Abstract
Hickory (Carya cathayensis Sarg.), an important nut-producing species in Southeastern China, has high economic value, but so far there has been no cultivar bred under species although it is mostly propagated by seeding and some elite individuals have been found. It has been found recently that this species has a certain rate of apomixis and poor knowledge of its genetic background has influenced development of a feasible breeding strategy. Here in this paper we first release SSR (Simple sequence repeat) markers developed in this species and their transferability to other three species of the same genus, Carya. A total of 311 pairs of SSR primers in hickory were developed based on sequenced cDNAs of a fruit development-associated cDNA library and RNA-seq data of developing female floral buds and could be used to distinguish hickory, C. hunanensis Cheng et R. H. Chang ex R. H. Chang et Lu, C. illinoensis K. Koch (pecan) and C. dabieshanensis M. C. Liu et Z. J. Li, but they were monomorphic in both hickory and C. hunanensis although multi-alleles have been identified in all the four species. There is a transferability rate of 63.02% observed between hickory and pecan and the markers can be applied to study genetic diversity of accessions in pecan. When used in C. dabieshanensis, it was revealed that C. dabieshanensis had the number of alleles per locus ranging from 2 to 4, observed heterozygosity from 0 to 0.6667 and expected heterozygosity from 0.333 to 0.8667, respectively, which supports the existence of C. dabieshanensis as a separate species different from hickory and indicates that there is potential for selection and breeding in this species.
Keywords: Carya cathayensis Sarg., SSR (Simple sequence repeat), C. hunanensis, C. illinoensis (pecan), C. dabieshanensis.
1. INTRODUCTION
Hickory (Carya cathayensis Sarg.) is an important non-timber species in the southeast of China in such provinces as Zhejiang and Anhui, the nut of which has high nutritional [1-5] and economical values [6]. A high economic return based on an annual output value of US$0.413 billion (US$1 = RMB¥6.30) has driven not only local farmers but also others in other provinces to develop hickory plantation on a large scale, but so far there has been no cultivar bred in this species.
Although this species is monoecious with separate male and female flowers, it has been discovered in hickory in recent years a poor polymorphism at the DNA level [7], a high percentage (15.7%) of and significant LD (linkage disequilibrium) using AFLP (Amplified fragment length polymorphism) markers [7] and RAPD (Random amplified polymorphic DNAs) markers [8]. It is reported that the recombination fraction between the markers determines the proportion in which the LD between two markers decays with generation for a random-mating population [9]. And limited recombination is also responsible for a significant LD [10, 11] and a recombination rate is important in predicting levels of LD in a population [12]. Eldon reported that a high LD might be associated with a phenomenon that the offspring of a single individual replace an intermediate fraction of the population [12]. Therefore it was inferred that there might be apomictic phenomenon in hickory, which was later embryologically confirmed [13].
So far co-dominant molecular markers have not been developed in hickory and dominant markers are still used in the genetic study of this species like other tree species [8]. Recently it has been suggested that it is feasible to study apomixis based on linkage analysis [14] although large-fragment linkage with no crossover has been reported in apomictic species [15-17]. However, linkage analysis is based on polymorphic markers. SSR (Simple sequence repeat), a co-dominant marker, has been reported in the classification of cultivars and accessions of Kentucky bluegrass (Poa pratensis L.), a facultative apomictic grass species [18], which implies that SSR markers might be useful in the genetic study of hickory. Here in this paper, it is reported for the first time the development of SSR markers in hickory and their transferability to other species of Carya. The specific objectives were to test the feasibility of SSR markers to be used in the future study of apomixis in hickory based on statistical genetics.
2. METHODS
2.1. Sources of SSR-containing Sequences and Primer Design
ESTs (Expressed sequence tags) of a fruit development-associated cDNA library (GenBank: JN786116 - JN786290) and RNA-seq data (http://www.cls.zju.edu.cn/binfo/ hickory/) of developing and differentiating female floral buds of hickory were analyzed using SSRHunter 1.3 for identification of SSR-characterized sequences. Then SSR primers were designed using Primer Premier 5.0, which satisfied the following parameters set: 100 to 300 bp fragments to be amplified, a primer length of 18 – 25 bp, a Tm of 45 - 65℃ with a difference no more than 5℃ between a forward and a backward primers, a GC content of 40 – 60% and avoidance of formation of a secondary structure. The designed primers were synthesized by Sangon Biotech (Shanghai) Co. Ltd.
2.2. Genomic DNA Extraction and Testing of the Primers Designed
Genomic DNAs of 32 individual hickory trees from natural populations in Daoshi Town, Henglu Township, Shunxi Town, Tuankou Town, Longgang Town and Maxiao Township of Lin’an, Anji County and Chun’an County of Zhejiang Province as well as She County and Ningguo City of Anhui Province were extracted from young but fully open leaves by a modified CTAB (Cetyltrimethyl ammonium bromide) method [19], then measured in optical density (OD) with ND-1000 Spectrophotometer V3.3 (NanoDrop Technologies, Inc.), and electrophoresed in 1% agarose gel for quality examination and dilution. The same method was applied to isolation of DNAs of pecan, C. dabieshanensis and C. hunanensis. Leaf samples of 12 accessions were collected from a pecan germplasm-collecting garden in Yvhang of Zhejiang Province. Twenty-nine samples from seedlings of C. dabieshanensis originated from a natural population in Jiuzihe Town, Luotian County of Hubei Province and that in Yvtan Village, Tiantangsai Town, Jinzhai County of Anhui Province were used in the experiment. Fourteen samples of C. hunanensis were from Dabaozi Township, Jinzhou County of Hunan Province.
Diluted DNA was amplified with the designed primers by a method modified by Song et al. [20]. PCR products were separated in 1% Argrose gel to examine whether these primers were working or not. Repeated amplification was conducted for the primers with no amplification product to confirm they were not applicable. For those applicable primers, if their amplification products fell in the range of fragment size initially designed, amplified fragments were purified with EZ-10 Spin Column DNA Gel Extraction Kit (Bio Basic Inc.) and then ligated to a T-vector following the description of pGEM®-T Easy Vector System I (Promega), followed by transformation using competent cells of DH5α in Escherichia coli prepared by a calcium chloride method. Positive clones selected using blue/white selection after growing on an IPTG (Isopropyl-1-thio-β-D-galactoside)/X-gal-added plate were cultured overnight at 37℃ in a liquid LB (Lysogeny broth) medium and the culture was used as a template for amplification. Then the overnight suspension-cultured E. coli was sent to Sangon Biotech (Shanghai) Co. Ltd. for sequencing of inserts to confirm the existence of SSRs if the size of inserts amplified was similar to that of the initial PCR fragment.
2.3. Screening of Primer Pairs in Hickory and Transferability of SSR Primers Developed from C. cathayensis to Other Species of the Same Genus
Those primers were thought to be applicable if their amplified products contained the same simple sequence repeat motifs by sequencing as those observed in sequences initially obtained from cDNA sequencing and RNA-seq data. Then these pairs of primers were used to amplify and separate DNAs of six hickory samples for screening of primers in terms of polymorphism.
The SSR primers that have been confirmed to be applicable to amplification in hickory were used to amplify DNAs of pecan, C. dabieshanensis and C. hunanensis to see whether these four species could be separated by SSR markers. Cloning and sequencing as described above were conducted to confirm the separation of the four species and existence of multiple alleles. For pecan, all the primers were tested and their amplification products were sequenced for validation of SSR motifs.
2.4. Comparison Between Hickory and C. dabieshanensis
Thirty-two samples of hickory and 29 samples of C. dabieshanensis were amplified with 30 pairs of SSR primers. The data obtained were then analyzed using POPGENE version 1.32 for observed number of alleles (Na), effective number of alleles (Ne), Shannon's Information index (I), observed heterozygosity (Ho), and expected heterozygosity (He).
3. RESULTS
3.1. Identification and Characterization of SSRs
Forty-three SSR-characterized loci were identified from the cDNAs sequenced of a fruit development-associated cDNA library, in which four types of di-nucleotide repeat motifs, mainly TC/GA, were found in 26 sequences (accounting for 60.5% of the total), seven types of tri-nucleotide repeat motifs, mainly GCA/TGC, found in 15 sequences (34.9%), and only one tetra-nucleotide repeat (CATG/ CATG) identified in two sequences (4.6%) [20]. A total of 1575 sequences was found out of 52274 contigs to contain SSRs from the RNA-seq data of developing female floral buds. Finally 1629 SSR loci were screened, out of which forty-nine sequences were found to have two SSR loci, five sequences three loci, and one sequence four loci. 871 loci (53.34%) were characterized by di-nucleotide repeats, followed by 543 loci (33.56%) containing tri-nucleotide repeats. There were 175 types of repeat motifs comprising of 6 types of di-nucleotide repeats with AG/CT and GA/TC dominant, 33 types of tri-nucleotide repeats mainly characterized by GAA/TTC, 30, 40 and 64 types of tetra-, penta- and hexa-nucleotide repeats, and one type each of hepta- and octa-nucleotide repeats.
3.2. Development and Testing of SSR Primers in Hickory
With SSR locus-containing sequences, 704 pairs of primers were designed as prescribed initially in parameters set. Then they were used to amplify three genomic DNAs extracted from young leaves of hickory trees. As a result, 272 pairs of primers had no PCR products and 393 pairs of primers had amplified fragments meeting the initial requirements of primer design, accounting for 58.66% of all the primers designed. Finally, it was confirmed by sequencing that a total of 311 pairs of SSR primers was developed from hickory (Appendix 1) based on the fact that their PCR products contained SSRs.
3.3. Screening of Primer Pairs in Hickory and Transferability of SSR Primers Developed to Other Species of the Same Genus
With DNAs from six randomly selected samples of hickory, all the primer pairs were tested to screen for polymorphism. Unfortunately, only monomorphism was observed among these six DNAs.
Forty-five pairs of primers developed were used to amplify four DNAs samples (each of hickory, pecan, C. hunanensis and C. dabieshanensis of Carya). Of these species tested, only does pecan have cultivars. Among these primers, 36 pairs (Cc2, Cc9, Cc13, Cc19, Cc31, Cc33, Cc35, Cc78, Cc137, Cc139, Cc140, Cc156, Cc175, Cc176, Cc183, Cc185, Cc187, Cc188, Cc191, Cc193, Cc195, Cc196, Cc197, Cc199, Cc200, Cc209, Cc221, Cc222, Cc229, Cc236, Cc245, Cc258, Cc283, Cc304, Cc306 and Cc309) were confirmed to create polymorphism in these four species, nine pairs (Cc12, Cc52, Cc177, Cc178, Cc179, Cc194, Cc198, Cc218 and Cc252) were not applicable for no amplification product and 23 pairs (Cc2, Cc19, Cc31, Cc33, Cc78, Cc137, Cc139, Cc140, Cc183, Cc191, Cc193, Cc196, Cc197, Cc199, Cc200, Cc209, Cc221, Cc229, Cc245, Cc283, Cc304, Cc306 and Cc309) could distinguish pecan from other three species, which means that primers developed have to be screened for their use in the study of multiple species of Carya. In addition, DNA fragments amplified with six pairs of primers were sequenced and proven to contain corresponding SSR motifs, which suggested that the primers developed from hickory are transferable in these species of the same genus (Table 1). Moreover, it had been also revealed by sequencing of PCR products that multi-alleles existed in these four species (Table 2). Similar results were also observed by Grauke et al. in pecan [21, 22].
Table 1.
Sequencing of PCR products amplified with different pairs of primers in four species of Carya.
| Species | PCR Fragment Length (bp) / Repeat Motif | |||||
|---|---|---|---|---|---|---|
| Cc195 | Cc197 | Cc199 | Cc200 | Cc221 | Cc222 | |
| C. cathayensis | 236 / (TGTTGG)4 | 125 / (GA)14 | 156 / (CT)23 | 244 / (TC)13 | 152 / (AGAAA)4 | 159 / (TC)13 |
| C. illinoensis | 244 / (TGTTGG)2 | 92 / (GA)5 | 156 / (CT)23 | 236 / (TC)6 | 152 / (AGAAA)4 | 157 / (TC)11 |
| C. hunanensis | 238 / (TGTTGG)3 | 122 / (GA)19 | 148 / (CT)19 | 240 / (TC)12 | 152 / (AGAAA)4 | 161 / (TC)14 |
| C. dabieshanensis | 241 / (TGTTGG)5 | 131 / (GA)17 | 148 / (CT)19 | 244 / (TC)13 | 142 / (AGAAA)4 | 155 / (TC)11 |
Table 2.
Multi-alleles expressed in SSR markers in different species of Carya.
| Species | Sample No. - Fragment No. | Primer | Repeat Motif | Length of PCR Fragments (bp) |
|---|---|---|---|---|
| C. hunanensis | S1-1 | Cc197 | (GA)5 | 114 |
| S1-2 | (GA)19 | 122 | ||
| S2-1 | Cc222 | (TC)11 | 155 | |
| S2-2 | (TC)14 | 161 | ||
| C. dabieshanensis | S1-1 | Cc222 | (TC)14 | 161 |
| S1-2 | (TC)13 | 159 | ||
| C. illinoensis | S1-1 | Cc193 | (TGG)5 | 244 |
| S1-2 | (TGG)4 | 244 | ||
| S2-1 | Cc194 | (CAT)5 | 233 | |
| S2-2 | (CAT)6 | 236 | ||
| S3-1 | Cc222 | (TC)11 | 157 | |
| S3-2 | (TC)17 | 167 | ||
| S3-3 | (TC)21 | 175 | ||
| S3-4 | (TC)27 | 187 | ||
| C. cathayensis | S1-1 | Cc195 | (TGTTGG)4 | 200 |
| S1-2 | (TGTTGG)5 | 230 | ||
| S2-1 | Cc58 | (TTC)6 | 111 | |
| S2-2 | (TTC)8 | 118 | ||
| S3-1 | Cc131 | (CT)11 | 298 | |
| S3-2 | (CT)43 | 389 |
More DNAs samples of each species (12 samples of pecan, 29 samples in C. dabieshanensis, 32 samples of hickory and 14 samples in C. hunanensis) were used in amplification with 26 pairs of primers (Cc2, Cc12, Cc19, Cc78, Cc175, Cc176, Cc177, Cc178, Cc179, Cc183, Cc185, Cc187, Cc188, Cc191, Cc195, Cc197, Cc209, Cc218, Cc221, Cc222, Cc245, Cc258, Cc283, Cc304, Cc306 and Cc309). As a result, polymorphism occurred in C. illinoensis and C. dabieshanensis, which was not the case in either hickory or C. hunanensis. Fourteen pairs of primers (Cc19, Cc176, Cc177, Cc179, Cc185, Cc187, Cc191, Cc195, Cc197, Cc222, Cc229, Cc258, Cc306 and Cc309) were found to be polymorphic among the accessions of pecan tested.
Since pecan has been used in the reciprocal cross with hickory to study apomixis [14, 23], sequencing of PCR fragments of pecan DNAs amplified with all the SSR primers developed from hickory was conducted. As a result, 276 pairs of primers were amplifiable but finally 196 pairs of primers were confirmed to have SSRs contained in their PCR products (Appendix 1), out of which 121 pairs could be used to distinguish hickory from pecan.
3.4. Comparison Between Hickory and C. dabieshanensis
So far only hickory and C. dabieshanensis are applied in the production of nuts, both of which have no cultivar so far and mostly propagated by seeding. The main producing area of both species is located in the areas of Tianmu Mountain bordering Zhejiang and Anhui. C. dabieshanensis, a species identified in the 1980s [24] and mostly growing in a wild habitat [25], has been increasingly used in recent 10 years because of its high economic value to farmers in Anhui and widely extended there except hickory, which is not the case in Zhejiang where only hickory trees are planted. The nut of C. dabieshanensis is larger than that of hickory [25, 26].
Based on the analysis with 30 pairs of primers, it suggested that there was monomorphism in hickory but it was not true in C. dabieshanensis. In C. dabieshanensis, eight pairs of SSR primers revealed polymorphism (Table 3; Fig. 1), which indicated that these two species differed from each other in genetic background. In both species, eight amplified loci had 15 alleles obtained from hickory and 22 from C. dabieshanensis. In C. dabieshanensis, the number of alleles per locus (Na) ranged from 1 to 4 with an average of 2.75 and the even effective number of alleles (Ne) was 2.26. An average Shannon Information index (I) of 0.75 was detected. The observed heterozygosity (Ho) and expected heterozygosity (He) ranged from 0.345 to 1 and 0 to 0.752, respectively (Table 3).
Table 3.
Results of eight SSR markers screened in 29 samples of C. dabieshanensis.
| Locus | Naa | Neb | Ic | Hod | Hee |
|---|---|---|---|---|---|
| Cc19 | 3.0000 | 2.5837 | 1.0152 | 0.6552 | 0.6237 |
| Cc197 | 3.0000 | 2.6159 | 1.0250 | 1.0000 | 0.6286 |
| Cc221 | 2.0000 | 1.0351 | 0.0871 | 0.0345 | 0.0345 |
| Cc222 | 3.0000 | 1.8512 | 0.7105 | 0.6429 | 0.4682 |
| Cc245 | 1.0000 | 1.0000 | 0.0000 | 0.0000 | 0.0000 |
| Cc258 | 2.0000 | 1.3993 | 0.4597 | 0.3448 | 0.2904 |
| Cc304 | 4.0000 | 3.8244 | 1.3632 | 1.0000 | 0.7519 |
| Cc309 | 4.0000 | 3.7798 | 1.3569 | 1.0000 | 0.7483 |
| Mean | 2.7500 | 2.2612 | 0.7522 | 0.5847 | 0.4432 |
Number of alleles.
Number of effective alleles.
Shannon Information index.
Observed heterozygosity.
Expected heterozygosity.
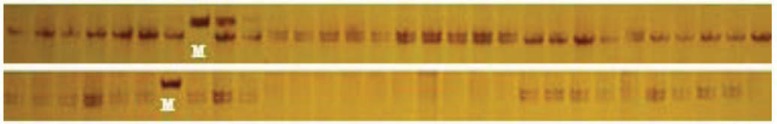
Fig. (1).
Electrophoresis profile of DNAs in C. dabieshanensis amplified with SSR primers developed from hickory (Primers: Cc258 (up) and Cc245 (down); M: marker (up: 200 bp and down: 300bp).
4. DISCUSSIONS
SSR markers are co-dominant, which, in theory, would be high in the rate of polymorphism in hickory that is propagated by seeding, but they are monomorphic. It has been embryologically confirmed that hickory is an apomictic species with a certain rate of outcross [13, 23]. Apomixis was reported to be regulated by a single master regulatory gene or by a gene complex comprising of several tightly linked genes that were located in a recombinationally suppressed region [15-17], which was observed in Oryza sativa [15, 27], Paspalum simplex [28], Tripsacum dactyloides [29], et al. When compared with their sexual relatives, apomictic species had cosegregated fragments that had a length of 15 – 40 cM [15], which could be regarded as a singly inherited Mendelian trait [30]. That means that apomictic species have large gene-linked fragments in a chromosome. Since development of EST-derived SSR markers is based on expressed genes and amplified SSR-containing fragments are relatively short as compared with dominant markers, it is quite possible in hickory that there is monomorphism tested with SSR markers, but this is different from the result obtained with dominant markers, in which polymorphism to different extents was observed [7, 8].
For transferability of SSR markers, it is quite possible and common among species of the same genus or even across genera of the same family [31-34] for conservation of SSR loci [35]. Similar results are obtained and described in this paper, which is that the SSR markers developed from hickory are transferable to C. dabieshanensis, C. hunanensis and pecan. Apomixis is also found in pecan [23] and a transferability rate of 63.02% is observed between hickory and pecan here. In addition, it seems that types of mechanism responsible for apomixis in hickory and pecan are different for the fact that there is monomorphism in hickory tested by SSR markers but polymorphism among accessions of pecan, the latter of which is similar to the case in Kentucky bluegrass [18]. Moreover, testing of SSR markers developed from hickory in C. dabieshanensis supports the existence of C. dabieshanensis as a separate species different from hickory for its polymorphism, which is smilar to the result obtained with RAPD markers [27, 36].
ACKNOWLEDGEMENTS
This work is supported by grants “Linkage analysis-based genetic study of apomitic hickory (Carya cathayensis Sarg.) (31370678)” and “Mechanism of apomixis in Carya catyayensis and its biological significance in breeding system (31100229)” from NSFC (Natural Science Foundation of China), a grant (2011R50030) from the Program for Zhejiang Leading Team of S&T Innovation, grants (LZ13C160002 and Y3110311) from the NSF of Zhejiang Province, China and a grant (2013AA102605) from "863" Program of China.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
APPENDIX
Appendix 1.
Characteristics of 311 SSR markers developed in Carya cathayensis.
| Locus | Primer sequences (5’-3’) | Repeat motif | Ta (°C) | Size (bp) | Applicability in pecan |
|---|---|---|---|---|---|
| Cc1 | F: ATGAGGGTTATGGAGGACA | (TA)9 | 51.8 | 173 | Y (Yes) |
| R: TGCGATAAGTGGCATTCAC | |||||
| Cc2 | F: ATGAGGGTTATGGAGGACA | (AT)10 | 55.4 | 188 | Y |
| R: GGTAGCCGGACAAACAGA | |||||
| Cc3 | F: ACGCTCCATCCGTAGTTG | (TGC)6 | 53.1 | 281 | Y |
| R: GAGTCTCATTCGCCAAGC | |||||
| Cc4 | F: GCTCCAAGCGAAAGTCAAGT | (TC)6 | 56.7 | 176 | N (No) |
| R: TCATAAACCAACGCCAAAGA | |||||
| Cc5 | F: CAAATCGAGCTGTTGGTTC | (TA)7 | 55.4 | 140 | Y |
| R: GTACTCCTGCTTGCTCCTTA | |||||
| Cc6 | F: ATCTGGGCATAGGTAGCA | (ATT)5 | 51.6 | 241 | Y |
| R: ATCGGAATAGTTCCTTTCAT | |||||
| Cc7 | F: AGCCAATTCCATATCCCAAGC | (TA)5 | 55.3 | 143 | Y |
| R: GACATACCAAACCGTTCTGC | |||||
| Cc8 | F: CGCCTGTATGTCACCCTT | (TA)5 | 55.4 | 159 | Y |
| R: AGAGTCGCTTGGGTTTATC | |||||
| Cc9 | F: CAAACGACGGAGTCAACA | (AG)11(AGA) 8 | 52.0 | 164 | N |
| R: ACCCGAAGTGGTTCAAAT | |||||
| Cc10 | F: GCTTCGGCTTCTTCTGCA | (TC)5 | 57.3 | 265 | Y |
| R: GGATACCCGGAGTCTTACGT | |||||
| Cc11 | F: ACAACAAGCAATACACCCAA | (ATT)9 | 50.1 | 199 | Y |
| R: CTAATCCAAGTAAGCCTCAA | |||||
| Cc12 | F: CAAATCAGTTCAGACACTCCCCCTC | (TC)5 | 63.8 | 262 | Y |
| R: AGTTCTCGGTGCGTGCTTGTAGTAT | |||||
| Cc13 | F: CCCAAGATCCAGTACAACG | (TC)14 | 51.3 | 262 | N |
| R: AATGCCACCGATTCTTG | |||||
| Cc14 | F: ATTGCTGTTGCCACTGAG | (CTG)5 | 48.7 | 322 | Y |
| R: ATGTCATCATCGGACTCTT | |||||
| Cc15 | F: CTGTAACTGCAAAAGACC | (AG)12 | 45.0 | 137 | Y |
| R: AGGCTATCTCATACCACC | |||||
| Cc16 | F: TGCCTTCTCCAGCTCATG | (AT)5 | 57.3 | 274 | N |
| R: CAACCCTAGCAATCTACGAC | |||||
| Cc17 | F: AAGTTTGGCTTTGTGAGG | (CATG)5 | 50.3 | 137 | N |
| R: ACAAACGGTATGGATCGC | |||||
| Cc18 | F: CTTTCCTGCCATACTCAC | (AG)9 | 45.9 | 116 | Y |
| R: ATCGTCCTCTTTACATCC | |||||
| Cc19 | F: AAACCTTGGCATAGTCATTTGAGA | (TC)8 | 60.1 | 96 | Y |
| R: GCTTTGTCAACTTTGTTTTGGGTGT | |||||
| Cc20 | F: ACAAGAAGAAGAAGCGAG | (AG)5 | 46.8 | 116 | Y |
| R: CTTCCTCTTTCTGCCTCT | |||||
| Cc21 | F: GCTTCGGCTTCTTCTGCA | (TC)5 | 57.0 | 311 | N |
| R: GATGCCAAGGCGATTTCA | |||||
| Cc22 | F: GGGCAAAGCACAAACTCC | (AT)4 | 57.0 | 166 | Y |
| R: TATCGGCGGTGTATCAGG | |||||
| Cc23 | F: ACTTCGGCTTCTTCTGCA | (TC)5 | 55.0 | 271 | Y |
| R: GAATACCCGGAGTCTTACGT | |||||
| Cc24 | F: AAGTGGAACACGAGGAGC | (GAG)6 | 52.7 | 277 | Y |
| R: TGCTTGTTCACTGCCTTT | |||||
| Cc25 | F: TGCTCCTTCCAAGAACATAA | (AGA)6 | 53.3 | 177 | Y |
| R: CGTCGCAATGAGAATGTTT | |||||
| Cc26 | F: ATTACCGAAGCGGATTAGA | (GAA)6 | 53.3 | 148 | Y |
| R: GTGGAAGAACCCGTATGTT | |||||
| Cc27 | F: AGAAGAGGAGCAACGAACC | (CAA)7 | 53.9 | 261 | Y |
| R: CATAATGCCAAACCGAAAC | |||||
| Cc28 | F: TGCTGGGAATTTGGAGAC | (CAG)7 | 53.3 | 256 | Y |
| R: TGACAAGGCGATTATGGTT | |||||
| Cc29 | F: GAAAGATGTTTGGGAGGAA | (AG)11 | 53.3 | 183 | Y |
| R: GCTTCCAAGTTCCAACCC | |||||
| Cc30 | F: ACCAGTTTGGAGGAAGACA | (AGA)6 | 53.7 | 209 | Y |
| R: AAGATTAGCCATCCCATACA | |||||
| Cc31 | F: TAGTCAGAGGCTCGAAGAAA | (TC)9 | 52.5 | 72 | N |
| R: TGCTATTACCCAGAAACCAT | |||||
| Cc32 | F: AGAACTGCGATTCGTTATTC | (TCT)9 | 53.7 | 151 | Y |
| R: ACAACCTTCCTCAGCCAC | |||||
| Cc33 | F: TCGCTCGGACTCAACTCT | (CT)11 | 53.7 | 165 | Y |
| R: ATCCACCATCTTCTCCTTTT | |||||
| Cc34 | F: GCCATAGATTTCTAATCCTCTT | (AG)9 | 54.5 | 207 | Y |
| R: CAAACCCAGCCGACATAA | |||||
| Cc35 | F: GGCAATCTACCTCCATCAC | (AT)10 | 55.0 | 276 | N |
| R: TAGCAACGTGCTCCATTC | |||||
| Cc36 | F: GCTTCATCACCGCCTTCT | (TCT)6 | 54.0 | 255 | Y |
| R: CGGAGCAACAATCCCTAA | |||||
| Cc37 | F: AGGGCATTTGGCTGGTTT | (TA)8 | 55.0 | 220 | Y |
| R: AAGGAGCTTGCAGAAGTTGA | |||||
| Cc38 | F: CAAGGGCAACCAGAAACA | (GCA)7 | 55.0 | 201 | Y |
| R: GATGGCTACGAAGGAGAAGA | |||||
| Cc39 | F: CGGTCTCAAACCCAAATC | (CTT)6 | 55.0 | 235 | Y |
| R: AATTATACTCCTCCACCACAAG | |||||
| Cc40 | F: TGCAGGTTGCTTTCTTCG | (TCT)12 | 52.2 | 167 | Y |
| R: GGATCTGAGGCATTTGTAAG | |||||
| Cc41 | F: TCTGAGTTGTTGTGCGTCTG | (CT)9 | 54.7 | 156 | Y |
| R: AGCGGTTGGTGCGATTTA | |||||
| Cc42 | F: TACGAGGAACACCAAAGC | (AG)15 | 55.0 | 143 | N |
| R: TCGAGACATGAATCTAACCC | |||||
| Cc43 | F: GACCCATTTGCTTCTGCT | (CAC)6 | 55.0 | 170 | Y |
| R: CAGAGGGAAAGTCCAGGTT | |||||
| Cc44 | F: GAGGCTGCGAATCGTCTG | (AAC)6 | 55.0 | 265 | Y |
| R: TTGATTGAGGAAGGAGGG | |||||
| Cc45 | F: GAGGTGGTGGTGGTGGTGGT | (GGT)7 | 55.4 | 214 | Y |
| R: ATTGCCGTTCCTCGCTTTT | |||||
| Cc46 | F: TGGCTCCTTGCCTGTATTT | (AAT)6 | 55.4 | 296 | Y |
| R: CTCTCTTTGTCTGTGTCTGTCTCTG | |||||
| Cc47 | F: ATGAGCGTAGGGCATGTAA | (CTG)6 | 58.0 | 194 | Y |
| R: CAACCAACGGCGGTGATA | |||||
| Cc48 | F: CCTTCGTCGTCTTCATCTT | (CT)8 | 55.4 | 135 | Y |
| R: TGTGCCTCTGTGACCTCC | |||||
| Cc49 | F: GTTACGACCGAGATAGATAGAAG | (GAG)6 | 53.6 | 116 | Y |
| R: TGATAAACGACGAGGACAAG | |||||
| Cc50 | F: CAATCTGTTGCTTCCGTCTT | (GA)17 | 55.8 | 169 | Y |
| R: TGCCTCCTCATTCATACTCC | |||||
| Cc51 | F: CATCTGCTCCACCCAAAC | (TCT)6 | 56.1 | 165 | Y |
| R: ATTCATACGCTGTACCATCAC | |||||
| Cc52 | F: CAGGTTGAATGTGGGAGC | (TCT)7 | 56.4 | 377 | Y |
| R: TAGCCACTGTCTTTCCTGTATT | |||||
| Cc53 | F: ATCCCACCTTTAAGATACAACC | (ATAC)6 | 56.4 | 153 | Y |
| R: TATGCGACGGAAGCGAAC | |||||
| Cc54 | F: GTAGTGGACGCAGCAAGA | (TC)9 | 51.7 | 207 | Y |
| R: TCGTAGGAGCACGGAGTT | |||||
| Cc55 | F: GTGGGCTAGGTGGAGGAT | (CTT)9 | 53.9 | 173 | Y |
| R: AACCAGGGAGACGGACAA | |||||
| Cc56 | F: CTTCACAACGGAGCGAGCCT | (AG)10 | 63.3 | 355 | N |
| R: GCAATGCCACGCCATCAAA | |||||
| Cc57 | F: GGGAAGCCTCCTCTGCTAT | (AG)14 | 56.5 | 143 | Y |
| R: GTGCTGGTTGGATTTGGTG | |||||
| Cc58 | F: CTCCGTGGCTCCTCTATC | (TTC)6 | 57.6 | 111 | N |
| R: TCTCCTTCTGGGAGGTTCT | |||||
| Cc59 | F: CTGCTGCCGACAACTGCT | (TC)14 | 55.8 | 140 | N |
| R: CGAGAATGTATCCCGCTATG | |||||
| Cc60 | F: TCCTCATCGTCGCCACCT | (TC)9 | 57.4 | 229 | N |
| R: CGTCACATAGATGAAGAAAGTTACC | |||||
| Cc61 | F: CTTCCTCCCTCGTTGCCTCAT | (ATC)6 | 59.6 | 131 | Y |
| R: TCCACCACCGCCTCCAAA | |||||
| Cc62 | F: GGACCAGCGATTCTTAGGC | (TCT)6 | 59.7 | 161 | Y |
| R: CTCCGAATCTCCTCGCTTG | |||||
| Cc63 | F: GACGAAGTTGATGGAGGCAATG | (GAT)6 | 61.8 | 222 | Y |
| R: TCGCAGGTGGCTGAGGTG | |||||
| Cc64 | F: GGGGATGAACGGCCAGGAT | (CGG)6 | 61.9 | 140 | Y |
| R: ACCCACGGTCACGCCCACTA | |||||
| Cc65 | F: CGCAGTCACTCACTCGTTT | (TC)16 | 51.8 | 140 | Y |
| R: CAAGTTTGGAGGTTTGTTCT | |||||
| Cc66 | F: AAAGCCAACCCCAAAATC | (TTC)4 | 52.7 | 200 | Y |
| R: CTCCATGAGCACCATCCA | |||||
| Cc67 | F: TGCTTCCCGTGTAGATGA | (TGG)6 | 53.3 | 300 | N |
| R: AAGTCCCAGGAATCAAAGA | |||||
| Cc68 | F: TTCAAAAATCCACCTGCCAA | (CTT)6 | 53.7 | 189 | Y |
| R: CGGTATCGTTCAAATCCTCG | |||||
| Cc69 | F: TCAATAAAGCATTCCGTCCA | (TC)11 | 53.7 | 153 | Y |
| R: ACTGAACCATTTCCTCGTAGC | |||||
| Cc70 | F: AAAGGTTCCATCGGCATAAA | (AGA)8 | 53.7 | 143 | Y |
| R: CAGTGTAGGGGATTCCTGTAGC | |||||
| Cc71 | F: GAACTCCGCTTGTGTTTCATA | (TC)13 | 53.7 | 121 | Y |
| R: CATCAAATTCCAACCCCTAA | |||||
| Cc72 | F: GCTAAAACCTCTCCTGAACCA | (TC)9 | 53.7 | 133 | Y |
| R: CCACAAATGAAAGAATCCCA | |||||
| Cc73 | F: TTTTCTGATGCTTTGCTTG | (TCCTCT)4 | 53.7 | 196 | Y |
| R: GGAGACTGTTCCGATGGTGT | |||||
| Cc74 | F: GGGACGACGAGGAGAAAGAC | (TC)9 | 54.6 | 265 | Y |
| R: AAAGCCGCAGACGCAAA | |||||
| Cc75 | F: AACCGCCATTGCCATTC | (AG)7 | 54.6 | 237 | Y |
| R: CAACCGCCCTTTCCTTTT | |||||
| Cc76 | F: TCAAGCCAAGGATCACCA | (TCT)6 | 55.0 | 231 | Y |
| R: GCCACTGCCAGGACTCTAT | |||||
| Cc77 | F: GAAGCCTACTCCTACAGCGA | (TAT)4 | 55.0 | 411 | Y |
| R: CACCAACCAAAGCCATCA | |||||
| Cc78 | F: ACATCCGAGCATCACCAC | (TC)11 | 55.4 | 148 | Y |
| R: TCAGATTCCGAACAACGAC | |||||
| Cc79 | F: ACGAGGCACAAACATACAG | (TTC)7 | 55.4 | 131 | Y |
| R: GTTGACAGTGGTGGACAGAG | |||||
| Cc80 | F: ATTTGGGGTCTTTCTGGGT | (AG)8 | 55.4 | 164 | Y |
| R: GGCTGTTCTTATTTGTCGCA | |||||
| Cc81 | F: TACACTCCAAGAAGGAAAGGC | (GAG)5 | 55.4 | 278 | Y |
| R: TTGTCGTGGGTTCATTTGC | |||||
| Cc82 | F: CCCACCCAACTGCTCTACCT | (CTT)6 | 55.4 | 136 | Y |
| R: TCGCCATTCACAACGGATT | |||||
| Cc83 | F: CAATGGAGGAGTGAGAGCG | (AGA)6 | 55.4 | 189 | Y |
| R: TTCGTCGGAAAATGGAGAC | |||||
| Cc84 | F: CTGGTTGGTCTTTCGGTCA | (AT)11 | 55.4 | 285 | Y |
| R: AAGCCCTTCTCGTTTCACA | |||||
| Cc85 | F: CTTAGAGCCCAGGTGAATG | (GCA)7 | 55.8 | 243 | Y |
| R: GCTATCCTTCTTTCGGTTTC | |||||
| Cc86 | F: TCCTGCCCCTGGTGATT | (TC)10 | 55.8 | 193 | Y |
| R: TTGCTGGATTGTCAGTTTGG | |||||
| Cc87 | F: TCAAAGAAGCAAAGCACCGA | (AAGA)5 | 55.8 | 196 | Y |
| R: CCGAGCCAATGAATAGAATGTC | |||||
| Cc88 | F: AGCCTGGGGAAAAGGAACA | (TTC)6 | 55.8 | 170 | Y |
| R: GGGTTTGGTTGTCTTATTGG | |||||
| Cc89 | F: GAAGGTGAAACAGTTGAAGCAG | (TC)9 | 55.8 | 173 | Y |
| R: GAATCTCATTTTTGGTCGGC | |||||
| Cc90 | F: CTAACTTCCTCCTCTTCGTCTTC | (TCGTCC)2 | 55.8 | 102 | Y |
| R: GGATTATTTCGTGTGCGTGA | |||||
| Cc91 | F: GTACATTTGATGCCGTTTCG | (GA)9 | 55.8 | 214 | Y |
| R: ACTTGTTAAGCCCTCCTCGT | |||||
| Cc92 | F: TGCCAAGAATGAATGGGAAG | (GAA)6 | 55.8 | 383 | Y |
| R: GAGAGGAAGCAAAGACAAAGGTT | |||||
| Cc93 | F: ATTTGAATGCCAGAAGAGCG | (GAA)6 | 55.8 | 179 | Y |
| R: CTGAGGAGAAGCCATACCCA | |||||
| Cc94 | F: AATGATCTGGGTATCTAGCGTC | (TTTCT)6 | 55.8 | 127 | Y |
| R: CTTGGTTGAGTTGGGTTTTG | |||||
| Cc95 | F: AACTGGCTCACTTTCTTCTGG | (ATG)6 | 55.8 | 194 | Y |
| R: CAACATCCTGCTTATTTCCG | |||||
| Cc96 | F: TGGGGATTGAGAAGAGCAGT | (GA)10 | 55.8 | 128 | Y |
| R: AATGGAAGAACCGAACAGCA | |||||
| Cc97 | F: GGGATTATTTGTTTGCCGTCA | (TC)17 | 56.1 | 205 | Y |
| R: GAGAGAAGAAGGGCTGGAGGA | |||||
| Cc98 | F: AGGAACTGAAGGGGCTAAAGG | (GGTTT)4 | 56.1 | 260 | Y |
| R: AAAACAGAATGCGTCGGAGAA | |||||
| Cc99 | F: AGGAAACTGACGGCGAGAC | (AACC)5 | 56.1 | 179 | Y |
| R: GGAATGAACGACTTTGGATGA | |||||
| Cc100 | F: GTAGAGCCATCCACATCAAAGT | (TTTTATTT)3 | 56.1 | 206 | Y |
| R: CCGAAACGATAAGGAAAAGAG | |||||
| Cc101 | F: GACGAAGATGGTGATGTGGTA | (TGA)4 | 56.1 | 167 | Y |
| R: TGCTGGGTTTGAACAGAAGTA | |||||
| Cc102 | F: TCTTAGGAGCAAGCCCCA | (TA)9 | 56.1 | 155 | Y |
| R: GCATTCTTTATTACAGCAGCG | |||||
| Cc103 | F: GGAGGTGACATAGCACAGGC | (AG)9 | 56.1 | 151 | Y |
| R: ACCAACCAAGAAACAATAGCG | |||||
| Cc104 | F: ATCACATCCTCGCAGACCTAC | (CT)9 | 56.1 | 312 | Y |
| R: CTACCAAATGGCACATCAACA | |||||
| Cc105 | F: TCACCATCTCAGTCACTCCGTC | (GGAA)5 | 56.1 | 239 | Y |
| R: CGAATCAAAAAAACCCTCCCT | |||||
| Cc106 | F: TCGTTTATGGTCAATGCCTCT | (GA)16 | 56.1 | 259 | Y |
| R: GCAGTCACTCACTCGTTTCCT | |||||
| Cc107 | F: TGATTTTTAGCAGCAGAAGTCG | (GTC)6 | 56.4 | 187 | N |
| R: AGAGCCACAGCCTCGTTCA | |||||
| Cc108 | F: ACTTCCACCACCATCATCCA | (AAG)7 | 56.1 | 181 | Y |
| R: CTTGTTGGGTATCATTCAGCAT | |||||
| Cc109 | F: AATAAGCCCTTCTCGTTTCACA | (AT)10 | 56.4 | 278 | Y |
| R: TTTCGGTCAGACCTACAGCG | |||||
| Cc110 | F: TCATCAATGGGCGGTAGC | (AGCC)5 | 56.4 | 298 | Y |
| R: TCAACAAACTGACTGGCAGTAA | |||||
| Cc111 | F: CGCACCAATCCAACAGG | (GAAGG)5 | 56.4 | 147 | Y |
| R: CAAGAGTGAGTTCAAACAAAGC | |||||
| Cc112 | F: AAGACGACGCTGTATCCATTAT | (TA)10 | 56.4 | 180 | N |
| R: CCTTTCTCCCTTTCACTTTCC | |||||
| Cc113 | F: CCTCTGTTATCGCCGTTATCT | (GA)10 | 56.4 | 273 | Y |
| R: AAAAGTTTACCAGAAGGCAGTG | |||||
| Cc114 | F: CTACAGAAGCAGCAGGGGA | (AGA)7 | 56.6 | 163 | Y |
| R: CAATCGCATACAAAGTCTCAGTT | |||||
| Cc115 | F: GTCCTTCTCCACCAAATCCTG | (AAG)8 | 57.0 | 189 | Y |
| R: CCGCCTGTTGCCTTTCA | |||||
| Cc116 | F: GTCCGCAACCGACGAAA | (GAG)6 | 57.0 | 206 | Y |
| R: CAGAACCTAAATCGCCCACA | |||||
| Cc117 | F: CCCAACCCGTAACAGCA | (GAG)6 | 57.0 | 127 | Y |
| R: GAGAAGGGCTCCAGAAGGT | |||||
| Cc118 | F: GGGCTCGCAGAAGATACTCAC | (CT)10 | 57.3 | 313 | Y |
| R: TTTCCGCTTGGCACCTCA | |||||
| Cc119 | F: CCTTCTCATCCAAATCCCTC | (AGG)7 | 57.3 | 389 | Y |
| R: CGAATGCCCTCCACAACT | |||||
| Cc120 | F: GGTTCTTCTTCCCAGCGA | (TC)12 | 57.3 | 202 | Y |
| R: ACCGAAAAGCACCCACAC | |||||
| Cc121 | F: GCCGATTTGATTCCACCC | (TTC)7 | 57.3 | 207 | Y |
| R: GGCTAAACTTCTCTCTTCCCATT | |||||
| Cc122 | F: ACGGGATACCTCCGCAAA | (CCA)7 | 57.3 | 440 | Y |
| R: ATCGTCGCAGTGGCTACAGT | |||||
| Cc123 | F: GGGAGGAAGCGGTGAAAA | (AGG)5 | 57.3 | 268 | Y |
| R: AGAGGGATTGGATGTGGGTAA | |||||
| Cc124 | F: TGGGTTCCCTGCCTTCTA | (GAC)6 | 57.3 | 231 | Y |
| R: CGAGTATCCCTCACGCAAA | |||||
| Cc125 | F: AAACCCTCGCTGTTCATCG | (TTC)7 | 57.6 | 235 | Y |
| R: TCAACCCAATCCGTATCCG | |||||
| Cc126 | F: CTGGGGTTGGGATTGTGTT | (AG)10 and (AGA) 6 | 57.6 | 214 | Y |
| R: GTCTTCATCGTCGTCGTTTG | |||||
| Cc127 | F: CAAGTTGTAGTTGTGGGAGGC | (CAC)7 | 57.6 | 218 | Y |
| R: AGATTGTGATTGGGAGGGC | |||||
| Cc128 | F: ATGAGGTATTCGGCGTAGC | (AAAC)5 | 57.6 | 303 | N |
| R: GGGGGTTCGCGGTTCTTATTT | |||||
| Cc129 | F: TTGGGGTGGTTTCTTGGTC | (GAT)6 | 57.6 | 297 | Y |
| R: CTGCGTGTGTGGGAGTTTG | |||||
| Cc130 | F: CAATCTAAACATCACTCACTCCC | (CT)11 | 57.6 | 603 | Y |
| R: AGAACACCACCAAGCGAAC | |||||
| Cc131 | F: ACCACCGAAGCACTACCTCAC | (CT)11 | 57.6 | 297 | N |
| R: AATACGATGGGTCCAGCGA | |||||
| Cc132 | F: GAGAAGAGCAGTATCTGTGTCCG | (GA)10 | 57.6 | 396 | Y |
| R: AGCGTATCCTCGCAGCAAT | |||||
| Cc133 | F: TTCCCTCGCCTTGCTTTCT | (AG)10 | 57.6 | 156 | Y |
| R: AAGTCAATCGGTGTGGTGCTC | |||||
| Cc134 | F: CCTCTGCTATCGCCCAAAA | (AG)11 | 57.6 | 345 | Y |
| R: TGCCCTCTTCTTCGTGCTC | |||||
| Cc135 | F: ATAGATAAGGGAGGGGGCAT | (AAAG)5 | 57.8 | 183 | Y |
| R: GCAAAACACAGGTATCGGGT | |||||
| Cc136 | F: GTGGTAGTGGCGGAGGAAAT | (GGT)5 | 57.8 | 197 | Y |
| R: CTGAAAAGGGGATGGGGATA | |||||
| Cc137 | F: GGCTTCGTTTGTTGTAGGCA | (AG)9 | 57.8 | 168 | Y |
| R: ATGGGGACTGTGATTCTGGAGT | |||||
| Cc138 | F: ACAAGAAGTCGGAGCCCAC | (TC)10 | 57.8 | 193 | Y |
| R: GATTTGCCAAAGACGAGAGG | |||||
| Cc139 | F: TTTCGCTCTCGTTTCTCAGG | (AG)10 | 57.8 | 105 | Y |
| R: CTCTCTTGTTCCCGCTACCC | |||||
| Cc140 | F: CTGGAGTGAGTCAGTATTGGCT | (GAAAA)4 | 57.8 | 226 | Y |
| R: CGATTGCTATACAGGGGTCA | |||||
| Cc141 | F: CTCACTGTCAAAAGTCTCAATCGT | (ATT)7 | 57.8 | 288 | Y |
| R: TAACCCAACCACCTCCACAA | |||||
| Cc142 | F: GGGTGGAGGATACAGGAAAA | (CT)10 | 57.8 | 168 | Y |
| R: CACATTAGGAGGTAGTGTTTATTGG | |||||
| Cc143 | F: TGGCGGTAGTTGTTGTTGATG | (TAG)5 and (GAA) 6 | 57.8 | 149 | Y |
| R: CCAAAACCCCAGAACCCTAA | |||||
| Cc144 | F: TGTAGAAGGTATTGGACTGAAGG | (TCA)7 | 57.8 | 123 | Y |
| R: AGATGAAGCAGGTGATGACG | |||||
| Cc145 | F: ACAGTGAGGGCAAATGAAGG | (AAG)7 | 57.8 | 127 | Y |
| R: TGACCCAAGCACGATTATGAC | |||||
| Cc146 | F: GAGAAGGATGTGGTGTCACTGTT | (GAT)6 | 57.8 | 291 | Y |
| R: CCTCGCCAAAAAGGTCTACA | |||||
| Cc147 | F: CTTTCGGGTTTGGAGGGTAA | (GA)8 | 57.8 | 118 | Y |
| R: CGGCAGGCACAATCTTATCA | |||||
| Cc148 | F: AACGACCTTGAACTCCAGCA | (CTCCT)4 | 57.8 | 92 | Y |
| R: TGAGATGACCTGAGGCGAGA | |||||
| Cc149 | F: CCTTCTCCTTCCGTCAAACA | (TC)14 | 57.8 | 239 | Y |
| R: AACGATAGAGTGGCTGCTGG | |||||
| Cc150 | F: GACCACCAGAACTCCCCAAT | (AAG)6 | 57.8 | 275 | Y |
| R: ATCCACAACAAAGGACGCAG | |||||
| Cc151 | F: AGGAGCAGAGCAATATCCAAGT | (GAA)6 | 57.8 | 223 | Y |
| R: GGTTCTTCCGCAGTTGTTCT | |||||
| Cc152 | F: CAACACCTTCCACTTCCAGC | (GAAGA)4 | 57.8 | 250 | Y |
| R: GACCCTGAAAACAATGCGAC | |||||
| Cc153 | F: GCCAAAGAGAAGAAACCCAGA | (TCT)6 | 58.0 | 191 | Y |
| R: AGAAGCAGTGTTGAAGGTGAGG | |||||
| Cc154 | F: GTAATGCCGATGAGACTGAGA | (GAAGAG)4 | 58.0 | 195 | Y |
| R: CTTCGCTACCTGGCTTGTC | |||||
| Cc155 | F: ACCACACTCCGACACCTCTT | (TC)8 | 58.0 | 221 | Y |
| R: CACCCAAATACTTCCTCCTTC | |||||
| Cc156 | F: CGTTACTGTTGTTGCGGTGAA | (TGG)6 | 58.0 | 247 | Y |
| R: ACCTCTGACCCGACCTCTAAAC | |||||
| Cc157 | F: ACCCTAACTGCCCTTCTTCC | (CT)9 | 58.0 | 269 | Y |
| R: CGTCTTCTTATCGGTAATCGG | |||||
| Cc158 | F: TCCACTGCTGACTCCACTTCT | (TCT)6 | 58.0 | 123 | N |
| R: ACCCGTAAATACTACCAACCG | |||||
| Cc159 | F: GGGCTGGGACTGGTTATAGT | (CCTCG)4 | 58.0 | 249 | Y |
| R: ATTCTCTCGTGAGATTGAGGC | |||||
| Cc160 | F: GGAGGGCACAAGCAAGAGT | (GAT)6 | 58.0 | 191 | Y |
| R: CTGGAGACCAAACAACAAACC | |||||
| Cc161 | F: GATTGTCCTCGTGTTCGTCG | (AGC)7 | 58.0 | 210 | Y |
| R: TTCGTGAGTGGCTGAAGTGTT | |||||
| Cc162 | F: CCGAGCACAAGACAAAAACTC | (GAA)6 | 58.0 | 258 | Y |
| R: CATATCCGTCACCATCCTCTG | |||||
| Cc163 | F: TACCTCAGCCAACCACTCC | (CAG)6 | 58.2 | 111 | Y |
| R: CAAATCCTCACACTGTCACTCA | |||||
| Cc164 | F: GAGCATTTTGTGTCATAAAGGAGAG | (AAT)7 | 58.2 | 272 | Y |
| R: ATGGATTTTAGAGGTGAGCGTG | |||||
| Cc165 | F: GCTACCAGTCTCACCGTTATCG | (GAA)6 | 58.2 | 286 | Y |
| R: TCCTTTTTCAACTCCTCCCTCT | |||||
| Cc166 | F: GGAGAAAGAAGACGAGGAGAGA | (CGAG)5 | 58.2 | 104 | N |
| R: AGTTCAATCCCACTATCCCAGT | |||||
| Cc167 | F: CTCGTTCGTAGTCTCAGCCTC | (CCCTAA)4 | 58.2 | 292 | Y |
| R: CCTTCCTCATAGTCGTCAAATC | |||||
| Cc168 | F: TGAGAGCAGAACTTGAACCAGA | (TC)9 | 58.2 | 192 | Y |
| R: TAGAGAAACGAGCCAGAGCG | |||||
| Cc169 | F: CTGCTCCTTCACTCCACTCA | (GCA)6 | 58.2 | 128 | Y |
| R: TCTCCTCCCTAACAATAACACC | |||||
| Cc170 | F: GCTTTACAGATGTCTCAAGGATTC | (CAGCAA)4(CAACAG) 4 | 58.4 | 249 | Y |
| R: GCCTCTGAAACTGGGTATGC | |||||
| Cc171 | F: CGAGACTTTGGTTGAGTGTGAA | (GAT)6 | 58.4 | 252 | Y |
| R: GTTTGCCTACACGATGACAGTTA | |||||
| Cc172 | F: GGAAGGAAGAGTATTGCGAAGAT | (GAA)6 | 58.4 | 197 | Y |
| R: GATACTACCCCGACTGCTCTGAC | |||||
| Cc173 | F: CCCTTGAAAACCTTGATCTCTGTA | (TC)8 | 58.6 | 231 | Y |
| R: TGTGATAAATGATGGGGGATGCC | |||||
| Cc174 | F: ATGAGGGGGAGGATGGACT | (GAT)7 | 58.6 | 154 | N |
| R: GCATTAAACAAACTACCACCACTC | |||||
| Cc175 | F: ACTGTGTGATGGGTGAAGATAGATT | (GTTTTC)4 | 58.6 | 279 | Y |
| R: GAAAACAAGACTGGAAGAAGGAAC | |||||
| Cc176 | F: CAGTGGGATGAGACGAATGATG | (TA)9 | 58.6 | 286 | Y |
| R: GACAAATACTAAAAGCAAGGGTGG | |||||
| Cc177 | F: AGGAACAGCACACAGAACCC | (TTC)6 | 58.7 | 118 | Y |
| R: TGTGATGTAGATAGAAACCTGGAAC | |||||
| Cc178 | F: TTCCCCTCCTCCTTCCTCA | (GCT)6 | 59.7 | 277 | Y |
| R: GCAAAACAGCGGTGGTCAG | |||||
| Cc179 | F: TGACATCACTCCGCACTCTC | (CCA)6 | 59.7 | 279 | Y |
| R: GCTCTCTTGGGTGTTGACG | |||||
| Cc180 | F: AAGCCCTCCTTTACCTGCC | (GCG)9 | 59.7 | 298 | N |
| R: CGAACTTACCATCCGCCAC | |||||
| Cc181 | F: ACACAAGCCCCAGCACATC | (TC)10 | 59.7 | 152 | Y |
| R: AGCCAGGAGCAGCACCAG | |||||
| Cc182 | F: CAAAGACCTAAGCCCGAGAG | (ATGAGC)4 | 59.7 | 329 | Y |
| R: ATGGCGTGCCCTAAGAGTC | |||||
| Cc183 | F: CGAAGCAGTTATCTGGGCAC | (CAT)6 | 59.9 | 227 | Y |
| R: CGGGTGAAAACTTGGTCTCTC | |||||
| Cc184 | F: CCAACAAAGCCCCAGAGAAC | (CT)10 | 59.9 | 216 | Y |
| R: AGCAGAGATGGAGAGAAGCGA | |||||
| Cc185 | F: GGTCACGGTCTTGGGCTTAT | (GAA)6 | 59.9 | 290 | Y |
| R: GGTCAACTCGGGTGTTTTCC | |||||
| Cc186 | F: CTCTCCAGTCCCAACCTCAG | (CGCCTC)3 | 59.9 | 281 | Y |
| R: TGAAGTAGGAGCAGGCGTCT | |||||
| Cc187 | F: GATCCCTAGTTCCATGCTCG | (TGC)6 | 59.9 | 301 | Y |
| R: ATCTCTACCTCAGTCACCCACA | |||||
| Cc188 | F: GGTCTGAACTCCCCACCATAG | (CCA)6 | 59.9 | 264 | Y |
| R: CGTCTCCATACGGAACCTCA | |||||
| Cc189 | F: CCATCTTCTACCCCCTACCAT | (TC)13 | 60.0 | 200 | Y |
| R: CAGCCTACCTACACTTTCTTCG | |||||
| Cc190 | F: AGTCTCCCTTCAGCCCTTCC | (CT)9 | 60.2 | 327 | Y |
| R: CTCATCTTTCTGCGTCTACTTGC | |||||
| Cc191 | F: CATCTTCCTCTAACTGATCCCTC | (GA)8 | 60.2 | 201 | Y |
| R: GACACTGTTGGTGGTTCTGTATC | |||||
| Cc192 | F: GAACCACTCTTCACCCAGCAC | (GA)15 | 61.9 | 273 | Y |
| R: GAGGATAACAGTCAGCAGCGG | |||||
| Cc193 | F: GCGAAAAATGGCGGAAT | (TGG)8 | 52.2 | 251 | Y |
| R: CACACAGTCAGAGACGCAAAA | |||||
| Cc194 | F: TGCCCTTTTCATTATCGCT | (CAT)6 | 53.2 | 236 | Y |
| R: AGGAGGTAGTTGGCTATCTGTAA | |||||
| Cc195 | F: AAGCAAACCCGTCAAACAA | (TGTTGG)4 | 53.2 | 236 | Y |
| R: AACCGAAGACCCAGAACCA | |||||
| Cc196 | F: AGACGCTGAGAACGCCTTT | (TC)7 | 53.2 | 183 | Y |
| R: TTTCCAATTCCGATTCCCT | |||||
| Cc197 | F: CTCCCAGTGGACGAACAGA | (GA)14 | 53.2 | 125 | Y |
| R: CCCAAATCAGAATCGCAAA | |||||
| Cc198 | F: CTTTGTTGTTGAATCCCCAT | (TGGTCG)5 | 53.7 | 173 | Y |
| R: TCCTTATCCTCACACTGCTTG | |||||
| Cc199 | F: AAAAAAAGGAATCGCCCATC | (CT)23 | 53.7 | 156 | Y |
| R: GCCGAGCCTCCAAGAGTAGA | |||||
| Cc200 | F: AGCCCCGAAGATACAGACA | (TC)13 | 53.7 | 244 | Y |
| R: AACAAACGAAGCAAAAGCAG | |||||
| Cc201 | F: CAAGATAACAAAAGCCAGCA | (AG)9 | 53.7 | 122 | Y |
| R: AAGGTGAAACAGTTGAAGCAG | |||||
| Cc202 | F: TTCAACTTCCTCCGATTCAA | (TC)13 | 53.7 | 208 | Y |
| R: GGCAACTACCAAGCATACGA | |||||
| Cc203 | F: TTTTCCAAGGATGATGCTCT | (AGC)4 | 53.7 | 241 | N |
| R: GTTTGCGGGAGTAATCGTAT | |||||
| Cc204 | F: ACCGCAAAATACTGAAAAGG | (AG)6 | 53.7 | 163 | Y |
| R: TGTGAGGGAGGGGAAGAA | |||||
| Cc205 | F: CTCCTTACTCCACGGATTTGA | (AAT)7 | 53.7 | 163 | Y |
| R: GGGGGGTTTTTTTCTTCTTT | |||||
| Cc206 | F: GTTCTGTAGCAAAGAGTGTCGG | (CTT)6 | 54.6 | 285 | Y |
| R: AAAATCTCGGGCGGTCA | |||||
| Cc207 | F: GGGAAGACTGGGAAGTGAGT | (TCT)9 | 54.6 | 180 | Y |
| R: TGGTTTCATTCGGTGGC | |||||
| Cc208 | F: TTCGCACTTGCCCAACA | (TTC)6 | 54.6 | 287 | N |
| R: ATAATAGTCTCCCAACCAAACG | |||||
| Cc209 | F: GGGTCGGTGCTTGATTTT | (CGG)6 | 55.0 | 294 | Y |
| R: CTCCAAACAAAGCCCACA | |||||
| Cc210 | F: AACTGGAAAAGGCACGGA | (GAA)8 | 55.0 | 294 | N |
| R: GAAACCCCAATCGCAGAA | |||||
| Cc211 | F: TAAAACCCTAAACCCTCGC | (TTC)7 | 55.4 | 240 | Y |
| R: TGAGAATCGGGGAAGAGAA | |||||
| Cc212 | F: AACAATCTGCTGCCAAAGG | (TA)12 | 55.4 | 239 | Y |
| R: GTCCAATAGGTAAATACACTCGG | |||||
| Cc213 | F: GGATTATTTCGTGTGCGTG | (GGACGA)2 | 55.4 | 102 | N |
| R: CTAACTTCCTCCTCTTCGTCTT | |||||
| Cc214 | F: AAAAAAGGGGTTAGCAGTGG | (AGC)6 | 55.4 | 264 | N |
| R: ATAAATACACCTCGGCGGA | |||||
| Cc215 | F: TCGGTATGGGTTTCCTTGA | (AG)9 | 55.4 | 157 | Y |
| R: AGTGGGTGGTGTTGGTTTG | |||||
| Cc216 | F: AAGGGCAATACCCCGTC | (CCA)6 | 55.4 | 211 | Y |
| R: AAGAATAGAGGCGTGGCAA | |||||
| Cc217 | F: TGAAGCCAGATTTTCGGAC | (GAA)6 | 55.4 | 258 | Y |
| R: AACTCATCTCCGCCTCTCTTA | |||||
| Cc218 | F: ATGAGCCAAGTCCGAGAAA | (GAT)6 | 55.4 | 257 | Y |
| R: TTGTATCCCAGGTCAAATGC | |||||
| Cc219 | F: TAGAAGCCATTCCAACACTCA | (AGAC)5 | 55.4 | 138 | N |
| R: TAACCTCGCCATAACCCAT | |||||
| Cc220 | F: CGAGGAACACCAAAGCAAA | (AG)12 | 55.4 | 285 | N |
| R: ATACGGACACTTCACAAACCC | |||||
| Cc221 | F: ATCTTGCGAAATCGGGACT | (AGAAA)4 | 55.8 | 152 | Y |
| R: CAGCGTTTAGAAGGGAGGG | |||||
| Cc222 | F: TAAGGAGCAGGTGCGGAAG | (TC)13 | 55.8 | 159 | Y |
| R: AAGACAACAGTATGTAGGTAAGGGC | |||||
| Cc223 | F: TTCTCTTCATTCCCATCCTG | (CAG)9 | 55.8 | 229 | N |
| R: TGCCTGAGTGGTCGCTT | |||||
| Cc224 | F: GAGCGAGAAAGTAGAGCCAA | (TTC)6 | 55.8 | 134 | Y |
| R: GTCACGCCTAAAACCAAAGA | |||||
| Cc225 | F: AACCCTAATGACGGTATCCA | (GAGAG)4 | 55.8 | 226 | Y |
| R: AACGACCTTGAACTCCAGC | |||||
| Cc226 | F: ACTTTGCCTACTCACAGCCC | (AAG)6 | 55.8 | 296 | Y |
| R: CCTTTTCCACACCGATAACA | |||||
| Cc227 | F: TAAACCCGTCTCTAACATCCG | (AAG)8 | 55.8 | 209 | Y |
| R: TTCCGTCGTTCACTTCACAA | |||||
| Cc228 | F: ATCCCTCTCCACCTCTATCTCT | (GAA)7 | 55.8 | 192 | Y |
| R: TTCTTTCCTAACCTCCCCTT | |||||
| Cc229 | F: ATCCCTTTGTTCCATCACCA | (TC)20 | 55.8 | 145 | Y |
| R: AAGAAACCCACTTCCTCATACC | |||||
| Cc230 | F: CGAAACTGAAACCGACCAAA | (TA)10 | 55.8 | 136 | Y |
| R: CCAGACAACCAGCGACCTTA | |||||
| Cc231 | F: GAAAAGGTTCCATCGGCATA | (AGA)8 | 55.8 | 145 | Y |
| R: CAGTGTAGGGGATTCCTGTAGC | |||||
| Cc232 | F: TACTTCGGTGATTTGATAGCG | (CGTTTC)6 | 56.1 | 143 | Y |
| R: TTTGGGATAGACAAGACAACG | |||||
| Cc233 | F: ACTCAAACGCTATCACTTCCA | (AAG)6 | 56.1 | 145 | Y |
| R: CTCCTCTCCTGTTCATGCTCT | |||||
| Cc234 | F: CTGAACATCATAGACAAACCGT | (GAA)6 | 56.1 | 276 | Y |
| R: TCTCTCTCTTAATCCATCTGCTC | |||||
| Cc235 | F: AGTGGAGCACACCAGAAGG | (CTG)6 | 56.1 | 360 | Y |
| R: TGACCAGATTGAGTGACGAAA | |||||
| Cc236 | F: CCTCTATTCCTTTGCTTTTCC | (AT)9 | 56.1 | 250 | N |
| R: TCTTCGTGCGTGTATGTGAG | |||||
| Cc237 | F: TCTCTCCGTGTGTTTTTGAAG | (CAACAC)4 | 56.1 | 249 | Y |
| R: GGTGATGGGTCTGAAAGGTT | |||||
| Cc238 | F: AGACAAACACAGCGAAACAAC | (AGG)7 | 56.1 | 243 | Y |
| R: CCCATCACCGTCACCAA | |||||
| Cc239 | F: GGCATAGTGAACCTCAACCA | (GAA)6 | 56.1 | 160 | Y |
| R: CGCTTATCTTTTTTCTCCCTC | |||||
| Cc240 | F: TCACCATCTCAGTCACTCCGTC | (GGAA)5 | 56.1 | 239 | Y |
| R: CGAATCAAAAAAACCCTCCCT | |||||
| Cc241 | F: ACCTGCTTGGCTTTTACGATA | (TGG)5 | 56.1 | 208 | Y |
| R: CACACCTTGACCAGAACGG | |||||
| Cc242 | F: TCGGTATCGTTCAAATCCTCG | (AGA)6 | 56.1 | 186 | Y |
| R: AAAATCCACCTGCCAAATCTG | |||||
| Cc243 | F: GAGCCCATTTGCTTCTATTTG | (TTC)6 | 56.1 | 258 | Y |
| R: CTTTCCATCATTACCGAACATC | |||||
| Cc244 | F: ATCCCTCAAAATACTCGGTCA | (TTC)6 | 56.1 | 154 | Y |
| R: AATACTGCGGAGCCAACTAAT | |||||
| Cc245 | F: CTTGCTGCTTGCTTCTGG | (ATTTT)6 | 56.1 | 189 | Y |
| R: ACTAATACGGTGGTTGATGGA | |||||
| Cc246 | F: TGATGACGCTGTACTTGCC | (GA)11 | 56.1 | 225 | N |
| R: GACTCCGATTTGAGATTTCCT | |||||
| Cc247 | F: GAAATACTACCGTGGAAATGGA | (AGG)7 | 56.3 | 219 | Y |
| R: AGAGGGATTGGATGTGGGT | |||||
| Cc248 | F: ATCTTGGAAATGGCACTCTGTA | (GGA)6 | 56.3 | 222 | Y |
| R: TTCTTTGAGATGGAGAGGGAG | |||||
| Cc249 | F: GAGCCTGGAAGATATGCGA | (CAT)6 | 56.3 | 132 | Y |
| R: AAGGGAGAAGATACTTTTGGTG | |||||
| Cc250 | F: CATTGCTTCCGTAGATTCTGTT | (TC)12 | 56.3 | 118 | Y |
| R: GACCCGCTTCTTGTTATTCC | |||||
| Cc251 | F: CTCCTTACTGCCTTTGTTGACT | (TC)11 | 56.3 | 128 | Y |
| R: TCTTCTGGGGTTTCTTACTTTC | |||||
| Cc252 | F: GTCGTCTGCTCGTGTAAATAAA | (GTTTTC)4 | 56.3 | 246 | Y |
| R: AACAAGACTGGAAGAAGGAACA | |||||
| Cc253 | F: GGGGCTCGCAGAAGATACT | (CT)11 | 57.0 | 193 | Y |
| R: GTTTTCCGCTTGGCACC | |||||
| Cc254 | F: AGCCCTTCGCCCATAAC | (TGG)7 | 57.0 | 185 | Y |
| R: GAACTGCGTGTCAAATCCTC | |||||
| Cc255 | F: GGAGATTGCCCCGTTTG | (AAG)6 | 57.0 | 84 | N |
| R: TTGTCCACAGAATCGTCAGC | |||||
| Cc256 | F: GGAACGAAGCCCACCTAT | (AC)10 | 57.3 | 162 | Y |
| R: AATCCTGATGACCCCCTG | |||||
| Cc257 | F: CCACGGAAGACGGCTATT | (GA)10 | 57.3 | 132 | Y |
| R: CAGAATCCTGCTGCGACTC | |||||
| Cc258 | F: AGAGACGAAGCGGGTTGA | (GAAGGT)4 | 57.3 | 183 | Y |
| R: TCTCTCATAAAGCCTGTGCC | |||||
| Cc259 | F: CCCCAATCCCATCTCTGT | (CT)11 | 57.3 | 244 | Y |
| R: AAACGAGGCAGTCGGTCT | |||||
| Cc260 | F: TCTCAAACCGACCCACCA | (CTT)6 | 57.3 | 377 | Y |
| R: AACTCTTCTCTCTCCCTTTCTCTTC | |||||
| Cc261 | F: GCAGAGCCAAAGAAGCCA | (GTG)6 | 57.3 | 252 | Y |
| R: CCTGTAAGACGCCATTCCA | |||||
| Cc262 | F: CGCAAAGCGATCCCATCT | (GA)11 | 57.3 | 271 | Y |
| R: TGCCAGGTGAGCACGGTAT | |||||
| Cc263 | F: GAAATACCAACGCACACCC | (CT)10 | 57.6 | 138 | Y |
| R: CCTATGCTTCACTCCCCAA | |||||
| Cc264 | F: GTAACGACTCACCAACTTCAACA | (GA)12 | 57.6 | 173 | Y |
| R: GGGGGATTTTCCTCTGCTA | |||||
| Cc265 | F: ATTCCGCTATTACGGTGGC | (CT)16 | 57.8 | 199 | Y |
| R: ACCGCAATAGAGTTTTGGGAC | |||||
| Cc266 | F: GGTATTCGGCGTAGCCATTC | (AAAC)5 | 57.8 | 299 | N |
| R: GGGGGTTCTCGGTTCTTATTT | |||||
| Cc267 | F: CAACGCTGGTTCCTCTTATG | (AGA)6 | 57.8 | 270 | Y |
| R: TCCACGACCGACAACTTCT | |||||
| Cc268 | F: TTTGGGTTCTGTGACTCTTGG | (CT)14 | 57.8 | 199 | Y |
| R: CGTCCCCCTTTTTCTTTTATC | |||||
| Cc269 | F: AAGCCGCACCTCACCTTTTA | (GTT)6 | 57.8 | 296 | Y |
| R: GCTGTCAGATTCCCGTCCAC | |||||
| Cc270 | F: TTATCGGCAACCACCACACT | (TC)8 | 57.8 | 234 | Y |
| R: TCACCCAAATACTTCCTCCTTC | |||||
| Cc271 | F: TGCCGACAACTGCTTCTCTA | (TC)17 | 57.8 | 107 | N |
| R: CTAACCCCACCTTCAACACC | |||||
| Cc272 | F: CTGGATTAGTCGCTTGGCAA | (AG)3C(AG) 15 | 57.8 | 183 | Y |
| R: TGAAGGTTTCGGTTAGGTGGT | |||||
| Cc273 | F: ACGGAGGAGGGTGATGAGT | (ATT)7 | 57.8 | 211 | Y |
| R: GTAACGCCATGAGCTTCAAC | |||||
| Cc274 | F: TTTGAGGGACTTGCTGCTGA | (ATT)6 | 57.8 | 233 | Y |
| R: GGACTTTGCTTTGTGACCATCTA | |||||
| Cc275 | F: CATAGCCCCCTCTTCTCGT | (CT)10 | 57.8 | 266 | N |
| R: TTACTTGCCCTCACACCACA | |||||
| Cc276 | F: CCATTTTTCCGAGGAGTTGC | (TCG)8 | 57.8 | 238 | Y |
| R: TTGATGTCTGTGCCTGATAACG | |||||
| Cc277 | F: ACACGACCAAGATACTCATTCACA | (CCA)6 | 57.8 | 289 | N |
| R: CCACGCCATAATATCGCTCA | |||||
| Cc278 | F: TTCTCTTCTGGCACTCCTCTG | (ATTTG)4 | 57.8 | 270 | N |
| R: GTCCCAACCCAACCATAAGT | |||||
| Cc279 | F: TGCCGTTTGGAGACCATAAG | (CT)11 | 57.8 | 279 | Y |
| R: ACCTGGGACATACCGTGACA | |||||
| Cc280 | F: ACCCACTGGACAAAGAAACC | (CTT)7 | 57.8 | 183 | Y |
| R: AGTGAAATGTAGGAGGACGAGA | |||||
| Cc281 | F: GACTTGTGAAGAACTCGGCA | (GAA)6 | 57.8 | 127 | Y |
| R: TCTGTGAGACACGCACTAACC | |||||
| Cc282 | F: GGATTTTCTCACTGGCGGTA | (TAG)6 | 57.8 | 145 | Y |
| R: ACTTCAAGGCTCCTCAAGATTC | |||||
| Cc283 | F: GGAACATTAGCAGCGGAGAC | (ACT)7 | 57.8 | 336 | Y |
| R: GACAACAAACGCCATAGCCT | |||||
| Cc284 | F: ATCCTATGCTCCAATGCCAG | (CAG)3 | 57.8 | 124 | Y |
| R: CCTCACTGTCACTCAACTTCA | |||||
| Cc285 | F: CTAACCAGGGAGACGGACAA | (AAG)9 | 57.8 | 249 | Y |
| R: GAAACTGAGCGATGAAGGGA | |||||
| Cc286 | F: GCAATACGGACACGGTAGG | (AACAC)2 | 57.8 | 334 | Y |
| R: ATTCACAGTGCGTCAAGGTC | |||||
| Cc287 | F: GACATCATCAGCGGGGTAG | (CT)10 | 57.8 | 267 | Y |
| R: AGAGCCACTCGTTTCCATTC | |||||
| Cc288 | F: GGAGGCACAAAAATCCAAACTAC | (GTTGCT)4 | 58.0 | 238 | Y |
| R: ATTGACAACTTTCTGCGACCG | |||||
| Cc289 | F: CAGTGTTTGTCAAAGAGGAGAGA | (TTC)6 | 58.0 | 216 | Y |
| R: GAGTTGGTTTGGGTGGTTCTA | |||||
| Cc290 | F: ACCGCCCTTCTACTGATTCTT | (AG)9 | 58.0 | 297 | Y |
| R: CTCACCATTCTCTCTTCGCTG | |||||
| Cc291 | F: CGAGACTTTGGTTGAGTGTGA | (GAT)6 | 58.0 | 254 | Y |
| R: GAGTTTGCCTACACGATGACA | |||||
| Cc292 | F: AGGGTTTTGATTGGACAGACG | (CAAA)5 | 58.0 | 184 | Y |
| R: AGCGAATGGGAGGAGATAAGAT | |||||
| Cc293 | F: CGGGACGACTATGAAGGTTTT | (TC)11 | 58.0 | 104 | Y |
| R: CAGTTGGATACAGATAGACGCAGA | |||||
| Cc294 | F: CTACCCCATACTCCAAACCAT | (TCT)7 | 58.0 | 300 | Y |
| R: CAGCCTTCTACAAAGCAGTCAT | |||||
| Cc295 | F: CATCCCATACACACAGCACAAT | (TTC)6 | 58.2 | 169 | Y |
| R: GAGCGTTGTCGTCCTTTGG | |||||
| Cc296 | F: TCAGGACACCGATACATACGC | (TGG)5 | 58.4 | 293 | Y |
| R: CCTTCTCATCCACTCAAAACACT | |||||
| Cc297 | F: TTCATCTTCCTCTAACTGATCCCT | (GA)9 | 58.6 | 208 | Y |
| R: TGTGACACTGTTGGTGGTTCTG | |||||
| Cc298 | F: GCAGCAATAGAAGAAGAAACAACC | (CCT)6 | 58.6 | 171 | Y |
| R: GATAAGGATGATGCGGTCGTG | |||||
| Cc299 | F: GTCAGAGGCTTTGGGTTCC | (GGA)7 | 59.7 | 204 | Y |
| R: GCATTCGGAGTCTACCACCT | |||||
| Cc300 | F: TCCTCCTCGTGCTTCGTCT | (AGA)8 | 59.7 | 329 | Y |
| R: TACTCCTTCCGTCCCCACTT | |||||
| Cc301 | F: CGCACACACTGACACATCCA | (CT)9 | 59.7 | 121 | Y |
| R: CGACGAAGACGACGGGTTA | |||||
| Cc302 | F: CGCATCCAGGAATCACACC | (TC)9 | 59.7 | 222 | N |
| R: TGAGCACGAAGAGAACGCC | |||||
| Cc303 | F: GAAGGAGTACGAGGAGTAGCAG | (AAG)7 | 59.8 | 183 | Y |
| R: GGTCAACGCCACCATCA | |||||
| Cc304 | F: AACACTACGCTCAGCAACTGTG | (CAC)6 | 59.8 | 262 | Y |
| R: GGGTTCACCTTCCAATCCAG | |||||
| Cc305 | F: GGGCTCTCCTTTTCCTTGTC | (AAG)6 | 59.8 | 267 | Y |
| R: CACCAGTAACCCCTCCTATCTC | |||||
| Cc306 | F: CCAACAAAGCCCCAGAGAAC | (CT)10 | 59.8 | 214 | Y |
| R: AGCAGAGATGGAGAGAAGCGA | |||||
| Cc307 | F: GCTCTTGGCTCTTCGCTTCT | (CTT)6 | 59.8 | 126 | Y |
| R: GACATCTTGCTCCACTGACACAC | |||||
| Cc308 | F: CGAAGCAGTTATCTGGGCAC | (CAT)6 | 59.8 | 227 | Y |
| R: CGGGTGAAAACTTGGTCTCTC | |||||
| Cc309 | F: AGACAAGGATTGAGAGGTGGAG | (GA)10 | 60.0 | 260 | Y |
| R: AGTTGGGTCGGAATAGTGAGC | |||||
| Cc310 | F: GTCCGTGAGTGAGGGTTAGG | (CTT)6 | 60.1 | 265 | Y |
| R: AGTTGGGTCGGAATAGTGAGC | |||||
| Cc311 | F: GCTGCTCCTCTGCTTCTTGG | (AGG)6 | 60.2 | 251 | Y |
| R: TTCCTTCTACTGGACCGTTCATC |
REFERENCES
- 1.Qi Y, Weng L, Wang Z. Progress in studies on chemical ingredients and their pharmacological activities. Chinese J. Clin. Hosp. Pharm. 2010;30 (19):1682–1685. [Google Scholar]
- 2.Lu H, Yang H, Bi Y, Liang S, Mei J. Physicochemical properties and fatty acid composition of hickory oil. China Oils and Fats. 2010;35 (5):73–76. [Google Scholar]
- 3.Shen J, Xiao RZ, Chen Z, Chen Q. Study on the physicochemical properties and oxidative stability of cold-pressed Carya cathayensis Sarg oil. Journal of the Chinese Cereals and Oils Association. 2012;27 (4):64–73. [Google Scholar]
- 4.Zhang P, Zhong H, Yao X, Wang K, Wang Y, Chang J. Oil yield and fatty acid composition of nuts of four species in Carya family. Jiangxi. Nongye. Daxue. Xuebao. 2012;34 (3):499–504. [Google Scholar]
- 5.Jia X, Wang C, Mo Z, Li Y, Guo Z, Qiao Y. Comparison on fruit quality of pecan and Zhejiang walnut. Tianjin Agricultural Sciences. 2013;19 (3):28–31. [Google Scholar]
- 6.Hu W. Forest industries with some characteristics have inflated farmers’ wallets. Forestry in Anhui. 2010;(1):21. [Google Scholar]
- 7.Shen L, Guo J, Li Y, Zeng Y. Population structure and genetic diversity of Carya cathayensis revealed by AFLP analysis. J. Fujian Forestry Sci. .Technol. 2009;36 (4):84–86. [Google Scholar]
- 8.Li Y, Li Y, Wu S, Han K, Wang Z, Hou W, Zeng Y, Wu R. Estimation of multilocus linkage disequilibria in diploid populations with dominant markers. Genetics. 2007;176:1–11. doi: 10.1534/genetics.106.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates: Sunderland MA. 1998 [Google Scholar]
- 10.Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Oefner PJ, Stahl EA, Weigel D. The extent of linkage disequilibrium in Arabidopsis thaliana. Nature Genetics. 2002;30:190–193. doi: 10.1038/ng813. [DOI] [PubMed] [Google Scholar]
- 11.Falconer DS, Mackay TF, editors. Longman Group Ltd: Essex UK. 1996. Introduction to Quantitative Genetics. [Google Scholar]
- 12.Eldon B, Wakeley J. Linkage disequilibrium under skewed offspring distribution among individuals in a population. Genetics. 2008;178:1517–1532. doi: 10.1534/genetics.107.075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Wang ZJ, Jin SH, Xia GH, Huang YJ, Huang JQ. A pattern of unique embryogenesis occurring via apomixis in Carya cathayensis. Biologia Plantarum. 2012;56 (4):620–627. [Google Scholar]
- 14.Hou W, Shen L, Li Y, Pang XM, Zeng YR, Wu RL. A model for linkage analysis with apomixis. Theor. Appl. Genet. 2011;123 (5):681–691. doi: 10.1007/s00122-011-1618-4. [DOI] [PubMed] [Google Scholar]
- 15.Grossniklaus U, Nogler G, van Dijk P. How to avoid sex: the genetic control of gametophytic apomyxis. Plant Cell. 2001;13:1491–1998. doi: 10.1105/tpc.13.7.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roche D, Chen Z, Hanna WW, Ozias AP. Non-Mendelian transmission of an apospory-specific genomic region in a reciprocal cross between sexual pearl millet (Pennisetum glaucum) and an apomictic F1 (P glaucum × P squamulatum). Sex. Plant Reprod. 2001;13(4):217–223. [Google Scholar]
- 17.Labombarda P, Busti A, Caceres ME, Pupilli F, Arcioni S. An AFLP marker tightly linked to apomixis reveals hemizygosity in a portion of the apomixis-controlling locus in Paspalum simplex. Genome. 2002;45(3):513–519. doi: 10.1139/g02-014. [DOI] [PubMed] [Google Scholar]
- 18.Honig JA, Averello V, Bonos SA, Meyer WA. Classification of Kentucky bluegrass (Poa pratensis L. cultivars and accessions based on microsatellite (simple sequence repeat) markers. HortScience. 2012;47 (9):1356–1366. [Google Scholar]
- 19.Guo J, Chi W , Zhao X, Wang L, Zeng Y, Zhang Q. An AFLP system for Carya cathayensis. J. Zhejiang Forestry College. 2008;25 (4):532–537. [Google Scholar]
- 20.Song S, Huang Y, Dong L, Huang Y, Zeng Y, Hu Z. Simple sequence repeat (SSR) markers in Carya cathayensis. J. Zhejiang A &F University. 2012;29 (4):622–633. [Google Scholar]
- 21.Grauke LJ, Mendoza-Herrera MA, Binzel ML. Plastid microsatellite markers in Carya. Acta Horticulturae. 2010;859:237–246. [Google Scholar]
- 22.Grauke LJ, Iqbal MJ, Reddy AS. Thompson TE: Developing microsattelite DNA markers in pecan. J. Am. Soc. Hortic. Sci. 2003;128 (3):374–380. [Google Scholar]
- 23.Zeng Y, Hou W, Song S, Feng S, Shen L, Xia G, Wu R. A statistical design for testing apomictic diversification through linkage analysis. Brief. Bioinform. 2012;15(2):306–318. doi: 10.1093/bib/bbs080. [DOI] [PubMed] [Google Scholar]
- 24.Li Z. Cultivation and Processing in C Cathayensis. China Agricultural Science and Technology Press: Beijing. 2003 [Google Scholar]
- 25.Wang Z, Huang Y, Guo C, Huang J, Wang H. RAPD analysis on genetic diversity of Carya dabieshanensis populations. Journal of Plant Ecology (formerly Acta Phytoecologica Sinaca): 2006;30 (3):534–538. [Google Scholar]
- 26.Li Q, Du Z C. dabieshanensis. Forest Science and Technology. 2001;12:26–27. [Google Scholar]
- 27.Pupilli F, Labombarda P, Caceres ME, Quarin CL, Arcioni S. The chromosome segment related to apomixes in Paspalum simplex is homoeologous to the telomeric region of the long arm of rice chromosome 12. Mol. Breed. 2001;8:53–61. [Google Scholar]
- 28.Caceres ME, Matzk F, Busti A, Pupilli F, Arcioni S. Apomixis and sexuality in Paspalum simplex: characterization of the mode of reproduction in segregating progenies by different methods. Sex. Plant Reprod. 2001;14:201–206. doi: 10.1007/s00497-001-0109-1. [DOI] [PubMed] [Google Scholar]
- 29.Grimanelli D, Leblanc O, Espinosa E, Perotti E, González de León D, Savidan Y. Mapping diplosporous apomixis in tetraploid Tripsacum: one gene or several genes. Heredity. 1998;80:33–39. doi: 10.1046/j.1365-2540.1998.00263.x. [DOI] [PubMed] [Google Scholar]
- 30.Bicknell RA, Borst NK, Koltunow AM. Monogenic inheritance of apomixis in two Hieracium species with distinct developmental mechanisms. Heredity. 2000;84:228–237. doi: 10.1046/j.1365-2540.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- 31.Garcia RAV, Rangel PN, Brondani C, Martins WS, Melo LC, Carneiro MS, Borba TCO, Brondani RPV. The characterization of a new set of EST-derived simple sequence repeat (SSR) markers as a resource for the genetic analysis of Phaseolus vulgaris. BMC Genetics. 2011;12:41–54. doi: 10.1186/1471-2156-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S, Huguet T, Huyghe C. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol. 2003;3:9–27. doi: 10.1186/1471-2229-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaitieng B, Kaga A, Tomooka N, Isemura T, Kuroda Y, Vaughan DA. Development of a black gram (Vigna mungo (L. Hepper) linkage map and its comparison with an azuki bean (Vigna angularis (Willd.) Ohwi and Ohashi) linkage map. Theoretical and Applied Genetics. 2006;113:1261–1269. doi: 10.1007/s00122-006-0380-5. [DOI] [PubMed] [Google Scholar]
- 34.Yu K, Park SJ, Poysa V. Abundance and variation of microsatellite DNA sequences in beans (Phaseolus and Vigna). Genome. 1999;42:27–34. [Google Scholar]
- 35.Métais I, Hamon B, Jalouzot R, Peltier D. Structure and level of genetic diversity in various bean types evidenced with microsatellite markers isolated from a genomic enriched library. Theor. Appl. Genet. 2002;104:1346–1352. doi: 10.1007/s00122-002-0901-9. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, Zhang B, Wang Z, Guo C. RAPD analysis on genetic relationship among species in genus Carya. J. Southwest Forestry College. 2003;23(4):1–3, 11. [Google Scholar]