Abstract
N-acetylgalactosaminyltransferase (GalNAc)-type (mucin-type) O-glycosylation is an abundant and highly diverse modification of proteins. This type of O-glycosylation is initiated in the Golgi by a large family of up to 20 homologous polypeptide GalNAc-T isoenzymes that transfer GalNAc to Ser, Thr and possibly Tyr residues. These GalNAc residues are then further elongated by a large set of glycosyltransferases to build a variety of complex O-glycan structures. What determines O-glycan site occupancy is still poorly understood, although it is clear that the substrate specificities of individual isoenzymes and the repertoire of GalNAc-Ts in cells are key parameters. The GalNAc-T isoenzymes are differentially expressed in cells and tissues in principle allowing cells to produce unique O-glycoproteomes dependent on the specific subset of isoforms present. In vitro analysis of acceptor peptide substrate specificities using recombinant expressed GalNAc-Ts has been the method of choice for probing activities of individual isoforms, but these studies have been hampered by biological validation of actual O-glycosylation sites in proteins and number of substrate testable. Here, we present a systematic analysis of the activity of 10 human GalNAc-T isoenzymes with 195 peptide substrates covering known O-glycosylation sites and provide a comprehensive dataset for evaluating isoform-specific contributions to the O-glycoproteome.
Keywords: GALNT, ISOGlyP, MALDI-TOF, NetOGlyc4.0, SimpleCell
Introduction
N-acetylgalactosaminyltransferase (GalNAc)-type (mucin-type) O-glycosylation (here after simply O-glycosylation) is arguably the most abundant and complex protein posttranslational modification, which is highly differentially regulated in cells and tissues both with respect to where O-glycans are attached but also with respect to glycan structures (Ten Hagen et al. 2003; Bennett et al. 2012). Unique for this type of protein glycosylation is the large number of polypeptide GalNAc-T isoenzymes, up to 20, that initiate and control sites of O-glycosylation. These isoenzymes have different substrate specificities, are differentially expressed in cells and the repertoire of isoenzymes in a given cell is believed to orchestrate the O-glycoproteome produced. Furthermore, GalNAc-Ts may be distributed differently in the Golgi stacks (Rottger et al. 1998) and may relocalize to ER in cancer cells (Gill et al. 2010). Site-specific O-glycosylation is important for modulating protein function (Schjoldager and Clausen 2012) and deficiencies in individual GalNAc-T isoforms are known to cause diseases (Topaz et al. 2004; Fakhro et al. 2011). However, detailed insight into the specific functions of individual isoenzymes and their fine acceptor substrate specificities and contribution to the O-glycoproteome is still limited.
Most of our current knowledge of the functions of GalNAc-Ts is derived from in vitro enzyme assays with short peptide acceptor substrates. These studies have been limited by the ability to express active homogeneous recombinant enzymes (several isoenzymes have been difficult to express such as GalNAc-T8, T9, T18, T19 and T20) and by the limited number of diverse acceptor peptides characterized in the literature based on native O-glycoproteins and O-glycosites. Today only 15 of the known GalNAc-T isoforms have been expressed, purified and analyzed in various detail (Bennett et al. 2012), typically using a rather limited set of peptide substrates selected mainly from mucins or mucin-like O-glycoproteins with high density of O-glycosylation in PST rich sequences (Ten Hagen et al. 2003; Bennett et al. 2012). Major progress has been achieved using random peptide libraries as substrates in identifying sequence features such as positions of Pro and other residues relative to the acceptor site that can distinguish the acceptor substrate specificities of GalNAc-T isoforms (Gerken et al. 2011). Another area of progress has come from novel strategies for proteome-wide discovery of O-glycoproteins and O-glycosites (Halim et al. 2012; Steentoft et al. 2013). We recently used the so-called “SimpleCell (SC)” strategy with 12 human cell lines from different organs and identified almost 3000 glycosites derived from over 600 O-glycoproteins (Steentoft et al. 2013). The SC strategy involves precise genetic engineering where the COSMC gene encoded a private chaperone for the Core1 C1GalT1 synthase is knocked out to produce cells incapable of elongating O-glycans. As a result the truncated Tn (GalNAcα1-O-Ser/Thr) or STn (NeuAcα2-6GalNAcα1-O-Ser/Thr) O-glycans are homogenously expressed in most cells (Steentoft et al. 2011). While the knockout of COSMC and truncation of O-glycans may have effects on cells including altered expression of proteins, we have compared the O-glycoproteomes of wildtype K562 and K562 SCs and found reasonable overlap given the experimental conditions involved (Steentoft et al. 2011), suggesting that O-glycoproteome data from SCs represent the capacity of cells for O-glycosylation. Thus, the SC O-glycoproteome datasets now provide access to a greater variety of in vivo utilized acceptor substrates for design and testing of GalNAc-T substrate specificities.
Analyses of GalNAc-T activities using in vitro assays with peptide acceptor substrates have provided substantial insight into the nature of acceptor sites and specificities of GalNAc-T isoforms. One of the first comprehensive studies by the Tabak group (O'Connell et al. 1992) analyzed the specificity of bovine colostrum GalNAc-T activity with a peptide sequence derived from human von Willebrand factor using systematic substitutions of amino acids adjacent to the Thr acceptor site. This study demonstrated that residues from −6 to +5 flanking the Thr acceptor site are important for assessment of the in vitro glycosylation as well as the essential effect of adjacent Pro residues in position +3 and positive effect in positions −1, +1, +2 and +4. Other studies have shown that Ser acceptor sites are less efficiently used compared to Thr sites in vitro although the same flanking sequences are required (O'Connell and Tabak 1993; Wang et al. 1993; Gerken et al. 2006). An in vivo study with short sequence motifs in a reporter construct has also demonstrated the positive effect of Pro as well the negative effect of charged residues by substitution of residues adjacent to the acceptor site (Nehrke et al. 1996). A more systematic analysis of adjacent residue preferences using random peptide libraries provided a quantitative tool to explore the general rules of O-GalNAc glycosylation, and this has revealed that the GalNAc-T preference motif primarily locates at positions −3 to +3 of the acceptor site (Gerken et al. 2008, 2011). This strategy has further shown that Pro residues (particularly at +1 and +3) are a major determining factor driving most GalNAc-T isoform substrate preferences (Gerken et al. 2011). In addition, unique N-terminal substrate determinants have been identified by this approach that tend to differentiate the different GalNAc-T isoforms (Gerken et al. 2011). For example, Pro is highly preferred by GalNAc-T2 at position −1, while Ile, Val and Pro are only moderately favored at this position for GalNAc-T1 (Gerken et al. 2006).
In vitro analyses have also identified specific peptide substrates for several GalNAc-T isoforms (Bennett et al. 2012). The first in vitro analyses of multiple GalNAc-T isoforms (GalNAc-T1, T2 and T3) with peptides derived from the mucin 1 (MUC1) tandem repeat showed different preferences for three of the five potential substrate sites in the repeat (Wandall et al. 1997). Besides, GalNAc-T3 and later T6 were shown to have unique substrate specificity for a peptide derived from the V3-loop of HIVIIIB gp120 (Bennett et al. 1996; Bennett, Hassan, Mandel et al. 1999), and this was later confirmed by in vivo analysis using a reporter protein (Nehrke et al. 1998). Unique substrates for GalNAc-T4 were found in the MUC1 tandem repeat where two substrate sites not utilized by other GalNAc-T isoforms tested were specifically glycosylated by GalNAc-T4 (Bennett et al. 1998). GalNAc-T4 also appears to be the only isoform capable of glycosylating Thr57 of P-selectin-ligand-glycoprotein-1 (Bennett et al. 1998), which produces the P-selectin ligand. More recently, studies of human diseases associated with GalNAc-transferase gene (GALNT) genes have led to identification of unique substrates for GalNAc-T3 [Thr178 in fibroblast growth factor 23 (FGF23)] (Kato et al. 2006) and GalNAc-T2 with [Thr226 in Angiopoietin-like Protein 3 (ANGPTL3)] (Schjoldager et al. 2010), which have both been validated in cell systems and shown to affect protein functions by regulating proprotein processing. Similarly, unique substrates for GalNAc-T1 were identified in the mouse galnt1 knockout in osteopontin and bone sialoprotein (Miwa et al. 2010).
Another feature of some GalNAc-Ts is their unique substrate specificity toward prior GalNAc-glycosylated glycopeptide substrates. This was first demonstrated with GalNAc-T4, where the glycosylation of two sites in the MUC1 tandem repeat was shown to be exclusively performed by GalNAc-T4, but only following prior glycosylation by other isoforms at one or more of the other three sites in the repeat (Hassan et al. 2000). This finding led to the discovery of the function of the putative lectin domain found in the C-terminal end of most GalNAc-Ts (Hazes 1996; Imberty et al. 1997). It has since been shown that the lectin domain has binding specificity for GalNAc and modulates the catalytic function of the enzyme (Hassan et al. 2000; Tenno et al. 2002; Wandall et al. 2007; Pedersen et al. 2011). Such lectin domain-directed glycopeptide glycosylation has been demonstrated for several other GalNAc-Ts, with some isoforms directing glycosylation N-terminal (∼7–12 residues) of a prior site of glycosylation, others directing C-terminal and some even showing equal N- and C-terminal preference (Wandall et al. 2007; Raman et al. 2008; Pedersen et al. 2011; Gerken et al. 2013). Several GalNAc-T isoforms have nearly exclusive specificities for GalNAc-glycopeptide substrates such as GalNAc-T7 (Bennett, Hassan, Hollingsworth et al. 1999) and GalNAc-T10 (Cheng et al. 2002; Raman et al. 2008). GalNAc-T10 has been shown to typically glycosylate acceptor sites immediately N terminal to O-GalNAc glycosites (Raman et al. 2008; Perrine et al. 2009). This preference is best explained by direct binding of the catalytic site to the GalNAc-glycopeptide substrate (Kubota et al. 2006; Raman et al. 2008).
A major limitation in past studies has been access to a large unbiased set of validated in vivo utilized O-glycosites from diverse O-glycoproteins. The O-glycosite dataset derived from our SC strategy provides such a discovery platform (Steentoft et al. 2013). We previously presented a preliminary analysis of 181 peptide substrates derived from 131 O-glycoproteins tested with eight GalNAc-T isoforms (Steentoft et al. 2013). Here, we present a more comprehensive analysis of 195 (162 of the above 181 peptides included) peptide substrates derived from randomly selected O-glycoproteins in vitro glycosylated with 10 GalNAc-T isoforms. The results suggest that the major players in human O-glycosylation are GalNAc-T1, T2 and T3, while other isoforms contribute with surprisingly few novel functions and instead appear to be involved in partly overlapping functions.
Results
In order to expand our knowledge on the individual functions of GalNAc-T isoforms, we generated 195 synthetic ∼20mer peptides designed to cover 327 O-glycosites identified in 148 O-glycoproteins by the SC strategy (Steentoft et al. 2013). Our panel of peptide substrates represents a selection of O-linked glycoproteins, which is biased against mucin and mucin-like sequences. This is because the SC glycoproteomics strategy tends to select against heavily O-glycosylated mucin domains, due to limitation in our proteolytic digestion strategies to produce suitable size O-glycopeptides for MS sequencing (Steentoft et al. 2013). The panel of peptides was tested by in vitro enzyme reactions using time-course product development assays monitored by MALDI-TOF MS, which enables semi-quantitative evaluation of the rate of reactions as well as the number of GalNAc residues incorporated (Bennett et al. 1998; Hassan et al. 2000). A preliminary single time-point analysis of a subset of the peptides (162) against eight GalNAc-Ts (T1, T2, T3, T5, T11, T12, T14 and T16) was presented previously (Steentoft et al. 2013). Here, we have expanded the peptide panel and analyzed additional isoforms (GalNAc-T4 and T7) with time-course assays [4 h and overnight (ON)]. Figure 1 summarizes the characteristics of peptide substrates and GalNAc-T isoforms analyzed.
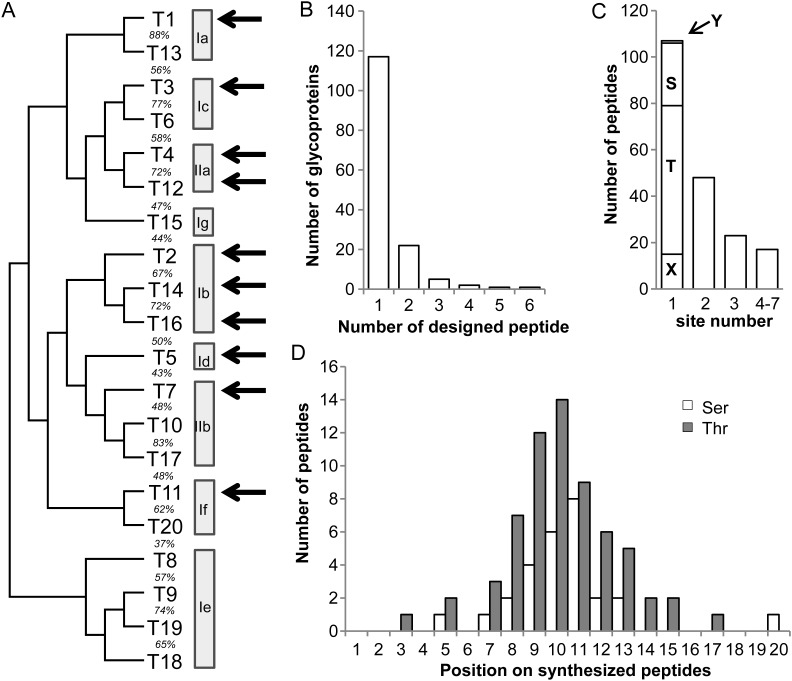
Fig. 1.
Selection and design of ∼20 mer peptide substrates based on the SC O-glycoproteome. One hundred and ninety-five peptides covering 327 SC strategy identified glycosites were selected from 148 glycoproteins and used as in vitro substrates for 10 recombinant GalNAc-Ts. (A) Human GalNAc-T phylogenetic tree and subfamilies based on Bennett et al. (2012). Recombinant human GalNAc-Ts used in this study are shown by arrows representing GalNAc-T isoforms from seven out of nine subfamilies. (B) The number of synthesized peptides designed per selected proteins. (C) The number of SC identified O-glycosites covered per synthesized peptide (both ambiguous and unambiguous sites included). Peptides containing only one site were further divided: Y, 1 Tyr site; S, 27 Ser sites; T, 64 Thr sites and X, 15 ambiguous sites. (D) The position of identified O-glycosites on synthesized peptides. Only the 91 peptides containing one unambiguous Ser or Thr SC identified glycosite were analyzed.
Selection and design of peptide substrates based on the SC O-glycoproteome
A total of 148 O-glycoproteins identified in 12 human cancer cell lines by the SC strategy (Steentoft et al. 2013) were selected for analysis (Supplementary data, Figure S1, Supplementary data, Table S1A). Most of the selected O-glycoproteins were only identified in a single cell line; however, a representative subset of O-glycoproteins expressed in more than one cell line was also included (Supplementary data, Figure S1A and Supplementary data, Table S1A, Columns I and J). Of the selected O-glycoproteins, 52 had only one identified O-glycosylation site, while others had up to 31 sites identified from the SC O-glycoproteome (Supplementary data, Figure S1B). A total of 195 ∼20mer peptide substrates were selected for synthesis (Figure 1B, Supplementary data, Table S1A). Peptide substrates were selected using one or more of the following criteria: (i) peptides contained a single (or multiple) O-glycosylation site with either Thr, Ser, or Tyr as acceptor sites (Figure 1C); (ii) acceptor sites preferably located within the center or at least three residues away from either terminal of the peptide (except for four peptides, Supplementary data, Table S1A, Column A) (Figure 1D) and (iii) acceptor sites located in regions of proteins with predicted unstructured nature (110 peptides) as well as within regions predicted to have structural constraints, i.e., in annotated folded domains of proteins (62 peptides) (Supplementary data, Table S1A, Column H). The latter being of interest as O-glycosylation is thought to predominantly occur in the Golgi after protein folding, and therefore peptides covering O-glycosites positioned in folded domains may potentially behave differently in the in vitro glycosylation assays.
Expression and purification of GalNAc-T isoforms
One or more members of seven of the nine GalNAc-T subfamilies (Figure 1A) previously shown to be functionally active were selected for study. The GalNAc-Ts were expressed as secreted proteins in insect cells and purified to apparent homogeneity as evaluated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Supplementary data, Figure S2). GalNAc-T1, having similar substrate activities with GalNAc-T13 (Bennett et al. 2012) (unpublished), was selected from subfamily Ia. Three members from subfamily Ib (GalNAc-T2, T14 and T16) were selected since detailed comparisons of their substrate specificities are lacking (Wang et al. 2003; Raman et al. 2012). From subfamily Ic GalNAc-T3 was chosen since it has almost identical substrate specificity with GalNAc-T6 (Bennett, Hassan, Mandel et al. 1999). GalNAc-T5 and T11 constituting subfamilies Ig and If, respectively, were also included (Bennett, Hassan, Hollingsworth et al. 1999; Schwientek et al. 2002). GalNAc-T4 and T12 from subfamily IIa exhibiting peptide as well as GalNAc-glycopeptide substrate specificities were included (Bennett et al. 1998; Hassan et al. 2000; Guo et al. 2002), and GalNAc-T7 reported to exhibit exclusive GalNAc-glycopeptide specificity from subfamily IIb was also selected (Bennett, Hassan, Hollingsworth et al. 1999). All the purified GalNAc-Ts used in these studies were tested and shown to be active against control peptide substrates (Supplementary data, Table S1G).
In vitro GalNAc-T time-course assays monitored by MALDI-TOF MS
A summary of MALDI-TOF analysis of 4 h and ON glycosylation reactions with the peptide panel (195 peptides) using the 10 GalNAc-Ts (T1, T2, T3, T4, T5, T7, T11, T12, T14 and T16) is presented in Supplementary data, Table S1B. A semi-quantitative scoring system, described in Supplementary data, Figure S3, was used to convert the MALDI-TOF MS peak intensity data into a measure of both the relative extent of GalNAc glycosylation [i.e., (+), +, ++ and +++] and the number of sites of GalNAc incorporated (1, 2, 3, …) into each acceptor peptide. This scoring ignored minor peaks (intensity of glycopeptide peak <5% of the intensity of the peptide peak) to disregard false-positive assignment of products. The results showed that 122 of 195 (63%) peptides served as substrate for one or more GalNAc-Ts after 4 h and 146 (75%) after ON incubation (Supplementary data, Table S1A, Columns F and G). Of the 107 peptides containing a single SC identified O-glycosite 55 (51%) were glycosylated ON with one GalNAc residue, 16 (15%) with more than one GalNAc residue, while 36 (34%) were not glycosylated. Of the 88 peptides with more than one SC identified glycosite 44 (50%) were glycosylated with one GalNAc residue, 31 (35%) with more than one, while 13 (15%) were not glycosylated (Supplementary data, Figure S4).
Most SC identified sites were located within protein sequences containing other Ser/Thr/Tyr residues, thus most peptide substrate designs also include one or more Ser/Thr/Tyr residues not identified as O-glycosites. Several peptide substrates were glycosylated with more than one GalNAc residue, but we did not sequence the glycosylation products and did not determine the actual site(s) of incorporation of GalNAc residues due to the extensive efforts required. Thus, the analyses of the in vitro glycosylation data are under the assumption that incorporation primarily occur at the sites identified by the SC approach. Analysis of peptides with single SC identified O-glycosites (92 peptides) under these premises revealed that Thr acceptor sites were glycosylated in 68% cases ON (54% at 4 h), which was slightly better compared to Ser sites with 52% glycosylated ON (41% at 4 h) (Figure 2A). This is in agreement with previous reports showing that Thr residues are better acceptors than Ser (O'Connell and Tabak 1993; Wang et al. 1993; Elliott et al. 1994; Gerken et al. 2002).
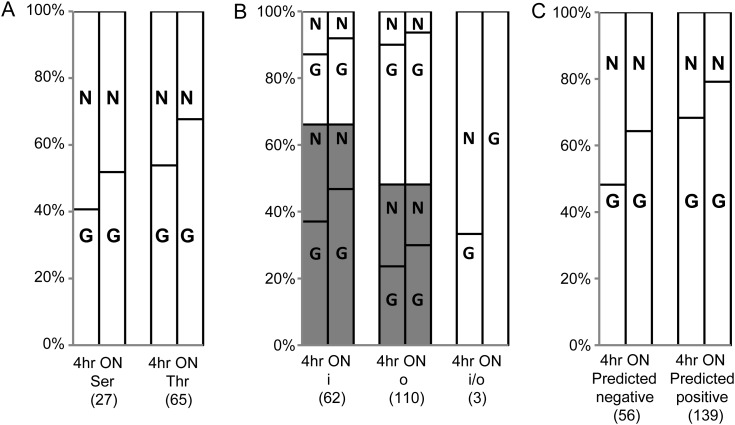
Fig. 2.
Analyses of in vitro peptide substrate preference. In vitro glycosylation of peptide substrates by all tested GalNAc-Ts were analyzed. (A) In vitro preference of Ser or Thr residues as acceptor sites. In vitro glycosylation of 92 peptides containing one unambiguously identified Thr/Ser site (peptide from Apo-CIII with one ambiguous, but known Thr glycosite included) were analyzed. Twenty-seven peptides contain Ser glycosites while 65 have Thr glycosites. (B) Analysis of the relationship between in vitro substrate preference and their location in proteins. 175 peptides annotated by the Conserved Domain Database were analyzed (Supplementary data, Table S1A, Column H). These peptides include 62 peptides located in an annotated folded domain (i), 110 peptides located outside a folded domain (o) and three peptides at the i/o interface (i/o), and they were further divided into two subsets, peptides with single (black bar) or multiple (white bar) identified glycosites. (C) Comparison of in vitro glycosylation and NetOGlyc4.0 prediction. All 195 synthetic peptides were analyzed to evaluate the prediction ability of NetOGlyc4.0. These peptides are divided into 56 negative-predicted peptides and 139 positive-predicted peptides. The number of nonglycosylated or glycosylated peptides was demonstrated as percentage. N: in vitro nonglycosylated; G: in vitro glycosylated. 4 h: in vitro glycosylation in 4 h (Supplementary data, Table S1A, Column D); ON: in vitro glycosylation ON (Supplementary data, Table S1A, Column E); Number in brackets: number of synthetic peptides.
Perhaps interestingly, we found that when analyzing glycosylation of single SC identified glycosites about 71% ON (56% at 4 h) of the peptides derived from folded domain structures served as substrates for GalNAc-Ts, while 62% ON (49% at 4 h) of the peptides derived from unstructured regions of proteins served as substrates (Figure 2B). But for sequences with multiple SC identified glycosites a reverse tendency was demonstrated.
Finally, we evaluated the performance of neutral network prediction of mucin type GalNAc O-glycosylation sites (NetOGlyc) to predict whether peptides served as substrates for glycosylation, and 79% ON (68% at 4 h) of the peptides predicted to be glycosylated in fact served as substrates for the GalNAc-Ts, which is a slightly higher proportion compared with the peptides not predicted to be glycosylated where 64% ON (48% at 4 h) could be glycosylated (Figure 2C).
Probing GalNAc-T isoform specific functions
The three best characterized GalNAc-Ts, GalNAc-T1–T3, glycosylated 135 of 195 (69%) peptides showing both overlapping substrate specificities as well as apparent unique substrate specificities (Figure 3A). Sequence logo-plots of the peptides with single identified O-glycosites are shown in Supplementary data, Figure S5. Both the overall and individual (for GalNAc-T1–T3) in vitro glycosylated peptides demonstrate the commonly observed Pro preferences at positions −1 and +3 of the acceptor site as demonstrated previously (Briand et al. 1981; Wilson et al. 1991; O'Connell et al. 1992; Nehrke et al. 1996; Gerken et al. 2006, 2011).
Fig. 3.
Substrate selectivity and overlap analyses of GalNAc-T isoforms. ON glycosylation data was used for analysis with the 4 h glycosylation data in brackets. (A) Substrate overlap between GalNAc-T1, T2 and T3. (B) Substrate overlap between GalNAc-T1-3 and the remaining GalNAc-Ts. Eleven peptides were specifically glycosylated by GalNAc-T4, T5, T7, T11, T12, T14 and T16. (C) Substrate overlap among subfamily Ib including GalNAc-T2, T14 and T16. (D) Substrate overlap between GalNAc-T4 and T12 (subfamily IIa).
Collectively, the seven remaining GalNAc-Ts (T4, T5, T7, T11, T12, T14 and T16) glycosylated 103 (53%) peptides, and surprisingly only 11 of these peptides were not substrates for GalNAc-T1, T2 or T3 after ON reaction (Figure 3B). After ON incubation, the number of glycosylated peptide substrates for GalNAc-T4, T5, T7, T11, T12, T14 and T16 were 29, 14, 49, 31, 27 and 45, respectively, compared with 86 for GalNAc-T1, 84 for T2 and 107 for T3. Overall, GalNAc-T1–T3 showed considerably higher catalytic efficiency with the identified peptide substrates compared with the other tested GalNAc-T isoforms (both 4 h and ON reactions). Selected specific and preferred peptide substrates for GalNAc-T1, T2, T3 and the other GalNAc-Ts are shown in Supplementary data, Table S1C–S1F.
The three members of the close homologous subfamily Ib (GalNAc-T2, T14, T16) showed substantial peptide substrate overlap (Figure 3C). Interestingly, GalNAc-T2 was found to have a higher extent of glycosylation and wider substrate selection than T14 and T16. In contrast, only partial overlap was found between the two members from subfamily IIa (GalNAc-T4 and T12) on nonglycosylated peptide substrates (Figure 3D).
Comparisons with ISOGlyP predictions
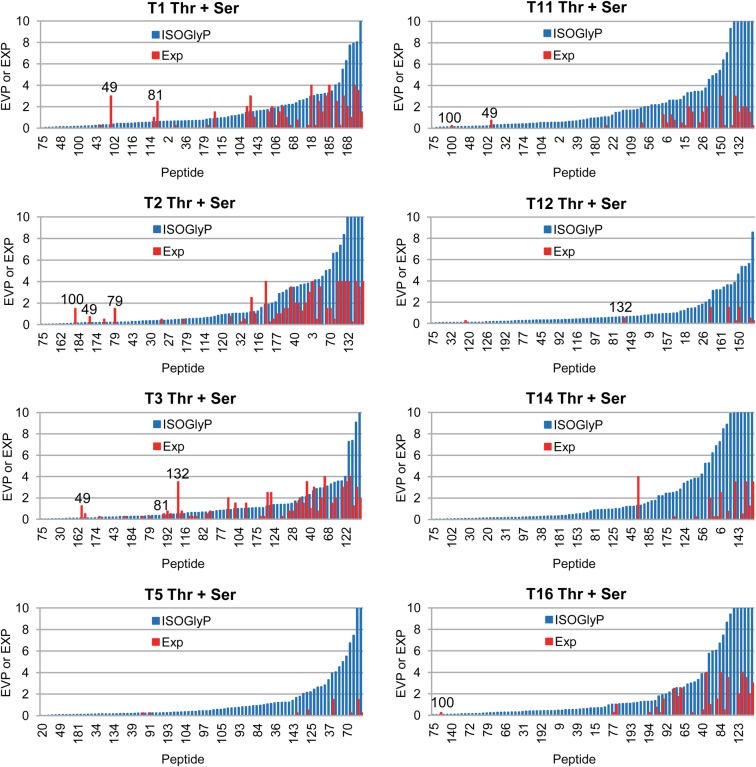
The isoform specific O-glycosylation prediction (ISOGlyP) web based isoform specific predictor of O-glycosylation is based on the glycosylation of random peptides substrates containing only one Thr acceptor flanked by 3–5 random positions plus flanking GAGA residues (Gerken et al. 2006, 2008, 2011; Perrine et al. 2009). The result of the ISOGlyP analysis of a given sequence is an enhancement value product (EVP), which is thought to represent the propensity (or rate) of a site for glycosylation, with a value of 1 being neutral, values <1 inhibitory and values >1 enhanced for glycosylation. For optimal comparison, we therefore restricted the comparison of our in vitro glycosylation data to peptide substrates containing only single unambiguous SC identified sites. We also excluded the few peptide substrates where the SC identified site was not positioned centrally (Supplementary data, Table S2A, Column A). Calculation of the ISOGlyP EVP values and comparison to the experimentally observed glycosylation data (Supplementary data, Table S2A) was performed under the assumption that the SC identified glycosite was the same utilized in the in vitro assay. The plots of the EVP value and relative experimental glycosylation are given in Figure 4. The ISOGlyP EVP values reasonably predict glycosylation by the corresponding GalNAc-Ts, i.e., peptides with low EVP values are not glycosylated while those with the high values are more likely to be glycosylated. Interestingly, the most active GalNAc-Ts (T1–T3) that glycosylate the most peptides show the best correlations. One might speculate that by using a greater amount of enzymes, thus leading to higher extent of glycosylation the correlation with the less active GalNAc-Ts would improve. Nevertheless, if the ISOGlyP EVP value is low the likelyhood of glycosylation is also very low. Presently we do not understand why high ISOGlyP EVP values do not fully predict glycosylation, which we are presently pursuing in further studies. Interestingly, there were five peptides where the ISOGlyP value was very low (significantly <1) and the observed peptide highly glycosylated (Thr peptides 49 (DPM3), 79 (GLT8D1), 132 (MRC2), Ser peptides 81 (GPA33) and 100 (KIAA0494), Supplementary data, Table S2A, Figure 4). An analysis of the peptide sequence revealed that there was another Thr or Ser residue in the peptide which had an EVP value of >1 (Supplementary data, Table S2B) suggesting that the alternative site would be glycosylated instead.
Fig. 4.
Comparison of GalNAc-T isoform substrate preference with the ISOGlyP EVP. The in vitro glycosylation (red bars) of 90 peptides containing a single unambiguously identified Thr or Ser glycosite (Supplementary data, Table S2A) (peptide derived from Apo-CIII with an ambiguous but known glycosite included) are compared with the site's EVP value obtained by ISOGlyP (http://isoglyp.utep.edu/) for GalNAc-T1, T2, T3, T5, T11, T12, T14 and T16 (blue bars). As the in vitro glycosites for synthetic peptides were not defined, the in vivo identified glycosites were considered as the acceptor sites and used for EVP value calculation. The in vitro glycosylation data was converted as follows: (+), 0.5; +, 1; ++, 2; +++, 3; ++++, 4 and the average of the 4 h and ON glycosylation data plotted. Peptides were arranged according to EVP values and EVP values >10 were truncated. Five peptides (49, 79, 81, 100, 132, Supplementary data, Table S2A for sequences) with very low EVP values at in vivo Ser/Thr glycosites, but glycosylated by some isoforms and containing alternative Ser/Thr sites with EVP values >1 were labeled in the plot.
Discussion
Here, we have performed a comprehensive analysis of multiple GalNAc-Ts with a large panel of peptides designed from a set of O-glycoproteins identified from our SC O-glycoproteome strategy (Steentoft et al. 2013). We found that: (i) the majority of peptides served as substrates for one or more GalNAc-T isoforms; (ii) the most broadly expressed isoforms, GalNAc-T1–T3, glycosylated by far the majority of these peptide acceptor substrates; (iii) the isoforms with more restricted expression, GalNAc-T4, T5, T11, T12, T14, T16, generally glycosylated a minor subset of the peptides and only contributed a few novel substrates; (iv) the GalNAc-glycopeptide preferring isoforms, GalNAc-T4, T7 and T12, were indeed found to glycosylate a subset of peptides and (v) we did not identify substantial general features that characterize peptide substrates differentially glycosylated between GalNAc-T isoforms. Several examples have demonstrated how in vitro O-glycosylation correlates and predicts in vivo function of GalNAc-T isoforms (Wandall et al. 1997; Nehrke et al. 1998; Kato et al. 2006; Schjoldager et al. 2010, 2012). The present data further supports this idea and suggests that if anything the experimental in vitro peptide assay may under report the actual in vivo glycosylation. The results provide a baseline for further studies of the functions of the many GalNAc-T isoforms and their contribution to regulation of the large differential O-glycoproteome of cell.
In vitro analysis of properties of the GalNAc-Ts depends on relevant peptide substrates, and except for the random peptide substrate approach (Gerken et al. 2006, 2011), only a limited number of peptide substrates have been analyzed with multiple GalNAc-T isoforms (Bennett et al. 2012). Here, we used a large panel of peptides designed from over 140 O-glycoproteins identified by the SC strategy (Steentoft et al. 2013). The selected peptide substrates differ from those used in the past by having single or isolated O-glycosylation sites rather than primarily having highly clustered sites typical of mucins and mucin-like domains (Ten Hagen et al. 2003; Bennett et al. 2012) (Figure 1). A potential drawback with the SC strategy is that it does not currently enable assessment of stoichiometry of identified O-glycosites. Thus, some of the glycosites identified may represent low occupancy sites because they serve as poor substrates for O-glycosylation in cells. A classical example of this is the single Ser O-glycosylation site (121-PPDAASAAPLR-131) in erythropoietin, which is only partially utilized in CHO cells (Delorme et al. 1992). Interestingly, in vitro glycosylation of different peptides covering this site are very poor substrates for all the GalNAc-Ts tested so far (Wang et al. 1993; Wandall et al. 1997; Schwientek et al. 2002). In line with this only 75% of the peptides covering in vivo identified O-glycosites indeed served as substrates for in vitro O-glycosylation (Figure 3). An alternative explanation may be that other GalNAc-T isoforms not tested in the present study would glycosylate these peptides, but we did test at least one member of all GalNAc-T subfamilies. The only exceptions were GalNAc-T15 from subfamily Ig (Cheng et al. 2004) and the Ie subfamily of four isoforms that so far have not been found to be active (Bennett et al. 2012). Another explanation may be that short peptides do not always mimic acceptor substrate sites that depend on conformation in, e.g., folded domains of proteins. We recently found that GalNAc-T11 is the only isoform capable of glycosylating the low-density lipoprotein receptor (LDLR) in its ligand-binding region, where the short linker regions between the LDLR class A repeats (found in LDLR and related receptors) contain a highly conserved O-glycosite (Steentoft et al. 2013; Pedersen et al. 2014) (Supplementary data, Figure S6). Interestingly, none of the peptides derived from these sites tested in the present in vitro assay served as efficient substrates for GalNAc-T11 (peptide 105, 107 from LRP1 and peptide 108 from LRP8 given in Supplementary data, Table S1B) (Pedersen et al. 2014). Given that the Class A repeats are structurally similar to EGF repeats containing three intradomain disulphide bridges and that the linker regions consist of only four amino acids with the O-glycosites positioned −1 to the first cysteine residue, we reasoned that our short model peptides may not present the substrate sites in the proper conformation for in vitro glycosylation. However, we found that GalNAc-T11 in fact glycosylates a recombinant Escherichia coli expressed LDLR reporter protein in vitro and confirmed this in vivo using GALNT11 knock out cells (Pedersen et al. 2014). It is possible that other isoforms exhibit substrate specificities that have unique structural constraints.
Our study confirmed that the broadly expressed GalNAc-T1–T3 isoforms are the major contributors to the O-glycoproteome. We did not test the close subfamily homologs of GalNAc-T1 and T3, GalNAc-T13 and T6, respectively, but we previously showed that GalNAc-T3 and T6 have almost identical substrate specificities on a small peptide panel (Bennett, Hassan, Mandel et al. 1999) and a survey of a large set of peptide substrates showed in preliminary studies that GalNAc-T1 and T13 also shared similar substrate specificities (unpublished), which was additionally demonstrated by the random peptide strategy (Gerken and Revoredo, unpublished). The latter findings are in disagreement with a previous report using a limited set of peptides (Zhang et al. 2003). We did test all members of subfamily Ib, including GalNAc-T2, T14, and T16, and observed that GalNAc-T14 and T16 showed similar but more restricted substrate specificities compared to GalNAc-T2 (Figure 4C). Similar glycosylation motifs for these three isoforms have been obtained with the random peptide approach (Gerken and Revoredo, unpublished). GalNAc-T11 is classified in subfamily If together with GalNAc-T20, but the T20 isoform has not been demonstrated to function as a GalNAc-T and it has been proposed to represent a pseudo gene (Bennett et al. 2012; Raman et al. 2012). However, recent studies do suggest a role for GalNAc-T20 in male infertility (Takasaki et al. 2014).
GalNAc-Ts with selective or exclusive GalNAc-glycopeptide substrate specificities are classified in subfamily IIa (GalNAc-T4 and T12) and IIb (GalNAc-T7 and T10), respectively (Bennett et al. 2012). GalNAc-T4 glycosylated a small number of peptides as reported previously (Bennett et al. 1998; Hassan et al. 2000), while T12 showed distinct but partially overlapping peptide substrate specificity with T4 (Figure 3D). We did not further assay and compare GalNAc-glycopeptide substrate specificities of these isoforms since the majority of substrate sites studied represents isolated sites, but it appears that both GalNAc-T4 and T12 have important functions against glycopeptide substrates and these differ (unpublished). The analysis of GalNAc-T7 from subfamily IIb, whose members (GalNAc-T7, T10 and T17) are reported to have strict preference for glycopeptides (Bennett, Hassan, Hollingsworth et al. 1999; Cheng et al. 2002; Raman et al. 2012), demonstrated that T7 in fact catalyzed partial incorporation of GalNAc into 26 peptide substrates, although the time-course assay suggested that this activity was relatively low. Further studies are needed to explore this, but our findings would suggest that members of IIb similar to members of IIa may have unique and quite restricted peptide substrate specificities beyond their glycopeptide activities. Noteworthy was that several of the substrates identified for GalNAc-T7 were not glycosylated by any of the other isoforms tested. Interestingly, the random peptide substrate studies of GalNAc-T10 show that this GalNAc-T possesses a broad range of specificity and lacks the typical Pro enhancements although with significantly reduced activity (Perrine et al. 2009).
A few Tyr glycosites with no obvious consensus surrounding residues have been identified in the SC O-glycoproteome (Steentoft et al. 2013; Vakhrushev et al. 2013), and we hypothesized that these are also catalyzed by the GalNAc-Ts. The site from NUCB2 (peptide 144, HDQLEAQKLEYHQVIQQMEQK) included in our study could not be glycosylated by any of the GalNAc-Ts tested (Supplementary data, Table S1B). Two other Tyr glycosites from EGFLAM (RQLNINGALYVGGMKEIALH) and APP (Aβ) (DAEFRHDSGYEVHHQKLV) (Supplementary data, Figure S6) (Halim et al. 2011) also did not serve as substrates for any isoforms tested (data not shown). Currently, we do not know how efficiently these Tyr sites are glycosylated in vivo, but the Tyr glycosite identified in Aβ was found with low stoichiometry in Aβ isolated from human cerebrospinal fluid (CSF).
Congenital deficiencies in GALNT genes have been found to be a direct cause of disease (Topaz et al. 2004; Fakhro et al. 2011) or be associated with diseases (Kathiresan et al. 2008; Willer et al. 2008) demonstrating that some GalNAc-T isoforms serve nonredundant essential functions. Since GalNAc-Ts are differentially expressed in cells and tissues (Bennett et al. 2012), functional redundancy has to be evaluated in the context of the repertoire of GalNAc-T isoforms expressed. We previously showed that familial tumoral calcinosis (FTC) (Topaz et al. 2004) is caused by loss of GalNAc-T3 mediated glycosylation of Thr178 in a proprotein processing inactivation site of FGF23, which is produced in osteoblasts (Kato et al. 2006). Similarly GalNAc-T2 is required for glycosylation of Apo-CIII and ANGPTL3 produced in liver (Schjoldager et al. 2010, 2012). In both cases other GalNAc-T isoforms can compensate these functions, but such isoforms are not expressed in the cell types producing these substrate proteins. It is, therefore, important to analyze functions of GalNAc-Ts in the context of the repertoire of isoforms expressed in relevant cells. To this end, we are employing isogenic cell lines with selective loss of a single GalNAc-T isoform in the so-called SC strategy. In a first preliminary study, we were able to validate the unique function of GalNAc-T2 with Apo-CIII and ANGPTL3 in HepG2 liver cells using a differential glycoproteomics approach (Schjoldager et al. 2012).
In summary, this study provides the first comprehensive in vitro analysis of the peptide substrate specificities of a large number of the GalNAc-T isoforms and suggests that three isoforms (GalNAc-T1–T3) are major contributors to formation of the overall O-glycoproteome in most cell lines studied to date, while a number of other isoforms (GalNAc-T4, T5, T7, T11, T12, T14 and T16) appear to be minor contributors with more overlapping than unique functions. Future studies of differential O-glycoproteomes in model cell systems or animals are needed to further evaluate nonredundant functions of the many GalNAc-T isoforms, but the in vitro studies as reported here clearly provide a valuable discovery platform for identifying GalNAc-T isoform-specific functions and potential roles in disease.
Materials and methods
Expression and purification of GalNAc-Ts
Soluble truncated recombinant GalNAc-Ts were expressed as secreted proteins in High five insect cells (Vester-Christensen et al. 2013). Enzymes were purified by either SP-sepharose (Sigma-Aldrich, St. Louis, MO, USA) followed by MonoS (5/50 GL, GE Healthcare, Europe GmbH, Denmark) (GalNAc-T1–T3), or by Ni-NTA agarose (Invitrogen, Carlsbad, CA, USA) followed by MonoQ/S (5/50 GL, GE Healthcare) (GalNAc-T4, T5, T7, T11, T12, T14 and T16). SP-sepharose chromatography was performed after incubation of the culture medium with Amberlite IRA96 (Sigma-Aldrich) and dilution (3×) in 25 mM Bis-Tris (pH 6.0, 10 mM NaCl). The SP-sepharose was extensively washed with 20 column volumes (CVs) 25 mM Bis-Tris (pH 6.0, 10 mM NaCl) and the enzymes eluted with a linear gradient of NaCl from 10 mM to 1 M in 10 CVs. Ni-NTA chromatography was performed after prior dialysis of culture medium against 50 mM Tris (pH 8.0, 150 mM NaCl) at 4°C. The Ni-NTA column was extensively washed with 40–50 CV 25 mM Tris (pH 8.0, 300 mM NaCl, 10 mM Imidazole) and enzymes eluted with 5 CVs elution buffer (25 mM Tris, pH 8.0, 300 mM NaCl, 250 mM Imidazole). Further purification was performed using MonoQ or MonoS using an ÄKTA™ fast protein liquid chromatography (GE Healthcare) interfaced by the UNICORN 4.12 control software. Purified enzymes were analyzed by SDS-PAGE (4–12% Bis-Tris gradient gel, Invitrogen) Coomassie staining and quantified by ImageJ with bovine serum albumin as standards. Western blots with anti-His or specific monoclonal antibodies to individual enzymes were used for confirmation.
In vitro O-GalNAc glycosylation assays
A panel of 195 peptides containing glycosites identified by the SC O-glycoproteome strategy (Steentoft et al. 2013) was designed and custom synthesized (NeoBioSci and Schafer-N). In vitro glycosylation assays were performed as product development assays in 25 μL buffer (25 mM cacodylic acid sodium, pH 7.4, 10 mM MnCl2, 0.25% Triton X-100), 2 mM UDP-GalNAc (Sigma), 10 μg of acceptor peptides and 0.1 μg of purified enzyme at 37°C. For time-course evaluation 1 μL of the reaction mixtures were taken at 4 h and ON, and analyzed by MALDI-TOF MS. Positive control peptide substrates (Supplementary data, Table S1G) used for different GalNAc-T isoforms were: Muc1a (GalNAc-T1); IgA-h (GalNAc-T2, T3 and T11); Muc7 (GalNAc-T4); Muc2-1b (GalNAc-T5); Muc2-1a-Tn (GalNAc-T7); Pomc (GalNAc-T12) and Act8 (GalNAc-T14 and T16).
MALDI-TOF MS analysis
Evaluation of incorporation of GalNAc residues into peptide substrates was performed by MALDI-TOF MS. One microliter of reaction mixture was diluted with 9 μL 0.1% TFA/H2O and 1 μL mixed with 1 μL of 10 mg/mL 2,5-dihydrobenzoic acid dissolved in ACN/H2O (7:3). Acquisition of MS spectra was performed on MALDI-TOF instruments, Voyager-DE™ (Applied Biosystems, Foster City, CA, USA) operating at an accelerating voltage of 20 kV in the linear mode, and with Bruker Autoflex (Bruker Daltonik GmbH, Bremen, Germany) using the Bruker Flex Control 3.4 software. Spectra were recorded in the positive ion mode and the raw spectra were processed by the Voyager Data Explorer 5.1 and Bruker FlexAnalysis 3.4 software.
Data analysis
The NetOGlyc4.0 predictor (Steentoft et al. 2013) (http://www.cbs.dtu.dk/services/NetOGlyc/) was used to evaluate O-glycosites included in the study. The predictor ISOGlyP (Gerken et al. 2006, 2008, 2011; Perrine et al. 2009) (http://isoglyp.utep.edu/) was utilized to evaluate GalNAc-T isoform specificity of substrate sites. The Conserved Domain Database (Marchler-Bauer et al. 2013) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) was used for prediction of general protein folded domain composition and analysis of spacial relationships of O-glycosites and peptide locations on proteins.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the Carlsberg Foundation, the Danish Research Councils, a program of excellence from the University of Copenhagen, the Danish National Research Foundation (DNRF107), the National Institutes of Health (R01 CA078834 to T.A.G.) and a scholarship from the China Scholarship Council (to Y.K.).
Abbreviations
ACN, acetonitrile; ANGPTL3, angiopoietin-like Protein 3; APOC3, Apo-CIII, Apolipoprotein C-III; BSA, bovine serum albumin; DHB, 2,5-dihydroxybenzoic acid; EVP, enhancement value product; FGF23, fibroblast growth factor 23; FPLC, fast protein liquid chromatography; FTC, familial tumoral calcinosis; GalNAc, N-acetyl-D-galactosamine; GalNAc-T, UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase; GALNT, GalNAc-transferase gene; ISOGlyP, isoform specific O-glycosylation prediction; LDLR, low-density lipoprotein receptor; MALDI-TOF, matrix assisted laser desorption/ionization time-of-flight; MS, mass spectrometry; MUC1, mucin 1; NetOGlyc, neutral network prediction of mucin type GalNAc O-glycosylation sites; ON, overnight; SC, SimpleCell; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; Ser, serine; TFA, trifluoroacetic acid; Thr, threonine; Tris, tris(hydroxymethyl)aminomethane; Tyr, tyrosine; UDP, uridine diphosphate.
Supplementary Material
References
- Bennett EP, Hassan H, Clausen H. cDNA cloning and expression of a novel human UDP-N-acetyl-alpha-D-galactosamine. Polypeptide N-acetylgalactosaminyltransferase, GalNAc-T3. J Biol Chem. 1996;271:17006–17012. doi: 10.1074/jbc.271.29.17006. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Hollingsworth MA, Clausen H. A novel human UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. 1999;460:226–230. doi: 10.1016/s0014-5793(99)01268-5. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Mandel U, Hollingsworth MA, Akisawa N, Ikematsu Y, Merkx G, van Kessel AG, Olofsson S, Clausen H. Cloning and characterization of a close homologue of human UDP-N-acetyl-alpha-D-galactosamine:Polypeptide N-acetylgalactosaminyltransferase-T3, designated GalNAc-T6. Evidence for genetic but not functional redundancy. J Biol Chem. 1999;274:25362–25370. doi: 10.1074/jbc.274.36.25362. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, van Kessel AG, et al. Cloning of a human UDP-N-acetyl-alpha-D-Galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J Biol Chem. 1998;273:30472–30481. doi: 10.1074/jbc.273.46.30472. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand JP, Andrews SP, Jr, Cahill E, Conway NA, Young JD. Investigation of the requirements for O-glycosylation by bovine submaxillary gland UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosamine transferase using synthetic peptide substrates. J Biol Chem. 1981;256:12205–12207. [PubMed] [Google Scholar]
- Cheng L, Tachibana K, Iwasaki H, Kameyama A, Zhang Y, Kubota T, Hiruma T, Kudo T, Guo JM, Narimatsu H. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T15. FEBS Lett. 2004;566:17–24. doi: 10.1016/j.febslet.2004.03.108. [DOI] [PubMed] [Google Scholar]
- Cheng L, Tachibana K, Zhang Y, Guo J, Kahori Tachibana K, Kameyama A, Wang H, Hiruma T, Iwasaki H, Togayachi A, et al. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T10. FEBS Lett. 2002;531:115–121. doi: 10.1016/s0014-5793(02)03399-9. [DOI] [PubMed] [Google Scholar]
- Delorme E, Lorenzini T, Giffin J, Martin F, Jacobsen F, Boone T, Elliott S. Role of glycosylation on the secretion and biological activity of erythropoietin. Biochemistry. 1992;31:9871–9876. doi: 10.1021/bi00156a003. [DOI] [PubMed] [Google Scholar]
- Elliott S, Bartley T, Delorme E, Derby P, Hunt R, Lorenzini T, Parker V, Rohde MF, Stoney K. Structural requirements for addition of O-linked carbohydrate to recombinant erythropoietin. Biochemistry. 1994;33:11237–11245. doi: 10.1021/bi00203a020. [DOI] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Jamison O, Perrine CL, Collette JC, Moinova H, Ravi L, Markowitz SD, Shen W, Patel H, Tabak LA. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J Biol Chem. 2011;286:14493–14507. doi: 10.1074/jbc.M111.218701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Raman J, Fritz TA, Jamison O. Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J Biol Chem. 2006;281:32403–32416. doi: 10.1074/jbc.M605149200. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Revoredo L, Thome JJ, Tabak LA, Vester-Christensen MB, Clausen H, Gahlay GK, Jarvis DL, Johnson RW, Moniz HA, et al. The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J Biol Chem. 2013;288:19900–19914. doi: 10.1074/jbc.M113.477877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen KG, Jamison O. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase ortholog pairs. Glycobiology. 2008;18:861–870. doi: 10.1093/glycob/cwn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Zhang J, Levine J, Elhammer A. Mucin core O-glycosylation is modulated by neighboring residue glycosylation status. Kinetic modeling of the site-specific glycosylation of the apo-porcine submaxillary mucin tandem repeat by UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases T1 and T2. J Biol Chem. 2002;277:49850–49862. doi: 10.1074/jbc.M205851200. [DOI] [PubMed] [Google Scholar]
- Gill DJ, Chia J, Senewiratne J, Bard F. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J Cell Biol. 2010;189:843–858. doi: 10.1083/jcb.201003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JM, Zhang Y, Cheng L, Iwasaki H, Wang H, Kubota T, Tachibana K, Narimatsu H. Molecular cloning and characterization of a novel member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family, pp-GalNAc-T12. FEBS Lett. 2002;524:211–218. doi: 10.1016/s0014-5793(02)03007-7. [DOI] [PubMed] [Google Scholar]
- Halim A, Brinkmalm G, Ruetschi U, Westman-Brinkmalm A, Portelius E, Zetterberg H, Blennow K, Larson G, Nilsson J. Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc Natl Acad Sci USA. 2011;108:11848–11853. doi: 10.1073/pnas.1102664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim A, Nilsson J, Ruetschi U, Hesse C, Larson G. Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol Cell Proteomics. 2012;11:M111.013649. doi: 10.1074/mcp.M111.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H, Reis CA, Bennett EP, Mirgorodskaya E, Roepstorff P, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J, Clausen H. The lectin domain of UDP-N-acetyl-D-galactosamine: Polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J Biol Chem. 2000;275:38197–38205. doi: 10.1074/jbc.M005783200. [DOI] [PubMed] [Google Scholar]
- Hazes B. The (QxW)3 domain: A flexible lectin scaffold. Protein Sci. 1996;5:1490–1501. doi: 10.1002/pro.5560050805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imberty A, Piller V, Piller F, Breton C. Fold recognition and molecular modeling of a lectin-like domain in UDP-GalNac:polypeptide N-acetylgalactosaminyltransferases. Protein Eng. 1997;10:1353–1356. doi: 10.1093/protein/10.12.1353. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem. 2006;281:18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- Kubota T, Shiba T, Sugioka S, Furukawa S, Sawaki H, Kato R, Wakatsuki S, Narimatsu H. Structural basis of carbohydrate transfer activity by human UDP-GalNAc: Polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10) J Mol Biol. 2006;359:708–727. doi: 10.1016/j.jmb.2006.03.061. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa HE, Gerken TA, Jamison O, Tabak LA. Isoform-specific O-glycosylation of osteopontin and bone sialoprotein by polypeptide N-acetylgalactosaminyltransferase-1. J Biol Chem. 2010;285:1208–1219. doi: 10.1074/jbc.M109.035436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrke K, Hagen FK, Tabak LA. Charge distribution of flanking amino acids influences O-glycan acquisition in vivo. J Biol Chem. 1996;271:7061–7065. doi: 10.1074/jbc.271.12.7061. [DOI] [PubMed] [Google Scholar]
- Nehrke K, Hagen FK, Tabak LA. Isoform-specific O-glycosylation by murine UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-T3, in vivo. Glycobiology. 1998;8:367–371. doi: 10.1093/glycob/8.4.367. [DOI] [PubMed] [Google Scholar]
- O'Connell BC, Hagen FK, Tabak LA. The influence of flanking sequence on the O-glycosylation of threonine in vitro. J Biol Chem. 1992;267:25010–25018. [PubMed] [Google Scholar]
- O'Connell BC, Tabak LA. A comparison of serine and threonine O-glycosylation by UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase. J Dent Res. 1993;72:1554–1558. doi: 10.1177/00220345930720120401. [DOI] [PubMed] [Google Scholar]
- Pedersen JW, Bennett EP, Schjoldager KT, Meldal M, Holmer AP, Blixt O, Clo E, Levery SB, Clausen H, Wandall HH. Lectin domains of polypeptide GalNAc transferases exhibit glycopeptide binding specificity. J Biol Chem. 2011;286:32684–32696. doi: 10.1074/jbc.M111.273722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NB, Wang S, Narimatsu Y, Yang Z, Halim A, Schjoldager KT, Madsen TD, Seidah NG, Bennett EP, Levery SB, et al. Low Density Lipoprotein Receptor Class A Repeats Are O-Glycosylated in Linker Regions. J Biol Chem. 2014;289:17312–17324. doi: 10.1074/jbc.M113.545053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine CL, Ganguli A, Wu P, Bertozzi CR, Fritz TA, Raman J, Tabak LA, Gerken TA. Glycopeptide-preferring polypeptide GalNAc transferase 10 (ppGalNAc T10), involved in mucin-type O-glycosylation, has a unique GalNAc-O-Ser/Thr-binding site in its catalytic domain not found in ppGalNAc T1 or T2. J Biol Chem. 2009;284:20387–20397. doi: 10.1074/jbc.M109.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Fritz TA, Gerken TA, Jamison O, Live D, Liu M, Tabak LA. The catalytic and lectin domains of UDP-GalNAc:polypeptide alpha-N-Acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J Biol Chem. 2008;283:22942–22951. doi: 10.1074/jbc.M803387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Guan Y, Perrine CL, Gerken TA, Tabak LA. UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferases: Completion of the family tree. Glycobiology. 2012;22:768–777. doi: 10.1093/glycob/cwr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottger S, White J, Wandall HH, Olivo JC, Stark A, Bennett EP, Whitehouse C, Berger EG, Clausen H, Nilsson T. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J Cell Sci. 1998;111(Pt 1):45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- Schjoldager KT, Clausen H. Site-specific protein O-glycosylation modulates proprotein processing - deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim Biophys Acta. 2012;1820:2079–2094. doi: 10.1016/j.bbagen.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Schjoldager KT, Vakhrushev SY, Kong Y, Steentoft C, Nudelman AS, Pedersen NB, Wandall HH, Mandel U, Bennett EP, Levery SB, et al. Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc Natl Acad Sci USA. 2012;109:9893–9898. doi: 10.1073/pnas.1203563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Vester-Christensen MB, Bennett EP, Levery SB, Schwientek T, Yin W, Blixt O, Clausen H. O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: Possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J Biol Chem. 2010;285:36293–36303. doi: 10.1074/jbc.M110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwientek T, Bennett EP, Flores C, Thacker J, Hollmann M, Reis CA, Behrens J, Mandel U, Keck B, Schafer MA, et al. Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J Biol Chem. 2002;277:22623–22638. doi: 10.1074/jbc.M202684200. [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KT, Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- Takasaki N, Tachibana K, Ogasawara S, Matsuzaki H, Hagiuda J, Ishikawa H, Mochida K, Inoue K, Ogonuki N, Ogura A, et al. A heterozygous mutation of GALNTL5 affects male infertility with impairment of sperm motility. Proc Natl Acad Sci USA. 2014;111:1120–1125. doi: 10.1073/pnas.1310777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hagen KG, Fritz TA, Tabak LA. All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- Tenno M, Saeki A, Kezdy FJ, Elhammer AP, Kurosaka A. The lectin domain of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 1 is involved in O-glycosylation of a polypeptide with multiple acceptor sites. J Biol Chem. 2002;277:47088–47096. doi: 10.1074/jbc.M207369200. [DOI] [PubMed] [Google Scholar]
- Topaz O, Shurman DL, Bergman R, Indelman M, Ratajczak P, Mizrachi M, Khamaysi Z, Behar D, Petronius D, Friedman V, et al. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat Genet. 2004;36:579–581. doi: 10.1038/ng1358. [DOI] [PubMed] [Google Scholar]
- Vakhrushev SY, Steentoft C, Vester-Christensen MB, Bennett EP, Clausen H, Levery SB. Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol Cell Proteomics. 2013;12:932–944. doi: 10.1074/mcp.O112.021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester-Christensen MB, Bennett EP, Clausen H, Mandel U. Generation of monoclonal antibodies to native active human glycosyltransferases. Methods Mol Biol. 2013;1022:403–420. doi: 10.1007/978-1-62703-465-4_30. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Hassan H, Mirgorodskaya E, Kristensen AK, Roepstorff P, Bennett EP, Nielsen PA, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J, et al. Substrate specificities of three members of the human UDP-N-acetyl-alpha-D-galactosamine:Polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J Biol Chem. 1997;272:23503–23514. doi: 10.1074/jbc.272.38.23503. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Irazoqui F, Tarp MA, Bennett EP, Mandel U, Takeuchi H, Kato K, Irimura T, Suryanarayanan G, Hollingsworth MA, et al. The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc: Lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology. 2007;17:374–387. doi: 10.1093/glycob/cwl082. [DOI] [PubMed] [Google Scholar]
- Wang Y, Agrwal N, Eckhardt AE, Stevens RD, Hill RL. The acceptor substrate specificity of porcine submaxillary UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is dependent on the amino acid sequences adjacent to serine and threonine residues. J Biol Chem. 1993;268:22979–22983. [PubMed] [Google Scholar]
- Wang H, Tachibana K, Zhang Y, Iwasaki H, Kameyama A, Cheng L, Guo J, Hiruma T, Togayachi A, Kudo T, et al. Cloning and characterization of a novel UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase, pp-GalNAc-T14. Biochem Biophys Res Commun. 2003;300:738–744. doi: 10.1016/s0006-291x(02)02908-x. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IB, Gavel Y, von Heijne G. Amino acid distributions around O-linked glycosylation sites. Biochem J. 1991;275(Pt 2):529–534. doi: 10.1042/bj2750529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iwasaki H, Wang H, Kudo T, Kalka TB, Hennet T, Kubota T, Cheng L, Inaba N, Gotoh M, et al. Cloning and characterization of a new human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, designated pp-GalNAc-T13, that is specifically expressed in neurons and synthesizes GalNAc alpha-serine/threonine antigen. J Biol Chem. 2003;278:573–584. doi: 10.1074/jbc.M203094200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.