Abstract
Aims: A recent study conducted in mice reported that liver-specific knockout of tumor suppressor Pten augments nuclear factor (erythroid-derived 2)-like 2 (NRF2) transcriptional activity. Here, we further investigated how phosphatase and tensin homolog deleted on chromosome 10 (PTEN) controls NRF2 and the relevance of this pathway in human carcin ogenesis. Results: Drug and genetic targeting to PTEN and phosphoproteomics approaches indicated that PTEN leads to glycogen synthase kinase-3 (GSK-3)-mediated phosphorylation of NRF2 at residues Ser335 and Ser338 and subsequent beta-transducin repeat containing protein (β-TrCP)-dependent but Kelch-like ECH-associated protein 1 (KEAP1)-independent degradation. Rescue experiments in PTEN-deficient cells and xerographs in athymic mice indicated that loss of PTEN leads to increased NRF2 signature which provides a proliferating and tumorigenic advantage. Tissue microarrays from endometrioid carcinomas showed that 80% of PTEN-negative tumors expressed high levels of NRF2 or its target heme oxygenase-1 (HO-1). Innovation: These results uncover a new mechanism of oncogenic activation of NRF2 by loss of its negative regulation by PTEN/GSK-3/β-TrCP that may be relevant to a large number of tumors, including endometrioid carcinomas. Conclusion: Increased activity of NRF2 due to loss of PTEN is instrumental in human carcinogenesis and represents a novel therapeutic target. Antioxid. Redox Signal. 21, 2498–2514.
Introduction
Nuclear factor (erythroid-derived 2)-like 2 (NRF2, also called NFE2L2), a master regulator of cell tolerance to stress (12, 54), exhibits a dark side insofar as it is frequently activated during tumor promotion and progression (2, 13, 19, 26, 43). In normal cells, NRF2 activity is kept low under homeostatic basal conditions by the E3 ubiquitin ligase adapter Kelch-like ECH-associated protein 1 (KEAP1), which binds to DLG and ETGE motifs in the Neh2 domain of NRF2, leading to its ubiquitination by a Cullin-3/Rbx1 E3 ligase (22). The constitutive activation of NRF2 in lung, breast, head and neck, ovarian, endometrial, and prostate cancers has been connected with somatic mutations in the DLG or ETGE motifs of the NRF2-Neh2 domain or with somatic mutations and epigenetic modification throughout KEAP1 (15, 24, 46, 48, 50, 60) [an updated review is provided in Ref. (26)]. Thus, genetic changes result in abnormally high NRF2 protein levels and expression of the antioxidant response element (ARE)-driven genes that produces increased resistance to oxidative, proteostatic, and inflammatory stresses (54) and metabolic reprogramming (23).
Innovation.
This study uncovers a new mechanism of inhibition of transcription factor nuclear factor (erythroid-derived 2)-like 2 (NRF2) by phosphatase and tensin homolog deleted on chromosome 10 (PTEN), which requires the glycogen synthase kinase-3/beta-transducin repeat-containing protein (GSK-3/β-TrCP) axis and is independent of Kelch-like ECH-associated protein 1 (KEAP1). The clinical relevance of this innovation is demonstrated in endometrioid tumors lacking PTEN. In addition, the study advances the possibility to inhibit NRF2 to prevent tumorigenesis associated with PTEN loss.
However, it is intriguing that somatic mutations in KEAP1 or NRF2 coding genes are found in only 10%–20% of certain tumors, while the NRF2 signature seems to be more frequently upregulated in cancer. This fact suggests that other mechanisms besides loss of NRF2 repression by KEAP1 may contribute to carcinogenesis. We have previously reported that Glycogen Synthase kinase-3 (GSK-3) phosphorylates NRF2 (33, 34). Based on site-directed mutagenesis and two-dimensional (2D) polyacrylamide gel electrophoresis, the sites of phosphorylation were predicted to be located in the sequence DSGIS, but definitive proof by a proteomic approach is lacking. GSK-3 phosphorylation creates a phosphosite in the Neh6 domain that is recognized by SCF/β-TrCP, an E3 ligase adapter which leads to Cullin-1/Rbx1-mediated NRF2 ubiquitination and its subsequent degradation. Since GSK-3 is inhibited by phosphorylation at its N-terminal pseudosubstrate domain (Ser21 in GSK-3α and Ser9 in GSK-3β) by the protein Ser/Thr kinase AKT, it is possible that NRF2 might be upregulated in tumors through activation of AKT and permanent inactivation of GSK-3. Indeed, the participation of phosphatidylinositol 3 kinase (PI3K) and its downstream effector AKT in the activation of NRF2 has been reported in several studies (21, 37), and the ability of PI3K to metabolically reprogram tumor cells through NRF2 activation has also been described (23). Consistent with this, a very recent study has reported that Phosphatase and Tensin Homolog deleted on chromosome 10 (PTEN), which is a phosphatase that counteracts PI3K activity, participates in repression of NRF2 activity (55). In that study, liver-specific disruption of Keap1 and Pten in mice augmented NRF2 activity, leading to cholangiocyte expansion. It is, therefore, essential to determine the mechanistic connection between PTEN and NRF2 and its relevance to human carcinogenesis in those cases where somatic mutations disrupting KEAP1/NRF2 have not been reported.
The suppressor gene PTEN is one of the most commonly mutated genes in human cancers (47, 53). It is both a lipid phosphatase that eliminates phosphatidylinositol 3,4,5-trisphosphate generated by PI3K and a protein phosphatase which self-dephosphorylates at Thr366, and both activities appear to be relevant in invasion (57). A multitude of post-translational modifications such as phosphorylation, ubiquitination, acetylation, and redox modifications fine tune its catalytic activity (10, 49). Thus, PTEN is inhibited by oxidation (17) or S-sulfhydration (8) of the redox-sensitive cysteine Cys124 located at the catalytic center and by S-nitrosylation of the alosteric Cys83 residue (28). In some cancers, PTEN enzymatic activity has been reported to be inactivated through these cysteine alterations (16). Two cancer types in which PTEN presents a high frequency of mutations are prostate and endometrioid carcinoma. In this study, we have characterized the mechanistic connection between PTEN and NRF2 in human tumor cell lines of both origins and have further explored the impact of this connection in tissue microarrays (TMA) of endometrioid carcinomas, where inactivating PTEN mutations occur in 61% and its expression is severely reduced in 97% of cases (25, 35, 56). Our results uncover a general mechanism of participation of NRF2 in carcinogenesis for the vast majority of human tumors, where PTEN function is lost.
Results
The antioxidant tert-butylhydroquinone regulates PI3K/AKT/GSK-3β
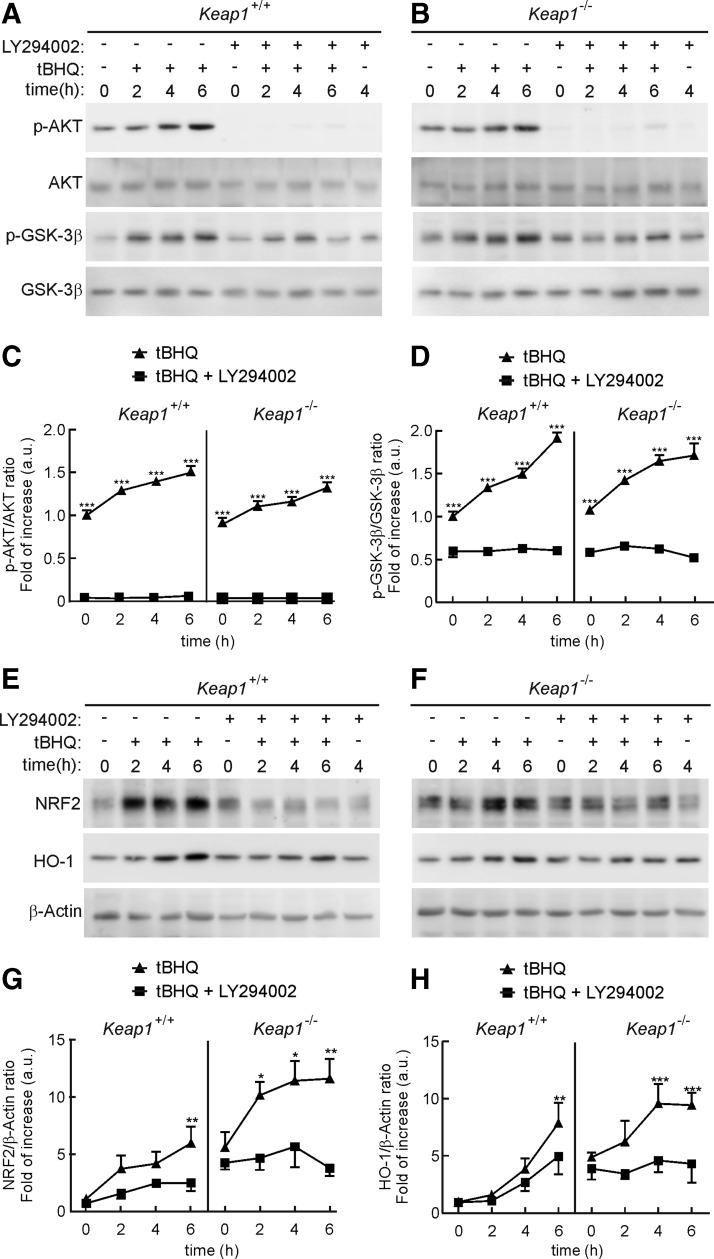
Tert-butylhydroquinone (tBHQ) is widely used to activate NRF2 through inhibition of KEAP1, but we challenged this assumption under conditions of KEAP1 deficiency. Mouse embryonic fibroblasts (MEFs) from wild-type (Keap1+/+) or KEAP1-knockout (Keap1−/−) mice were treated with 10 μM tBHQ for the times indicated in Figure 1A–D. Treatment with tBHQ produced a two-fold increase in the levels of active AKT (phospho-AKTSer473) and inactive GSK-3β (phospho-GSK-3βSer9) proteins in both MEF cell lines. This effect was totally abrogated when cells were pretreated with the PI3K inhibitor LY294002 (30 μM, 15 min). In parallel, as shown in Figure 1E–H, tBHQ increased NRF2 protein abundance not only in wild-type MEFs, as expected, but also in KEAP1-deficient MEFs. Moreover, at 6 h after the addition of tBHQ, both MEF lines displayed higher levels of the prototypic ARE-regulated gene product heme oxygenase-1 (HO-1). Pretreatment with the PI3K inhibitor LY294002 (30 μM, 15 min) prevented the tBHQ-induced increase in NRF2 protein levels and HO-1 in both wild-type and mutant MEFs. Therefore, our results provide evidence of a KEAP1-independent mechanism by which this compound induces NRF2-target genes that correlate with activation of the PI3K/AKT pathway and inhibition of GSK-3β.
FIG. 1.
tBHQ regulates AKT/GSK-3β activity and increases NRF2 and HO-1 in a KEAP1-independent manner. MEFs from wild-type (Keap1+/+) or KEAP1-deficient (Keap1−/−) littermates were maintained in low-serum for 16 h, and then treated with LY294002 (30 μM) or DMSO as vehicle for 15 min before the tBHQ treatment (10 μM). (A, B) Immunoblots with specific antibodies as indicated in the panels. (C, D) Densitometric analysis of p-AKT and p-GSK-3β protein levels of representative blots from (A, B). (E, F) Immunoblots with specific antibodies as indicated in the panels. (G, H) Densitometric analysis of NRF2 and HO-1 protein levels from representative blots from (E, F). For (C, D) and (G, H), data are mean±SEM (n=3). Statistical analysis was performed with two-way ANOVA followed by Bonferroni post-hoc test. *p<0.05, **p<0.01, and ***p<0.001 versus the group at 0 min. ANOVA, analysis of variance; GSK-3, glycogen synthase kinase-3; KEAP1, Kelch-like ECH-associated protein 1; MEFs, mouse embryonic fibroblasts; NRF2, nuclear factor (erythroid-derived 2)-like 2; tBHQ, tert-buthylhydroquinone.
PTEN inhibition increases NRF2 protein levels and activates the expression of its target genes
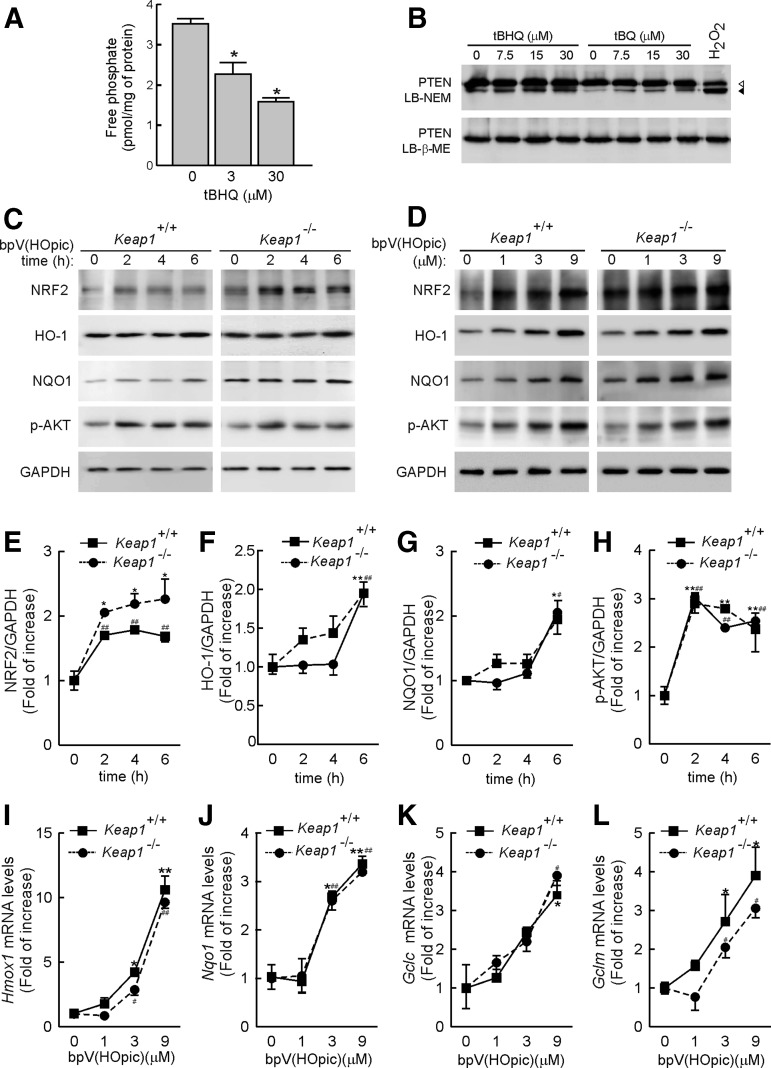
We hypothesized that tBHQ might activate NRF2 by inhibiting PTEN, which negatively controls the PI3K/AKT pathway, and, thus, indirectly activates GSK-3. tBHQ, and its oxidized form tert-butylbenzoquinone (tBQ), stimulated an oxidative band shift of PTEN in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), consistent with its oxidation (6, 17, 20), that correlated with inhibition of its catalytic activity (Fig. 2A, B). As a more selective approach, Keap1+/+ and Keap1−/− MEFs were incubated with the PTEN inhibitor dipotassium bisperoxo (5-hydroxypyridine-2-carboxyl) oxovanadate [bpV(HOpic), 3 μM] for the times indicated in Figure 2C and E–H; loss of PTEN activity was confirmed by demonstrating that bpV(HOpic) increased phospho-AKTSer473 levels. We found that inhibition of PTEN for 2 h by bpV(HOpic) led to an increase in NRF2 protein levels in both Keap1+/+ and Keap1−/− MEFs, which lasted for at least 6 h. In addition, HO-1 and NAD(P)H quinone oxidoreductase 1 (NQO1) protein levels were also increased after a 6 h incubation with bpV(HOpic). This effect was more noticeable in a dose-response curve after a 6 h incubation with bpV(HOpic) (Fig. 2D). Further, as shown in Figure 2I–L, inhibition of PTEN for 6 h resulted in a 2- to 30-fold induction of the NRF2-regulated genes Hmox1 (encoding HO-1), Nqo1, glutamate-cysteine ligase catalytic (Gclc), and glutamate-cysteine ligase modifier (Gclm). Overall, these results indicate that PTEN inhibits NRF2 function in a KEAP1-independent manner.
FIG. 2.
Pharmacological inhibition of PTEN induces the NRF2 signature. (A) In vitro phosphatase assay with immunocomplexes from HEK293T cells transfected with HA-tagged PTEN. After 24 h from transfection, HA-PTEN was immunoprecipitated using anti-HA antibody. Inmunocomplexes were treated in vitro with the indicated concentrations of tBHQ or vehicle (DMSO, 0.01%) and then submitted to an in vitro phosphatase assay. Graph depicts PTEN activity as free phosphate measured by malachite green assay. Data indicate mean±SEM (n=3). Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test. *p<0.05 versus the vehicle-treated group. (B) tBHQ and tBQ induce oxidant modification of PTEN. HEK293T cells were treated for 2 h with the indicated concentrations of tBHQ, tBQ, or 2 mM hydrogen peroxide as control. Cell extracts were prepared in lysis buffer containing 50 mM N-ethylmaleimide (LB-NEM) or 2 mM β-mecaptoethanol (LB-β-ME) and resolved in 10% SDS-PAGE. Empty and filled arrowheads point the respective reduced and the oxidized forms of PTEN after treatments. (C) Time-dependent effect of PTEN inhibition on the NRF2 signature. Keap1+/+ and Keap1−/− MEFs were treated with 3 μM bpV(HOpic) for the indicated time points. Whole-protein lysates were then immunoblotted with specific antibodies as indicated in the panels. (D) Dose-dependent effect of PTEN inhibition on NRF2 signature. Keap1+/+ and Keap1−/− MEFs were treated with the indicated concentrations of bpV(HOpic) for 6 h. (E–H) Densitometric analysis of NRF2, HO-1, NQO1, and p-AKT protein levels of representative blots from (C). (I–L), Keap1+/+ and Keap1−/− MEFs were treated for 6 h with the indicated doses of bpV(HOpic). The mRNA levels of Hmox1, Nqo1, Gclc, and Gclm were determined by qRT-PCR and normalized by β-Actin levels. Data are mean±SEM (n=3). Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test. *p<0.05; **p<0.01 points Keap1+/+ MEFs versus groups at 0 min. #p<0.05; ##p<0.01 points Keap1−/− MEFs versus groups at 0 min. bpV(HOpic), dipotassium bisperoxo (5-hydroxypyridine-2-carboxyl) oxovanadate; NQO1, NAD(P)H quinone oxidoreductase 1; PTEN, phosphatase and tensin homolog deleted on chromosome 10; qRT-PCR, quantitative reverse transcriptase-polymerase chain reaction; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; tBQ, tert-butylbenzoquinone.
PTEN activation impairs NRF2 activity
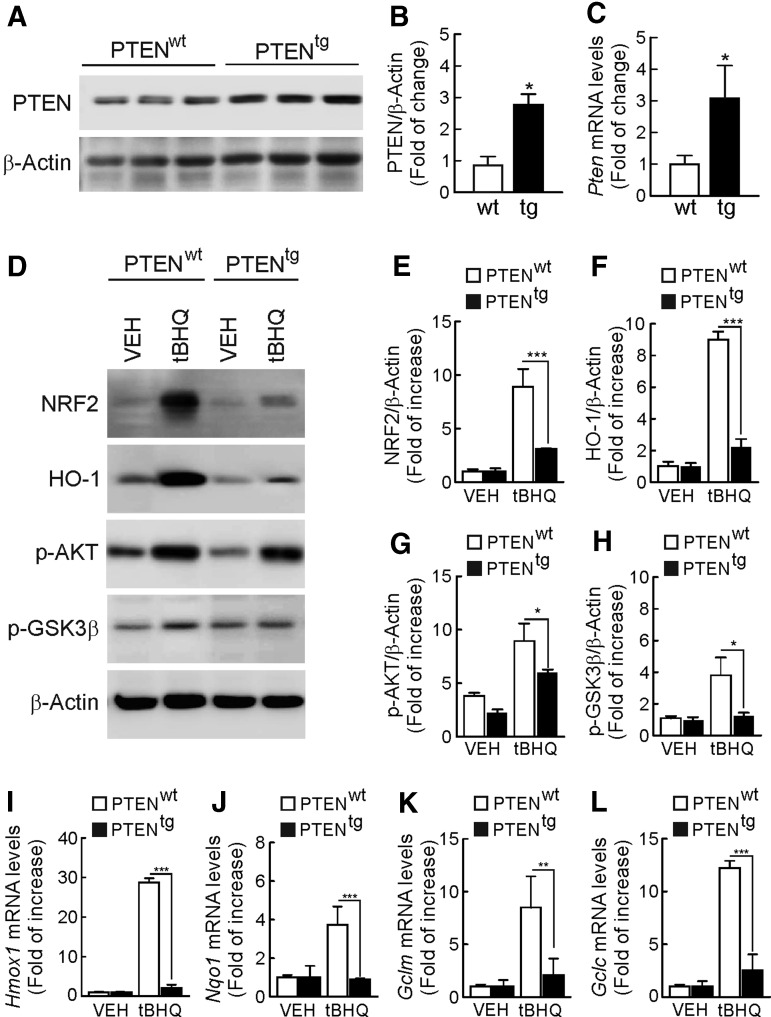
To gain further support for the results obtained with bpV(HOpic), we used MEFs from transgenic mice that overexpress PTEN (PTENtg) by ∼3-fold relative to control littermates (PTENwt) (30) (Fig. 3A–C). The PTENtg MEFs exhibited impaired PI3K/AKT activity under basal conditions or on treatment with tBHQ (Fig. 3D, G–H). Thus, tBHQ (10 μM, 6 h) increased NRF2 and HO-1 protein levels by 10-fold in PTENwt MEFs, while this induction was drastically reduced in PTENtg MEFs (Fig. 3D–F). In addition, Figure 3I–L shows that expression of the NRF2-regulated genes Hmox1, Nqo1, Gclc, and Gclm was significantly impaired in PTENtg MEFs.
FIG. 3.
MEFs from PTENtg mice exhibit impaired activation of NRF2 in response to tBHQ. (A) PTEN protein levels in MEFs from PTENwt and PTENtg transgenic mice. Each lane corresponds to MEFs prepared from different mice. (B) Densitometric quantification of representative blots from (A). (C) Determination of PTEN mRNA levels by qRT-PCR. Data are mean±SEM (n=3). Statistical analysis was performed with a Student's t-test. *p<0.05 versus PTENwt MEFs. (D) Immunoblots of PTENwt and PTENtg MEFs treated with 10 μM of tBHQ for 6 h. (E–H) densitometric quantification of NRF2, HO-1, p-AKT, and p-GSK-3β protein levels of representative blots from (D). (I–L) mRNA levels of Hmox1, Nqo1, Gclc, and Gclm were determined by qRT-PCR and normalized by β-Actin levels. Data are mean±SEM (n=3). Statistical analysis was performed with two-way ANOVA followed by Bonferroni post-hoc test. *p<0.05; **p<0.01; and ***p<0.001 versus PTENwt MEFs.
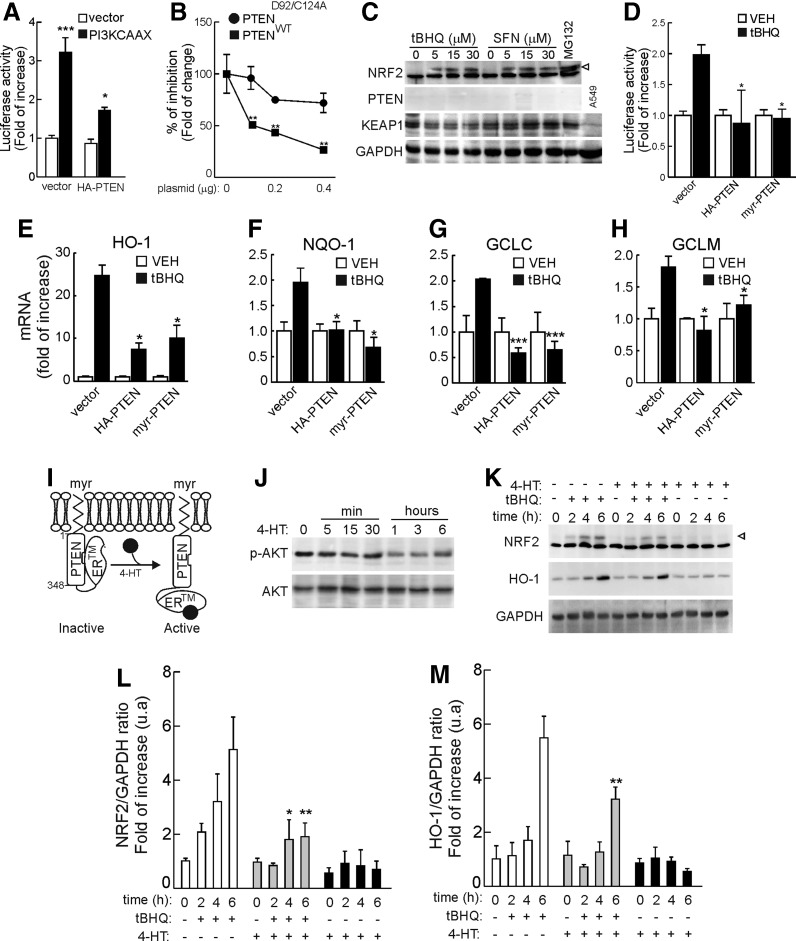
To further analyze the impact that PTEN makes on NRF2 activity without the interference of the endogenous phosphatase, we performed complementation experiments in the PTEN-null human prostate carcinoma cell line PC-3. These tumor cells exhibit KEAP1 activity (27, 59, 60), and we have established that they express KEAP1 and NRF2 and that they are responsive to the KEAP1 modulators tBHQ and sulforaphane (Fig. 4A). First, we conducted ARE-driven luciferase reporter assays in PC-3 cells in which PTEN activity was rescued with ectopically expressed versions of the phosphatase. In Figure 4B, PC-3 cells were transfected with plasmids for the NRF2 firefly luciferase reporter ARE-LUC, pTK-Renilla luciferase (as an internal control), an active version of p110αPI3K (PI3K-CAAX), and either empty vector or expression vector for wild-type HA-PTEN. Under these conditions, PI3K upregulated the ARE-LUC reporter activity while HA-PTEN attenuated this effect. In addition, in Figure 4C, we found that the PI3K-CAAX-mediated activation of ARE-LUC was dose dependently diminished by expression of wild-type PTEN but not by the inactive mutant HA-PTEND92A/C124A. Similarly, in Figure 4D, we found that the ability of tBHQ to induce ARE-LUC reporter activity was prevented by expression of wild-type PTEN or a membrane-anchored, active, myristoylated version of PTEN (myr-PTEN). These results, showing that ectopic expression of active PTEN inhibits ARE-driven luciferase activity, complement those obtained with MEFs from PTENtg mice.
FIG. 4.
Rescue of PTEN expression restrains NRF2 activation. (A) The KEAP1/NRF2 axis is functional in prostate carcinoma PC-3 cells. PC-3 cells were maintained in low-serum medium for 16 h and then submitted to tBHQ, sulforaphane (SFN), or MG132 (30 μM) as control, for 6 h. Whole-protein lysates were then immunoblotted with specific antibodies as indicated in the panels. As a negative control for KEAP1 expression, we introduced one lane from human A549 cells that are deficient in KEAP1. The anti-human NRF2 antibody recognizes a strong unspecific band just below NRF2. The arrowhead points to the NRF2 band (apparent MW of 110 kDa). (B) PC-3 cells were co-transfected with ARE-LUC, pTK-Renilla as control vector, and PI3K-CAAX plus either empty vector or expression vector for wild-type HA-PTEN. (C) PC-3 cells were transfected as in (B) but with increasing amounts of HA-PTEND92A/C124A or wild-type HA-PTEN, as indicated. For (B, C), after 24 h, luciferase activity was determined. Data are mean±SEM (n=3). Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test. **p<0.01 versus groups at the same concentration points. (D) PC-3 cells were co-transfected with ARE-LUC, pTK-Renilla as control vector, and either wild-type HA-PTEN or active myr-PTEN. After transfection, cells were maintained in low-serum conditions supplemented with 3 μM tBHQ. After 16 h, luciferase activity was determined. Data are mean±SEM (n=6). Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test. *p<0.05 versus PC-3-pcDNA3.1 treated with tBHQ. (E–H) PC-3 cells were transiently transfected with empty vector (pcDNA3.1), wild-type HA-PTEN, or myr-PTEN. After 16 h in low-serum medium, mRNA levels of HMOX1, NQO1, GCLC, and GCLM were determined by qRT-PCR. Data are mean±SEM (n=3). Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test. *p<0.05; ***p<0.001 versus PC-3-pcDNA3.1 treated with tBHQ. (I) scheme showing the regulation of myr-PTEN-ER* by 4-HT. (J) induction of myr-PTEN-ER* activity by 1 μM 4-HT as determined by time-dependent reduction of p-AKT levels. (K) PC-3 cells were transiently transfected with myr-PTEN-ER*. After 16 h, cells were preincubated with 4-HT for 1 h and then submitted to tBHQ (15 μM) for the indicated time points. Arrowhead points to the NRF2 band (apparent MW of 110 kDa). (L, M) Densitometric quantification of NRF2 and HO-1 protein levels in representative blots from (K). Data are mean±SEM. Statistical analysis was performed with two-way ANOVA followed by Bonferroni post-hoc test. *p<0.05; **p<0.01 versus tBHQ-treated PC3 cells. 4-HT, 4-hydroxytamoxifen; ARE, antioxidant response element; PI3K, phosphatidylinositol 3 kinase; MW, molecular weight.
We next examined the expression of endogenous NRF2-target genes in PC-3 cells under basal conditions and after rescue with expression vectors for wild-type HA-PTEN or myr-PTEN. After 16 h in low-serum medium (1% fetal bovine serum), PC-3 transfected cells were treated with 10 μM tBHQ for 6 h (Figure 4E–H). Induction by tBHQ of Hmox1, Nqo1, Gclc, and Gclm was greatly attenuated in cells expressing wild type or myr-PTEN. In additional experiments, we generated a conditionally active chimera, myr-PTEN-ER* (Fig. 4I) in which the C-terminus of myr-PTEN was replaced with a mutated version of the hormone-binding domain from the estrogen receptor (residues 281 to 599) that is sensitive to 4-hydroxytamoxifen (4-HT). This fusion protein is expressed in an inactive form and becomes activated in the presence of 4-HT, thus enabling chemical genetic activation of PTEN. Figure 4J shows a time-dependent activation of myr-PTEN-ER* by addition of 1 μM 4-HT as determined by progressive loss of p-AKT. Interestingly, as shown in Figure 4K–M, when PC-3 cells were transfected with myr-PTEN-ER* and stimulated with 4-HT (1 μM), the tBHQ induction of NRF2 and HO-1 protein levels decreased gradually while 4-HT had a negligible effect in the absence of this construct. Taken together, these results indicate that PTEN inhibits NRF2 function.
PTEN targets the Neh6 domain of NRF2
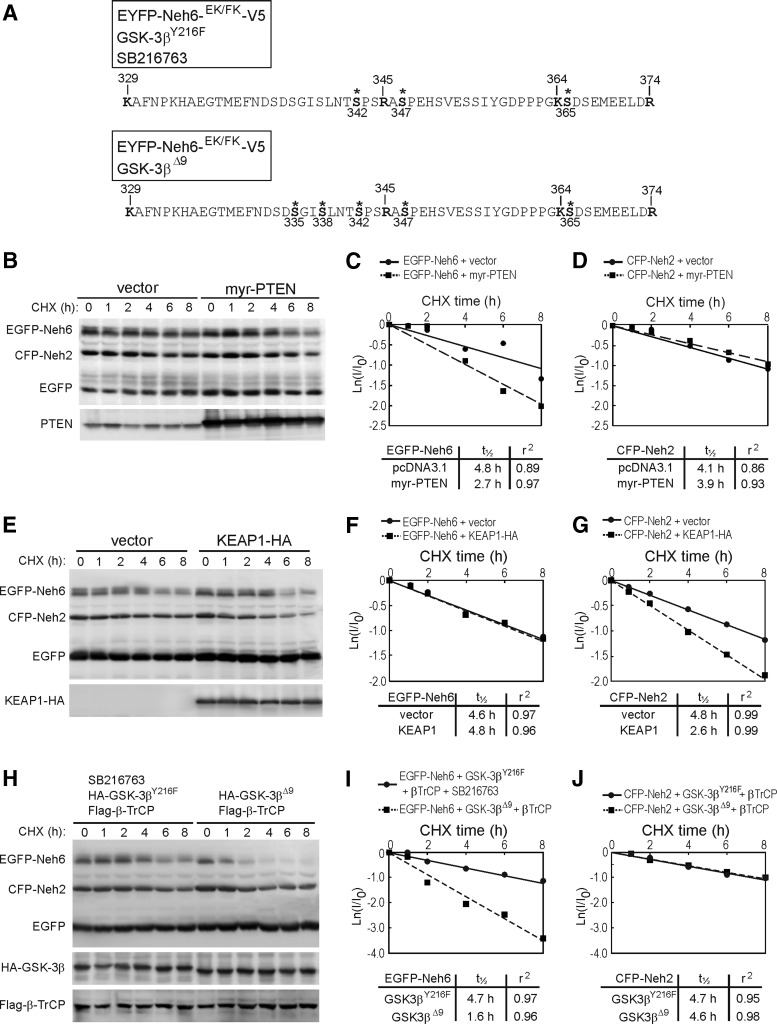
We have previously predicted that GSK-3β phosphorylates two Ser residues within the Neh6 domain of NRF2 (33, 34), but we still lack a formal proof based on phosphoproteomics analysis. We generated a fusion protein comprising enhanced yellow fluorescent protein (EYFP) and residues 317 to 372 of mouse NRF2 (EYFP-mNRF2(317–372)-V5) that contained the putative sites for GSK-3 phosphorylation. In addition, we introduced two point mutations, E329K and F364K (EYFP-Neh6-EK/FK-V5), to enable smaller peptides to be generated after trypsin digestion, thereby facilitating the phosphoproteomics study. HEK293T cells were transfected with EYFP-Neh6-EK/FK-V5 under conditions where GSK-3 activity was modulated. Inactivation of GSK-3 was achieved by co-transfection with a hypomorphic version of GSK-3β that retains only residual activity (GSK-3βY216F) and incubation throughout the experiment with the GSK-3 inhibitor SB216763 (10 μM). Activation of GSK-3 was achieved by co-transfection with active GSK-3βΔ9, lacking the first 9 N-terminal residues which prevented pseudosubstrate-negative regulation. After exposure to these forms of GSK-3β, the EYFP-Neh6-EK/FK-V5 protein was resolved by SDS-PAGE, gel slices were trypsinized, and the digests were subjected to mass spectrometry analysis using liquid chromatography tandem mass spectrometry (LC-MS/MS). As shown in Figure 5A, Ser342 and Ser347 were phosphorylated regardless of GSK-3β activity, as expected from our previous work (33, 34), and we identified an additional phosphosite at Ser365. However, when cells were co-transfected with EYFP-Neh6-EK/FK-V5 and GSK-3βΔ9, we identified two additional phosphoresidues, Ser335 and Ser338. For details, see Supplementary Tables S3–S12 (Supplementary Data are available online at www.liebertpub.com/ars). These results confirm by phosphoproteomics our previous predictions based on side-directed mutagenesis and 2D-polyacrylamide gel electrophoresis (33, 34).
FIG. 5.
PTEN targets the Neh6 degradation domain of NRF2. (A) GSK-3β phosphorylates NRF2 at Ser335 and Ser338. HEK293T were transfected with fusion protein EYFP-Neh6EK/FK-V5 under conditions of GSK-3 inhibition (co-transfection of hypomorphic GSK-3βY216 mutant plus incubation with 10 μM SB216763) and activation (co-transfection of active GSK-3βΔ9). MG132 (30 μM) was added to prevent protein degradation. Mass spectrometry analysis identified five phosphorylated Ser residues at positions 335, 338, 342, 347, and 365 (indicated by *) but phosphorylation of Ser335 and Ser338 was found only under conditions of GSK-3 stimulation. Annotated mass spectra of the identified phosphorylated peptides are provided in Supplementary Tables S3–S12. (B) PTEN induces the degradation of NRF2 at the level of its Neh6 phosphodegron. HEK293T cells were co-transfected with expression vectors for CFP-Neh2, EGFP-Neh6, EGFP, and either empty vector or myr-PTEN. Then, cells were maintained in low-serum for 16 h and pulse-chased with 100 μg/ml CHX. Whole-protein lysates were immunoblotted with anti-GFP antibody (upper blot) or anti-PTEN antibody (lower blot) showing over-expression of myr-PTEN. (C, D) Determination of half lives. Graphs depict the natural logarithm of the relative protein levels of the indicated proteins as a function of CHX chase time. Protein half life was determined using the linear part of the degradation curves. (E) CFP-Neh2 but not EGFP-Neh6 is modulated by KEAP1. HEK293T cells were co-transfected with expression vectors for CFP-Neh2, EGFP-Neh6, and EGFP and either vector or HA-tagged KEAP-1. Then, the cells were maintained in low-serum medium for 16 h and pulse-chased with 100 μg/ml CHX at the indicated time points. (F, G) Determination of half lives. (H) EGFP-Neh6 but not CFP-Neh2 is modulated by GSK-3β/β-TrCP. HEK293T cells were co-transfected with expression vectors for CFP-Neh2, EGFP-Neh6, EGFP, Flag-β-TrCP, and either HA-tagged GSK-3βY216F (hypomorphic version of GSK-3β that retains only residual activity, plus incubation in 10 μM SB216763) or GSK-3βΔ9 (constitutively active version) and pulse-chased with CHX. (I, J) Determination of half lives. β-TrCP, beta-transducin repeat containing protein; CFP, cyan fluorescent protein; CHX, cycloheximide; EGFP, enhanced green fluorescent protein; EYFP, enhanced yellow fluorescent protein.
These phosphoserines create a binding site for beta-transducin repeat-containing protein (β-TrCP) in the Neh6 domain of NRF2, and we, therefore, sought to determine whether PTEN could contribute to this inhibitory pathway. We constructed chimeric proteins made of cyan fluorescent protein (CFP) fused to the Neh2 (CFP-Neh2) and enhanced green fluorescence protein (EGFP) fused to Neh6 (EGFP-Neh6) degrons, which are regulated by KEAP1 or GSK-3β/β-TrCP, respectively. In Figure 5E–J, we show that the half lives of the CFP-Neh2 and EGFP-Neh6 chimeric proteins are diminished by the co-expression of KEAP1 and GSK-3β/β-TrCP, validating these proteins for selective regulation by each respective pathway. HEK293T cells were co-transfected with expression vectors for CFP-Neh2, EGFP-Neh6, and EGFP as internal control. In addition, cells were co-transfected with empty plasmid or myr-PTEN. Then, the cells were pulse-chased with cycloheximide (CHX, 100 μg/ml) for the indicated times. As shown in Figure 4B–D, EGFP was stable for the entire length of the experiment, whereas EGFP-Neh6 and CFP-Neh2 had shorter half lives of about 4.8 and 4.1 h, respectively. However, in the presence of myr-PTEN, only the EGFP-Neh6 turn-over was accelerated, with a half life of about 2.7 h. These results indicate that PTEN targets the Neh6 degron of NRF2.
A dominant-negative NRF2 mutant inhibits growth of PTEN-deficient tumor cells
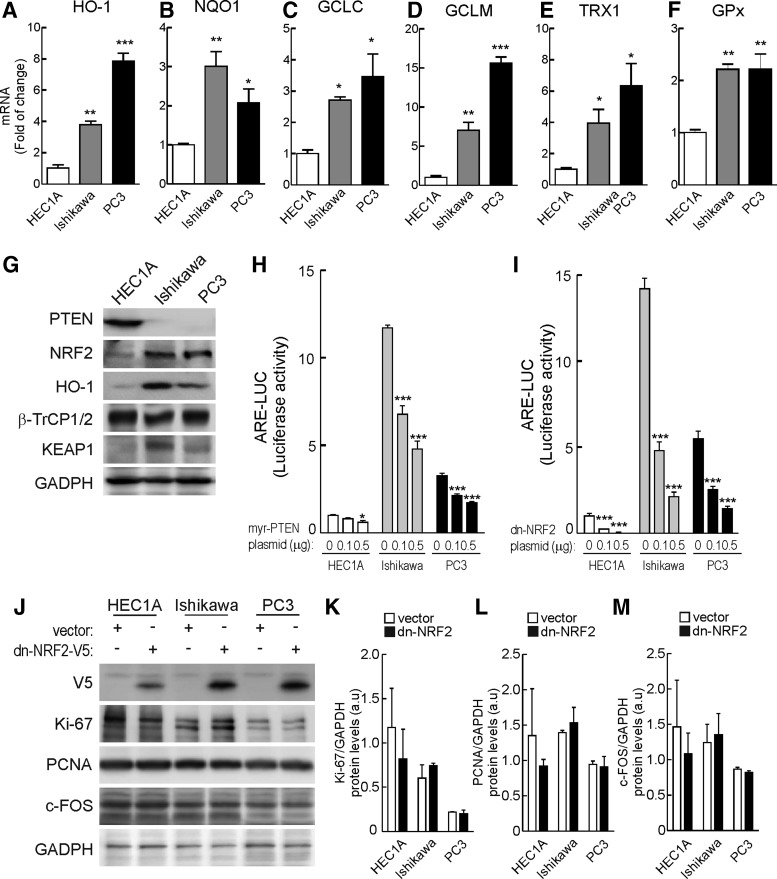
To study the balance between PTEN and NRF2 activity in tumorigenesis, first we characterized the NRF2 signatures in three tumor cell lines: HEC1A and Ishikawa are endometrioid carcinoma cell lines that express and lack PTEN, respectively, and PC-3 is the prostate cancer cell line which lacks PTEN. As shown in Figure 6A–F, the two cell lines lacking PTEN, Ishikawa and PC-3, exhibited increased messenger RNA levels of six NRF2-regulated genes coding HO-1, NQO1, GCLC, GCLM, thioredoxin reductase 1 (TRX1), and glutathione peroxidase (GPX). At the protein level (Fig. 6G), the cell lines lacking PTEN exhibited increased NRF2 and HO-1 while β-TrCP levels were roughly similar. KEAP1 levels were higher in the PTEN-deficient cells, consistent with the reported observation that the promoter of KEAP1 contains NRF2-regulated AREs (3). Moreover, in the three cell lines, but more evident in the PTEN-deficient ones, transfection of active myr-PTEN or a dominant-negative form of NRF2 that lacks the transactivation domain (dn-NRF2; residues 408 to 597 in the mouse sequence) dose dependently reduced the activity of an ARE-luciferase reporter (Fig. 6H, I). For the experiments related to cell proliferation and tumorigenesis described later on, it was important to determine whether dn-NRF2 would affect proliferation of these tumor cell lines. We analyzed protein levels of three markers of proliferation, namely Ki-67, PCNA, and c-FOS. As shown in Figure 6J–M, they were similar at 16 h after transfection of dn-NRF2. Considering that PC-3 cells have been previously used in the context of electrophile regulation of KEAP1 (Fig. 4) (27, 59) and that sequence analysis demonstrated their KEAP1 is no mutated (60), we decided to use this cell line for future experiments.
FIG. 6.
PTEN deficiency increases the NRF2-signature. (A) After 16 h in low-serum medium, whole-cell lysates from HEC1A (endometrial tumor cell line expressing wild-type PTEN), Ishikawa (endometrial tumor cell line null for PTEN expression), or PC-3 (prostate tumor cell line null for PTEN expression) were immunoblotted with the indicated antibodies. (B–G) mRNA levels of HMOX1, NQO1, GCLC, GCLM, TRX1, and GPX were determined by qRT-PCR. Data are mean±SEM (n=3). Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test. *p<0.05; **p<0.01; and ***p<0.001 versus HEC1A cells. (H, I) HEC1A, Ishikawa, and PC-3 cells were co-transfected with ARE-LUC, pTK-Renilla as control vector, and increasing amounts of myr-PTEN or dn-NRF2 as indicated. After 24 h, luciferase activity was determined. Data are mean±SEM (n=3). Statistical analyses were performed with two-way ANOVA followed by Bonferroni post-hoc test. *p<0.05; **p<0.01; ***and p<0.001 versus control cells. (J) HEC1A, Ishikawa, and PC-3 were transfected with dn-NRF2-V5 plasmid. After 24 h, whole-cell lysates were immunoblotted with specific antibodies as indicated in the panels. (K–M) Densitometric quantification of Ki-67, PCNA, and c-FOS protein levels, respectively, of representative blots from (J). Data are mean±SEM. TRX1, thioredoxin reductase 1.
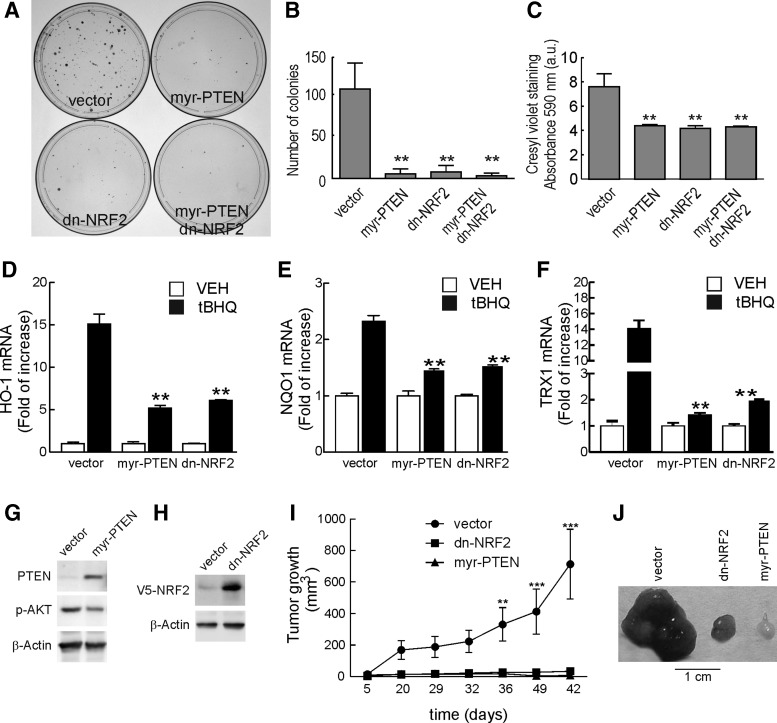
Then, we performed colony-forming assays using PC-3 cells that had been transfected with expression plasmids for myr-PTEN, dn-NRF2, or the empty vector (pcDNA 3.1) as control. Twenty-four hours after transfection, the PC-3 cells were cultured for 4 weeks in the presence of 0.5 mg/ml G418. As shown in Figure 7A, overexpression of either myr-PTEN or dn-NRF2 or both together strongly reduced the appearance of crystal violet-stained colonies when compared with control plates, as assessed by number (Fig. 7B) or optical absorbance (Fig. 7C). These results indicate that not only the proliferative activity of PC-3 cells is tightly dependent on the loss of the tumor suppressor PTEN but also that dn-NRF2 abolishes that proliferative advantage. Compared with the results of short-term transfection of dn-NRF2 (Fig. 6J–M), these results suggest that dn-NRF2 is not toxic by itself but sensitizes against the high proliferation rate which is expected in PTEN-deficient tumor cells. A similar idea has been previously suggested for KRAS-, BRAF-, and MYC-transformed cells (4).
FIG. 7.
NRF2 inhibition or restoration of PTEN expression reduces the tumorigenic capacity of PC-3 cells. (A) PC-3 cells were transfected with pcDNA3.1 (vector), myr-PTEN, or dn-NRF2-V5, as indicated. After 24 h from transfection, cells were treated for 4 weeks with 0.5 mg/ml of G418. Then, plates were fixed and stained with cresyl violet solution. The plates shown are representative of three different experiments. (B) Number of stained colonies. (C) Densitometric quantification of crystal violet staining at λ=590 nm. Data are mean±SEM (n=3). Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test. **p<0.01 versus PC-3-pcDNA3.1. (D–F) Characterization of NRF2 signature in PC-3 stably transfected cell lines. PC-3-pcDNA3.1, PC-3-myr-PTEN, and PC-3-dn-NRF2 were maintained in low-serum medium. After 16 h, cells were treated with tBHQ (10 μM) for 6 h and mRNA levels of HMOX1, NQO1, and TRX1 were determined by qRT-PCR and normalized by β-Actin. Data are mean±SEM (n=3). Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test. **p<0.01 versus PC-3-pcDNA3.1 treated with tBHQ. (G) Whole-protein lysates from PC-3 stably transfected with pcDNA3.1 (vector) or myr-PTEN were immunoblotted with specific antibodies as indicated in the panels. (H) Whole-protein lysates from PC-3 stably transfected with pcDNA3.1 (vector) or dn-NRF2-V5 were immunoblotted with specific antibodies as indicated in the panels. (I) 3x106 of PC-3 pcDNA3.1, PC-3-myr-PTEN, or PC-3-dnNRF2 cells were resuspended in 100 μl of PBS and transplanted subcutaneously into the left flank of the mouse. Tumor volume was measured twice at week. Data are mean±SEM (n=8). Statistical analysis was performed with two-way ANOVA followed by Bonferroni post-hoc test. **p<0.01; ***p<0.001 versus PC-3-pcDNA3.1. (J) Representative tumors from PC-3 pcDNA3.1, PC-3-myr-PTEN, and PC-3-dnNRF2 removed after 42 days from injection. PBS, phosphate-buffered saline.
Inhibition of NRF2 impaired PC-3 tumorigenesis in nude mice
We analyzed whether myr-PTEN or dn-NRF2 might abrogate the tumorigenic capacity of these cells. We generated three lines of PC-3 cells stably transfected with myr-PTEN, dn-NRF2-V5, or empty vector (pcDNA3.1). According to the low growth rate observed in the colony-forming assays, production of the myr-PTEN and dn-NRF2-V5 cells took several more weeks than those with control vector. Figure 7G and H show the expression of myr-PTEN and dn-NRF2-V5 in these cells, respectively. Overexpression of myr-PTEN reduced p-AKT basal levels compared with control cells (Fig. 7G). Treatment of these rescued cells with 10 μM tBHQ for 6 h increased the mRNA levels of HO-1, NQO1, and TRX1 in the PC-3-pcDNA3.1 line but overexpression of myr-PTEN and dn-NRF2-V5 attenuated transcriptional activity of these genes (Fig. 7D–F). Then, we injected 3×106 of PC-3-myr-PTEN, PC-3-dn-NRF2-V5, or PC-3-pcDNA3.1 cells in the left flank of nude mice, and monitored tumor appearance after an additional injection of matrigel in the right flank. In these experiments, the PC-3-pcDNA3.1 cells formed tumors of about 1 cm in diameter in about 6 weeks after Matrigel injection (Fig. 7I, J). By contrast, PC-3-myr-PTEN and, more importantly, PC-3-dn-NRF2-V5 cells either did not develop tumors or they were very small (less than 2 mm in diameter). These results indicate that PC-3 cells require both loss of PTEN and NRF2 expression in order to exhibit tumorigenicity.
Absence of PTEN correlates with high NRF2 and HO-1 protein levels in human endometrioid tumors
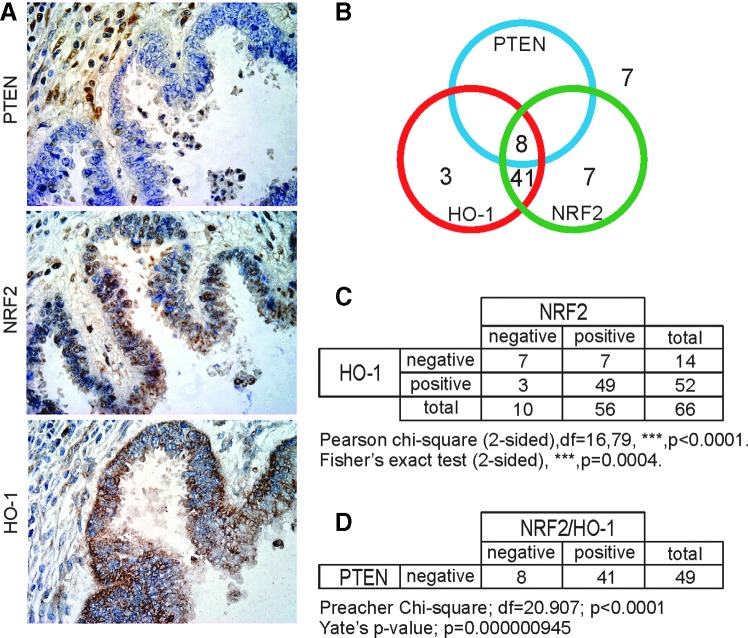
Loss of PTEN and overexpression of NRF2 are frequently found in endometrioid cancer. To analyze whether there is a causative connection between PTEN and NRF2 in these tumors, we analyzed their levels in TMA of paraffin-wax-embedded endometrioid tumors from 66 patients. We also analyzed the abundance of HO-1 protein as a reporter of NRF2 transcriptional activity. Core tissues, 0.6 mm in diameter, were arrayed in duplicate on each slice to reinforce the anatomopathological characterization. The staining results for PTEN, NRF2, and HO-1 were scored by three independent observers. Figure 8A shows an example of an abnormal endometrioid gland with three 4-μm thick sections that were negative for PTEN expression in the tumor, but positive for the surrounding cells, while being positive for NRF2 and HO-1. As shown in the Venn diagram of Figure 8B, 58 tumors (88%) were negative for PTEN expression, providing further evidence that PTEN loss is frequent in this tumor type. Moreover, 49 tumors (74%) exhibited co-expression of NRF2 and HO-1 as expected from the fact that NRF2 induces the expression of HO-1. Most importantly, 41 of the 58 PTEN-negative tumors (80.4%) were positive for both NRF2 and HO-1 staining (Fig. 8C). A comparison of the predicted and observed frequencies of biologically relevant samples by Preacher's Chi-square analysis indicated that the observed frequency in PTEN versus (NRF2+HO-1) expression is significant with regard to the expected distribution: Preacher Chi-square, df=20.907, ***p<0.0001 (Fig. 8D). Taken together, the correlation between PTEN and NRF2/HO-1 expression might be considered an emerging marker to diagnose and monitor endometrioid cancers.
FIG. 8.
PTEN deficiency correlates with elevated NRF2 and HO-1 protein levels in human endometrioid tumors. (A) Immunohistochemistry performed in 4-μm-thick sections of human endometrial tumor. (B) Venn diagram showing evaluation and quantification of PTEN, NRF2, and HO-1 expression in tissue microarrays from 66 tumors of a cohort of patients with endometrioid cancer. (C) Contingency analysis of NRF2 and HO-1 expression in the 66 tumors using Pearson chi-square test followed by Fisher exact test in 66 patients. Pearson chi-square analysis indicates a statistical difference between observed distribution of NRF2 and HO-1 with a Fisher's p-value of 0.0004. (D) Analysis of PTEN expression in the 49 tumors that were positive for NRF2 and HO-1 determined in (C). Preacher's chi-square analysis indicates a statistical difference between observed distribution of NRF2 and HO-1 in the PTEN-negative tumors and the expected fair distribution with a Yate's, p-value of 9.4e-7. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Discussion
Since the initial discovery of NRF2 as a master regulator of cytoprotective genes (12), a large number of studies have sought to understand its influence on many pathophysiological processes (9). At least two lines of research have provided an insight into how NRF2 stability is regulated: redox regulation by KEAP1 and kinase regulation though GSK-3/β-TrCP. Both are intimately connected, because electrophiles alter the balance between kinase and phosphatase activities. Thus, we reported that the diterpene carnosol (21) and nordihydroguayaretic acid (37) activate signaling pathways, including PI3K/AKT, and lead to NRF2 activation. Considering that PTEN has a redox-sensitive cysteine in its catalytic center, we speculated that redox cycling compounds could inhibit its phosphatase activity, therefore leading to signaling events that could impact NRF2 phosphorylation and stability. In support of this, a biotinylated derivative of the synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), one of the most potent NRF2 inducers, has been reported to bind directly to Cys124 within the catalytic center and inhibit its lipid phosphatase activity in vitro (32). Also, the phytosterol guggulsterone has been suggested to induce HO-1 expression through PTEN-targeted activation of NRF2 (1). In the case of tBHQ, the diphenolic ring of this molecule can cycle between alcohol and keto groups and is, therefore, capable of behaving as an antioxidant (tBHQ) or a pro-oxidant (tBQ), respectively, as demonstrated in studies by Dinkova-Kostova and colleagues (5, 58). In fact, a previous report showed that PTEN is sensitive to tBHQ, although the study focused on histone modification that might participate in upregulation of the ARE (41). Consistent with this hypothesis, we found that PTEN activity was inhibited by tBHQ both in vitro and in vivo. These findings are of great transcendence, because most likely many compounds that have been claimed to target NRF2 though KEAP1 will also inhibit PTEN and, hence, their mechanism of action should be revisited. It is conceivable that drug alterations of PTEN, either directly through adduct formation or indirectly through other cysteine modifications, might inhibit not only its lipid phosphatase but also its protein phosphatase activity, which according to (57) is responsible for autodephosphorylation of residue Thr336. This autodephosphorylation mechanism provides a feed-forward loop of its lipid phosphatase activity. Indeed, mutation of this site makes lipid phosphatase activity sufficient for PTEN to inhibit invasion. Further studies will be required to determine how electrophilic compounds alter either of these two activities, but for the purpose of this study it seems clear that at least the lipid phosphatase activity is crucial because it connects with upregulation of GSK-3 and subsequent downregulation of NRF2.
The highly selective PTEN inhibitor bpV(HOpic) also upregulated NRF2, even in KEAP1-deficient cells, and conversely, MEFs from transgenic mice overexpressing PTEN exhibited lower basal and tBHQ-induced levels of NRF2 and its target genes. Moreover, active myr-PTEN antagonized the upregulation of ARE by PI3K-CAAX. All these data suggest that PTEN serves as an effective negative regulator of NRF2. These mechanistic results are in agreement with a recent study (55). Here, specific knockout of mouse Keap1 and Pten in liver led to the conclusion that PTEN participates in the regulation of NRF2, at least in part, through a KEAP1-independent manner. This study also reported the inhibition of GSK-3 in the Pten-deficient livers, building on our previous reports indicating that this kinase inhibits NRF2 (33, 34, 38, 39, 42) but the mechanistic connection between GSK-3 and NRF2 was not studied. We have further characterized this inhibitory function by phosphoproteomic identification of the two residues, Ser335 and Ser338 in the mouse sequence that are specifically phosphorylated by GSK-3. These two residues lie in the Neh6 degradation domain of NRF2 (DSGIS) and confirm our previous predictions based on site-directed mutagenesis.
The role of NRF2 as a survival factor for highly proliferating and tumorigenic cells is being increasingly recognized (4). However, apart from some tumors with somatic mutations identified in the interface between NRF2 and KEAP1, it is not known how NRF2 is upregulated. It was, therefore, interesting to determine the relevance of this PTEN/NRF2 axis in tumorigenesis, because the PTEN tumor suppressor is lost in the vast majority of human cancers. To this end, we chose the HEC1A endometrioid cells, as a control with normal PTEN, and Ishikawa and PC-3 cells derived from endometrioid and prostate cancer, respectively, both of which lack PTEN. Importantly, these cells retain an electrophile-responsive KEAP1/NRF2 axis, and in the case of PC-3 cells, KEAP1 has been sequenced and found to be unaltered (60). In these cells, we found increased NRF2 levels and activation of an ARE-LUC reporter, but expression of active myr-PTEN abrogated this effect. Moreover, overexpression of a dominant negative version of NRF2 dramatically abolished their proliferative and tumorigenic capacity as determined in colony-forming assays and xenograft growth in athymic mice, respectively. These results are consistent with the data of Mitsuishi and colleagues, who proposed that upregulation of NRF2 in mice with sustained PI3K-AKT signaling triggers a metabolic switch which enables tumor growth (23). Our data add to that study by showing that loss of PTEN and subsequent inhibition of GSK-3 lead to upregulation of NRF2.
It should be noted, however, that although our study demonstrates a mechanistic and functional connection between PTEN and NRF2, other additional ways of NRF2 activation in tumorigenesis may also take place. An increasing number of reports indicate that NRF2 provides a protective advantage to tumor cells against the oxidative stress which is produced under high proliferation rates (51). For instance, DeNicola et al. (4) showed that oncogene-induced NRF2 transcription promotes the detoxification of reactive oxygen species and tumorigenesis. On the other hand, NRF2-knockout mice have a normal development, indicating that NRF2-loss is not toxic by itself, but their tumorigenic capacity is impaired due to the need of NRF2 to sustain high proliferative rates (44) and this may be independent of PTEN, which is normal in these mice. So, most likely, ablation of NRF2 activity will prevent a high rate of proliferation as this is accompanied by increased production of reactive oxygen species. Consistent with this, dn-NRF2 did not alter parameters of proliferation but prevented the high proliferation rate and tumorigenesis of PTEN-deficient cells in nude mice.
Validation of the inverse relationship between NRF2 and loss of PTEN was undertaken in endometrioid carcinoma microarrays. Both NRF2 and its downstream target HO-1 were upregulated in the majority (80%) of the PTEN-negative tumors. These observations suggest a crucial role of NRF2/HO-1 in the development of at least this tumor type. Our results are essentially consistent with those reported in endometrial serous carcinoma, by Zhang and colleagues (14). These workers also reported that inhibition of NRF2 expression by overexpressing KEAP1 sensitized both the endometrioid tumor cell line SPEC-2 and SPEC-2-derived xenografts to chemotherapeutic agents. Here, we found that overexpression of an active version of PTEN led to a similar lack of tumorigenic response in the prostate cancer cell line PC-3. Therefore, both the KEAP1/NRF2 and the newly described PTEN/GSK-3/NRF2 pathways provide a molecular basis to target NRF2 inhibition as anticancer therapy.
Experimental Procedures
Cell culture and reagents
Keap1−/− and Keap1+/+ MEFs were provided by Dr. Ken Itoh (Center for Advanced Medical Research, Hirosaki University Graduate School of Medicine, Hirosaki, Japan). MEFs from wild-type (PTENwt) and transgenic PTEN (PTENtg) mice were obtained as described in (31). MEFs and Human embryonic kidney 293T (HEK293T) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 80 μg/ml gentamicin. PC-3 cells null for PTEN expression were provided by Dr. Ines Diaz Laviada (Faculty of Medicine, University of Alcala de Henares, Spain) and maintained in Roswell Park Memorial Institute Medium supplemented with 10% fetal bovine serum and 80 μg/ml gentamicin. Endometrial cell lines, HEC1A and Ishikawa, were provided by Dr. Gema Moreno (Department of Biochemistry, Faculty of Medicine, Autonomous University of Madrid, Spain) and were maintained in DMEM supplemented with 10% fetal bovine serum and 80 μg/ml gentamicin. Transient transfections were performed with TransFectin lipid reagent from Bio-Rad. tBHQ, tBQ, MG132, and LY294002 were from Sigma-Aldrich. CHX was from Boehringer Mannheim. bpV(HOpic) was from Millipore. SFN was from LKT laboratories.
Plasmids
pEF-HA-PTEN, pEF-HA-PTEND92A/C124A, and pEF-myr-PTEN plasmids were provided by Dr. Andrew Chan (The Mount Sinai School of Medicine, NY). pTG6600-P110α-PI3K-CAAX was provided by Dr. Silvio Gutkind (NIH, Bethesda, MD). The vectors pCGN-HA-GSK-3βΔ9 and pCGN-HA-GSK-3βY216F were provided by Dr. Akira Kikuchi (Department of Biochemistry, Faculty of Medicine, Hiroshima University). pcDNA3-Flag-β-TrCP2 was provided by Dr. Tomoki Chiba (Department of Molecular Biology, University of Tsukuba, Japan). pcDNA3.1-KEAP1-HA was generated by replacement of the V5-tag of pcDNA3.1-KEAP1-V5 (Biomedical Research Institute, Ninewells Hospital and Medical School, University of Dundee, Scotland, United Kingdom) for an HA-tag using the following primers: 5′-TCGAGCGATCGTCGACATGTACCCATACGATGTTCCAGATTACGCTTCTAGATATCAAG-3′ (forward), and 5′-CCGGCTTGATATCTAGAAGCGTAATCTGGAACATCGTATGGGTACATGTCGACGATCGC-3′ (reverse). Vectors pCFP-Neh2 (2–101) and pEGFP-Neh6 (306–483) were generated as described in (37). Plasmid encoding EYFP-Neh6-EK/FK-V5 containing the mutations E329K and F364K was generated with QuikChange® Lightning Site-Directed Mutagenesis Kit (Agilent). Expression vector for myr-PTEN-ER* was generated by cloning the mutated estrogen receptor (40) (BluescriptKST-ER*) in frame to the 5′-end of myr-PTEN (pEF-myr-PTEN) plasmid.
Immunoblotting
Immunoblots were performed as described in (37). The primary antibodies used are described in Supplementary Table S1. Membranes were analyzed using the appropriate peroxidase-conjugated secondary antibodies. Proteins were detected by enhanced chemiluminescence (GE Healthcare).
Analysis of mRNA levels
Total RNA extraction, reverse transcription, and quantitative polymerase chain reaction (PCR) were done as detailed elsewhere (36). Primer sequences are shown in Supplementary Table S2. Data analysis was based on the ΔΔCT method with normalization of the raw data to housekeeping genes (Applied Biosystems). All PCRs were performed in triplicate.
Luciferase assays
Transient transfections of HEC1A, Ishikawa and PC-3 cells were performed with the expression vectors for pTK-renilla (Promega) and ARE-LUC (a gift of Dr. J. Alam, Department of Molecular Genetics, Ochsner Clinic Foundation) as previously described (11) using Lipofectamine 2000 reagent (Invitrogen).
Phosphoproteomics
Coomassie Blue-stained SDS-PAGE gel bands were digested with trypsin as previously described (45). Peptides were extracted from gel pieces as described by Sprenger and Horrevoets (52). Phosphorylated peptides were enriched using TiO2 chromatography according to (18). Before LC-MS/MS analysis, phosphopeptides were desalted on microcolumns packed with POROS R3 RP material as previously described (7). Purified phosphopeptide fractions were analyzed by nanoflow liquid chromatography (RP HPLC, EASY-nanoLC; Thermo Fisher Scientific) coupled via a nano-electrospray ion source to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Briefly, samples were re-suspended in 5 μl solvent A (0.1% FA) and loaded onto the analytical column using intelligent flow control at 260 bar. The analytical column consisted of a 17–18 cm×75 μm i.d. fused silica capillary with a laser-pulled emitter packed in house with Reprosil Pur C18 AQ, 3 μm (Dr. Maisch, Germany). Peptides were eluted in a linear gradient of 0%–34% solvent B (95% ACN 0.1% FA) over 30 min at a flow rate of 250 nl/min. Mass spectra were generated in the positive ion mode using data-dependent acquisition. Survey scans in the m/z range 300–1650 were measured in the Orbitrap at a resolution of 30,000. The top 3 most intense precursor ions were selected for dual fragmentation using both collision-induced dissociation-multistage activation (CID-MSA) and higher collision energy dissociation (HCD). For CID-MSA 35, normalized collision energy was used and fragment ion spectra were acquired in the LTQ. For HCD, normalized collision energy was 45 and fragment ion spectra were acquired in the Orbitrap at a 7500 resolution. Raw data were processed by ProteomeDiscoverer software version 1.4 (Thermo Fisher Scientific). The Mascot searches were performed using an in-house Mascot server (version 2.3.02; Matrix Science). The search parameters were as follows: peptide mass tolerance 10 ppm; fragment match tolerance 0.8 Da (CID mode), 20 mmu (HCD mode); fixed modifications: carbamidomethylation of cysteine residues; variable modifications: phosphorylation of Ser/Thr/Tyr, oxidation of Met; enzyme: trypsin, two miss-cleavages allowed. Percolator tool within the ProteomeDiscoverer software was used for estimating false discovery rate for peptide identification. A cut-off value of peptide rank 1 and high confidence and 1% false discover rate were chosen. Phosphorylation sites probabilities were calculated using phosphoRS node built in the ProteomeDiscoverer software. Only sites with the highest probability were selected.
Colony-forming assay
PC-3 were seeded on 24-well plates (100,000 cells per well), cultured for 16 h, and transfected using Lipofectamine 2000 (Invitrogen) with different indicated expression plasmids. The day after transfection, each well was transferred to a p100-plate and treated with 0.5 mg/ml of G418 in completed medium during 4 weeks until colony number was scored. Cells were fixed with 4% paraformaldehide solution and for 30 min, washed with phosphate-buffered saline (PBS), stained with 1% of crystal violet solution (Sigma-Aldrich) during 1 h, washed twice with PBS and water, and then dried. For absorbance quantification, plates were treated with 10% acetic acid during 30 min and the absorbance from the crystal violet staining was measured at 560 nm. Parallel experiments were used to obtain stably transfected PC-3 cells to be used in tumorigenic assays.
Phosphatase activity assay
After 24 h of transfection, HA-PTEN was immunoprecipitated as indicated in Ref. (33). A lysate from nontransfected cells was incubated only with G protein to control for nonspecific binding (data not shown). The complexes were harvested by centrifugation and washed three times with lysis buffer, twice with water, and once with TBS. Immunocomplexes were pretreated with the indicated doses of tBHQ or with vehicle (DMSO) for 30 min. PTEN activity was measured as indicated in Ref. (29).
Tumor xenografts in athymic nude mice
All experiments were authorized and supervised by the ethics committee for Research of the Autonomous University of Madrid following institutional, Spanish, and European guidelines. PC-3-pcDNA3.1, PC-3-myr-PTEN, and PC-3-dn-NRF2-V5 stable cell lines were used to induce tumor xenografts in 7-week-old athymic nude female mice (Charles River). Cells were harvested, washed, counted, and resuspended in PBS. 3×106 viable cells resuspended in 100 μl of PBS were transplanted subcutaneously into the left flank of the mouse. Mice were monitored twice weekly until each animal developed one tumor in the area of the cell injection. After 3 weeks of the cell injection, tumors were not noticeable and we reinforced tumor growth by injecting 7 mg/ml of Matrigel (basement membrane extracellular matrix; Trevigen). Tumor volume and body weight were measured during the period of investigation. Tumor volumes (V) were determined by the formula V=L×W2×0.5, with L being the longest cross-section and W being the shortest.
Immunohistochemistry and TMA
Four-μm thick sections of paraffin-embedded samples of endometrioid tissue from patients treated at the Hospital Universitario La Paz (Madrid, Spain) were arrayed in a collection of 5 TMA slides. Taken together, these slides encompassed 108 TMA cores; however, a total of 42 cores were missing or damaged. This results in a total of 66 valid TMA cores. TMA slices were deparaffinized and rehydrated in water, after which antigen retrieval was carried out by incubation in 1 mM EDTA, 0.05% Tween 20, and pH 8.0 at 50°C for 45 min. Endogenous peroxidase and nonspecific antibody reactivity was blocked with peroxidase blocking reagent (Dako) at room temperature for 15 min. The sections were then incubated for 60–90 min at 4°C with the corresponding primary antibodies indicated in Supplementary Table S1. Detection was performed with Envision Plus Detection System (Dako). Negative controls were used with goat serum replacing the primary antibody. The slides were mounted with DPX mountant (VWR International). We considered a PTEN-negative tumor when staining was absent in the epithelial cells (cytosol and nucleus) and present in the stromal cells of the same tumor.
Image analyses and statistics
Different band intensities corresponding to immunoblot detection of protein samples were quantified using MCID software (MCID). Analysis of variance one-way, two-way, and Student's t-test were used to assess differences between groups. A p-Value of <0.05 was considered significant. Unless otherwise indicated, all experiments were performed at least three times with similar results. The values presented in the graphs are the means of at least three samples. Results are expressed as mean±SEM. For the analysis of TMA, a Chi-square test was used. To analyze NRF2 and HO-1 contingency, we performed Pearson chi-square test followed by a Fisher exact test. To determine PTEN distribution among NRF2/HO-1-positive tumors, we performed a chi-square test.
Supplementary Material
Abbreviations Used
- β-TrCP
beta-transducin repeat-containing protein
- 4-HT
4-hydroxytamoxifen
- ARE
antioxidant response element
- bpV(HOpic)
dipotassium bisperoxo (5-hydroxypyridine-2-carboxyl) oxovanadate
- CDDO
2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid
- CFP
cyan fluorescent protein
- CHX
cycloheximide
- CID-MSA
collision-induced dissociation-multistage activation
- DMEM
Dulbecco's modified Eagle's medium
- EGFP
enhanced green fluorescent protein
- EYFP
enhanced yellow fluorescent protein
- Gclc
mouse gene encoding the catalytic subunit of γ-glutamyl cysteine ligase
- Gclm
mouse gene encoding the modulator subunit of γ-glutamyl cysteine ligase
- GSK-3
glycogen synthase kinase-3
- HCD
higher collision energy dissociation
- Hmox1
mouse gene coding heme oxygensase-1
- HO-1
heme oxygenase-1
- KEAP1
Kelch-like ECH-associated protein 1
- MEFs
mouse embryonic fibroblasts
- Nef2l2
mouse gene coding NRF2
- Nqo1
mouse gene encoding NQO1
- NQO1
NAD(P)H quinone oxidoreductase 1
- NRF2
nuclear factor (erythroid-derived 2)-like 2
- PI3K
phosphatidylinositol 3 kinase
- PTEN
phosphatase and tensin homolog deleted on chromosome 10
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- tBHQ
tert-butylhydroquinone
- tBQ
tert-butylbenzoquinone
- TMA
tissue microarrays
- TRX1
thioredoxin reductase 1
Acknowledgments
This study was supported by the European Cooperation in Science and Research (COST Action BM1203/EU-ROS) and by SAF2013-43271-R of the Spanish Ministry of Economy and Competitiveness. The authors thank Prof. John D. Hayes (Ninewells Hospital and Medical School, Dundee) for a critical reading of this article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Almazari I, Park JM, Park SA, Suh JY, Na HK, Cha YN, Surh YJ. Guggulsterone induces heme oxygenase-1 expression through activation of Nrf2 in human mammary epithelial cells: PTEN as a putative target. Carcinogenesis 33: 368–376, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519–525, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, and Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res 40: 7416–7429, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, and Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinkova-Kostova AT, and Wang XJ. Induction of the Keap1/Nrf2/ARE pathway by oxidizable diphenols. Chem Biol Interact 192: 101–106, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Gebremedhin D, Terashvili M, Wickramasekera N, Zhang DX, Rau N, Miura H, and Harder DR. Redox signaling via oxidative inactivation of PTEN modulates pressure-dependent myogenic tone in rat middle cerebral arteries. PLoS One 8: e68498, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gobom J, Nordhoff E, Mirgorodskaya E, Ekman R, and Roepstorff P. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom 34: 105–116, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Greiner R, Palinkas Z, Basell K, Becher D, Antelmann H, Nagy P, and Dick TP. Polysulfides link H2S to protein thiol oxidation. Antioxid Redox Signal 19: 1749–1765, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes JD. and Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39: 199–218, 2014 [DOI] [PubMed] [Google Scholar]
- 10.Heneberg P. Reactive nitrogen species and hydrogen sulfide as regulators of protein tyrosine phosphatase activity. Antioxid Redox Signal 20: 2191–2209, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Innamorato NG, Rojo AI, Garcia-Yague AJ, Yamamoto M, de Ceballos ML, and Cuadrado A. The transcription factor Nrf2 is a therapeutic target against brain inflammation. J Immunol 181: 680–689, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, and Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236: 313–322, 1997 [DOI] [PubMed] [Google Scholar]
- 13.Jaramillo MC. and Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27: 2179–2191, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, and Zhang DD. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res 70: 5486–5496, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, Eom HS, Yoo NJ, and Lee SH. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol 220: 446–451, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Kitagishi Y. and Matsuda S. Redox regulation of tumor suppressor PTEN in cancer and aging (Review). Int J Mol Med 31: 511–515, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Kwon J, Lee SR, Yang KS, Ahn Y, Kim YJ, Stadtman ER, and Rhee SG. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci U S A 101: 16419–16424, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, and Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics 4: 873–886, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Lau A, Villeneuve NF, Sun Z, Wong PK, and Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res 58: 262–270, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SR, Yang KS, Kwon J, Lee C, Jeong W, and Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem 277: 20336–20342, 2002 [DOI] [PubMed] [Google Scholar]
- 21.Martin D, Rojo AI, Salinas M, Diaz R, Gallardo G, Alam J, De Galarreta CM, and Cuadrado A. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J Biol Chem 279: 8919–8929, 2004 [DOI] [PubMed] [Google Scholar]
- 22.McMahon M, Thomas N, Itoh K, Yamamoto M, and Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem 281: 24756–24768, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, and Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22: 66–79, 2012 [DOI] [PubMed] [Google Scholar]
- 24.Muscarella LA, Parrella P, D'Alessandro V, la Torre A, Barbano R, Fontana A, Tancredi A, Guarnieri V, Balsamo T, Coco M, Copetti M, Pellegrini F, De Bonis P, Bisceglia M, Scaramuzzi G, Maiello E, Valori VM, Merla G, Vendemiale G, and Fazio VM. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics 6: 710–719, 2011 [DOI] [PubMed] [Google Scholar]
- 25.Mutter GL, Lin MC, Fitzgerald JT, Kum JB, Baak JP, Lees JA, Weng LP, and Eng C. Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. J Natl Cancer Inst 92: 924–930, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Na HK. and Surh YJ. Oncogenic potential of Nrf2 and its principal target protein heme oxygenase-1. Free Radic Biol Med 67: 353–365, 2014 [DOI] [PubMed] [Google Scholar]
- 27.Nair S, Barve A, Khor TO, Shen GX, Lin W, Chan JY, Cai L, and Kong AN. Regulation of Nrf2- and AP-1-mediated gene expression by epigallocatechin-3-gallate and sulforaphane in prostate of Nrf2-knockout or C57BL/6J mice and PC-3 AP-1 human prostate cancer cells. Acta Pharmacol Sin 31: 1223–1240, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Numajiri N, Takasawa K, Nishiya T, Tanaka H, Ohno K, Hayakawa W, Asada M, Matsuda H, Azumi K, Kamata H, Nakamura T, Hara H, Minami M, Lipton SA, and Uehara T. On-off system for PI3-kinase-Akt signaling through S-nitrosylation of phosphatase with sequence homology to tensin (PTEN). Proc Natl Acad Sci U S A 108: 10349–10354, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odriozola L, Singh G, Hoang T, and Chan AM. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J Biol Chem 282: 23306–23315, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Ortega-Molina A, Efeyan A, Lopez-Guadamillas E, Munoz-Martin M, Gomez-Lopez G, Canamero M, Mulero F, Pastor J, Martinez S, Romanos E, Mar Gonzalez-Barroso M, Rial E, Valverde AM, Bischoff JR, and Serrano M. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab 15: 382–394, 2012 [DOI] [PubMed] [Google Scholar]
- 31.Palmero I. and Serrano M. Induction of senescence by oncogenic Ras. Methods Enzymol 333: 247–256, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Pitha-Rowe I, Liby K, Royce D, and Sporn M. Synthetic triterpenoids attenuate cytotoxic retinal injury: cross-talk between Nrf2 and PI3K/AKT signaling through inhibition of the lipid phosphatase PTEN. Invest Ophthalmol Vis Sci 50: 5339–5347, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, and Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31: 1121–1133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rada P, Rojo AI, Evrard-Todeschi N, Innamorato NG, Cotte A, Jaworski T, Tobon-Velasco JC, Devijver H, Garcia-Mayoral MF, Van Leuven F, Hayes JD, Bertho G, and Cuadrado A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol Cell Biol 32: 3486–3499, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risinger JI, Hayes AK, Berchuck A, and Barrett JC. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res 57: 4736–4738, 1997 [PubMed] [Google Scholar]
- 36.Rojo AI, Innamorato NG, Martin-Moreno AM, De Ceballos ML, Yamamoto M, and Cuadrado A. Nrf2 regulates microglial dynamics and neuroinflammation in experimental Parkinson's disease. Glia 58: 588–598, 2010 [DOI] [PubMed] [Google Scholar]
- 37.Rojo AI, Medina-Campos ON, Rada P, Zuniga-Toala A, Lopez-Gazcon A, Espada S, Pedraza-Chaverri J, and Cuadrado A. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: Role of glycogen synthase kinase-3. Free Radic Biol Med 52: 473–487, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Rojo AI, Rada P, Egea J, Rosa AO, Lopez MG, and Cuadrado A. Functional interference between glycogen synthase kinase-3 beta and the transcription factor Nrf2 in protection against kainate-induced hippocampal cell death. Mol Cell Neurosci 39: 125–132, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Rojo AI, Sagarra MR, and Cuadrado A. GSK-3beta down-regulates the transcription factor Nrf2 after oxidant damage: relevance to exposure of neuronal cells to oxidative stress. J Neurochem 105: 192–202, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Rojo AI, Salinas M, Martin D, Perona R, and Cuadrado A. Regulation of Cu/Zn-superoxide dismutase expression via the phosphatidylinositol 3 kinase/Akt pathway and nuclear factor-kappaB. J Neurosci 24: 7324–7334, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakamoto K, Iwasaki K, Sugiyama H, and Tsuji Y. Role of the tumor suppressor PTEN in antioxidant responsive element-mediated transcription and associated histone modifications. Mol Biol Cell 20: 1606–1617, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salazar M, Rojo AI, Velasco D, de Sagarra RM, and Cuadrado A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem 281: 14841–14851, 2006 [DOI] [PubMed] [Google Scholar]
- 43.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, and Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 45: 860–867, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Satoh H, Moriguchi T, Takai J, Ebina M, and Yamamoto M. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res 73: 4158–4168, 2013 [DOI] [PubMed] [Google Scholar]
- 45.Shevchenko A, Wilm M, Vorm O, and Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858, 1996 [DOI] [PubMed] [Google Scholar]
- 46.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, and Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A 105: 13568–13573, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simpson L. and Parsons R. PTEN: life as a tumor suppressor. Exp Cell Res 264: 29–41, 2001 [DOI] [PubMed] [Google Scholar]
- 48.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, and Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 3: e420, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh G. and Chan AM. Post-translational modifications of PTEN and their potential therapeutic implications. Curr Cancer Drug Targets 11: 536–547, 2011 [DOI] [PubMed] [Google Scholar]
- 50.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN, Minna JD, Stewart DJ, and Wistuba II. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 16: 3743–3753, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sporn MB. and Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12: 564–571, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sprenger RR. and Horrevoets AJ. Proteomic study of caveolae and rafts isolated from human endothelial cells. Methods Mol Biol 357: 199–213, 2007 [DOI] [PubMed] [Google Scholar]
- 53.Stebbing J, Lit LC, Zhang H, Darrington RS, Melaiu O, Rudraraju B, and Giamas G. The regulatory roles of phosphatases in cancer. Oncogene 33: 939–953, 2014 [DOI] [PubMed] [Google Scholar]
- 54.Suzuki T, Motohashi H, and Yamamoto M. Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol Sci 34: 340–346, 2013 [DOI] [PubMed] [Google Scholar]
- 55.Taguchi K, Hirano I, Itoh T, Tanaka M, Miyajima A, Suzuki A, Motohashi H, and Yamamoto M. Nrf2 enhances cholangiocyte expansion in Pten-deficient livers. Mol Cell Biol 34: 900–913, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tashiro H, Blazes MS, Wu R, Cho KR, Bose S, Wang SI, Li J, Parsons R, and Ellenson LH. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res 57: 3935–3940, 1997 [PubMed] [Google Scholar]
- 57.Tibarewal P, Zilidis G, Spinelli L, Schurch N, Maccario H, Gray A, Perera NM, Davidson L, Barton GJ, and Leslie NR. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Sci Signal 5: ra18, 2012 [DOI] [PubMed] [Google Scholar]
- 58.Wang XJ, Hayes JD, Higgins LG, Wolf CR, and Dinkova-Kostova AT. Activation of the NRF2 signaling pathway by copper-mediated redox cycling of para- and ortho-hydroquinones. Chem Biol 17: 75–85, 2010 [DOI] [PubMed] [Google Scholar]
- 59.Xu C, Yuan X, Pan Z, Shen G, Kim JH, Yu S, Khor TO, Li W, Ma J, and Kong AN. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther 5: 1918–1926, 2006 [DOI] [PubMed] [Google Scholar]
- 60.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, and Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther 9: 336–346, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.