMAA | DAB | D5DAB | AEG | BMAA | DAB | D5DAB | AEG | 88/102 | 76/102 | 101/102 | 74/102 | |
| Standards | 11.25 ± 0.08 | 13.27 ± 0.10 | 13.34 ± 0.11 | 15.47 ± 0.11 | 0.9999 | 0.9998 | 0.9996 | 0.9996 | 13.1 ± 0.8 | 12 ± 0.8 | 60.3 ± 2 | 14.2 ± 0.8 |
| 73110 a | 11.42 ± 0.05 | 13.39 ± 0.08 | 13.47 ± 0.08 | 15.50 ± 0.13 | 0.9997 | 0.9995 | 0.9995 | 0.9995 | 13.0 ± 0.1 | 12.0 ± 0.4 | 58.8 ± 2 | 14.7 ± 1 |
| 73110 b | 11.12 ± 0.04 | 13.03 ± 0.04 | 13.08 ± 0.04 | 15.17 ± 0.09 | 0.9998 | 0.9973 | 0.9992 | 0.9990 | 12.6 ± 1 | 11.5 ± 1 | 58.9 ± 4 | 14.3 ± 1 |
| Oyster a | 11.42 ± 00.5 | 13.41 ± 0.06 | 13.44 ± 0.07 | 15.55 ± 0.06 | 0.9999 | 0.9991 | 0.9997 | 0.9993 | 12.4 ± 1 | 11.3 ± 1 | 58.6 ± 1 | 14 ± 0.4 |
| Oyster b | 11.06 ± 0.04 | 12.82 ± 0.09 | 12.88 ± 0.10 | 15.08 ± 0.14 | 0.9986 | 0.9807 | 0.9993 | 0.9915 | 12.0 ± 0.5 | 10.5 ± 0.4 | 58.4 ± 2 | 14.4 ± 0.6 |
| Mussel a | 11.30 ± 0.07 | 13.14 ± 0.07 | 13.17 ± 0.07 | 15.46 ± 0.11 | 0.9997 | 0.9983 | 0.9996 | 0.9995 | 12.8 ± 0.9 | 11.5 ± 0.9 | 59.4 ± 0.6 | 14.6 ± 0.3 |
| Mussel b | 10.91 ± 0.03 | 12.53 ± 0.05 | 12.53 ± 0.05 | 15.14 ± 0.11 | 0.9975 | 0.9498 | 0.9990 | 0.9919 | 12.0 ± 0.3 | 10.5 ± 0.4 | 58.5 ± 0.7 | 14.4 ± 0.4 |
RT, retention time; a samples spiked after extraction of free amino acids; b samples spiked after extraction of total amino acids.
2.2. Characterization of the Analytical Procedure
2.2.1. Recoveries of SPE Clean-Up and Remaining Matrix Effects
SPE recoveries and matrix effects were assumed to have a significant impact on the analysis of BMAA and isomers. We aimed to screen cyanobacteria, but also mollusks, for the presence of BMAA, DAB and AEG. After the selection of the Plexa PCX cartridge, recoveries of the SPE clean-up with representative matrices (cyanobacteria, oyster and mussel) and the remaining matrix effects were assessed for extraction procedures of both free and total BMAA and isomers (Table 1).
Table 1.
Recoveries (n = 3) of the solid phase extraction (SPE) clean-up (spiked solutions at 30 ng/mL) and remaining matrix suppression of the three matrices after the extraction of free and total BMAA and isomers. Matrix effects were evaluated as explained in Section 3.5.
| Extraction | Matrix | Recoveries of SPE Clean-Up (mean% ± SD) | Matrix Effect (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BMAA | DAB | D5DAB | AEG | BMAA | DAB | D5DAB | AEG | ||
| Free analytes | Cyanobacteria | 61.1 ± 4.9 | 51.0 ± 5.6 | 61.0 ± 1.6 | 48.1 ± 5.5 | 8.8 | 5.6 | 4.7 | 9.3 |
| Oyster | 56.1 ± 4.5 | 64.5 ± 4.5 | 65.4 ± 4.5 | 63.1 ± 3.8 | 7.3 | 0.9 | −3.5 | 0.2 | |
| Mussel | 63.8 ± 3.1 | 79.0 ± 6.8 | 75.6 ± 5.7 | 73.3 ± 2.1 | 5.3 | −6.3 | 1.2 | 0.7 | |
| Total analytes | Cyanobacteria | 67.6 ± 3.3 | 63.1 ± 1.9 | 63.4 ± 7.5 | 59.5 ± 1.0 | 3.8 | 1.9 | 4.2 | −9 |
| Oyster | 71.0 ± 2.0 | 76.4 ± 9.5 | 81.1 ± 4.1 | 74.0 ± 6.4 | 7.5 | 3.5 | 3 | 3.6 | |
| Mussel | 64.3 ± 8.0 | 65.0 ± 8.1 | 73.1 ± 6.5 | 64.3 ± 0.9 | 8.7 | 12.1 | 15.7 | −5.5 | |
Bond Elut® Plexa PCX is a mixed-mode SPE sorbent, which contains a highly polar polymeric cation-exchange resin with strong cation exchange functionalities. According to the manufacturer [48], Plexa PCX removes neutral and acidic interferences from the matrix and concentrates basic analytes, like BMAA and isomers. Ion suppression is reduced, because the highly polar, hydroxylated polymer surface is entirely amide-free and does not provide binding sites for endogenous species, such as proteins and lipids.
Recoveries of BMAA and isomers are globally within the same range (≥60%). Surprisingly, recoveries of analytes from cyanobacterial matrix were on average lower than those from the two more complex mollusk matrices. The cation-exchange functionality did not suffer from the higher content of amino acids in samples after acid hydrolysis, since similar recoveries were observed for the extraction of both free and total amino acids. A decrease of SPE recoveries due to the presence of matrix components can be seen in comparison with recoveries obtained with standards, as shown by Kasprzyk-Hordern [49]. Recoveries obtained for BMAA and DAB are comparable to those reported by other groups using Oasis® MCX cartridges and HILIC-MS methods to analyze cyanobacteria (>70% for Li et al. [45] and Combes et al. [24], but >80% for Kubo et al. [21]). Overall, the recoveries of the methods reported in the literature for the quantification of BMAA in cyanobacteria are generally better without SPE clean-up; however, matrix effects have not always been evaluated [19,20]. We could not find any information on SPE recoveries of underivatized analytes in mollusk matrices.
The matrix effects of BMAA and especially its isomers are not well documented. Li et al. [45] have reported no effect on BMAA, but DAB suppression with hydrolyzed cyanobacterial matrix, while strong matrix effects (>30%) were still present after SPE clean-up of cyanobacteria and biofilms in the procedure of Combes et al. [24]. Due to previous studies in our laboratory [50], matrix effects caused by mollusks were assumed to be higher than those caused by microalgae. However, this was not the case for this analyte-matrix combination.
After our SPE clean-up protocol, the remaining matrix effects, i.e., signals suppression or enhancement in the electrospray interface of LC-MS/MS, were relatively low for the three underivatized matrices (less than 15% of loss after extraction of both free and total amino acids). This suggests that our method was not affected by the strong signal suppression due to BMAA reactivity (i.e., metal adduct formation), as recently reported by Glover et al. [51] for the analysis of underivatized BMAA. Despite somewhat lower recoveries than reported elsewhere, the proposed SPE protocol seems to be very effective at cleaning cyanobacteria and mollusk matrices, while reproducibly extracting BMAA and isomers. For example, a strong matrix effect was observed for DAB in mussel matrix without the SPE clean-up step, while peak shapes and retention times were better and closer to the standards after SPE clean-up (Figure 6).
Figure 6.
The effect of the SPE clean-up on mussel matrix spiked with BMAA and AEG after extraction of free amino acids (DAB naturally present in the sample). The black line corresponds to the mass spectral transition m/z 119 > 102. (A) Spiked mussel extracted with TCA 0.1 M and injected in this solvent, while (B) the same sample subjected to PCX SPE clean-up and injected in ACN/water/FA. The shift in retention times compared to other chromatograms is due to the non-use of the guard column in (B) and due to the injection solvent, matrix effects and the non-use of a guard column in (A).
Among the internal standards that have been used, deuterated-BMAA, i.e., D3BMAA [22,25], and 5+BMAA [52] are more pertinent to accurately quantify samples by applying a corrective factor. In the absence of commercially available deuterated-BMAA (or otherwise isotopically-labeled BMAA), a group has recently validated a procedure to quantify underivatized BMAA and DAB using D3DAB as the internal standard [24]. As a result, we chose D-2,4-diaminobutyric acid-2,3,3,4,4-2D5 dihydrochloride (D5DAB) as the internal standard. D5DAB was expected to have recoveries and matrix effects similar to DAB and presumably similar to BMAA and AEG. This hypothesis was confirmed since concentrations obtained after correction with D5DAB recovery are within a 15% error range (20% for free AEG in cyanobacteria) and not overestimated. Therefore, a corrective factor based on D5DAB recovery could be applied to more accurately quantify BMAA and isomers in cyanobacteria, oyster and mussel matrices.
2.2.2. LC-MS/MS Performance: Linearity, LOD, LOQ and Repeatabilites (RT, R2 and Ion Ratios)
The calibration curves for BMAA, DAB and AEG showed good linearity (R2 > 0.9996) over the concentration range 1–500 ng/mL. However, a five-point calibration curve ranging from 1 to 100 ng/mL was classically used to quantify samples. The equations for the linear regressions were: y = 42,414x − 6703 for BMAA, y = 41,400x − 15,604 for DAB, y = 66,882x − 44,981 for AEG and y = 1252x − 585 for D5DAB.
The limit of detection (LOD) was defined as the lowest concentration giving a signal-to-noise (S/N) ratio of three (standard deviation of noise = σ; S/N = 3 σ) for the four qualitative mass spectral transitions of BMAA and DAB. The limit of quantification (LOQ) was calculated using the general transition m/z 119 > 102. Because the area of the peak at m/z 102 was more than seven-times higher than the peaks at m/z 76 and 74, the LOQ (with S/N > 10, σ = 3) was defined as equal to the LOD.
The LOQ was 3 ng/mL for standards and equivalent to 15 pg on the column. The LOQs within matrices were calculated by spiking freeze-dried material subjected to the extraction of both free and total analytes, and they were 0.225 µg/g dry weight (DW) of cyanobacteria and 0.15 µg/g DW of shellfish. However, since no clean mollusk matrices were found (especially for DAB and BMAA after extraction of total amino acids), LOQs for mollusks were estimated based on matrix effects. The difference of LOQs observed between cyanobacteria and mollusk matrices relied on the amount of freeze-dried material extracted (respectively, 10 and 15 mg).
While having an optimized selectivity, the instrument and method sensitivity are among the best reported for HILIC methods [19,20,22,24,45], but inferior to those reported for derivatized analytes in cyanobacterial samples [25,35]. The LOQ we obtained for cyanobacteria was better than the LOQ reported in the procedure of Combes et al. [24], possibly due to higher matrix effects in their HILIC-MS/MS method. For mollusk matrices, no LOQ of underivatized BMAA and isomers is reported. Nevertheless, LOQs of 1.7 µg/g DW and 0.15 µg/g wet weight were reported for derivatized BMAA with a gastropod [53] and a mussel matrix [54].
Thanks to an effective clean-up step and optimized gradient elution, we obtained a sensitive and selective method to quantify BMAA, DAB and AEG in cyanobacteria and mollusk matrices.
The repeatability of retention times, the correlation coefficient (R2) and ion ratios of standards calibration curves (n = 33) and spiked matrices are shown in Table 2.
Even if no clean matrices were found to assess the matrix effects, the strategy adopted (see Section 3.5) allowed for the good linearity of spiked matrices. Indeed, R2 > 0.998 with extraction of free amino acids and R2 > 0.99 (with some exceptions) after extraction of total amino acids were observed. The lower (R2 ≤ 0.98) coefficient of determination for DAB (oyster and mussel) came from the natural high content of this isomer in the mollusk matrices used.
Despite the presence of a relatively low matrix effect, the retention times of analytes and the ion ratios were rather stable. The good repeatability obtained (i.e., relative standard deviations less than 1% and 10% for RT and ion ratios, respectively) made this HILIC-MS/MS method suitable for the accurate quantification of BMAA and isomers in cyanobacteria and mollusk matrices, after extraction of both free and bound BMAA and its isomers.
2.3. Screening of BMAA and Isomers in Cyanobacteria
2.3.1. Kinetics of Growth and Production of BMAA and Isomers
As the production of BMAA by cyanobacteria has been suggested to be “a function of growth condition and/or life cycle stages” [7], the kinetics of growth and toxin production in batch cultures were made with two non-axenic strains that had previously been reported to produce BMAA: Leptolyngbya PCC 73110 [12,25] and Nostoc CCMP 2511 [36]. The Nostoc strain was reported to produce BMAA by methods lacking in selectivity (e.g., FLD), while Leptolyngbya was recently quantified with a highly selective MS/MS method. Biomass was regularly monitored, and cells were harvested between seven to 111 days of growth. The extraction of free analytes was performed on all samples, while the extraction of free plus bound (=total) analytes was carried out when freeze-dried material was available (Day 13 for both Nostoc CCMP 2511 and Day 40 for Leptolyngbya PCC73110 (Figure 7).
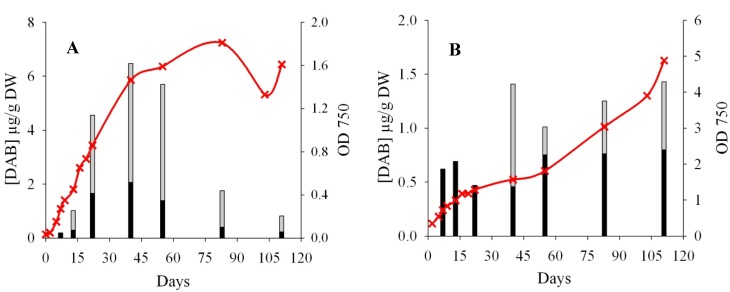
Figure 7.
Growth curves (red line, cross symbols) and free (dark histograms) and bound (grey histograms) DAB concentration for (A) Nostoc CCMP 2511 and (B) Leptolyngbya PCC 73110.
Neither free nor bound BMAA were found in any of the two strains, at any time of the growth curve. However, these strains are known to be BMAA producers, and the reported concentrations are above our LOQ [25,36]. For the Nostoc strain, the use of a non-selective method could have led to misidentification and/or overestimation of BMAA in previous studies [18]. Nevertheless, Jiang et al. have reported 0.73 µg/g DW of BMAA in the Leptolyngbya PCC73110 thanks to a highly selective and validated MS/MS method involving the derivatization of samples [25]. The conditions of the culture that we and their group applied are really close [12], and the selectivity of both methods drastically reduces the risk of misidentification. It should be noted that this group had performed a total extraction of BMAA after six days of growth, while we could only perform total extraction after 40 days of growth (due to limited freeze-dried material).
The fact that “cyanobacteria in culture display a fluctuating BMAA production”, as suggested by this same group [30], may explain why we could not find BMAA in our Leptolyngbya culture, if the production is highly variable. Further trials were made with this strain using different sources of nitrogen in the culture medium and different culture durations; however, BMAA was never found in our work, while AEG was sometimes detected (data not shown).
Interestingly DAB, a possible neurotoxic isomer of BMAA [23], was detected in all samples analyzed. As observed for BMAA [7], no correlation between the free and bound form of DAB was observed, but the bound form seemed to be more important. For example, free DAB from LOQ to 2 µg/g was reported for CCMP 2511, while the total form ranged from 0.82 to 5.8 µg/g DW. The kinetic of DAB production (both free and bound) by CCMP 2511 seemed to follow the kinetics of growth (Figure 7A). On the contrary, DAB production by the other strain seemed more stable and independent of growth. The two kinetics of DAB production were different, and the concentrations found varied by up to one order of magnitude; however, DAB was always detected in these two strains during the kinetic experiment.
2.3.2. Screening of Others Lab-Cultured Cyanobacteria
We screened eight other cyanobacterial strains for the production of BMAA and isomers (Table 3). Unlike Banack et al. [28], we could not find AEG in Synechocystis sp. PCC 6803 and Nostoc PCC 7120 extracts, but trace levels (from LOD to 1.5 µg/g DW of total AEG) were sometimes found in Leptolyngbya PCC 73110, Synechococcus elongatus CCAP1479/1B and Nostoc endophytum CCAP1453/14 (data not shown).
Table 3.
The content of free and total DAB (µg/g DW) in ten strains of cyanobacteria. When more than one discrete sample was screened (different medium of culture or days of growth), the minimum and the maximum concentrations are reported.
| Cyanobacterial Strain | Free DAB (µg/g DW) | Total DAB (µg/g DW) |
|---|---|---|
| Leptolyngbya PCC 73110 a | <LOD–2.69 | 1.01–1.57 |
| Nostoc CCMP 2511/ CMMED01 a | <LOD–2.27 | 0.82–12.48 |
| Microcystis PCC 7806 a | <LOD | <LOD |
| Nostoc PCC 7120 a | <LOD | * |
| Nostoc PCC 7107 a | 2.12–7.2 | * |
| Symploca PCC 8002 a | 0.3 | 0.43 |
| Synechocystis sp.PCC 6803 | <LOD | <LOD |
| Synechococcus elongatus CCAP1479/1B | 0.23–2.71 | 0.4–3.52 |
| Calothrix crustacea CCAP1410/9 | 0.6–0.92 | 6.95–14.53 |
| Nostoc endophytum CCAP1453/14 | 0.34–0.55 | 6.78–7.52 |
* Not analyzed for total DAB; <LOD, a peak corresponding to the transition 119 > 102 was detected at a retention time close to D5DAB, but was not quantifiable (<0.225 µg/g); a strain reported to be a BMAA producer.
Besides the three strains above, four more cyanobacteria were screened, as they had been previously reported to produce BMAA by the non-highly selective FLD [7] or selective MS/MS method with derivatization [29]. Finally, more than 60 samples corresponding to ten strains were analyzed with the HILIC-MS/MS method that we have optimized. No BMAA was found, while at least trace amounts of DAB were observed in all samples (except for PCC6803). The lack of selectivity of FLD is now well admitted, so the high concentrations of BMAA [7] reported with this detection technique cannot be fully considered. However, DAB production by cyanobacteria had already been reported, in Calothrix (<50 µg/g) in 2008 [22], then in 16 cyanobacterial samples (<0.83 µg/g) in 2010 [20] and in trace amounts in 2012 [45].
In these cases, HILIC methods were used, but DAB was also observed after derivatization of Leptolyngbya PCC73110 [12]. Once again, the bound form of DAB seemed to predominate. The concentrations we obtained here for DAB varied by a factor of 10 to 15 within the same strain, yet they still were within the concentration ranges reported before.
Krüger et al. [17] have raised a possible substance conversion into DAB during the hydrolysis of the sample preparation procedure, but we did detect free DAB with our procedure to extract free amino acids in which no acid hydrolysis of the samples was performed.
Little is known about BMAA and DAB production by cyanobacteria. The only work focusing on the factors that influence BMAA production showed that nitrogen starvation resulted in the production of BMAA [55]. However, the methodological approach and conclusion of this study were subsequently criticized [18]. This same group has recently reaffirmed a link between the content of BMAA and nitrogen availability in a field study of cyanobacterial blooms [56]. They have also suggested that “at high nitrogen level (>40 µM), BMAA production is suppressed” [56]. However, this is not confirmed by the publication of Jiang et al. [25] in which BMAA was reported in lab-cultured Leptolyngbya PCC 73110 grown in BG11 (Blue-Green 11) medium that contains a very high nitrate concentration. We also could not confirm this hypothesis with our lab-cultured cyanobacteria. Indeed, we grew cultures of Nostoc CCMP 2511 and Leptolyngbya PCC 73110 in media without any nitrogen source and did not observe the appearance of BMAA production in those strains that had been reported to produce BMAA before.
Now, highly selective methods exist, with and without a derivatization step [24,25], so the ability of cyanobacteria to produce BMAA and the factors controlling this production can be effectively assessed. In this attempt, only two strains (i.e., Leptolyngbya PCC73110 and Nostoc PCC7120) were confirmed to contain BMAA after derivatization of samples [25,29]. However, we could not find BMAA in these two strains with our selective HILIC-MS/MS method, maybe due to different conditions of culture [45], or because of the supposed brief nature of BMAA production by cyanobacteria [30], or due to the lack of sensitivity of our method. Nevertheless, we did detect DAB in almost all samples and sometimes AEG, which stresses the requirement of highly selective methods to accurately quantify BMAA in cyanobacteria.
2.3.3. Screening of Mollusks of Thau Lagoon
Since the discovery of BMAA in 1967, the link between BMAA and neurodegenerative diseases, like amyotrophic lateral sclerosis (ALS), is still under debate [3,57,58]. The consumption of contaminated aquatic organisms/seafood is a possible pathway of human exposure to BMAA. Potential associations of BMAA found in aquatic organisms with sporadic ALS were recently hypothesized in Chesapeake Bay, Maryland, USA [59], and in Thau Lagoon, France [35], with methods using the derivatization of analytes.
We decided to screen BMAA and isomers in mollusks collected in the Thau Lagoon during summer, 2009, with the HILIC-MS/MS that we have developed.
The three isomers, BMAA, DAB and AEG, were found in mussels and oysters of this bivalve farming area (Figure 8). The samples were freeze-dried and stored at room temperature since 2009.
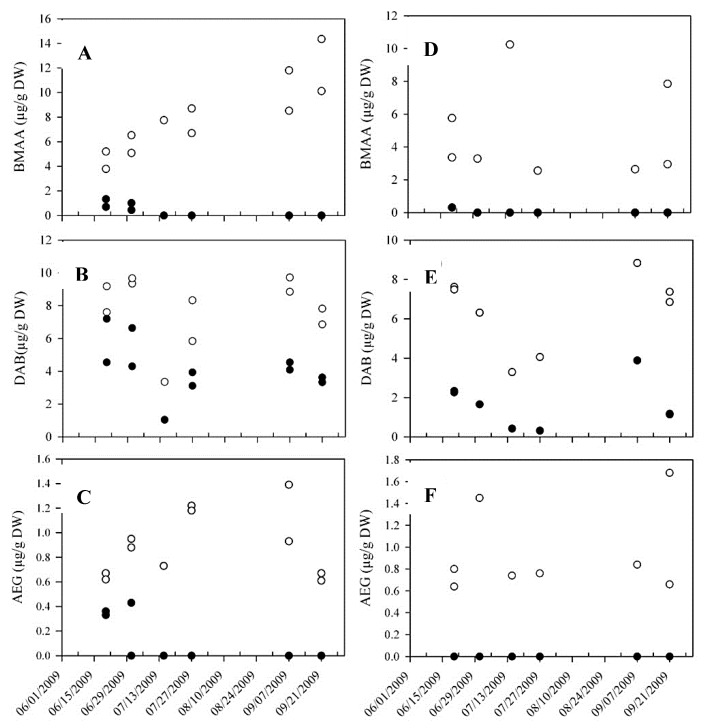
Figure 8.
(A–C) BMAA, DAB and AEG concentrations in free (black circles) and total form (empty circles) in mussels collected in 2009 and (D–F) in oysters. Mollusks were sampled in two of the three areas where shellfish were cultured in the lagoon and analyzed for both areas when material was available.
While both free BMAA and AEG were detected only up to July 1 (in six out to 19 samples for BMAA and in three of 19 samples for AEG), free DAB was always observed. However, after acid hydrolysis, all isomers were detected and quantified at higher concentrations. The concentrations of total BMAA in mussels showed a time-dependent increase during the summer of 2009, while they were more stable in oyster. The concentrations of total DAB were similar in the two mollusk matrices, between 3.4–9.7 and 3.3–8.8 µg/g DW in mussel and oyster, respectively. As for BMAA, the concentrations of total AEG in mussels increased between 20 June and 7 September. The higher content of BMAA and isomers found in mussels may be explained by their generally higher filtration activity compared to oysters [35].
During the summer, 2009, two phytoplankton blooms dominated by diatoms (e.g., Chaetoceros sp.) were observed at the beginning of July and at the end of August in the Thau Lagoon [60]. As diatoms were recently suggested to produce BMAA [30], the occurrence of such microalgae in addition to picocyanobacteria [35] could explain the presence of BMAA in mollusks during the summer, 2009. However, the production of BMAA by microalgae of the Thau Lagoon has not been studied so far.
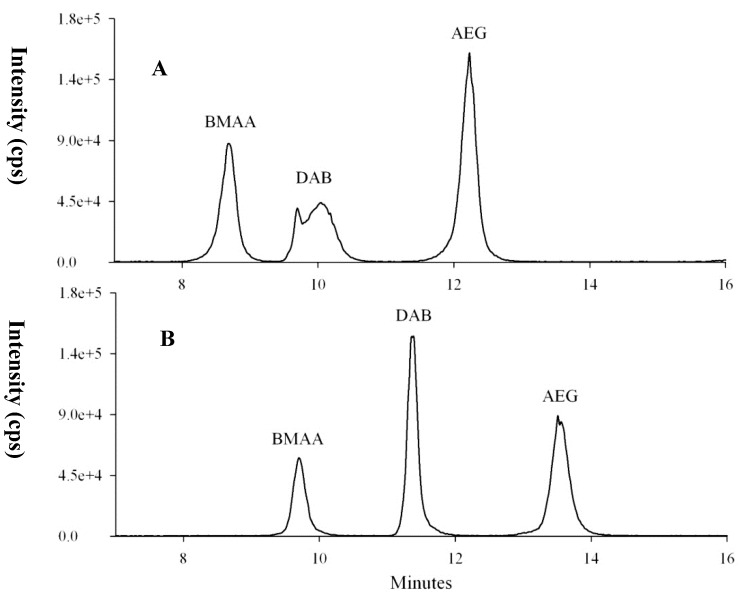
As no uncontaminated mussel or oyster samples could be found in the study area, we decided to spike the analytes into extracts of naturally contaminated samples. This exercise has allowed for the verification of retention times and ion ratios. No differences in retention time or ion ratios were detected between naturally-contaminated and spiked samples (Figure 9). It should be noted that after acid hydrolysis, an interfering compound giving a signal at the transition m/z 119 > 102 (corresponding to the loss of the NH3 group [20]) and eluting just before BMAA was observed for mollusk matrices (Figure 9). This unknown interference always eluted before BMAA, and the retention times of both compounds were reproducible. For example, mean retention times of 10.62 min ± 0.04 and 10.90 min ± 0.04, respectively, for the interference and BMAA in mussel matrix were obtained for seven replicates of the same sample injected twice and one replicate of four other mussel samples.
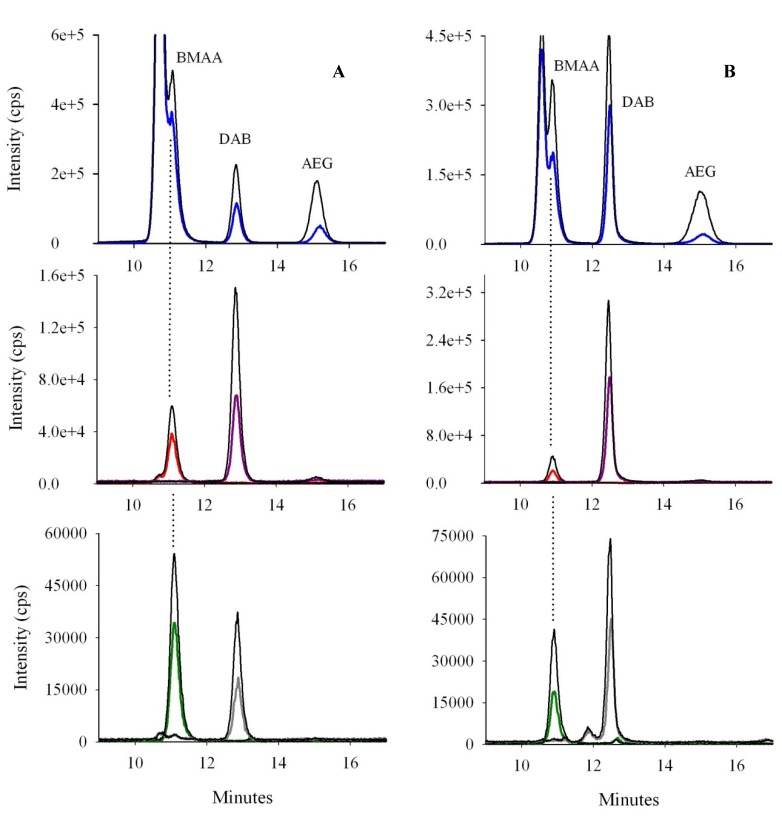
Figure 9.
Chromatograms of spiked (dark line) and non-spiked, naturally-contaminated (A) oyster and (B) mussel samples after performing the extraction for the determination of total BMAA and isomers. Colored lines represent mass spectral transitions m/z 119 to m/z 102 (blue, top graphs), 88 (red) and 101 (purple) in the middle graphs and 76 (green) and 74 (grey) in the bottom graphs.
Similarly, an interfering compound, partially resolved from BMAA, was reported by Jiang et al. [27] for cyanobacteria and mollusks from the Baltic Sea, after the derivatization of analytes. Nevertheless, retention time, specific mass spectral transitions (m/z 119 to m/z 88 and 76) and ion ratios (88/119 and 76/119) allowed us to unambiguously identify BMAA in hydrolyzed mollusk matrices. Moreover, the chromatograms of spiked oyster and mussel matrix clearly showed that the areas of mass spectral transitions corresponding to BMAA (and other isomers) are higher than in the non-spiked samples (Figure 9), thereby confirming that BMAA, DAB and AEG were present in these matrices. This compound could potentially interfere with the quantification of BMAA. However, as no interfering compound was detected with the mass spectral transitions m/z 119 to m/z 88 and 76, we quantified BMAA in parallel using either the highest transition (which has the interfering compound eluting at the front) or the two specific smaller transitions, and the concentrations obtained with either calculation method were within an error range of 10%–15%. As this error is in the range of the overall method variability, we decided to report quantification with the same transition as for the other isomers for the purpose of consistency.
The presence of BMAA isomers (DAB and AEG in this study plus BAMA in another study [27]) and interfering compounds within the same samples demonstrates the need for highly selective methods to accurately quantify those molecules in biological matrices.
Our results confirmed the presence of BMAA and isomers in mollusks of the Thau Lagoon, but with a HILIC-MS/MS method without the derivatization of samples. The concentrations we obtained here are similar to those that Masseret et al. have reported [35] for total BMAA in mussels, but higher for total DAB in mussels and both total BMAA and DAB in oysters. However, the application of a corrective factor is not mentioned in their study.
Apart from Thau Lagoon, BMAA and isomers were also reported in other ecosystems, after the derivatization of samples. If we consider the most selective methods that have been employed, BMAA was severally detected in mollusks coming from the Baltic Sea [12], in mussels and oysters either in very low [13] or at similar concentrations than what we obtained (0.27–1.16 µg/g wet weight corresponding to about 1.3–5.8 µg/g DW of mussel, if wet mussels are assumed to contain 80% water [54]). BMAA was also reported in oysters originating from the southeastern United States (6.8–46.9 µg/g DW) [53], in mollusks of Lake Taihu, China (mean of 3.21 µg/g DW) [15] and in cockles collected on Portuguese coasts (up to 0.434 µg/g DW) [32]. Finally, the presence of BMAA in mollusks of South Florida waters should be considered despite the use of a non-selective LC-FLD method [14].
Concerning BMAA isomers, BAMA and DAB in Swedish oysters [12,27], DAB in blue crabs [59] and DAB and AEG in Portuguese cockles [32] were observed. Interestingly, Lage et al. [32] mentioned that “these two isomers were observed along with BMAA in nearly all samples”. Masseret et al. [35] also found both BMAA and DAB in all mollusks samples of the Thau Lagoon. When BMAA isomers are considered, they are frequently detected in filter-feeding shellfish mollusk matrices. Recently, they were also reported in shark cartilage dietary supplements with both the FLD and MS/MS detection methods after the derivatization of samples [61].
Nevertheless, as far as we know, this is the first time that BMAA and isomers have been reported in mollusk matrices without the derivatization of samples, suggesting that HILIC-MS/MS methods are suitable to quantify these analytes in complex matrices.
While BMAA toxicity has been well studied [2] and DAB was reported to be a neurotoxic non-proteinogenic amino acid [62], the toxicity of AEG is unknown [25]. The incorporation of BMAA into proteins remains to be “indisputably proven” [63], especially because it is suggested to be involved in BMAA bioaccumulation [5]. Further work is required to understand the bioaccumulation pathway of BMAA in higher trophic levels of ecosystems, as observed in Guam or in the Baltic Sea.
3. Experimental Section
3.1. Chemicals and Reagents
β-N-methylamino-l-alanine hydrochloride (BMAA, B107) and trichloroacetic acid (TCA, 33731) were purchased from Sigma-Aldrich, France, while N-2-aminoethylglycine (AEG, A1153) and 2,4-diaminobutyric acid dihydrochloride (DAB, D0083) were obtained from TCI, Belgium. d-2,4-diaminobutyric acid-2,3,3,4,4-2D5 dihydrochloride (D5DAB), used as the internal standard, was purchased from CDN isotopes (CIL, Sainte-Foy-La-Grande, France).
Methanol (MeOH) and acetonitrile (ACN) were obtained as HPLC grade solvents from JT Baker. Water for analysis was supplied by a Milli-Q integral 3 system (Millipore). Solutions of formic acid (FA, 33015), hydrochloric acid 37% (HCl, 258148) and ammonium hydroxide (221228), all reagent grade, were purchased from Sigma-Aldrich.
Calibrant stock solutions (10 µg/mL) of BMAA, DAB, AEG and D5DAB were prepared in Milli-Q water and stored at 4 °C. As observed by Combes et al., 2013 [24], no degradation of stock solutions was observed after more than 6 months in this storage condition.
3.2. Samples
3.2.1. Cultures of Cyanobacteria
Ten non-axenic species of cyanobacteria, six of which are reported to be BMAA producers, were cultured for the screening of BMAA and isomers. Leptolyngbya PCC 73110 and Synechocystis PCC 6803 were maintained in BG11 medium [64]. Nostoc PCC 7120 and PCC 7107 were cultured in BG110 (nitrogen free). Microcystis PCC 7806 was cultured in modified BG11, BG110 with the addition of 2 mM NaNO3 and 10 mM NaHCO3 , and Symploca PCC 8002 in a mixture of BG11 and modified L1 medium, salinity adjusted to 27, no vitamins added (1:12, v/v). Nostoc CCMP 2511 was cultured in modified L1 medium, with four-times the classic NaNO3 concentration of L1 medium [65]. Synechococcus elongatus CCAP1479/1B was grown in Conway 10%, seawater diluted 3.33-times with Milli-Q water before adding the Conway elements [66] without silica. Calothrix crustacea CCAP1410/9 was cultured in F/2 medium [67] and Nostoc endophytum CCAP1453/14 in Conway medium (without silica).
The Milli-Q water used to prepare BG11 and modified BG11 medium was supplied by a Milli-Q integral 3 system (Millipore, Saint-Quentin-Yvelines, France), while filter-sterilized seawater at a salinity of 35 was used for seawater culture media. All cultures were maintained under sterile conditions at 22 °C under a 16:8 h light/dark cycle at a light intensity of 50–60 μmol/m2/s, except Synechococcus elongatus, Calothrix crustacean and Nostoc endophytum (20 °C, 12:12 h light/dark cycle, light intensity of 300 μmol/m2/s). Batch cultures were grown to establish growth factors and toxin production for the strains Leptolyngbya PCC 73110 and Nostoc CCMP 2511. Biomass was regularly monitored by the measure of DO750 nm (Jenway spectrophotometer 6705 series), and cyanobacteria were sampled after a growth period of 7, 13, 22, 40, 55, 83 and 111 days. All other strains were maintained in batch culture. Cyanobacterial cells were harvested via centrifugation at 4000× g for 30 min at 4 °C. The supernatant was carefully discarded, and the resulting pellet was freeze-dried, homogenized and stored at room temperature until further processing.
3.2.2. Collection of Field Samples
During summer, 2009, mussels (Mytilus galloprovincialis) and oysters (Crassostrea gigas) were sampled in the Mediterranean Lagoon of Thau, France, from two sites (Bouzigues and Marseillan; see Pernet et al. [60]). Whole flesh aliquots were freeze-dried, stored at room temperature and homogenized with a Microtron MB 550™ blender (Kinematica) before extraction.
3.3. Sample Preparation
3.3.1. Extraction
Aliquots of freeze-dried material (10 mg for cyanobacteria and 15 mg for mollusks) were weighted in a 1.5 mL Eppendorf® tube, and both 0.5 g of 200 µm glass beads and 750 µL of trichloroacetic acid (TCA) 0.1 M were added. Extraction was performed with a mixer mill (Retsch MM400, Germany) for 30 min at 30 Hz. Deuterated DAB (D5DAB, concentrations of 30 or 50 ng/mL for the extraction of free and total analytes) was added before extraction as an internal standard. Tubes were centrifuged at 13,000× g after grinding to precipitate beads and debris.
For the extraction of free BMAA and isomers, the supernatant was collected and filtered through a 0.22-µm filter (Nanosep® MF, Pall, Mexico) to remove particulate material. The volume was adjusted to 1 mL with TCA 0.1 M before SPE clean-up.
For the extraction of total analytes, the supernatant was collected after grinding and evaporated to dryness under a stream of nitrogen. The residue was dissolved in 600 µL HCl 6 M. Acid hydrolysis was performed in a Thermomixer® comfort (Eppendorf, Fisher Scientific, Illkirch, France) for 24 h at 99 °C to release the bound form of BMAA and isomers from proteins. HCl was dried under a stream of nitrogen, and the residue was dissolved in 1 mL of TCA 0.1 M before SPE clean-up.
3.3.2. SPE Clean-Up
Two kinds of polymeric cation-exchange sorbents (60 mg/3cc cartridges), Bond Elut® Plexa PCX (Agilent Technologies, VWR, France) and Oasis® MCX (Waters, France), were compared with the recoveries of standards. Then, SPE clean-up with the Plexa PCX cartridge was carried out according to Combes et al. [24] with some modifications. The mixed-mode sorbents were conditioned with 2 mL of methanol (MeOH) followed by 1 mL of TCA 0.1 M. Then, 1 mL of sample was loaded onto the cartridge, which was consecutively rinsed with 1 mL of HCl 0.1 M and 2 mL of MeOH. After allowing the cartridges to dry, analytes were eluted with 3 + 1 mL of MeOH/NH4OH (93:7, v/v). The eluate was evaporated to dryness under a stream of nitrogen at 40 °C. Finally, the residue was dissolved in a mixture of ACN/water (63:37, v/v), both containing 0.1% FA, before injection.
3.4. Analysis by LC-MS/MS
Analysis by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) was performed on a Ultra Fast Liquid Chromatograhy (UFLC) (model Nexera, Shimadzu, Champs-sur-Marne, France) coupled to a triple-quadrupole mass spectrometer (5500 QTRAP, AB Sciex). Chromatography was performed with a ZIC®-HILIC column (150 × 2.1 mm, 5 μm, Merck Sequant®, Fontenay-sous-Bois, France) and a TSK gel amide 80 guard column (2 × 10 mm, 5 μm).
Mobile phases were Milli-Q water (Mobile Phase A) and ACN (Mobile Phase B), both containing 0.1% formic acid. The flow rate was 0.2 mL/min, and the injection volume was 5 μL. The column temperature was 30 °C, while samples were kept at 4 °C. The linear gradient elution started with 37% of Mobile Phase A, rising to 55% over 18 min, held for 2 min, then decreased to 37% of Mobile Phase A for 3 min and held for 15 min to equilibrate the system. In order to reduce the quantity of impurities entering the MS system, the flow of the first five minutes was directed to the waste container.
To optimize the chromatographic resolution of BMAA and its isomers, a gradient elution was developed thanks to a 23 factorial design. Resolution was calculated as 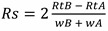 with Rt the retention time and w the width at the base of the common mass spectral transition m/z 119 > 102. Results were analyzed and interpreted using STATGRAPHICS Centurion XV software.
with Rt the retention time and w the width at the base of the common mass spectral transition m/z 119 > 102. Results were analyzed and interpreted using STATGRAPHICS Centurion XV software.
The LC-MS/MS system was used in positive ion mode with multiple reaction monitoring (MRM) detection. The most intense and common transition m/z 119 > 102 was used to quantify BMAA, DAB and AEG, while the transition m/z 124 > 47 was used to quantify D5DAB, because the other transitions (e.g., m/z 124 > 106) suffered from a high background signal.
To increase selectivity, two specific mass spectral transitions of BMAA (m/z 119 > 88 and 119 > 76) and DAB (m/z 119 > 101 and 119 > 74) and the ion ratios 88/102 and 76/102 for BMAA and 101/102 and 74/102 for DAB were also monitored. A sample was considered positive if the specific transitions were detectable and the variability of ion ratios observed in samples was less than 20%. Classically, the accepted variability is between 10% and 20% for ratios of this height of order. As we observed differences of less 10% for all analytes, we obtained an acceptable variability of ion ratios between standards and samples.
The ESI interface was operated using the following parameters: curtain gas 20 psi; temperature 600 °C; Gas 1 40 psi; Gas 2 60 psi; ion spray voltage 5500 V. For detection, the parameters are reported in Table 4.
Table 4.
Parameters used for optimal detection of BMAA and isomers with the 5500 QTRAP mass spectrometer.
| Transition (m/z) | EP (V) | DP (V) | CE (eV) | CXP (V) |
|---|---|---|---|---|
| 119 > 102 | 10 | 81 | 13 | 12 |
| 124 > 47 | 71 | 21 | 22 | |
| 119 > 88 | 66 | 17 | 10 | |
| 119 > 76 | 66 | 17 | 10 | |
| 119 > 101 | 86 | 11 | 8 | |
| 119 > 74 | 86 | 19 | 8 |
EP, entrance potential; DP, declustering potential; CE, collision energy; CXP, collision cell exit potential.
BMAA and isomers were quantified using a five-point calibration curve of pure standards constructed by dilution series (1–100 ng/mL) of stock solutions in a mixture of ACN and water (63:37, v/v, both containing 0.1% FA). A corrective factor derived from D5DAB recovery was applied to quantify more accurately BMAA, DAB and AEG. The software Analyst 1.5.1 was used to analyze the acquired data.
3.5. Recoveries and Matrix Effects
Recoveries of the SPE clean-up step and remaining matrix effects were estimated with one cyanobacterial and two shellfish matrices after extraction of both free and total BMAA and isomers. It has to be noted that no clean matrices were found (particularly after acid hydrolysis). As a compromise, a naturally-contaminated matrix was analyzed in parallel with and without spiking, and the peak areas of the non-spiked samples were subtracted from the peak areas of the spiked samples.
The Leptolyngbya PCC73110 (grown 40 days in conditions reported in Section 3.2.1) was chosen as the cyanobacterial matrix. The two shellfish matrices, mussel (M. galloprovincialis) and oyster (C. gigas), were sampled in 2009 in the Thau Lagoon where BMAA and DAB had been reported [35].
Recoveries of the SPE clean-up protocol with the Plexa PCX cartridge were evaluated with these three matrices with the extraction of both free and total analytes (see Section 2.2.1). Matrices were spiked in triplicate just before SPE with a mixture of BMAA and isomers at a final concentration of 30 ng/mL.
The remaining matrix effects were also assessed for both free and total extraction procedures. Before injection, the dried matrices subjected to extraction procedures were dissolved in a spiked ACN/water mixture to establish three-point standard curves (3, 10, 50 ng/mL). The slope of the spiked matrix curves was compared to the slope of standard curves in solvent to estimate the matrix effects.
3.6. Statistical Analysis
Statistical analyses were carried out with SigmaPlot 12.5 (Systat Software Inc., Chicago, IL, USA).
4. Conclusions
We present here a highly selective HILIC-MS/MS method to confidently identify and accurately quantify underivatized BMAA and isomers (DAB and AEG) in cyanobacterial and shellfish samples, in both free and total form. Thanks to an optimized gradient elution and an effective SPE clean-up step, good sensitivity (LOQ of 0.22 and 0.15 µg/g DW for cyanobacteria and mollusks, respectively) and low matrix effects were obtained. The use of D5DAB as the internal standard allowed for correction and accurate quantification of BMAA and isomers in all samples. This reliable method was applied to screen ten species of cyanobacteria. Neither free nor bound BMAA were found in our cultures. Nevertheless, DAB, a neurotoxic isomer of BMAA, was commonly detected. The HILIC-MS/MS method was also used to screen mollusks collected in 2009 in the Thau Lagoon. BMAA was identified in all shellfish samples after acid hydrolysis, and the concentrations of total BMAA were similar to those reported by Masseret et al. [35] in mussels. Additionally, we identified free BMAA in some samples during the month of June, 2009; however, no free BMAA was detected in either mussels or oysters after July 1, 2009. The significance of the general occurrence of DAB and to a lesser extent AEG in different matrices (cyanobacteria and mollusks) should be assessed further. Research is currently underway in our laboratory to pinpoint phytoplankton organisms producing BMAA in the Thau Lagoon. Furthermore, we intend to study the spatio-temporal distribution of BMAA and isomers over a longer time period and in other areas to provide data for risk evaluation.
Acknowledgments
This study was carried out under the RISALTOX project (Ifremer) and co-funded by the Regional Council of the “Pays de la Loire”. The authors would like to thank all of the members of the laboratory Phycotoxins at the Atlantic Centre of Ifremer for their help and advice during this study.
Author Contributions
D.R. developed the method, performed the analysis of samples and wrote the manuscript. Z.A. initiated the topic and, with V.Se. and L.B., supervised the thesis of DR. Z.A. and V.Se. contributed to the experimental designs, data interpretation and reviewed the manuscript. L.B. gave advice on cyanobacterial cultures. V.Sa. (chemical analysis) and M.B. (culture of cyanobacteria) provided technical assistance. E.A. performed the mollusk sampling in the Thau Lagoon. P.H. contributed to the literature search, data interpretation and proofreading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Vega A., Bell E.A. α-Amino-β-methylaminopropionic acid, a new amino acid from seeds of Cycas circinalis. Phytochemistry. 1967;6:759–762. doi: 10.1016/S0031-9422(00)86018-5. [DOI] [Google Scholar]
- 2.Chiu A.S., Gehringer M.M., Welch J.H., Neilan B.A. Does α-amino-β-methylaminopropionic acid (BMAA) play a role in neurodegeneration? Int. J. Environ. Res. Public Health. 2011;8:3728–3746. doi: 10.3390/ijerph8093728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karamyan V.T., Speth R.C. Animal models of BMAA neurotoxicity: A critical review. Life Sci. 2008;82:233–246. doi: 10.1016/j.lfs.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Cox P.A., Banack S.A., Murch S.J. Biomagnification of cyanobacterial neurotoxins and neurodegenerative disease among the Chamorro people of Guam. Proc. Natl. Acad. Sci. USA. 2003;100:13380–13383. doi: 10.1073/pnas.2235808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murch S.J., Cox P.A., Banack S.A. A mechanism for slow release of biomagnified cyanobacterial neurotoxins and neurodegenerative disease in Guam. Proc. Natl. Acad. Sci. USA. 2004;101:12228–12231. doi: 10.1073/pnas.0404926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murch S.J., Cox P.A., Banack S.A., Steele J.C., Sacks O.W. Occurrence of β-methylamino-l-alanine (BMAA) in ALS/PDC patients from Guam. Acta Neurol. Scand. 2004;110:267–269. doi: 10.1111/j.1600-0404.2004.00320.x. [DOI] [PubMed] [Google Scholar]
- 7.Cox P.A., Banack S.A., Murch S.J., Rasmussen U., Tien G., Bidigare R.R., Metcalf J.S., Morrison L.F., Codd G.A., Bergman B. Diverse taxa of cyanobacteria produce β-N-methylamino-l-alanine, a neurotoxic amino acid. Proc. Natl. Acad. Sci. USA. 2005;102:5074–5078. doi: 10.1073/pnas.0501526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metcalf J.S., Banack S.A., Lindsay J., Morrison L.F., Cox P.A., Codd G.A. Co-occurrence of β-N-methylamino-l-alanine, a neurotoxic amino acid with other cyanobacterial toxins in British waterbodies, 1990–2004. Environ. Microbiol. 2008;10:702–708. doi: 10.1111/j.1462-2920.2007.01492.x. [DOI] [PubMed] [Google Scholar]
- 9.Esterhuizen M., Downing T.G. β-N-methylamino-l-alanine (BMAA) in novel South African cyanobacterial isolates. Ecotoxicol. Environ. Safety. 2008;71:309–313. doi: 10.1016/j.ecoenv.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Johnson H.E., King S.R., Banack S.A., Webster C., Callanaupa W.J., Cox P.A. Cyanobacteria (Nostoc commune) used as a dietary item in the Peruvian highlands produce the neurotoxic amino acid BMAA. J. Ethnopharmacol. 2008;118:159–165. doi: 10.1016/j.jep.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Li A., Tian Z., Li J., Yu R., Banack S.A., Wang Z. Detection of the neurotoxin BMAA within cyanobacteria isolated from freshwater in China. Toxicon. 2010;55:947–953. doi: 10.1016/j.toxicon.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 12.Spáčil Z., Erkisson J., Jonasson S., Rasmussen U., Ilag L.L., Bergman B. Analytical protocol for identification of BMAA and DAB in biological samples. Analyst. 2010;135:127–132. doi: 10.1039/b921048b. [DOI] [PubMed] [Google Scholar]
- 13.Jonasson S., Eriksson J., Berntzon L., Spáčil Z., Ilag L.L., Ronnevi L.-O., Rasmussen U., Bergman B. Transfer of a cyanobacterial neurotoxin within a temperate aquatic ecosystem suggests pathways for human exposure. Proc. Natl. Acad. Sci. USA. 2010;107:9252–9257. doi: 10.1073/pnas.0914417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brand L.E., Pablo J., Compton A., Hammerschlag N., Mash D.C. Cyanobacterial blooms and the occurrence of the neurotoxin, beta-N-methylamino-l-alanine (BMAA), in South Florida aquatic food webs. Harmful Algae. 2010;9:620–635. doi: 10.1016/j.hal.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y., Chen Q., Chen X., Wang X., Liao X., Jiang L., Wu J., Yang L. Occurrence and transfer of a cyanobacterial neurotoxin beta-methylamino-l-alanine within the aquatic food webs of Gonghu Bay (Lake Taihu, China) to evaluate the potential human health risk. Sci. Total Environ. 2014;468:457–463. doi: 10.1016/j.scitotenv.2013.08.064. [DOI] [PubMed] [Google Scholar]
- 16.Pablo J., Banack S.A., Cox P.A., Johnson T.E., Papapetropoulos S., Bradley W.G., Buck A., Mash D.C. Cyanobacterial neurotoxin BMAA in ALS and Alzheimer’s disease. Acta Neurol. Scand. 2009;120:216–225. doi: 10.1111/j.1600-0404.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- 17.Kruger T., Oelmuller R., luckas B. The origin of β-N-methylamino-l-alanine (BMAA): Cycads and/-or cyanobacteria? J. Endocytobiosis Cell Res. 2012;22:29–36. [Google Scholar]
- 18.Faassen E.J. Presence of the neurotoxin BMAA in aquatic ecosystems: What do we really know? Toxins. 2014:1109–1138. doi: 10.3390/toxins6031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faassen E.J., Gillissen F., Lürling M. A Comparative Study on Three Analytical Methods for the Determination of the Neurotoxin BMAA in Cyanobacteria. PLoS One. 2012;7:e36667. doi: 10.1371/journal.pone.0036667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krüger T., Mönch B., Oppenhäuser S., Luckas B. LC–MS/MS determination of the isomeric neurotoxins BMAA (β-N-methylamino-l-alanine) and DAB (2,4-diaminobutyric acid) in cyanobacteria and seeds of Cycas revoluta and Lathyrus latifolius. Toxicon. 2010;55:547–557. doi: 10.1016/j.toxicon.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Kubo T., Kato N., Hosoya K., Kaya K. Effective determination method for a cyanobacterial neurotoxin, β-N-methylamino-l-alanine. Toxicon. 2008;51:1264–1268. doi: 10.1016/j.toxicon.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 22.Rosen J., Hellenas K.E. Determination of the neurotoxin BMAA (beta-N-methylamino-l-alanine) in cycad seed and cyanobacteria by LC-MS/MS (liquid chromatography tandem mass spectrometry) Analyst. 2008;133:1785–1789. doi: 10.1039/b809231a. [DOI] [PubMed] [Google Scholar]
- 23.Banack S.A., Downing T.G., Spácil Z., Purdie E.L., Metcalf J.S., Downing S., Esterhuizen M., Codd G.A., Cox P.A. Distinguishing the cyanobacterial neurotoxin β-N-methylamino-l-alanine (BMAA) from its structural isomer 2,4-diaminobutyric acid (2,4-DAB) Toxicon. 2010;56:868–879. doi: 10.1016/j.toxicon.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Combes A., El Abdellaoui S., Sarazin C., Vial J., Mejean A., Ploux O., Pichon V., Group B. Validation of the analytical procedure for the determination of the neurotoxin β-N-methylamino-l-alanine in complex environmental samples. Anal. Chim. Acta. 2013;771:42–49. doi: 10.1016/j.aca.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L., Johnston E., Åberg K.M., Nilsson U., Ilag L. Strategy for quantifying trace levels of BMAA in cyanobacteria by LC/MS/MS. Anal. Bioanal. Chem. 2013;405:1283–1292. doi: 10.1007/s00216-012-6550-1. [DOI] [PubMed] [Google Scholar]
- 26.Banack S.A., Metcalf J.S., Spacil Z., Downing T.G., Downing S., Long A., Nunn P.B., Cox P.A. Distinguishing the cyanobacterial neurotoxin beta-N-methylamino-l-alanine (BMAA) from other diamino acids. Toxicon. 2011;57:730–738. doi: 10.1016/j.toxicon.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Jiang L., Aigret B., Borggraeve W., Spacil Z., Ilag L. Selective LC-MS/MS method for the identification of BMAA from its isomers in biological samples. Anal. Bioanal. Chem. 2012;403:1719–1730. doi: 10.1007/s00216-012-5966-y. [DOI] [PubMed] [Google Scholar]
- 28.Banack S.A., Metcalf J.S., Jiang L., Craighead D., Ilag L.L., Cox P.A. Cyanobacteria produce N-(2-aminoethyl) glycine, a backbone for peptide nucleic acids which may have been the first genetic molecules for life on Earth. PLoS One. 2012;7:e49043–e49043. doi: 10.1371/journal.pone.0049043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berntzon L., Erasmie S., Celepli N., Eriksson J., Rasmussen U., Bergman B. BMAA Inhibits Nitrogen Fixation in the Cyanobacterium Nostoc sp.PCC 7120. Mar. Drugs. 2013;11:3091–3108. doi: 10.3390/md11083091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang L., Eriksson J., Lage S., Jonasson S., Shams S., Mehine M., Ilag L.L., Rasmussen U. Diatoms: A novel source for the neurotoxin BMAA in aquatic environments. PLoS One. 2014;9:e84578. doi: 10.1371/journal.pone.0084578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L., Ilag L.L. Detection of endogenous BMAA in dinoflagellate (Heterocapsa triquetra) hints at evolutionary conservation and environmental concern. PubRaw Sci. 2014;1:1–8. [Google Scholar]
- 32.Lage S., Costa P.R., Moita T., Eriksson J., Rasmussen U., Rydberg S.J. BMAA in shellfish from two Portuguese transitional water bodies suggests the marine dinoflagellate Gymnodinium catenatum as a potential BMAA source. Aquat. Toxicol. 2014;152:131–138. doi: 10.1016/j.aquatox.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 33.Dunlop R.A., Cox P.A., Banack S.A., Rodgers K.J. The Non-Protein Amino Acid BMAA Is Misincorporated into Human Proteins in Place of l-Serine Causing Protein Misfolding and Aggregation. PLoS One. 2013;8:e75376. doi: 10.1371/journal.pone.0075376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen S.A. Analytical techniques for the detection of alpha-amino-beta-methylaminopropionic acid. Analyst. 2012;137:1991–2005. doi: 10.1039/c2an16250d. [DOI] [PubMed] [Google Scholar]
- 35.Masseret E., Banack S., Boumédiène F., Abadie E., Brient L., Pernet F., Juntas-Morales R., Pageot N., Metcalf J., Cox P., et al. The French Network on A.L.S.: Investigation, dietary bmaa exposure in an amyotrophic lateral sclerosis cluster from Southern France. PLoS One. 2013;8:e83406. doi: 10.1371/journal.pone.0083406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banack S., Johnson H., Cheng R., Cox P. Production of the Neurotoxin BMAA by a Marine Cyanobacterium. Mar. Drugs. 2007;5:180–196. doi: 10.3390/md504180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eriksson J., Jonasson S., Papaefthimiou D., Rasmussen U., Bergman B. Improving derivatization efficiency of BMAA utilizing AccQ-Tag® in a complex cyanobacterial matrix. Amino Acids. 2009;36:43–48. doi: 10.1007/s00726-007-0023-4. [DOI] [PubMed] [Google Scholar]
- 38.Lawrenz E., Fedewa E., Richardson T. Extraction protocols for the quantification of phycobilins in aqueous phytoplankton extracts. J. Appl. Phycol. 2011;23:865–871. doi: 10.1007/s10811-010-9600-0. [DOI] [Google Scholar]
- 39.Serive B., Kaas R., Berard J.-B., Pasquet V., Picot L., Cadoret J.-P. Selection and optimisation of a method for efficient metabolites extraction from microalgae. Bioresource Technol. 2012;124:311–320. doi: 10.1016/j.biortech.2012.07.105. [DOI] [PubMed] [Google Scholar]
- 40.Ravn H., Anthoni U., Christophersen C., Nielsen P.H., Oshima Y. Standardized extraction method for paralytic shellfish toxins in phytoplankton. J. Appl. Phycol. 1995;7:589–594. doi: 10.1007/BF00003947. [DOI] [Google Scholar]
- 41.Zendong Z., Herrenknecht C., Abadie E., Brissard C., Tixier C., Mondeguer F., Séchet V., Amzil Z., Hess P. Extended evaluation of polymeric and lipophilic sorbents for passive sampling of marine toxins. Toxicon. 2014 doi: 10.1016/j.toxicon.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Chambers E., Wagrowski-Diehl D.M., Lu Z., Mazzeo J.R. Systematic and comprehensive strategy for reducing matrix effects in LC/MS/MS analyses. J. Chromatogr. B. 2007;852:22–34. doi: 10.1016/j.jchromb.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 43.Gerssen A., McElhinney M.A., Mulder P.P.J., Bire R., Hess P., de Boer J. Solid phase extraction for removal of matrix effects in lipophilic marine toxin analysis by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2009;394:1213–1226. doi: 10.1007/s00216-009-2790-0. [DOI] [PubMed] [Google Scholar]
- 44.Kilcoyne J., Fux E. Strategies for the elimination of matrix effects in the liquid chromatography tandem mass spectrometry analysis of the lipophilic toxins okadaic acid and azaspiracid-1 in molluscan shellfish. J. Chromatogr. A. 2010;1217:7123–7130. doi: 10.1016/j.chroma.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 45.Li A.F., Fan H., Ma F.F., McCarron P., Thomas K., Tang X.H., Quilliam M.A. Elucidation of matrix effects and performance of solid-phase extraction for LC-MS/MS analysis of b-N-methylamino-l-alanine (BMAA) and 2,4-diaminobutyric acid (DAB) neurotoxins in cyanobacteria. Analyst. 2012;137:1210–1219. doi: 10.1039/c2an15887f. [DOI] [PubMed] [Google Scholar]
- 46.Scott P.M., Niedzwadek B., Rawn D.F.K., Lau B.P.Y. Liquid chromatographic determination of the cyanobacterial toxin b-N-methylamino-l-alanine in algae food supplements, freshwater fish, and bottled water. J. Food Prot. 2009;78:1769–1773. doi: 10.4315/0362-028x-72.8.1769. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida T. Peptide separation by Hydrophilic-Interaction Chromatography: A review. J. Biochem. Biophys. Methods. 2004;60:265–280. doi: 10.1016/j.jbbm.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Agilent Technologies Bond Elut Plexa PCX. [(accessed on 13 November 2014)]. Available online: http://www.chem.agilent.com/en-US/products-services/Columns-Sample-Preparation/Sample-Preparation/Solid-Phase-Extraction-%28SPE%29/Bond-Elut-Plexa-PCX/Pages/default.aspx.
- 49.Kasprzyk-Hordern B., Dinsdale R.M., Guwy A.J. The effect of signal suppression and mobile phase composition on the simultaneous analysis of multiple classes of acidic/neutral pharmaceuticals and personal care products in surface water by solid-phase extraction and ultra performance liquid chromatography-negative electrospray tandem mass spectrometry. Talanta. 2008;74:1299–1312. doi: 10.1016/j.talanta.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 50.Jauffrais T., Herrenknecht C., Séchet V., Sibat M., Tillmann U., Krock B., Kilcoyne J., Miles C., McCarron P., Amzil Z., et al. Quantitative analysis of azaspiracids in Azadinium spinosum cultures. Anal. Bioanal. Chem. 2012;403:833–846. doi: 10.1007/s00216-012-5849-2. [DOI] [PubMed] [Google Scholar]
- 51.Glover W.B., Liberto C.M., McNeil W.S., Banack S.A., Shipley P.R., Murch S.J. Reactivity of β-methylamino-l-alanine in Complex Sample Matrixes Complicating Detection and Quantification by Mass Spectrometry. Anal. Chem. 2012;84:7946–7953. doi: 10.1021/ac301691r. [DOI] [PubMed] [Google Scholar]
- 52.Downing S., Contardo-Jara V., Pflugmacher S., Downing T.G. The fate of the cyanobacterial toxin beta-N-methylamino-l-alanine in freshwater mussels. Ecotoxicol. Environ. Safety. 2014;101:51–58. doi: 10.1016/j.ecoenv.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 53.Christensen S.J., Hemscheidt T.K., Trapido-Rosenthal H., Laws E.A., Bidigare R.R. Detection and quantification of beta-methylamino-l-alanine in aquatic invertebrates. Limnol. Oceanogr. Methods. 2012;10:891–898. doi: 10.4319/lom.2012.10.891. [DOI] [Google Scholar]
- 54.Salomonsson M.L., Hansson A., Bondesson U. Development and in-house validation of a method for quantification of BMAA in mussels using dansyl chloride derivatization and ultra performance liquid chromatography tandem mass spectrometry. Anal. Methods UK. 2013;5:4865–4874. doi: 10.1039/c3ay40657a. [DOI] [Google Scholar]
- 55.Downing S., Banack S.A., Metcalf J.S., Cox P.A., Downing T.G. Nitrogen starvation of cyanobacteria results in the production of β-N-methylamino-l-alanine. Toxicon. 2011;58:187–194. doi: 10.1016/j.toxicon.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 56.Scott L.L., Downing S., Phelan R.R., Downing T.G. Environmental modulation of microcystin and beta-N-methylamino-l-alanine as a function of nitrogen availability. Toxicon. 2014;87:1–5. doi: 10.1016/j.toxicon.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 57.Banack S.A., Caller T.A., Stommel E.W. The cyanobacteria derived toxin beta-N-methylamino-l-alanine and amyotrophic lateral sclerosis. Toxins. 2010;2:2837–2850. doi: 10.3390/toxins2122837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee M., McGeer P.L. , Weak BMAA toxicity compares with that of the dietary supplement beta-alanine. Neurobiol. Aging. 2012;33:1440–1447. doi: 10.1016/j.neurobiolaging.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 59.Field N.C., Metcalf J.S., Caller T.A., Banack S.A., Cox P.A., Stommel E.W. Linking beta-methylamino-l-alanine exposure to sporadic amyotrophic lateral sclerosis in Annapolis, MD. Toxicon. 2013;70:179–183. doi: 10.1016/j.toxicon.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 60.Pernet F., Malet N., Pastoureaud A., Vaquer A., Quéré C., Dubroca L. Marine diatoms sustain growth of bivalves in a Mediterranean lagoon. J. Sea Res. 2012;68:20–32. doi: 10.1016/j.seares.2011.11.004. [DOI] [Google Scholar]
- 61.Mondo K., Broc Glover W., Murch S.J., Liu G., Cai Y., Davis D.A., Mash D.C. Environmental neurotoxins beta-N-methylamino-l-alanine (BMAA) and mercury in shark cartilage dietary supplements. Food Chem. Toxicol. 2014;70:26–32. doi: 10.1016/j.fct.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 62.Weiss J.H., Christine C.W., Choi D.W. Bicarbonate Dependence of Glutamate Receptor Activation by Beta-N-methylamino-l-alanine—Channel recording and study with related-compounds. Neuron. 1989;3:321–326. doi: 10.1016/0896-6273(89)90256-0. [DOI] [PubMed] [Google Scholar]
- 63.Okle O., Stemmer K., Deschl U., Dietrich D.R. (l)-BMAA induced er stress and enhanced caspase 12 cleavage in human neuroblastoma sh-sy5y cells at low nonexcitotoxic concentrations. Toxicol. Sci. 2013;131:217–224. doi: 10.1093/toxsci/kfs291. [DOI] [PubMed] [Google Scholar]
- 64.Stanier R.Y., Kunisawa R., Mandel M., Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol. Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guillard R.R.L., Hargraves P.E. Stichochrysis-immobilis is a diatom, not a chrysophyte. Phycologia. 1993;32:234–236. [Google Scholar]
- 66.Tompkins J., DeVille M.M., Day J.G., Turner M.F. Catalogue of Strains. Titus Wilson & Son lmtd; Ambleside, UK: 1995. Culture collection of algae and protozoa; p. 204. [Google Scholar]
- 67.Guillard R.L. Culture of phytoplankton for feeding marine invertebrates. In: Smith W., Chanley M., editors. Culture of Marine Invertebrate Animals. Springer; New York, NY, USA: 1975. pp. 29–60. [Google Scholar]