Abstract
The first transgenic pigs were generated for agricultural purposes about three decades ago. Since then, the micromanipulation techniques of pig oocytes and embryos expanded from pronuclear injection of foreign DNA to somatic cell nuclear transfer, intracytoplasmic sperm injection-mediated gene transfer, lentiviral transduction, and cytoplasmic injection. Mechanistically, the passive transgenesis approach based on random integration of foreign DNA was developed to active genetic engineering techniques based on the transient activity of ectopic enzymes, such as transposases, recombinases, and programmable nucleases. Whole-genome sequencing and annotation of advanced genome maps of the pig complemented these developments. The full implementation of these tools promises to immensely increase the efficiency and, in parallel, to reduce the costs for the generation of genetically engineered pigs. Today, the major application of genetically engineered pigs is found in the field of biomedical disease modeling. It is anticipated that genetically engineered pigs will increasingly be used in biomedical research, since this model shows several similarities to humans with regard to physiology, metabolism, genome organization, pathology, and aging.
Key words: : binary transposon, disease model, domestic animal, humanized pig, large animal model, programmable nuclease
Introduction
Progress in reproductive techniques and gene transfer methods nowadays allows targeted modifications of the porcine genome, although the overall success rates are still low.1–4 A bottleneck for porcine transgenesis is the lack of authentic embryonic stem (ES) cells, which are suitable for blastocyst complementation experiments.5,6 Recently, the first attempts to generate porcine induced pluripotent stem (iPS) cells have been published7–10; however, the potential of current porcine iPS cells to contribute to chimera formation in blastocyst complentations,11 or to become reprogrammed by somatic cell nuclear transfer (SCNT),12 seems to be limited. Despite these bottlenecks, the recent employment of active transgenesis techniques based on ectopic enzymes, such as transposases, recombinases, and programmable nucleases, allows the generation of pig models at unprecedented pace.4
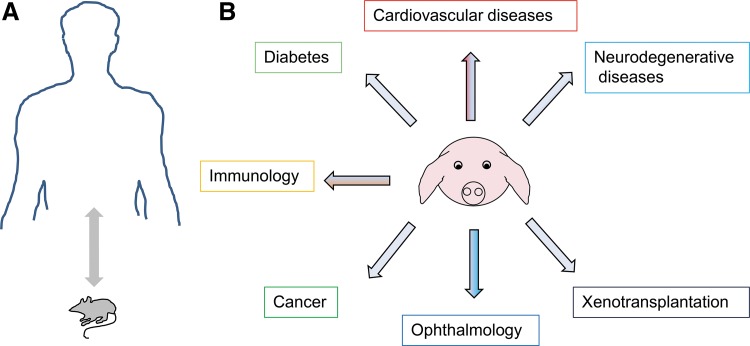
Nowadays, at least 90% of genetically modified pigs are generated for biomedical studies. Sequencing and annotation of the pig genome are important milestones to accelerate the generation of transgenic models. Since physiology, anatomy, pathology, genome organization, body weight, and life span of pigs and minipigs are more similar to those of humans, the domesticated pig represents an alternative biomedical model to rodents for specific human diseases.3,6 This article briefly discusses the current progress of transgenic pig models for cancer, cardiovascular diseases, diabetes, neurodegenerative diseases, ophthalmology, and xenotransplantation (Fig. 1), and future potential of this large animal model. Excellent comprehensive reviews regarding transgenesis in pigs in general and recent developments of innovative molecular tools, such as transposons, recombinases, and programmable nucleases, can be found elsewhere.3,4,13,14
FIG. 1.
Transgenic pigs as models for human diseases. (A) Rodent models differ in a number of critical parameters, such as size, life span, genome organization, physiology, and metabolism, from humans. (B) The domesticated pig and its minipig varieties seem to be relevant models for specific human diseases.
Porcine Biomedical Models
Cystic fibrosis
Cystic fibrosis (CF) is an autosomal recessive genetic disorder caused by mutations in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR) that lead to pancreatic insufficiency, inadequate hydration of mucous secretion, and development of intestinal, hepatic, and lung disease.15,16 The most prevalent cystic fibrosis mutation is a deletion of the phenylalanine at amino acid position 508 (Δ508).17
The efforts to develop a CFTR mouse model have not resulted in the appearance of the CF symptoms in this species.18 Importantly, genetically modified minipigs carrying either ΔF508, or CFTR knockout genotypes develop highly similar disease phenotypes as human patients (Table 1).19 This includes meconium ileus, defective chloride transport, early focal biliary cirrhosis, and pancreatic destruction.19–21
Table 1.
Selected Pig and Minipig Models for Biomedical Research
| Model | Comment | Reference |
|---|---|---|
| Xenotransplantation | Knockout of N-glycolylneuraminic acid and alpha-galactosyltransferase | Kwon et al.94 |
| Expression of tumor necrosis factor ligand | Klose et al.95 | |
| Expression of human leukocyte antigen | Weiss et al.96 | |
| PERV knockdown | Dieckhoff et al.79 | |
| Expression of human A20 (anti-apoptotic gene) | Oropeza et al.97 | |
| Cystic fibrosis | Knockout of cystic fibrosis transmembrane conductance receptor | Rogers et al.19 |
| Expression of mutated CFTR protein | Ostedgaard et al.21 | |
| Intestinal CFTR expression in CF pigs | Stoltz et al.98 | |
| Diabetes model | Expression of mutated hepatocyte nuclear factor-1 | Umeyama et al.39 |
| Expression of mutated insulin 2 | Renner et al.41 | |
| Immunology | Knockout of light chain | Ramsoondar et al.99 |
| Knockout of joining gene cluster | Mendicino et al.100 | |
| Knockout of interleukin-2 receptor subunit gamma | Watanabe et al.64 | |
| Knockout of RAG1/2 | Huang et al.65 | |
| Expression of hCD46/HLA-E and knockout GGTA1 | Bongoni et al.74 | |
| Neurodegenerative diseases | Expression of mutated huntingtin with polyglutamine tract | Yang et al.49 |
| Expression of mutated human amyloid precursor protein | Kragh et al.44 | |
| Expression of PSEN1M146I | Jakobsen et al.46 | |
| Expression of HTT protein (124Q) with human HD promoter | Baxa et al.50 | |
| Cancer | Knockout of BRCA1 gene | Luo et al.26 |
| Expression of mutated p53 | Sieren et al.25 | |
| Expression of mutated APC | Flisikowska et al.23 | |
| Ophthalmology | Introduced deletion in ELOVL4 gene | Sommer et al.60 |
| Expression of mutated rhodopsin gene | Scott et al.56 | |
| Cardiovascular diseases | Expression of mutated proprotein convertase subtilisin/kexin type 9 | Al-Mashhadi et al.29 |
PERV, porcine endogenous retrovirus; CFTR, cystic fibrosis transmembrane conductance regulator; GGTA1, galactosyl transferase1 gene; HTT, huntingtin protein; HD, Huntington's disease; BRCA1, breast cancer 1 gene; APC, adenomatous polyposis coli; ELOVL4, elongation of very long chain fatty acids protein 4.
Porcine cancer models
Cancer is a group of more than 100 diseases caused by uncontrolled growth of cells. Different types of cancer are driven by genetic instability or epigenetic alterations of DNA repair genes, such as mutations in tumor suppressor genes and oncogenes. Although genetically engineered mouse models have significantly contributed to cancer biology,22 they still have significant limitations in their usefulness for modeling human cancer due to the differences of biology of human and mice.23
The first transgenic pig model for the Li Fraumeni syndrome was created by gene targeting of the tumor suppressor gene TP53 in mesenchymal stem cells (MSCs) and subsequently using these cells in somatic cell nuclear transfer (SCNT).24 A latent R167H mutation in the p53 orthologous gene of the human oncogene mutant R175H was created with conditional gene activation system by the Cre-lox recombinase system. The strategy used by Leuchs et al.24 resulted in 10-fold greater p53-R167H expression in homozygous MSCs similar to the expression status of the mutant human allele. However, no viable pigs were obtained with the homozygous mutation. Fifteen heterozygous piglets are currently observed for signs of tumor formation.
Recently, a successful porcine model for the development of Li Fraumeni syndrome has been generated and resulted in homozygous pigs expressing TP53-R167H. Lymphomas and osteogenic tumor formations were observed and confirmed by histopathological evaluation, computed tomography, and magnetic resonance imaging.25
Adenomatous polyposis coli (APC) is a tumor suppressor gene, and mutations may result in colorectal cancer such as familial adenomatous polyposis (FAP). Mutated genes from human patients generally contain premature stop codons between amino acids 1061–1309. Similar to FAP patients, the APC pigs exhibit dysplastic adenomas and abnormal crypt foci,23 making them a useful model for longitudinal studies of disease progression, development of early diagnosis, and potential interventions.
A breast cancer 1 gene (BRCA1) knockout model for mammary carcinogenesis in porcine has been developed by targeting exon 12 using adeno-associated virus (AAV)–mediated gene targeting and SCNT.26 The protein product of the BRCA1 participates in DNA repair and checkpoint control processes.27 Since the homology between human and pig BRCA1 proteins (74%) is higher than the homology between human and murine proteins (58%), it is anticipated that cancer pathology and response to potentials drugs can be investigated by using this porcine model. Although heterozygous BRCA1 knockout pigs were produced successfully, development of tumorigenesis was not observed because of the early mortality of the piglets.26
Cardiovascular diseases
The domestic pig has been recognized as an excellent model for the human cardiovascular system due to similarities in coronary anatomy, physiology of heart, and blood flow.28,29 Transgenic studies with relevance to cardiovascular diseases have been developed in pig models. Employing the Sleeping Beauty (SB) transposon system, minipigs with liver-specific expression of a mutant human proprotein convertase subtilisin/kexin type 9 (PCSK9) gene were generated.29 The transgenic pigs displayed reduced hepatic low-density lipoprotein (LDL) receptor levels, impaired LDL clearance, hypercholesterolemia, and spontaneous development of progressive atherosclerotic lesions.29
Promising results were obtained in transgenic pigs that carry a fatty acid desaturase (FAD2) gene from spinach (Spinacia oleracea) under the control of an adipocyte P2 promoter to increase linoleic acid level.30 Transgenic Yucatan pigs that express higher levels of endothelial nitric oxide synthase (eNOS) which regulates vascular function and structure by generating nitric oxide (NO) have been generated to investigate the regulation of vasodilation.31 Since vascular homeostasis by NO signaling is closely related to hydrogen peroxide (H2O2), which modulates the eNOS activity,32 the role of endothelium-derived H2O2 on cardiovascular disease has been investigated by the generation of transgenic minipigs that overexpress the human endothelial catalase.14,33
Peroxisome proliferator-activated receptor-γ (PPAR-γ) has been seen as a target for therapeutic invention of cardiovascular disease due to its role on fatty acid uptake by adipocytes.34 Effects of PPAR-γ have been investigated with a new strategy for the production of transgenic pig models using zinc-finger nuclease technology following SCNT.35 These pig models promise to be useful in understanding of the human cardiovascular diseases and may serve as a model for testing new therapeutic strategies.
Diabetes
Diabetes mellitus (DM) is a group of metabolic disorders resulting from deficiency or ineffectiveness of endogenous insulin, which leads to an increase in blood glucose levels. Long-term hyperglycemia results in life-threatening consequences with dysfunction and failure of kidneys, heart, nerves, and blood vessels.36 Type I DM is also called juvenile diabetes, which is characterized by low production of insulin, while type II DM results from the resistance to insulin.
A transgenic pig model for type II DM was produced based on impaired glucose-dependent insulinotropic polypeptide (GIP) receptor using lentiviral transgenesis.37 The role of GIP in DM was demonstrated by a transgenic pig model, which showed significantly reduced oral glucose tolerance.38 Significant reduction in number and mass of β-cells were obtained in these transgenic pigs.37
A transgenic pig model expressing a dominant-negative hepatocyte nuclear factor 1α (HNF-1α), responsible for progressive reduction in insulin secretion was produced.39,40 Although 12 of the 22 live born piglets died before weaning, 4 lived up to 196 days. Surviving piglets exhibited nonfasting blood glucose levels over 200 mg/dl. Diabetic characteristics include immature renal development and abnormal Langerhans islet formation in which high glucose level and poor insulin secretion were detected by histochemical analysis.40 More recently, a transgenic pig model for permanent neonatal diabetes, which has been associated with mutations in the insulin gene (INS) in humans,41 was generated by the introduction of an orthologous mutation in the pig genome.42 The transgenic pigs showed hyperglycemia soon after birth. A decrease in body weight (41%) and β-cell mass (72%) was observed at the age of 4.5 months. The production of INSC94Y transgenic pigs having a stable diabetic phenotype might become an attractive model for insulin treatment studies.
Neurodegenerative diseases
Alzheimer's disease (AD) is the most common multifactorial neural disorder characterized by progressive loss of memory and neurodegeneration in specific brain regions such as pyramidal cells in the cortex.42 Causative mutations have been identified in presenilin genes 1 and 2 (PSEN1, PSEN2), tau gene, and the amyloid precursor protein (APP) gene leading to increased production of hyper-phosphorylated tau in tangles and amyloid β-peptide (Aβ) in plaques, which in turn results in synaptic damage and neuropathy.43 A genetically modified pig model for AD was produced with a cDNA construct containing the Swedish mutation of human APP gene (APPsw).44 Although transgene expression was detected in brain tissues, Aβ peptide accumulation at symptomatic levels and appearance of a clinical consequences were yet not observed. At 1–2 years of age, the APPsw-carrying AD minipigs were analyzed by the spontaneous object recognition test (SORT) as a measure of memory. However, no significant differences were found in age-matched controls.45 It is estimated that AD symptoms may develop with increasing age of the animals.44
Recently, another AD pig model carrying PSEN1M146I mutation was developed. Therefore, a recombinase-mediated cassette exchange (RMCE) for targeted transgenesis and SCNT were employed.46 Up to now, no AD-like behavioral symptoms have been observed in these animals; however, onset of symptoms may take more time. Potentially, these models may offer the possibility for developing early diagnosis of AD.
Huntington's disease (HD) is an autosomal dominant, progressive neurodegenerative disorder characterized by expansion of a tri-nucleotide (CAG) repeat in the huntingtin gene (HTT) that results in extended amino-terminal poly-glutamine tracts ranging from 36 to 120 repeats. The mutated huntingtin protein misfolds, leading to aggregate accumulation in neurons that affects the transmission of neurotransmitters and are suspected to induce apoptosis.47,48 Transgenic HD minipigs that express amino-terminal mutant HTT (105Q) under the control of the cytomegalovirus enhancer and chicken β-actin promoter were successfully generated via SCNT. The authors suggested that high levels of mutant HTT expression lead to early death phenotype of the carrier animals. Out of one litter, three of the five piglets died within 3 days and the fourth died at the age of 25 days. The fifth and vital piglet showed the lowest expression level of mutant HTT.49 Whereas genetic mouse models often failed to replicate overt neurodegeneration and apoptosis, histological analyses of brains sections from these piglets showed clear signs of apoptotic neurons with activated caspase-3 and DNA fragmentation.49
Baxa et al.50 recently reported the successful generation of transgenic HD minipigs using a lentiviral-based vector carrying human amino-terminal truncated HTT gene containing 145 repeats of CAG/CAA codons under the control of human HTT promoter. Although stable HTT expression was obtained in brain tissues, no motor deficits developed in this pig model so far.
Ophthalmology
Retinitis pigmentosa (RP) is an inherited retinal disease that causes loss of central vision due to loss of light-sensitive photoreceptor neurons. Numerous mutations in various genes and loci can result in RP in humans, but the majority of autosomal dominant forms of RP cases are related to mutations in the rhodopsin (RHO) gene. Unlike laboratory rodents or dogs, swine is an appropriate choice as an RP model, since it has morphological and functional similarities, including similar medium- and short-wavelength cone photoreceptors and spatial distribution of rod photoreceptors.51,52 Transgenic pigs expressing P347L mutation in the RHO gene created by pronuclear microinjection showed similar progressive cone photoreceptor degeneration and loss of function in cone-mediated vision as humans.53 Expression of the most common human P23H mutation in the rhodopsin gene was developed in a minipig model via SCNT and resulted in successful mirroring of the human phenotype of RP.54 Recently, Fernandez de Castro et al.55 created a pig model carrying P23H mutation that lacks rod function. Independent of the rod, the cone function developed normally for the first 30 days and then slowly declined. The same group has recently reported the effects of RHOP23H expression on rod and cone photoreceptor in pig embryos. This resulted in an abnormal morphological and functional development of rod photoreceptors, but no change in cone photoreceptors was observed related to the cone dysfunction.56 Therefore, this large animal model may be useful in the therapeutic intervention to rescue cone function in RP patients.
A pig model for a cone–rod dystrophy (CORD) characterized by the progressive loss of cone cells was developed recently by using lentiviral transgenesis. It has been reported that guanylate cyclase 2D gene (GUCY2D) mutations are a frequent cause for autosomal dominant CORD.57,58 The pigs expressing a GUCY2D mutation can be useful to understand the pathophysiology of the disease.59
Truncation mutations in the elongation of very long chain fatty acids protein 4 (ELOVL4) gene cause macular dystrophy. Photoreceptor topography in the pig retina is more similar to that in humans as it includes cone-rich, macula-like area centralis, whereas mice lack a macular. Transgenic pigs expressing disease-causing ELOVL4 mutations were generated by pronuclear DNA injection and SCNT.60 A detailed analysis showed photoreceptor loss, disorganized inner and outer segments, and diminished electroretinography responses, suggesting that the transgenic pigs mirror macular degeneration and provide an unique model for therapeutic interventions.
Immunology
The first successful immunodeficient pig model was generated by homologous recombination in porcine fetal fibroblasts resulting in a interleukin-2 receptor subunit gamma (IL2RG) deletion.61 IL2RG is responsible for growth and maturation of T cells and NK cells via signaling through different interleukins.62 The resulting IL2RG heterozygous male pigs showed deficiency of T and NK cells, and lacked thymus, similar to human SCID, which is a genetic disorder characterized by malfunctioning of T- and B-lymphocytes.63 Watanabe et al.64 generated IL2RG knockout pigs by zinc-finger nuclease technology, which showed an identical phenotype with above-mentioned pig model.
Subsequently, SCID pig models were established by TALENs targeting RAG1 and RAG2 in porcine fetal fibroblasts, followed by SCNT. Bi-allelic mutant RAG1/2 piglets showed the absence of mature B and T lymphocytes in their lymphoid organs, which was the result of the deficit in V(D)J recombination. Compared to wild type, heterozygous RAG2 piglets showed similar development and sexual maturity, whereas the RAG1 or RAG2 homozygous piglets were characterized by a higher death rate.65 Another SCID pig model carrying RAG2 bi-allelic mutations has been generated resulting in piglets that lacked B- and T-cells, but possessed NK-cells, providing an alternative to immunocompromised mice for the assessment of tumorigenicity.66 Since the immune reactions of pigs and humans are quite similar, the RAG2 knockout pigs may help to understand the phenotype of human SCID patients and to evaluate the efficacy and safety of cell transplantations.66 However, long-term maintenance of severely immunodeficient pigs would require housing under pathogen-free conditions, which currently is not available in most experimental pig facilities.
Xenotransplantation
Xenotransplantation seems to be one option to close the growing gap between demand and availability of appropriate organs for human patients with acute or chronic organ failure.67 Genetic engineering of pigs allows to overcome immunological hurdles for xenotransplantation and to prevent transmission of porcine pathogens to human recipients.
A major obstacle to successful porcine-to-primate xenotransplantation is caused by hyperacute rejection resulting from preformed antibodies that recognize a galactose-α1,3-galactose (α-Gal) carbohydrate.68 The synthesis of α-Gal epitopes is catalyzed by the alpha 1,3 galactosyl transferase1 gene (GGTA1) found in all mammals except old world monkeys, apes, and humans.69 The production of GGTA1 knockout animals significantly increased the survival of xenotransplants, maximal survival rates of up to 3–6 months have been achieved with porcine GGTA1 knockout organs (kidney or heart) transplanted to baboons, and hyperacute rejections were greatly reduced in the grafts.70,71 The hyperacute rejection can be further reduced by simultaneous expression of human complement regulatory proteins such as CD46, CD55, or CD59 combined with a GGTA1 deletion. A more advanced pig line carrying deletions of two xenoantigens, α-Gal and N-glycolylneuraminic acid, has been produced, and showed reduced humoral activity compared to a GGTA1 knockout alone.72 In addition to the immunosuppressive approach, graft survival is considered to depend on coagulation dysregulation, leading to the development of thrombotic microangiopathy of the graft, consumptive coagulopathy of the recipient, or both.73
Recently, Bongoni et al.74 generated GGTA1 heterozygous knockout/hC46/HLA-E transgenic pigs to explore the effect of hCD46/HLA-E expression on complement deposition, endothelial activation, and NK cell interaction. Ex vivo xenoperfusion of the transgenic pig limbs with human blood resulted in reduction of accumulation of C3b/c, C4b/c, and C6 and a reduced up-regulation of the proinflammatory cell adhesion molecules E-selectin and VCAM-1.74 Although the contribution of the HLA-E expression was not specified due to the double mutation, hCD46-expressing transgenic pigs lacking the α-Gal epitope resulted in reduced complement activation during ex vivo lung perfusions.75 Accordingly, these models will likely represent useful tools to develop strategy for xenotransplantation. A recent study describing the xenotransplantation of multitransgenic porcine islets into diabetic cynomolgus monkeys, however, did not demonstrate long-term correction of insulin and blood glucose levels.76
Extensive research has been conducted to reduce the potential risk of zoonotic pathogen transmission to human patients of a xenotransplantation; a particular emphasis has been put on porcine endogenous retrovirus (PERV).77,78 RNA interference (RNAi) is a promising method for knocking down the PERV expression. The efficacy of RNAi to reduce PERV expression has been demonstrated in cloned piglets by targeting the PERV genes encoding capsid protein, reverse transcriptase, and envelope glycoprotein.79–82 It seems likely that these improvements will be helpful to increase the biosecurity of potential porcine xenotransplants. However, recent data suggest that other common opportunistic pathogens, like cytomegalovirus (CMV), influence xenograft survival in nonhuman primates and need to be controlled in a xenotransplantation scenario.83,84
Conclusions and Perspectives
Rapidly increasing numbers of biomedical pig models were published in recent years (Table 1). The development of advanced methods for genetic engineering provided the basis for this recent movement. Advanced methodological improvements include the establishment of hyperactive transposases, recombinases, and programmable endonucleases. Together with more accurate genome data, precise genetic modifications became feasible. In own experiments, we explored and established the SB transposon system for porcine transgenesis. The in ovo transposition turned out to be a highly efficient method resulting in success rates of up to 60% transgenic piglets per litter.85,86 Importantly, the monomeric SB transposons seem to escape epigenetic regulation, and faithful promoter-dependent expression patterns were obtained (Fig. 2). Using the chimeric CAGGS promoter, an ubiquitous expression in somatic and germline cells was found.85,87,88 Induced PS cells derived from these animals showed a robust expression of the reporter (Fig. 2) and will be instrumental for vital cell tracking studies, for example, to determine the fate of iPS cells in blastocyst complementation, to analyze the contribution to chimera formation, and to assess fetal cell chimerism in the pig.89 The robust reporter expression will also allow to track transplanted cells in pig models of innovative cell therapies. In combination with a recombinase, such as the Cre/loxP system, the SB-transposon-tagged genomic loci can be readdressed, thereby enabling targeted integration of functional transgenes into pretested genomic loci (Fig. 3).
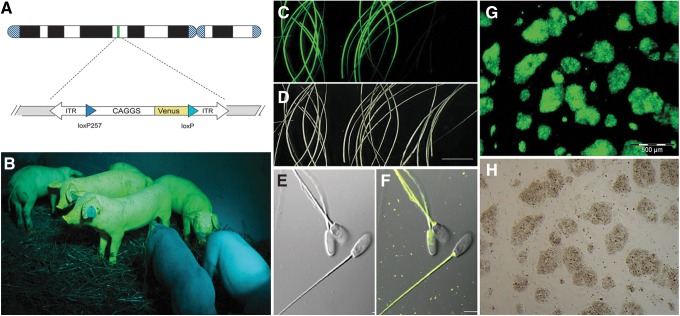
FIG. 2.
Fluorescent reporter transposon pigs as tool for vital cell tracking. (A) Schematic depiction of Sleeping Beauty (SB)–Venus transposon.85 (B) Transposon transgenic pigs, generated by cytoplasmic injection of plasmids, carrying a Venus reporter fluorescent green under specific excitation. Wild-type animals (front) appear bluish due to reflected and scattered excitation light.87 (C) Amazingly, the Venus reporter is also deposited in the hair, and two samples from transgenic (left and middle) and a hair sample from a wild-type pig (right) are shown under epifluorescence. (D) Same view as in (C) showing under darkfield conditions. Scale bar=1 cm. (E) Spermatozoa from a reporter transposon boar shown under differential interference contrast (DIC) and (F) as overlay of specific fluorescence and DIC images. The spermatozoa loaded with the Venus protein and allow tracking the fate of paternal proteins after fertilization.87 Scale bar=5 μm. (G) Porcine induced pluripotent stem cell colonies derived from an SB–Venus animal shown under Venus fluorescence conditions, and (H) brightfield view.
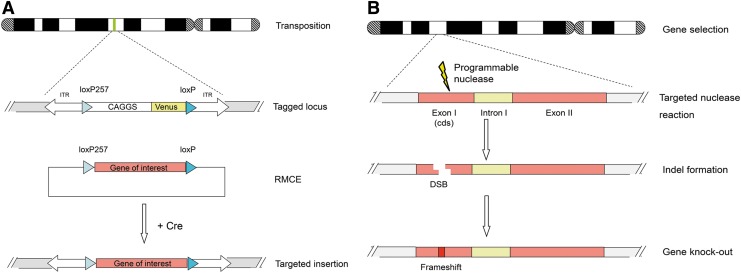
FIG. 3.
Genetic engineering of the pig genome. (A) Additive genome engineering by transposase-catalyzed integration. Subsequently, a targeted replacement of the reporter against a transgene of choice is possible via Cre recombinase-mediated cassette exchange (RMCE).4,85 (B) Subtractive genome engineering by a programmable nuclease approach. A designer nuclease, ZFN, TALEN, or CRISPR/Cas9, is directed against a protein-coding exon. The CRISPR/Cas9 system seems to combine the efficiency of ZFNs and TALENs with a much simpler design principle, as target-site selection is determined solely by base complementarity to the guide RNA. Indel formation after faulty repair will likely result in a frameshift mutation and a functional gene knockout. In principle, this mechanism can be coupled with homologous recombination.
The development of programmable endonucleases, like ZFNs, TALENs, and CRISPR/Cas nucleases, will complement this toolbox for genetic engineering of the pig (Fig. 3).13,14,34,90–92 It is anticipated that authentic pluripotent cells of the pig will be generated in the near future. Thus, genetic engineering in the pig will become a routine tool for the generation of relevant humanized pig models. The most obvious application of transgenic pigs will be for disease models and biomedical therapies, which are not well reflected in small rodent models. The expected progress for pig transgenesis (increase of success rates and decreasing costs) will make the pig an attractive complementary model for advanced approaches in biomedical and pharmaceutical research.
Pig models allow longitudinal studies of disease initiation and progression, thus allowing for improved diagnosis methods. The ongoing breeding of miniature pig strains, such as Göttingen, NIH and Sinclair minipigs, and the even smaller microminipigs,93 will contribute to a broader acceptance of this animal model. Miniaturized pigs will be easier to implement in existing infrastructures and will fit in current schedules for pharmacological testing of potential drug substance.
Genetically modified pigs will complement and extend disease modeling in transgenic mice, in particular for diseases with slowly progressing pathophysiology. The examples of biomedical pig models discussed here demonstrate that the technology for genetic engineering in this species caught up to the possibilities already available in the mouse. In the near future, the biomedical and pharmaceutical research with genetically engineered pigs will have a significant impact for molecular and cellular understanding of disease development, early diagnosis, and therapeutic interventions, and will establish the “laboratory pig” as valuable large mammalian model species. Moreover, pig models will allow the preclinical assessment of novel cell therapies to restore diseased tissues and organs, and to test the prospects of genetic correction of mutated alleles.
Abbreviations Used
- AAV
adeno-associated virus
- Aβ
amyloid β peptide
- AD
Alzheimer's disease
- APC
adenomatous polyposis coli (gene)
- APP
amyloid precursor protein
- BRCA
breast cancer (gene)
- C3, C4, C6
complement components
- CAGGS
CMV enhancer, chicken beta-actin promoter
- CD46, CD55, CD59
complement proteins
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane regulator
- CMV
cytomegalovirus
- CORD
cone–rod dystrophy
- Cre
causes recombination
- CRISPR
clustered regularly interspaced short palindromic repeats
- DIC
differential interference contrast
- DM
diabetes mellitus
- eNOS
endothelial nitric oxide synthase
- ELOV4
elongation of very long chain fatty acids protein 4
- ES
embryonic stem (cell)
- FAD2
fatty acid desaturase 2 (gene)
- FAP
familiar adenomatous polyposis
- GGTA1
alpha 1,3 galactosyltransferase 1
- GIP
glucose-dependent insulinotropic polypeptide
- GUCY2D
guanylate cyclase 2D (gene)
- HD
Huntington's disease
- HLA-E
class I histocompatibility antigen, alpha chain E
- HNF-1α
hepatocyte nuclear factor 1α
- HTT
huntingtin (gene)
- IL2RG
interleukin-2 receptor subunit gamma
- Indel
insertion of deletion of bases
- INS
insulin
- iPS
induced pluripotent stem (cell)
- ITR
inverted terminal repeats
- LDL
low-density lipoprotein
- MSC
mesenchymal stem cell
- NK cell
natural killer cell
- NO
nitric oxide
- PCSK9
protein convertase subtilisin/kexin type 9 (gene)
- PERV
porcine endogenous retrovirus
- PNI
pronuclear DNA injection
- PPAR-γ
peroxisome proliferator-activated receptor-γ
- PSEN
presenilin
- RAG
recombination activating gene
- RHO
rhodopsin
- RMCE
recombinase mediated cassette exchange
- RNAi
RNA interference
- RP
retinitis pigmentosa
- SB
Sleeping Beauty (transposon)
- SCID
severe combined immunodeficiency
- SCNT
somatic cell nuclear transfer
- SORT
spontaneous object recognition test
- TALEN
transactivator-like element nuclease
- TP35
tumor protein 53
- VCAM1
vascular cell adhesion molecule 1
- Venus
variant of yellow fluorescent protein
- ZFN
zinc finger nuclease
Acknowledgments
Financial support by the Deutsche Forschungsgemeinschaft and the Erasmus program (University of Istanbul) is gratefully acknowledged.
Author Disclosure Statement
The authors declare no conflicts of interest.
References
- 1.Clark J, Whitelaw B. A future for transgenic livestock. Nat Rev Genet. 2003;4:825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robl JM, Wang Z, Kasinathan P, Kuroiwa Y. Transgenic animal production and animal biotechnology. Theriogenology. 2007;67:127–133 [DOI] [PubMed] [Google Scholar]
- 3.Whyte JJ, Prather RS. Genetic modifications of pigs for medicine and agriculture. Mol Reprod Dev. 2011;78:879–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrels W, Ivics Z, Kues WA. Precision genetic engineering in large mammals. Trends Biotechnol. 2012;7:386–393 [DOI] [PubMed] [Google Scholar]
- 5.Hall VJ. Early development of the porcine embryo: the importance of cell signalling in development of pluripotent cell lines. Reprod Fertil Dev. 2012;25:94–102 [DOI] [PubMed] [Google Scholar]
- 6.Nowak-Imialek M, Niemann H. Pluripotent cells in farm animals: state of the art and future perspectives. Reprod Fertil Dev. 2012;25:103–128 [DOI] [PubMed] [Google Scholar]
- 7.Esteban MA, Xu J, Jang J, et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2011;284:17634–17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezashi T, Telugu B, Alexenko AP, et al. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci USA. 2011;106:10993–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Chen J, Ren J, et al. Generation of pig-induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol. 2011;1:46–54 [DOI] [PubMed] [Google Scholar]
- 10.Kues WA, Herrmann D, Barg-Kues B, et al. Derivation and characterization of sleeping beauty transposon-mediated porcine induced pluripotent stem cells. Stem Cells Dev. 2013;22:124–135 [DOI] [PubMed] [Google Scholar]
- 11.West FD, Terlouw SL, Kwon DJ, et al. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2011;19:1211–1220 [DOI] [PubMed] [Google Scholar]
- 12.Fan N, Chen J, Shang , et al. Piglets cloned from induced pluripotent stem cells. Cell Res. 2013;23:162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334 [DOI] [PubMed] [Google Scholar]
- 14.Prather RS, Lorson M, Ross JW. Genetically engineered pig models for human diseases. Annu Rev Anim Biosci. 2013;1:203–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boucher RC. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur Respir J. 2004;23:146–158 [DOI] [PubMed] [Google Scholar]
- 16.Veit G, Bossard F, Goepp J, et al. Proinflammatory cytokine secretion is suppressed by TMEM16A or CFTR channel activity in human cystic fibrosis bronchial epithelia. Mol Biol Cell. 2012;23:4188–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du K, Lukacs GL. Cooperative assembly and misfolding of CFTR domains in vivo. Mol Biol Cell. 2009;20:1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bragonzi A. Murine models of acute and chronic lung infection with cystic fibrosis pathogens. Int J Med Microbiol. 2010;300:584–593 [DOI] [PubMed] [Google Scholar]
- 19.Rogers CS, Stoltz DA, Meyerholz DK, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321:1837–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers CS, Hao Y, Rokhlina T, et al. Production of CFTR null and ΔF508 heterozygous pigs by AAV-mediated gene targeting and somatic cell nuclear transfer. J Clin Invest. 2008;118:1571–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostedgaard LS, Meyerholz DK, Chen JH, et al. The ΔF508 mutation causes CFTR misprocessing and cystic fibrosis-like disease in pigs. Sci Transl Med. 2011;3:74ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheon D-J, Orsulic S. Mouse models of cancer. Annu Rev Pathol. 2011;6:95–119 [DOI] [PubMed] [Google Scholar]
- 23.Flisikowska T, Merkl C, Landmann M, et al. A porcine model of familial adenomatous polyposis. Gastroenterology 2012;143:1173–1175 [DOI] [PubMed] [Google Scholar]
- 24.Leuchs S, Saalfrank A, Merkl C, et al. Inactivation and inducible oncogenic mutation of p53 in gene targeted pigs. PLoS ONE. 2012;7:e43323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sieren JC, Meyerholz DK, Wang XJ, et al. Development and translational imaging of a TP53 porcine tumorigenesis model. J Clin Invest. 2014;124:4052–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, Li J, Liu Y, et al. High efficiency of BRCA knockout using rAAV-mediated gene targeting: developing a pig model for breast cancer. Transgenic Res. 2011;20:975–988 [DOI] [PubMed] [Google Scholar]
- 27.Deng CX, Wang RH. Roles of BRCA1 in DNA damage repair: a link between development and cancer. Hum Mol Genet. 2003;12:113R–123R [DOI] [PubMed] [Google Scholar]
- 28.Turk JR, Laughlin MH. Physical activity and atherosclerosis: which animal model? Can J Appl Physiol. 2004;29:657–683 [DOI] [PubMed] [Google Scholar]
- 29.Al-Mashhadi RH, Sørensen CB, Kragh PM, et al. Familial hypercholesterolemia and atherosclerosis in cloned minipigs created by DNA transposition of a human PCSK9 gain-of-function mutant. Sci Transl Med. 2013;5:166ra1. [DOI] [PubMed] [Google Scholar]
- 30.Saeki K, Matsumoto K, Kinoshita M, et al. Functional expression of a Delta12fatty acid desaturase gene from spinach in transgenic pigs. Proc Natl Acad Sci USA. 2004;101:6361–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao YH, Yong HY, Murphy CN, et al. Production of endothelial nitric oxide synthase (eNOS) over-expressing piglets. Transgenic Res. 2006;15:739–750 [DOI] [PubMed] [Google Scholar]
- 32.Thomas SR, Chen K, Keaney JF., Jr Hydrogen peroxide activates endothelial nitric-oxide synthase through coordinated phosphorylation and dephosphorylation via a phosphoinositide 3-kinase-dependent signaling pathway. J Biol Chem. 2002;277:6017–6024 [DOI] [PubMed] [Google Scholar]
- 33.Whyte JJ, Samuel M, Mahan E, et al. Vascular endothelium-specific overexpression of human catalase in cloned pigs. Transgenic Res. 2011;20:989–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholls SJ, Uno K. Peroxisome proliferator-activated receptor (PPARa/g) agonists as a potential target to reduce cardiovascular risk in diabetes. Diabetes Vasc. Dis. Res. 2012;9:89–94 [DOI] [PubMed] [Google Scholar]
- 35.Yang D, Yang H, Li W, et al. 2011. Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21:979–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2013;36:S67–S74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renner S, Fehlings C, Herbach N, et al. Glucose intolerance and reduced proliferation of pancreatic beta-cells in transgenic pigs with impaired glucose-dependent insulinotropic polypeptide function. Diabetes. 2010;59:1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyunt O, Wu JY, McGown IN, et al. Investigating maturity onset diabetes of the young. Clin Biochem Rev. 2009;30:67–74 [PMC free article] [PubMed] [Google Scholar]
- 39.Umeyama K, Watanabe M, Saito H, et al. 2009. Dominant-negative mutant hepatocyte nuclear factor 1alpha induces diabetes in transgenic-cloned pigs. Transgenic Res. 2009;18:697–706 [DOI] [PubMed] [Google Scholar]
- 40.Støy J, Steiner DF, Park S-Y, et al. Clinical and molecular genetics of neonatal diabetes due to mutations in the insulin gene. Rev Endocr Metab Disord. 2010;11:205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renner S, Braun-Reichhart C, Blutke A, et al. Permanent neonatal diabetes in INS(C94Y) transgenic pigs. Diabetes. 2013;62:1505–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;6:1204–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh DM, Klyubin I, Shankar GM, et al. The role of cell-derived oligomers of Abeta in Alzheimer's disease and avenues for therapeutic intervention. Biochem Soc Trans. 2005;33:1087–1090 [DOI] [PubMed] [Google Scholar]
- 44.Kragh PM, Nielsen AL, Li J, et al. 2009. Hemizygous minipigs produced by random gene insertion and handmade cloning express the Alzheimer's disease-causing dominant mutation APPsw. Transgenic Res. 2009;18:545–558 [DOI] [PubMed] [Google Scholar]
- 45.Sondergaard LV, Ladewig J, Dagnaes-Hansen F, et al. Object recognition as a measure of memory in 1–2 years old transgenic minipigs carrying the APPsw mutation for Alzheimer's disease. Transgenic Res. 2012;21:1341–1348 [DOI] [PubMed] [Google Scholar]
- 46.Jakobsen JE, Johansen MG, Schmidt M, et al. Generation of minipigs with targeted transgene insertion by recombinase-mediated cassette exchange (RMCE) and somatic cell nuclear transfer (SCNT). Transgenic Res. 2013;22:709–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan J, Yankner BA. Apoptosis in the nervous system. Nature 2000;407:802–809 [DOI] [PubMed] [Google Scholar]
- 48.Rubinsztein DC, Carmichael J. Huntington's disease: molecular basis of neurodegeneration. Expert Rev Mol Med. 2003;5:1–21 [DOI] [PubMed] [Google Scholar]
- 49.Yang D, Wang CE, Zhao B, et al. Expression of Huntington's disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Hum Mol Genet. 2010;19:3983–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baxa M, Hruska-Plochan M, Juhas S, et al. A transgenic minipig model of Huntington's disease. J Huntingtons Dis 2013;2:47–68 [DOI] [PubMed] [Google Scholar]
- 51.Chandler MJ, Smith PJ, Samuelson DA, et al. Photoreceptor density of the domestic pig retina. Vet Ophthalmol. 1999;2:179–184 [DOI] [PubMed] [Google Scholar]
- 52.Gerke CG, Hao Y, Wong F. Topography of rods and cones in the retina of the domestic pig. Hong Kong Med J. 1995;1:302–308 [Google Scholar]
- 53.Petters RM, Alexander CA, Wells KD, et al. Genetically engineered large animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa. Nat Biotechnol. 1997;15:965–970 [DOI] [PubMed] [Google Scholar]
- 54.Ross JW, Fernandez de Castro JP, Zhao J, et al. Generation of an inbred miniature pig model of retinitis pigmentosa. Investig Ophthalmol Vis Sci. 2012;53:501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez de Castro JP, Scott PA, James W, et al. Cone photoreceptors develop normally in the absence of functional rod photoreceptors in a transgenic swine model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014;55:2460–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott PA, Fernandez de Castro JP, Kaplan HJ, et al. A Pro23His mutation alters prenatal rod photoreceptor morphology in a transgenic swine model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014;28:2452–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitiratschky VB, Wilke R, Renner AB, et al. Mutation analysis identifies GUCY2D as the major gene responsible for autosomal dominant progressive cone degeneration. Invest Ophthalmol Vis Sci. 2008;49:5015–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelsell RE, Gregory-Evans K, Payne AM, et al. Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum Mol Genet. 1998;7:1179–1184 [DOI] [PubMed] [Google Scholar]
- 59.Kostic C, Lillico SG, Crippa SV, et al. Rapid cohort generation and analysis of disease spectrum of large animal model of cone dystrophy. PLoS ONE. 2013;8:e71363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sommer JR, Estrada JL, Collins EB, et al. Production of ELOVL4 transgenic pigs: a large animal model for Stargardt-like macular degeneration. Br J Ophthalmol. 2011;95:1749–1754 [DOI] [PubMed] [Google Scholar]
- 61.Suzuki S, Iwamoto M, Saito Y, et al. Il2rg gene-targeted severe combined immunodeficiency pigs. Cell Stem Cell. 2012;10:753–758 [DOI] [PubMed] [Google Scholar]
- 62.Belizário JE. Immunodeficient mouse models: an overview. Open Immunol J. 2009;2:79–85 [Google Scholar]
- 63.Burg M, Gennery AR. Educational paper: the expanding clinical and immunological spectrum of severe combined immunodeficiency. Eur J Pediatr. 2011;170:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watanabe M, Nakano K, Matsunari H, et al. Generation of interleukin-2 receptor gamma gene knockout pigs from somatic cells genetically modified by zinc finger nuclease-encoding mRNA. PLoS ONE. 2013;8:e76478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang J, Guo X, Fan N, et al. RAG1/2 Knockout pigs with severe combined immunodeficiency. J Immunol. 2014;193:1496–1503 [DOI] [PubMed] [Google Scholar]
- 66.Lee K, Kwon DN, Ezashi T, et al. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc Natl Acad Sci USA. 2014;111:7260–7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang YG, Sykes M. Xenotransplantation: current status and a perspective on the future. Nat Rev Immunol. 2007;7:519–531 [DOI] [PubMed] [Google Scholar]
- 68.Klymiuk N, Aigner B, Brem G, et al. Genetic modification of pigs as organ donors for xenotransplantation. Mol Reprod Dev. 2010;77:209–221 [DOI] [PubMed] [Google Scholar]
- 69.Galili U. The alpha-gal epitope and the anti-Gal antibody in xenotransplantation and in cancer immunotherapy. Immunol Cell Biol. 2005;83:674–686 [DOI] [PubMed] [Google Scholar]
- 70.Kuwaki K, Tseng YL, Dor FJ, et al. Heart transplantation in baboons using alpha1,3-galactosyltransferase gene-knockout pigs as donors: initial experience. Nat Med. 2005;11:29–31 [DOI] [PubMed] [Google Scholar]
- 71.Yamada K, Yazawa K, Shimizu A, et al. Marked prolongation of porcine renal xenograft survival in baboons through the use of α1, 3-galactosyltransferase gene-knockout donors and the cotransplantation of vascularized thymic tissue. Nat Med. 2005;11:32–34 [DOI] [PubMed] [Google Scholar]
- 72.Lutz AJ, Li P, Estrada JL, et al. Double knockout pigs deficient in N-glycolylneuraminic acid and galactose a-1,3-galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation. 2013;20:27–35 [DOI] [PubMed] [Google Scholar]
- 73.Iwase H, Ekser B, Hara H, et al. Regulation of human platelet aggregation by genetically modified pig endothelial cells and thrombin inhibition. Xenotransplantation. 2014;21:72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bongoni AK, Kiermeir D, Jenni H, et al. Complement dependent early immunological responses during ex vivo xenoperfusion of hCD46/HLA-E double transgenic pig forelimbs with human blood. Xenotransplantation. 2014;21:230–243 [DOI] [PubMed] [Google Scholar]
- 75.Burdorf L, Stoddard T, Zhang T, et al. Expression of human CD46 modulates inflammation associated with GalTKO lung xenograft injury. Am J Transplant. 2014;14:1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bottino R, Wijkstrom M, van der Windt DJ, et al. Pig-to-monkey islet xenotransplantation using multi-transgenic pigs. Am J Transplant. 2014;14:2275–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Switzer W, Michler M, Shanmugam RE, et al. Lack of cross-species transmission of porcine endogenousretrovirus infection to nonhuman primate recipients of porcine cells, tissues, or organs. Transplantation. 2001;71:959–965 [DOI] [PubMed] [Google Scholar]
- 78.Irgang M, Sauer IM, Karlas A, et al. Porcine endogenous retroviruses: no infection in patients treated with a bioreactor based on porcine liver cells. J Clin Virol. 2003;28:141–154 [DOI] [PubMed] [Google Scholar]
- 79.Dieckhoff B, Petersen B, Kues WA, et al. Knockdown of porcine endogenous retrovirus (PERV) expression by PERV-specific shRNA in transgenic pigs. Xenotransplantation. 2008;15:36–45 [DOI] [PubMed] [Google Scholar]
- 80.Ramsoondar J, Vaught T, Ball S, et al. Production of transgenic pigs that express porcine endogenous retrovirus small interfering RNAs. Xenotransplantation. 2009;16:164–180 [DOI] [PubMed] [Google Scholar]
- 81.Chung HC, Nguyen VG, Kim HJ, et al. Inhibition of porcine endogenous retrovirus in PK15 cell line by efficient multitargeting RNA interference. Transpl Int. 2014;27:96–105 [DOI] [PubMed] [Google Scholar]
- 82.Li ZG, Lie GB, Pan MX, et al. Knockdown of porcine endogenous retroviruses by RNA interference in chinese experimental miniature pig fibroblasts. Transplant Proc. 2013;45:748–755 [DOI] [PubMed] [Google Scholar]
- 83.Yamada K, Tasaki M, Sekijima M, et al. Porcine cytomegalovirus infection is associated with early rejection of kidney grafts in a pig to baboon xenotransplantation model. Transplantation. 2014;98:411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sekijima M, Waki S, Sahara H, et al. Results of life-supporting galactosyltransferase knockout kidneys in cynomolgus monkeys using two different sources of galactosyltransferase knockout swine. Transplantation. 2014;98:419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garrels W, Mates L, Holler S., et al. Germline transgenic pigs by Sleeping Beauty transposition in porcine zygotes and targeted integration in the pig genome. PLoS ONE 2011;6:e23573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ivics Z, Garrels W, Mátés L, et al. Germline transgenesis in pigs by cytoplasmic microinjection of Sleeping Beauty transposons. Nat Protoc. 2014;9:810–827 [DOI] [PubMed] [Google Scholar]
- 87.Garrels W, Holler S, Taylor U, et al. 2011. Genotype-independent transmission of transgenic fluorophore protein by boar spermatozoa. PLoS ONE. 2011;6:e27563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garrels W, Holler S, Cleve N, et al. Assessment of fecundity and germ line transmission in two transgenic pig lines produced by sleeping beauty transposition. Genes (Basel). 2012;3:615–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garrels W, Holler S, Taylor U, et al. Assessment of fetal cell chimerism in transgenic pig lines generated by sleeping beauty transposition. PLoS ONE. 2014;9:e96673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whyte JJ, Zhao J, Wells KD, et al. Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Mol Reprod Dev. 2011;78:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hauschild J, Petersen B, Santiago Y, et al. generation of a biallelic knockout in pigs using zinc-finger-nucleases. Proc Natl Acad Sci USA. 2011;108:12013–12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hai T, Teng F, Guo R, et al. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaneko N, Itoh K, Sugiyama A, Izumi Y. Microminipig, a non-rodent experimental animal optimized for life science research: preface. J Pharmacol Sci. 2011;115:112–114 [DOI] [PubMed] [Google Scholar]
- 94.Kwon DN, Lee K, Kang MJ, et al. Production of biallelic CMP-Neu5Ac hydroxylase knock-out pigs. Sci Rep. 2013;3:1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klose R, Kemter E, Bedke T, et al. Expression of biologically active human TRAIL in transgenic pigs. Transplantation. 2005;80:222–230 [DOI] [PubMed] [Google Scholar]
- 96.Weiss EH, Lilienfeld BG, Müller S, et al. HLA-E/human beta2-microglobulin transgenic pigs: protection against xenogeneic human anti-pig natural killer cell cytotoxicity. Transplantation. 2009;87:35–43 [DOI] [PubMed] [Google Scholar]
- 97.Oropeza M, Petersen B, Carnwath JW, et al. Transgenic expression of the human A20 gene in cloned pigs provides protection against apoptotic and inflammatory stimuli. Xenotransplantation. 2009;16:522–534 [DOI] [PubMed] [Google Scholar]
- 98.Stoltz DA, Rokhlina T, Ernst SE, et al. Intestinal CFTR expression alleviates meconium ileus in cystic fibrosis pigs. J Clin Invest. 2013;123:2685–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ramsoondar J, Mendicino M, Phelps C, et al. Targeted disruption of the porcine immunoglobulin kappa light chain locus. Transgenic Res. 2011;20:643–653 [DOI] [PubMed] [Google Scholar]
- 100.Mendicino M, Ramsoondar J, Phelps C, et al. Generation of antibody- and B cell-deficient pigs by targeted disruption of the J-region gene segment of the heavy chain locus. Transgenic Res. 2011;20:625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]