Abstract
Induced pluripotent stem cells (iPSCs) are reprogrammed from somatic cells via ectopic gene expression and, similarly to embryonic stem cells (ESCs), possess powerful abilities to self-renew and differentiate into cells of various lineages. However, the neural differentiation potency of iPSCs remains unknown. In this study, we demonstrated the neural differentiation ability of iPSCs compared with ESCs using an retinoic acid (RA) induction system. The neural differentiation efficiency of iPSCs was obviously lower than that of ESCs. Retinoic acid receptor-α (RARα) was critical in the RA-induced neural differentiation of iPSCs, and the effect of RARα was confirmed by applying a specific RARα antagonist ER50891 to ESCs. These findings indicate that iPSCs do not possess the complete properties that ESCs have.
Introduction
Embryonic stem cells (ESCs) are derived in vitro from the inner cell mass of blastocysts and possess unlimited self-renewal ability and pluripotency to differentiate into various cell types of all three germ layers (Evans and Kaufman, 1981). These characteristics of ESCs provide a promising resource for studying the mechanisms of pluripotency, lineage commitment, and cell fate specification and allow their application to disease modeling, drug screening, and cell-based therapy. Although ESCs possess powerful properties, it is difficult to apply them to autologous cell transplantation because of immune and ethical issues. To address these problems, somatic cells were reprogrammed via ectopic expression of Oct4, Sox2, Klf4, and cMyc to derive induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006). These iPSCs, similarly to ESCs, exhibit an unlimited proliferation ability and are pluripotent and germ-line competent (Okita et al., 2007). Although the transgenes present in iPSCs raised concerns regarding their clinical application, these cells represent an unlimited source for cell therapy with clearly reduced immune rejection events (Araki et al., 2013). On the basis of these powerful characteristics, differentiated gene-targeted autologous iPSCs have served as therapeutic cells for clinical treatment (Deyle et al., 2012). However, although iPSCs undergo differentiation programs, the differentiation efficiency of iPSCs remains obscure.
All-trans retinoic acid (ATRA), which is a metabolic product of vitamin A, is a well-known and important morphogen that induces stem cell differentiation into various cell lineages, especially a neural lineage (Maden, 2007; Rhinn and Dolle, 2012). After binding to nuclear retinoic acid receptors (RARs) and coordinating with retinoid X receptors (RXRs), the RA–RAR–RXR complex binds to functional retinoic acid response elements (RAREs) to activate downstream genes. Thus, RA triggers a downstream signaling that is involved in the maintenance of adult neurons and neural stem cells and induces axon outgrowth and nerve regeneration (Corcoran and Maden, 1999; Corcoran et al., 2000; Corcoran et al., 2002). In previous studies, ATRA was used to induce neural differentiation from ESCs in vitro; the derived neurons exhibited functional neuronal properties (Bain et al., 1995; Fraichard et al., 1995).
In this study, we used RA as an inducer to promote in vitro neural differentiation, and we compared the neural differentiation potency of iPSCs with that of ESCs. We observed that iPSCs were able to differentiate into neurons and glial cells, albeit with a lower differentiation efficiency. We found that the expression of RARα in iPSCs was one of the major factors that attenuated the RA effects of neural differentiation. Our results indicate that iPSCs are capable of yielding differentiated cells but with lower neural differentiation efficiency.
Materials and Methods
Cultivation of mouse ESCs and iPSCs
The mouse ESC line AB1 from 129S7/SvEvBrd mice was kindly provided by Dr. You-Tzung Chen (Graduate Institute of Clinical Genomics, National Taiwan University, Taipei, Taiwan) (McMahon and Bradley, 1990). The D3 line from 129S2/SvPas mice was purchased from the American Cell Type Collection, and the iPS-MEF-Ng-20D-17 mouse iPSC line from RF8 mouse ESCs from 129S4 mice was generously provided by Dr. Shinya Yamanaka (Center for iPS Cell Research and Application, Kyoto University, Kyoto, Japan) (Okita et al., 2007). ESCs and iPSCs were maintained on tissue-culture dishes (Corning, Corning, NY, USA) in the presence of gamma-irradiated mouse embryonic fibroblasts in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 15% Knockout Serum Replacement (KSR), 1% GlutaMAX, 1% Minimum Essential Medium (MEM) nonessential amino acids (NEAA), 1% antibiotic–antimycotic (all from Invitrogen, Carlsbad, CA, USA), 0.2 mM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), and 1000 U/mL of ESGRO Leukemia Inhibitory Factor (LIF) (Millipore, Billerica, MA, USA). All cultures were kept at 37°C in a 5% CO2 humidified air incubator.
Neural induction
For embryoid body (EB) formation, ESCs and iPSCs were detached and dissociated into single cells with 0.25% trypsin–EDTA (Invitrogen) and resuspended in EB medium (ESC medium without ESGRO LIF) at a density of 5×104 cells/mL. Hanging drops plated onto lids of nonadherent bacterial Petri dishes were cultured for 4 days; each drop contained 1000 cells in 20 μL of EB medium. After 4 days of hanging drop culture, EBs were formed and transferred to new Petri dishes in fresh EB medium containing 1 μM RA (Sigma-Aldrich). EBs were cultured for an additional 4 days, and the medium was changed every 2 days (4−/4+ protocol) (Bain et al., 1995). For treatment with the RARα antagonist ER50891, this drug was added into EB medium at day 4 to day 8 of differentiation, and the medium was changed every day. After 4 days of RA induction, EBs were collected and plated on Matrigel (Becton Dickinson, Palo Alto, CA, USA)-coated tissue culture dishes in serum-free neural induction medium, which consists of a 1:1 mixture of DMEM/F12 supplemented with N2 and neurobasal medium supplemented with B27 (all from Invitrogen). The medium was replaced every 2 days until 8 days of adherent culture.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 10 min at room temperature, washed with phosphate-buffered saline (PBS) (Invitrogen) three times, and permeabilized for 15 min in 0.1% Triton X-100/PBS. After washing with PBS three times, cells were blocked in PBS containing 4% normal goat serum for 30 min. Primary antibodies were diluted in blocking buffer and applied for 1 h at room temperature. After washing three times with PBS, the cells were incubated for 1 h at room temperature with secondary antibodies and the nuclear-staining dye Hoechst 33342 (1:1000; Sigma-Aldrich). After washing with PBS, cells were mounted in Crystal Mount (Biomeda, Foster City, CA, USA), and images were obtained with a Leica TCS SP5 laser confocal microscope (Leica, Wetzlar, Germany). The primary antibodies used were as follows: Mouse anti-Oct3/4 (1:100; Santa Cruz, Santa Cruz, CA, USA), goat anti-Sox1 (1:400; Santa Cruz), mouse anti-Nestin (1:400; Becton Dickinson), rabbit anti-Tuj1 (1:1000; Sigma-Aldrich), mouse anti-glial fibrillary acidic protein (GFAP; 1:400; Sigma-Aldrich), and mouse anti-2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase; 1:400; Millipore, Billerica, MA, USA). The secondary antibodies used were as follows: Alexa Fluor 488-conjugated donkey anti-mouse immunoglobulin G (IgG), Alexa Fluor 594-conjugated donkey anti-rabbit IgG, and Alexa Fluor 594-conjugated donkey anti-goat IgG (1:400; Invitrogen).
Quantitative RT–PCR
Total RNA was isolated using the TRI Reagent (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. First-strand cDNA was synthesized from 2 μg of total RNA using ThermoScript RT (Invitrogen) with oligo(dT) primers. Quantitative PCR was performed with FastStart SYBR Green Master (Roche, Mannheim, Germany), and signals were detected on an Mx3000P QPCR System (Agilent Technologies, Santa Clara, CA, USA). The relative expression level was calculated using the Ct−ΔΔ method and normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The experimental results contained three biological repeats, and each biological repeat was assayed with three technical triplicates. Each experiment was performed more than three times with same trend. The primer sequences used were as follows: Oct4, forward (F), 5′-GAAGCAGAAGAGGATCACCTTG-3′, reverse (R), 5′-TTCTTAAGGCTGAGCTGCAAG-3′ (129 bp); Sox1, F, 5′-ATACCGCAATCCCCTCTCAG-3′, R, 5′-ACAACATCCGACTCCTCTTCC-3′ (167 bp); Nestin, F, 5′-GGTCACTGTCGCCGCTACTC-3′, R, 5′-CGGACGTGGAGCACTAGAGAA-3′ (77 bp); Tuj1, F, 5′-TAGACCCCAGCGGCAACTAT-3′, R, 5′-GTTCCAGGTTCCAAGTCCACC-3′ (127 bp); CYP26a1, F, 5′-TTCTGCAGATGAAGCGCAGG-3′, R, 5′-TTTCGCTGCTTGTGCGAGGA-3′ (211 bp); Raldh2, F, 5′-TTGCAGATGCTGACTTGGAC-3′, R, 5′-TCTGAGGACCCTGCTCAGTT-3′ (201 bp); RARα, F: 5′-CTTCTGACTGTGGCTGCTTG-3′, R, 5′-CTCTTCGGAACTGCTGCTCT-3′ (232 bp); RARβ, F, 5′-GGACCTTGAGGAACCAACAA-3′, R, 5′-GAATGTCTGCAACAGCTGGA-3′ (375 bp); RARγ, F, 5′-AGGTCACCAGAAATCGATGC-3′, R, 5′-CTGGCAGAGTGAGGGAAAAG-3′(212 bp); RXRα, F, 5′-CTTTGACAGGGTGCTAACAGAGC-3′, R, 5′-ACGCTTCTAGTGACGCATAACACC-3′ (173 bp); RXRβ, F, 5′-TCAACTCCACAGTGTCGCTC-3′, R, 5′-TAAACCCCATAGTGCTTGCC-3′ (175 bp); RXRγ, F, 5′-TTCTTCAAAAGGACCATCAGG-3′, R, 5′-CGTTCATGTCACCGTAGGATTCT-3′ (289 bp); Fgf4, F, 5′-CTACTGCAACGTGGGCATC-3′, R, 5′-TCGGTAAAGAAAGGCACACC-3′ (201 bp); and Fgf8, F, 5′-CATCAGCGCGAGGTGCACTTC–3′, R: 5′–CGTGAAGGGCGGGTAGTTGAG-3′ (99 bp).
Western blotting
Cells were collected and lysed in Cell Lysis Buffer (Cell Signaling Technology, Danvers, MA, USA) supplemented with a protease inhibitor cocktail (Roche). After centrifugation at 14,000 rpm for 30 min, the supernatant was collected, and the protein concentration was measured using the Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA, USA). Protein samples (30 μg) in 1× sample buffer were boiled for 10 min, resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Subsequently, blots were blocked with 5% nonfat milk in Tris-buffered saline with 0.1% Tween-20 (TBST) for 1 h, and incubated with primary antibodies in blocking buffer overnight at 4°C. After washing three times with TBST, the blots were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies in blocking buffer for 1 h at room temperature. The detection of signals was performed using the Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA). The primary antibodies used were as follows: Mouse anti-Oct3/4 (1:1000; Santa Cruz), rabbit anti-phosphor-Erk1/2 (1:1000; Cell Signaling Technology), rabbit anti-Erk1/2 (1:1000; Cell Signaling Technology), and mouse anti-GAPDH (1:5000; Sigma-Aldrich). The secondary antibodies used were as follows: HRP-conjugated rabbit anti-mouse IgG and HRP-conjugated goat anti-rabbit IgG (1:2000; Sigma-Aldrich).
Flow cytometry
Cultured cells were trypsinized into individual cells, fixed in 4% paraformaldehyde for 10 min, and washed three times with PBS. Subsequently, cells were permeabilized with 0.1% Triton X-100/PBS for 15 min, washed in PBS three times, and blocked in PBS containing 2% bovine serum albumin (BSA) for 30 min. The primary antibodies were diluted in blocking buffer and applied for 1 h at room temperature. After three washes in PBS, cells were incubated with secondary antibodies for 1 h at room temperature. Finally, cells were washed three times in PBS and used for sorting. Fluorescence-activated cell sorting (FACS) analysis was performed using a BD FACSAria II Cell Sorter (Becton Dickinson), and data were analyzed by the BD FACSDiva Software (Becton Dickinson). The primary antibodies used were mouse anti-Oct3/4 (1:100; Santa Cruz) and goat anti-Sox1 (1:100; Santa Cruz). The secondary antibodies used were Alexa Fluor 488-conjugated donkey anti-mouse IgG and Alexa Fluor 594-conjugated donkey anti-goat IgG (all at 1:200; all from Invitrogen). Each result was obtained from at least 1×106 cells in total for one antibody staining.
Statistical analyses
The statistical data were analyzed by the SigmaPlot 10.0 software (Systat Software, San Jose, CA, USA). An analysis of variance (ANOVA) test was used to compare the dynamic changes of gene expression at different time points in one cell type, and Student's t-test was used to compare the difference of gene expression between two cell types at one time point. A p value <0.05 was considered significant. All data were shown as the mean±standard error of the mean (SEM).
Results
iPSCs differentiated into neural-lineage cells, similar to ESCs but with some differences
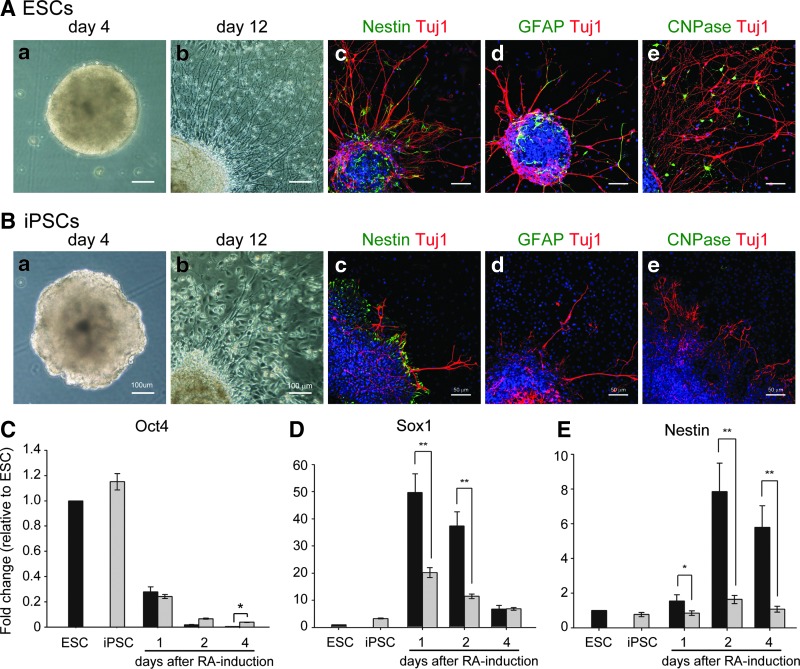
To understand the neural differentiation ability of iPSCs, cells were subjected to differentiation according to the procedure of in vitro neural differentiation. Undifferentiated cells were detached from feeder cells and cultured in the suspension condition with EB medium, to form EBs (Fig. 1Aa, Ba). The EBs from ESCs looked like spherical cell aggregates with a smooth surface (Fig. 1Aa); EBs from iPSCs also formed spherical aggregates, albeit with a rough and irregular surface (Fig. 1Ba). After 4 days of treatment with RA and plating onto cell culture dishes, neurite outgrowth from both types of EBs was observed in adherent cultures (Fig. 1Ab, Bb). Immunocytochemical analyses revealed that Nestin-positive neural-progenitor cells (Fig. 1Ac, Bc), Tuj1-positive neurons (Fig. 1Ac–e, Bc–e), GFAP-positive glial cells (Fig. 1Ad, Bd), and CNPase-positive oligodendrocytes (Fig. 1Ae, Be) were present in differentiated ESCs and iPSCs. Interestingly, the number of Nestin-positive neural-progenitor cells, Tuj1-positive neurons, GFAP-positive glial cells, and CNPase-positive oligodendrocytes derived from ESCs was apparently greater than that of those derived from iPSCs; moreover, the length of Tuj1-positive neurites derived from ESCs was obviously longer than that of those derived from iPSCs (Fig. 1Ac–e, Bc–e). These observations indicate that these two types of stem cells differentiated into neural lineage cells, with some differences.
FIG. 1.
Neural differentiation of ESCs and iPSCs. Both ESCs and iPSCs formed EBs (Aa and Ba). ESC-derived EBs looked like spherical aggregates with a smooth surface (Aa); in contrast, iPSCs formed spherical aggregates with a rough surface (Ba). After plating EBs for 8 days, ESC- and iPSC-derived neurite outgrowth was observed by phase-contrast microscopy (Ab and Bb). Nestin-positive neural progenitors and Tuj1-positive neurons were detected in ESC- and iPSC-differentiated cells by immunocytochemical analysis using confocal microscopy (Nestin, Ac and Bc; Tuj1, Ac–e and Bc–e). GFAP-positive astrocytes and CNPase-positive oligodendrocytes were detected in ESC-differentiated cells (Ad and Ae), but were hardly found in iPSC-differentiated cells, as assessed by immunocytochemical analysis using confocal microscopy (Bd and Be). Nuclear staining with Hoechst 33342 was shown (Ac–e and Bc–e). Oct4 was rapidly downregulated in both ESCs and iPSCs after RA treatment (C). Sox1 expression in ESCs and iPSCs was first increased at day 1 after RA treatment, with expression levels that were markedly lower in iPSCs than they were in ESCs (D). Nestin expression in ESCs and iPSCs was rapidly increased at day 2 after RA treatment, with expression levels that were markedly lower in iPSCs compared with ESCs (E). Tuj1 expression in ESC- and iPSC-derived cells was upregulated after RA treatment, with expression levels that were significantly higher in ESC-derived cells than they were in iPSC-derived cells. Error bars represent the mean±SEM. (*) P<0.05, (**)P<0.01.
To clarify whether there was a difference in early neural differentiation between ESCs and iPSCs, we examined the mRNA expression of a pluripotent gene (Oct4) and of early neural genes (Sox1 and Nestin) by quantitative PCR at days 1, 2, and 4 after RA treatment. Oct4 mRNA expression decreased dramatically in both RA-treated ESCs and iPSCs as assessed by ANOVA test (Fig. 1C). This observation suggests that RA promotes the differentiation of ESCs and iPSCs, with no obvious difference in the Oct4 mRNA expression level between ESCs and iPSCs, which implies that the differentiating program started in both types of cells. Subsequently, we focused on the neural differentiation potency of ESCs and iPSCs after RA treatment. During rapid downregulation of Oct4 expression, upregulation of the Sox1 and Nestin mRNAs was observed in RA-treated cells by quantitative PCR as assessed by ANOVA test (Fig. 1D, E). Sox1 expression in ESCs and iPSCs was first increased at day 1 after RA treatment, and then was decreased gradually after day 2. Although the temporal expression pattern of iPSCs was similar to that of ESCs, the Sox1 mRNA expression level in iPSCs was significantly lower than that observed in ESCs at days 1, 2, and 4 after RA treatment (Fig. 1D). Similar to (but with some differences from) the Sox1 expression pattern, the expression of the Nestin mRNA was upregulated after RA-induced Sox1 expression as assessed by ANOVA test; moreover, the level of expression of Nestin in iPSCs was significantly lower than that observed in ESCs, similar to Sox1 expression (Fig. 1E). These findings suggest that iPSCs possess a weaker neural differentiation potency than do ESCs.
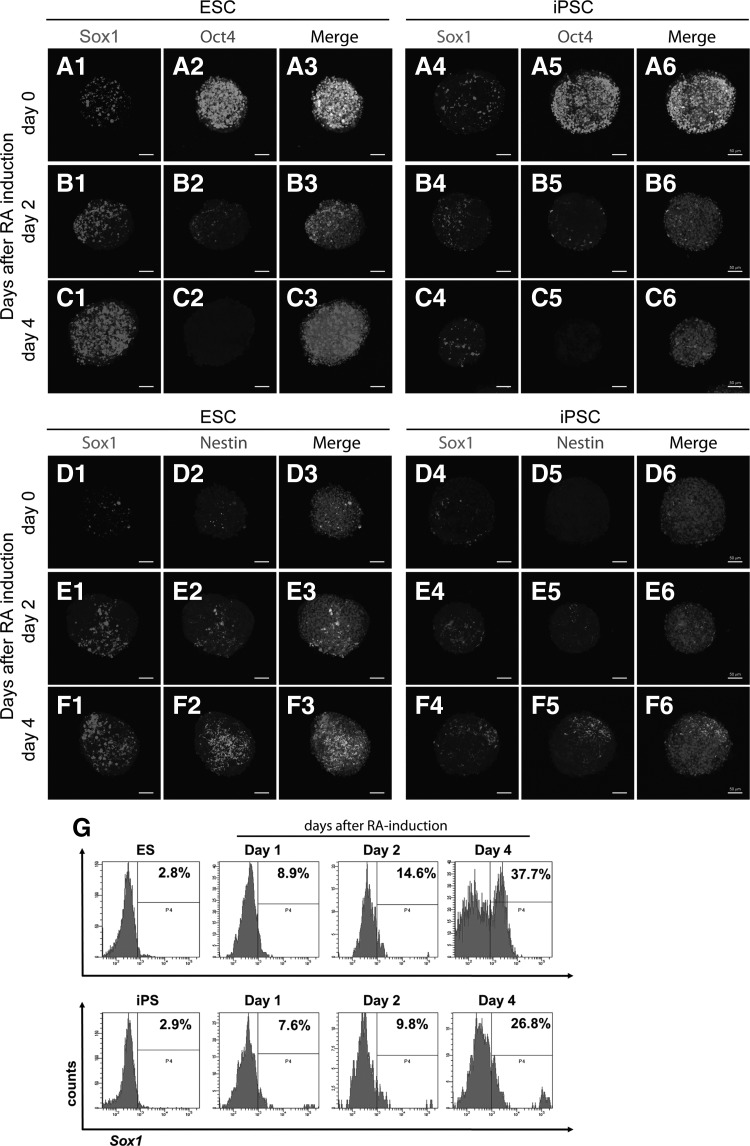
To confirm the temporal protein expression pattern, differentiating ESCs and iPSCs were subjected to examination of Oct4, Sox1, and Nestin expression by immunostaining analysis at days 0, 2, and 4 after RA treatment. Similar to what was found for the RNA expression patterns, Oct4-positive cells were decreased readily after RA treatment in both ESCs and iPSCs as assessed by ANOVA test (Fig. 2A–C). Conversely, Sox1- and Nestin-positive cells were clearly enriched after RA treatment (Sox1, Fig. 2A–F; Nestin, Fig. 2D–F). Similar to the results obtained in the RNA analysis (Fig. 1), the population of Sox1- and Nestin-positive cells was obviously smaller in differentiating iPSCs compared with differentiating ESCs. The low Sox1-positive cell proportion detected in differentiating iPSCs was confirmed by flow cytometric analysis, in which undifferentiated cells were used as the parallel controls (Fig. 2G). Taken together, these results indicate that, although the order and timing of early neural differentiation in iPSCs resembled those observed in ESCs, the expression levels of early neural genes in iPSCs were significantly lower than those detected in ESCs.
FIG. 2.
Detection of the expression of pluripotent and neural markers during early neural differentiation by immunocytochemical analysis. During early neural differentiation, the pluripotent marker Oct4 was rapidly downregulated after RA treatment (A2, A5, B2, B5, C2, and C5). Cells that were positive for the neural markers Nestin and Sox1 were enriched after RA treatment in both ESCs and iPSCs (Nestin, D2, D5, E2, E5, F2, and F5; Sox1, A1, A4, B1, B4, C1, C4, D1, D4, E1, E4, F1, and F4). However, the number of Nestin-positive cells derived from iPSCs (D5, E5, and F5) was obviously lower than that derived from ESCs (D2, E2, and F2), and the number of Sox1-positive cells derived from iPSCs (A4, B4, C4, D4, E4, and F4) was obviously lower than that derived from ESCs (A1, B1, C1, D1, E1, and F1). Nuclei were stained with Hoechst 33342 and shown in merged images. Scale bar, 50 μm. (G) Flow cytometry analysis of RA-treated mouse ESCs and iPSCs revealed that the proportion of Sox1-positive cells increased largely in ESCs and iPSCs, and that the proportion of Sox1-positive cells in ESC cultures was greater than that observed in iPSC cultures. Undifferentiated cells were used as the control.
RA-Fgf/Erk signaling was attenuated in iPSCs
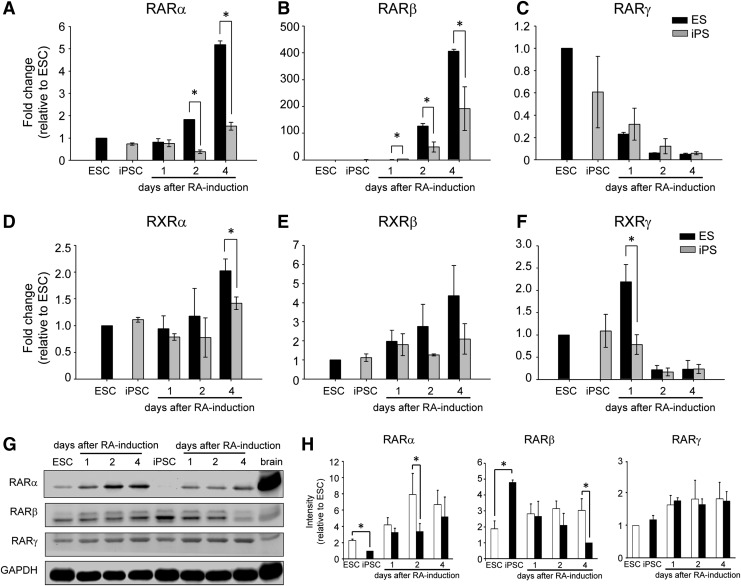
To elucidate the mechanism underlying this difference in neural differentiation potency between iPSCs and ESCs, we first examined the RA signaling pathway. In this signaling pathway, RA located in the extracellular region penetrates the membrane and travels to the nucleus to bind RARs. Subsequently, the complex functions as a transcription activator. Thus, the expression pattern of RARs was first examined by qRT-PCR analysis. Among the RARs and RXRs, the increased patterns of RARα, RARβ, RXRα, and RXRβ expression in ESCs and those of RARα, RARβ, and RXRα in iPSCs were found after RA treatment as assessed by ANOVA test (Fig. 3A, B, D, E). In contrast, the decreased pattern of RARγ and RXRγ expression ESCs and that of RARγ expression in iPSCs were observed during early neural induction (Fig. 3C, F). Notably, at the early stage of RA-induced differentiation, the expression of RARα, RARβ, RXRα, and RXRγ was apparently different between ESCs and iPSCs, and the levels of these RARs in iPSC cultures were markedly lower than those in ESC cultures (Fig. 3A, B, D, F). However, although downregulation of RARγ and upregulation of RXRβ were observed, respectively, there was no significant difference in this regard between ESCs and iPSCs (Fig. 3C, E).
FIG. 3.
Expression of RA nuclear receptors during early neural differentiation Screening of the RNA expression of RARs (RARα, RARβ, and RARγ) and RXRs (RXRα, RXRβ, and RXRγ) by qRT-PCR showed that RARγ and RXRγ exhibited a rapid decrease during early neural induction, whereas RARα, RARβ, RXRα, and RXRβ were upregulated in ESC culture after RA treatment (A–F). Western blot analysis revealed that the expression of RARα was induced after RA treatment in ESCs and iPSCs (G). Using ImageJ software to quantify protein expression, we showed that the level of induction of the RARα protein in ESCs was significantly higher than that detected in iPSCs (H). The error bar represents the mean±SEM. (*) P<0.05.
Because the change of RAR expression levels was apparent in iPSCs and ESCs, the cells were further subjected to western blot analysis to examine the protein expression pattern. Undifferentiated cells and mouse brain were used in parallel as controls. Similar to what was found regarding the RNA and protein expression pattern, the expression of the RARα protein was obviously increased after RA treatment, as assessed by ANOVA test (Fig. 3G, H). Notably, RARα protein was significantly lower than that detected in differentiating ESCs, not only in undifferentiated but also differentiating iPSCs at day 2 after RA treatment (Fig. 3H). The protein expression levels of RARβ and RARγ were not increased or decreased, respectively, which was not consistent to the RNA expression pattern (Fig. 3G, H). These results suggest that the expression of RARs in iPSCs was aberrant, and the expression of RARα was attenuated from the undifferentiated cell stage.
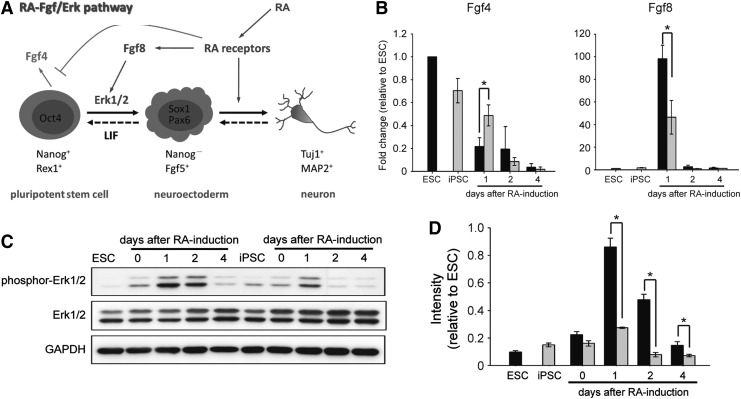
According to previous investigations on RA-induced neural differentiation (Kunath et al., 2007; Li et al., 2006; Lu et al., 2009; Stavridis et al., 2007; Stavridis et al., 2010), fibroblast growth factor (Fgf)/extracellular signal-regulated kinase (Erk) signaling may cooperate with RA signaling and play an important role in promoting the transition from a pluripotent state to neural lineage commitment in RA-induced early neural induction. In the presence of RA, Fgf8-Erk1/2 signaling was activated and Fgf4 signaling was weakened, as shown in Figure 4A. To test whether Fgf8 and Fgf4 signaling were affected in RA-treated ESCs and iPSCs, differentiating cells were subjected to an analysis of the RNA expression of Fgf4 and Fgf8. The results of qRT-PCR showed that Fgf4 was downregulated after RA treatment and that the expression of Fgf8 peaked at day 1 after RA treatment in both ESCs and iPSCs by ANOVA test (Fig. 4B). Further examination of the Fgf8 downstream effector phosphor-Erk1/2 by western blot analysis showed that the phosphor-Erk1/2 protein was readily upregulated after RA treatment and responded faithfully to the expression of Fgf8 (Fig. 4C).
FIG. 4.
Expression of RA-Fgf/Erk signaling during early neural differentiation Schematic diagram of RA-Fgf/Erk signaling in early neural differentiation (A). The screening of the RNA expression of Fgf4 and Fgf8 by qRT-PCR showed that the expression of Fgf4 was readily decreased during the differentiation process and the expression of Fgf8 was increased (as a surge) at day 1 after RA treatment (B). Western blot analysis revealed that phosphor-Erk1/2 was upregulated after RA treatment, and then downregulated in differentiating ESCs and iPSCs (C). Using the ImageJ software to quantify protein expression, we showed that the ratio of phosphor-Erk1/2 intensity to total Erk1/2 intensity was increased to a greater extent in ESCs compared with iPSCs at days 1, 2, and 4 after RA treatment (D). Error bars represent the mean±SEM. (*) P<0.05.
These results indicated that an effect of RA on Fgf4 and Fgf8 signaling was present and functional in these two pluripotent cells. However, the tendency for Fgf4 downregulation in differentiating iPSCs was slower than that observed in differentiating ESCs, and the level of Fgf8 and its downstream effector phosphor-Erk1/2 were obviously lower in iPSCs compared with ESCs (Fig. 4B, C). Taken together, these results suggest that RARs, especially RARα, and its downstream effectors were attenuated in iPSCs.
RARα was a critical cause of impairment of RA signaling in iPSCs
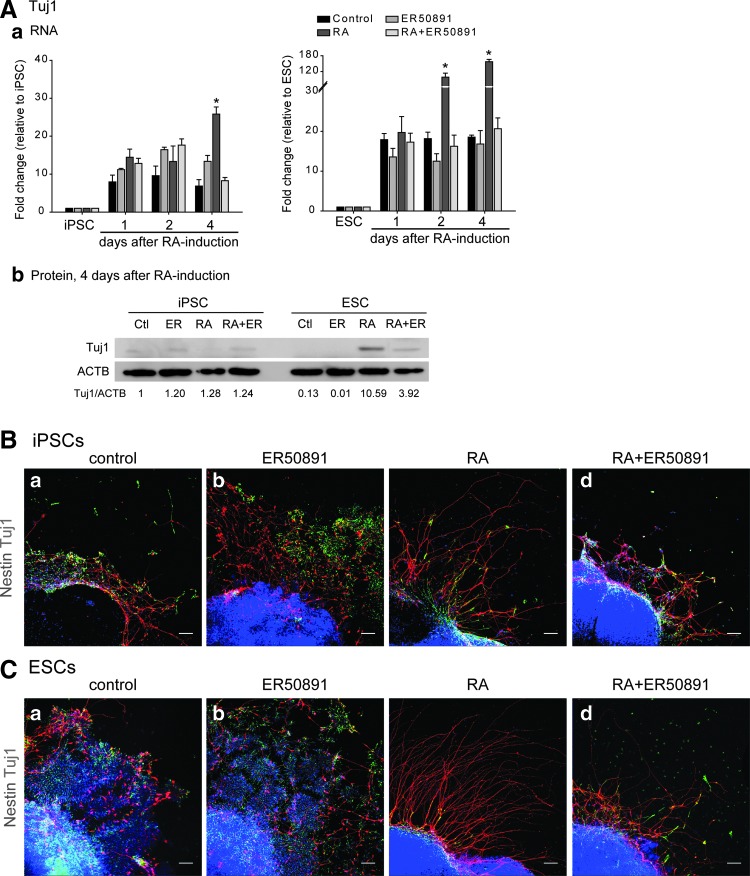
Our screening revealed that the expression of RARs was lower in iPSCs and that downstream SOX1 expression and RA-Fgf/Erk signaling were downregulated, indicating that RARs might be the cause of the weakened RA signaling in iPSCs. Because the expression pattern in RNA analysis and protein analysis of RARα was consistent whereas that of RARβ was not (Fig. 3A, B, G, H), we next examined whether RARα is a critical factor in the impairment of the neural differentiation ability of iPSCs. To this end, we used the RARα antagonist ER50891 to prevent RARα signaling in RA-treated ESCs and iPSCs. To examine whether the antagonist yielded side effects or even cytotoxicity, ESCs and iPSCs were only treated with ER50891 in parallel as the control (Fig. 5A, Bb, Cb). The screening of the expression of the neural differentiation effector Tuj1 by qRT-PCR analysis and western blot analysis showed that this effector was induced by RA treatment in both ESCs and iPSCs (Fig. 5A). However, its expression was delayed and was obviously lower in iPSCs compared with ESCs as assessed by ANOVA test (Fig. 5A).
FIG. 5.
Deficient RARα pathway impaired neural differentiation At day 16, Tuj1 RNA expression was increased after RA treatment in both ESCs and iPSCs, and its expression was downregulated in the presence of the RARα antagonist ER50891, as assessed by qRT-PCR analysis (Aa) and western blot analysis (Ab). Immunocytochemical analysis showed that Nestin- and Tuj1-immunoreactive cells were detected in all conditions of iPSCs (B) and ESCs (C). Long processes were observed easily under RA treatment in iPSCs (Bc) and ESCs (Cc); however, the presence of the RARα antagonist ER50891 caused profound loss of long processes in both ESCs (Bd) and iPSCs (Cd). Cells without RA or ER50891 treatment (Ba and Ca) and with only ER50891 treatment (Bb and Cb) were used as controls. Scale bar, 50 μm. Error bars represent the mean±SEM. (*) P<0.05.
In addition, the presence of the RARα antagonist ER50891 reversed the effect of RA significantly in both ESCs and iPSCs (Fig. 5A). Further examination of Nestin and Tuj1 protein expression by immunostaining showed that the presence of RA induced long Tuj1-positive processes in differentiating ESCs and iPSCs compared with that observed in untreated and ER50891-treated control cells (Fig. 5Ba–c, Ca–c). Moreover, the addition of the RARα antagonist ER50891 to RA treatment inhibited the RA effect significantly in both differentiating ESCs and iPSCs (Fig. 5Bd, Cd); this observation was consistent with the findings obtained in the qRT-PCR analysis (Fig. 5Aa). Taken together, these results suggest that deficient RARα expression was a critical cause of impairment of the neural differentiation ability of iPSCs.
Discussion
Previous studies have reported that iPSCs achieve pluripotency and provide beneficial opportunities in regenerative medicine, with reduced immune responses and ethical issues (Araki et al., 2013; Deyle et al., 2012). However, the differentiation efficiency of iPSCs remains obscure. The aim of the current study was to improve our understanding of the neural differentiation ability of iPSCs. Our results showed that iPSCs were capable of differentiating into neural lineages, including neurons and glial cells, via the known RA induction pathway. However, the efficiency of neural differentiation in iPSCs was significantly lower than that of ESCs. Further screening of molecules involved in the RA pathway revealed that RARα may serve as an important deficient cue in iPSCs that attenuates their neural differentiation efficiency. Our combined observations suggest that iPSCs exhibited a low differentiation efficiency.
In our study and previous investigations, RA was used to induce ESC and iPSC differentiation into neural lineage cells (Fig. 1) (Bain et al., 1995; Fraichard et al., 1995). It has been reported that RA binds to RAR and forms a transcriptional complex with RXR (Bastien and Rochette-Egly, 2004). This RA–RAR–RXR complex recognizes RAREs to modulate downstream gene expression (Maden, 2007; Rhinn and Dolle, 2012). For instance, the RA–RAR–RXR complex binds to the RARE located in the Oct4 promoter to suppress the expression of the Oct4 pluripotent gene (Okazawa et al., 1991; Pikarsky et al., 1994). Conversely, this complex activates downstream neural lineage genes and signaling pathways to promote neural differentiation (Freemantle et al., 2002).
These findings suggest that the presence of RARs and RXRs in cells is important for RA signal pathways. In previous studies, the activation of RARα and RARβ was required for neural differentiation and proliferation in vitro and in vivo (Goncalves et al., 2005; Goncalves et al., 2009), and these molecules were upregulated by RA-induced neural differentiation in ESCs (Lu et al. 2009). Moreover, RARα participated in the differentiation process of neurons and glial cells (Chandran et al., 2003; Goncalves et al., 2005). Consistent with previous studies, we found that RARα was important for the neural differentiation process and that deficiency of RARα expression in iPSCs may be a critical defect that leads to their impaired neural-differentiation ability.
Currently, study on the comparison of human ESCs and iPSCs showed that during neural differentiation both human ESCs and iPSCs followed the same neural developmental time course and genetic regulation program whereas iPSCs showed significantly reduced efficiency and variability between lines (Hu et al., 2010). In line with previous reports, our study found that, although the neural differentiation potency of iPSCs was significantly lower than that of ESCs, both differentiation progressions followed the same temporal program. For example, pluripotent genes were downregulated and neural genes were upregulated (Figs. 1C–E, 2G, 5A, and Fig. S1) (Supplementary Data are available at www.liebertpub.com/cell/). Unlike the human system, the mouse iPSCs we used were germ-line transmission (Okita et al., 2007), i.e., the characteristics of mouse iPSCs were very close to those of ESCs in at least general body formation and gonad environment. However, we still observed similar results as in human studies.
The discrepancy of between ESCs and iPSCs might be a result of several possibilities. One is incomplete reprogramming of iPSC generation. Incomplete reprogamming causes epigenetic unevenness. It has been demonstrated that genomic methylation, which is an epigenetic modification of DNA, in iPSCs might influence gene expression and the propensity of cell fate commitment (Hu et al., 2010; Kim et al., 2010). This property, so-called “epigenetic memory,” in iPSCs is caused by residual DNA methylation of donor cells with incomplete erasure during reprogramming, rather than by the presence of the transgenes, and might explain the lower efficiency of neural differentiation of iPSCs compared with the ESCs observed here. During reprogramming, demethylation is a late phenomenon that probably occurs passively (Mikkelsen et al., 2008), and the origin of somatic cells might affect the efficiency and fidelity of reprogramming to pluripotency (Aoi et al., 2008; Kim et al., 2010; Maherali et al., 2008; Miura et al., 2009).
Interestingly, the differences between iPSCs and ESCs may not be apparent in the pluripotent state and may appear only after differentiation (Kim et al., 2010). Here, although we used germ line–competent iPSCs, we still obtained consistent results, which might have resulted from residual epigenetic markers that are retained in specific loci of the genome and influence the cell differentiation fate (Doi et al., 2009; Kim et al., 2010). Our results may also support this suggestion. Thus, to completely reprogram somatic cells into “ground-state” pluripotent stem cells was important for further application (Silva et al., 2008). Previous studies have suggested several methods for improving reprogramming, such as serial reprogramming steps (Chin et al., 2009; Hanna et al., 2007; Kim et al., 2010; Wernig et al., 2008) or applying demethylating agents (Chiu and Blau, 1985; Eden et al., 1998). In doing so, patient-specific cells might be able to reprogram fully into ground-state pluripotent stem cells without a propensity for differentiation. In this way, iPSCs might be more suitable for drug screening and cell therapy.
Supplementary Material
Acknowledgments
This work was supported by the Stem Cell Priority Grants from the National Science Council, Taiwan (NSC-100-2321-B-002-075 and NSC-101-2321-B-002-038). Facilities were provided by grants from the Ministry of Education, Taiwan, to the Center of Genomic Medicine in National Taiwan University. We would like to thank Dr. You-Tzung Chen for the kind gift of AB1 mouse ESCs and RIKEN, Japan, for the mouse iPSCs.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Aoi T., Yae K., Nakagawa M., Ichisaka T., Okita K., Takahashi K., Chiba T., and Yamanaka S. (2008). Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 321, 699–702 [DOI] [PubMed] [Google Scholar]
- Araki R., Uda M., Hoki Y., Sunayama M., Nakamura M., Ando S., Sugiura M., Ideno H., Shimada A., Nifuji A., and Abe M. (2013). Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature 494, 100–104 [DOI] [PubMed] [Google Scholar]
- Bain G., Kitchens D., Yao M., Huettner J.E., and Gottlieb D.I. (1995). Embryonic stem cells express neuronal properties in vitro. Dev. Biol. 168,342–357 [DOI] [PubMed] [Google Scholar]
- Bastien J., Rochette-Egly C. (2004). Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328, 1–16 [DOI] [PubMed] [Google Scholar]
- Chandran S., Kato H., Gerreli D., Compston A., Svendsen C.N., and Allen N.D. (2003). FGF-dependent generation of oligodendrocytes by a hedgehog-independent pathway. Development 130, 6599–6609 [DOI] [PubMed] [Google Scholar]
- Chin M.H., Mason M.J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J., Khvorostov I., Ott V., Grunstein M., Lavon N., Benvenisty N., Croce C.M., Clark A.T., Baxter T., Pyle A.D., Teitell M.A., Pelegrini M., Plath K., and Lowry W.E. (2009). Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5,111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.P., and Blau H.M. (1985). 5-Azacytidine permits gene activation in a previously noninducible cell type. Cell 40, 417–424 [DOI] [PubMed] [Google Scholar]
- Corcoran J., and Maden M. (1999). Nerve growth factor acts via retinoic acid synthesis to stimulate neurite outgrowth. Nat. Neurosci. 2,307–308 [DOI] [PubMed] [Google Scholar]
- Corcoran J., Shroot B., Pizzey J., and Maden M. (2000). The role of retinoic acid receptors in neurite outgrowth from different populations of embryonic mouse dorsal root ganglia. J. Cell Sci. 113 (Pt 14), 2567–2574 [DOI] [PubMed] [Google Scholar]
- Corcoran J., So P.L., Barber R.D., Vincent K.J., Mazarakis N.D., Mitrophanous K.A., Kingsman S.M., and Maden M. (2002). Retinoic acid receptor beta2 and neurite outgrowth in the adult mouse spinal cord in vitro. J. Cell Sci. 115(Pt 19), 3779–3786 [DOI] [PubMed] [Google Scholar]
- Deyle D.R., Khan I.F., Ren G., Wang P.R, Kho J., Schwarze U., and Russell D.W. (2012). Normal collagen and bone production by gene-targeted human osteogenesis imperfecta iPSCs. Mol. Ther. 20,204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A., Park I.H., Wen B., Murakami P., Aryee M.J., Irizarry R., Herb B., Ladd-Acosta C., Rho J., Loewer S., Miller J., Schlaeger T., Daley G.Q., and Feinberg A.P. (2009). Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41,1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S., Hashimshony T., Keshet I., Cedar H., and Thorne A.W. (1998). DNA methylation models histone acetylation. Nature 394, 842. [DOI] [PubMed] [Google Scholar]
- Evans M.J., and Kaufman M.H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- Fraichard A., Chassande O., Bilbaut G., Dehay C., Savatier P., and Samarut J. (1995). In vitro differentiation of embryonic stem cells into glial cells and functional neurons. J. Cell Sci. 108 (Pt 10), 3181–3188 [DOI] [PubMed] [Google Scholar]
- Freemantle S.J., Kerley J.S., Olsen S.L., Gross R.H., and Spinella M.J. (2002). Developmentally-related candidate retinoic acid target genes regulated early during neuronal differentiation of human embryonal carcinoma. Oncogene 21, 2880–2889 [DOI] [PubMed] [Google Scholar]
- Goncalves M.B., Boyle J., Webber D.J., Hall S., Minger S.L., and Corcoran J.P. (2005). Timing of the retinoid-signalling pathway determines the expression of neuronal markers in neural progenitor cells. Dev. Biol. 278,60–70 [DOI] [PubMed] [Google Scholar]
- Goncalves M.B., Agudo M., Connor S., McMahon S., Minger S.L., Maden M., and Corcoran J.P. (2009). Sequential RARbeta and alpha signalling in vivo can induce adult forebrain neural progenitor cells to differentiate into neurons through Shh and FGF signalling pathways. Dev. Biol. 326,305–313 [DOI] [PubMed] [Google Scholar]
- Hanna J., Wernig M., Markoulaki S., Sun C.W., Meissner A., Cassady J.P., Beard C., Brambrink T., Wu L.C., Townes T.M., and Jaenisch R. (2007). Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318, 1920–1923 [DOI] [PubMed] [Google Scholar]
- Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A. and Zhang S.C. (2010). Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 107,4335–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I., Yabuuchi A., Takeuchi A., Cunniff K.C., Hongguang H., McKinney-Freeman S., Naveiras O., Yoon T.J., Irizarry R.A., Jung N., Seita J. H.anna P. M.urakami R. J.aenisch R. W.eissleder S. H. Orkin I. L. Weissman J., Feinberg A.P., and Daley G.Q. (2010). Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., and Smith A. (2007). FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134, 2895–2902 [DOI] [PubMed] [Google Scholar]
- Li Z., Theus M.H., and Wei L. (2006). Role of ERK 1/2 signaling in neuronal differentiation of cultured embryonic stem cells. Dev. Growth Differ. 48,513–523 [DOI] [PubMed] [Google Scholar]
- Lu J., Tan L., Li P., Gao H., Fang B., Ye S., Geng Z., Zheng P., and Song H. (2009). All-trans retinoic acid promotes neural lineage entry by pluripotent embryonic stem cells via multiple pathways. BMC Cell Biol. 10,57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M. (2007). Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci. 8,755–765 [DOI] [PubMed] [Google Scholar]
- Maherali N., Ahfeldt T., Rigamonti A., Utikal J., Cowan C., and Hochedlinger K. (2008). A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3,340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A.P., and Bradley A. (1990). The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62, 1073–1085 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., and Meissner A. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Okada Y., Aoi T., Okada A., Takahashi K., Okita K., Nakagawa M., Koyanagi M., Tanabe K., Ohnuki M., Ogawa D., Ikeda E., Okano H., and Yamanaka S. (2009). Variation in the safety of induced pluripotent stem cell lines. Nat. Biotechnol. 27,743–745 [DOI] [PubMed] [Google Scholar]
- Okazawa H., Okamoto K., Ishino F., Ishino-Kaneko T., Takeda S., Toyoda Y., Muramatsu M., and Hamada H. (1991). The oct3 gene, a gene for an embryonic transcription factor, is controlled by a retinoic acid repressible enhancer. EMBO J. 10,2997–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., and Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- Pikarsky E., Sharir H., Ben-Shushan E., and Bergman Y. (1994). Retinoic acid represses Oct-3/4 gene expression through several retinoic acid-responsive elements located in the promoter-enhancer region. Mol. Cell. Biol. 14,1026–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M., and Dolle P. (2012). Retinoic acid signalling during development. Development 139, 843–858 [DOI] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., and Smith A. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6,e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavridis M.P., Lunn J.S., Collins B.J., and Storey K.G. (2007). A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development 134, 2889–2894 [DOI] [PubMed] [Google Scholar]
- Stavridis M.P., Collins B.J., and Storey K.G. (2010). Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation. Development 137, 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Wernig M., Lengner C.J., Hanna J., Lodato M.A., Steine E., Foreman R., Staerk J., Markoulaki S., and Jaenisch R. (2008). A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 26,916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.