Abstract
In this study, we selected 181 nematode glycogenes that are orthologous to human glycogenes and examined their RNAi phenotypes. The results are deposited in the Caenorhabditis elegans Glycogene Database (CGGDB) at AIST, Tsukuba, Japan. The most prominent RNAi phenotypes observed are disruptions of cell cycle progression in germline mitosis/meiosis and in early embryonic cell mitosis. Along with the previously reported roles of chondroitin proteoglycans, glycosphingolipids and GPI-anchored proteins in cell cycle progression, we show for the first time that the inhibition of the functions of N-glycan synthesis genes (cytoplasmic alg genes) resulted in abnormal germline formation, ER stress and small body size phenotypes. The results provide additional information on the roles of glycoconjugates in the cell cycle progression mechanisms of germline and embryonic cells.
Keywords: alg genes, germline cell division, glycogene, oligosaccharyltransferase, RNAi
Introduction
Carbohydrates play important roles in development and morphogenesis, in immunity and in medicine. The roles of carbohydrates in cell–cell interactions and host–pathogen interactions are typical examples of the key roles of carbohydrates in living organisms (Committee on Assessing the Importance and Impact of Glycomics and Glycosciences 2012). Thus, studying the roles of glycoconjugates and conducting a functional analysis of their synthesizing enzymes are essential for our understanding of the nature of life. One approach to studying the role of carbohydrates in organisms is assessing the phenotypes caused by inhibiting the functions of glycogenes, which are the genes involved in the synthesis, degradation, modification or recognition of carbohydrates. The nematode Caenorhabditis elegans is an ideal organism for this functional glycomics approach because the inhibition of gene function via RNAi and deletion mutagenesis is quick and easy (Ahringer 2006; Kutscher and Shaham 2014). All of the organism's cell lineages and its detailed neural wiring patterns (connectome) have been described in studies starting in the 1980s (White et al. 1986; Varshney et al. 2011; Jarrell et al. 2012), and its development can be traced at a single cellular level using differential interference contrast (DIC) microscopy because of its transparent body. Among >21,000 protein-coding genes, ∼60–80% of the genes are presumed to be human gene orthologs (Shaye and Greenwald 2011; Lui and Colaiacovo 2013). Thus, studying the function of human orthologs in the simple nematode organism is a powerful strategy to shed light on the roles of orthologous genes in humans. With these definite advantages in mind, we have been studying the roles of carbohydrate-related genes in the nematode Caenorhabditis elegans since the 1990s. RNAi and deletion mutagenesis have been intensively used, and various new roles of C. elegans glycogenes have been reported from our laboratory (see below).
The GlycoGene Database (GGDB, AIST, Japan) is an Internet database of various glycogenes, including glycosyltransferases, sulfotransferases and nucleotide-sugar transporters (http://jcggdb.jp/rcmg/ggdb/). Detailed annotations, links to other databases and orthologs of the human genes (Mus musculus, Rattus norvegicus, Drosophila melanogaster, C. elegans, Saccharomyces cerevisiae and Arabidopsis thaliana) are listed if identified. All up-to-date information on human glycogenes is summarized in the GGDB. Researchers can browse the list of glycogenes in their own web browser and can effectively retrieve essential information on the glycogenes in which they are interested, such as the substrate specificities and gene expressions (Narimatsu 2004; Togayachi et al. 2008).
In parallel with constructing the GGDB, we searched for glycogenes in the C. elegans genome using the same in silico cloning strategy developed for the GGDB (Togayachi et al. 2006). For all selected C. elegans putative glycogenes, we performed RNAi to examine the roles of glycogenes in development and morphogenesis. Several of the results have been published from our laboratory, including the roles of chondroitin synthesis in cytokinesis (Mizuguchi et al. 2003; Izumikawa et al. 2004), GlcCer synthesis in cell division and oocyte formation (Nomura et al. 2011) and GPI-anchor synthesis in germline formation (Murata et al. 2012). In this study, we report the RNAi phenotypes of the glycogenes listed in the database and briefly describe our new C. elegans Glycogene Database (CGGDB) and its entries. In addition to the abovementioned groups of genes, we show that the RNAi-mediated inhibition of the genes involved in N-glycan synthesis resulted in abnormal germline formation, an ER stress phenotype (unfolded protein response: UPR) and small body size (Sma) phenotype. These phenotypes were predominately caused by the RNAi of the genes acting at the cytoplasmic side of the ER and not of the genes acting at ER-lumen side. The results strongly suggest that N-glycan synthesis plays key roles in germline formation and early embryogenesis, along with other glycogenes in the nematode C. elegans.
Results
The selection of putative glycogenes in the nematode C. elegans
To conduct a functional glycomics study to identify the roles of glycogenes in the nematode C. elegans, we selected human glycogene orthologs in the worm genome. Obtaining the human glycogene sequences listed in our Glycogene Database (GGDB) (Narimatsu 2004; Togayachi et al. 2006; Togayachi et al. 2008), we used the basic local alignment search tool (BLAST) and a suite of Profile Hidden Markov Model tools program using the profile hidden Markov Model (HMM) to identify orthologs of these genes in the worm genome. The DXD motif was searched in the translated gene sequences because it interacts with the phosphate groups of the sugar nucleotide donor through the coordination of a divalent cation such as Mn2+. The transmembrane domain, which is often localized at the N-terminal end, is identified as a region that contains 18–22 hydrophobic amino acid residues by SOSUI (Hirokawa et al. 1998; Togayachi et al. 2007). Then, we classified these putative orthologs by comparing their homology to the human glycogenes, referring to the Cazy GT Database (Cazy; Campbell et al. 1997; Coutinho et al. 2003). A sequence set for each GT family of the Cazy GT Database was used as an MEME query (Bailey and Elkan 1994), and we obtained six motifs in the GT families (gg_motif; M1–M6 in the Supplementary data, Tables SI and SII). The presence of each motif was searched in each putative glycogene, and the motif was used to classify worm glycogene families. Positive amino acids (K/R) are typically abundant in the inner side of the transmembrane region of the glycosyltransferase (positive-inside rule) (von Heijne 1989); therefore, the presence of the K/R region at the N-terminal side was also used as an indication of a glycosyltransferase. The information from the OrthoList (Shaye and Greenwald 2011; Greenwald Lab 2013) was also examined and used to verify annotations (Table I; Supplementary data, Tables SI and SII). In total, we selected 181 genes, including glycosyltransferases, sulfation-related genes and GPI-anchor synthesis genes. For brevity, all of the selected genes are listed and described concisely in Supplementary data, Tables SI and SII. The list of these in silico-selected genes can be used for the further verification of the enzymatic activities and/or functions of the gene products and will be a valuable resource to clarify the roles of glycogenes in living organisms.
Table I.
Examples of C. elegans glycogenes with observed RNAi phenotypes
| Family | Human ortholog | C. elegans ortholog | RNAi phenotype | hsp-4::GFP fluorescence |
|---|---|---|---|---|
| ALG | ||||
| Cytoplasmic ALG (GnT) | ALG7 | Y60A3A.14 | Sma (100%, n = 33) | Increased (C, FM) |
| ALG13 | R10D12.12 | WT | Increased (C, FM) | |
| ALG14 | M02B7.4 | Oocyte morphology variant (35%, n = 20) | Increased (C, FM) | |
| Cytoplasmic ALG (ManT) | ALG1 | T26A5.4 | Emb (33%, n = 6) | Normal (C, FM) |
| ALG2 | F09E5.2 | Sma, oocyte morphology variant (83%, n = 12) | Increased (C, FM) | |
| ALG2 | bus-8 (T23F2.1) | Sma, Skd, Dpy (100%, n = 15) | Normal (C, FM) | |
| ALG11 | B0361.8 | Sma, oocyte morphology variant (76%, n = 50) | Increased (C, FM) | |
| Dol-P-Man synthesis | DPM1 | dpm-1 (Y66H1A.2) | Sma, oocyte morphology variant (78%, n = 32) | Increased (C, FM) |
| Dol-P-Glc synthesis | ALG5 | H43I07.3 | WT | Increased (C, FM) |
| OST | ||||
| WBP1, DDOST, OST48 beta subunit | ostb-1 (T09A5.11) | Sma, no oocytes (83%, n = 24) | Increased (FM) | |
| DAD1, OST2 | dad-1 (F57B10.10) | Sma, oocyte morphology variant (89%, n = 29) | Increased (FM) | |
| STT3 | stt-3 (T12A2.2) | Sma, oocyte morphology variant (95%, n = 20) | Increased (FM) | |
| GAG | CSS1, CSS2, CSS3, CSGlcAT, CSGalNAcT1, CSGalNAcT2 | sqv-5 (T24D1.1) | Emb (83%, n = 12) | Normal (C) |
| CSS2 | mig-22 (PAR2.4) | Emb (100%, n = 13) oocyte morphology variant | Normal (C) | |
| XylT | XYLT1, XYLT2 | sqv-6 (Y50D4C.4) | Emb (45%, n = 20) | Normal (C) |
| GlcAT | B3GAT3, B3GAT2 | sqv-8 (ZK1307.5) | Emb (55%, n = 20) | Normal (C) |
| SulfoT | HS2ST1 | hst-2 (C34F6.4) | Oocyte morphology variant (50%, n = 20) | Normal (C) |
| HS3ST1 | hst-3.1 (F40H3.5) | Oocyte morphology variant (50%, n = 10) | Normal (C) | |
| Others (see text for references): hpo-3 (F33D11.9), hpo-4 (T05E11.6), F38E1.9, C27A12.9, Y48E1B.2, T22C1.3, T09B4.1, dpm-3 (F28D1.11), cgt-1 (T06C12.10), cgt-2 (F20B4.6), cgt-3 (F59G1.1), pps-1 (T14G10.1), chs-1 (T25G3.2), chs-2 (F48A11.1), sqv-2 (Y110A2AL.14), sqv-3 (R10E11.4), pad-2 (K10G9.3), gly-2 (C55B7.2), gly-4 (Y116F11B.12) and D2045.9 | ||||
ALG, asparagine-linked glycosylation genes; C, COPASTM Biosort; Dpy, dumpy body; Emb, embryonic lethal; FM, fluorescence microscope; GAG, glycosaminoglycan synthesis gene; OST, oligosaccharyltransferase complex; Skd, skiddy; Sma, small body size; WT, wild type.
RNAi screening of all the selected C. elegans glycogenes
To knockdown each glycogene, the feeding RNAi technique was used for N2 (wild type) animals and for a strain coexpressing the mCherry membrane probe (mCherry DNA fused to a PH domain) and GFP-tagged β-tubulin marker. The OD70 and AZ244 strains contain the mCherry membrane probe and the GFP-tagged β-tubulin marker, respectively; thus, we crossed the OD70 strain to the AZ244 strain to visualize the germ cell membrane and tubulin simultaneously. The strain is very useful for visualizing germline cells in vivo (Nomura et al. 2011; Murata et al. 2012). In this study, the phenotypes were monitored in P0, F1 and F2 animals, and especially close attention was paid to the formation of germline cells and early embryogenesis. Using a transgenic worm that had the hsp-4::GFP transgene (SJ4005), the ER stress response was also monitored for each RNAi treatment. The HSP-4 is a homolog of the human chaperone and ER stress sensor GRP78/BiP. Observations under a fluorescence microscope and/or COPAS TM (Complex Object Parametric Analysis and Sorting) Biosort analysis (Pulak 2006; Dejima et al. 2009) was used for the experiments. By using the COPASTM Biosort, we can measure the fluorescence intensity and the body size of the worms simultaneously. All results are listed in Supplementary data, Tables SI and SII, and the same data can be found in our CGGDB. For several of the 181 genes, deletion alleles were isolated and examined for phenotypes. For example, RNAi treatment of the rib-1 or rib-2 genes involved in heparan sulfate (HS) synthesis or the piga-1 gene involved in the first step of GPI-anchor synthesis resulted in less evident phenotypes, and severe phenotypes were observed only in the deletion alleles (Morio et al. 2003; Kitagawa et al. 2007; Murata et al. 2012).
Abnormal phenotypes observed in our RNAi studies
Of the selected 181 glycogenes, remarkable RNAi phenotypes were observed in 38 different genes. Table I lists all the glycogenes that showed severe phenotypes under RNAi treatments. The results of functional glycomics analyses from our laboratory and others identified several important roles of glycans in development and morphogenesis. Figures 1 and 2 show examples of abnormal cell division and abnormal germline phenotypes observed in our RNAi-treated animals.
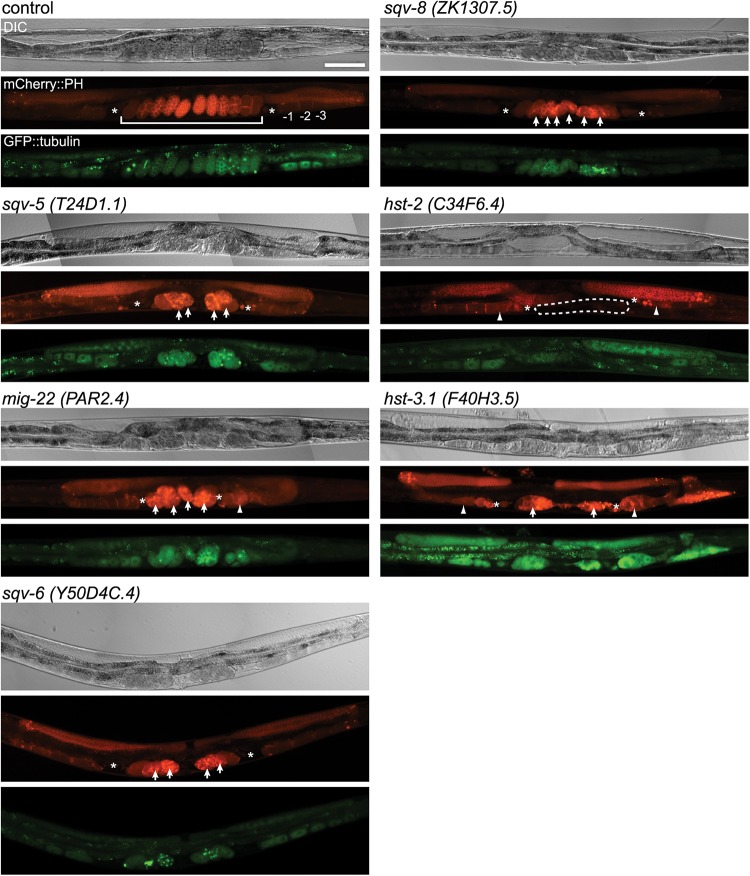
Fig. 1.
RNAi-mediated knockdown of several glycosyltransferases and sulfotransferases showed germline phenotypes. In each set of photographs, the upper panel shows the Nomarski DIC micrograph, the middle panel shows the PH domain detected by mCherry (red) and the bottom panel shows tubulin detected by gfp (green). In the mCherry::PH panel, the numbers indicate the normal oocytes, where −1 is the most proximal, and the brackets indicate the normal uterus (control); arrow: embryonic lethal (sqv-5, mig-22, sqv-6, sqv-8 and hst-3.1); arrowhead: oocyte morphology variant (mig-22, hst-2 and hst-3.1); dashed line: no eggs in uterus (hst-2), asterisks; spermatheca, scale bar: 100 μm.
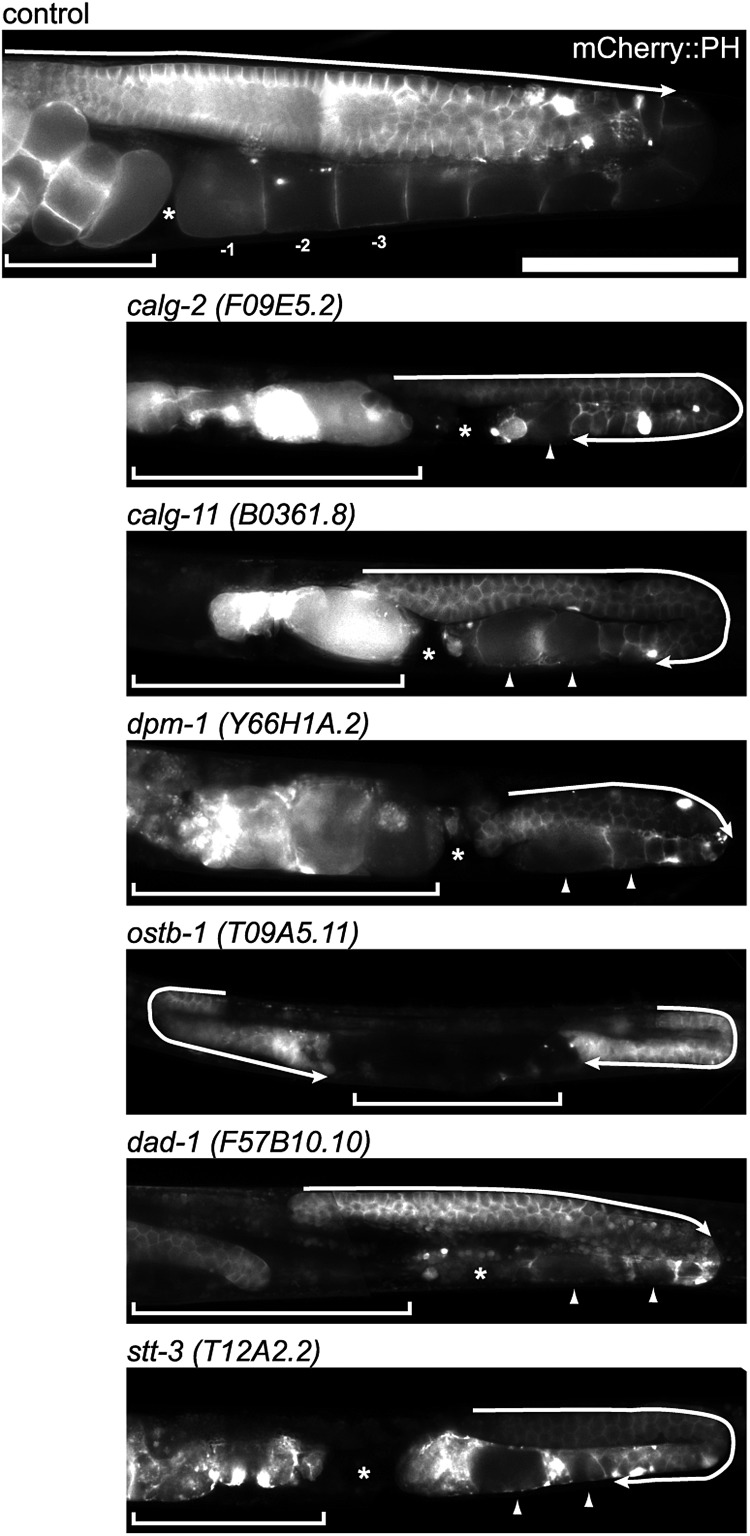
Fig. 2.
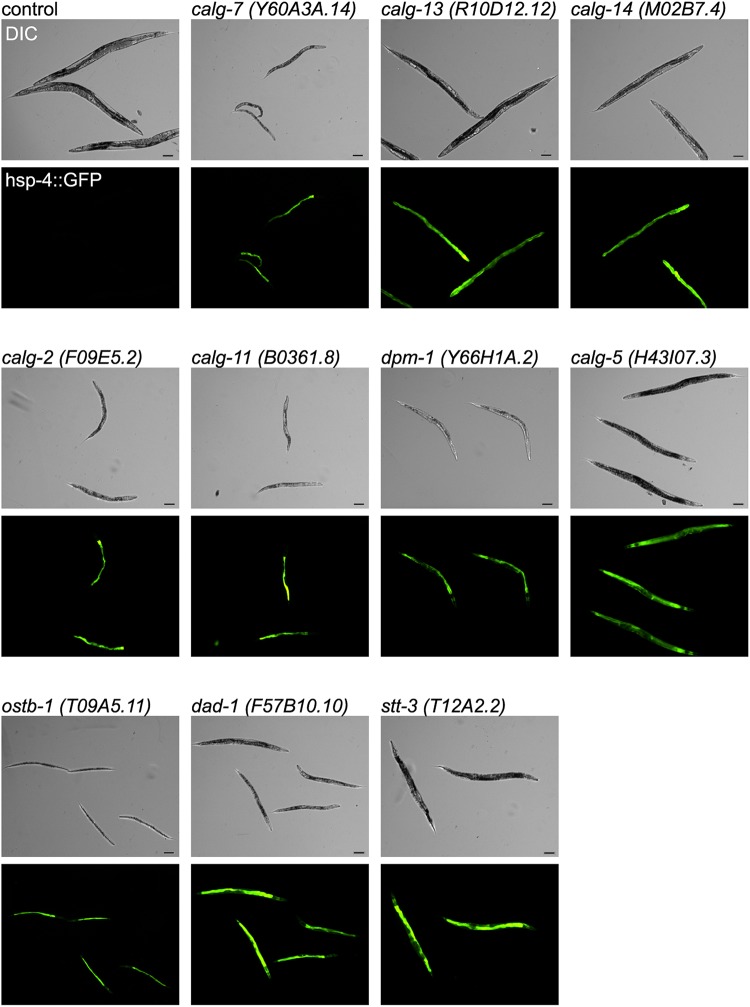
RNAi-inactivation of glycosyltransferases involved in LLO synthesis showed germline phenotypes. The germline of the control shows the normal syncytium end (arrow). The knockdown of calg-2 and calg-11 resulted in the extension of the syncytial region over the loop of gonad (arrow). The oocyte number decreased, and the oocyte morphology became variant (arrowhead). dpm-1, dad-1 and stt-3 RNAi also produced abnormal oocytes (arrowheads), and ostb-1 and stt-3 RNAi resulted in expanded syncytium. The inhibition of ostb-1 resulted in no oocytes, a very short gonad and expanded syncytium (arrow) phenotypes. The RNAi of these glycosyltransferases involved in LLO (lipid-linked oligosaccharides) synthesis all showed a small gonad phenotype compared with the control. The PH domain was detected by mCherry. The brackets indicate the uterus region. The asterisk indicates the spermatheca. Scale bar: 100 μm.
The inhibition of chondroitin synthesis results in abnormal cytokinesis and abnormal distal tip cell migration, which affects germline formation and early embryonic cell division (Hwang, Olson and Brown 2003; Hwang, Olson and Esko 2003; Mizuguchi et al. 2003; Izumikawa et al. 2004; Suzuki et al. 2006; Pedersen et al. 2013). The inhibition of the genes involved in chondroitin proteoglycan synthesis, including chondroitin synthase (sqv-5), chondroitin polymerizing factor (mig-22), xylosyltransferase essential for GAG synthesis (sqv-6) (Hwang, Olson and Brown 2003; Hwang, Olson and Esko 2003) and glucronyltransferase (sqv-8) (Bulik et al. 2000), all showed cytokinesis defects in the fertilized eggs (lethality) as described by the previous studies, and, in several cases, showed oocyte formation abnormality (Figure 1, mig-22 arrow head) which is a novel phenotype found in this study. Figure 1 shows that many abnormal eggs accumulated in the uteri of GAG-related gene-deficient animals.
RNAi-mediated inhibition of 2-O-sulfotransferase hst-2 and 3-O-transferase hst-3.1, which are sulfotransferases that are essential for HS sulfation (Figure 1), resulted in abnormal oocyte formation. Although extensive studies have been carried out on abnormal neuronal phenotypes and DTC migration defect under inhibition of these sulfotransferases (Turnbull et al. 2003; Kinnunen et al. 2005; Bülow et al. 2008; Bhattacharya et al. 2009; Tecle et al. 2013), to our knowledge, this is the first report on the germline defects caused by inhibition of these genes. In hst-2 RNAi-treated animals, the proximal oocytes were abnormal, and no eggs were found in the uterus (50%, n = 20). Oocyte morphology was abnormal in hst-3.1-defective animals (RNAi, 50%, n = 10), and sterility was observed in both groups of animals (Figure 1).
In the nematode, HS synthesis is mediated by the rib-1 and rib-2 genes. The deletion of these genes results in abnormal embryogenesis after gastrulation (e.g., abnormal ventral/epidermal enclosure formation) and abnormal axon guidance (Kitagawa et al. 2007). 3′-Phosphoadenosine 5′-phosphosulfate (PAPS) is a universal metabolic donor of sulfate groups (high-energy sulfate), and the inhibition of PAPS synthase pps-1 (Dejima et al. 2006) or PAPS transporters (pst-1 and pst-2) results in HS-deficient phenotypes and phenotypes that can be found under the inhibition of protein tyrosine sulfation (Bhattacharya et al. 2009; Dejima et al. 2010), indicating that sulfation plays essential roles in synthesizing sulfated HS proteoglycans and tyrosine-sulfated proteins (Kinnunen et al. 2005; Dejima et al. 2006; Mizuguchi et al. 2009; Dejima et al. 2010; Kalis et al. 2010). The inhibition of GlcCer synthesis also results in both abnormal cytokinesis and abnormal oocyte formation (Nomura et al. 2011) besides larval arrest possibly due to defects in digestive tract cells (Marza et al. 2009). GPI-anchor synthesis is also essential to oocyte formation, and without GPI-anchored protein synthesis, no oocytes are formed (Murata et al. 2012).
The RNAi of oligosaccharyltransferase (OST) genes, conducted by our group (ostb-1, stt-3 and dad-1; Figure 2) and others (for ostb-1, see Bonner et al. 2013; for ribo-1, see Stevens and Spang 2013), showed various abnormal phenotypes, including ER stress and embryonic lethality. These results indicate that the synthesis of N-glycans may play essential roles in meiosis and/or mitosis.
Glycogenes involved in N-glycan synthesis are essential for meiosis and/or mitosis
To examine whether the glycogenes involved in N-glycan synthesis also have roles in mitosis or meiosis, we determined the RNAi of the N-glycan-related genes in the selected glycogenes (see Figure 3 and Table I; Supplementary data and the glycogene list, Tables SI and SII) and examined the phenotypes. The RNAi-mediated inhibition of gly- genes coding enzymes such as GalNAcT1, T2, T10 and T11 or glct- genes (beta3GlcAT) showed no obvious phenotypes in oogenesis (mitosis and meiosis) or in early embryonic cell division (mitosis) (Supplementary data, Tables SI and SII). In contrast, the RNAi of the genes involved in LLO (lipid-linked oligosaccharides)-mediated glycan synthesis or the attachment of LLO to proteins resulted in severe phenotypes. In Figure 2, typical RNAi phenotypes of genes involved in LLO synthesis are depicted as observed in our experiments. The RNAi of calg-2 (C. elegans ALG2 ortholog), calg-11, dpm-1, ostb-1, dad-1 and stt-3 showed severe phenotypes in oocyte formation. In this study, the names of genes whose official names have not been assigned in the C. elegans community are represented as calg-2 (i.e., C. elegans human ALG2 gene ortholog); this name is composed of “c”, meaning “C. elegans”, followed by a gene name indicative of human ortholog of the gene.
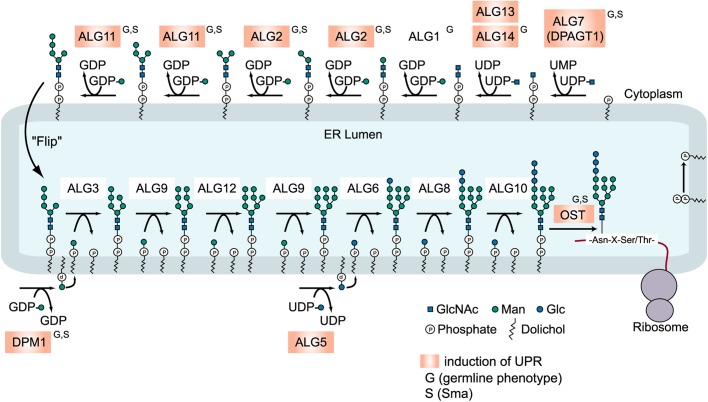
Fig. 3.
Synthesis of dolicol-P-P-GlcNAc2Man9Glc3 in the ER of eukaryotes. A model for the eukaryotic biosynthetic pathway of lipid-linked oligosaccharides (LLOs) and its transfer to proteins. The glycogenes that showed an increase of hsp-4::GFP expression by RNAi are highlighted in orange, and the genes showing small body size (S) or germline phenotype (G) are indicated. ostb-1, dad-1 and stt-3 code subunits of OST.
Figure 3 presents the eukaryotic N-glycan synthesis pathway and summarizes abnormalities found in this study. The inhibition of calg-7 or calg-14 also resulted in similar phenotypes (data not shown). The RNAi of calg-2 resulted in the extension of the germline syncytial region across the loop of the gonad (Figure 2, arrow). Few oocytes were formed, and the oocyte size was small or abnormal (arrowhead). No fertilized eggs were found in the uterus. The RNAi of the calg-11 ortholog also showed an extension of the germline syncytial region across the loop of the gonad (arrow), and the oocyte number decreased. The oocytes were abnormally swollen (arrowheads), and the abnormal eggs were found in the uterus. The inhibition of dpm-1, which transfers mannose to dolichol lipid, also severely affected oocyte formation (Figure 2, arrowhead), and no cellular compartment of the oocytes was observed. No fertilized eggs were formed in the animal. The inhibition of ostb-1, which is a subunit of OST, resulted in no oocyte formation. The length of the syncytial region before the loop region was abnormally short, and the syncytial region extended over the loop region. No cellularization of oocytes was observed. The inhibition of dad-1, which is also a subunit of OST, resulted in abnormal oocyte formation (arrowheads). The number of oocytes was decreased, and no fertilized eggs were found. The inhibition of stt-3, which is a potentially active subunit of OST, resulted in abnormal oocytes (Figure 2, arrowheads), as observed in dpm-1 RNAi-treated animals. The syncytial region also extended over the loop region (arrow), and no fertilized eggs were formed (Figure 2). In all of the RNAi-treated animals shown in Figure 2, the gonads were small and poorly formed, and fertilization was defected.
We performed a detailed analysis of the calg-7 gene functions, which catalyzes the first step of N-glycan synthesis with RNAi and deletion mutant analysis, and the results will be published in a separate study. The inhibition of bus-8, which is another putative ALG2 ortholog, resulted in no abnormality in germline formation; however, the Sma or weak dumpy (Dpy) phenotype, and skiddy phenotypes (Yook and Hodgkin 2007; Partridge et al. 2008) were observed (see below). The RNAi-mediated inhibition of cytoplasmic alg genes resulted in abnormal germline formation; however, the RNAi of ER-lumen alg genes (calg-3, calg-9, calg-12, calg-6, calg-8 and calg-10) did not result in abnormal phenotypes (Figure 3). Thus, abnormal germline phenotypes were observed under the RNAi-mediated knockdown of cytoplasmic alg genes or OST genes.
Inhibition of cytoplasmic alg genes but not ER-lumen alg genes resulted in ER stress/Sma phenotype
The inhibition of the alg7 gene with tunicamycin induces the UPR due to the accumulation of abnormal non-glycosylated proteins in the ER (Hulsmeier et al. 2011). It is also reported that mild dose of tunicamycin (3 μg/mL) results in Sma phenotype in C. elegans (Struwe et al. 2009). Thus, we next examined whether the inhibition of glycogenes causes the ER stress response and Sma phenotype.
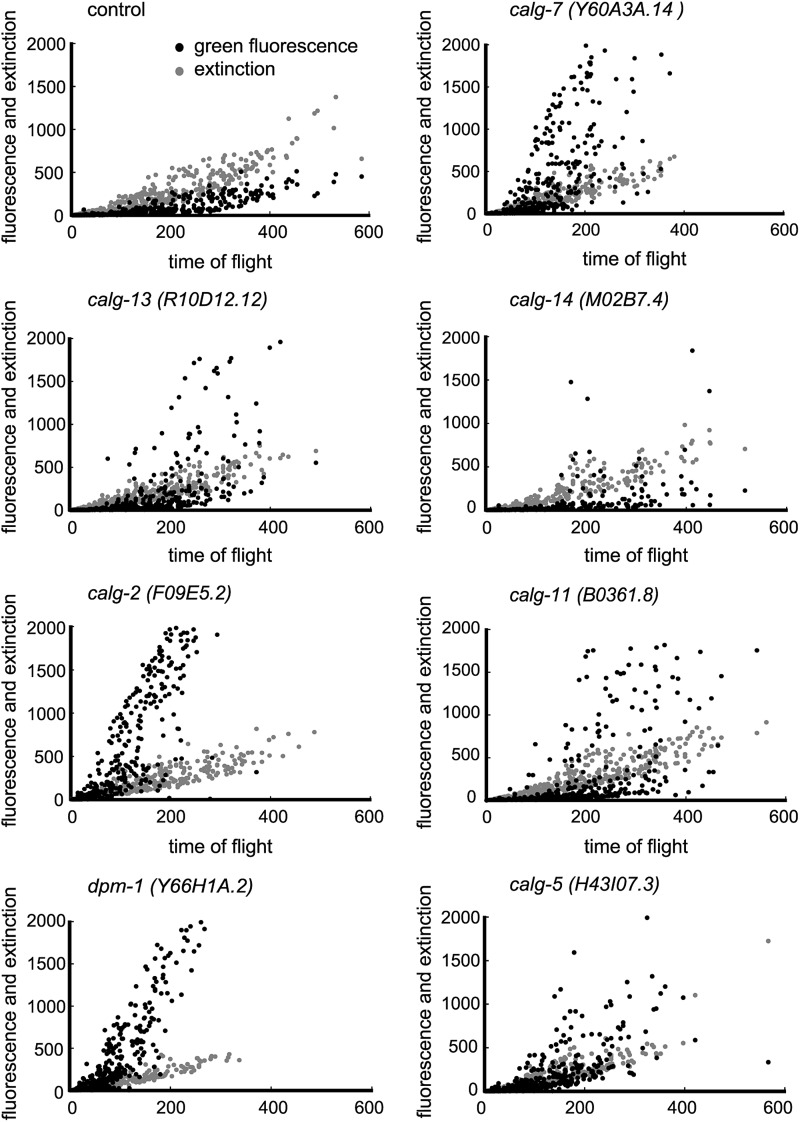
To monitor UPR, transgenic animals containing hsp-4::GFP (see above) were RNAi treated for each glycogene. The inhibition of calg-7, calg-13, calg-14, calg-2 and calg-11 resulted in the increase of hsp-4 transgene expression (the COPASTM Biosort analyses and fluorescence micrographs are shown in Figures 4 and 5). The dpm-1 and calg-5 genes act at the cytoplasmic side of the ER, and the RNAi of these genes also resulted in the UPR (Figure 4). The RNAi of calg-1, which is a cytoplasmic alg gene, did not result in the UPR (Figure 3, Supplementary data, Figures S1 and S2). Thus, we detected the UPR in RNAi-treated animals of most of the cytoplasmic alg genes. The inhibition of OST genes such as ostb-1, dad-1 and stt-3 also resulted in the UPR (Figure 5). In sharp contrast to these results, the inhibition of the ER-lumen alg genes including calg-3, calg-9, calg-12, calg-6, calg-8 and calg-10 resulted in no stress response (Supplementary data, Figures S1 and S2).
Fig. 4.
RNAi of glycosyltransferases involved in LLO synthesis at the cytoplasmic side of ER resulted in the strong induction of hsp-4::GFP. The black dots represent hsp-4::GFP fluorescence, and the gray dots represent extinction, which provides a measurement of optical density of the organism. COPASTM Biosort was used for the analyses. X-axis: time of flight (length of each organism); Y-axis: intensity.
Fig. 5.
RNAi of C. elegans cytoplasmic alg gene orthologs involved in LLO synthesis and RNAi of OST subunit genes induced strong GFP fluorescence under fluorescence microscopy. In each of the micrographs, the upper panel shows the Nomarski DIC micrograph, and the bottom panel shows the fluorescence of the hsp-4::gfp (green). Scale bar: 100 μm.
The inhibition of bus-8 did not result in the UPR (Supplementary data, Figures S1 and S2). The inhibition of uggt-1 and uggt-2 (Buzzi et al. 2011), which are ER quality-control genes, resulted in no stress response, and the inhibition of ogt-1, which is an O-linked GlcNAc transferase ortholog, resulted in no stress response at all (Supplementary data, Tables SI and SII). As expected (Hulsmeier et al. 2011), inhibition of glycogenes acting at Golgi (e.g., genes involved in GAG synthesis) resulted in no stress response (Supplementary data, Figure S1). Only alg genes and the dpm-1 gene, which act at the cytoplasmic side of the ER, showed the UPR after RNAi-mediated knockdown. The RNAi-mediated inhibition of no other genes caused the UPR.
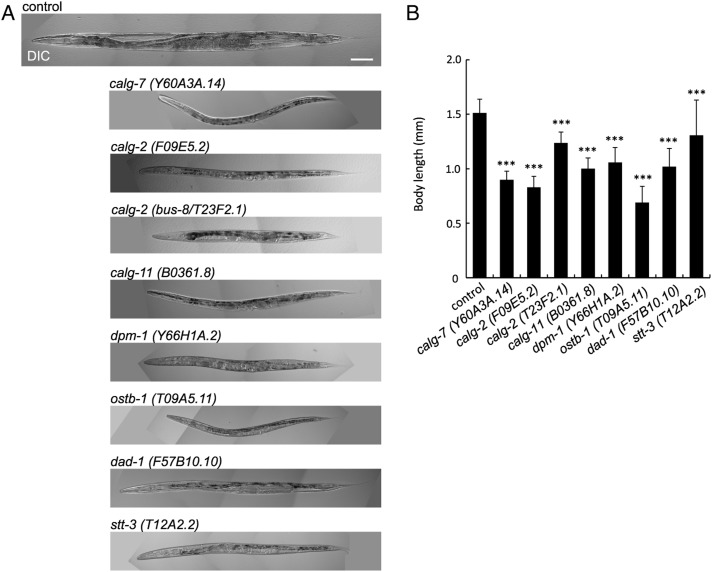
In our systematic RNAi study of glycogenes, we noted that the Sma phenotype (Sma phenotype) was found in animals treated with RNAi against LLO synthesis genes. Figure 6 shows the Sma phenotype observed in the RNAi-treated animals that had calg-7, calg-2, bus-8, calg-11, dpm-1, ostb-1, dad-1 and stt-3 gene orthologs. The extent of the body size reduction was comparable with the previously reported Sma gene phenotype of the nematode. The Sma phenotype was only observed under the inhibition of cytoplasmic alg gene orthologs or OST genes (Figure 3 shows the cytoplasmic/ER-lumen alg genes and OSTs). Our inspection of the previously reported RNAi experiments revealed that the Sma phenotype was reported for ribo-1 OST subunit, but no Sma RNAi phenotype has been reported for other N-glycan synthesis genes.
Fig. 6.
RNAi-mediated knockdown of glycosyltransferases involved in LLO synthesis showed small (Sma) phenotype. (A) The RNAi-mediated inactivation of LLO synthesis genes resulted in small body size. Scale bar: 100 μm. (B) Body length of RNAi-treated animals. The error bar indicates the standard deviation. Control n = 28, calg-7 (Y60A3A.14) n = 33, calg-2 (F09E5.2) n = 20, calg-2 (bus-8/T23F2.1) n = 13, calg-11 (B0361.8) n = 21, dpm-1 (Y66H1A.2) n = 30, ostb-1 (T09A5.11) n = 25, dad-1 (F57B10.10) n = 36 and stt-3 (T12A2.2) n = 26. ***P ≤ 0.001.
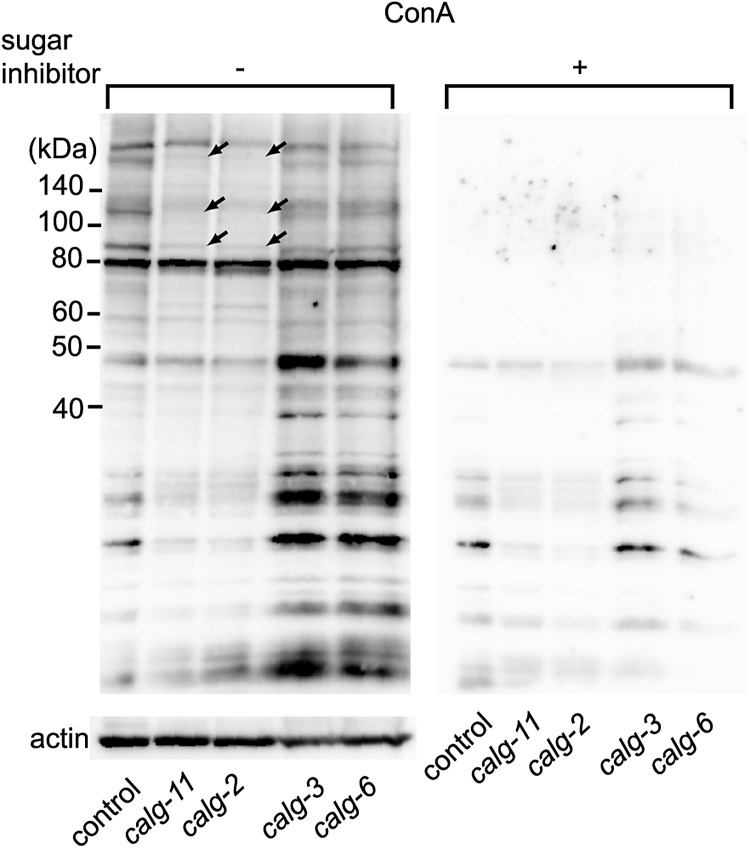
Figure 7 shows the reduction of ConA staining of total proteins of calg-2 or calg-11 RNAi-treated animals after SDS–PAGE. ConA recognizes mannose rich N-glycans including “core” trimannosides found in LLOs (Naismith and Field 1996). ConA staining was only reduced in cytoplasmic alg RNAi-treated animals (calg-11 and calg-2 in Figure 7), and no reduction of staining was observed in ER-lumen alg RNAi-treated animals (calg-3 and calg-6 in Figure 7). This finding indicates that the synthesis of LLO is reduced in cytoplasmic calg-11 or calg-2 RNAi-treated animals, and the observed phenotypes result from the reduction of N-glycan synthesis.
Fig. 7.
Knockdown of calg-11 or calg-2 shows reduction of ConA staining. ConA staining was decreased in the RNAi-mediated inhibition of cytoplasmic alg genes (calg-2 and calg-11) compared with the control. The arrows indicate the bands showing strong reduction. This reduction was not detected under the inhibition of ER-lumen alg genes (calg-3 and calg-6). The blotted membrane of the same samples shown at the right panel was incubated with sugar inhibitor plus HRP-conjugated ConA (Plus sign), and the left membrane was incubated with HRP-conjugated ConA in the absence of the sugar inhibitor (minus sign), followed by ECL detection. The bottom panel shows the anti-actin staining of the same lanes of the same samples as a control.
In summary, the germline abnormal phenotypes, body size small phenotype and UPR phenotype after RNAi were specific for the cytoplasmic alg genes but not for the ER-lumen alg genes. Among the 181 glycogenes examined, only the genes involved in LLO synthesis at the cytoplasmic side showed these phenotypes, indicating that the formation of the LLO core structure is essential for normal germline formation and development and morphogenesis.
Construction of CGGDB
The results of the RNAi-induced germline phenotypes of the C. elegans glycogenes are listed in our newly constructed CGGDB (http://jcggdb.jp/cggdb/). The C. elegans genes orthologous to human glycogenes are listed in alphabetical order of the gene family name, and the human orthologous gene name is indicated. Clicking the JCEG ID (Japan C. elegans Glycogene ID) opens a new window, and detailed information on each gene is listed. The NCBI gene link, the human ortholog information in the GGDB/OrthoList, the results of transmembrane domain prediction and the presence of the DXD motif, the presence of unique motifs conserved among glycosyltransferases (gg_motif), and the presence of the K/R region are also indicated. Users are referred to the bibliography with PubMed links for further sources of information. The coding DNA sequence and protein sequence can also be retrieved through the links on the page. The WormBase link and a direct link to the in situ database NEXTDB (if available) are also present in the page, and the user can see the in situ images with one click.
Previously reported RNAi phenotypes and allele phenotypes are also collected in the database, enabling the user to access all of the gene phenotypes quickly. By clicking the Expression tab, the user can find a list of the cell names in C. elegans, and clicking the cell name opens a new window showing the glycogenes expressed in that particular cell. The phenotype tab represents various phenotypes associated with the glycogenes, and clicking a phenotype opens a new window that lists all of the glycogenes associated with the phenotype. Other C. elegans glycogenes that are not orthologous to human glycogenes are also listed and tested for RNAi phenotypes, and the results will be included in a future version of our CGGDB. By navigating through our CGGDB, users can grasp the up-to-date information on particular C. elegans glycogenes to apply to their own studies.
Discussion
In his excellent review article on the glycogenes of C. elegans, Prof. Schachter listed 148 genes that are relevant to glycosylation, including putative glycosyltransferases, glycosidases and nucleotide-sugar transporters (Schachter 2004; Berninsone 2006). His review compiled RNAi information as well as mutant phenotypes and has been an essential reference for studying C. elegans glycogenes. Independently, we used an in silico cloning approach that was developed to construct our GlycogeneDatabase and listed all of the C. elegans glycogenes that are orthologous to human glycogenes. The current version of the CGGDB does not include glycosidases and sugar transporters, although we plan to add these genes to a future version of the database. Further verification of the catalytic activity of the in silico-selected genesis (e.g., by expressing the genes in vitro) is labor intensive; however, the RNAi-mediated knockdown of each glycogene function is simple in C. elegans. Thus, we used the selected gene list to examine the roles of C. elegans glycogenes systematically. As described above, we found new roles of glycogenes in early cell division, early embryogenesis, axon guidance, innate immunity (Ideo et al. 2009) and germline formation. Owing to the transparency of its body, the development of germline and embryonic cell division in C. elegans can be easily examined in vivo. Detailed knowledge on the molecular mechanisms of C. elegans germline formation has been accumulated, including the molecular mechanisms of germline stem cell niche induction and maintenance, stem cell to oocyte formation, cell cycle progression of oocytes and oocyte-embryo transition (Schedl 2013). Thus, we have paid special attention to the germline phenotypes and early embryogenesis in our study.
In this study, by examining the germline phenotype of various glycogene RNAi-treated animals, we found that the genes involved in N-glycan synthesis are also essential in the germline development and egg formation of C. elegans. The inhibition of N-glycan synthesis genes, including cytoplasmic alg genes and dpm-1, resulted in abnormal germline formation, UPR and Sma phenotype; however, the inhibition of any ER-lumen alg gene did not result in these phenotypes. The germline abnormal, UPR and Sma phenotypes were also observed in the OST gene RNAi-treated animals. A possibility that RNAi efficiency to ER-lumen alg genes is generally lower to cytoplasmic alg genes appears to be less probable because our RNAi against other ER-lumen expressed genes (e.g., OST genes and GPI-anchor synthesis gene -PIGV, PIGO, PIGK, GAA1 and PIGU orthologs) showed severe phenotypes. These results indicate that the “core trimannoside” formation and attachment of LLO to proteins play essential roles in germline formation and early embryonic cell division. No single RNAi treatment of the genes that are involved in the N-glycan chain elongation/modification that occurs at the ER-lumen side resulted in these germline phenotypes. In several cases, the core trimannoside can be modified by the addition of GlcNAc, followed by several different glycosyltransferases and glycosidases, to form fucose-modified oligosaccharides (Schiller et al. 2012). These unique structures may compensate for the reduction of orthodox N-glycan structures resulting from ER-lumen alg RNAi. Thus, N-glycan core trimannoside synthesis occurring at the cytoplasmic side before flipping to the ER side or the addition of LLO to proteins by OST appear to be indispensable, but the addition of monosaccharides at the ER-lumen side appears to be dispensable. To confirm this finding, we need to analyze both ER-alg knockout animals and cytoplasmic alg knockout animals, and these studies are currently underway in our laboratory and will be published separately.
Previously published high-throughput RNAi screening on N2 animals failed to detect germline phenotypes for cytoplasmic alg genes, but in several RNAi experiments using the RNAi sensitive strain (rrf-3), phenotypes including lethality or Bt toxin hypersensitivity were reported for the calg-11, calg-13 and calg-14 genes, which are cytoplasmic alg genes. For the calg-8 gene, which is an ER-lumen alg gene, a lethal phenotype was reported in rrf-3 animals. The rrf-3 strain shows a high incidence of male phenotypes and a temperature-dependent decrease in brood size at 25°C; therefore, we excluded this strain from the present analysis (Simmer et al. 2002). Reduced brood size, partial embryonic lethality and larval arrest in F1 animals were reported for N2 calg-8 (RNAi) animals using the ORFeome RNAi sequence (Rual et al. 2004). Larval arrest and reduced brood size in F1 animals may be caused by the presence of abnormal glycosylated proteins in the later stages of development or by a difference in RNAi efficiency due to the different RNAi constructs used by our group and by Rual et al. To resolve this discrepancy, the use of calg-8 knockout animals and the quantitation of calg-8 mRNA are necessary.
The inhibition of N-glycan synthesis genes resulted in abnormal germline, UPR and Sma phenotypes. One possibility of causing these RNAi phenotypes is that the absence of several specific “N-glycosylated” proteins that are indispensable for germline formation and morphogenesis resulted in the phenotypes. N-glycosylated C. elegans proteins have been extensively studied by mass spectroscopy; therefore, the identification of major N-glycosylated proteins expressed in the germline is within the scope of the present technology (Kaji et al. 2003; Kaji et al. 2012). The identification of these proteins and the inhibition of the function of each N-glycosylation protein may shed light on this possibility. The other possibility is that the disturbance of gene networks responsible for germline formation and morphogenesis result from a reduction in the expression level of N-glycan synthesis genes, followed by UPR. The reduction of the gene functions induces the accumulation of abnormally glycosylated and/or non-glycosylated proteins, which results in UPR. In the absence of N-glycan synthesis gene function, no properly N-glycosylated proteins are formed. Therefore, the UPR cannot restore the abnormal ER state, which may be the cause of defects in germline cells and early embryonic cells. In this case, we can identify the major N-glycosylated proteins and UPR-associated proteins that have been affected (i.e., overexpressed and decreased proteins) in the germline. This type of analysis may help identify the roles of various normal proteins and stress–responsive proteins in germline formation and morphogenesis.
The cytoplasmic alg genes and ER-lumen alg genes are associated with various congenital disorders of glycosylation in humans. Through the in silico identification of alg gene orthologs followed by RNAi experiments of each gene, we found that RNAi is a very powerful method to induce various N-glycosylation-defective phenotypes including germline abnormalities, the UPR and the body size small phenotype. Tunicamycin, which is an inhibitor of ALG7 (DPAGT), induces no phenotypes in N2/rrf-3 animals at 2 μg/mL, while it induces Dpy, Unc, Sma and other phenotypes with low penetrance at 3 μg/mL. These phenotypes disappear at higher concentration of the drug. Lethality dominates at higher concentration (Struwe et al. 2009; Struwe and Warren 2010). Struwe et al. carried out genome-wide RNAi screening of rrf-3 worms to identify genes hypersensitive to a mild dose (2 μg/mL) of tunicamycin. They pinpointed 512 genes that show phenotypes under the drug-induced mild inhibition of N-glycosylation. In the presence of tunicamycin, RNAi of ER-lumen calg- genes (calg-3, -6, -8, -9, -10 and -12) showed phenotypes (Emb, Lvl, Lava, Gro, small brood or Ste), but no RNAi phenotypes were recorded without tunicamycin. No phenotypes were found in cytoplasmic calg- genes in their experiments. The 512 RNAi hypersensitive genes could be promising candidates for the phenotypes observed in our cytoplasmic alg gene RNAi experiments. With these previous studies, the RNAi-mediated inhibition of calg- genes described in this study could be a powerful way to study the molecular mechanisms of the onset and pathology of congenital disorders of glycosylation.
There are a lot of difference in glycans between humans and the nematode. For examples, no sialylated glycoconjugates are found in the nematode, and genes of neither sialyltransferases nor sialidases are found in the genome. By selecting C. elegans glycogenes orthologous to human glycogenes, we can exclude these non-conserved glycogenes, and only conserved glycogenes are depicted in our study. Most of the molecular mechanisms found in C. elegans are often conserved in higher organisms including humans; therefore, listing the human glycogene orthologs in C. elegans and using the gene list for the systematic inhibition of gene functions has a unique advantage for clarifying the roles of human glycogenes due to the various advantages of C. elegans over mammalian model organisms and cultured cells. We hope that our CGGDB will be a valuable source for researchers to use to discover the molecular mechanisms of human glycogene-associated diseases and their novel treatments.
Materials and methods
C. elegans strains
All worm strains were cultured at 20°C as previously described (Brenner 1974). The following strains were used: N2: wild type, OD70: ltIs44; [pie-1p-mCherry::PH(PLC1delta1) + unc-119(+)] (Kachur et al. 2008), AZ244: unc-119(ed3) III; ruIs57[pie-1::GFP::tubulin + unc-119(+)] (Praitis et al. 2001) and SJ4005: zcIs4[hsp-4p::gfp] V. Previously, we isolated a strain coexpressing the GFP-labeled β-tubulin marker and the mCherry membrane probe by mating OD70 to AZ244 (Nomura et al. 2011), and the strain was used throughout this study.
RNAi by feeding
The protocol for RNAi by feeding was based on previously described methods (Timmons et al. 2001; Akiyoshi et al. 2013). Caenorhabditis elegans RNAi library clones (Kamath and Ahringer 2003) were used for the RNAi of most of the glycogenes. Unreliable RNAi bacterial strains of the library (Qu et al. 2011) were omitted from our RNAi experiments. For several of the glycogenes, RNAi constructs were made from amplified cDNA or genomic DNA, as previously described (Mizuguchi et al. 2003). A fragment of cDNA was cloned into the L4440 (pPD129.36) vector, and the cloned plasmids were transformed into Escherichia coli HT115(DE3). A single colony of HT115(DE3) carrying the plasmids was cultured in LB medium containing 100 µg/mL carbenicillin for 8 h, and the E. coli was seeded onto NGM plates and incubated at 37°C overnight. The plates were treated with 2 mM isopropyl-β-d-thiogalactoside (IPTG; 100 µL/plate) and incubated at 37°C for 6 h to allow HT115 to express the double-stranded RNA. L4 hermaphrodites were transferred onto the feeding plates and maintained at 25°C. We observed the phenotypes of the F1 generation after 72 and 96 h. HT115 carrying the L4440 vector without any insert was used as a control.
Scoring RNAi phenotypes
For phenotypic characterization and the lethality assay, L4 hermaphrodites were transferred onto the plates, and dsRNA was introduced into the worms by feeding. The F1 and F2 progenies were further incubated for 130 h and were then observed by microscopy. The worms were picked at the late L4 stage from culture plates, allowed to lay eggs for 24 h, and then transferred to a new plate daily for the next 3 days. Larvae that failed to develop into L3 after 40 and 96 h were scored as larval arrest/lethal at 25°C. To measure body length, three L4 worms of N2 or RNAi-treated worms were put onto a 60-mm plate and fed for 96 h at 25°C before analysis. We collected 40 adult worms on a glass slide for each experiment, and heated the worms for 1 min at 60°C and measured their linear body lengths by ocular micrometer. To assess the germline and early embryonic phenotypes, the animals were observed with a full automatic microscope (Leica or Olympus) equipped using MetaMorph Software (version 6.1r5, Molecular Devices Corporation, CA) (see Microscopy).
Microscopy
DIC and fluorescent images were obtained using a Leica DMRXA full automatic microscope (Leica, Wetzlar, Germany) or Olympus BX51 fluorescence/DIC microscope (Tokyo, Japan) equipped with an MAC5000 controller system (Ludl Electronic Products Ltd., NY) as previously described (Mizuguchi et al. 2008). The acquired images were processed using MetaMorph software (version 6.1r5, Molecular Devices Corporation ). Confocal images were acquired with an LSM 510 META microscope (Carl Zeiss, Oberkochen, Germany). The worms were placed on an 8-well printed microscope glass slide (Matsunami Glass, Osaka, Japan) and anesthetized with M9 buffer containing 16 mM sodium azide.
Scoring UPR with COPASTM Biosort
Phsp-4::GFP fluorescence induction of the SJ4005 strain and time-of-flight of the populations of worms fed with control or glycogene dsRNA were measured using a COPAS (Complex Object Parametric Analysis and Sorting)™ Biosort dual color fluorescence sorting system (Union Biometrica, Holliston, MA) at 119 h at 25°C after placing five L4 worms following the manufacturer's instructions (Pulak 2006; Dejima et al. 2009). Further data analyses were performed using Excel 2003 software (Microsoft, Redmond, WA). A fluorescence dissecting microscope (Olympus SZX16) was also used to monitor the induction of the Phsp-4::GFP gene.
Lectin blotting and western blotting
In general, worms from three NGM agar plates were combined and used for blot analyses. The worms were frozen cracked in liquid nitrogen, and the samples were sonicated on ice in the solubilization buffer (7 M urea, 2 M thiourea, 4% CHAPS, 10 mM Tris–HCl, pH 8.0–8.5) in the presence of protease inhibitors (cOmplete ULTRA Tablets, Mini, EDTA-free, Roche Diagnostics, Basel, Switzerland). After incubation of the buffer at 4°C overnight, the samples were centrifuged at 18,000 × g at 4°C for 5 min, and the supernatant was collected and acetone precipitated at−20°C for 2 h. After centrifugation at 12,000 × g for 10 min, the pellet was collected, and after removing the acetone through drying, the pellet was solubilized in the SDS sample buffer. After SDS– PAGE (e-PAGEL 12.5%-E-T12.5L, ATTO, Tokyo, Japan), the gels were blotted onto PVDF membranes (Immobilon transfer membranes, pore size, 0.45 μm, Merck Millipore, Tokyo, Japan) using a standard semi-dry blotting protocol (ATTO). The blotted membranes were blocked with PBS-0.5% Tween 20 solution (PBS-T) for 30 min at room temperature followed by incubating the membrane with HRP (horseradish peroxidase-conjugated concanavalin A (EY LABORATORIES, INC., CA) (0.33 μg/mL in 0.5% PBS-T) with or without the sugar inhibitor (0.5 M α-methyl-d-mannoside, Sigma-Aldrich, Japan) for 1 h at room temperature. For actin immunoblotting, the PVDF membrane after blotting was incubated with anti-actin antibody (MAB1501, Merck Millipore, 1:4000 in PBS-T) for 1 h. After extensive washing, the membrane was incubated with HRP-conjugated anti-mouse IgG (NA931V, GE Healthcare Japan, 1:4000 in PBS-T) for 1 h at room temperature. The reactive protein bands were detected using an ECL chemiluminescence protein detection kit (Western Lighting ECL Pro, Perkin Elmer, MA) according to the manufacturer's protocol using LAS-3000 (FUJIFILM, Tokyo, Japan).
The selection of human glycogene orthologs using bioinformatics
To select glycogenes of the nematode (C. elegans) comprehensively, orthologous genes of human glycogenes were first collected. The set of genes registered in the GlycoGene Database (GGDB) was used to query the bioinformatics tools for a similarity search such as BLAST and HMM (Campbell et al. 1997; Kikuchi and Narimatsu 2006). To eliminate the genes that have homology on the site other than the enzymatic activity domain, the resultant putative glycogenes were confirmed to have homology between the glycosyltransferase domain and known glycosyltransferases. Subsequently, to select the glycogenes that do not have homology to human genes, the gene sequences of the nematode registered in the CAZy GT were collected and combined into the first dataset (those that have homology to human genes) (Coutinho et al. 2003). Finally, the collected putative genes were confirmed to possess the known characteristics of glycosyltransferases (e.g., DXD sequences and positive-inside rule), and this information was registered as annotation data into CGGDB (Kikuchi et al. 2003; Kikuchi and Narimatsu 2006).
Statistical analysis
Statistical analyses, including one-way ANOVA, Student's t-test and Holm's multiple comparison, were performed using the R statistical package (R version 2.11.0).
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Conflict of interest statement
None declared.
Funding
This work was supported by the Core Research for Evolutional Science and Technology (CREST) Program of the Japan Science and Technology Corp. (to KN) and by Grant-in-Aid for Scientific Research (C) (grant number 24570165) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to KN). The work was also supported by Mizutani Foundation Grant 120140 (to KN). The database construction work was supported by the grant for the integrated database project by the National Bioscience Database Center (NBDC), Japan Science and Technology Agency (JST) (to YS, TS, AT and HN).
Abbreviations
ANOVA, analysis of variance; COPAS, Complex Object Parametric Analysis and Sorting; CGGDB, C. elegans Glycogene Database; DIC, differential interference contrast; Dpy, dumpy body; EGFP, enhanced green fluorescent protein; Emb, embryonic lethal; ER, endoplasmic reticulum; GAG, glycosaminoglycan; GGDB, Glycogene Database; GPI, glycosylphosphatidylinositol; Gro, slow growth; GT, glycosyltransferase; HMM, Hidden Markov Model; HS, heparan sulfate; LLO, lipid-linked oligosaccharides; Lva, larval arrest; Lvl, larval lethal; NEXTDB, The Nematode Expression Pattern DataBase; OST, oligosaccharyltransferase; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; PH, Pleckstrin-homology; Sma, small body size; Ste, sterile; Unc, uncoordinated; UPR, unfolded protein response
Supplementary Material
Acknowledgements
We thank our former colleagues from the NEDO GG project, Dr Norihiro Kikuchi and Dr Yeon-Dae Kwon for their knowledge on glycosyltransferases, which largely contributed to our current work. We acknowledge the Caenorhabditis Genetics Center supported by the NIH National Center for Research Resources (NCRR) for the strains used in this study.
References
- Ahringer J. Reverse Genetics. WormBook. 2006 doi:10.1895/wormbook.1.47.1. [Google Scholar]
- Akiyoshi S, Nomura KH, Nomura K. 2013. GlycoPOD RNAi protocols http://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t205 , last accessed July 14, 2014.
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Berninsone PM. Carbohydrates and glycosylation. WormBook. 2006 doi: 10.1895/wormbook.1.125.1. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.125.1, http://www.wormbook.org, last accessed July 14, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R, Townley RA, Berry KL, Bulow HE. The PAPS transporter PST-1 is required for heparan sulfation and is essential for viability and neural development in C. elegans. J Cell Sci. 2009;122:4492–4504. doi: 10.1242/jcs.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner MK, Han BH, Skop A. Profiling of the mammalian mitotic spindle proteome reveals an ER protein, OSTD-1, as being necessary for cell division and ER morphology. PLoS ONE. 2013;8:e77051. doi: 10.1371/journal.pone.0077051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik D, Wei G, Toyoda H, Kinoshita-Toyoda A, Waldrip W, Esko J, Robbins P, Selleck S. sqv-3 -7, and -8, a set of genes affecting morphogenesis in Caenorhabditis elegans, encode enzymes required for glycosaminoglycan biosynthesis. Proc Natl Acad Sci USA. 2000;97:10838–10843. doi: 10.1073/pnas.97.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow HE, Tjoe N, Townley RA, Didiano D, van Kuppevelt TH, Hobert O. Extracellular sugar modifications provide instructive and cell-specific information for axon-guidance choices. Curr Biol. 2008;18:1978–1985. doi: 10.1016/j.cub.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzi LI, Simonetta SH, Parodi AJ, Castro OA. The two Caenorhabditis elegans UDP-glucose: Glycoprotein glucosyltransferase homologues have distinct biological functions. PLoS ONE. 2011;6:e27025. doi: 10.1371/journal.pone.0027025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazy H. 2013. CAZy – Home http://www.cazy.org/ , last accessed July 14, 2014.
- Committee on Assessing the Importance and Impact of Glycomics and Glycosciences, Board on Chemical Sciences and Technology, Board on Life Sciences, Division on Earth and Life Studies, National Research Council. Transforming glycoscience: A roadmap for the future. The National Academies Press; 2012. [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Dejima K, Murata D, Mizuguchi S, Nomura KH, Gengyo-Ando K, Mitani S, Kamiyama S, Nishihara S, Nomura K. The ortholog of human solute carrier family 35 member B1 (UDP-galactose transporter-related protein 1) is involved in maintenance of ER homeostasis and essential for larval development in Caenorhabditis elegans. FASEB J. 2009;23:2215–2225. doi: 10.1096/fj.08-123737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima K, Murata D, Mizuguchi S, Nomura KH, Izumikawa T, Kitagawa H, Gengyo-Ando K, Yoshina S, Ichimiya T, Nishihara S, et al. Two Golgi-resident 3’-phosphoadenosine 5’-phosphosulfate transporters play distinct roles in heparan sulfate modifications and embryonic and larval development in Caenorhabditis elegans. J Biol Chem. 2010;285:24717–24728. doi: 10.1074/jbc.M109.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima K, Seko A, Yamashita K, Gengyo-Ando K, Mitani S, Izumikawa T, Kitagawa H, Sugahara K, Mizuguchi S, Nomura K. Essential roles of 3’-phosphoadenosine 5’-phoshosulfate synthase in embryonic and larval development of the nematode Caenorhabditis elegans. J Biol Chem. 2006;281:11431–11440. doi: 10.1074/jbc.M601509200. [DOI] [PubMed] [Google Scholar]
- Greenwald Lab. 2013. OrthoList http://www.greenwaldlab.org/ortholist/ , last accessed July 14, 2014.
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Hulsmeier AJ, Welti M, Hennet T. Glycoprotein maturation and the UPR. Methods Enzymol. 2011;491:163–182. doi: 10.1016/B978-0-12-385928-0.00010-9. [DOI] [PubMed] [Google Scholar]
- Hwang H, Olson S, Brown J, Esko J, Horvitz H. The Caenorhabditis elegans genes sqv-2 and sqv-6, which are required for vulval morphogenesis, encode glycosaminoglycan galactosyltransferase II and xylosyltransferase. J Biol Chem. 2003;278:11735–11738. doi: 10.1074/jbc.C200518200. [DOI] [PubMed] [Google Scholar]
- Hwang H, Olson S, Esko J, Horvitz H. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 2003;423:439–443. doi: 10.1038/nature01634. [DOI] [PubMed] [Google Scholar]
- Ideo H, Fukushima K, Gengyo-Ando K, Mitani S, Dejima K, Nomura K, Yamashita K. A Caenorhabditis elegans glycolipid-binding galectin functions in host defense against bacterial infection. J Biol Chem. 2009;284:26493–26501. doi: 10.1074/jbc.M109.038257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa T, Kitagawa H, Mizuguchi S, Nomura KH, Nomura K, Tamura J, Gengyo-Ando K, Mitani S, Sugahara K. Nematode chondroitin polymerizing factor showing cell-/organ-specific expression is indispensable for chondroitin synthesis and embryonic cell division. J Biol Chem. 2004;279:53755–53761. doi: 10.1074/jbc.M409615200. [DOI] [PubMed] [Google Scholar]
- Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW. The connectome of a decision-making neural network. Science. 2012;337:437–444. doi: 10.1126/science.1221762. [DOI] [PubMed] [Google Scholar]
- Kachur TM, Audhya A, Pilgrim DB. UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C-elegans. Dev Biol. 2008;314:287–299. doi: 10.1016/j.ydbio.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- Kaji H, Shikanai T, Sasaki-Sawa A, Wen H, Fujita M, Suzuki Y, Sugahara D, Sawaki H, Yamauchi Y, Shinkawa T, et al. Large-scale identification of N-glycosylated proteins of mouse tissues and construction of a glycoprotein database, GlycoProtDB. J Proteome Res. 2012;11:4553–4566. doi: 10.1021/pr300346c. [DOI] [PubMed] [Google Scholar]
- Kalis AK, Kroetz MB, Larson KM, Zarkower D. Functional genomic identification of genes required for male gonadal differentiation in Caenorhabditis elegans. Genetics. 2010;185:523–535. doi: 10.1534/genetics.110.116038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kikuchi N, Kwon YD, Gotoh M, Narimatsu H. Comparison of glycosyltransferase families using the profile hidden Markov model. Biochem Biophys Res Commun. 2003;310:574–579. doi: 10.1016/j.bbrc.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Kikuchi N, Narimatsu H. Bioinformatics for comprehensive finding and analysis of glycosyltransferases. Biochim Biophys Acta. 2006;1760:578–583. doi: 10.1016/j.bbagen.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Kinnunen T, Huang Z, Townsend J, Gatdula MM, Brown JR, Esko JD, Turnbull JE. Heparan 2-O-sulfotransferase, hst-2, is essential for normal cell migration in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:1507–1512. doi: 10.1073/pnas.0401591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H, Izumikawa T, Mizuguchi S, Dejima K, Nomura KH, Egusa N, Taniguchi F, Tamura J, Gengyo-Ando K, Mitani S, et al. Expression of rib-1, a Caenorhabditis elegans homolog of the human tumor suppressor EXT genes, is indispensable for heparan sulfate synthesis and embryonic morphogenesis. J Biol Chem. 2007;282:8533–8544. doi: 10.1074/jbc.M611107200. [DOI] [PubMed] [Google Scholar]
- Kutscher LM, Shaham S. Forward and Reverse Mutagenesis in C. elegans. WormBook. 2014. doi:10.1895/wormbook.1.167.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui DY, Colaiacovo MP. Meiotic development in Caenorhabditis elegans. Adv Exp Med Biol. 2013;757:133–170. doi: 10.1007/978-1-4614-4015-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marza E, Simonsen KT, Faergeman NJ, Lesa GM. Expression of ceramide glucosyltransferases, which are essential for glycosphingolipid synthesis, is only required in a small subset of C. elegans cells. J Cell Sci. 2009;122:822–833. doi: 10.1242/jcs.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi S, Dejima K, Nomura K, Murata D, Nomura K. Functional glycomics at the level of single cells: Studying roles of sugars in cell division, differentiation, and morphogenesis with 4d microscopy. In: Taniguchi N, Suzuki A, Ito Y, Narimatsu H, Kawasaki T, Hase S, editors. Japan: Springer; 2008. pp. 290–294. [Google Scholar]
- Mizuguchi S, Dejima K, Nomura K. Sulfation and related genes in Caenorhabditis elegans. Trends Glycosci Glycotechnol. 2009;21:179–191. [Google Scholar]
- Mizuguchi S, Uyama T, Kitagawa H, Nomura K, Dejima K, Gengyo-Ando K, Mitani S, Sugahara K, Nomura K. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature. 2003;423:443–448. doi: 10.1038/nature01635. [DOI] [PubMed] [Google Scholar]
- Morio H, Honda Y, Toyoda H, Nakajima M, Kurosawa H, Shirasawa T. EXT gene family member rib-2 is essential for embryonic development and heparan sulfate biosynthesis in Caenorhabditis elegans. Biochem Biophys Res Commun. 2003;301:317–323. doi: 10.1016/s0006-291x(02)03031-0. [DOI] [PubMed] [Google Scholar]
- Murata D, Nomura KH, Dejima K, Mizuguchi S, Kawasaki N, Matsuishi-Nakajima Y, Ito S, Gengyo-Ando K, Kage-Nakadai E, Mitani S, et al. GPI-anchor synthesis is indispensable for the germline development of the nematode Caenorhabditis elegans. Mol Biol Cell. 2012;23:982–995. doi: 10.1091/mbc.E10-10-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith JH, Field RA. Structural basis of trimannoside recognition by concanavalin A. J Biol Chem. 1996;271:972–976. doi: 10.1074/jbc.271.2.972. [DOI] [PubMed] [Google Scholar]
- Narimatsu H. Construction of a human glycogene library and comprehensive functional analysis. Glycoconj J. 2004;21:17–24. doi: 10.1023/B:GLYC.0000043742.99482.01. [DOI] [PubMed] [Google Scholar]
- Nomura KH, Murata D, Hayashi Y, Dejima K, Mizuguchi S, Kage-Nakadai E, Gengyo-Ando K, Mitani S, Hirabayashi Y, Ito M, et al. Ceramide glucosyltransferase of the nematode Caenorhabditis elegans is involved in oocyte formation and in early embryonic cell division. Glycobiology. 2011;21:834–848. doi: 10.1093/glycob/cwr019. [DOI] [PubMed] [Google Scholar]
- Partridge FA, Tearle AW, Gravato-Nobre MJ, Schafer WR, Hodgkin J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev Biol. 2008;317:549–559. doi: 10.1016/j.ydbio.2008.02.060. [DOI] [PubMed] [Google Scholar]
- Pedersen ME, Snieckute G, Kagias K, Nehammer C, Multhaupt HA, Couchman JR, Pocock R. An epidermal microRNA regulates neuronal migration through control of the cellular glycosylation state. Science. 2013;341:1404–1408. doi: 10.1126/science.1242528. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol. 2006;351:275–286. doi: 10.1385/1-59745-151-7:275. [DOI] [PubMed] [Google Scholar]
- Qu W, Ren C, Li Y, Shi J, Zhang J, Wang X, Hang X, Lu Y, Zhao D, Zhang C. Reliability analysis of the Ahringer Caenorhabditis elegans RNAi feeding library: A guide for genome-wide screens. BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-170. 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter H. Protein glycosylation lessons from Caenorhabditis elegans. Curr Opin Struct Biol. 2004;14:607–616. doi: 10.1016/j.sbi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Schedl T. Germ cell development in C. elegans (advances in experimental medicine and biology) New York: Springer; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller B, Hykollari A, Yan S, Paschinger K, Wilson IB. Complicated N-linked glycans in simple organisms. Biol Chem. 2012;393:661–673. doi: 10.1515/hsz-2012-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. OrthoList: A compendium of C. elegans genes with human orthologs. PLoS ONE. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Stevens J, Spang A. N-Glycosylation is required for secretion and mitosis in C. elegans. PLoS ONE. 2013;8:e63687. doi: 10.1371/journal.pone.0063687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe WB, Hughes BL, Osborn DW, Boudreau ED, Shaw KMD, Warren CE. Modeling a congenital disorder of glycosylation type I in C. elegans: A genome-wide RNAi screen for N-glycosylation-dependent loci. Glycobiology. 2009;19:1554–1562. doi: 10.1093/glycob/cwp136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe WB, Warren CE. High-throughput RNAi screening for N-glycosylation dependent loci in Caenorhabditis Elegans. Methods Enzymol. 2010;480:477–493. doi: 10.1016/S0076-6879(10)80021-7. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Toyoda H, Sano M, Nishiwaki K. Chondroitin acts in the guidance of gonadal distal tip cells in C-elegans. Dev Biol. 2006;300:635–646. doi: 10.1016/j.ydbio.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Tecle E, Diaz-Balzac CA, Bulow HE. Distinct 3-O-sulfated heparan sulfate modification patterns are required for kal-1-dependent neurite branching in a context-dependent manner in Caenorhabditis elegans. G3 (Bethesda) 2013;3:541–552. doi: 10.1534/g3.112.005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Dae K, Shikanai T, Narimatsu H. A database system for glycogenes (GGDB) In: Taniguchi N, Suzuki A, Ito Y, Narimatsu H, Kawasaki T, Hase S, editors. Experimental Glycoscience Glycobiology. Japan: Springer; 2008. pp. 423–425. [Google Scholar]
- Togayachi A, Sato T, Narimatsu H. Comprehensive enzymatic characterization of glycosyltransferases with a beta3GT or beta4GT motif. Methods Enzymol. 2006;416:91–102. doi: 10.1016/S0076-6879(06)16006-1. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Sato T, Iwai T, Narimatsu H. Cloning and characterization of β1,3-glycosyltransferase family with a β3GT motifs. Trends Glycosci Glycotechnol. 2007;19:29–40. [Google Scholar]
- Turnbull J, Drummond K, Huang Z, Kinnunen T, Ford-Perriss M, Murphy M, Guimond S. Heparan sulphate sulphotransferase expression in mice and Caenorhabditis elegans. Biochem Soc Trans. 2003;31:343–348. doi: 10.1042/bst0310343. [DOI] [PubMed] [Google Scholar]
- Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol. 2011;7:e1001066. doi: 10.1371/journal.pcbi.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Yook K, Hodgkin J. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2007;175:681–697. doi: 10.1534/genetics.106.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.