Abstract
Since the time of Darwin, biologists have sought to understand the origins and maintenance of life’s diversity of form. However, the nature of the exact DNA mutations and molecular mechanisms that result in morphological differences between species remains unclear. Here, we characterize a nonsynonymous mutation in a transcriptional coactivator, limb bud and heart homolog (lbh), which is associated with adaptive variation in the lower jaw of cichlid fishes. Using both zebrafish and Xenopus, we demonstrate that lbh mediates migration of cranial neural crest cells, the cellular source of the craniofacial skeleton. A single amino acid change that is alternatively fixed in cichlids with differing facial morphologies results in discrete shifts in migration patterns of this multipotent cell type that are consistent with both embryological and adult craniofacial phenotypes. Among animals, this polymorphism in lbh represents a rare example of a coding change that is associated with continuous morphological variation. This work offers novel insights into the development and evolution of the craniofacial skeleton, underscores the evolutionary potential of neural crest cells, and extends our understanding of the genetic nature of mutations that underlie divergence in complex phenotypes.
Keywords: adaptive radiation, evo-devo, coding mutation, neural crest cell
Introduction
Vertebrates demonstrate astounding diversity in facial structures due to intense selective pressure to exploit divergent ecological niches. Although alterations in gene expression have been associated with natural variation in facial morphology in a number of systems (Abzhanov et al. 2004, 2006; Albertson et al. 2005; Wu et al. 2006; Brugmann et al. 2010; Roberts et al. 2011; Powder et al. 2012; Schoenebeck et al. 2012), the specific DNA changes that underlie craniofacial evolution remain largely unknown due to the challenges of identifying causative mutations that contribute to phenotypic variation in complex traits. Most of the craniofacial skeleton in vertebrates is derived from neural crest cells (NCCs) (Noden 1978; Schilling et al. 1996; Chai et al. 2000; Jiang et al. 2002; Kague et al. 2012), a multipotent cell population whose derivatives are so widespread that it has been referred to as the fourth germ layer (Hall 2010). Enormous evolutionary potential has also been ascribed to the NCCs, as they have been shown to contain species-specific patterning information for the traits that they give rise to (Schneider and Helms 2003; Tucker and Lumsden 2004).
An extreme example of natural craniofacial variation is found in the cichlid fishes of the East African rift-valley lakes, which have undergone elaborate modifications of their oral jaws, contributing to their explosive adaptive radiation (Danley and Kocher 2001; Albertson and Kocher 2006; Cooper et al. 2010). Maylandia zebra (MZ) and Labeotropheus fuelleborni (LF), two species used in this study, represent opposite ends of a major ecomorphological axis which distinguishes species that, respectively, forage in the water column (i.e., pelagic) from those that feed from the rocky substrate (i.e., benthic) (Albertson et al. 2005; Cooper et al. 2010). Variation in jaw length relative to the postorbital region of the head (Cooper et al. 2010) is integral to these alternate feeding strategies; all other things equal, the longer mandible in MZ (fig. 1a) allows for more rapid jaw movement as the animal plucks food from the water column, whereas the shortened mandible of LF (fig. 1a) confers greater force as the animal shears attached algae from the substrate (Albertson et al. 2005). We previously identified a major effect quantitative trait locus (QTL) on linkage group 19 for this adaptive trait (Albertson et al. 2005; Cooper et al. 2011). Here, we extend these genetic mapping data to implicate a causative mutation and developmental mechanism mediating adaptive variation in jaw length. We integrate methods from both population genetics and experimental embryology, and leverage the experimental advantages of several animal systems in order to draw explicit connections between genotype and phenotype. In all, this work contributes a more comprehensive understanding of how the craniofacial skeleton develops and evolves.
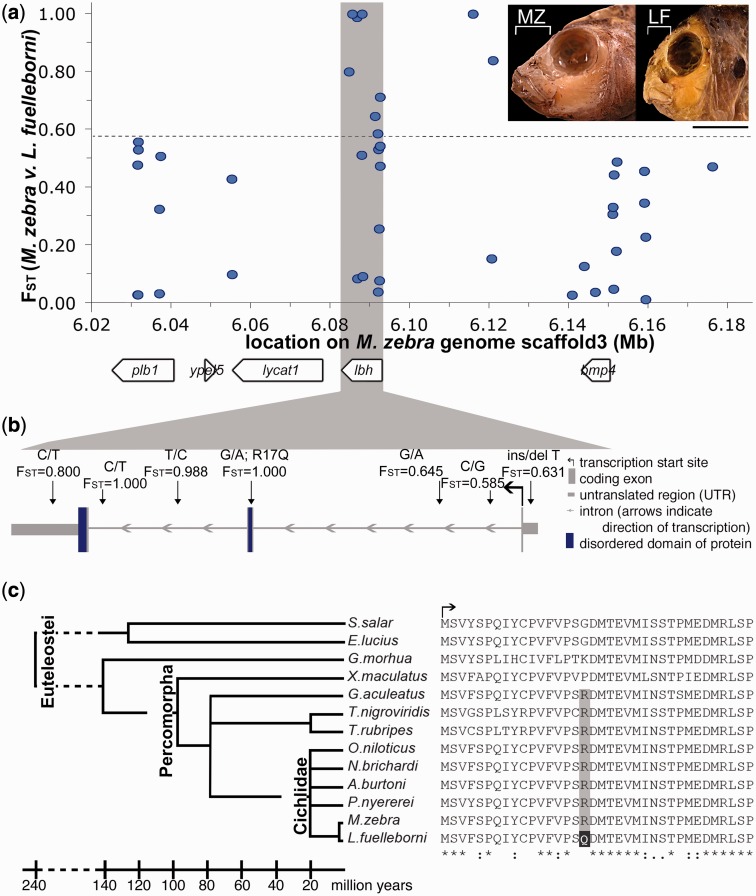
Fig. 1.
Derived nonsynonymous mutation in lbh is alternatively fixed between cichlids with different craniofacial morphologies. (a) Population level genetic diversity (FST) for SNPs within a QTL for relative mandible length (white bracket on inset) between MZ and LF. Dashed line indicates experimental FST threshold (0.569) between these species (Mims et al. 2010). Gene predictions indicated by white boxes. (b) Schematic of lbh, with highly differentiated SNPs indicated. SNPs listed as on sense transcript. (c) Protein alignment for the N-terminal of Lbh across selected members of Teleostei. Scale bar = 1 cm.
Results and Discussion
Fine Mapping of a Craniofacial Locus Implicates Limb Bud and Heart Homolog
The QTL for relative jaw length maps to an approximately 6 cM interval on LG19 (Cooper et al. 2011), which corresponds to approximately 5 Mb of sequence on scaffold 3 of the Lake Malawi cichlid genome (see Materials and Methods for specifics). The gene bmp4 is within this region, and we have previously found that variation in bmp4 expression is associated with the development of species-specific mandible shapes (Albertson et al. 2005). In addition, the gene limb bud and heart homolog (lbh), which is expressed in the developing pharyngeal arches in mouse (Briegel and Joyner 2001), is adjacent to bmp4. We therefore focused our resequencing efforts within and around these two genes (∼160 kb in total, though we cannot rule out the possibility that other genes may also contribute to this QTL). In order to characterize polymorphisms within this region, we selectively sequenced a panel of wild-caught MZ and LF animals (fig. 1a and supplementary table S1, Supplementary Material online) to identify single nucleotide polymorphisms (SNPs) that were outliers for FST. Given the genetic homogeneity of cichlids, this is an appropriate and efficient method of identifying candidate loci for species-level divergence in this group (Loh et al. 2008; Roberts et al. 2011; Albertson et al. 2014).
Although allelic variation within and immediately around bmp4 does not segregate with jaw shape, highly differentiated SNPs between MZ and LF (FST >0.569, a genome-wide empirical threshold for genetic divergence between LF and MZ [Mims et al. 2010]) were found in two other regions of this interval (fig. 1a), indicating they may be involved in the phenotypic divergence between these two species. One of these regions resides in a 52-kb interval downstream of bmp4 and upstream of lbh (fig. 1a and supplementary table S1, Supplementary Material online). Given the differential expression of bmp4 in cichlids (Albertson et al. 2005) and that bmp genes in general (DiLeone et al. 1998; Portnoy et al. 2005; Chandler et al. 2007; Guenther et al. 2008; Pregizer and Mortlock 2009), and bmp4 specifically (Chandler et al. 2009), are known to have tissue specific distal enhancer elements up to 150 kb away, it is possible that this region may transcriptionally regulate bmp4. It is also possible that it may be involved in the regulation of lbh, both lbh and bmp4, or neither gene.
The second region with outlier FST values spans the lbh gene (fig. 1a). Specifically, we identified seven SNPs within the lbh gene that are highly differentiated between MZ and LF. Six of these SNPs are noncoding (fig. 1b), and could represent changes in cis-regulation of lbh between LF and MZ. Seventh SNP is a nonsynonymous change within the coding sequence that is alternatively fixed between MZ and LF (FST = 1.00). The derived allele in LF encodes an R>Q change in a region of Lbh that is necessary for its transcriptional co-activator activity (fig. 1b, amino acid17 of tilapia ENSONIP00000001722) (Ai et al. 2008). This region is also highly disordered (Al-Ali et al. 2010) (and thus lacks a stable tertiary structure), which is thought to confer conformational flexibility, allowing Lbh to bind to a range of other proteins (Dunker et al. 2008).
Aside from the change in LF, this amino acid residue is conserved over 80 My of teleost evolution (fig. 1c and supplementary fig. S1, Supplementary Material online). Because Lbh is a disordered protein, we are unable to infer what specific effects this amino acid (aa) substitution may have on protein structure. However, the R>Q change in LF results in a loss of charge, and is predicted to affect protein function based on both PolyPhen-2 (Adzhubei et al. 2010) and SIFT (Ng and Henikoff 2003) protein prediction algorithms (0.863 and 0.01, respectively; PolyPhen scores approaching 1 are not tolerated; SIFT scores less than 0.05 are also considered not tolerated). Despite amino acid changes in Esox lucius (R>G), Salmo salar (R>G), and Gadus morhua (R>K), these same algorithms predict that the function of this residue is largely conserved over 240 My of teleost evolution. The R>G change in pike and salmon yield PolyPhen-2 scores of 0.0 and SIFT scores of 0.21, which suggests that this amino acid change does not impact protein function. The PolyPhen-2 score for the R>K change in cod is 0.155 (tolerated), and the SIFT score is 0.02 (not tolerated). Within percomorph teleosts, the only other species with an amino acid change at this residue that is predicted to disrupt protein function is the platyfish, Xiphophorus maculatus (PolyPhen-2 score = 0.863, SIFT score = 0.04). Head shape in the platyfish is not overtly similar to that of LF (supplementary fig. S1, Supplementary Material online). However, relative to other teleosts, platyfish demonstrate marked differences in NCC development and migration (Sadaghiani and Vielkind 1989), and thus any roles for Lbh in platyfish craniofacial development may be quite different from those in other percomorphs. It is also important to note that outside of cichlids, amino acid identity in Lbh drops to below 90%, which means that protein structure and function could be quite different in these other lineages. It is therefore most notable that the amino acid change in LF is predicted to alter protein function relative to other cichlids, where the remainder of the protein is highly conserved.
Although most of the noncoding SNPs in lbh segregate in other cichlid species, the derived coding allele is exclusively and always found in LF, even when its closest ecomorphological (e.g., short jawed, biting mode of feeding) counterparts (Cooper et al. 2010) are considered (supplementary table S2, Supplementary Material online). Given that LF defines one end of continuous craniofacial variation among East-African rift lake cichlids (Cooper et al. 2010), we hypothesized that lbh, and particularly the R>Q coding mutation, may have contributed to this extreme, adaptive mandible morphology.
Lbh Is a Novel Regulator of NCC Development
Since this gene is relatively uncharacterized, we next sought to determine the function of lbh during craniofacial development. In order to investigate lbh in a broad phylogenetic context, we utilized cichlids as well as two experimental models of NCC development, zebrafish and Xenopus laevis. At the pharyngula stage in both cichlids and zebrafish, lbh expression is widespread across the head, including within the cranial NCCs (fig. 2a and supplementary fig. S2, Supplementary Material online). However, in Xenopus, lbh expression in the head is limited to migratory cranial NCCs (fig. 2b), which is similar to its expression in mouse (Briegel and Joyner 2001), suggesting there may have been a restriction of lbh expression in the tetrapod lineage. Given its expression in NCCs (fig. 2a and b) and correlation with the evolution of a NCC derivative in cichlids (fig. 1), we hypothesized that lbh is a novel regulator of NCC development.
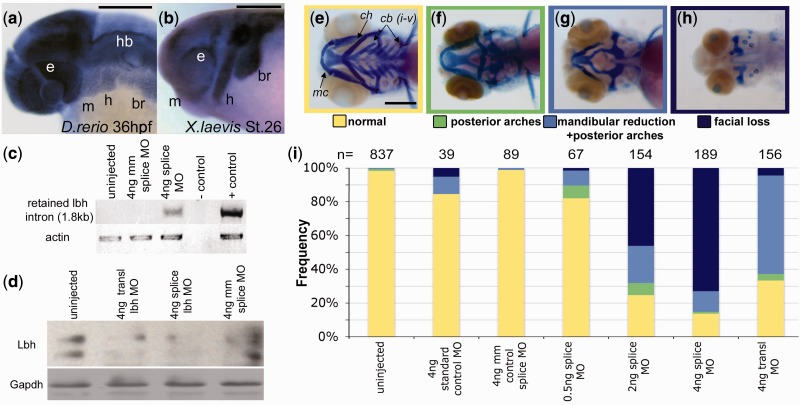
Fig. 2.
lbh is expressed in cranial NCCs and depletion of Lbh results in severe facial abnormalities. Whole-mount in situ hybridization of lbh in (a) zebrafish and (b) Xenopus. Reverse transcription-PCR (c) confirms intron retention and Western blot (d) confirms Lbh depletion following morpholino (MO) injection. Alcian blue and Alizarin red staining of (e) wild-type zebrafish at 96 hpf. Morphants have three classes of defects: (f) malformation of the ceratohyal (ch) and ceratobranchials (cb) only, (g) defects extend to include hypoplasia of Meckel’s cartilage (mc), and (h) complete loss of NCC-derived craniofacial structures. Stacked bar graphs (i) summarizing phenotypes observed following MO injections. br, branchial streams; e, eye; h, hyoid stream; hb, hindbrain; m, mandibular stream; mm, mismatch. Scale bars = 200 µm.
To test this hypothesis in vivo, we depleted Lbh in zebrafish using both translation- and splice-blocking morpholinos (MOs) (fig. 2c and d). Though morphants exhibited defects in other NCC derivatives such as melanocytes, MZ and LF exhibit extensive overlap in pigmentation, as both species are generally characterized by dark vertical bars on top of light blue bodies (Konings 2001). Moreover, both species exhibit considerable geographic variation in pigmentation levels (i.e., degree/strength of barring) (Ribbink et al. 1983). Alternatively, mandible length consistently and markedly differs between these two species. We therefore focused on the chondrogenic/osteogenic NCC lineage, and assessed effects of the MOs on the craniofacial skeleton. Craniofacial abnormalities (fig. 2e–h) were observed in a MO dose-dependent manner, with the majority of morphants (n = 98/156 for translation-blocking, n = 161/189 for splice-blocking) exhibiting malformation of the mandible or complete loss of all NCC-derived facial structures (Kague et al. 2012) at the highest MO dose (fig. 2g–i). Our most severe morphants (fig. 2h) were remarkably similar to genetic mutants with a complete loss of NCCs (e.g., foxd3;tfap2a [Arduini et al. 2009; Wang et al. 2011]). Although this defect was predominantly observed only in splice-blocking morphants, the effect was specific to NCC derived cartilages. Morphants exhibiting this “facial loss” still possessed mesodermally derived cartilaginous aspects of the neurocranium (Kague et al. 2012), suggesting that Lbh is not acting at the level of chondrogenesis.
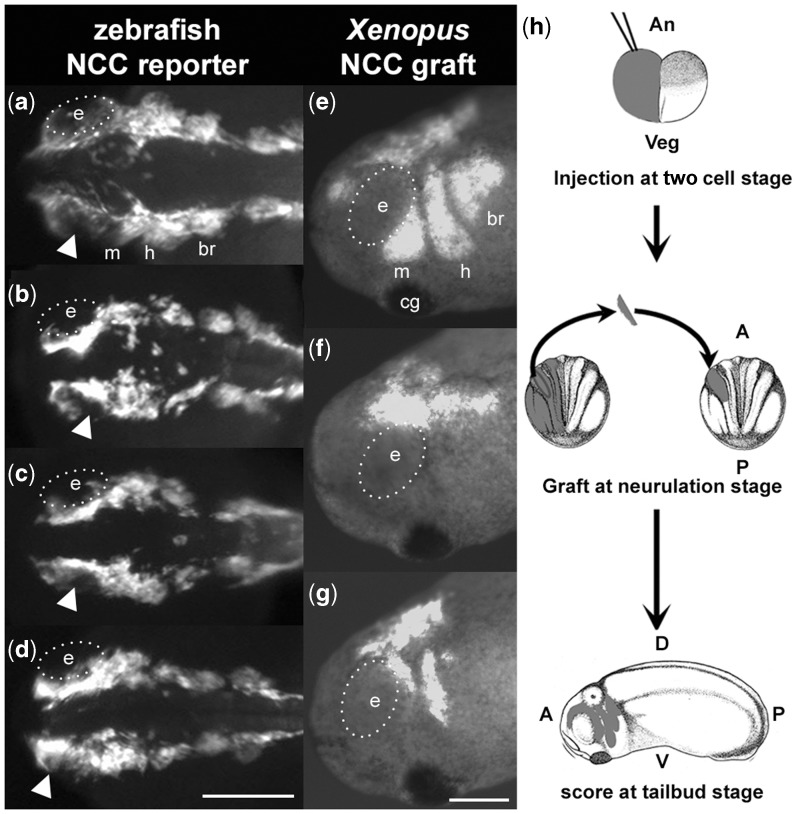
To identify the cellular mechanism through which lbh regulates NCC development, we conducted MO knockdowns in a zebrafish NCC reporter line, Tg(-7.2sox10:eGFP) (Hoffman et al. 2007), and observed defects in cranial NCC development (fig. 3a–d and supplementary fig. S3 and movie S1, Supplementary Material online). Although NCC induction appears to be unaffected, NCCs in morphants are disorganized and remain in the dorsal margins of the embryos (fig. 3a–d), failing to migrate ventrally (supplementary fig. S3 and movie S1, Supplementary Material online) into the pharyngeal arches (n = 113/118 morphants vs. n = 0/128 uninjected controls). In particular, anterior NCCs accumulate above the eye in lbh morphants (see arrowheads in fig. 3a–d, and arrows in supplementary fig. S3, Supplementary Material online). To mitigate potential secondary effects and determine cell autonomy of NCC defects, we utilized Xenopus, which allowed us to graft morphant cranial NCCs onto wild-type embryos (fig. 3h). Xenopus morphant experiments confirmed that depletion of Lbh results in cell-autonomous defects in NCC migration, particularly in the mandibular streams (n = 5/20 show complete inhibition of migration [e.g., fig. 3f] and n = 8/20 show reduced/defective migration [e.g., fig 3g] vs. n = 0/17 controls [e.g., fig. 3e]), and commensurate defects in craniofacial cartilages (supplementary fig. S4, Supplementary Material online). Expression of the NCC specifier slug (Sauka-Spengler and Bronner-Fraser 2008) was unaffected in morphants (supplementary fig. S5, Supplementary Material online), confirming that lbh acts during NCC migration, but not during induction and formation of migratory NCCs.
Fig. 3.
Depletion of Lbh results in cell-autonomous defects in NCC migration. (a–d) Dorsal views of uninjected (a) or lbh morphant (b–d) NCC reporter zebrafish [Tg(-7.2sox10:EFGP)] at 24 hpf. Arrowheads indicate accumulation of NCCs above the eye in morphants relative to controls. Lateral views of st.24 Xenopus with control (e) or morphant (f–g) NCC grafts. (h) Schematic of Xenopus NCC grafting experiment. Eye (e) is outlined with white dotted line. A, anterior; An, animal pole; br, branchial streams; cg, cement gland; D, dorsal; h, hyoid stream; m, mandibular stream; P, posterior; V, ventral; Veg, vegetal pole. Scale bars = 200 µm.
Alternate Forms of Cichlid Lbh Have Differential Effects on NCC Development
Given its genetic association with an evolutionary adaptation, lbh morphant phenotypes, and rarity of coding mutations that affect animal morphology (see Discussion), we next examined whether there might be a functional consequence of the single R>Q amino acid mutation identified in cichlids. For this experiment we used both zebrafish (supplementary fig. S6, Supplementary Material online) and Xenopus (fig. 4 and supplementary fig. S7, Supplementary Material online) systems. Over expression of either MZ or LF morphs of Lbh in zebrafish led to dramatic developmental defects, which is not surprising given that injections were performed at the one-cell stage and thus ectopic expression of Lbh affected all cells at all stages of development. Nevertheless, we detected statistically significant differences in the frequency and severity of defects depending on which form was injected, with the LF variant yielding defects that were consistently more severe than that of MZ (supplementary fig. S6, Supplementary Material online). These data suggest that Lbh morphs have differential activity, and that the single amino acid change in LF may be functionally relevant.
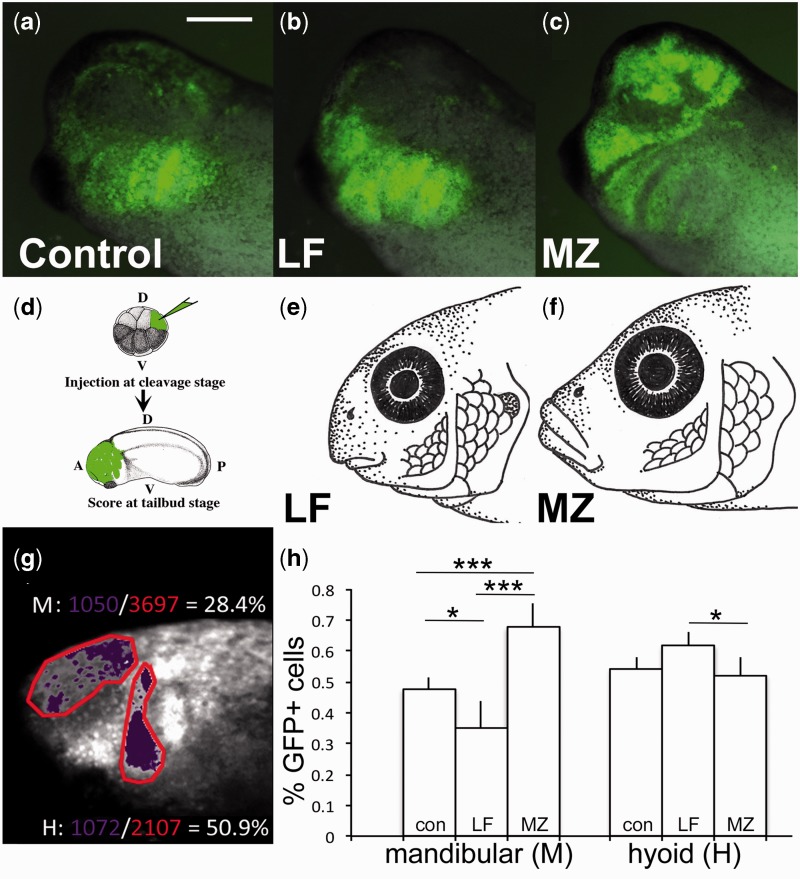
Fig. 4.
Single amino acid mutation in LF and MZ Lbh differentially regulates NCC migration. (d) Targeted injections in Xenopus of (a) control, (b) LF lbh mRNA, and (c) MZ lbh mRNA shows alternate patterns of NCC migration consistent with adult (e) LF and (f) MZ phenotypes. (g–h) Pixel density analysis of NCC migration: Percent of GFP-positive cells (purple) per stream area (red) were calculated for the anterior portion of the mandibular stream (M) and hyoid stream (H). (h) NCCs migrate more anteriorly following injection of MZ lbh (n = 16) compared with LF lbh (n = 16) or controls (n = 19). P values based on Tukey’s HSD. *P < 0.05, ***P < 0.001. Scale bar = 200 µm.
To avoid the confounding secondary effects associated with injecting at the one-cell stage, we again turned to the Xenopus model. Coexpression of GFP with either MZ or LF lbh mRNA in Xenopus using both NCC grafting (fig. 3h) and targeted injections (fig. 4d) resulted in alternate and statistically significant NCC behaviors. First, embryos expressing LF Lbh showed a significant decrease in overall NCC migration relative to those with MZ Lbh (30% vs. 59% of embryos exhibiting NCC migration, respectively, P < 0.001) (supplementary fig. S7a, Supplementary Material online), demonstrating differential potency of Lbh variants. Second, for embryos in which NCC migration was observed, alternate patterns of migration were noted. Whereas LF Lbh resulted in NCC migration that was biased to more posterior migratory streams (fig. 4b and h and supplementary fig. S7b, Supplementary Material online), the MZ Lbh morph increased the amount of migratory NCCs in the mandibular stream, particularly in the anterior portion of this stream over the developing eye (68% vs. 35% GFP+ cells for MZ and LF, respectively, P < 0.001) (fig. 4c and h and supplementary fig. S7b, Supplementary Material online). Given that NCCs are induced properly in lbh loss of function experiments (fig. 3 and supplementary figs. S3 and S5, Supplementary Material online), we hypothesize that animals expressing either LF or MZ Lbh begin with a similar pool of premigratory NCCs that is then differentially partitioned between anterior and posterior arches. Further experiments are necessary to explore this hypothesis, and to determine whether observed differences in NCC patterning might also be due to changes in cell survival and/or proliferation.
Nevertheless, these different NCC behaviors in response to alternate forms of the Lbh protein are highly consistent with adult phenotypes (fig. 4e–f). Specifically, LF possess jaws (i.e., mandibular arch derivatives) that are extremely reduced relative to other cichlid species, as well as compared to the rest of the LF pharyngeal skeleton (i.e., hyoid and branchial arch derivatives). In other words, it is the preorbital region of the skull in LF, which is derived from anterior NCCs, that distinguishes it from other cichlid species (Cooper et al. 2010, 2011; Parsons et al. 2014). Given these observations, we returned to the cichlid system in order to test the hypothesis that differences in relative mandible length observed in adult cichlids could be traced to differences in NCC development.
Manipulation of Lbh in the Lab Mimics Natural Variation in Early Craniofacial Morphogenesis
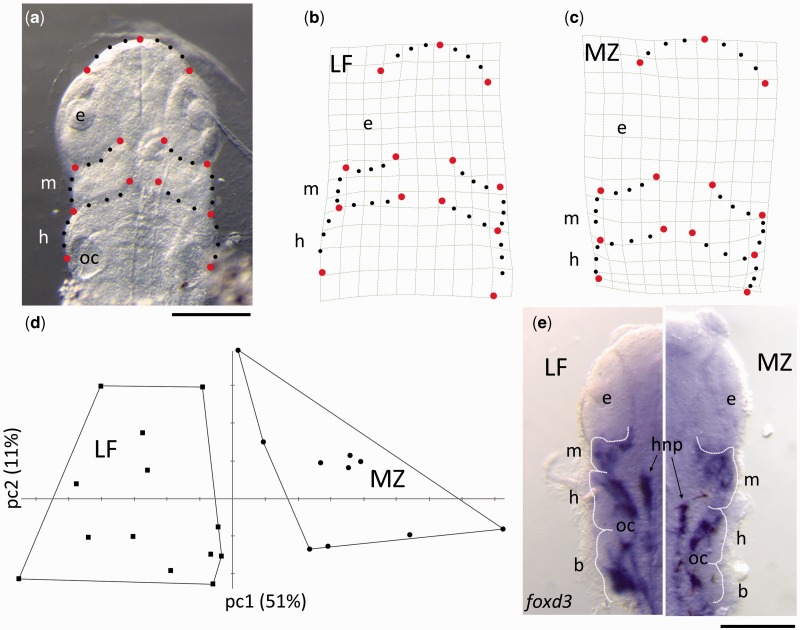
The results of the functional analysis in Xenopus suggest that the LF (i.e., short jaw) form of Lbh will drive more NCCs to posterior arches, whereas the MZ (i.e., long jaw) form will result in relatively more NCCs going to the anterior arch compared to posterior arches. Not only is this prediction consistent with differences in adult morphologies between LF and MZ (fig. 4e–f), it is also consistent with variation in the relative size of the pharyngeal arches within LF and MZ embryos (fig. 5). Specifically, geometric morphometric analysis demonstrates that LF and MZ embryos significantly differ (P < 0.001) along the primary axis of shape variation (i.e., PC1, 51% of the total variation in the data set) at stages when NCC migration has just completed (fig. 5d). Notably, this axis describes differences in the relative proportions of the developing pharyngeal arches. In MZ, the mandibular arch is expanded in size relative to the hyoid arch (fig. 5c). Alternatively, in LF, the mandibular arch is smaller relative to the hyoid arch (fig. 5b). This pattern of pharyngeal arch shape variation is also reflected in patterns of NCC development (shown by in situ hybridization for foxd3, a marker of NCC development [Sauka-Spengler and Bronner-Fraser 2008]) in similarly staged embryos. Within LF embryos, NCCs appear to be evenly distributed among the three arches, whereas in MZ more foxd3 positive cells are found in the mandibular arch relative to the posterior arches (fig. 5e).
Fig. 5.
Pharyngeal arch morphology foreshadows differences in adult head/jaw shape. Geometric morphometric analysis of pharyngula stage (st.13) cichlids using both landmark and semilandmark (e.g., curve) data (a) shows differences in the relative size of arches with LF and MZ embryos consistent with functional analysis of Lbh in Xenopus. Deformation grids (b and c) characterize shape variation along the primary axis of variation (pc1), which clearly distinguishes LF and MZ embryos (d). LF embryos are characterized by a relatively small mandibular (m) arch compared with the hyoid (h) arch (b). Alternatively, comparably staged MZ embryos possess relatively larger mandibular arches as compared to the hyoid arch (c). Differences in arch shape are also reflected in patterns of NCC development, as illustrated by whole-mount in situ hybridization using a probe for foxd3, a NCC marker (e). The anterior extent of hindbrain neural precursor cells (hnp) approximates the division between the mandibular and hyoid arch, and the otic capsule (oc) marks the junction between the hyoid and branchial arch. e, eye. Scale bars = 200 µm.
These embryonic patterns correlate with the adult phenotypes in these fishes (fig. 4e–f), where LF shows a dramatic shortening of the preorbital (mandibular arch derived) region of the face relative to MZ (Cooper et al. 2010). Differences in mandible length were previously shown to be evident between LF and MZ at the initial stages of chondrogenesis (Albertson et al. 2005). Here, we show that these differences can be traced to even earlier stages in craniofacial development. Taken together, data from 1) the functional analyses of alternate Lbh alleles in Xenopus, 2) the analysis of embryonic head shapes, and 3) differences in adult cichlid jaw lengths suggest that the amino acid polymorphism in Lbh may act to shift patterns of NCC migration in LF and MZ. Thus, differences in adult jaw lengths may be due, at least in part, to differences in the size of the founding population of NCCs from which specific sets of craniofacial bones arise and elongate.
Variation in this initial patterning event is likely elaborated at later stages of cichlid development through the differential activity of other genes (Albertson et al. 2005; Roberts et al. 2011; Parsons et al. 2014) and cellular events (e.g., apoptosis, proliferation, differentiation), leading to the production of species-specific differences in adult head shapes. This is consistent with developmental theory, which posits that development is a hierarchical process wherein each ontogenetic event builds upon (and is influenced by) the phenotype established from previous events (Hallgrimsson et al. 2009). It is also consistent with empirical findings in other systems. For example, recent comparative studies in avians show that initial differences in the allocation of postmigratory NCCs are combined with later differences in proliferation to result in species-specific mandible lengths (Fish et al. 2014). Moreover, in humans the pathology of many congenital facial dysmorphologies (e.g., CHARGE and Goldenhar syndromes) has been traced to primary defects in NCC migration (Hall 2010). Thus, a primary goal of craniofacial biology across systems is to identify the developmental origins for natural and clinical variation in facial form.
Roles for cis-Regulatory versus Coding Mutations in Morphological Evolution
For over 40 years (Britten and Davidson 1969; King and Wilson 1975; Jacob 1977) evolutionary biologists have debated the relative importance of coding and cis-regulatory mutations in evolution (Wray et al. 2003; Carroll et al. 2005; Hoekstra and Coyne 2007; Wray 2007; Stern and Orgogozo 2008; Wagner and Lynch 2008; Wittkopp and Kalay 2012). Current theory (Gilbert 2003; Wray et al. 2003; Carroll et al. 2005; Wray 2007; Wittkopp and Kalay 2012) suggests that, rather than coding changes, continuous variation in animal morphology is due to cis-regulatory mutations that result in quantitative, temporal, and/or spatial alterations in gene expression. Although there are examples of coding changes that regulate the evolution of animal morphology, it is notable that many, if not most, of these seem to mediate discrete shits in categorical phenotypes, for instance Oca2 in cavefish albinism (Protas et al. 2006) and EphB2 in the presence/absence of pigeon feather head crests (Shapiro et al. 2013). By contrast, mutations implicated in continuous (i.e., quantitative) phenotypic variation are predominantly cis-regulatory, including optix in Heliconius butterfly wing patterning (Reed et al. 2011) and shavenbaby in Drosophila trichome patterning (McGregor et al. 2007).
The amino acid change in Lbh therefore represents a rare example of a coding mutation that is associated with continuous, adaptive morphological variation. This mutation is also notable considering its potential for pleiotropy, which is often cited for the paucity of coding mutations that underlie variation in complex morphologies (Carroll et al. 2005; Wray 2007; Stern and Orgogozo 2008; Wagner and Lynch 2008). For instance, it occurs in a transcriptional regulator (Ai et al. 2008), and thus has the potential to influence the expression of many target genes. In addition, the mutation occurs in a gene necessary for the early development of NCCs, and a cell population that contributes to the development of a wide spectrum of tissues and organs (Hall 2010). It remains to be determined if Lbh has pleiotropic effects in cichlids, but given the widespread anatomical differences (e.g., craniofacial, fins, and heart) between LF and MZ, this represents an interesting area of future investigation.
Modular Protein Structure as a Target of Selection
Although modularity of cis-regulatory regions is often cited in morphological evolution (Wray et al. 2003; Carroll et al. 2005; Wray 2007; Stern and Orgogozo 2008; Wagner and Lynch 2008; Wittkopp and Kalay 2012), it has also been proposed that modules within coding regions, such as those that mediate protein–protein interactions (e.g., disordered domains and short linear motifs), may also have a high degree of evolutionary plasticity (Wagner and Lynch 2008). Limited empirical support for this comes from the comparison of protein function in plants (Bartlett and Whipple 2013), distantly related insect orders (Galant and Carroll 2002), and placental and marsupial mammals (Brayer et al. 2011). As the coding mutation in Lbh occurs in a disordered domain (Al-Ali et al. 2010) and the derived allele results in the loss of a predicted short linear motif (Mooney et al. 2012), this study represents the first empirical support of this theory with respect to microevolutionary change in animals. These studies set the stage for future investigations of protein evolution across phylogenetic levels in order to determine how prevalent this phenomenon may be during morphological evolution. In all, such work will contribute to a more comprehensive understanding of the genetic nature of evolutionary divergence in a wide spectrum of taxa.
Materials and Methods
Genetic Analysis
We previously mapped a QTL for relative lower jaw length (i.e., mechanical advantage) to an approximately 6 cM region on cichlid linkage group 19 (Albertson et al. 2005; Cooper et al. 2011). When anchored to the Malawi cichlid genome (Metriaclima [i.e., Maylandia] zebra v.0, which is available here: http://cichlid.umd.edu/cichlidlabs/kocherlab/bouillabase.html, last accessed September 2014), this interval corresponds to approximately 5 Mb of sequence on scaffold 3 (3.2–8.1 Mb). Bmp4 was perceived to be an excellent candidate for underlying this QTL. In addition, we noted that lbh was adjacent to bmp4. Based on its expression in the developing pharyngeal arches in mouse (Briegel and Joyner 2001), we considered lbh to be another strong candidate for this QTL. We therefore focused our resequencing efforts on the genomic interval around these two genes. DNA polymorphisms were identified and genotyped in wild-caught MZ and LF from five distinct populations along the southern shore of Lake Malawi (up to n = 10 per population) using Applied Biosystems Big Dye Terminator chemistry and an ABI-3730 Sequencer or, as applicable, restriction enzyme digestion and gel electrophoresis. In order to identify SNPs, 1–2 kb fragments were fully sequenced in at least two LF and two MZ, all representing different populations, in approximately 10 kb intervals, given that linkage disequilibrium around other craniofacial and pigmentation loci in cichlids appears to be approximately 15 kb (Roberts et al. 2011; Albertson et al. forthcoming). Once the coding polymorphism was identified in lbh, sequencing resolution was expanded to include a contiguous 10 kb fragment, including all untranslated regions (UTRs), exons, and introns of lbh. Additional genotyping was conducted with equal representation from all cichlid populations. Primers used available upon request. FST calculations were performed using GenePop (http://genepop.curtin.edu.au/, last accessed September 2014). The empirical threshold for outlier status of FST values was 0.569, which is based on an examination of genomic divergence among LF and MZ across multiple populations (Mims et al. 2010). The lbh gene on MZ scaffold 3 (Broad v.0) was annotated based on Ensembl gene annotation for Nile tilapia (Oreochromis niloticus, ENSONIG00000001365.1), another member of the Cichlidae family, and MZ expressed sequence tags (ESTs) (Cichlid Genome Database, http://bouillabase.org, last accessed September 2014). Additional genotyping of a subset of lbh SNPs was conducted on fish in the genus Tropheops, which exhibit a continuum of trophic phenotypes roughly between MZ and LF (Roberts et al. 2011), and were wild-caught in the same region of Lake Malawi (Tropheops microstoma, T. gracilior, T. intermediate, T. tropheops, T. “red cheek,” T. “orange chest,” T. sp chinyamwezi). Sequences for MZ and LF lbh cDNA have been deposited to GenBank under accessions KJ593150 and KJ593151, respectively.
Protein Conservation
Conservation of the amino acid change in cichlids was assessed across selected members of Teleostei, with phylogeny derived from (Near et al. 2012). Gene sequences were identified by BLAST to GenBank (Salmo salar [salmon] and Esox lucius [Northern pike]), Ensembl (Gadus morhua [cod], Xiphophorus maculatus [platyfish], Gasterosteus aculeatus [stickleback], Tetraodon nigroviridis [tetraodon], Takifugu rubripes [fugu], and Oreochromis niloticus [Nile tilapia]), and the Cichlid Genome Database (Astatotilapia burtoni, Neolamprologus brichardi, and Pundamilia nyererei). Sequences were confirmed using EST libraries as available. DNA sequences were translated computationally and aligned using ClustalW. The effect of the amino acid change on predicted linear motifs was assessed using SLiMPred0.9 (Mooney et al. 2012).
Animals
All animals were maintained and used according to guidelines and protocols approved by the Institutional Animal Care and Use Committee at the University of Massachusetts Amherst. Cichlids (MZ and LF) and zebrafish (Danio rerio) were maintained and bred on separate filtration systems at 28.5 °C in a 14 h light/10 h dark cycle. Cichlid species were collected from Lake Malawi and reared in 40-gallon glass aquaria. Embryos (all F1- or F2-derived from wild-caught stocks) were obtained by natural matings, collected from mouth-brooding females, and staged according to Fujimura and Okada (2007). Wild-type (EKW) or NCC reporter line Tg(-7.2sox10:EFGP) (Hoffman et al. 2007) zebrafish embryos were collected by natural mass matings (Westerfield 2000) and staged according to Kimmel et al. (1995). Xenopus laevis embryos were obtained by in vitro fertilization as described in (Cousin et al. 2000) and staged according to Nieuwkoop and Faber (1967).
In Situ Hybridizations
Riboprobes were designed from ENSDARG00000087377 (zebrafish lbh), NM_001088038 (Xenopus lbh), and NM_001088649 (Xenopus slug). Cichlid lbh and foxd3 coding regions used for probe design were identified by BLAST of tilapia genes ENSONIG00000001365.1 and ENSONIG00000021415.1, respectively, to the MZ genome (Broad v.0) on the Cichlid Genome Database. Probes were prepared and whole-mount in situ hybridization was performed as previously described for cichlids (Albertson et al. 2005), zebrafish (Albertson and Yelick 2005), and Xenopus (Cousin et al. 2000).
Lbh Cloning and mRNA Expression
Full-length lbh was amplified by polymerase chain reaction (PCR) from both MZ and LF whole head cDNA using oligonucleotides containing a Flag tag, and cloned into the pCS2+ vector at the BamHI and EcoRI sites. Sequencing was performed to ascertain that no errors were introduced during PCR. Capped mRNA was synthesized using SP6 RNA polymerase on DNA linearized with NotI as described in Cousin et al. (2000).
Zebrafish Injections and Skeletal Staining
Antisense morpholino oligomers (MO) (Gene Tools, LLC) were designed to the translational start site (5′-CATCACGTCAGTCATCACGGCTACA-3′) or splice acceptor site (5′-AGATCTGAAGACGAGCACAGAGACA-3′) of zebrafish lbh. The standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) and a splice acceptor MO with five mismatches (mm) (5′-AGATGTCAACACGACCACACAGACA-3′) were used as negative controls. All morpholinos were injected at 0.5–4 ng/embryo. For lbh mRNA overexpression, each embryo was injected with 10 pg of either LF or MZ lbh mRNA plus 4 ng splice-blocking zebrafish MO, included to deplete endogenous Lbh. Embryos were injected with 1–3 nl MO or mRNA with 0.05% phenol red tracer (Sigma-Aldrich) at the yolk/blastoderm interface of one- to two-cell embryos. Each experimental treatment was repeated on 2–11 (mean = 5) different mass matings. To assess the effects of lbh overexpression, embryos were verified to be developing normally at 90% epiboly to bud stages (9–10 h postfertilization [hpf]) and phenotypically classified at 24 hpf. Skeletal effects were assessed at 96 hpf using Alcian blue and Alizarin red to stain for cartilage and bone, respectively, as previously described (Walker and Kimmel 2007). Analysis of NCC migration were conducted in Tg(-7.2sox10:EFGP) zebrafish reporter lines using the splice-blocking lbh MO. Movies were taken from the 3- to 5-somite stage to approximately 20–22 somites, with pictures taken every 3 min and movies reconstructed at five frames per second.
Reverse Transcription PCR
Either whole zebrafish embryos (n = 20 per stage from four-cell stage to 24 hpf) or tissue rostral to the eye (n = 20 per stage from 36 hpf to 1 month) were collected in TRIzol reagent (Invitrogen), and total RNA was extracted per manufacturer’s instructions. Genomic DNA was removed with DNase (Invitrogen) and cDNA was synthesized using random primers and the High Capacity cDNA Reverse Transcription Kit (Invitrogen). Following degradation of rRNA, 300 ng cDNA was used as PCR template for lbh (5′-CAGCACCGATGGATGATATG-3′ and 5′-TTCTGCCGATCCTCATCTTC-3′) and internal control actb2 (5′-TCAGCCATGGATGATGAAAT-3′ and 5′-GGTCAGGATCTTCATGAGGT-3′); both of these PCR products span an intron. Retention of the 1.8 kb lbh intron was assessed using primers sitting entirely within the intron (5′-CATCCTCACCCGACTGAAAT-3′ and 5′-CACGCTCATGATTCACACCT-3′) using RNA and cDNA prepared as above from 6 hpf embryos (n = 60).
Western Blotting
Protein was extracted from dechorionated 24 hpf embryos (n = 20) in 1 × Modified Barth Solution (MBS), 1% Triton X-100, 2 mM EDTA, and 1X Protease and Phosphatase Inhibitor Cocktail (ThermoFisher). Lbh was immunoprecipitated with 1µg of rabbit polyclonal antibody (pAb) (AbCam) coupled on protein A-agarose beads (Pierce) overnight at 4 °C. Samples were run in nonreducing Laemmli buffer on a 15% polyacrylamide gel, transferred on a PVDF membrane, and the portion of the membrane containing proteins above 37 kDa (including the immunoglobulins) was cut out. The remainder of the membrane containing Lbh was blotted using the Lbh pAb (AbCam) and an HRP coupled donkey antimouse pAb (Jackson lab) as previously described (Neuner et al. 2009). A loading control blot was carried out on one embryo equivalent on the same samples after immunoprecipitation using GAPDH antibody (Millipore).
Xenopus Morpholino Injections and Grafting
Embryos were injected at either the two- or eight-cell stage as previously described (Cousin et al. 2012), and all experiments were repeated on 3–4 different fertilizations. Briefly, for targeted injections (fig. 4d), 250 pg of GFP mRNA and 500 pg of either LF or MZ lbh mRNA was injected in one blastomere at the eight-cell stage. For grafting experiments (fig. 3h), one cell of a two-cell stage embryo was injected with: 1) 25 ng of X. laevis lbh translation-blocking morpholino (5′-ATACAGACATTAAGTCCAGAGGAGC-3′) with 200 pg of GFP mRNA or 2) 250 pg of GFP mRNA with 500 pg of either LF or MZ lbh mRNA. Control grafting embryos were injected with 200–250 pg of GFP mRNA. At st.15, when the neural folds are visible, cranial NCC were unilaterally grafted onto a host whose cranial NCC had been removed using the procedure described in Cousin et al. (2012).
Migration and Pixel Density Analysis
For assessing effects of lbh overexpression, each embryo was scored for presence or absence of fluorescent NCCs in the mandibular, hyoid, and/or branchial arches at the end of NCC migration, between st.25 and 28. Embryos were scored as “present” regardless of whether NCC had migrated partway or completely to the ventral edge of the embryo.
For only those embryos that had targeted injections and were scored as “present” as described above, pixel density analysis following (Roberts et al. 2011) was used to assess patterns of NCC migration. In brief, the percentage of GFP-positive cells was quantified and compared between the anterior portion of the mandibular migratory stream and the hyoid stream for control, LF-Lbh injected, and MZ-Lbh injected animals. For all animals, high-quality digital images of st.25–28 Xenopus embryos in the lateral view were imported into Adobe Photoshop CS3. Using the lasso tool, the anterior mandibular (i.e., above the eye) and hyoid streams were outlined, and the total number of pixels counted within each stream. Distinct streams were specified using a combination of GFP-positive expression and anatomical landmarks (e.g., eye and hatching gland). Next, using Select > Color Range, we used a standard set of green pixels to estimate the number of GFP-positive pixels within each pharyngeal arch. The ratio between GFP-positive and total number of pixels was used to estimate and compare the amount NCCs within each arch. Significance was assessed using ANOVA and Tukey’s HSD test in R.
Geometric Morphometrics
Three day (st.13) cichlid embryos were fixed, positioned in 80% glycerol, and photographed using a Leica DFC450 camera mounted to a Leica MF15 stereomicroscope. All embryos were carefully stage matched and sampled from multiple families. The final sample sizes were n = 13 for LF and n = 10 for MZ. All photos were taken in the ventral view such that pharyngeal arches could be visualized. Landmarks and semilandmarks were collected from photos using the software tpsdig2 (Rohlf 2010). Semilandmarks were collected as series of curves that defined the shape of the rostral aspect of the embryo as well as the shape of the first two pharyngeal arches. These data were reduced to three evenly spaced landmarks per curve and subsequently defined as semilandmarks using the software tsputil (Rohlf 2010). The program tpsrelw (Rohlf 2010) was used to superimpose landmarks and semilandmarks using a chord-distance (Procrustes distance) based “sliders” method, generate partial warps from landmark data, and perform a principal component analysis (PCA) on these variables. Data are presented for this PCA.
Supplementary Material
Supplementary tables S1 and S2, figures S1–S7 and movie S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the Karlstrom lab for technical assistance, J. Bennett for fish maintenance, and past and present members of the Albertson lab, UMass Amherst Behavior and Morphology group, and Drs D. Alfandari, C. Babbitt, and M. Bartlett for helpful discussions. Thanks too to Dr J. Sylvester for helpful discussions about cichlid NCC biology. Sequences for MZ and LF lbh cDNA have been deposited to GenBank under accessions KJ593150 and KJ593151, respectively. This work was supported by NSF Career IOS-1054909 (R.C.A.), NIH/NIDCR 1F32DE023707 (K.E.P.), NIH/NIDCR DE016289 (H.C.), UMass Healey Endowment Grant (H.C.), UMass Commonwealth Honors College Fellowship (G.M.). K.E.P., H.C., and R.C.A. conceived and designed experiments. K.E.P., H.C., and G.M. performed the experiments. K.E.P., H.C., and R.C.A. analyzed the data. K.E.P., H.C., and R.C.A. wrote the manuscript.
References
- Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai J, Wang Y, Tan K, Deng Y, Luo N, Yuan W, Wang Z, Li Y, Wang Y, Mo X, et al. A human homolog of mouse Lbh gene, hLBH, expresses in heart and activates SRE and AP-1 mediated MAPK signaling pathway. Mol Biol Rep. 2008;35:179–187. doi: 10.1007/s11033-007-9068-4. [DOI] [PubMed] [Google Scholar]
- Al-Ali H, Rieger ME, Seldeen KL, Harris TK, Farooq A, Briegel KJ. Biophysical characterization reveals structural disorder in the developmental transcriptional regulator LBH. Biochem Biophys Res Commun. 2010;391:1104–1109. doi: 10.1016/j.bbrc.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Kocher TD. Genetic and developmental basis of cichlid trophic diversity. Heredity. 2006;97:211–221. doi: 10.1038/sj.hdy.6800864. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: the molecular basis of alternate feeding strategies. Proc Natl Acad Sci U S A. 2005;102:16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Yelick PC. Roles for fgf8 signaling in left-right patterning of the visceral organs and craniofacial skeleton. Dev Biol. 2005;283:310–321. doi: 10.1016/j.ydbio.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Powder KE, Hu Y, Coyle KP, Roberts RB, Parsons KJ. Genetic basis of continuous variation in the levels and modular inheritance of pigmentation in cichlid fishes. Mol Ecol. 2014 doi: 10.1111/mec.12900. Advance Access published September 18, 2014, doi: 10.1111/mec.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduini BL, Bosse KM, Henion PD. Genetic ablation of neural crest cell diversification. Development. 2009;136:1987–1994. doi: 10.1242/dev.033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett ME, Whipple CJ. Protein change in plant evolution: tracing one thread connecting molecular and phenotypic diversity. Front Plant Sci. 2013;4:382. doi: 10.3389/fpls.2013.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayer KJ, Lynch VJ, Wagner GP. Evolution of a derived protein-protein interaction between HoxA11 and Foxo1a in mammals caused by changes in intramolecular regulation. Proc Natl Acad Sci U S A. 2011;108:E414–420. doi: 10.1073/pnas.1100990108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel K, Joyner A. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol. 2001;233:291–304. doi: 10.1006/dbio.2001.0225. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Powder KE, Young NM, Goodnough LH, Hahn SM, James AW, Helms JA, Lovett M. Comparative gene expression analysis of avian embryonic facial structures reveals new candidates for human craniofacial disorders. Hum Mol Genet. 2010;19:920–930. doi: 10.1093/hmg/ddp559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: molecular genetics and the evolution of animal design. 2nd ed. Malden (MA): Blackwell; 2005. [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chandler KJ, Chandler RL, Mortlock DP. Identification of an ancient Bmp4 mesoderm enhancer located 46 kb from the promoter. Dev Biol. 2009;327:590–602. doi: 10.1016/j.ydbio.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3' enhancer located 156.3 kilobases from the promoter. Mol Cell Biol. 2007;27:2934–2951. doi: 10.1128/MCB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WJ, Parsons K, McIntyre A, Kern B, McGee-Moore A, Albertson RC. Bentho-pelagic divergence of cichlid feeding architecture was prodigious and consistent during multiple adaptive radiations within African rift-lakes. PLoS One. 2010;5:e9551. doi: 10.1371/journal.pone.0009551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WJ, Wernle J, Mann K, Albertson RC. Functional and genetic intergration in the skulls of Lake Malawi Cichlids. Evol Biol. 2011;38:316–334. [Google Scholar]
- Cousin H, Abbruzzese G, McCusker C, Alfandari D. ADAM13 function is required in the 3 dimensional context of the embryo during cranial neural crest cell migration in Xenopus laevis. Dev Biol. 2012;368:335–344. doi: 10.1016/j.ydbio.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H, Gaultier A, Bleux C, Darribere T, Alfandari D. PACSIN2 is a regulator of the metalloprotease/disintegrin ADAM13. Dev Biol. 2000;227:197–210. doi: 10.1006/dbio.2000.9871. [DOI] [PubMed] [Google Scholar]
- Danley PD, Kocher TD. Speciation in rapidly diverging systems: lessons from Lake Malawi. Mol Ecol. 2001;10:1075–1086. doi: 10.1046/j.1365-294x.2001.01283.x. [DOI] [PubMed] [Google Scholar]
- DiLeone RJ, Russell LB, Kingsley DM. An extensive 3' regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics. 1998;148:401–408. doi: 10.1093/genetics/148.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Fish JL, Sklar RS, Woronowicz KC, Schneider RA. Multiple developmental mechanisms regulate species-specific jaw size. Development. 2014;141:674–684. doi: 10.1242/dev.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Okada N. Development of the embryo, larva and early juvenile of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae). Developmental staging system. Dev Growth Differ. 2007;49:301–324. doi: 10.1111/j.1440-169X.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Opening Darwin's black box: teaching evolution through developmental genetics. Nat Rev Genet. 2003;4:735–741. doi: 10.1038/nrg1159. [DOI] [PubMed] [Google Scholar]
- Guenther C, Pantalena-Filho L, Kingsley DM. Shaping skeletal growth by modular regulatory elements in the Bmp5 gene. PLoS Genet. 2008;4:e1000308. doi: 10.1371/journal.pgen.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BK. The neural crest and neural crest cells in vertebrate development and evolution. New York: Springer; 2010. [Google Scholar]
- Hallgrimsson B, Jamniczky H, Young NM, Rolian C, Parsons TE, Boughner JC, Marcucio RS. Deciphering the palimpsest: studying the relationship between morphological integration and phenotypic covariation. Evol Biol. 2009;36:355–376. doi: 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol. 2007;308:679–691. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Kague E, Gallagher M, Burke S, Parsons M, Franz-Odendaal T, Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS One. 2012;7:e47394. doi: 10.1371/journal.pone.0047394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Konings A. Malawi cichlids in their natural habitat. El Paso (TX): Cichlid Press; 2001. [Google Scholar]
- Loh YH, Katz L, Mims M, Kocher TD, Yi SV, Streelman JT. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;8:R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- Mims MC, Darrin Hulsey C, Fitzpatrick BM, Streelman JT. Geography disentangles introgression from ancestral polymorphism in Lake Malawi cichlids. Mol Ecol. 2010;19:940–951. doi: 10.1111/j.1365-294X.2010.04529.x. [DOI] [PubMed] [Google Scholar]
- Mooney C, Pollastri G, Shields DC, Haslam NJ. Prediction of short linear protein binding regions. J Mol Biol. 2012;415:193–204. doi: 10.1016/j.jmb.2011.10.025. [DOI] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A. 2012;109:13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner R, Cousin H, McCusker C, Coyne M, Alfandari D. Xenopus ADAM19 is involved in neural, neural crest and muscle development. Mech Dev. 2009;126:240–255. doi: 10.1016/j.mod.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) 2nd ed. Amsterdam: North-Holland; 1967. [Google Scholar]
- Noden DM. The control of avian cephalic neural crest cytodifferentiation. I. Skeletal and connective tissues. Dev Biol. 1978;67:296–312. doi: 10.1016/0012-1606(78)90201-4. [DOI] [PubMed] [Google Scholar]
- Parsons KJ, Trent Taylor A, Powder KE, Albertson RC. Wnt signalling underlies the evolution of new phenotypes and craniofacial variability in Lake Malawi cichlids. Nat Commun. 2014;5:3629. doi: 10.1038/ncomms4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy ME, McDermott KJ, Antonellis A, Margulies EH, Prasad AB, Program NCS, Kingsley DM, Green ED, Mortlock DP. Detection of potential GDF6 regulatory elements by multispecies sequence comparisons and identification of a skeletal joint enhancer. Genomics. 2005;86:295–305. doi: 10.1016/j.ygeno.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Powder KE, Ku Y-C, Brugmann SA, Veile RA, Renaud NA, Helms JA, Lovett M. A cross-species analysis of microRNAs in the developing avian face. PLoS One. 2012;7:e35111. doi: 10.1371/journal.pone.0035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregizer S, Mortlock DP. Control of BMP gene expression by long-range regulatory elements. Cytokine Growth Factor Rev. 2009;20:509–515. doi: 10.1016/j.cytogfr.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, Jiggins CD, Chamberlain NL, Kronforst MR, Chen R, et al. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333:1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- Ribbink AJ, Marsh AC, Ribbink CC, Sharp BJ. A preliminary survey of the cichlid fishes of rocky habitats in Lake Malawi. S Afr J Zool. 1983;18:149–310. [Google Scholar]
- Roberts RB, Hu Y, Albertson RC, Kocher TD. Craniofacial divergence and ongoing adaptation via the hedgehog pathway. Proc Natl Acad Sci U S A. 2011;108:13194–13199. doi: 10.1073/pnas.1018456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf FJ. Department of Ecology and Evolution, State University of New York at Stony Brook. 2010. TPSdig v2.16, TPSutil v1.47, and TPSrelw v1.49. Available from: http://life.bio.sunysb.edu/morph/ [Google Scholar]
- Sadaghiani B, Vielkind JR. Neural crest development in Xiphophorus fishes: scanning electron and light microscopic studies. Development. 1989;105:487–504. [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell. Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Schilling T, Piotrowski T, Grandel H, Brand M, Heisenberg C, Jiang Y, Beuchle D, Hammerschmidt M, Kane D, Mullins M, et al. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development. 1996;123:329–344. doi: 10.1242/dev.123.1.329. [DOI] [PubMed] [Google Scholar]
- Schneider RA, Helms JA. The cellular and molecular origins of beak morphology. Science. 2003;299:565–568. doi: 10.1126/science.1077827. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Hutchinson SA, Byers A, Beale HC, Carringon B, Faden DL, Rimbault M, Decker B, Kidd JM, Sood R, et al. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 2012;8:e1002849. doi: 10.1371/journal.pgen.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Kronenberg Z, Li C, Domyan ET, Pan H, Campbell M, Tan H, Huff CD, Hu H, Vickrey AI, et al. Genomic diversity and evolution of the head crest in the rock pigeon. Science. 2013;339:1063–1067. doi: 10.1126/science.1230422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Lumsden A. Neural crest cells provide species-specific patterning information in the developing branchial skeleton. Evol Dev. 2004;6:32–40. doi: 10.1111/j.1525-142x.2004.04004.x. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Lynch VJ. The gene regulatory logic of transcription factor evolution. Trends Ecol Evol. 2008;23:377–385. doi: 10.1016/j.tree.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Wang WD, Melville DB, Montero-Balaguer M, Hatzopoulos AK, Knapik EW. Tfap2a and Foxd3 regulate early steps in the development of the neural crest progenitor population. Dev Biol. 2011;360:173–185. doi: 10.1016/j.ydbio.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) Eugene: University of Oregon Press; 2000. [Google Scholar]
- Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Wu P, Jiang TX, Shen JY, Widelitz RB, Chuong CM. Morphoregulation of avian beaks: comparative mapping of growth zone activities and morphological evolution. Dev Dyn. 2006;235:1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.